Many transferable quinolone resistance mechanisms have been identified in Gram-negative bacteria. The plasmid-encoded 65-amino-acid-long ciprofloxacin-modifying enzyme CrpP was recently identified in Pseudomonas aeruginosa isolates. We analyzed a collection of 100 clonally unrelated and multidrug-resistant P. aeruginosa clinical isolates, among which 46 were positive for crpP-like genes, encoding five CrpP variants conferring variable levels of reduced susceptibility to fluoroquinolones.
KEYWORDS: Pseudomonas aeruginosa, CrpP, fluoroquinolones, ciprofloxacin
ABSTRACT
Many transferable quinolone resistance mechanisms have been identified in Gram-negative bacteria. The plasmid-encoded 65-amino-acid-long ciprofloxacin-modifying enzyme CrpP was recently identified in Pseudomonas aeruginosa isolates. We analyzed a collection of 100 clonally unrelated and multidrug-resistant P. aeruginosa clinical isolates, among which 46 were positive for crpP-like genes, encoding five CrpP variants conferring variable levels of reduced susceptibility to fluoroquinolones. These crpP-like genes were chromosomally located as part of pathogenicity genomic islands.
INTRODUCTION
Bacterial resistance to quinolones and fluoroquinolones mainly results from decreased outer membrane permeability, overexpression of efflux pumps, and/or amino acid substitutions in the sequences of the chromosomally encoded topoisomerases that are the targets of these antibiotics (1). However, a series of acquired and transferable mechanisms have been identified in Gram-negative bacteria, including quinolone target protection conferred by Qnr-type pentapeptide proteins, enzymatic inactivation of the antibiotic through the action of the AAC(6′)-Ib-cr acetyltransferase (coresistance to aminoglycosides), and plasmid-encoded efflux pumps, such as OqxAB or QepA (2, 3). Nevertheless, the latter horizontally transferred resistance determinants have rarely been identified in Pseudomonas aeruginosa, in which resistance to fluoroquinolones is mainly driven by mutations in topoisomerases, efflux overproduction, and outer membrane permeability defects (4–6).
Recently, a plasmid-encoded 65-amino-acid-long ciprofloxacin-modifying enzyme, CrpP (renamed CrpP1 here for clarity), was identified in a single clinical P. aeruginosa isolate from Mexico (7). The crpP1 gene was located in a 123-kb conjugative plasmid (pUM505) coharboring virulence and heavy-metal resistance genes (8). CrpP confers reduced susceptibility (7.5-fold) to ciprofloxacin (CIP) once produced in Escherichia coli J53-3. In P. aeruginosa, this enzyme confers decreased susceptibility (4-fold decrease of MIC values) to CIP, norfloxacin, and moxifloxacin but only when transferring the crpP1-positive pUM505 natural plasmid, since expression of crpP1 from a recombinant plasmid once transformed in P. aeruginosa did not have any significant effect. Very recently, homologues of the crpP1 gene that shared low-level nucleotide identity (corresponding proteins being 10% to 43% identical) were identified among Enterobacterales from Mexico, with corresponding proteins exhibiting conserved residues Thr8, Asp9, Lys33, Gly34, and Cys40; these amino acids are possibly key residues in respect to the CrpP activity (9). It was demonstrated that CrpP1 exhibits ATP-dependent phosphorylation activity toward CIP, explaining the decreased antibacterial activity of this antibiotic (7). Considering the single report of the crpP1 gene in P. aeruginosa, our aim was to evaluate the dissemination of this novel resistance determinant.
A collection of 100 P. aeruginosa isolates recovered in France and Switzerland during 2000 to 2015 were selected for this study, including extended-spectrum β-lactamase (n = 17) and carbapenemase (n = 73) producers. A PCR approach was used for screening the crpP gene among these isolates, using designed primers crpP-F (5′-AGAGCGGGATCGATCAGAAAT-3′) and crpP-R (5′-ACGAGGTGCAGTGTGTCAAA-3′), followed by sequencing of the corresponding amplicons (Microsynth, Balgach, Switzerland). A total of 46 isolates were positive for crpP-like genes, and 5 CrpP variants were identified, namely, CrpP1 (referring to the sequence previously reported from Mexico [7]) and 4 additional variants exhibiting few amino acid substitutions: CrpP2 (Lys4Arg and Gly7Asp), CrpP3 (Gly7Asp), CrpP4 (Gly7His, Phe16Tyr and Ile26Leu), and CrpP5 (Lys4Arg, Gly7Asp and Phe55Tyr).
Multilocus sequencing typing (MLST) was performed, and sequence type assignation was done by using the MLST database for P. aeruginosa (https://pubmlst.org/paeruginosa/). Sequence typing of all the crpP-positive isolates showed the most prevalent types to be ST235, ST111, ST233, and ST273 (24%, 17.4%, 6.5%, and 4.3%, respectively).
To evaluate the impact of the different CrpP enzymes on quinolone susceptibility, a series of cloning and expression experiments were performed. The crpP-like genes were cloned into shuttle vector pUCp24 using primers crpP-XbaI-F (5′-TCTAGAAGAGCGGGATCGATCAGAAAT-3′) and crpP-SacI-R (5′-GAGCTCACGAGGTGCAGTGTGTCAAA-3′) and then transformed in both E. coli TOP10 and P. aeruginosa PAO1 that did not harbor any crpP gene, as described (10). MICs were determined for the different recombinant strains by the broth microdilution method using cation-adjusted Mueller-Hinton (MH) broth. This showed similar decreased susceptibility to CIP for the E. coli strains producing CrpP2, CrpP3, CrpP4, and CrpP5 but not CrpP1. Indeed, CrpP1 did not decrease the susceptibility of E. coli TOP10 to CIP, even though it was reported to do so in a previous study that used E. coli J53-3 as the recipient strain (7). Of note, CrpP1 did not have as much effect on quinolones as on the other variants tested. In particular, production of CrpP1 in E. coli TOP10 did not decrease the susceptibility to levofloxacin (LEV) and CIP. MIC values were 7.5-fold higher upon production of CrpP2, CrpP3, and CrpP5 and 4-fold higher with CrpP4. However, only a 2-fold increase in MIC of CIP was observed for the P. aeruginosa recombinant strain producing CrpP2, but MICs remained unchanged for the other P. aeruginosa recombinant strains. Instead, a 3.75-fold increase in the MIC of LEV was observed once CrpP2, CrpP3, CrpP4, and CrpP5 were produced in E. coli isolates, although no change was observed with CrpP1 (Table 1). The MIC of LEV was increased by 2-fold once CrpP2, CrpP3, and CrpP5 were produced in P. aeruginosa. P. aeruginosa PAO1isolates expressing CrpP2, CrpP3, and CrpP4 were resistant to LEV according to the 2020 EUCAST breakpoint value for resistance (>1 μg/ml).
TABLE 1.
MICs of fluoroquinolones for the P. aeruginosa and E. coli recombinant strains examined in this study
| Antibiotic | MIC (μg/ml) in: |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Escherichia coli TOP10 |
Pseudomonas aeruginosa PAO1 |
|||||||||||
| TOP10 | CrpP variant |
PAO1 | CrpP variant |
|||||||||
| CrpP1 | CrpP2 | CrpP3 | CrpP4 | CrpP5 | CrpP1 | CrpP2 | CrpP3 | CrpP4 | CrpP5 | |||
| Ciprofloxacin | 0.002 | 0.002 | 0.015 | 0.015 | 0.008 | 0.015 | 0.25 | 0.25 | 0.5 | 0.25 | 0.25 | 0.25 |
| Levofloxacin | 0.004 | 0.004 | 0.015 | 0.015 | 0.015 | 0.015 | 1 | 1 | 2 | 2 | 2 | 1 |
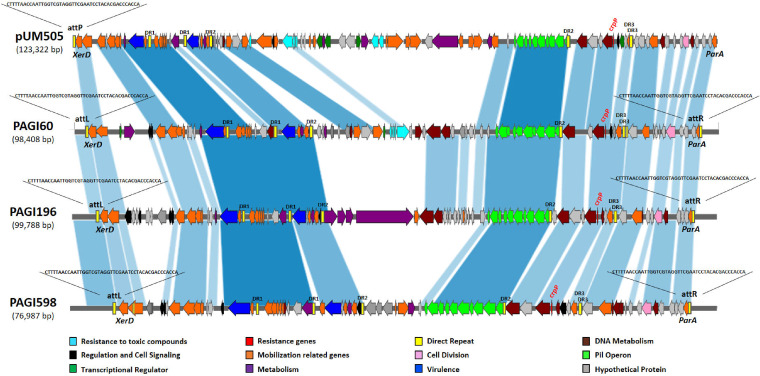
To identify the genetic location of the crpP-like genes in the positive isolates, we performed whole-genome sequencing (WGS) on 7 of the 46 crpP-positive isolates as representatives of the different ST types. DNAs were extracted by use of a DNeasy blood and tissue kit using the QIAcube apparatus (Qiagen). Genomic libraries were assessed using a Nextera XT library preparation kit (Illumina, Inc., San Diego, CA), and sequencing was performed using an Illumina MiniSeq system with 300-bp paired-end reads and a coverage of 50 times. The generated FastQ data were compiled and analyzed using the CLC Genomic Workbench (version 7.5.1; CLC Bio, Aarhus, Denmark). Reads were assembled de novo with automatic bubble and word size, and contigs with a minimum contig length of 800 nucleotides were generated using the mapping-mode map reads back to contigs. Analysis of WGS data showed that the crpP-like genes were always located on the chromosome of the corresponding isolates within large fragments (∼70 to 100 kb) bracketed by two direct repeats named attL and attR of 45 bp each. At both extremities of this whole fragment, tyrosine recombinase (XerD) and ParA protein-encoding genes were identified, respectively. These two proteins are known to be linked to the mobilization and integration, respectively, of pathogenicity genomic islands (PAGI) (11). The presence of these genes together with the presence of the attL and attR direct repeats bracketing the entire DNA fragment strongly suggested that the crpP genes were located into chromosomal PAGI (Fig. 1). The PAGIs presented one conserved region, encompassing the crpP gene, the pil operon, a series of virulence genes, and mobilization-related genes. In addition, they actually contained a variable region, in which the same genes encoding resistance to toxic compounds as originally reported in plasmid pUM505 were detected in two isolates (R60 and R90).
FIG 1.
Sequence comparison between pUM505 and PAGIs (60, 196, and 598). Arrows and arrowheads indicate open reading frames and their direction of transcription. Shading between bars indicates portions of sequences that align with each other. The map was drawn using Kablammo software (http://kablammo.wasmuthlab.org/).
To evaluate whether the PAG1-like structures are functional in terms of mobilization, PCR assays were performed to determine their putative circularization. Outward primers designed from the sequences located at the extremities of the structures, i.e., PAGIact-F (5′-ATTTCCCTACACCACCCTTG-3′) and PAGIact-R (5′-TTCGAGCAAAGAGTGCTGTT-3′), were used for that purpose. Positive PCR results were obtained, showing that a circular form of these PAGI-like elements was present and confirming their functionality as mobile structures.
Several features identified here were also found in the sequence of plasmid pUM505. In pUM505, a single copy of the attL sequence was present (corresponding sequence, 15,649 to 15,693 bp); and the parA gene (14,687 to 15,553 bp) and xerD integrase gene (16,030 to 17,331 bp), both encoding proteins necessary for the mobilization of pathogenic islands, were also identified. Furthermore, because the so-called pUM505 plasmid did not harbor any replicase gene, we hypothesize that plasmid pUM505 was actually a product of the circularized form of this newly described pathogenic island in the original description (8).
In summary, we report the spread of crpP-like genes in a large series of P. aeruginosa isolates from Europe. CrpP-like enzymes, despite not conferring clinical resistance per se but only reduced susceptibility to fluoroquinolones once produced in E. coli isolates, may be considered transferable mechanisms of fluoroquinolone resistance that may further be transferred to other strains. Acquisition of these genes found in clonally unrelated P. aeruginosa strains was likely mediated by the acquisition of a PAGI-like element. While this work was in progress, a study performed by Ruiz (12) revealed that CrpP-like-encoding genes were frequently identified within P. aeruginosa genomes according to an in silico analysis of sequences available from GenBank, which is in line with our observations.
Interestingly, we show here that susceptibility to fluoroquinolone molecules might vary slightly depending on the nature of the CrpP protein, but without conferring a high level of resistance. Supporting our data, modifications in conserved amino acids within the CrpP protein sequence were recently reported to affect their respective enzymatic activities (13). Furthermore, apart from their impact on fluoroquinolone susceptibility, acquisition of the PAGI-like elements in P. aeruginosa is of significant concern, considering that many of them coharbor genes encoding resistance to heavy metals and encode genes for virulence factors (14, 15).
Accession number(s).
The sequences of PAGIs have been deposited in the GenBank database under accession numbers MT074669 (PAGI-R60), MT074670 (PAGI-R90), MT074671 (PAGI-R104), MT074672 (PAGI-R135), MT074673 (PAGI-R184), MT074674 (PAGI-R196), and MT074675 (PAGI-R598). GenBank accession numbers for CrpP variants are WP_033179079 (CrpP1), WP_031644039 (CrpP2), WP_003109353 (CrpP3), WP_071533909 (CrpP4) and WP_071580329 (CrpP5).
ACKNOWLEDGMENTS
This work was partly supported by the University of Fribourg, the Swiss National Science Foundation (project PNR72-40AR40_173686), and INSERM, Paris, France.
REFERENCES
- 1.Hooper DC, Jacoby GA. 2016. Topoisomerase inhibitors: fluoroquinolone mechanisms of action and resistance. Cold Spring Harb Perspect Med 6:a025320. doi: 10.1101/cshperspect.a025320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martinez-Martinez L, Pascual A, Jacoby GA. 1998. Quinolone resistance from a transferable plasmid. Lancet 351:797–799. doi: 10.1016/S0140-6736(97)07322-4. [DOI] [PubMed] [Google Scholar]
- 3.Ruiz J, Pons MJ, Gomes C. 2012. Transferable mechanisms of quinolone resistance. Int J Antimicrob Agents 40:196–203. doi: 10.1016/j.ijantimicag.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 4.Rehman A, Patrick WM, Lamont IL. 2019. Mechanisms of ciprofloxacin resistance in Pseudomonas aeruginosa: new approaches to an old problem. J Med Microbiol 68:1–10. doi: 10.1099/jmm.0.000873. [DOI] [PubMed] [Google Scholar]
- 5.El-Badawy MF, Alrobaian MM, Shohayeb MM, Abdelwahab SF. 2019. Investigation of six plasmid-mediated quinolone resistance genes among clinical isolates of pseudomonas: a genotypic study in Saudi Arabia. Infect Drug Resist 12:915–923. doi: 10.2147/IDR.S203288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamed SM, Elkhatib WF, El-Mahallawy HA, Helmy MM, Ashour MS, Aboshanab K. 2018. Multiple mechanisms contributing to ciprofloxacin resistance among Gram negative bacteria causing infections to cancer patients. Sci Rep 8:12268. doi: 10.1038/s41598-018-30756-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chávez-Jacobo VM, Hernández- Ramírez KC, Romo-Rodríguez P, Pérez-Gallardo RV, Campos-García J, Gutiérrez-Corona JF, García-Merinos JP, Meza-Carmen V, Silva-Sánchez J, Ramírez-Díaz MI. 2018. CrpP is a novel ciprofloxacin-modifying enzyme encoded by the Pseudomonas aeruginosa pUM505 plasmid. Antimicrob Agents Chemother 62:e02629-17. doi: 10.1128/AAC.02629-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramírez-Díaz MI, Díaz-Magaña A, Meza-Carmen V, Johnstone L, Cervantes C, Rensing C. 2011. Nucleotide sequence of Pseudomonas aeruginosa conjugative plasmid pUM505 containing virulence and heavy-metal resistance genes. Plasmid 66:7–18. doi: 10.1016/j.plasmid.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 9.Chavez-Jacobo VM, Hernández- Ramírez KC, Silva-Sanchez J, Garza-Ramos U, Barrios-Camacho H, Ortiz-Alvarado R, Cervantes C, Meza-Carmen V, Ramírez-Díaz MI. 2019. Prevalence of the crpP gene conferring decreased ciprofloxacin susceptibility in enterobacterial clinical isolates from Mexican hospitals. J Antimicrob Chemother 74:1253–1259. doi: 10.1093/jac/dky562. [DOI] [PubMed] [Google Scholar]
- 10.Ortiz de la Rosa JM, Nordmann P, Poirel L. 2019. ESBLs and resistance to ceftazidime/avibactam and ceftolozane/tazobactam combinations in Escherichia coli and Pseudomonas aeruginosa. J Antimicrob Chemother 74:1934–1939. doi: 10.1093/jac/dkz149. [DOI] [PubMed] [Google Scholar]
- 11.Qiu X, Gurkar AU, Lory S. 2006. Interstrain transfer of the large pathogenicity island (PAPI-1) of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 103:19830–19835. doi: 10.1073/pnas.0606810104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruiz J. 2019. CrpP, a passenger or a hidden stowaway in the Pseudomonas aeruginosa genome? J Antimicrob Chemother 74:3397–3399. doi: 10.1093/jac/dkz316. [DOI] [PubMed] [Google Scholar]
- 13.Chávez-Jacobo VM, Garcia Merinos JP, López Y, Meza-Carmen V, Ramírez-Diaz MI. 2020. Identification of essential residues for ciprofloxacin resistance of ciprofloxacin-modifying enzyme (CrpP) of pUM505. Microbiology doi: 10.1099/mic.0.000889. [DOI] [PubMed] [Google Scholar]
- 14.Hernández-Ramírez KC, Chávez-Jacobo VM, Valle-Maldonado MI, Patiño-Medina JA, Díaz-Pérez SP, Jácome-Galarza IE, Ortiz-Alvarado R, Meza-Carmen V, Ramírez-Díaz MI. 2017. Plasmid pUM505 encodes a toxin-antitoxin system conferring plasmid stability and increased Pseudomonas aeruginosa virulence. Microb Pathog 112:259–268. doi: 10.1016/j.micpath.2017.09.060. [DOI] [PubMed] [Google Scholar]
- 15.Hernández-Ramírez KC, Reyes-Gallegos RI, Chávez-Jacobo VM, Díaz-Magaña A, Meza-Carmen V, Ramírez-Díaz MI. 2018. A plasmid-encoded mobile genetic element from Pseudomonas aeruginosa that confers heavy metal resistance and virulence. Plasmid 98:15–22. doi: 10.1016/j.plasmid.2018.07.003. [DOI] [PubMed] [Google Scholar]