Abstract
Protein kinase B/Akt is a serine/threonine kinase that links receptors coupled to the PI3K lipid kinase to cellular anabolic pathways. Its activity in cells is controlled by reversible phosphorylation and an intramolecular lipid‐controlled allosteric switch. In this review, I outline the current progress in understanding Akt regulatory mechanisms, define three models of Akt activation in cells, and highlight how intramolecular allosterism cooperates with cell‐autonomous mechanisms to control Akt localization and activity and direct it toward specific sets of substrates in cells.
Keywords: protein kinase Akt, signaling
Abbreviations
- AGC
kinases, a group of kinases related to protein kinases A, ‐G and ‐C
- BNID
bionumbers record identification number
- DFG motif
a conserved motif containing aspartic acid – phenylalanine – glycine residues
- FRET
Förster's resonant energy transfer
- PDK1
phosphatidylinositol‐dependentkinase 1
- PIF
PDK1‐interacting fragment
- PI3K
phosphatidylinositol‐3‐kinase
- PI(3,4)P2
phosphatidylinositol‐3,4‐bisphosphate
- PI(3,4,5)P3
phosphatidylinositol‐3,4,5‐trisphosphate
- mTOR
mammalian target of rapamycin
1. AKT: AN INTRODUCTION
Akt belongs to the AGC family of serine/threonine kinases.1, 2 In mammalian cells, Akt is expressed as three isozymes (Akt1, Akt2, and Akt3), which display high degree of sequence identity and partially overlapping substrate specificities. Functionally, Akt controls cell survival, proliferation, DNA repair, and anabolic processes (reviewed in Reference 3) through phosphorylation of multiple (>300) substrates4, 5, 6 in response to activation of the phosphatidylinositol‐3 kinase (PI3K)‐coupled growth factor receptors.
Structurally, Akt contains the N‐terminal lipid‐binding pleckstrin homology (PH) domain, the bilobal kinase core domain (KD), and the C‐terminal hydrophobic extension, also known as the hydrophobic motif (HM), typical for the AGC kinases (Figure 1a). Like all eukaryotic protein kinases, the catalytic activity of Akt is controlled through alignment of the conserved residues in the regulatory (R‐) and catalytic (C‐) hydrophobic “spines,”7 linking the N‐ and C‐lobes of the kinase domain (Figure 1b). In the active conformation, proper alignment of the residues comprising the C‐ and R‐spines ensures optimal architecture of the active site for ATP binding and phosphate transfer onto the bound substrate molecule. Conversely, disassembly of either spine inactivates the kinases, allowing them to act as molecular switches. In Akt, the assembly and disassembly of the hydrophobic spines are controlled through posttranslational modifications and allosterically, through conformational rearrangements induced by binding of the phosphatidylinositol lipids PI(3,4,5)P3 or PI(3,4)P2 to the PH domain. Here, we will consider these mechanisms separately and then discuss how they contribute to Akt (in)activation in cells.
Figure 1.
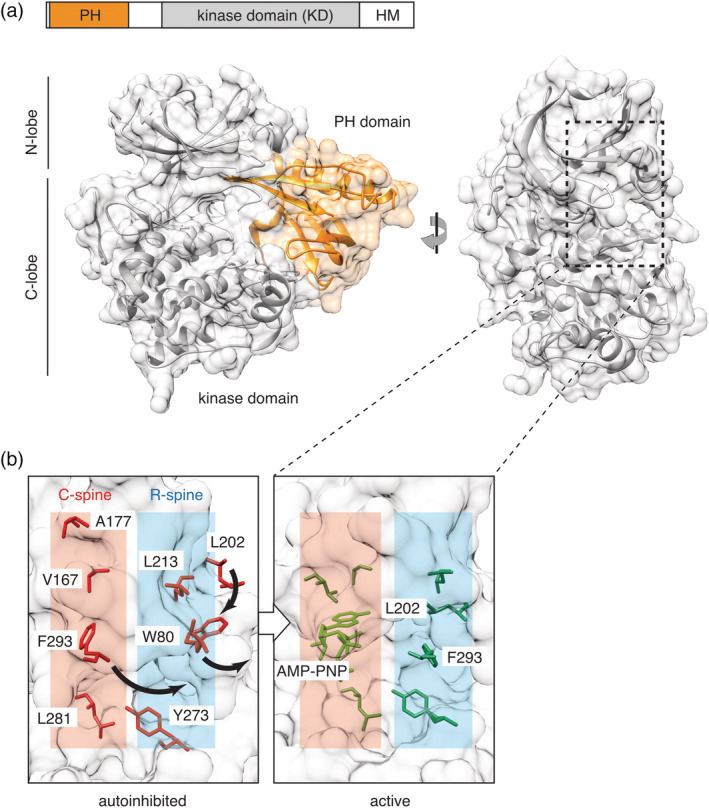
Akt structure and activation. (a) In the autoinhibited conformation, stabilized by the allosteric inhibitor VIII (PDB: 3o96), the lipid‐binding pleckstrin homology (PH) domain (orange) forms an extensive allosteric interface with the C‐lobe of the kinase domain (KD, gray). In the rotated view, the PH domain is hidden for clarity. Location of the C‐ and R‐spines connecting the N‐ and C‐lobes of the kinase domain is shown by dotted rectangle. (b) Akt phosphorylation on T308/S473 and allosteric mechanism cooperate to activate the kinase by aligning residues in the conserved hydrophobic spines. In the autoinhibited conformation (PDB: 3o96), W80 and F293 occupy the corresponding positions of L202 and nucleotide in the C‐ and R‐spines of the active form (PDB: 1o6k). Residues comprising the C‐ and R‐spines are highlighted; numbering corresponds to human Akt1 isozyme
1.1. Control of Akt activity by PTMs
Many posttranslational modifications have been reported for Akt (reviewed in References 8 and 9), yet the activity in vitro and in cells requires phosphorylation of two critical residues: T308 in the activation loop by PDK1 (here and henceforth residue numbering corresponds to human Akt1 isozyme) and S473 in the C‐terminal hydrophobic extension by the mTOR complex 2 (mTORC2).
Phosphorylation of T308 activates the kinase by promoting ordering of the activation loop and alignment of the residues in both R‐ and C‐spines, thereby enabling ATP binding and phosphate transfer. Crystal structures of the unphosphorylated and phosphorylated Akt kinase domains10, 11 showed that phosphorylation results in flipping of F293 of the DFG motif out of the C‐spine, which enables accommodating the adenine ring of the ATP and simultaneously restoring the R‐spine (Figure 1b, active). This conformational rearrangement is accompanied by repositioning of K179 and D292, which coordinate the ATP phosphates, closure of the β1/β2 (“glycine‐rich”) loop and ordering of the αC helix and the activation loop, thereby completing the kinase core and poising it for catalysis.
Many AGC kinases are regulated by intramolecular binding of HM to a hydrophobic pocket (“PIF pocket”) on the N‐lobe of the kinase domain.2 This binding helps recruit the PDK1 kinase, which phosphorylates the activation loop, and stabilizes the C‐ and R‐spines in the active conformation. This “PIF pocket” mechanism is common for many AGC kinases and was also proposed for Akt. Indeed, the in vitro activity of Akt monophosphorylated on T308 is only a fraction of the maximal, and phosphorylation of S473 in the HM or presence of the peptides mimicking the latter was often reported to increase Akt in vitro activity ten‐ to hundred‐fold.11, 12, 13, 14
How exactly S473 phosphorylation affects Akt activity, however, is unclear. Crystal structures of the unphosphorylated and T308‐phosphorylated kinase domain lacking the HM are nearly identical,10, 11 suggesting that engagement of the PIF pocket could stabilize the active conformation. However, recent studies using protein semisynthesis13 and genetic code expansion14, 15 demonstrated that while phosphorylation of S473 increased activity of T308‐phosphorylated Akt in vitro, the former site is essentially dispensable for the activity in cells.14, 15 Indeed, mutation of S473 or genetic ablation of mTORC2 components have only minor effect on cellular Akt phosphorylation and activity.12, 16, 17, 18, 19
What is the reason for the discrepancy between Akt behavior in cells and in vitro? The apparent dependence of Akt activity upon S473 phosphorylation in vitro could simply reflect the higher sensitivity of the in vitro assays and be irrelevant for Akt regulation in cells. Indeed, intracellular concentration of Akt substrates is likely to be well below the K M (BNID 103954, 101958),20 so that the rate of substrate phosphorylation would be limited by substrate availability rather than activity of the kinase. Alternatively, HM could contribute to Akt regulation in cells via additional mechanisms. Thus, phosphorylation of S473 was proposed to help protect Akt from dephosphorylation by cellular phosphatases,14, 21 although to which extent this is due to the PIF pocket occupancy or Akt membrane binding (discussed in the following) is currently unclear. It is also possible that engagement of the PIF pocket could help stabilize the active conformation of the kinase and/or modulate Akt substrate specificity in cells19; future studies should help gauge contribution of the HM‐dependent mechanisms to cellular Akt activity.
Several recent studies have identified two additional phosphorylation sites in Akt C‐terminal extension: S477 and T479.6, 22 Residues surrounding S477 (but not T479) partially resemble a Cdk2 consensus sequence, and its phosphorylation preceded that of S473,6, 22 prompting the authors to suggest that S477 primes Akt to phosphorylation by mTORC2. An alternative model13 proposed that S477 and T479 interact with the PH‐KD linker, helping destabilize the autoinhibitory interface (see in the following), though the authors' own data argue against this model, showing that phosphorylation of S477 and T479 increased K M for ATP to millimolar (3.7 mM) range. The exact role of the newly discovered C‐terminal phosphorylation sites in Akt regulation as well as the structural insight into their role in the context of full‐length Akt remain to be established.
Several other posttranslational modifications of Akt were reported, including Akt phosphorylation by both serine/threonine (Tbk1 and Ikkε) and tyrosine (Ack1, Src, and Ptk6) kinases (reviewed in References 8, 9, and 23) as well as proline hydroxylation by the oxygen‐dependent hydroxylase EglN1.24 While being interesting phenomenologically, the mechanisms and specific role of these PTMs in the control of Akt activity in cells await further characterization.
1.2. Allosteric control of Akt by lipid binding
It has long been established that the PH domain is required for Akt activation in cells. Indeed, Akt PH domain binds PI(3,4,5)P3 and PI(3,4)P2, thereby recruiting Akt to cellular membranes, where it is phosphorylated. In line with this, PI3K inhibition or mutations in the PH domain interfering with lipid binding all prevent Akt activation and phosphorylation of its substrates. However, deletion of the PH domain also dramatically increased Akt activity in vitro,13, 14, 18, 25, 26 suggesting that it could also act as an allosteric site.
The critical evidence for the allosteric function of the PH domain came from the only full‐length Akt crystal structure in complex with the inhibitor VIII.27 In this structure, the bound inhibitor promoted formation of an extensive (>1,500 Å2) interface between the PH and the C‐lobe of the kinase domain, with the PH domain essentially masking the active site. This structure clearly corresponds to the inactive conformation, with F293 occupying the ATP site in the C‐spine and the position of L202 in the R‐spine being occupied by the bulky W80 side chain (Figure 1b, autoinhibited). Akt activation, therefore, minimally requires displacement of W80 by L202 in the R‐spine, which is likely coupled to binding of lipids to the PH domain (see in the following), and F293 swinging out of the nucleotide binding site in the C‐spine, coupled to Akt phosphorylation on T308/S473 (Figure 1b). PH domain also partially occludes the substrate‐binding groove, which becomes fully exposed only when the allosteric interface is destabilized. The structure provided the first empirical evidence for the hypothetical autoinhibited “PH‐in” conformation, which was originally proposed as the opposite to the experimentally observed “PH‐out” species of the monomolecular FRET‐based Akt reporter28, 29 and firmly established the role of the PH domain in Akt allosteric regulation, as originally proposed by Stokoe et al.25
The structure of Akt in complex with the allosteric inhibitor highlighted two important points. First, it identified the key residues involved in maintaining the allosteric interface. Thus, E17 and K14 in the PH domain form the corresponding ionic bridges with R273 and D323 on the C‐lobe of the kinase domain. Mutation of key residues at the allosteric interface (E17, W80, D323, and D325) rendered Akt insensitive to allosteric inhibitors and was sufficient for kinase activation and cellular transformation.30 Indeed, the charge‐reversal mutation E17K resulted in increased Akt membrane binding and constitutive activity, drove cellular transformation, and was observed in patients with Proteus syndrome, megalencephaly, as well as colorectal, ovarian, and breast cancers.31, 32 Similarly, D323H mutation was identified in urinary carcinoma.33 The clear correlation between the mutations at the allosteric interface and their transformation potential underscore the importance of the autoinhibitory mechanism in restricting cellular Akt activity.
Second, the structure demonstrated that the allosteric interface included the residues in the PH domain that directly bind PI(3,4,5)P3 and PI(3,4)P2 lipids. This suggested that lipid binding and autoinhibited Akt conformation are mutually exclusive. This prediction was recently directly tested by biochemical measurements, demonstrating that PI(3,4,5)P3 or PI(3,4)P2 binding is indeed required for allosteric activation of the T308/S473‐phosphorylated Akt.18 This allosteric effect of the PH domain occupancy on Akt activity is likely coupled to W80 displacement by L202 in the R‐spine and changes in binding of the peptide substrates,13, 18 though T308 phosphorylation remains critical for ATP binding. Together, these data suggested that in cells active kinase is primarily associated with cellular membranes.
Interestingly, membrane binding might also help protect Akt from dephosphorylation. Earlier studies as well as the more recent reports demonstrated that ATP and ATP‐competitive analogs stabilize hyperphosphorylated, membrane‐bound Akt,14, 21, 34, 35, 36, 37, 38 suggesting that ATP‐bound Akt is less sensitive to dephosphorylation. Remarkably, ATP analogs did not induce hyperphosphorylation of the non‐membrane‐bound, cytosolic Akt35, 36; the effect was only observed in membrane‐targeted myristoylated form of Akt or in the presence of growth factors, recruiting full‐length Akt to the membranes, indicating that ATP‐competitive Akt inhibitors trap the kinase in the active, PI(3,4,5)P3‐bound conformation. In line with this notion, ATP analog induced membrane accumulation of the endogenous Akt.35 Furthermore, inhibition of upstream kinases only partially reduced phosphorylation of the membrane‐recruited Akt,39 demonstrating that membrane‐bound Akt is at least partially protected from dephosphorylation. Conversely, dissociation from the PI(3,4,5)P3/PI(3,4)P2‐containing membranes promoted desphosphorylation of the wild‐type, but not allosteric mutant of Akt,18 suggesting that re‐formation of the allosteric interface between the PH and kinase domains makes Akt a better substrate for phosphatases.
While accumulating evidence demonstrates that both phosphorylation and lipid binding play complementary roles in the allosteric control of Akt activity, several important questions remain unanswered. Thus, it is not clear whether membrane binding precedes Akt phosphorylation by PDK1 and/or mTORC2 and whether it can indeed stabilize the phosphorylated form of the kinase. Structures of the full‐length autoinhibited apo‐form as well as lipid‐bound active kinase would help better understand the molecular mechanisms of Akt allosterism.
1.3. Other Akt activation mechanisms
Several additional mechanisms were proposed to contribute to Akt activation in cells. Thus, reversible K63 ubiquitination of the lysine residues at the allosteric interface (K8, K14, and K20) was shown to induce Akt hyperactivation, even in the absence of growth factors or PI3K activity.40, 41, 42, 43 Consistently, Akt allosteric interface mutants E17K and E49K are hyperubiquitinated, hyperphosphorylated and correlate with human cancers.31, 40, 41, 44 Deficiency of the corresponding E3 ubiquitin ligases TRAF6, Skp2, and NEDD4 impaired Akt activation and breast cancer progression, whereas their overexpression correlated with Akt hyperactivation and tumorigenesis and mediated resistance to PI3K inhibitors in aggressive breast cancer.40, 41, 43, 45 Conversely, deficiency of the deubiquitinating enzyme CYLD promoted cell survival and growth of prostate tumors in mice.42 Interestingly, lysine acetylation appears to inhibit Akt activation by interfering with PI(3,4,5)P3 binding and membrane translocation,46 although the data on Akt acetylation and its functional role are scarce. Together, these reports clearly demonstrate that posttranslational modifications in the PH domain interfere with the allosteric mechanism, locking Akt in the active PH‐out conformation.
Several reports indicated that certain accessory proteins, such as Hsp90/Cdc37 chaperones,47 protooncogene product Tcl148, 49, 50 or nucleophosmin B2351, 52 induce increased Akt phosphorylation, most likely by protecting it from inactivation by phosphatases. Interestingly, Tcl1 binding site was mapped at the surface of the PH domain opposite to the lipid binding site, suggesting that it could potentially interfere with the Akt allosteric mechanism.50 Further biochemical and structural studies would be required to address the specific mechanisms by which accessory proteins affect Akt activity.
2. CELLULAR CONTROL OF AKT ACTIVITY
While many important insights into Akt regulatory mechanisms were obtained in vitro, it is essential to consider the relevance of these mechanisms in the complex cellular environment. Indeed, in cells both upstream and downstream lipid and protein kinases and phosphatases contribute to the kinetics and extent of Akt activation. In the following, we shall consider the relevance of the cell‐autonomous factors in relation to Akt‐intrinsic regulatory mechanisms outlined above. Such consideration allows to isolate three popular models of Akt regulation inside cells and formulate the biochemical criteria for Akt activity at subcellular compartments. We shall then discuss how these criteria define Akt activity and phosphorylation of its substrates in cells. Many of the results mentioned in the following were obtained using various Akt biosensors; interested readers are referred to an excellent comprehensive review on the topic,53 which, among others, covers many Akt‐specific reporters.
2.1. Models of Akt activation in cells
In quiescent cells Akt localizes in the cell nucleus and cytosol. Growth factors and other stimuli which activate production of PI(3,4,5)P3 and PI(3,4)P2 result in PH domain‐dependent accumulation of Akt at the respective membranes. This accumulation is transient, typically peaking around 2–5 min after stimulation; however, phosphorylation of both T308 and S473 is sustained for at least 2–3 hr post‐stimulation.18, 28, 54, 55, 56 Combined, these observations gave rise to a classical, or diffusive model of Akt activation (Figure 2a). According to this model, following the transient PI(3,4,5)P3 binding and phosphorylation by membrane‐associated PDK1 and mTORC2, Akt dissociates from the membranes and diffuses throughout the cell interior in its active form, phosphorylating its numerous substrates in the cytosol and nucleus until it is eventually inactivated by dephosphorylation.
Figure 2.
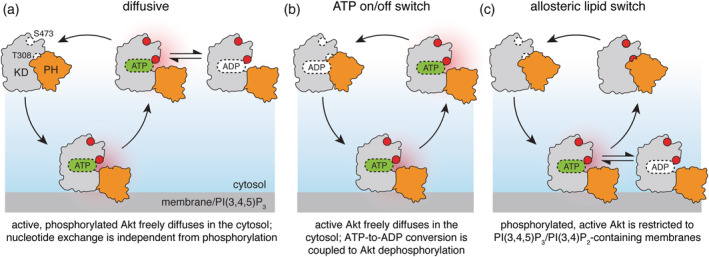
Models of intracellular Akt activation cycle. For all models, Akt activation requires binding to cellular membranes, containing PI(3,4,5)P3 and/or PI(3,4)P2 phosphoinositide lipids, accompanied by Akt phosphorylation on T308 and S473 (open and red‐filled circles) by membrane‐bound PDK1 and mTORC2 (not shown). PH domain is shown in orange, kinase domain in gray; red halo refers to catalytically active Akt. According to the diffusive model (a), phosphorylated, active Akt may dissociate from the membrane and diffuse in the cytosol phosphorylating the substrates (not shown) through multiple rounds of catalysis. An extension of the diffusive model, ATP on/off switch (b), links Akt dephosphorylation with the exchange of ATP for ADP during a single round of phosphate transfer onto the substrate. The allosteric lipid switch model (c) proposes that only membrane‐bound Akt is both phosphorylated and active, phosphorylating the substrates (not shown) in multiple rounds of catalysis. Dissociation from the membrane results in formation of the autoinhibited conformation and promotes rapid Akt dephosphorylation in the cytosol
Based on the fact that ATP‐competitive inhibitors induce paradoxical hyperphosphorylation of Akt in cells, Lin et al. have proposed an elegant extension of the diffusive model.37 According to their ATP on/off switch model (Figure 2b), ATP‐bound Akt is protected from dephosphorylation, as it diffuses through the cell. Substrate phosphorylation and the concomitant ATP‐to‐ADP conversion change Akt conformation such that it becomes a better substrate for cellular phosphatases and is therefore rapidly inactivated. Unlike the diffusive model, which neither imposed any restriction of Akt activity nor explicitly linked nucleotide exchange to Akt phosphorylation state, the ATP on/off switch limits kinase activity to a single round of catalysis, linking the model to empirical data demonstrating that Akt activity is closely coupled to PI(3,4,5)P3 and PI(3,4)P2 dynamics. This model was, however, challenged by the finding that Akt kinase‐inactive mutant that retains ATP binding capacity was dephosphorylated with the same kinetics as the wild type.18
While both models accounted for the existing empirical data, the subsequent phosphoproteomic analysis4, 6 showed that Akt substrates display distinct kinetics of phosphorylation, incompatible with the distributive kinetics implied by the freely diffusing Akt. Furthermore, free diffusion at ~12 μm2/s (Reference 18) would allow active Akt to reach any substrate in a typical cell well under 3 s; yet, the FRET reporters indicated much slower (t ½ 3÷5 min) kinetics of Akt activation in the cytosol and in the nucleus,28, 57 suggesting that active Akt is not freely diffusing inside cells.
Central to the diffusive model is the question of how long phosphorylated Akt persists in the cytosol following dissociation from PI(3,4,5)P3 or PI(3,4)P2. Akt activity in cells is limited by serine/threonine phosphatases dephosphorylating T308 and S473, such as PP2A and PHLPP1/2 (reviewed in Reference 58). Comparison of the kinetics of PI(3,4,5)P3 depletion and Akt inactivation in cells showed that Akt dephosphorylation is rate‐limited by its membrane dissociation.18 Furthermore, it was demonstrated that this effect was mediated by the allosteric mechanism described above and that active kinase in cells displayed the diffusive properties of a proximal membrane‐bound protein. The resulting allosteric lipid switch model (Figure 2c) suggests that in addition to phosphorylation of T308, Akt activity requires binding to PI(3,4,5)P3 or PI(3,4)P2, thereby limiting kinase activity to a subset of subcellular membrane compartments.
This conclusion has important implications for the control of Akt kinase activity and substrate phosphorylation in cells, suggesting that active kinase is exclusively associated with cellular membranes. The allosteric switch ensures that Akt activity remains proportional to the activity of the upstream regulators (lipid kinases and phosphatases) and enables to fine‐tune its activity inside cells according to the identity and lifetime of its lipid ligands. This latter prediction was recently supported empirically by an elegant imaging study showing that distinct intracellular localization and kinetics of PI(3,4,5)P3 and PI(3,4)P2 lipids dictate differential recruitment and activation profiles of Akt isozymes.59 Specifically, PI(3,4,5)P3 appears to acutely recruit Akt1 and −3 at the plasma membrane, whereas PI(3,4)P2 was responsible for the sustained activation of Akt2 at both the plasma membrane and early endosomes.
The allosteric switch model was challenged by a report proposing that dual S477/T479 phosphorylation helps destabilize the inhibitory interface.13 While the difference with the Ebner et al study could be accounted for by the authors using the soluble rather than membrane‐embedded PI(3,4,5)P3 as well as the saturating Mg2+ concentrations, the lack of exhaustive characterization of the proposed mechanism in cells is perhaps the most critical deficiency of this report, which prevents discrimination of the two models. The two papers, however, have set the high bar for the quantitative biochemical and cellular characterization of Akt activation mechanisms, which future studies will need to match.
Although the allosteric switch currently appears to be the most exhaustive model to‐date, providing both a molecular rationale for the empirical data and clues about the mechanisms to bypass the requirement for lipid binding, many questions remain unanswered. Thus, neither model satisfyingly addresses coupling of lipid binding to regulation of ATP/ADP exchange. Furthermore, the phosphatases responsible for Akt inactivation apparently play an active role in the intracellular control of Akt, yet the details of their regulation are only beginning to emerge. The ultimate test for the allosteric switch model will be the direct demonstration of the distinct phosphorylation kinetics for the membrane‐bound and cytosolic substrates, which is currently missing. Finally, mechanisms bypassing the allosteric switch (see discussion in the following) and helping protect Akt from inactivation remain to be characterized.
2.2. Akt activity at cellular membrane compartments
The observations outlined above allow us to formulate biochemical criteria for Akt activation at subcellular compartments: they should contain either PI(3,4,5)P3 or PI(3,4)P2 and display activities of the activating kinases upstream of Akt. Both plasma membrane and endosomal subpopulations satisfy these criteria: PI(3,4,5)P3 is primarily generated upon growth factor stimulation by class I PI3K in the inner leaflet of plasma membrane, membrane ruffles, clathrin‐coated pits and APPL1‐positive early endosomes. Another Akt‐activating lipid, PI(3,4)P2, is produced primarily through PI(3,4,5)P3 dephosphorylation60 or by phosphorylation of PI(4)P by class II PI3K in the endosomal vesicles (reviewed in Reference 61).
The second biochemical criterion of Akt activation is presence of the upstream kinases, PDK1 and mTORC2, at the subcellular membranes. While similar lipid binding specificity of the PDK1 and Akt PH domain ensures that both kinases are effectively co‐recruited to PI(3,4,5)P3‐ and PI(3,4)P2‐containing membranes, intracellular localization of mTORC2 until recently remained unclear. Initial immunofluorescence studies reported diffuse reticular staining of various mTORC2 components (reviewed in Reference 62); yet, they provided no insight on the activity of the complex in cells. Biochemical fractionation detected mTORC2 activity in the rough ER and crude mitochondrial fractions,63, 64 raising questions of how Akt is recruited to these compartments, which are not known to contain PI(3,4,5)P3 or PI(3,4)P2. An alternative hypothesis65 proposed that mTORC2 could be recruited to and activated by PI(3,4,5)P3 at the plasma membrane. Using a compartment‐specific probe for the endogenous mTORC2 activity, it was shown that both the localization and activity of mTORC2 at the plasma membrane are PI3K‐independent and that phosphorylation of Akt in response to growth factors is exclusively triggered by its membrane recruitment.39 However, it is important to keep in mind that mTORC2 is not absolutely required for Akt activity, though it augments phosphorylation of several Akt‐specific substrates, such as FoxO transcription factors, Tsc2, PRAS40, and AS160.19, 66
Localization of Akt activity to subcellular membranes may help target it toward specific sets of substrates as well as account for the distinct kinetics of Akt substrate phosphorylation.4, 6 Indeed, knockdown of the early endosomal protein Appl1 in zebrafish interfered with Akt‐mediated phosphorylation of Gsk3β, but not Tsc2.67 In agreement with the zebrafish study, local Akt activation on early endosomes led to Gsk3β phosphorylation and inactivation followed by relief of an autoinhibitory phosphorylation of the Gsk3β substrate dynamin1 and the corresponding increase in clathrin‐mediated endocytosis.68, 69, 70 Consistently, in PI3KC2γ knockout mice, which display markedly reduced level of PI(3,4)P2 on early endosomes, phosphorylation of both Gsk3β and its substrate glycogen synthase was reduced, whereas other Akt substrates, such as Tsc2 or FoxO transcription factors, were not affected.71 Similarly, phosphorylation of the lysosomal Akt substrate GFAP was shown to inhibit chaperone‐mediated autophagy.72 Furthermore, Akt phosphorylation on S473 could differentially regulate glucose transporters Glut1 and Glut4 in different cell types,19, 73 suggesting that compartment‐specific phosphorylation dynamics could help define functional outcomes of Akt signaling. These examples demonstrate that cellular membranes could both contribute to local Akt activation kinetics and guide its activity toward compartment‐specific substrates.
2.3. The enigma of nuclear Akt
In most cells and cell lines, Akt shuttles between the cytosol and nucleoplasm.74, 75, 76, 77, 78 As the potential functional roles of nuclear Akt have been discussed in detail elsewhere,79 here, we will concentrate on the question of whether nuclear Akt is catalytically active.
The typical argument for Akt being active in the nucleus is based on three major assumptions79: (a) Akt accumulates in the nucleus in response to different stimuli, (b) nucleus contains PI3K, PDK1, and mTORC2, all necessary to phosphorylate Akt, and (c) many Akt substrates reside in the nucleus, at least transiently. While each of these claims is sufficiently supported by multiple reports, the conjecture that Akt phosphorylates its substrates while in the nucleus is open for debate. Several questions must be addressed before making any definitive conclusions regarding the activity of nuclear Akt.
The first question is whether Akt can be phosphorylated while in the nucleus or if it is imported in the active form from the cytosol. Discriminating between these two scenarios is important, as they involve alternative mechanisms of Akt activation. The first scenario, where phosphorylated Akt is imported into the nucleus from the cytosol, is traditionally supported by the delayed phosphorylation of Akt FRET reporters in the nucleus57, 80 and several imaging studies of Akt localization in fixed cells.28, 29, 74, 75, 81 Yet, the evidence regarding the dependence of Akt nuclear accumulation on its phosphorylation state is at best contradictory.81, 82 More importantly, this scenario implies existence of a factor or factors that protect Akt from dephosphorylation by cellular phosphatases and bypasses the PI(3,4,5)P3 requirement to relieve Akt allosteric autoinhibition in the nucleus. One such potential factor could be Tcl1, which was shown to induce activation and nuclear accumulation of Akt.48, 49 Interaction with Hsp90 and/or Cdc37 also protected Akt from dephosphorylation,47, 83 suggesting that chaperones could contribute to accumulation of active, phosphorylated Akt in the nucleus. Yet, whether phosphorylated active Akt is imported into the nucleus from the cytosol and if additional factors help protect it against inactivation by cytosolic phosphatases is currently unclear.
An alternative scenario with Akt being phosphorylated in the nucleus requires mechanisms to transduce the growth factor signal from the plasma membrane or trigger PI3K‐independent nuclear resident kinases capable of activating Akt. One exciting model includes phosphorylation of S473 in the HM and recruitment of PDK1 via the PIF pocket mechanism resulting in Akt activation independent from PI3K.26 According to this report, UV‐induced DNA damage resulted in PI3K‐independent, DNA‐PK‐sensitive activation of Akt and phosphorylation of its substrates FoxO3a and Gsk3β. Furthermore, AktS473D “phosphomimetic” mutant was efficiently phosphorylated by PDK1 and could also phosphorylate its substrate Gsk3β (but not Tsc2) in vitro even in the absence of PI(3,4,5)P3‐containing vesicles.26 While the recent biochemical work provided strong evidence against the utility of “phosphomimetic” substitutions in Akt,15 it is tempting to speculate that a similar mechanism could indeed operate upon DNA damage and contribute to Akt activation in response to genotoxic drugs.84 Whether such mechanism contributes to accumulation of phosphorylated Akt in response to PI3K activation remains unclear.
The second popular argument for nuclear Akt activity is teleological: many Akt substrates are annotated as nuclear, so they are more likely to be phosphorylated by a nuclear‐localized kinase. This, however, could also be used as a counterargument: phosphorylation of an archetypal Akt substrate, FoxO family of transcription factors, serves to prevent their nuclear localization. More importantly, several recent proteomic studies have shown that most of Akt substrates annotated as nuclear in fact shuttle between nucleus and the cytoplasm,85, 86, 87 thereby challenging the simplistic, yet popular model that nuclear Akt substrates are phosphorylated in the nucleus. While one should not exclude such possibility, determination of the exact site of Akt activity under different conditions warrants further studies.
Nuclear accumulation of Akt could contribute to cellular transformation or reflect its hyperactivation, such as observed upon Tcl1 overexpression in T cell leukaemia or upon interference with the Akt allosteric mechanism (e.g., in E17K or D323H transforming mutants). Indeed, nuclear accumulation of Akt correlates with certain types of breast cancer78 and could be linked to the transformation potential of Tcl1.88 The potential roles of nuclear Akt in resistance of cancer cells to DNA‐damaging drugs84 and in promoting cell survival52 further highlights its physiological relevance and underscores the importance of further studies to unravel the mechanisms of Akt activation in the nucleus and its role in DNA repair and cellular transformation.
In summary, accumulation of active Akt in the nucleus requires particular mechanisms to induce and sustain its phosphorylation, which are likely to be context‐specific. The mere presence of the kinase in the nucleus does not warrant its activity and could equally reflect passive equilibration or sequestration of Akt inside cells. Localization of Akt in the nucleus could help restrict its substrate specificity and induce context‐specific cellular responses.
3. OUTLOOK
Three decades of Akt research have enormously contributed to the biochemical and molecular understanding of Akt regulation. Yet, despite the recent progress in methods for monitoring lipid and Akt dynamics in live cells, many questions remain unanswered. Thus, the atomic structure of the unphosphorylated as well as lipid‐bound full‐length Akt is still lacking. Such structures would improve our understanding of how the C‐terminal phosphorylation contributes to Akt activation or stabilization of the active conformation and shed light on the exact mechanism of Akt allosteric activation by lipid binding. Furthermore, direct demonstration of the distinct (distributive versus processive) phosphorylation kinetics of the cytosolic and membrane‐bound substrates by the membrane‐bound Akt is still lacking; whether the allosteric lipid switch indeed guides Akt toward specific substrates and/or affects processivity of substrate phosphorylation by Akt also remains unknown. Examination of the alternative mechanisms helping bypass the requirement for lipid binding, especially in the context of Akt activity in the nucleus, awaits further studies. Careful characterization of such mechanisms may help harness the therapeutic potential of the nuclear pool and develop new routes of interfering with Akt activity, orthogonal to its function in the control of anabolic processes and glucose homeostasis. Finally, examination of the spatially separated Akt pools with distinct regulatory mechanisms is a conceptually exciting paradigm for both biochemists and cell biologists, and elucidating the molecular mechanisms underlying this concept will help understand how biochemical architecture helps determine the fidelity of cellular signaling.
Yudushkin I. Control of Akt activity and substrate phosphorylation in cells. IUBMB Life. 2020;72:1115–1125. 10.1002/iub.2264
REFERENCES
- 1. Pearce LR, Komander D, Alessi DR. The nuts and bolts of AGC protein kinases. Nat Rev Mol Cell Biol. 2010;11:9–22. [DOI] [PubMed] [Google Scholar]
- 2. Leroux AE, Schulze JO, Biondi RM. AGC kinases, mechanisms of regulation and innovative drug development. Semin Cancer Biol. 2018;48:1–17. [DOI] [PubMed] [Google Scholar]
- 3. Manning BD, Toker A. AKT/PKB Signaling: Navigating the Network. Cell. 2017;169:381–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Humphrey SJ, Yang G, Yang P, et al. Dynamic adipocyte phosphoproteome reveals that Akt directly regulates mTORC2. Cell Metab. 2013;17:1009–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Toker A, Marmiroli S. Signaling specificity in the Akt pathway in biology and disease. Adv Biol Regul. 2014;55:28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Humphrey SJ, Azimifar SB, Mann M. High‐throughput phosphoproteomics reveals in vivo insulin signaling dynamics. Nat Biotechnol. 2015;33:990–995. [DOI] [PubMed] [Google Scholar]
- 7. Taylor SS, Kornev AP. Protein kinases: Evolution of dynamic regulatory proteins. Trends Biochem Sci. 2011;36:65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liao Y, Hung M‐C. Physiological regulation of Akt activity and stability. Am J Transl Res. 2010;2:19–42. [PMC free article] [PubMed] [Google Scholar]
- 9. Risso G, Blaustein M, Pozzi B, Mammi P, Srebrow A. Akt/PKB: One kinase, many modifications. Biochem J. 2015;468:203–214. [DOI] [PubMed] [Google Scholar]
- 10. Yang J, Cron P, Good VM, Thompson V, Hemmings BA, Barford D. Crystal structure of an activated Akt/protein kinase B ternary complex with GSK3‐peptide and AMP‐PNP. Nat Struct Biol. 2002;9:940–944. [DOI] [PubMed] [Google Scholar]
- 11. Yang J, Cron P, Thompson V, et al. Molecular mechanism for the regulation of protein kinase B/Akt by hydrophobic motif phosphorylation. Mol Cell. 2002;9:1227–1240. [DOI] [PubMed] [Google Scholar]
- 12. Biondi RM, Kieloch A, Currie RA, Deak M, Alessi DR. The PIF‐binding pocket in PDK1 is essential for activation of S6K and SGK, but not PKB. EMBO J. 2001;20:4380–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chu N, Salguero AL, Liu AZ, Chen Z, Dempsey DR, et al. Akt kinase activation mechanisms revealed using protein semisynthesis. Cell. 2018;174:897–907.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Balasuriya N, McKenna M, Liu X, Li SS‐C, O'Donoghue P. Phosphorylation‐dependent inhibition of Akt1. Genes. 2018;9:450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Balasuriya N, Kunkel MT, Liu X, et al. Genetic code expansion and live cell imaging reveal that Thr‐308 phosphorylation is irreplaceable and sufficient for Akt1 activity. J Biol Chem. 2018;293:10744–10756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jacinto E, Loewith R, Schmidt A, et al. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 2004;6:1122–1128. [DOI] [PubMed] [Google Scholar]
- 17. Guertin DA, Stevens DM, Thoreen CC, et al. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt‐FOXO and PKCα, but not S6K1. Dev Cell. 2006;11:859–871. [DOI] [PubMed] [Google Scholar]
- 18. Ebner M, Lučić I, Leonard TA, Yudushkin I. PI(3,4,5)P3 engagement restricts Akt activity to cellular membranes. Mol Cell. 2017;65:416–431.e6. [DOI] [PubMed] [Google Scholar]
- 19. Kearney AL, Cooke KC, Norris DM, et al. Serine 474 phosphorylation is essential for maximal Akt2 kinase activity in adipocytes. J Biol Chem. 2019;294:16729–16739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Milo R, Jorgensen P, Moran U, Weber G, Springer M. BioNumbers—the database of key numbers in molecular and cell biology. Nucleic Acids Res. 2010;38:D750–D753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chan TO, Zhang J, Tiegs BC, et al. Akt kinase C‐terminal modifications control activation loop dephosphorylation and enhance insulin response. Biochem J. 2015;471:37–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu P, Begley M, Michowski W, et al. Cell‐cycle‐regulated activation of Akt kinase by phosphorylation at its carboxyl terminus. Nature. 2014;508:541–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mahajan K, Mahajan NP. PI3K‐independent AKT activation in cancers: a treasure trove for novel therapeutics. J Cell Physiol. 2012;227:3178–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guo J, Chakraborty AA, Liu P, Gan W, Zheng X, et al. pVHL suppresses kinase activity of Akt in a proline‐hydroxylation‐dependent manner. Science. 2016;353:929–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stokoe D, Stephens LR, Copeland T, Gaffney PR, Reese CB, et al. Dual role of phosphatidylinositol‐3,4,5‐trisphosphate in the activation of protein kinase B. Science. 1997;277:567–570. [DOI] [PubMed] [Google Scholar]
- 26. Balzano D, Fawal M‐A, Velázquez JV, et al. Alternative activation mechanisms of protein kinase B trigger distinct downstream signaling responses. J Biol Chem. 2015;290:24975–24985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu W‐I, Voegtli WC, Sturgis HL, Dizon FP, Vigers GPA, Brandhuber BJ. Crystal structure of human AKT1 with an allosteric inhibitor reveals a new mode of kinase inhibition. PLoS ONE. 2010;5:e12913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Calleja V, Alcor D, Laguerre M, et al. Intramolecular and intermolecular interactions of protein kinase B define its activation in vivo. PLoS Biol. 2007;5:e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Calleja V, Laguerre M, Parker PJ, Larijani B. Role of a novel PH‐kinase domain interface in PKB/Akt regulation: Structural mechanism for allosteric inhibition. PLoS Biol. 2009;7:e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Parikh C, Janakiraman V, Wu W‐I, et al. Disruption of PH‐kinase domain interactions leads to oncogenic activation of AKT in human cancers. Proc Natl Acad Sci. 2012;109:19368–19373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Carpten JD, Faber AL, Horn C, et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448:439–444. [DOI] [PubMed] [Google Scholar]
- 32. Landgraf KE, Pilling C, Falke JJ. Molecular mechanism of an oncogenic mutation that alters membrane targeting: Glu17Lys modifies the PIP lipid specificity of the AKT1 PH domain. Biochemistry. 2008;47:12260–12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yi KH, Lauring J. Recurrent AKT mutations in human cancers: Functional consequences and effects on drug sensitivity. Oncotarget. 2015;7:4241–4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Han EK‐H, Leverson JD, McGonigal T, et al. Akt inhibitor A‐443654 induces rapid Akt Ser‐473 phosphorylation independent of mTORC1 inhibition. Oncogene. 2007;26:5655–5661. [DOI] [PubMed] [Google Scholar]
- 35. Okuzumi T, Fiedler D, Zhang C, Gray DC, Aizenstein B, et al. Inhibitor hijacking of Akt activation. Nat Methods. 2009;5:484–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chan TO, Zhang J, Rodeck U, et al. Resistance of Akt kinases to dephosphorylation through ATP‐dependent conformational plasticity. Proc Natl Acad Sci. 2011;108:E1120–E1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lin K, Lin J, Wu W‐I, Ballard J, Lee BB, et al. An ATP‐site on‐off switch that restricts phosphatase accessibility of Akt. Sci Signaling. 2012;5:ra37. [DOI] [PubMed] [Google Scholar]
- 38. Lu S, Deng R, Jiang H, Song H, Li S, et al. The Mechanism of ATP‐Dependent Allosteric Protection of Akt Kinase Phosphorylation. Structure. 2015;23:1725–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ebner M, Sinkovics B, Szczygieł M, Ribeiro DW, Yudushkin I. Localization of mTORC2 activity inside cells. J Cell Biol. 2017;216:343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yang W‐L, Wang J, Chan C‐H, Lee S‐W, Campos AD, et al. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science. 2009;325:1134–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fan C‐D, Lum MA, Xu C, Black JD, Wang X. Ubiquitin‐dependent regulation of phospho‐AKT dynamics by the ubiquitin E3 ligase, NEDD4‐1, in the insulin‐like growth factor‐1 response. J Biol Chem. 2013;288:1674–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang W‐L, Jin G, Li C‐F, Jeong YS, Moten A, et al. Cycles of ubiquitination and deubiquitination critically regulate growth factor‐mediated activation of Akt signaling. Sci Signaling. 2013;6:ra3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Clement E, Inuzuka H, Nihira NT, Wei W, Toker A. Skp2‐dependent reactivation of AKT drives resistance to PI3K inhibitors. Sci Signaling. 2018;11:eaao3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Askham JM, Platt F, Chambers PA, Snowden H, Taylor CF, Knowles MA. AKT1 mutations in bladder cancer: identification of a novel oncogenic mutation that can co‐operate with E17K. Oncogene. 2010;29:150–155. [DOI] [PubMed] [Google Scholar]
- 45. Chan C‐H, Li C‐F, Yang W‐L, et al. The Skp2‐SCF E3 ligase regulates Akt ubiquitination, glycolysis, herceptin sensitivity, and tumorigenesis. Cell. 2012;149:1098–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sundaresan NR, Pillai VB, Wolfgeher D, Samant S, Vasudevan P, et al. The deacetylase SIRT1 promotes membrane localization and activation of Akt and PDK1 during tumorigenesis and cardiac hypertrophy. Sci Signaling. 2011;4:ra46. [DOI] [PubMed] [Google Scholar]
- 47. Sato S, Fujita N, Tsuruo T. Modulation of Akt kinase activity by binding to Hsp90. Proc Natl Acad Sci U S A. 2000;97:10832–10837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pekarsky Y, Koval A, Hallas C, et al. Tcl1 enhances Akt kinase activity and mediates its nuclear translocation. Proc Natl Acad Sci U S A. 2000;97:3028–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Künstle G, Laine J, Pierron G, Kagami Si S‐I, Nakajima H, et al. Identification of Akt association and oligomerization domains of the Akt kinase coactivator TCL1. Mol Cell Biol. 2002;22:1513–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Auguin D, Barthe P, Royer C, et al. Structural basis for the co‐activation of protein kinase B by T‐cell leukemia‐1 (TCL1) family proto‐oncoproteins. J Biol Chem. 2004;279:35890–35902. [DOI] [PubMed] [Google Scholar]
- 51. Ahn J‐Y, Liu X, Cheng D, et al. Nucleophosmin/B23, a nuclear PI(3,4,5)P(3) receptor, mediates the antiapoptotic actions of NGF by inhibiting CAD. Mol Cell. 2005;18:435–445. [DOI] [PubMed] [Google Scholar]
- 52. Lee SB, Xuan Nguyen TL, Choi JW, et al. Nuclear Akt interacts with B23/NPM and protects it from proteolytic cleavage, enhancing cell survival. Proc Natl Acad Sci. 2008;105:16584–16589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Greenwald EC, Mehta S, Zhang J. Genetically encoded fluorescent biosensors illuminate the spatiotemporal regulation of signaling networks. Chem Rev. 2018;118:11707–11794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bruss MD, Arias EB, Lienhard GE, Cartee GD. Increased phosphorylation of Akt substrate of 160 kDa (AS160) in rat skeletal muscle in response to insulin or contractile activity. Diabetes. 2005;54:41–50. [DOI] [PubMed] [Google Scholar]
- 55. Galbaugh T, Cerrito MG, Jose CC, Cutler ML. EGF‐induced activation of Akt results in mTOR‐dependent p70S6 kinase phosphorylation and inhibition of HC11 cell lactogenic differentiation. BMC Cell Biol. 2006;7:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kubota H, Noguchi R, Toyoshima Y, et al. Temporal coding of insulin action through multiplexing of the AKT pathway. Mol Cell. 2012;46:820–832. [DOI] [PubMed] [Google Scholar]
- 57. Kunkel MT, Ni Q, Tsien RY, Zhang J, Newton AC. Spatio‐temporal dynamics of protein kinase B/Akt signaling revealed by a genetically encoded fluorescent reporter. J Biol Chem. 2005;280:5581–5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Newton AC, Trotman LC. Turning off AKT: PHLPP as a drug target. Annu Rev Pharmacol Toxicol. 2014;54:537–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liu S‐L, Wang Z‐G, Hu Y, Xin Y, Singaram I, et al. Quantitative lipid imaging reveals a new signaling function of phosphatidylinositol‐3,4‐bisphophate: Isoform‐ and site‐specific activation of Akt. Mol Cell. 2018;71:1092–1104.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Goulden BD, Pacheco J, Dull A, Zewe JP, Deiters A, et al. A high‐avidity biosensor reveals plasma membrane PI(3,4)P2 is predominantly a class I PI3K signaling product. J Cell Biol. 2018;265:jcb.201809026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Naguib A. Following the trail of lipids: Signals initiated by PI3K function at multiple cellular membranes. Sci Signaling. 2016;9:re4. [DOI] [PubMed] [Google Scholar]
- 62. Betz C, Hall MN. Where is mTOR and what is it doing there? J Cell Biol. 2013;203:563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Oh WJ, Wu C‐C, Kim SJ, Facchinetti V, Julien L‐A, et al. mTORC2 can associate with ribosomes to promote cotranslational phosphorylation and stability of nascent Akt polypeptide. EMBO J. 2010;29:3939–3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Betz C, Stracka D, Prescianotto‐Baschong C, Frieden M, Demaurex N, Hall MN. Feature Article: mTOR complex 2‐Akt signaling at mitochondria‐associated endoplasmic reticulum membranes (MAM) regulates mitochondrial physiology. Proc Natl Acad Sci. 2013;110:12526–12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Liu P, Gan W, Chin YR, et al. PtdIns(3,4,5)P3‐dependent activation of the mTORC2 kinase complex. Cancer Discov. 2015;5:1194–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Jacinto E, Facchinetti V, Liu D, et al. SIN1/MIP1 maintains rictor‐mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127:125–137. [DOI] [PubMed] [Google Scholar]
- 67. Schenck A, Goto‐Silva L, Collinet C, et al. The endosomal protein Appl1 mediates Akt substrate specificity and cell survival in vertebrate development. Cell. 2008;133:486–497. [DOI] [PubMed] [Google Scholar]
- 68. Reis CR, Chen P‐H, Srinivasan S, Aguet F, Mettlen M, Schmid SL. Crosstalk between Akt/GSK3β signaling and dynamin‐1 regulates clathrin‐mediated endocytosis. EMBO J. 2015;34:2132–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chen P‐H, Bendris N, Hsiao Y‐J, Reis CR, Mettlen M, et al. Crosstalk between CLCb/Dyn1‐Mediated Adaptive Clathrin‐Mediated Endocytosis and Epidermal Growth Factor Receptor Signaling Increases Metastasis. Dev Cell. 2017;40:278–288.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Schmid SL. Reciprocal regulation of signaling and endocytosis: Implications for the evolving cancer cell. J Cell Biol. 2017;216:2623–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Braccini L, Ciraolo E, Campa CC, et al. PI3K‐C2γ is a Rab5 effector selectively controlling endosomal Akt2 activation downstream of insulin signalling. Nat Commun. 2015;6:7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Arias E, Koga H, Diaz A, Mocholi E, Patel B, Cuervo AM. Lysosomal mTORC2/PHLPP1/Akt regulate chaperone‐mediated autophagy. Mol Cell. 2015;59:270–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Beg M, Abdullah N, Thowfeik FS, Altorki NK, McGraw TE. Distinct Akt phosphorylation states are required for insulin regulated Glut4 and Glut1‐mediated glucose uptake. Elife. 2017;6:e26896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Meier R, Alessi DR, Cron P, Andjelković M, Hemmings BA. Mitogenic activation, phosphorylation, and nuclear translocation of protein kinase Bbeta. J Biol Chem. 1997;272:30491–30497. [DOI] [PubMed] [Google Scholar]
- 75. Borgatti P, Martelli AM, Tabellini G, Bellacosa A, Capitani S, Neri LM. Threonine 308 phosphorylated form of Akt translocates to the nucleus of PC12 cells under nerve growth factor stimulation and associates with the nuclear matrix protein nucleolin. J Cell Physiol. 2003;196:79–88. [DOI] [PubMed] [Google Scholar]
- 76. Shiraishi I, Melendez J, Ahn Y, et al. Nuclear targeting of Akt enhances kinase activity and survival of cardiomyocytes. Circ Res. 2004;94:884–891. [DOI] [PubMed] [Google Scholar]
- 77. Mistafa O, Högberg J, Stenius U. Statins and ATP regulate nuclear pAkt via the P2X7 purinergic receptor in epithelial cells. Biochem Biophys Res Commun. 2008;365:131–136. [DOI] [PubMed] [Google Scholar]
- 78. Badve S, Collins NR, Bhat‐Nakshatri P, et al. Subcellular localization of activated AKT in estrogen receptor‐ and progesterone receptor‐expressing breast cancers: potential clinical implications. Am J Pathol. 2010;176:2139–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Martelli AM, Tabellini G, Bressanin D, et al. The emerging multiple roles of nuclear Akt. Biochimica et Biophysica Acta (BBA)—Mol Cell Res. 2012;1823:2168–2178. [DOI] [PubMed] [Google Scholar]
- 80. Mehta S, Zhang Y, Roth RH, et al. Single‐fluorophore biosensors for sensitive and multiplexed detection of signalling activities. Nat Cell Biol. 2018;20:1215–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Xuan Nguyen TL, Choi JW, Lee SB, et al. Akt phosphorylation is essential for nuclear translocation and retention in NGF‐stimulated PC12 cells. Biochem Biophys Res Commun. 2006;349:789–798. [DOI] [PubMed] [Google Scholar]
- 82. Saji M, Vasko V, Kada F, Allbritton EH, Burman KD, Ringel MD. Akt1 contains a functional leucine‐rich nuclear export sequence. Biochem Biophys Res Commun. 2005;332:167–173. [DOI] [PubMed] [Google Scholar]
- 83. Theodoraki MA, Kunjappu M, Sternberg DW, Caplan AJ. Akt shows variable sensitivity to an Hsp90 inhibitor depending on cell context. Exp Cell Res. 2007;313:3851–3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Brown KK, Montaser‐Kouhsari L, Beck AH, Toker A. MERIT40 is an Akt substrate that promotes resolution of DNA damage induced by chemotherapy. Cell Rep. 2015;11:1358–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wühr M, Güttler T, Peshkin L, et al. The nuclear proteome of a vertebrate. Curr Biol. 2015;25:2663–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Itzhak DN, Tyanova S, Cox J, Borner GH. Global, quantitative and dynamic mapping of protein subcellular localization. Elife. 2016;5:e16950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Thul PJ, Åkesson L, Wiking M, Mahdessian D, Geladaki A, et al. A subcellular map of the human proteome. Science. 2017;356:eaal3321. [DOI] [PubMed] [Google Scholar]
- 88. Teitell MA. The TCL1 family of oncoproteins: co‐activators of transformation. Nat Rev Cancer. 2005;5:640–648. [DOI] [PubMed] [Google Scholar]


