Abstract
Plant genomes contain two major classes of innate immune receptors to recognize different pathogens. The pattern recognition receptors perceive conserved pathogen‐associated molecular patterns and the resistance genes with nucleotide‐binding (NB) and leucine‐rich repeat (LRR) domains recognize specific pathogen effectors. The precise regulation of resistance genes is important since the unregulated expression of NB‐LRR genes can inhibit growth and may result in autoimmunity in the absence of pathogen infection. It was shown that a subset of miRNAs could target NB‐LRR genes and act as an important regulator of plant immunity in the absence of pathogens. Plants not only interact with pathogens, but they can also establish symbiotic interactions with microbes. Nitrogen‐fixing symbiotic interaction and nodule formation of legumes may also require the suppression of host defence to prevent immune responses. We found that upon symbiotic interactions, miRNAs repressing NB‐LRR expression are upregulated in the developing nodules of Medicago truncatula. Furthermore, we show that the suppression of the activity of the NB‐LRR genes targeted by these miRNAs is important during nodule development. Our results suggest that the downregulation of NB‐LRR resistance genes in the developing nodule produces a suitable niche that facilitates bacterial colonization and the development of an N‐fixing nodule.
Keywords: defence, Medicago truncatula, miRNA, NB‐LRR, nodulation, pathogen response, phasiRNA, receptor kinases, symbiosis
Short abstract
Nitrogen‐fixing symbiotic interaction and nodule formation of legumes require the suppression of host defenses by differential expression of NB‐LRR‐regulating miRNAs to prevent immune responses.
1. INTRODUCTION
Plants have evolved together with pathogens and during this evolution they developed several defence pathways to overcome pathogen infection. Plant genomes contain two major classes of innate immune receptors to recognize different pathogens (Chisholm, Coaker, Day, & Staskawicz, 2006; Jones & Dangl, 2006). The first line of defence comprises the pattern recognition receptors (PRRs) that recognize conserved pathogen‐associated molecular patterns (PAMPs) and activate PAMP‐triggered immunity (PTI). However, PTI can be suppressed by the pathogen, usually through secreted proteins called effectors. Plant genomes also contain large numbers of resistance (R) genes with nucleotide‐binding (NB) and leucine‐rich repeat (LRR) domains (Meyers, Kaushik, & Nandety, 2005), which function as a second line of defence, that recognize specific pathogen effectors and trigger resistance responses. As a consequence, R genes are protecting plants from diseases caused by devastating pathogens. However, the unregulated expression of NB‐LRR‐type resistance genes can inhibit growth and may result in autoimmunity in the absence of pathogen infection (Michael Weaver, Swiderski, Li, & Jones, 2006).
In addition to innate immune receptors, in plants and other eukaryotes, small RNA (sRNA) systems mediate gene silencing which is thought to have evolved as a defence system against viruses and other molecular intruders (Ding, 2010; Ding & Voinnet, 2007). sRNAs are also important gene expression regulators, involved in many developmental processes and stress responses. These molecules are generally 21‐ to 24‐nt‐long and based on their biogenesis we distinguish two main classes: microRNAs (miRNAs) and small interfering RNAs (siRNAs). miRNAs function in a post‐transcriptional manner by downregulating target mRNAs in a variety of cellular processes (Rogers & Chen, 2013). The large majority of the plant sRNAs are siRNAs and one class of siRNAs termed phased siRNAs (phasiRNAs) (Zhai et al., 2011) regulate protein‐coding genes in a similar fashion as miRNAs.
Plant genomes contain large numbers of NB‐LRR‐type resistance genes that recognize specific pathogen effectors and trigger resistance responses (Meyers, Kozik, Griego, Kuang, & Michelmore, 2003), however, the unregulated expression of NB‐LRR genes may be harmful to plant growth and to immune responses (Michael Weaver et al., 2006). It was shown that a subset of plant miRNAs (miR482/2118 superfamily, miR1507, miR2109) can target NB‐LRR genes and regulate plant immunity (Deng et al., 2018; Li et al., 2012; Shivaprasad et al., 2012; Su et al., 2018; Yang et al., 2015; Zhai et al., 2011). The miR482/2118 superfamily is the most ancient among the NB‐LRR‐regulating miRNAs and it emerged first in Gymnosperms, followed by extensive radiation in seed plants. This gene family is present in the sequenced legume genomes except in Lotus japonicus (Chávez Montes et al., 2014; de Vries, Kloesges, & Rose, 2015). The miR1507 and miR2109 (also known as miR5213 in Medicago truncatula) also target NB‐LRR genes and they are prevalent in Fabaceae (Taylor, Tarver, Hiscock, & Donoghue, 2014; Zhang, Xia, Kuang, & Meyers, 2016). Some of the NB‐LRR‐targeting miRNAs are capable of triggering the production of phasiRNAs from their cleaved target mRNAs and target a large number of NB‐LRR mRNAs by phasiRNAs (Zhai et al., 2011). Moreover, this regulation by miRNAs is proposed as a mechanism for reducing fitness costs associated with NB‐LRR genes by targeting them in the absence of pathogens (Shivaprasad et al., 2012). It was proposed that in the absence of pathogens, the NB‐LRR resistance genes are transcribed but cleaved by miRNAs/phasiRNAs keeping their translation at a very low level. During pathogen attack, the levels of miRNAs and phasiRNAs are reduced resulting in an increased level of resistance gene transcript (Shivaprasad et al., 2012). The NB‐LRR proteins are normally associated with effector‐triggered immunity (Jones & Dangl, 2006), however, if the NB‐LRR proteins are overexpressed (Bendahmane, Kanyuka, & Baulcombe, 1999), defence can also be induced independently of protein‐based recognition mechanisms. Therefore, the elevated levels of NB‐LRR proteins accelerate the activation of the defence pathways in non‐race‐specific defence against viral and bacterial pathogens (Shivaprasad et al., 2012).
Legumes establish symbiotic interactions with microbes, however, it is not fully understood how they differentiate between symbiotic or pathogenic microbial partners. Based on recent research achievements, symbiotic interaction and nodulation require the suppression of host defences to prevent immune responses (Yang, Tang, Gao, Krishnan, & Zhu, 2010). The M. truncatula genome encodes around 540 NB‐LRR genes, and more than 60% of them could be targeted by the NB‐LRR‐targeting miRNAs such as miR1507, miR2109, and miR2118 (member of the miR482 superfamily) or by the phasiRNAs that are produced from at least 114 phasiRNA producing loci by the action of these miRNAs (Zhai et al., 2011).
Legumes invite symbiotic soil bacteria, termed rhizobia to enter into their roots and induce the development of nitrogen‐fixing symbiotic nodules. It is a long‐lasting vital question of how the host plants differentiate between interacting beneficial and detrimental microbes. Remarkable similarities have been found in sensing both categories of microbes leading to activation of distinct signalling pathways and physiological responses (Cao, Halane, Gassmann, & Stacey, 2017; Gourion, Berrabah, Ratet, & Stacey, 2015; Zipfel & Oldroyd, 2017). Previous observations indicated the involvement of several miRNA families in nodule development (Combier et al., 2006; Lelandais‐Brière et al., 2009; Li, Deng, Wu, Subramanian, & Yu, 2010; Subramanian et al., 2008; Tsikou et al., 2018) and the regulation of R gene‐mediated immunity in a rhizobial strain‐specific manner was also reported (Yang et al., 2010).
Here we report, that the expression level of the NB‐LRR‐regulating miRNAs (miR1507, miR2109, and miR2118) is induced at the early phase of symbiotic interaction in M. truncatula and this induction is maintained in the symbiotic nodule, co‐localizing with symbiotic bacteria in colonized nodule cells. Furthermore, the target NB‐LRR mRNAs of these miRNAs are downregulated in the symbiotic nodules. We show that modification of the expression level of the NB‐LRR‐regulating miRNAs (either upregulation or downregulation) significantly changed the number of the symbiotic nodules on M. truncatula plants. These results indicate that nodulation requires the fine‐tuned differential regulation of NB‐LRR genes by miRNAs during symbiotic interactions to prevent the host's immune responses.
2. MATERIALS AND METHODS
2.1. Plant materials and growth conditions
Medicago truncatula Jemalong genotype was used in all experiments. Seeds were chemically scarified and sterilized with sulphuric acid as described in the M. truncatula handbook (Chabaud, Lichtenzveig, Ellwood, Pfaff, & Journet, 2006). Seeds were germinated with overnight incubation in the dark at room temperature on inverted agar plates (1% wt/vol agar in water) following 4–6‐day‐long cold treatment at 4°C. Seedlings for all experiments were planted in zeolite substrate (Geoproduct Kft., Mád, Hungary) perfused with nitrogen‐free Gibson medium (Gibson & Nutman, 1960) and were grown in a growth chamber under long‐day photoperiod (16 hr light and 8 hr darkness at 24°C) for 5 days before inoculation with rhizobia (Journet, De Carvalho‐Niebel, Andriankaja, Huguet, & Barker, 2006).
2.2. Viral inoculation of M. truncatula seedlings
Five‐day‐old M. truncatula cotyledons were infected with the sap of Nicotiana glutinosa infected with alfalfa mosaic virus (AMV) (Salamon, Sós‐Hegedűs, Gyula, & Szittya, 2018) as previously described (Kis, Salamon, Kis, & Szittya, 2017).
2.3. Treatment of M. truncatula seedlings with flg22
Roots of 7‐day‐old M. truncatula plants were immersed in 1 μM flg22 (Ezbiolab) solution diluted in Fahräeus medium (Barker et al., 2006) or in Fahräeus medium as a mock control for 6 hr. Then roots were washed with distilled water and incubated in Fahräeus medium for an additional 24 hr. RNA was isolated from roots and cDNA was synthesized for qPCR. As markers of the activation of plant defence responses, expression of PATHOGENESIS‐RELATED1 (PR1, Medtr2g435490) and PATHOGENESIS‐RELATED10 (PR10, Medtr2g035150) genes were tested by qPCR (Domonkos et al., 2017).
2.4. Bacterial strains and inoculation of M. truncatula roots
Five‐day‐old plants were infected with wild‐type Sinorhizobium (Ensifer) meliloti 1021 or its exoY mutant derivative defective in succinoglycan production (Reuber & Walker, 1993). Rhizobial strains were grown for 24–48 hr at 30°C in tryptone yeast (TY) medium supplemented with 6 mM CaCl2, and inoculations were carried out as described previously (Journet et al., 2006). In hairy root transformation experiments, Agrobacterium rhizogenes ARqua1 strain was used.
2.5. In situ hybridization
M. truncatula nodules inoculated either with wild‐type S. meliloti or S. meliloti exoY were collected at 4, 7, 14 days post‐inoculation (dpi) and 14 dpi, respectively. The nodules were fixed in a fixative solution of 4% (wt/vol) paraformaldehyde (Sigma) in phosphate‐buffered saline pH 7.4 (PBS) and processed for paraffin embedding as described previously (Várallyay & Havelda, 2011). Longitudinal sections of nodules (10 μm thick) were mounted to specific poly‐l‐lysine‐coated microscope slides (Leica). For all in situ hybridization, 5′ digoxigenin (DIG)‐labelled locked nucleic acid oligonucleotides (Exiqon) were used (Table S1). The stock solution of oligonucleotides was diluted to 20 pmol μl−1 concentrations, and 2 μl probe was used per slide in 150 μl hybridization solution. Hybridization was performed overnight at 50°C with each probe and slides were washed with 0.2× saline sodium citrate (SSC) buffer pH 7.0. Slides were incubated in Blocking I solution (0.5% vol/vol Blocking Reagent in 1× Tris‐buffered saline buffer pH 7.4, TBS, Roche) for 30 min and were transferred into Blocking II solution (1% vol/vol BSA in 1× TBS and 0.3% vol/vol Triton™ X‐100) for 15 min. The anti‐DIG solution was diluted in Blocking II solution (1:1,250), 150 μl solution was applied per slide and incubated for 90 min. After washing with 1× TBS, SB buffer containing nitro‐blue tetrazolium: 5‐bromo‐4‐chloro‐3′‐indolyl phosphate (1:50, vol/vol) was applied to develop the colour reaction (Medzihradszky, Schneitz, & Lohmann, 2014). The reaction was stopped after 12 hr by rinsing the slides with water.
2.6. Microscopy
In situ hybridized nodule sections were observed with ZEISS axiostar plus M light microscope and images captured using a euromex HD ultra 1080p digital camera. To analyse the presence of rhizobia in nodules, in situ sections were stained with 5 μM SYTO™ 13 in PBS (pH 7.4) for 20–60 min and were analysed with confocal laser scanning microscopy. Confocal laser scanning microscopy was performed using a Leica SP5AOBS confocal laser scanning microscope (Leica, Germany) on DMI6000 microscope base using HCX PL FLUOTAR ×5 dry objective with a numerical aperture of 0.15. SYTO™ 13 fluorescence was detected between 502 and 587 nm using 488 nm laser excitation. In situ hybridization signal of the same sample was detected using a Zeiss Axiocam MRc‐5 colour camera (Carl Zeiss MicroImaging GmbH, Germany) connected to the side port of the confocal microscope.
2.7. RNA extraction, northern hybridization
M. truncatula roots, nodules, leaves, or cotyledons were collected at different time points, and total RNA was isolated by using the phenol/chloroform extraction method (Szittya, Salamon, & Burgyán, 2000). Northern hybridization was performed essentially as previously described (Baksa et al., 2015). Briefly, 1 μg total RNA was separated in a 12% (wt/vol) polyacrylamide gel in 1× tris‐borate EDTA buffer containing 8 M urea and transferred to the neutral nylon membrane Hybond™‐NX (Amersham/GE Healthcare) by semi‐dry electroblotter for 45 min. RNA molecules were cross‐linked by 1‐ethyl‐3‐(3‐dimethylaminopropyl) carbodiimide chemical cross‐linking method at 60°C for 90 min (Pall & Hamilton, 2008). RNA blots were pre‐hybridized at 38°C for 1 hr with ULTRAhyb®‐oligo ultrasensitive hybridization buffer (Thermo Scientific). For miRNA detection, oligonucleotide probes complementary to miRNAs (Table S1) were end‐labelled with [γ32P] ATP (Perkin Elmer) for 1 hr at 37°C using T4 polynucleotide kinase (Thermo Scientific). Hybridization with labelled probes was performed at 38°C for 16–18 hr. Then blots were washed twice with 2× SSC buffer containing 0.1% (wt/vol) SDS at 38°C and exposed to PhosphorImager® screens (GE Healthcare).
2.8. RT‐qPCR assays
Total RNA samples were treated with DNase I (Thermo Scientific). The cDNA synthesis was performed with 500 ng total RNA by High‐Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer's instructions. For the RT‐qPCR, a Light Cycler 96 (Roche) machine and SYBR™ Select Master Mix (Applied Biosystems) were used. Data were analysed by LightCycler® 96 SW1.1 software (Roche). Relative expression levels were calculated by normalization against the expression of PTB (polypyrimidine tract‐binding‐like protein, Medtr3g090960). The miRNA cleavage sites in the target transcripts were predicted with the psRNATarget server (Dai & Zhao, 2011). Primers for the miRNA target genes were designed around the miRNA target sites using eprimer3, part of the EMBOSS software package (Rychlik, 1993).
2.9. Construct design and plant transformation
Overexpression constructs were generated by Gateway Technology (Life Technologies). For miRNA overexpression, constructs of miRNA precursors were amplified from M. truncatula genomic sequences with Phusion DNA Polymerase (Thermo Scientific) using the primers previously described (Fei, Li, Teng, & Meyers, 2015) (Table S1). miRNA precursors were inserted into pDONR201 vector by homologous recombination by BP clonase (Life Technologies), and sequences were verified. Destination clones were made by LR clonase II (Life Technologies) mediated recombination. The binary vector contained the 966‐bp‐long EF1α promoter (At1g07920—GTP binding Elongation factor Tu family protein) upstream the homologous recombination sites and DsRed fluorescent selection marker gene under the control of the Arabidopsis thaliana UBIQUITIN10 promoter. The base of the destination vector was the pKGW‐R vector (obtained from Plant Systems Biology, VIB‐Ghent University) in which the EF1α promoter was inserted between the SpeI and HindIII sites, the box containing the bacterial selection marker, chloramphenicol resistance, and the two homologous recombination sites (AttR1‐CmR‐ccDB‐AttR2) were inverted (pKGW‐R‐pEF1). We used short tandem target mimic (STTM) construct to decrease miR2118abc miRNA levels (Fei et al., 2015) and IPS‐based target mimicry construct (Franco‐Zorrilla et al., 2007) to decrease the miR2118a level (Table S1). in vitro synthesized STTM sequences (Integrated DNA Technologies) and the amplified nos terminator was cloned into SpeI and AatII site of pKGW‐R‐pEF1 plasmid, using infusion technology (In‐Fusion® HD cloning kit Clontech). For analysis of the expression of miRNA regulated NB‐LRR target genes, plants were treated with A. rhizogenes as described earlier (Boisson‐Dernier et al., 2001). After 6 days on Fahräeus agar medium, the transformed seedlings were transferred to Fahräeus agar complemented with kanamycin (25 μg ml−1) and cefotaxime (250 μg ml−1) for 2 weeks. Kanamycin was used as a selection marker for the vector and the constructs, while cefotaxime was used for eliminating Agrobacterium from the transformed hairy roots. The plants were kept sterile until the red fluorescent transformed roots were collected for RNA preparation and qPCRs were performed on NB‐LRR target genes.
For the quantification of infection events and nodule numbers, all constructs were introduced in M. truncatula Jemalong plants using A. rhizogenes hairy root transformation (Boisson‐Dernier et al., 2001). DsRed was the fluorescent selection marker of transformed hairy roots. Transformed plants were planted in zeolite substrate and inoculated with S. meliloti 1021 strain carrying the pXLGD4 plasmid constitutively expressing the lacZ reporter gene for counting infection events or S. meliloti 1021 (wt) for nodule number analysis. To quantify microcolonies and infection threads, at least 15 red fluorescent transformed roots per construct were fixed and stained with 0.1% (wt/vol) X‐Gal solution as described earlier (Domonkos et al., 2017). Blue‐stained microcolonies and infection threads were counted at 5 dpi. Numbers of infection events were normalized to root length (counts per mm of root).
Nodule number was counted at 4 weeks post‐inoculation (wpi) with S. meliloti 1021. Following quantification, the roots were individually frozen in liquid nitrogen and total RNA was isolated for northern analysis of sRNAs.
2.10. Statistical analysis
In the case of RT‐qPCR analysis and nodule number quantification, unpaired, one‐tailed t tests were performed to estimate statistical significance of the difference between the samples; infected plants were compared to mock‐treated ones or transgenic roots were compared to empty vector‐transformed roots. In the RT‐qPCR experiments, four independent biological replicates were measured with two technical repeats the mean of which was used in the statistical analysis. In the nodule number quantification, at least 45 (miRNA target mimicry lines) or 15 (miRNA overexpression lines) transgenic roots were counted. The statistical tests were performed in Microsoft Excel. Statistical significance of the observed difference was marked in the figures with asterisks according to the following categories: ***p ≤ .001; **p ≤ .01;*p ≤ .05, ns = not significant.
2.11. Promoter analysis
The pri‐miRNA sequences were identified by PatMan (Prüfer et al., 2008) using the mature miRNA sequences as the query patterns and the r5.0 version of the M. truncatula A17 annotated transcripts (Pecrix et al., 2018) as the database. Promoters of the five MIRNA genes were obtained by extracting the sequences 2 kb upstream of the transcription start site (TSS) of the identified transcripts using BEDTools v2.26.0 (Quinlan & Hall, 2010). Promoter analysis was conducted using the PLACE tool (Higo, Ugawa, Iwamoto, & Korenaga, 1999). The lists of motifs of the five MIRNA promoters were compared and drawn with an online Venn‐diagram drawing tool (http://bioinformatics.psb.ugent.be/webtools/Venn/). The selected common elements were plotted with a custom R script and further edited with Inkscape.
3. RESULTS AND DISCUSSION
3.1. Both pathogen infection and symbiotic interaction modulates the expression level of NB‐LRR‐regulating miRNAs
To test the effect of pathogen infection on the expression level of NB‐LRR‐regulating miRNAs (miR1507, miR2109, and miR2118) in legumes able to form symbiotic N‐fixing interaction, we infected M. truncatula plants with AMV (Salamon et al., 2018). The level of the NB‐LRR‐targeting miRNAs was monitored in the inoculated cotyledons of mock and virus‐infected plants with northern blots 4 dpi (Figure 1a). We found that similarly to previous reports in Solanaceae species (Deng et al., 2018; Li et al., 2012; Shivaprasad et al., 2012; Yang et al., 2015), the expression level of these three NB‐LRR‐regulating miRNAs decreased during AMV infection in M. truncatula cotyledons and the largest reduction was detected in the expression of miR2118.
Figure 1.
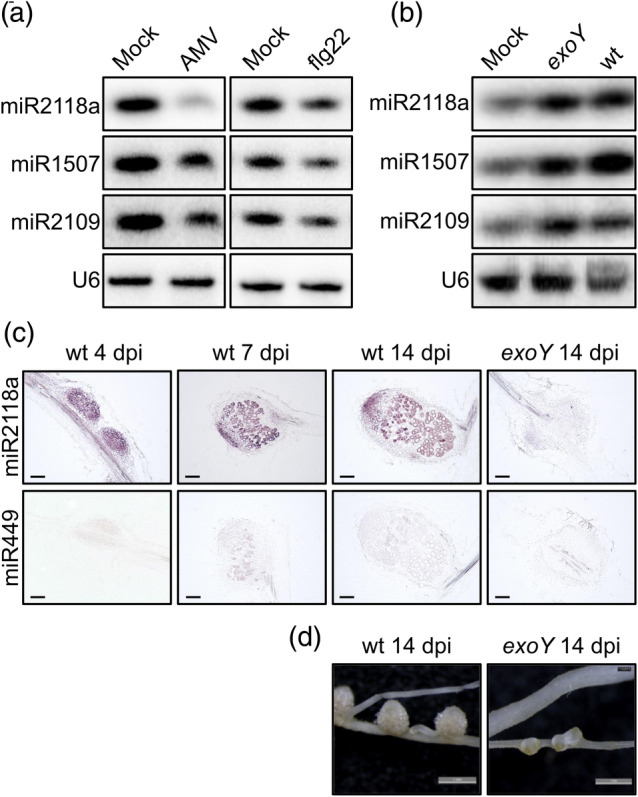
Expression of the NB‐LRR‐regulating miRNAs in Medicago truncatula roots and nodules during pathogen attack (a) and symbiosis (b, c). (a) Small RNA northern blot analysis showed the suppression of miR2118a, miR1507, and miR2109 expression in M. truncatula cotyledons 4 days post‐inoculation (dpi) with Alfalfa mosaic virus (AMV) compared to mock samples. The expression of the same miRNAs was analysed in M. truncatula seedlings 24 hr after treated with the synthetic flg22‐peptide for 6 hr. (b) Small RNA northern blot analysis of miR2118a, miR1507, and miR2109 in mock‐treated M. truncatula roots and roots inoculated with exoY mutant and wild‐type (wt) strains of Sinorhizobium meliloti at 1 dpi. The RNA was transferred to a membrane and probed with radiolabelled DNA oligonucleotides for miR2118a, miR1507, miR2109, and U6 snRNA as a loading control. (c) Spatial distribution of miR2118 in M. truncatula nodules. In situ hybridization of longitudinal sections of nodules either with 5′ digoxigenin (DIG)‐labelled LNA‐modified oligonucleotide probe for miR2118 or chicken miR449 as a negative control. Nodules were harvested at 4, 7, and 14 dpi with wt S. meliloti, and 14 dpi with exoY mutant of S. meliloti. (d) The images show the symbiotic phenotype of nodules induced by the wild‐type and the exoY mutant S. meliloti at 14 dpi. Scale bars: in panel c—100 μm; in panel d—1 mm
In plants, a 22‐amino acid sequence (flg22) of the conserved N‐terminal part of bacterial flagellin is a well‐characterized PAMP, known to activate PTI. Pre‐stimulation of plants with flg22 led to the activation of PTI and enhanced resistance against bacterial invaders (Zipfel et al., 2004). We tested the effect of the synthetic flg22 on the level of NB‐LRR‐regulating miRNAs (miR1507, miR2109, and miR2118). We immersed M. truncatula seedlings into media containing 1 μM flg22, and after incubation for 6 hr, the seedlings were transferred into a new media with no flg22. Following incubation for a further 24 hr, RNA was extracted from the seedlings and subjected to northern blot to analyse the expression level of the NB‐LRR‐regulating miRNAs. We found that the expression level of all three miRNAs decreased as a consequence of flg22 treatment (Figure 1a). This result shows that the activation of PTI with flg22 can trigger the plant defence signalling to suppress the level of NB‐LRR‐regulating miRNAs in M. truncatula.
The quick and precise regulation of plant immunity is not only paramount during pathogen attack, but it could also be very important during the recognition of beneficial microbes to establish symbiotic interaction with nitrogen‐fixing rhizobia. To test this hypothesis, we used the M. truncatula–Sinorhizobium meliloti symbiotic interaction as a model system to investigate the role of miRNAs in regulating NB‐LRR genes during symbiosis. In our experiments, we monitored the expression level of the three NB‐LRR‐regulating miRNAs at the early stage of the symbiotic interaction. Five‐day‐old M. truncatula seedlings were inoculated with wild‐type S. meliloti strain 1021 and RNA samples were collected from non‐inoculated roots (mock) and inoculated roots at 1 dpi. The collected RNA samples were subjected to sRNA northern blot, and the expression level of all three NB‐LRR‐regulating miRNAs was monitored (Figure 1b). We found that the basal expression of all three miRNAs increased at 1 dpi. During this experiment, we also inoculated M. truncatula seedlings with the exoY mutant derivative of S. meliloti deficient in succinoglycan (exopolysaccharide I) production (Reuber & Walker, 1993). The exoY mutant of S. meliloti is defective to induce infection thread formation and hence nodule primordia lacking rhizobia develop on the roots of M. truncatula (Jones et al., 2008). We collected RNA samples from non‐inoculated roots (mock) and inoculated roots at 1 dpi with the mutant rhizobia. The expression level of all three NB‐LRR‐regulating miRNAs was tested with sRNA northern blot. We found that the expression level of miR2118a, miR1507, and miR2109 was also increased in roots inoculated with S. meliloti exoY mutant, similarly what was found with wild‐type rhizobia (Figure 1b). These results indicate that the miRNAs regulating NB‐LRR genes could play a role during the establishment of symbiotic interaction with rhizobia by the post‐transcriptional downregulation of the NB‐LRR genes.
3.2. Promoters of Mtr‐MIR2118a,b,c, Mtr‐MIR2109, and Mtr‐MIR1507 genes contain both pathogen‐responsive and nodulation‐regulated motifs
The observed dual regulation of the miRNA levels raised the possibility that the promoters of these NB‐LRR‐regulating miRNAs respond both to pathogens and symbionts. We searched for known regulatory motifs in the promoters of the five miRNAs with PLACE (Higo et al., 1999) and found several sequence motifs that were associated either with pathogen response or nodulation, and other functions as well (Table S2). We wondered if there were motifs that were present in all the five promoters, therefore we analysed the sets of motifs and visualized the result in a Venn‐diagram (Figure 2a). There were 44 common sequence motifs present in the promoters of the five miRNAs, eight of which were associated either with pathogen‐response (i.e., W‐boxes) or nodulation (Table S2). For simplicity, we grouped these sequence motifs into two categories and visualized their location on the 2 kb sequence upstream to the TSS of the corresponding MIRNA genes (Figure 2b). The promoters of these MIRNA genes are diverse despite their similar mature miRNA sequences suggesting that they are regulated differently. For example, the promoter of the Mtr‐MIR2118a and Mtr‐MIR2118b genes contain many pathogen‐responsive motifs but only a few nodulation‐regulated motifs. This might explain our finding that the miR2118 level drops the most upon pathogen attack (Figure 1a) since the WRKY transcription factors that bind W‐boxes are often negative regulators of gene expression (Pandey & Somssich, 2009; Rushton, Somssich, Ringler, & Shen, 2010). Other motifs might be responsible for the integration of other external or internal signals, or for functions in tissues other than nodules.
Figure 2.
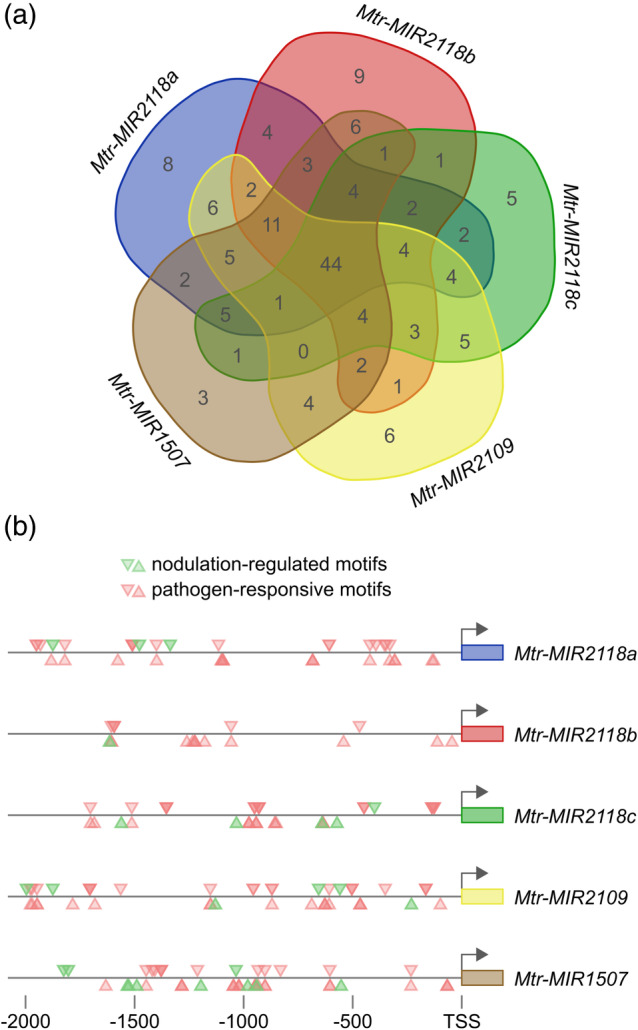
Promoter analysis of the NB‐LRR‐targeting MIRNA genes. Promoter analysis was conducted using the PLACE tool (Higo et al., 1999). The lists of motifs of the five MIRNA promoters were compared with each other using an online Venn‐diagram drawing tool (http://bioinformatics.psb.ugent.be/webtools/Venn/). There were 44 common elements in all the five promoters (a). From them, we selected the potential nodulation‐regulated motifs (NODCON2GM, OSE2ROOTNODULE) and the pathogen‐responsive elements (BIHD1OS, WBOXATNPR1, WRKY71OS, WBOXHVISO1, WBOXNTERF3, GT1GMSCAM4), and plotted them by position within the promoter (b). The downward‐ and upward‐pointing triangles mark the sites found on the forward and the reverse strands, respectively
3.3. The NB‐LRR‐targeting miRNAs are upregulated during symbiotic nodule development
The suppression of plant immunity could be important at two stages of the nitrogen‐fixing symbiosis. During the early events of the symbiotic interaction, leguminous plants control their defence responses by the rapid and transient activation of defence genes (Lohar et al., 2006). In addition, the involvement of an NB‐LRR disease protein controlling nodulation in a strain‐specific manner in soybean has been reported indicating the role of effector‐triggered immunity in the rhizobial symbiotic interaction (Yang et al., 2010). Also, during the later events, when the intracellular rhizobial invasion of nodule cells occurs, the regulation of plant immunity is also required. To monitor the presence of NB‐LRR‐targeting miRNAs during nodule development, we carried out in situ hybridizations of these miRNAs in developing nodules at different time points post‐inoculation with rhizobia.
We inoculated M. truncatula roots with wild‐type and exoY mutant S. meliloti and investigated the distribution of miR2118 in 4‐, 7‐, and 14‐day‐old nodules with in situ hybridization (Figures 1c and S1). At 4 dpi, strong expression of miR2118 was detected only in the symbiotic cells of nodule primordia. In the 7‐ and 14‐day‐old nodules, the miR2118 expression localized to the nodule meristem and the infected nodule cells. The presence of bacteria detected by the DNA‐binding SYTO™ 13 staining and the signal of miR2118 expression predominantly overlap in 7‐ and 14‐day‐old nodule sections indicating the elevated expression level of miR2118 in colonized nodule cells (Figure S1). This result also revealed that no signal of miR2118 expression was detected by in situ hybridizations in root tissue and non‐symbiotic cells of root nodules. Furthermore, we tested the expression pattern of miR2118 by in situ hybridization in M. truncatula nodules induced by the exoY mutant of S. meliloti. No expression of miR2118 was found in nodules devoid of any bacteria further suggesting the incidence of bacterial presence and miR2118 expression (Figure 1c). These results indicate that (i) cells colonized by rhizobia could have a lower NB‐LRR level than non‐infected cells and (ii) higher NB‐LRR gene expression level in non‐symbiotic nodule cells might block the colonization of these cells by rhizobia.
Next, we investigated the spatial distribution of all of the other NB‐LRR‐targeting miRNAs during different stages of nodule development by in situ hybridization. In 4‐day‐old nodules, the expression of all NB‐LRR‐targeting miRNAs (miR2118, miR2109, and miR1507) was detected and distributed uniformly in the developing nodules, and no expression of the miRNAs was detected in the non‐symbiotic tissues of the nodule (Figure S2a). In 7‐day‐old nodules, we were able to detect the expression of all three NB‐LRR‐targeting miRNAs (Figure S2b). Similarly, all three NB‐LRR‐targeting miRNAs were expressed in 14‐day‐old nitrogen‐fixing nodules, but they showed different spatial distribution. The miR2118 was evenly expressed in the nodule meristem and the different symbiotic zones of the 14‐day‐old nodules. The miR1507 expression showed a gradient expression predominantly localized to the meristem, the infection, and intermediate zones of the nodules, while a lower expression was found in the nitrogen fixation zone. The spatial expression pattern of miR2109 was mainly restricted to the apical part of the nodule (Figure S2c). We also investigated the spatial distribution of all of the other NB‐LRR‐targeting miRNAs in 14‐day‐old nodules elicited by the exoY mutant of S. meliloti and similarly to miR2118a, we did not detect the expression of any of these NB‐LRR‐targeting miRNAs in nodules devoid of rhizobia (Figure S2d). The nodules produced upon these infection processes are not able to fix nitrogen, because the exoY mutant S. meliloti bacteria are entrapped in the infection thread. Since the expression level of NB‐LRR‐targeting miRNAs is very low in these nodules lacking bacteria, the level of NB‐LRR proteins could be increased to a level that would contribute to the block of colonization of S. meliloti exoY mutant in these nodules.
3.4. miR2118, miR2109, and miR1507 silence NB‐LRR mRNAs
The M. truncatula genome encodes approx. 540 NB‐LRR genes and more than 60% of these genes can be targeted by the NB‐LRR‐targeting miRNAs or by the phasiRNAs that are produced by the action of the NB‐LRR‐targeting miRNAs (Zhai et al., 2011). The upregulation of all three NB‐LRR‐regulating miRNAs during symbiotic interactions in the nodules predicts that they decrease the level of the mRNAs of their target NB‐LRR genes. To confirm that the targeted NB‐LRR gene expressions are indeed downregulated, we selected two NB‐LRR genes targeted by each miRNA (miR2118, miR2109, and miR1507), respectively. We measured their expression level in symbiotic nodule by qRT‐PCR at 4, 7, and 14 dpi with S. meliloti. As it was expected, the expression level of all the six investigated miRNA‐targeted NB‐LRR gene was suppressed in the symbiotic nodules compared either to the symbiotically insensitive region of roots (Figure 3a) or to empty nodules elicited by S. meliloti exoY mutant (Figure 3b). However, the activity of NB‐LRR genes showed differential suppression and temporal variation (Figure 3). These results show that the three NB‐LRR‐regulating miRNAs control the expression level of the NB‐LRR genes and this regulation could contribute to the successful induction of symbiotic nodule development by facilitating the infection and the bacterial colonization of the root and the developing nodule tissue.
Figure 3.
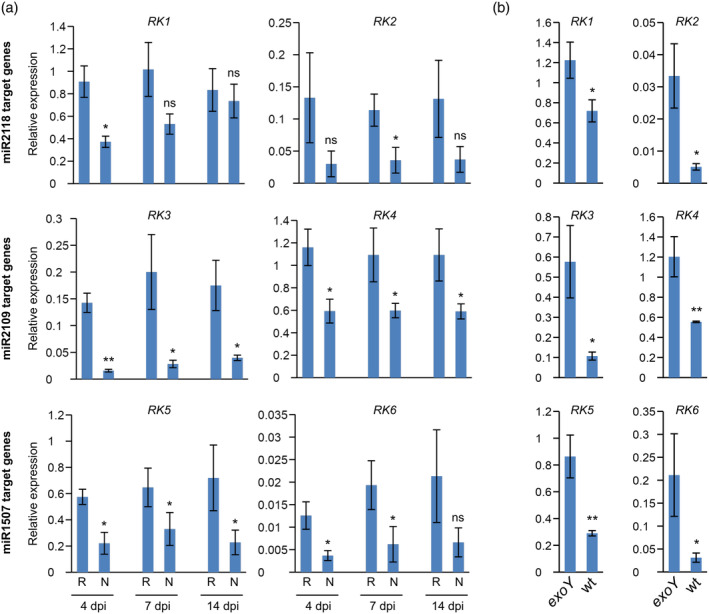
Expression of miRNA‐regulated NB‐LRR transcripts in Medicago truncatula nodules. (a) Roots were infected with Sinorhizobium meliloti and samples were taken from nodule‐less root parts (R) or nodules (N) at 4, 7, and 14 days post‐inoculation (dpi). (b) Roots were infected either with wt or exoY mutant S. meliloti. Samples were taken from nodules at 14 dpi. The expression levels were determined by RT‐qPCR. The expression level of target genes was normalized to the PTB (polypyrimidine tract‐binding‐like protein; Medtr3g090960.1) level in the same sample. RK1 = Medtr4g014990.1, RK2 = Medtr4g023400.1, RK3 = Medtr7g025250.1, RK4 = Medtr4g043630.1, RK5 = Medtr7g091110.1, RK6 = Medtr3g086070.1. The values are means of four independent experiments with SE. An unpaired, one‐tailed t test was performed to estimate statistical significance. ***p ≤ .001; **p ≤ .01; *p ≤ .05, ns = not significant
3.5. Symbiotic nodule number is regulated by the expression level of NB‐LRR‐regulating miRNAs
Our model predicts that if the reduced level of NB‐LRR gene expression is necessary to the formation and proper development of symbiotic nodules, the higher level of NB‐LRR gene expression will interfere with the process resulting in the reduction of nodule number. To test this hypothesis, we generated composite M. truncatula plants, using A. rhizogenes‐mediated hairy root transformation, expressing short sequences mimicking miRNA target sites (Franco‐Zorrilla et al., 2007; Yan et al., 2012) that lead to the degradation of targeted miR2118 and hence lowering the level of the NB‐LRR‐targeting miR2118. This observation was in line with a previous report (Fei et al., 2015) where the same short sequences mimicking miR2118 target sites (MIM2118) were reported to reduce the level of NB‐LRR‐targeting miRNAs. In MIM2118 transgenic roots, we checked the expression level of an NB‐LRR gene targeted by miR2118 and its expression level was elevated as expected (Figure S3a). Next, we examined the hairy roots of the composite plants for the characteristics of root growth and nodulation by S. meliloti. We also confirmed the downregulation of miR2118 in all of the transgenic roots by northern blot analysis (Figure 4a). We found that under our growing conditions, the lower level of miR2118 had no significant effects on root growth characteristics. To test the early infection events during the downregulation of miR2118, we counted the microcolonies (Figure S4a) and infection threads (Figure S4b) on MIM2118 transgenic roots at 5 dpi with S. meliloti. We found no significant differences in any early infection events compared to control roots. Nevertheless, when we scrutinized the nodule numbers in the transgenic hairy roots showing reduced miR2118 expression level 3 wpi with S. meliloti, we found a significant reduction in the number of mature nodules compared to the control roots transformed with empty vector (Figure 4b,c), irrespectively of which of the two mimicking constructs was expressed. The mature nodules were similar in size, infected with bacteria, and displayed normal morphology on both the MIM2118 transgenic and control roots (Figure S5). Based on these results, we concluded that the expression level of NB‐LRR‐regulating miR2118 correlated with the nodule number but had no effect on nodule morphology and rhizobial colonization.
Figure 4.
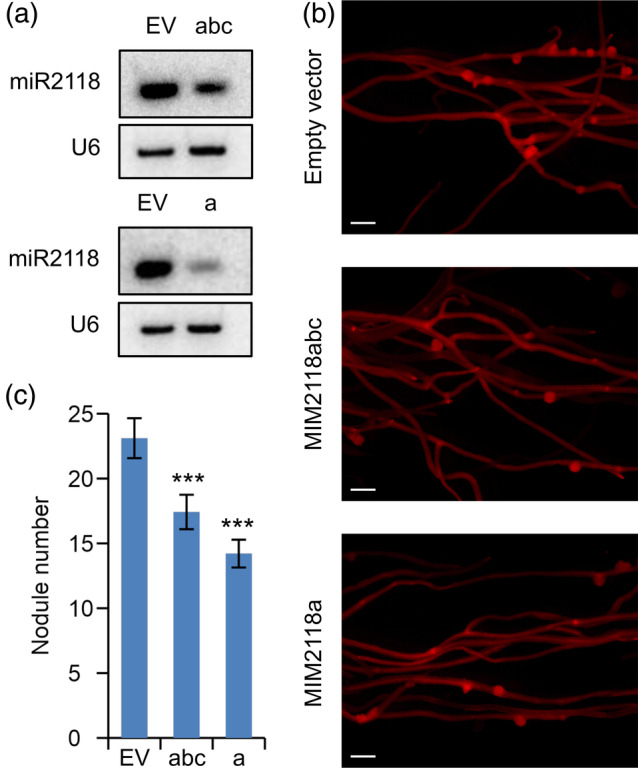
The decreased level of miR2118 induced by target mimicry constructs led to the reduction of nodule number on Medicago truncatula roots inoculated with Sinorhizobium meliloti. Target mimicry constructs reducing the expression level of miR2118a,b,c (MIM2118abc) and miR2118a (MIM2118a) respectively, controlled by the constitutively active Arabidopsis thaliana EF1α gene promoter were introduced into in M. truncatula roots using Agrobacterium rhizogenes‐mediated transformation. (a) Small RNA northern blot analysis of the same transgenic roots displayed in panel b showed a decreased abundance of mature miRNAs in transgenic lines at 3 weeks post‐inoculation (wpi). RNAs isolated from M. truncatula roots were separated on a 12% (wt/vol) polyacrylamide gel and subsequently transferred to a membrane and probed with radiolabelled DNA oligonucleotides for miR2118, and U6 snRNA as a loading control. (b) Representative images of nodulated transgenic roots 3 wpi with S. meliloti, transformed with MIM2118abc (abc) and MIM2118a (a) target mimicry constructs or with empty vectors (EV) identified by expressing the DsRed fluorescent protein. Scale bars: 1 mm. (c) Average nodule numbers of at least 45 transgenic roots were counted for each construct at 3 wpi with S. meliloti
To further confirm this conclusion, we tested the effect of symptomless virus infection on nodulation. We have shown that AMV infection of M. truncatula plants reduces the expression level of the NB‐LRR‐regulating miRNAs (Figure 1a). Furthermore, it has been previously demonstrated that the inhibition of the function of these miRNAs (miR2118, miR2109, and miR1507) leads to the upregulation of their target mRNAs (Fei et al., 2015). Based on these results, we speculated that AMV infection leads to the downregulation of the NB‐LRR‐regulating miRNAs that might cause the upregulation of its target NB‐LRR genes similarly to the target mimicry plants reported earlier (Fei et al., 2015). It is also important to mention that AMV infection, beside downregulating the NB‐LRR‐regulating miRNAs, does not induce any visible phenotypes of M. truncatula plants (Figure S6a). Therefore, the mock and the AMV infected plants were indistinguishable from each other by visual inspection. We infected five‐day‐old M. truncatula seedlings with AMV or mock inoculum, and after 7 days the plants were inoculated with S. meliloti. The nodule numbers of virus‐infected plants, verified by the presence of the viral AMV coat protein gene using northern blot (Figure S6b), were quantified at 7 and 10 dpi with S. meliloti. We found that virus‐infected plants developed a significantly reduced number of nodules compared to the mock‐inoculated control plants (Figure S7). This result might indicate that the downregulation of the NB‐LRR genes by miRNAs is important during nodule development.
To confirm this hypothesis, we generated the overexpressing constructs of the same miRNAs (miR2118OX, miR2109OX, and miR1507OX) that were shown previously (Fei et al., 2015) to increase the abundance of miR2118, miR2109, and miR1507 in M. truncatula roots, to test their effect on nodulation. We generated hairy root composite M. truncatula plants using A. rhizogenes‐mediated transformation system. Next, these plants were examined for root growth characteristics and nodulation by S. meliloti. We verified the overexpression of miRNAs in DsRed‐positive transgenic roots by northern blot analysis 4 wpi with rhizobia. The results confirmed that transgenic roots of the plants transformed by the three miRNA overexpressing constructs had significantly higher miRNA expression levels (Figure 5a). Under our growing conditions, overexpression of these miRNAs (miR2118, miR2109, and miR1507) did not cause any significant changes in the characteristics of root growth. Compared with empty vector transformed controls, the miRNA overexpressing transgenic lines showed no obvious differences in root length or lateral root density. In miR1507OX, miR2109OX, and miR2118OX transgenic roots we checked the expression level of NB‐LRR genes targeted by each of these miRNAs and their expression level was decreased, as it was expected (Figure S3). To test the early infection events during the overexpression of NB‐LRR‐regulating miRNAs, we quantified microcolonies and infection threads on the miRNA overexpressing (miR2118OX, miR2109OX, and miR1507OX) transgenic roots at 5 dpi with S. meliloti. We found significantly more microcolonies with all three miRNA overexpression constructs (Figure S4a) and a significantly increased number of infection threads on transgenic roots overexpressing miR2118 or miR2109 (Figure S4b) compared to empty vector transformed control roots. Next, we examined the nodule numbers in the transgenic hairy roots 4 weeks after S. meliloti inoculation. We found increased nodule numbers on transgenic hairy roots overexpressing miR2118, miR2109, and miR1507 compared to control empty vector transformed roots at 4 wpi with S. meliloti (Figure 5b,c). The mature nodules were similar in size, infected with bacteria, and morphologically looked normal in all three miRNA overexpressing transgenic and control roots (Figure S5). Our result is in line with a report showing that the overexpression of miR482, another member of the miR482/2118 superfamily increases soybean nodulation (Li et al., 2010). These results suggested that the NB‐LRR‐regulating miRNAs are important components of nodulation by regulating the expression level of NB‐LRR genes and, hence, promoting bacterial infection and development of nitrogen‐fixing symbiotic nodules.
Figure 5.
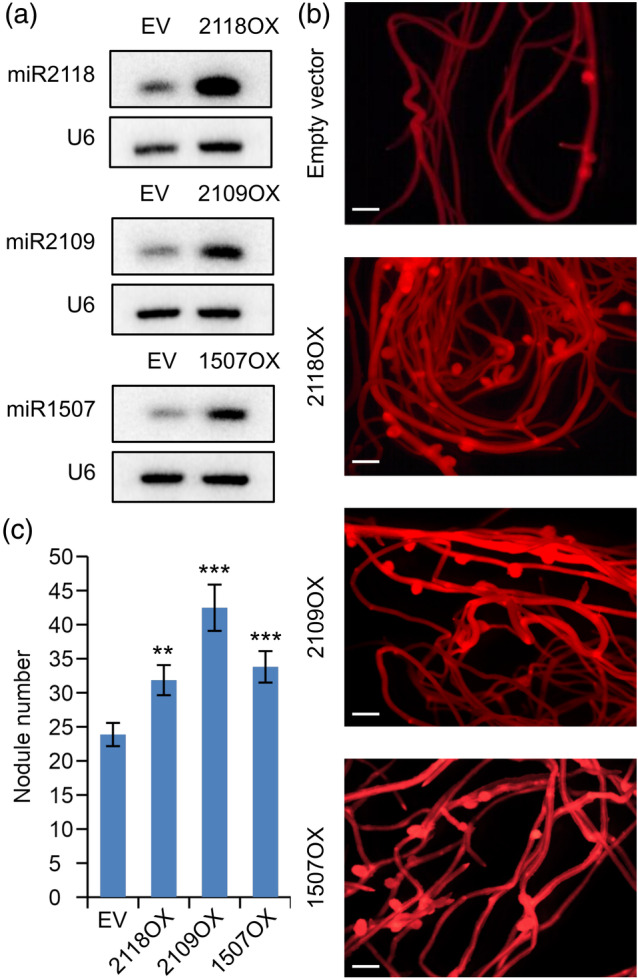
Constitutive overexpression of miRNAs in Medicago truncatula hairy roots generated with Agrobacterium rhizogenes‐mediated transformation resulted in an increased nodule number. NB‐LRR‐regulating miRNAs (miR2118, miR2109, and miR1507) were driven by the constitutive Arabidopsis thaliana EF1α gene promoter in M. truncatula transgenic roots. (a) Small RNA northern blot analysis of the same transgenic roots showed an increased abundance of mature miRNAs. RNAs isolated from M. truncatula transgenic roots were separated on a 12% (wt/vol) polyacrylamide gel. The RNA was transferred to a membrane and probed with radiolabelled DNA oligonucleotides for miR2118, miR2109, miR1507, and U6 snRNA as a loading control. (B) Representative images of nodulated transgenic roots, overexpressing miR2118 (2118OX), miR2109 (2109OX), and miR1507 (1507OX) or transformed with empty vector (EV), identified by expressing the DsRed fluorescent protein 4 weeks post‐inoculation (wpi) with Sinorhizobium meliloti. Scale bars: 1 mm. (c) Average nodule numbers of at least 15 transgenic roots for each construct were counted at 4 wpi with S. meliloti
4. CONCLUSIONS
The expression level of the three miRNA families (miR2118, miR2109, and miR1507) that regulate NB‐LRR mRNAs post‐transcriptionally is affected by the nature of plant–microbe interactions in legumes (see proposed model in Figure 6). In pathogen‐infected plants, the expression level of these NB‐LRR‐regulating miRNAs decreases and in line with previous findings (Li et al., 2012; Shivaprasad et al., 2012), this allows pathogen‐induced accumulation of NB‐LRR mRNAs which could result in elevated expression of NB‐LRR proteins and in the activation of defence responses. Interestingly, the interaction with the symbiotic microbial partner upregulates the expression level of the same set of miRNAs and thus affects the downregulation of the NB‐LRR defence genes in the developing nodule. This dual regulation may be attributed to the presence of both pathogen‐responsive and nodulation‐regulated sequence motifs in the promoter of the NB‐LRR‐regulating MIRNA genes (Figure 2b). The suppressed activity of these resistance genes facilitates both the infection and accommodation of legume roots by symbiotic bacteria finally resulting in the development of a higher number of nodules (Figure 6). The M. truncatula genome harbours more than 500 NB‐LRR genes and their post‐transcriptional regulation by miRNAs would reduce the fitness cost associated with these genes by keeping their basal level low in the absence of pathogen infection. The miRNA‐based post‐transcriptional regulation would allow the plant to fine‐tune the level of NB‐LRR transcripts depending on the character of the microbe interacting with the plant. Pathogen infection can decrease the basal level of the NB‐LRR‐regulating miRNAs resulting in efficient defence response. However, during symbiotic interaction, the increased expression level of the NB‐LRR‐regulating miRNAs lowers the transcript level of defence genes and therefore can provide a suitable niche to the symbiotic bacteria to infect the roots and colonize the developing nodules. Because the NB‐LRR‐targeting miRNAs can respond to both pathogens and symbiotic partners in M. truncatula, they could function as an elegant molecular switch to provide a fast and adequate response by increasing or decreasing the level NB‐LRR mRNAs according to the nature of plant–microbe interaction.
Figure 6.
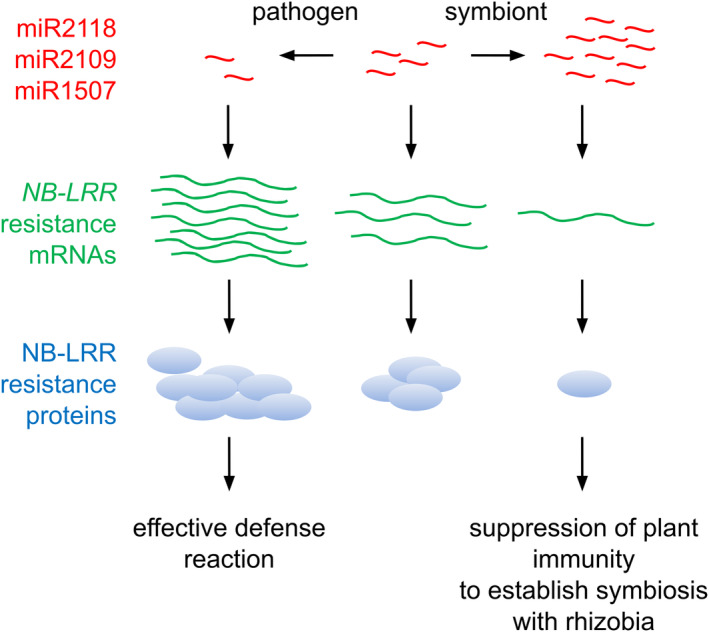
Model for the miRNA mediated regulation of NB‐LRR genes. The expression level of the three miRNA families (miR2118, miR2109, and miR1507) regulating NB‐LRR mRNAs post‐transcriptionally is modulated by the nature of plant–microbe interactions in legumes. In pathogen‐infected plants, the expression level of these miRNAs decreases advancing pathogen inducible expression of NB‐LRR proteins and thus activating defence mechanisms against pathogen attack. In contrast, the interaction with the nitrogen‐fixing symbiotic bacterial partner upregulates the expression level of the same set of miRNAs and resulting in the downregulation of the NB‐LRR defence genes. As a consequence, the plant's innate immunity is suppressed in the nodule during symbiosis. Red tildes represent miRNAs, green wavy lines represent NB‐LRR mRNAs, and light blue bubbles show NB‐LRR proteins
In M. truncatula, these miRNAs target at least 114 NB‐LRR genes that produce phasiRNAs that can target additional NB‐LRR genes and in this way, it is estimated that this regulating cascade can efficiently target at least 60% of the plant's NB‐LRR mRNA repertoire post‐transcriptionally (Fei et al., 2015; Zhai et al., 2011). The generation of phasiRNAs by these miRNAs, therefore, constitutes an efficient amplification system to modulate the expression of a large set of target mRNAs. By modulating the expression level of NB‐LRR‐regulating miRNAs, legume plants are able to fine‐tune their response to the microbial partner in a cost‐effective way. This mechanism along with other regulatory systems (Djordjevic, Mohd‐Radzman, & Imin, 2015; Imin, Patel, Corcilius, Payne, & Djordjevic, 2018; Mortier, Holsters, & Goormachtig, 2012) could contribute to the control of nodule numbers developed on legume roots. The spatial distributions of these miRNAs within the nodule furthermore indicate that they control the invasion of non‐symbiotic nodule cells by the symbiotic bacteria. However, the regulation of the NB‐LRR genes by a miRNA switch could have a fitness cost during special circumstances. Virus infection can trigger the accumulation of NB‐LRR mRNAs by downregulating the miRNAs, and as a consequence, the interaction with the symbiotic partner could be blocked or attenuated. In line with this hypothesis, a reduced number of developing nodules was observed on the roots of virus‐infected alfalfa (Medicago sativa), and these virus‐infected plants showed growth reduction when the nitrogen source was limiting (Ohki, Leps, & Hiruki, 1986).
Since miR482/2118 superfamily is present in many plant families (González, Müller, Baulcombe, & Puigdomènech, 2015), it is tempting to speculate that they might contribute to the regulation of both pathogenic and symbiotic interactions besides the legume family as well.
CONFLICT OF INTEREST
All authors declare that they have no conflict of interest. This article does not contain any studies with human participants or animals performed by any of the authors.
Supporting information
Table S1 Primers and probes used in this study.
Table S2 Results of promoter analysis.
Figure S1 Spatial distribution of miR2118 in Medicago truncatula nodules.
Figure S2 Spatial distribution of NB‐LRR‐regulating miRNAs in Medicago truncatula nodules inoculated with wild‐type (a)–(c) and exoY mutant (d) of Sinorhizobium meliloti.
Figure S3 Expression of miRNA‐regulated NB‐LRR transcripts in transgenic Medicago truncatula roots.
Figure S4 Quantification of infection events in miRNA overexpressing (OX) and miRNA target mimicry (MIM) Medicago truncatula hairy roots following inoculation with Sinorhizobium meliloti.
Figure S5 Nodulation phenotype of different transgenic Medicago truncatula hairy roots.
Figure S6 Mock and AMV infected Medicago truncatula plants at 14 and 31 dpi.
Figure S7 Downregulation of the expression of NB‐LRR‐targeting miRNAs leads to the repression of nodule number of Medicago truncatula infected with Sinorhizobium meliloti.
ACKNOWLEDGMENTS
The authors would like to thank Anikó Szigeti and Hajnalka Tolnainé Csákány for their valuable laboratory assistance during this project. We thank Ferhan Ayaydin (Cellular Imaging Laboratory, Biological Research Center, Szeged, Hungary) for performing confocal laser scanning microscopy. This work was supported by Hungarian National Research, Development and Innovation Office [K‐106068, K‐106170, K‐119701, K‐119652, K‐120300, K‐129547, K‐129171, PD‐116926]; A.S‐H. received the Bolyai Janos Fellowship (BO/00206/15) of the Hungarian Academy of Sciences.
Sós‐Hegedűs A, Domonkos Á, Tóth T, Gyula P, Kaló P, Szittya G. Suppression of NB‐LRR genes by miRNAs promotes nitrogen‐fixing nodule development in Medicago truncatula . Plant Cell Environ. 2020;43:1117–1129. 10.1111/pce.13698
Funding information Bolyai Janos Fellowship, Grant/Award Number: A.S‐H. received the Bolyai Janos Fellowship (BO/00206/15); Országos Tudományos Kutatási Alapok, Grant/Award Numbers: K‐106068; K‐106170; K‐119701; K‐119652; K‐129171, K‐129547, K‐120300, PD‐116926
Contributor Information
Péter Kaló, Email: kalo.peter@abc.naik.hu.
György Szittya, Email: szittya.gyorgy@abc.naik.hu.
REFERENCES
- Baksa, I. , Nagy, T. , Barta, E. , Havelda, Z. , Várallyay, É. , Silhavy, D. , … Szittya, G. (2015). Identification of Nicotiana benthamiana microRNAs and their targets using high throughput sequencing and degradome analysis. BMC Genomics, 16, 1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker, D. G. , Pfaff, T. , Moreau, D. , Groves, E. , Ruffel, S. , Lepetite, M. , … Journet, E.‐P. (2006). Growing M. truncatula: Choice of substrates and growth conditions In Mathesius U. & Journet E.‐P. (Eds.), Medicago truncatula handbook (pp. 1–26). Ardmore: The Samuel Roberts Noble Foundation. [Google Scholar]
- Bendahmane, A. , Kanyuka, K. , & Baulcombe, D. C. (1999). The Rx gene from potato controls separate virus resistance and cell death responses. The Plant Cell, 11, 781–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson‐Dernier, A. , Chabaud, M. , Garcia, F. , Bécard, G. , Rosenberg, C. , & Barker, D. G. (2001). Agrobacterium rhizogenes‐transformed roots of Medicago truncatula for the study of nitrogen‐fixing and endomycorrhizal symbiotic associations. Molecular Plant–Microbe Interactions: MPMI, 14, 695–700. [DOI] [PubMed] [Google Scholar]
- Cao, Y. , Halane, M. K. , Gassmann, W. , & Stacey, G. (2017). The role of plant innate immunity in the legume‐rhizobium symbiosis. Annual Review of Plant Biology, 68, 535–561. [DOI] [PubMed] [Google Scholar]
- Chabaud, M. , Lichtenzveig, J. , Ellwood, S. , Pfaff, T. , & Journet, E.‐P. (2006). Vernalization, crossings and testing for pollen viability In Mathesius U. & Journet E.‐P. (Eds.), Medicago truncatula handbook (pp. 1–13). USA: The Samuel Roberts Noble Foundation, Ardmore. [Google Scholar]
- Chávez Montes, R. A. , Rosas‐Cárdenas De, F. F. , De Paoli, E. , Accerbi, M. , Rymarquis, L. A. , Mahalingam, G. , … De Folter, S. (2014). Sample sequencing of vascular plants demonstrates widespread conservation and divergence of microRNAs. Nature Communications, 5, 1–15. [DOI] [PubMed] [Google Scholar]
- Chisholm, S. T. , Coaker, G. , Day, B. , & Staskawicz, B. J. (2006). Host‐microbe interactions: Shaping the evolution of the plant immune response. Cell, 124, 803–814. [DOI] [PubMed] [Google Scholar]
- Combier, J.‐P. , Frugier, F. , de Billy, F. , Boualem, A. , El‐Yahyaoui, F. , Moreau, S. , … Niebel, A. (2006). MtHAP2‐1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula . Genes & Development, 20, 3084–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, X. , & Zhao, P. X. (2011). psRNATarget: A plant small RNA target analysis server. Nucleic Acids Research, 39, W155–W159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries, S. , Kloesges, T. , & Rose, L. E. (2015). Evolutionarily dynamic, but robust, targeting of resistance genes by the miR482/2118 gene family in the Solanaceae. Genome Biology and Evolution, 7, 3307–3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, Y. , Wang, J. , Tung, J. , Liu, D. , Zhou, Y. , He, S. , … Li, F. (2018). A role for small RNA in regulating innate immunity during plant growth. PLoS Pathogens, 14, e1006756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, S.‐W. (2010). RNA‐based antiviral immunity. Nature Reviews. Immunology, 10, 632–644. [DOI] [PubMed] [Google Scholar]
- Ding, S.‐W. , & Voinnet, O. (2007). Antiviral immunity directed by small RNAs. Cell, 130, 413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djordjevic, M. A. , Mohd‐Radzman, N. A. , & Imin, N. (2015). Small‐peptide signals that control root nodule number, development, and symbiosis. Journal of Experimental Botany, 66, 5171–5181. [DOI] [PubMed] [Google Scholar]
- Domonkos, Á. , Kovács, S. , Gombár, A. , Kiss, E. , Horváth, B. , Kováts, G. Z. , … Kaló, P. (2017). NAD1 controls defense‐like responses in Medicago truncatula symbiotic nitrogen fixing nodules following rhizobial colonization in a BacA‐independent manner. Genes, 8, 387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei, Q. , Li, P. , Teng, C. , & Meyers, B. C. (2015). Secondary siRNAs from Medicago NB‐LRRs modulated via miRNA‐target interactions and their abundances. The Plant Journal: For Cell and Molecular Biology, 83, 451–465. [DOI] [PubMed] [Google Scholar]
- Franco‐Zorrilla, J. M. , Valli, A. , Todesco, M. , Mateos, I. , Puga, M. I. , Rubio‐Somoza, I. , … Paz‐Ares, J. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nature Genetics, 39, 1033–1037. [DOI] [PubMed] [Google Scholar]
- Gibson, A. H. , & Nutman, P. S. (1960). Studies on the physiology of nodule formation: With two figures in the text: A reappraisal of the effect of preplanting. Annals of Botany, 24, 420–433. [Google Scholar]
- González, V. M. , Müller, S. , Baulcombe, D. , & Puigdomènech, P. (2015). Evolution of NBS‐LRR gene copies among dicot plants and its regulation by members of the miR482/2118 superfamily of miRNAs. Molecular Plant, 8, 329–331. [DOI] [PubMed] [Google Scholar]
- Gourion, B. , Berrabah, F. , Ratet, P. , & Stacey, G. (2015). Rhizobium‐legume symbioses: The crucial role of plant immunity. Trends in Plant Science, 20, 186–194. [DOI] [PubMed] [Google Scholar]
- Higo, K. , Ugawa, Y. , Iwamoto, M. , & Korenaga, T. (1999). Plant cis‐acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Research, 27, 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imin, N. , Patel, N. , Corcilius, L. , Payne, R. J. , & Djordjevic, M. A. (2018). CLE peptide tri‐arabinosylation and peptide domain sequence composition are essential for SUNN‐dependent autoregulation of nodulation in Medicago truncatula . New Phytologist, 218, 73–80. [DOI] [PubMed] [Google Scholar]
- Jones, J. D. G. , & Dangl, J. L. (2006). The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Jones, K. M. , Sharopova, N. , Lohar, D. P. , Zhang, J. Q. , VandenBosch, K. A. , & Walker, G. C. (2008). Differential response of the plant Medicago truncatula to its symbiont Sinorhizobium meliloti or an exopolysaccharide‐deficient mutant. Proceedings of the National Academy of Sciences of the United States of America, 105, 704–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet, E.‐P. , De Carvalho‐Niebel, F. , Andriankaja, A. , Huguet, T. , & Barker, D. G. (2006). Rhizobial inoculation and nodulation of Medicago truncatula In Medicago truncatula handbook (pp. 1–6). Ardmore, USA: The Samuel Roberts Noble Foundation. [Google Scholar]
- Kis, S. , Salamon, P. , Kis, V. , & Szittya, G. (2017). Molecular characterization of a beet ringspot nepovirus isolated from begonia ricinifolia in Hungary. Archives of Virology, 162, 3559–3562. [DOI] [PubMed] [Google Scholar]
- Lelandais‐Brière, C. , Naya, L. , Sallet, E. , Calenge, F. , Frugier, F. , Hartmann, C. , … Crespi, M. (2009). Genome‐wide Medicago truncatula small RNA analysis revealed novel microRNAs and isoforms differentially regulated in roots and nodules. The Plant Cell, 21, 2780–2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F. , Pignatta, D. , Bendix, C. , Brunkard, J. O. , Cohn, M. M. , Tung, J. , … Baker, B. (2012). MicroRNA regulation of plant innate immune receptors. Proceedings of the National Academy of Sciences of the United States of America, 109, 1790–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Deng, Y. , Wu, T. , Subramanian, S. , & Yu, O. (2010). Misexpression of miR482, miR1512, and miR1515 increases soybean nodulation. Plant Physiology, 153, 1759–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohar, D. P. , Sharopova, N. , Endre, G. , Peñuela, S. , Samac, D. , Town, C. , … VandenBosch, K. A. (2006). Transcript analysis of early nodulation events in Medicago truncatula . Plant Physiology, 140, 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzihradszky, A. , Schneitz, K. , & Lohmann, J. U. (2014). Detection of mRNA expression patterns by nonradioactive in situ hybridization on histological sections of floral tissue In Riechmann J. L. & Wellmer F. (Eds.), Flower Development: Methods and Protocols. Methods in molecular biology (pp. 275–293). New York, NY: Springer New York. [DOI] [PubMed] [Google Scholar]
- Meyers, B. C. , Kaushik, S. , & Nandety, R. S. (2005). Evolving disease resistance genes. Current Opinion in Plant Biology, 8, 129–134. [DOI] [PubMed] [Google Scholar]
- Meyers, B. C. , Kozik, A. , Griego, A. , Kuang, H. , & Michelmore, R. W. (2003). Genome‐wide analysis of NBS‐LRR‐encoding genes in Arabidopsis. The Plant Cell, 15, 809–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael Weaver, L. , Swiderski, M. R. , Li, Y. , & Jones, J. D. G. (2006). The Arabidopsis thaliana TIR‐NB‐LRR R‐protein, RPP1A; protein localization and constitutive activation of defence by truncated alleles in tobacco and Arabidopsis. The Plant Journal, 47, 829–840. [DOI] [PubMed] [Google Scholar]
- Mortier, V. , Holsters, M. , & Goormachtig, S. (2012). Never too many? How legumes control nodule numbers. Plant, Cell & Environment, 35, 245–258. [DOI] [PubMed] [Google Scholar]
- Ohki, S. T. , Leps, W. T. , & Hiruki, C. (1986). Effects of alfalfa mosaic virus infection on factors associated with symbiotic N2 fixation in alfalfa. Canadian Journal of Plant Pathology, 8, 277–281. [Google Scholar]
- Pall, G. S. , & Hamilton, A. J. (2008). Improved northern blot method for enhanced detection of small RNA. Nature Protocols, 3, 1077–1084. [DOI] [PubMed] [Google Scholar]
- Pandey, S. P. , & Somssich, I. E. (2009). The role of WRKY transcription factors in plant immunity. Plant Physiology, 150, 1648–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecrix, Y. , Staton, S. E. , Sallet, E. , Lelandais‐Brière, C. , Moreau, S. , Carrère, S. , … Gamas, P. (2018). Whole‐genome landscape of Medicago truncatula symbiotic genes. Nature Plants, 4, 1017–1025. [DOI] [PubMed] [Google Scholar]
- Prüfer, K. , Stenzel, U. , Dannemann, M. , Green, R. E. , Lachmann, M. , & Kelso, J. (2008). PatMaN: Rapid alignment of short sequences to large databases. Bioinformatics, 24, 1530–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan, A. R. , & Hall, I. M. (2010). BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics, 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuber, T. L. , & Walker, G. C. (1993). Biosynthesis of succinoglycan, a symbiotically important exopolysaccharide of rhizobium meliloti. Cell, 74, 269–280. [DOI] [PubMed] [Google Scholar]
- Rogers, K. , & Chen, X. (2013). Biogenesis, turnover, and mode of action of plant microRNAs. The Plant Cell, 25, 2383–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton, P. J. , Somssich, I. E. , Ringler, P. , & Shen, Q. J. (2010). WRKY transcription factors. Trends in Plant Science, 15, 247–258. [DOI] [PubMed] [Google Scholar]
- Rychlik, W. (1993). Selection of primers for polymerase chain reaction. Methods in Molecular Biology (Clifton, N.J.), 15, 31–40. [DOI] [PubMed] [Google Scholar]
- Salamon, P. , Sós‐Hegedűs, A. , Gyula, P. , & Szittya, G. (2018). First report of the infection of alfalfa mosaic virus in Salvia sclarea in Hungary. Journal of Plant Pathology, 100(3), 608. [Google Scholar]
- Shivaprasad, P. V. , Chen, H.‐M. , Patel, K. , Bond, D. M. , Santos, B. A. C. M. , & Baulcombe, D. C. (2012). A microRNA superfamily regulates nucleotide binding site‐leucine‐rich repeats and other mRNAs. The Plant Cell, 24, 859–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, Y. , Li, H.‐G. , Wang, Y. , Li, S. , Wang, H.‐L. , Yu, L. , … Xia, X. (2018). Poplar miR472a targeting NBS‐LRRs is involved in effective defence against the necrotrophic fungus Cytospora chrysosperma . Journal of Experimental Botany, 69, 5519–5530. [DOI] [PubMed] [Google Scholar]
- Subramanian, S. , Fu, Y. , Sunkar, R. , Barbazuk, W. B. , Zhu, J.‐K. , & Yu, O. (2008). Novel and nodulation‐regulated microRNAs in soybean roots. BMC Genomics, 9, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szittya, G. , Salamon, P. , & Burgyán, J. (2000). The complete nucleotide sequence and synthesis of infectious RNA of genomic and defective interfering RNAs of TBSV‐P. Virus Research, 69, 131–136. [DOI] [PubMed] [Google Scholar]
- Taylor, R. S. , Tarver, J. E. , Hiscock, S. J. , & Donoghue, P. C. J. (2014). Evolutionary history of plant microRNAs. Trends in Plant Science, 19, 175–182. [DOI] [PubMed] [Google Scholar]
- Tsikou, D. , Yan, Z. , Holt, D. B. , Abel, N. B. , Reid, D. E. , Madsen, L. H. , … Markmann, K. (2018). Systemic control of legume susceptibility to rhizobial infection by a mobile microRNA. Science (New York, N.Y.), 362, 233–236. [DOI] [PubMed] [Google Scholar]
- Várallyay, E. , & Havelda, Z. (2011). Detection of microRNAs in plants by in situ hybridisation. Methods in Molecular Biology (Clifton, N.J.), 732, 9–23. [DOI] [PubMed] [Google Scholar]
- Yan, J. , Gu, Y. , Jia, X. , Kang, W. , Pan, S. , Tang, X. , … Tang, G. (2012). Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. The Plant Cell, 24, 415–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L. , Mu, X. , Liu, C. , Cai, J. , Shi, K. , Zhu, W. , & Yang, Q. (2015). Overexpression of potato miR482e enhanced plant sensitivity to Verticillium dahliae infection. Journal of Integrative Plant Biology, 57, 1078–1088. [DOI] [PubMed] [Google Scholar]
- Yang, S. , Tang, F. , Gao, M. , Krishnan, H. B. , & Zhu, H. (2010). R gene‐controlled host specificity in the legume‐rhizobia symbiosis. Proceedings of the National Academy of Sciences of the United States of America, 107, 18735–18740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai, J. , Jeong, D.‐H. , De Paoli, E. , Park, S. , Rosen, B. D. , Li, Y. , … Meyers, B. C. (2011). MicroRNAs as master regulators of the plant NB‐LRR defense gene family via the production of phased, trans‐acting siRNAs. Genes & Development, 25, 2540–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Xia, R. , Kuang, H. , & Meyers, B. C. (2016). The diversification of plant NBS‐LRR defense genes directs the evolution of MicroRNAs that target them. Molecular Biology and Evolution, 33, 2692–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel, C. , & Oldroyd, G. E. D. (2017). Plant signalling in symbiosis and immunity. Nature, 543, 328–336. [DOI] [PubMed] [Google Scholar]
- Zipfel, C. , Robatzek, S. , Navarro, L. , Oakeley, E. J. , Jones, J. D. G. , Felix, G. , & Boller, T. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature, 428, 764–767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Primers and probes used in this study.
Table S2 Results of promoter analysis.
Figure S1 Spatial distribution of miR2118 in Medicago truncatula nodules.
Figure S2 Spatial distribution of NB‐LRR‐regulating miRNAs in Medicago truncatula nodules inoculated with wild‐type (a)–(c) and exoY mutant (d) of Sinorhizobium meliloti.
Figure S3 Expression of miRNA‐regulated NB‐LRR transcripts in transgenic Medicago truncatula roots.
Figure S4 Quantification of infection events in miRNA overexpressing (OX) and miRNA target mimicry (MIM) Medicago truncatula hairy roots following inoculation with Sinorhizobium meliloti.
Figure S5 Nodulation phenotype of different transgenic Medicago truncatula hairy roots.
Figure S6 Mock and AMV infected Medicago truncatula plants at 14 and 31 dpi.
Figure S7 Downregulation of the expression of NB‐LRR‐targeting miRNAs leads to the repression of nodule number of Medicago truncatula infected with Sinorhizobium meliloti.


