The off-label use of third-generation cephalosporins (3GCs) during in ovo vaccination or vaccination of newly hatched chicks has been a common practice worldwide. CMY-2-producing Escherichia coli strains have been disseminated in broiler chicken production. The objective of this study was to determine the epidemiological linkage of blaCMY-2-positive plasmids among broilers both within and outside Japan, because the grandparent stock and parent stock were imported into Japan.
KEYWORDS: blaCMY-2, broiler, cephalosporin, off-label use, plasmids
ABSTRACT
The off-label use of third-generation cephalosporins (3GCs) during in ovo vaccination or vaccination of newly hatched chicks has been a common practice worldwide. CMY-2-producing Escherichia coli strains have been disseminated in broiler chicken production. The objective of this study was to determine the epidemiological linkage of blaCMY-2-positive plasmids among broilers both within and outside Japan, because the grandparent stock and parent stock were imported into Japan. We examined the whole-genome sequences of 132 3GC-resistant E. coli isolates collected from healthy broilers during 2002 to 2014. The predominant 3GC resistance gene was blaCMY-2, which was detected in the plasmids of 87 (65.9%) isolates. The main plasmid replicon types were IncI1-Iγ (n = 21; 24.1%), IncI (n = 12; 13.8%), IncB/O/K/Z (n = 28; 32.2%), and IncC (n = 22; 25.3%). Those plasmids were subjected to gene clustering, network analyses, and plasmid multilocus sequence typing (pMLST). The chromosomal DNA of isolates was subjected to MLST and single-nucleotide variant (SNV)-based phylogenetic analysis. MLST and SNV-based phylogenetic analysis revealed high diversity of E. coli isolates. The sequence type 429 (ST429) cluster harboring blaCMY-2-positive IncB/O/K/Z was closely related to isolates from broilers in Germany harboring blaCMY-2-positive IncB/O/K/Z. pST55-IncI, pST12-IncI1-Iγ, and pST3-IncC were prevalent in western Japan. pST12-IncI1-Iγ and pST3-IncC were closely related to plasmids detected in E. coli isolates from chickens in North America, whereas 26 IncB/O/K/Z types were related to those in Europe. These data will be useful to reveal the whole picture of transmission of CMY-2-producing bacteria inside and outside Japan.
INTRODUCTION
Third-generation cephalosporins (3GCs) are used as therapeutic agents for bacterial infectious diseases in humans. The World Health Organization (WHO) categorizes 3GCs as critically important antibiotics (1). The emergence and proliferation of 3GC-resistant bacteria in food-producing animals are global public health concerns (2).
The off-label use of ceftiofur (CTF) during in ovo vaccination or vaccination of newly hatched chicks has been a common practice in the poultry industry worldwide (3–6). Also, in Japan, the off-label use of CTF, which is not approved for broiler chicken and human use, was adopted at some hatcheries until it was voluntarily discontinued by farmers’ associations around March 2012 (3). The percentage of CTF-resistant Escherichia coli bacteria isolated from healthy broilers was around 3.0% in 2000 to 2002, which increased to approximately 10% in 2004. Subsequently, the percentage of CTF-resistant E. coli increased to between 15.0% and 20.0% in 2011 (3). However, the percentage of CTF-resistant E. coli decreased immediately after the off-label use of CTF was discontinued in 2012 (3). Hence, the off-label use of CTF was associated with cephalosporin resistance (7).
In E. coli, resistance to 3GC is mostly mediated by the extended-spectrum β-lactamases (ESBLs), or AmpC β-lactamases. AmpC β-lactamase-producing E. coli has been detected in food-producing animals, especially chickens (3, 8–11). Some researchers have reported that AmpC β-lactamase was found in Salmonella from chickens (12, 13). In fact, there was a report that the AmpC β-lactamase gene might be shared between E. coli and Salmonella (14). Moreover, AmpC β-lactamase-producing E. coli isolates were found at all levels of broiler production (15) and were isolated from chicken meat (16). Globally, the most common plasmid-mediated AmpC β-lactamase is CMY-2 (11). In Japan, blaCMY-2 was predominantly detected in 3GC-resistant E. coli isolates from healthy broilers (10). CMY-2, which can hydrolyze 3GC, is a plasmid-mediated AmpC β-lactamase belonging to class C of the Ambler classification. The blaCMY-2 gene is usually found on the transmissible plasmids of different replicon types (IncK, IncI1, IncA/C, IncF, IncI2, and IncL/M) (17–19).
In Japan, grandparent stocks and parent stocks have been imported from Europe and North America. Antibiotic-resistant bacteria may enter the broiler production chain due to transport and trade of eggs and chickens among many different countries (15). Furthermore, CMY-2-producing isolates were also detected in humans (17, 20). How CMY-2-producing bacteria from broilers in Japan are related to those in other countries and those from humans is still unknown.
In this study, to investigate the distribution and molecular characteristics of blaCMY-2-positive plasmids and to determine the global epidemiological linkage of blaCMY-2-positive plasmids, whole-genome sequence (WGS) analysis was performed for CMY-2-producing E. coli isolates from healthy broilers in Japan. Additionally, we compared the blaCMY-2-positive plasmid sequences with the plasmid sequences available in the public databases, including those of the human isolates.
RESULTS AND DISCUSSION
Pulsed-field gel electrophoresis with S1 nuclease (S1 PFGE) was performed to separate the chromosomal and plasmid DNAs of 132 3GC-resistant E. coli isolates from healthy broilers in Japan before the chromosomal and plasmid DNAs were individually analyzed using whole-genome sequencing. The total contig length of each plasmid was nearly in agreement with the estimated band size observed in the S1 PFGE, as in a previous study (21). Draft genome sequences of each plasmid were analyzed with the ABRicate program (see below), including the ResFinder and PlasmidFinder databases, to identify antimicrobial resistance genes and plasmid types.
The draft genome sequences of 132 E. coli isolates revealed that 87 (65.9%) isolates had blaCMY-2, and 28 isolates out of the 87 blaCMY-2-positive isolates carried other β-lactamase genes simultaneously. Another 44 isolates had the following β-lactamase genes other than blaCMY-2: blaTEM (n = 17), blaCTX-M group (blaCTX-M-1 [n = 7], blaCTX-M-2 [n = 13], blaCTX-M-9 [n = 4], and blaCTX-M-25 [n = 2]), blaSHV (n = 12), blaEC (n = 4), and blaOXA-21 (n = 1). Among the 44 isolates, 16 isolates had multiple β-lactamase genes. The remaining 1 isolate had no β-lactamase gene; however, we could not discover the reason. Carbapenemase genes were not detected in this study (see Table S1 in the supplemental material).
The blaCMY-2 gene was found in the following incompatibility groups: IncI1-Iγ (n = 21; 24.1%), IncI (n = 12; 13.8%), IncB/O/K/Z (n = 28; 32.2%), IncC (n = 22; 25.3%), IncI2 (n = 2; 2.3%), IncF (n = 1; 1.1%), p0111 (n = 1; 1.1%), and untypeable (n = 1; 1.1%). In this study, we refer to IncI1-Iγ and IncI as the IncI1 group. Most of the isolates harbored one blaCMY-2-positive plasmid, whereas two isolates (no. 23-Ec-C-50 and 24-Ec-C-171) harbored two blaCMY-2-positive IncB/O/K/Z and IncC plasmids and two blaCMY-2-positive IncC and IncC-IncFIB-IncFIC-IncX4 fusion plasmids, respectively.
Subsequently, the chromosomal DNAs of 82 isolates that had three major blaCMY-2-positive (IncI1 group, IncB/O/K/Z, and IncC) plasmids were analyzed with the multilocus sequence typing (MLST) program and compared with 131 blaCMY-2-positive publicly available strains and with 792 publicly available E. coli strains from Gallus gallus, chickens, or broilers within each sequence type (ST) cluster to discover the clonal dissemination. To investigate the features of our samples and the global linkage of blaCMY-2 plasmids, clustering analysis was performed for comparison with only our samples, and network analysis was performed to compare our samples with publicly available plasmids (IncI1 group, 133 plasmids; IncB/O/K/Z, 9 plasmids; IncC, 253 plasmids).
MLST and core genome single-nucleotide variant (SNV) phylogenetic analysis.
The STs were highly diverse among 82 E. coli isolates that had blaCMY-2-positive IncI1 group, IncB/O/K/Z, and IncC plasmids (see Table S2 in the supplemental material). These 82 isolates were assigned to 49 different STs, with 4 isolates exhibiting novel STs (ST9718, ST9719, ST9720, and ST9721). The STs were not dependent on the incompatibility type of the plasmid, district in Japan, or isolation year. This strongly suggested that blaCMY-2-positive plasmids are mainly transferred horizontally between E. coli isolates rather than by clonal spread of E. coli isolates harboring blaCMY-2-positive plasmids. These results agreed with those of earlier studies, which reported high ST strain diversity of CMY-2-producing E. coli isolates from broiler chickens (22, 23).
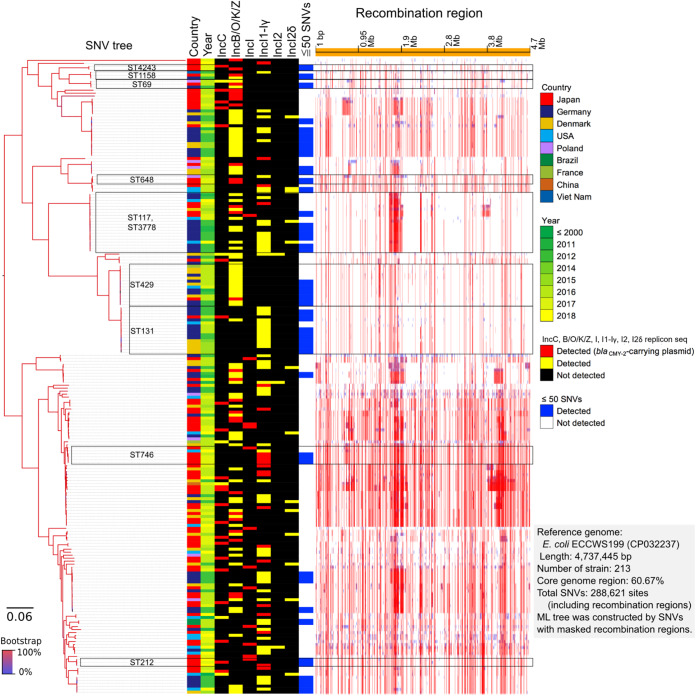
Furthermore, we identified 288,621 SNVs among the 213 blaCMY-2-positive E. coli isolates based on 60.67% of the core genome region in SNV-based phylogenetic analysis (Fig. 1). These results revealed several differences in the SNVs with the inclusion of various STs.
FIG 1.
Core genome SNV phylogenetic analysis. Eighty-two isolates were clustered with 131 publicly available blaCMY-2-positive E. coli genomes.
However, 20 isolates within the nine ST clusters (ST4243, ST1158, ST69, ST648, ST117, ST429, ST131, ST746, and ST212) exhibited differences in less than 50 SNVs with the blaCMY-2-positive strains isolated from Japan and other countries. Furthermore, the sequences of 20 isolates within the nine ST clusters were compared, cluster by cluster, with those of 792 publicly available E. coli isolates. A summary of the results, including sample numbers, total numbers of SNVs, and pairwise SNV counts in the nine ST clusters, is provided in Table S3 in the supplemental material. The SNV analysis based on each ST cluster revealed the presence of genetically closely related strains. Some strains in three STs out of nine revealed the differences in less than 50 SNVs between our strains (see Fig. S2 in the supplemental material). Two isolates (no. 23-Ec-C-57 and 23-Ec-C-110) assigned to ST648 differed by 33 SNVs. These two closely related isolates were collected from the Tohoku district, but not the same prefecture, in 2001. The 17-Ec-C-7 and 25-Ec-C-61 isolates assigned to ST117, which had differences in 50 SNVs, were collected from the Chugoku district in 2005 and 2013, respectively. Among the five ST746 isolates, two isolates (no. 19-Ec-C-93 and 19-Ec-C-96), which were collected from Kyushu, Okinawa district, in 2007, had differences in 11 SNVs. These results indicate that small-scale clonal spread also occurred in Japan.
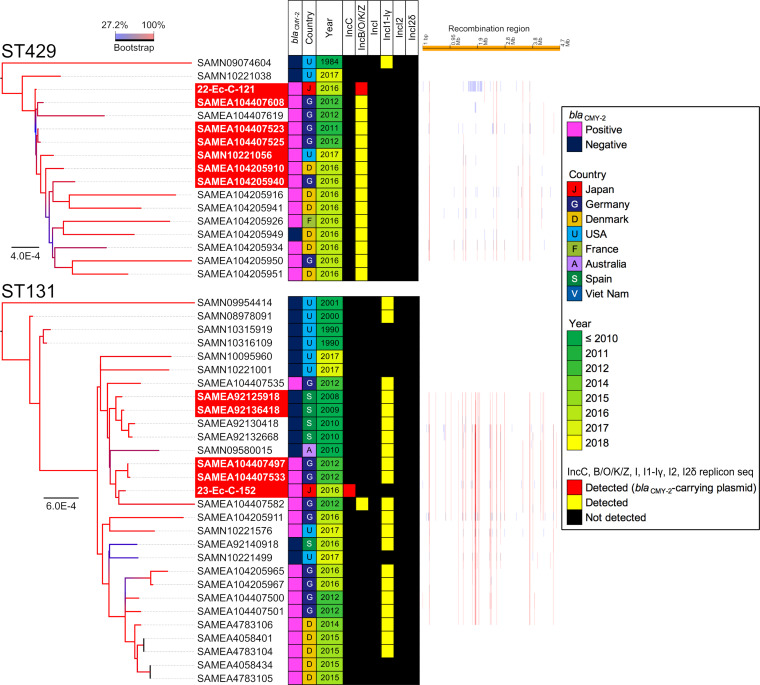
The isolates assigned to ST429 and ST131 and the isolates from other countries exhibited differences in less than 50 SNVs (see Fig. S2). The 22-Ec-C-121 isolate, isolated in 2010 and assigned to ST429, was closely related to isolates collected from Germany between 2011 (SAMEA104407523) and 2012 (SAMEA104407608 and SAMEA104407525) (Fig. 2). Additionally, the difference between 22-Ec-C-121 and SAMEA104407523 harboring a blaCMY-2-positive IncB/O/K/Z plasmid was only 7 SNVs. This demonstrated that a part of clone of CMY-2-producing E. coli isolates disseminated at broiler production in Japan could exist among the broilers produced in Europe around 2010. The 23-Ec-C-152 isolate, isolated in 2011 and assigned to ST131, and SAMEA104407497, isolated in Germany in 2012, had 36 SNVs. However, the plasmid types of the 23-Ec-C-152 and SAMEA104407497 isolates were different.
FIG 2.
Core genome SNV phylogenetic analysis. Two isolates assigned to ST429 and ST131 were compared with the blaCMY-2-positive or negative E. coli genomes.
IncI1 group.
We excluded one plasmid sample, p19C96-1, from hierarchical cluster analysis and plasmid network analysis for the following reason. The total read of p19C96-1 was twice as large as the band size of the plasmid, which might have been because a few types of plasmid that had the same size were extracted simultaneously when S1 PFGE was performed. Also, because the genome sequence of p19C96-1 was divided into seven contigs after de novo assembly, it was unknown which plasmid some contigs originated from.
Fourteen out of 21 blaCMY-2-IncI1-Iγ plasmid samples were identified as pST12, followed by untypeable (n = 5) and pST65 (n = 2). The majority of pST12-IncI1-Iγ plasmids (n = 11/14; 78.6%) were detected in isolates from western Japan. On the other hand, 12 samples identified as IncI showed sequence types pST55 (n = 11) and untypeable (n = 1). All but the pST55-IncI plasmids (n = 9/11; 81.8%) were detected in isolates from western Japan (see Table S2).
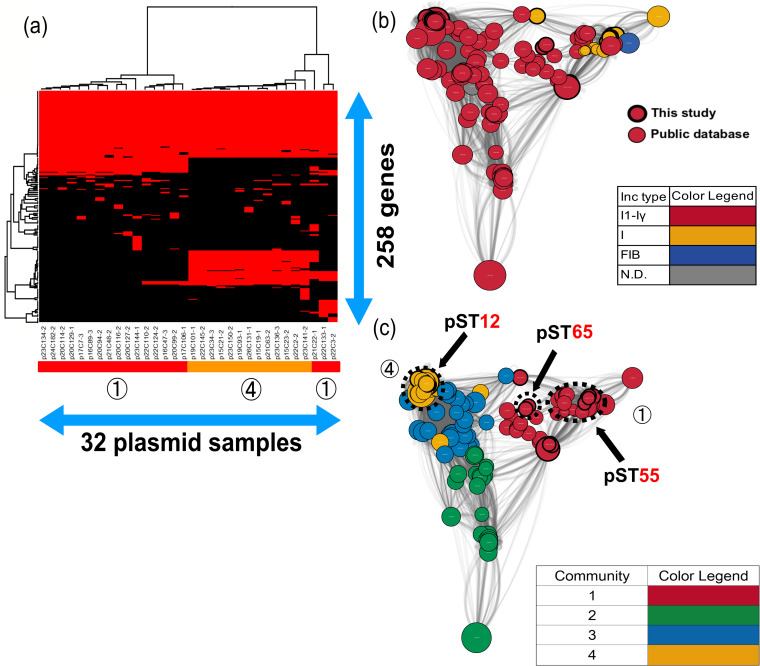
The gene structures of blaCMY-2-IncI1-Iγ and IncI were obviously different in comparative gene-clustering analyses (Fig. 3a). Interestingly, only pST12-IncI1-Iγ had ccdA and ccdB genes. The ccdA and ccdB genes encoded CcdA antitoxin and CcdB toxin, which provided a mechanism for control of the death of cells that had lost the plasmid carrying the genes (24). This system might contribute to the increase and persistence of blaCMY-2-IncI1-Iγ, not only in Japan, but also in other countries (Fig. 3b). Although the gene compositions of two pST65-IncI1-Iγ (p22C3-2 and p22C133-1) and one untypeable IncI (p21C22-1) plasmids were similar to that of pST12-IncI1-Iγ, the results of network analysis showed that pST65 plasmids were located between pST12-IncI1-Iγ and pST55-IncI, which suggested that those plasmids showed intermediate levels of evolution between IncI1-Iγ and IncI.
FIG 3.
Clustering analysis of the IncI1 group. (a) Thirty-two blaCMY-2-positive IncI1 group plasmids were clustered to determine the presence of 258 genes in the plasmids. Red indicates the plasmids that had the genes, whereas black indicates plasmids that lacked the genes. The numbers in circles and the color of the horizontal bar below the plasmid sample names represent community numbers and the color of each community in network analysis, respectively. (b and c) Network analysis of the IncI1 group. Thirty-two blaCMY-2-positive IncI1 group plasmid sequences were compared with 133 publicly available complete plasmid sequences. A color was given to each plasmid type (b) or community (c). The dotted circles represent each plasmid sample. Our samples are surrounded by thick black lines. The sizes of the circles indicate the sizes of the plasmids. When the constitutions of plasmids are similar, the distance between the plasmids is shorter.
pST55-IncI and pST12-IncI1-Iγ plasmids clustered in community 1 and community 4, respectively (Fig. 3c). In community 1, no publicly available sequences were detected in chickens, which indicated that pST55-IncI might not be so common among chickens in other countries. However, the relationship of the plasmids with those in other countries could not be denied due to one single-locus variant of pST2 (22). In contrast, all complete plasmids clustered in community 4 were detected in E. coli or Salmonella enterica isolates from humans (GenBank accession no. CP016865 and CP012929), turkeys (GenBank accession no. CP022064 and CP012936), and chickens (GenBank accession no. MG825376, CP016568, CP016522, CP012923, and CM004485) in America, Canada, Brazil, and China (see Data Set S1 in the supplemental material). In Europe, the most common plasmid ST was pST12, which was detected in E. coli isolates from humans, chickens, and chicken meat in Germany, Denmark, and Italy (22, 23, 25). The pST12 plasmids were associated with some serotypes of Salmonella among chickens in America (26). Additionally, Poppe et al. demonstrated that blaCMY-2-positive plasmids could be transferred between E. coli and Salmonella enterica serovar Newport through conjugation in the poultry intestinal tract (27). Thus, it is possible that blaCMY-2/IncI1/pST12 plasmids are shared between E. coli and Salmonella in different host species and in geographically different regions.
IncB/O/K/Z.
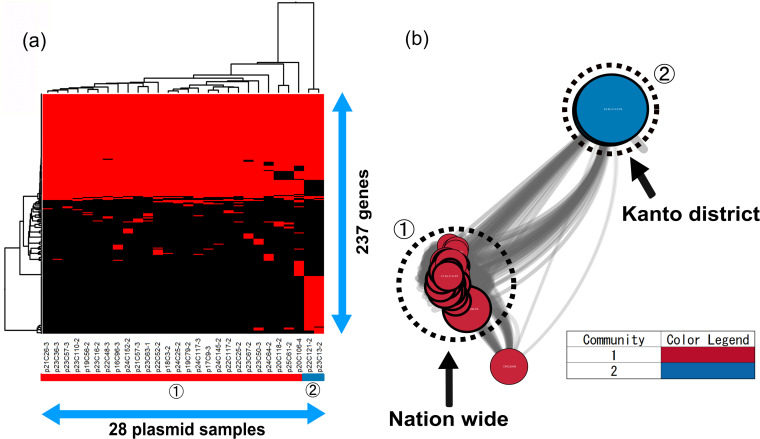
The blaCMY-2-positive IncB/O/K/Z plasmids, except two, had similar gene compositions in the gene-clustering analysis (Fig. 4a). In this study, although the plasmids were isolated from 2004 to 2013 throughout Japan, their frame structures had not changed for approximately 10 years, indicating that similar plasmids had been disseminated in Japan by recirculating in farms and repeated transmission. It was mentioned in other reports that blaCMY-2-positive IncK2 was highly preserved in chickens; it showed high stability in the absence of selective pressure in vitro (28), however, which seemed inconsistent with the fact that the resistance rate for 3GC in E. coli significantly decreased after the ban of off-label use. The plasmid stability system ParDEI provided a selection advantage for the plasmids (29). Indeed, our 26 blaCMY-2-positive IncB/O/K/Z plasmids had parE and pndA genes. Furthermore, the psiB and ssb genes, which are involved in conjugation, were carried by our plasmids. The results indicated that this type of blaCMY-2-positive IncB/O/K/Z plasmid spread in Japan while maintaining stability and that, in spite of the fact that our plasmid samples were derived from different farms nationwide and poultry of different ages, blaCMY-2-positive IncB/O/K/Z plasmids had a common ancestor.
FIG 4.
Clustering analysis of IncB/O/K/Z. (a) Twenty-eight blaCMY-2-positive IncB/O/K/Z plasmids were clustered to determine the presence of 237 genes in the plasmids. Red indicates the plasmids that had the genes, whereas black indicates plasmids that lacked the genes. The numbers in circles and the color of the horizontal bar below the plasmid sample names represent community numbers and the color of each community in network analysis, respectively. (b) Network analysis of IncB/O/K/Z. Twenty-eight blaCMY-2-positive IncB/O/K/Z plasmid sequences were compared with nine publicly available complete plasmid sequences. A color was given to each plasmid community. The dotted circles represent each plasmid sample. Our samples are surrounded by thick black lines. The sizes of the circles indicate the sizes of the plasmids. When the constitutions of plasmids are similar, the distance between the plasmids is shorter.
On the other hand, the remaining plasmids, p22C121-2 and p23C13-2, were detected from only the Kanto district, and they had tet(A), aac(3)-VIa, sul1, and aadA1 genes in addition to blaCMY-2 (see Table S2). Additionally, the plasmids carried emrE, which encodes the multidrug transporter EmrE. Also, the tyrosine recombinase genes xerC and xerD, which are involved in recombination (30), were carried on the plasmids. Moreover, two plasmids did not have the psiB and ssb genes but did have the cia gene, encoding bacteriocin, which was one of the stability mechanisms. This finding shows that the plasmids were preserved in a limited area without being propagated while escaping from some antimicrobials.
In this study, only 8.0% of publicly available IncB/O/K/Z plasmids were selected to be compared with our samples, indicating that our samples might have origins in Japan. However, in community 1, according to the results of network analysis, six plasmids (GenBank accession no. CP016548, KR905384, KR905385, KR905386, KR905387, and KR905389) had a close relationship with our samples (Fig. 4b). The plasmids were detected in E. coli isolates from chickens, chicken meat, and humans in the Netherlands and Switzerland. The majority of E. coli isolates from chickens harbored blaCMY-2-positive IncK plasmids in Denmark, the Netherlands, Norway, Sweden, and Finland (18, 22, 31–33). Furthermore, the plasmids from chickens, chicken meat, and humans in Switzerland were identical to each other, with more than 95% identity (34). blaCMY-2-positive IncB/O/K/Z plasmids could be an indicator of transmission between chickens and humans.
In community 2, none of the complete plasmids were extracted from the public database (see Data Set S2 in the supplemental material). Therefore, it was found that two plasmids clustered in community 2 had unique structures, suggesting that they originated in Japan.
IncC plasmid.
All the IncC plasmids had multidrug resistance genes, such as genes conferring resistance to β-lactam agents, aminoglycoside, amphenicol, quinolone, sulfonamide, trimethoprim, or tetracycline, as previously reported (35–38). Moreover, most of the IncC plasmids (17/23; 74%) were detected in the isolates from western Japan, like our IncI1 group (see Table S2).
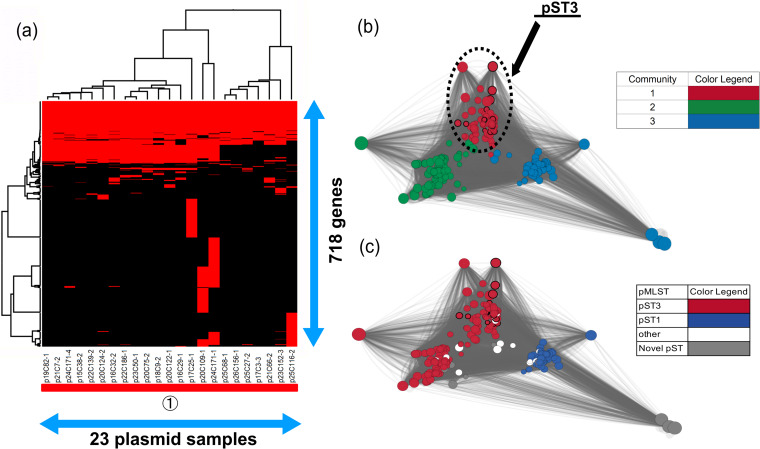
Almost all of the blaCMY-2-positive IncC plasmids (19/23; 82.6%) had both orf1832 and rhs1 genes, which type 1 IncC is known to have (39), indicating that type 1 IncC plasmids were disseminated in Japan. Furthermore, type 1 IncC plasmids were classified into type 1a, detected mainly in humans, and type 1b detected in humans and animals (39); however, we could not determine two types due to lack of part of a region that type 1a has specifically (39). Meanwhile, in spite of the fact that blaCMY-2-positive IncC plasmids, except for the plasmid p24C171-1, were identified as pST3, the gene structures of seven plasmids (p17C3-3, p21C66-2, p23C152-3, p25C27-2, p25C68-1, p25C116-2, and p26C156-1) were different from those of other samples (Fig. 5a). Those plasmids did not carry the ssb gene, and the type might differ from the genetic organization of the IncC plasmid provided in a previous report (39), which probably represented a new lineage originating from recombination, and further investigation of the evolution of IncC is necessary.
FIG 5.
Clustering analysis of IncC. (a) Twenty-three blaCMY-2-positive IncC plasmid sequences were clustered to determine the presence of 718 genes in the plasmids. Red indicates the plasmids that had the genes, whereas black indicates plasmids that lacked the genes. The numbers in circles and the color of the horizontal bar below the plasmid sample names represent community numbers and the color of each community in network analysis, respectively. (b and c) Network analysis of IncC. Twenty-three blaCMY-2-positive IncC plasmid sequences were compared with 253 publicly available complete plasmid sequences. A color was given to each plasmid community (b) or sequence type (c). The dotted circles represent each plasmid sample. Our samples are surrounded by thick black lines. The sizes of the circles indicate the sizes of the plasmids. When the constitutions of plasmids are similar, the distance between the plasmids is shorter.
In contrast, 95.5% of publicly available plasmids were selected to compare with our samples, and almost all the plasmids were identified as pST3 or pST1 (Fig. 5c), indicating that almost all IncA/C plasmids share a large number of gene regions (40). All publicly available blaCMY-2-positive IncA/C plasmids belonged to the same community as our samples, and those plasmids were detected in the following bacteria: Salmonella spp., Klebsiella pneumoniae, E. coli, Proteus mirabilis, Citrobacter freundii, Vibrio alginolyticus, Vibrio cholerae, and Aeromonas salmonicida (see Data Set S3 in the supplemental material). This was consistent with the results of earlier studies, which reported that IncA/C plasmids had a broad host range and were found in Enterobacteriaceae, Vibrio, and Aeromonas species (36, 37, 41, 42). This could be explained by changing the host’s methylation (43). The broad host range of IncC might make it difficult to understand IncC evolution, because interaction of the plasmid with various host DNAs increased (44). Additionally, large numbers of the plasmids were detected in E. coli and S. enterica isolates from cows, pigs, chickens, or turkeys in America and in E. coli, S. enterica, P. mirabilis, and K. pneumoniae isolates from pigs, chickens, and humans in China but not detected in Europe (see Data Set S3), consistent with previously published data (35).
The numbers of three major plasmid types and the 3GC resistance rate by year are shown in Fig. S1 in the supplemental material. The number of IncC plasmids was stable, whereas the number of IncI1 group and IncB/O/K/Z plasmids varied and was correlated with the 3GC resistance rate. Not only low fitness cost due to fewer resistance genes (45), but also the stability mechanism could be considered to understand why the IncI1 group and IncB/O/K/Z were the predominant types in Japan. Furthermore, the resistance rate for 3GC has been maintained at a low level since 2012 in Japan. Dame-Korevaar et al. reported that the sharp reduction of E. coli possessing blaCMY-2-IncA/C in a broiler parent flock occurred in the absence of antibiotics, but E. coli possessing blaCMY-2-IncA/C remained present in the environment (46), implying that E. coli possessing blaCMY-2-IncA/C could fail to disappear from farms unless sanitation was properly managed, even if antimicrobials were not used in hatcheries and farms. In addition, the multidrug resistance genes in the IncC plasmid might contribute to the stability of 3GC-resistant bacteria if antimicrobials were not properly used on farms.
The blaCMY-2-positive plasmids have been reported to play an important role in the transmission of 3GC-resistant Enterobacteriaceae isolated from humans and animals (13, 25, 31). Furthermore, some studies suggested that blaCMY-2-positive plasmids could be transmitted between food-producing animals and humans (14, 16, 17). In this study, network analysis revealed that the blaCMY-2-positive plasmids analyzed were in the same communities as the plasmids from human isolates abroad. Accordingly, further studies are needed that include the strains from humans in Japan.
This study has a limitation. We could not trace the countries from which the breeding companies imported the parent stocks and those to which the farms transported chicks. Elucidation of this epidemiological route may reveal the propagation of blaCMY-2-positive plasmids.
In conclusion, the pST55 and pST12-IncI1 group and pST3-IncC were more prevalent in western Japan, but we could not conclude that this was reflected in the transmission of those plasmids from overseas to certain farms owned by breeding companies or in the usage environment of antimicrobials in hatcheries. However, blaCMY-2-positive pST12-IncI1-Iγ and pST3-IncC plasmids were linked to those detected in E. coli and S. enterica isolates from chickens in North America, while blaCMY-2-positive IncB/O/K/Z plasmids were linked to chicken isolates in European countries. If transmission of the plasmids from overseas via imported chickens actually occurred, active monitoring of antimicrobial resistance for imported grandparent stocks and parent stocks will be needed. Taking the fact that grandparent stocks and parent stocks have been imported from Europe and America into consideration, our data revealed the linkage between blaCMY-2-positive plasmids in Japan and those in other countries.
MATERIALS AND METHODS
Bacterial strains.
In total, 1,756 E. coli samples were collected from fecal samples from healthy broiler chickens housed at different farms in each prefecture between 2002 and 2014 within the framework of the Japanese Veterinary Antimicrobial Resistance Monitoring System (JVARM) (47–50). One fecal sample per farm was collected, and two E. coli isolates were isolated from each sample using deoxycholate-hydrogen sulfide-lactose agar (Eiken Co., Ltd., Tokyo, Japan). The API 20E system (bioMérieux, Marcy I’Etoile, France) was used for the identification of E. coli.
Antimicrobial susceptibility testing.
The susceptibility of all E. coli isolates to ceftiofur was tested by agar dilution (2002 to 2009). The susceptibility of the E. coli isolates to cefotaxime was tested using a broth microdilution test (2010 to 2014) (Eiken, Japan), following the manufacturer’s instructions. The data were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) standards (2008, 2011, 2012, and 2013).
PFGE with S1 nuclease.
Of the 197 3GC-resistant E. coli isolates, 132 isolates were used for WGS analysis. This was because we selected one strain per fecal sample when two E. coli isolates were 3GC resistant and they were isolated from the same fecal sample. The genomic DNA of all 132 3GC-resistant E. coli isolates was subjected to PFGE with S1 nuclease (TaKaRa Bio, Japan) to separate the plasmid DNA and chromosomal DNA according to the method described by Barton et al. (51). The plugs of genomic DNA were incubated at 37°C for 30 min with 4.5 U of S1 nuclease. The digested DNA was electrophoresed in 1.0% SeaKem Gold agarose (Lonza Bio Co., Japan) under running conditions (2.0 to 25.0 s, 14°C, 6 V, and 18.5 h) generated by a CHEF-DR III. Electrophoresis was performed with 0.5× Tris-borate-EDTA (TBE) as a buffer. When degradation of DNA was observed, HEPES buffer was used to prevent it instead of 0.5× TBE buffer (52). The running conditions were modified by reducing the voltage to 5 V/cm when using HEPES buffer. The gel was stained with SYBR Gold nucleic acid gel stain (Life Technologies Co., USA) and visualized using a cyan LED transilluminator. The number and sizes of DNA bands predicted to be plasmids were determined, with visual confirmation. Subsequently, the gel fragments of chromosomal and plasmid DNA were excised and stored at −30°C until they were sequenced.
Whole-genome sequencing and sequence reconstruction.
The stored DNA was purified using a ZR-96 Zymoclean Gel DNA recovery kit (Zymo Research, USA). Next, the DNA samples were subjected to a tagmentation reaction and PCR amplification using a Nextera XT DNA sample preparation kit (Illumina, USA). The size of the prepared library was determined by electrophoresis. The samples were purified using the Wizard SV gel and PCR clean-up system (Promega, USA) and sequenced on an Illumina Miseq platform using a Miseq v3 reagent kit (Illumina, USA) with two 300-bp paired-end reads.
De novo assembly for each replicon separated by PFGE was performed with the A5-Miseq pipeline (53). Contig plasmid sequences of low read depth were excluded, as the sequences were constructed from a low-abundance DNA contamination of chromosomes and other plasmids. To confirm the exact plasmid size, the total lengths of final plasmid contigs were compared with the S1 PFGE fragment size. Gene prediction was performed with the Prodigal program (version 2.60) (54) and the BLASTP program (55) using the NCBI Protein Sequence Database (nr). The antimicrobial resistance genes and plasmid Inc types were detected with the ABRicate program (version 0.2) (https://github.com/tseemann/abricate) using the ResFinder 4.0 (56) and bacterial antimicrobial resistance reference gene database (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA313047) and the PlasmidFinder 2.1 (57) database, respectively. MLST of E. coli was performed with the MLST program (version 1.2) (https://github.com/tseemann/mlst/issues) against the MLST database (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli). Plasmid multilocus sequence typing (pMLST) was performed for plasmids belonging to the typeable groups, and the sequences were compared to the sequences deposited in the pMLST database (https://pubmlst.org/plasmid/).
Core genome SNV phylogenetic analysis.
The raw sequence data and assembled genome sequences of E. coli isolates from chickens were retrieved from the NCBI BioSample database. In total, 1,365 sample sequences were downloaded under the following parameters: “Escherichia coli” (organism) and “Gallus gallus” or “chicken” or “avian” (all fields). The submitted metadata (collection date and place) and sequencing data from the sequencing platform (Illumina paired-end raw reads or assembled sequences) were collected. Next, the low-depth data, organism prediction, and contamination check were verified. Then, the collection date and place information were provided, the data were registered in the SRA database and assembly database, and the data that had coverage read depths of more than 40 and the data identified as E. coli properly without contamination were selected. Finally, 792 samples were detected in total. The sequences of E. coli ECCWS199 (BioSample ID SAMN10023797), which had blaCMY-2 on pTB221 (GenBank accession no. NZ_CP032238), were also collected from the NCBI database as a complete genome sequence of blaCMY-2-positive E. coli isolated from chickens. For SNV analysis of only blaCMY-2-positive strains, the sequenced chromosomal short-read data (n = 82) and sequence data retrieved from the NCBI database (n = 131) were compared with the E. coli ECCWS199 complete chromosomal sequence (GenBank accession no. CP032237) using the BWA-MEM read-mapping program (58). All SNVs were extracted using SAMtools (59) and VarScan (v2.3.4) (60) software. The prophage SNVs and repeat regions predicted using the PHASTER (61) and NUCmer (62) programs, respectively, were excluded. The recombination regions were predicted using Gubbins (63). The SNVs in the recombination regions were masked. All extracted SNVs in core genome regions were concatenated as pseudosequences. Phylogenetic analysis was performed using the DNA approximately maximum-likelihood program (FastTree version2.1) (64). The SNV analysis within each ST cluster was performed not only for blaCMY-2-positive strains, but also for blaCMY-2-negative strains, using the method described above.
Plasmid comparative analysis.
A total of 14,731 complete plasmid sequences were retrieved from the NCBI nucleotide database and RefSeq database using the search keywords “complete sequence” and “plasmid.” The complete plasmid sequences were constructed. The sequences included 247 IncI1, 112 IncB/O/K/Z, and 265 IncA/C plasmids (September 2018). To perform the analysis under the same conditions against our plasmid sequence data, the genes in the complete plasmid sequences were predicted using the Prodigal program. Plasmid network analysis was performed as previously described (65, 66). Briefly, putative protein sequences of our draft plasmids and the complete plasmids deposited in the NCBI database were clustered using the UCLUST program (version 6.0.307). The following parameters were evaluated after sorting based on the amino acid sequence length, following the instructions accompanying the software: cluster_smallmem; id, 1.0; minsl, 0.9; minqt, 0.9; maxqt, 1.1; query_cov, 0.9; and target_cov, 0.9. These parameters indicated 100% amino acid sequence identity with at least 90% coverage and less than 10% length difference. We used the samples meeting the above criteria in network analysis. Plasmids sharing at least 40 homologous genes were connected as a network. A community was detected by the multilevel community method in the igraph library in R using the default parameter settings. Cytoscape version 3.2.0 was used to draw the plasmid network graph (67). The hierarchical cluster analysis and visualization were performed with presence and absence patterns of clustering genes using the heatmap.2 program of the gplot R package. The Simpson similarity coefficient and ward.D2 clustering method were used (68).
Data availability.
All raw short-read sequence data were deposited in the DNA Data Bank of Japan (BioSample IDs, SAMD00179258 to SAMD00179339; DRA accession no. DRA008702 and DRA008648). The assembled draft sequences of plasmids were deposited in the DDBJ/EMBL/GenBank database (accession no. LC501464 to LC501701).
Supplementary Material
ACKNOWLEDGMENTS
We thank the Pathogen Genomics Center, National Institute of Infectious Diseases, for technical assistance and the staffs of Livestock Hygiene Service Centers across Japan for providing E. coli isolates.
This study was partly supported by the Research Program on Emerging and Re-emerging Infectious Diseases of the Japan Agency for Medical Research and Development (grant 16fk0108305j0003).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.WHO-AGISAR. 2016. Critically important antimicrobials for human medicine, 5th revision World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Madec JY, Haenni M, Nordmann P, Poirel L. 2017. Extended-spectrum beta-lactamase/AmpC- and carbapenemase-producing Enterobacteriaceae in animals: a threat for humans?. Clin Microbiol Infect 23:826–833. doi: 10.1016/j.cmi.2017.01.013. [DOI] [PubMed] [Google Scholar]
- 3.Hiki M, Kawanishi M, Abo H, Kojima A, Koike R, Hamamoto S, Asai T. 2015. Decreased resistance to broad-spectrum cephalosporin in Escherichia coli from healthy broilers at farms in Japan after voluntary withdrawal of ceftiofur. Foodborne Pathog Dis 12:639–643. doi: 10.1089/fpd.2015.1960. [DOI] [PubMed] [Google Scholar]
- 4.Dutil L, Irwin R, Finley R, Ng LK, Avery B, Boerlin P, Bourgault AM, Cole L, Daignault D, Desruisseau A, Demczuk W, Hoang L, Horsman GB, Ismail J, Jamieson F, Maki A, Pacagnella A, Pillai DR. 2010. Ceftiofur resistance in Salmonella enterica serovar Heidelberg from chicken meat and humans. Emerg Infect Dis 16:48–54. doi: 10.3201/eid1601.090729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Webster P. 2009. The perils of poultry. CMAJ 181:21–24. doi: 10.1503/cmaj.091009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.EFSA Panel on Biological Hazards (BIOHAZ). 2011. Scientific opinion on the public health risks of bacterial strains producing extended-spectrum β-lactamases and/or AmpC β-lactamases in food and food-producing animals. European Food Safety Authority, Parma, Italy: https://efsa.onlinelibrary.wiley.com/doi/pdf/10.2903/j.efsa.2011.2322. [Google Scholar]
- 7.Webster P. 2009. Poultry, politics, and antibiotic resistance. Lancet 374:773–774. doi: 10.1016/s0140-6736(09)61578-6. [DOI] [PubMed] [Google Scholar]
- 8.Kojima A, Ishii Y, Ishihara K, Esaki H, Asai T, Oda C, Tamura Y, Takahashi T, Yamaguchi K. 2005. Extended-spectrum-beta-lactamase-producing Escherichia coli strains isolated from farm animals from 1999 to 2002: report from the Japanese Veterinary Antimicrobial Resistance Monitoring Program. Antimicrob Agents Chemother 49:3533–3537. doi: 10.1128/AAC.49.8.3533-3537.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kojima A, Asai T, Ishihara K, Morioka A, Akimoto K, Sugimoto Y, Sato T, Tamura Y, Takahashi T. 2009. National monitoring for antimicrobial resistance among indicator bacteria isolated from food-producing animals in Japan. J Vet Med Sci 71:1301–1308. doi: 10.1292/jvms.001301. [DOI] [PubMed] [Google Scholar]
- 10.Hiki M, Usui M, Kojima A, Ozawa M, Ishii Y, Asai T. 2013. Diversity of plasmid replicons encoding the blaCMY-2 gene in broad-spectrum cephalosporin-resistant Escherichia coli from livestock animals in Japan. Foodborne Pathog Dis 10:243–249. doi: 10.1089/fpd.2012.1306. [DOI] [PubMed] [Google Scholar]
- 11.Smet A, Martel A, Persoons D, Dewulf J, Heyndrickx M, Catry B, Herman L, Haesebrouck F, Butaye P. 2008. Diversity of extended-spectrum beta-lactamases and class C beta-lactamases among cloacal Escherichia coli isolates in Belgian broiler farms. Antimicrob Agents Chemother 52:1238–1243. doi: 10.1128/AAC.01285-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tiba-Casas MR, Camargo CH, Soares FB, Doi Y, Fernandes SA. 2019. Emergence of CMY-2-producing Salmonella Heidelberg associated with IncI1 plasmids isolated from poultry in Brazil. Microb Drug Resist 25:271–276. doi: 10.1089/mdr.2018.0044. [DOI] [PubMed] [Google Scholar]
- 13.Martin LC, Weir EK, Poppe C, Reid-Smith RJ, Boerlin P. 2012. Characterization of blaCMY-2 plasmids in Salmonella and Escherichia coli isolates from food animals in Canada. Appl Environ Microbiol 78:1285–1287. doi: 10.1128/AEM.06498-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winokur PL, Vonstein DL, Hoffman LJ, Uhlenhopp EK, Doern GV. 2001. Evidence for transfer of CMY-2 AmpC beta-lactamase plasmids between Escherichia coli and Salmonella isolates from food animals and humans. Antimicrob Agents Chemother 45:2716–2722. doi: 10.1128/AAC.45.10.2716-2722.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dierikx CM, van der Goot JA, Smith HE, Kant A, Mevius DJ. 2013. Presence of ESBL/AmpC-producing Escherichia coli in the broiler production pyramid: a descriptive study. PLoS One 8:e79005. doi: 10.1371/journal.pone.0079005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berg ES, Wester AL, Ahrenfeldt J, Mo SS, Slettemeas JS, Steinbakk M, Samuelsen O, Grude N, Simonsen GS, Lohr IH, Jorgensen SB, Tofteland S, Lund O, Dahle UR, Sunde M. 2017. Norwegian patients and retail chicken meat share cephalosporin-resistant Escherichia coli and IncK/blaCMY-2 resistance plasmids. Clin Microbiol Infect 23:407.e9–407.e15. doi: 10.1016/j.cmi.2016.12.035. [DOI] [PubMed] [Google Scholar]
- 17.Huijbers PM, Graat EA, Haenen AP, van Santen MG, van Essen-Zandbergen A, Mevius DJ, van Duijkeren E, van Hoek AH. 2014. Extended-spectrum and AmpC beta-lactamase-producing Escherichia coli in broilers and people living and/or working on broiler farms: prevalence, risk factors and molecular characteristics. J Antimicrob Chemother 69:2669–2675. doi: 10.1093/jac/dku178. [DOI] [PubMed] [Google Scholar]
- 18.Dierikx C, van der Goot J, Fabri T, van Essen-Zandbergen A, Smith H, Mevius D. 2013. Extended-spectrum-beta-lactamase- and AmpC-beta-lactamase-producing Escherichia coli in Dutch broilers and broiler farmers. J Antimicrob Chemother 68:60–67. doi: 10.1093/jac/dks349. [DOI] [PubMed] [Google Scholar]
- 19.Bortolaia V, Hansen KH, Nielsen CA, Fritsche TR, Guardabassi L. 2014. High diversity of plasmids harbouring blaCMY-2 among clinical Escherichia coli isolates from humans and companion animals in the upper Midwestern USA. J Antimicrob Chemother 69:1492–1496. doi: 10.1093/jac/dku011. [DOI] [PubMed] [Google Scholar]
- 20.Matsumura Y, Yamamoto M, Higuchi T, Komori T, Tsuboi F, Hayashi A, Sugimoto Y, Hotta G, Matsushima A, Nagao M, Takakura S, Ichiyama S. 2012. Prevalence of plasmid-mediated AmpC beta-lactamase-producing Escherichia coli and spread of the ST131 clone among extended-spectrum beta-lactamase-producing E. coli in Japan. Int J Antimicrob Agents 40:158–162. doi: 10.1016/j.ijantimicag.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 21.Kikuchi K, Lee K, Ueno H, Tomari K, Kobori S, Kaetsu A, Matsui M, Suzuki S, Sekizuka T, Kuroda M, Miyazaki M, Ohnishi M. 19 June 2019. Enterohaemorrhagic Escherichia coli O121:H19 acquired an extended-spectrum beta-lactamase gene during the development of an outbreak in two nurseries. Microb Genom 5 . doi: 10.1099/mgen.0.000278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen KH, Bortolaia V, Nielsen CA, Nielsen JB, Schonning K, Agerso Y, Guardabassi L. 2016. Host-specific patterns of genetic diversity among IncI1-Igamma and IncK plasmids encoding CMY-2 beta-lactamase in Escherichia coli isolates from humans, poultry meat, poultry, and dogs in Denmark. Appl Environ Microbiol 82:4705–4714. doi: 10.1128/AEM.00495-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pietsch M, RESET Study Group, Irrgang A, Roschanski N, Brenner Michael G, Hamprecht A, Rieber H, Kasbohrer A, Schwarz S, Rosler U, Kreienbrock L, Pfeifer Y, Fuchs S, Werner G. 2018. Whole genome analyses of CMY-2-producing Escherichia coli isolates from humans, animals and food in Germany. BMC Genomics 19:601. doi: 10.1186/s12864-018-4976-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bahassi EM, O'Dea MH, Allali N, Messens J, Gellert M, Couturier M. 1999. Interactions of CcdB with DNA gyrase. Inactivation of Gyra, poisoning of the gyrase-DNA complex, and the antidote action of CcdA. J Biol Chem 274:10936–10944. doi: 10.1074/jbc.274.16.10936. [DOI] [PubMed] [Google Scholar]
- 25.Accogli M, Fortini D, Giufre M, Graziani C, Dolejska M, Carattoli A, Cerquetti M. 2013. IncI1 plasmids associated with the spread of CMY-2, CTX-M-1 and SHV-12 in Escherichia coli of animal and human origin. Clin Microbiol Infect 19:E238–E240. doi: 10.1111/1469-0691.12128. [DOI] [PubMed] [Google Scholar]
- 26.Folster JP, Pecic G, McCullough A, Rickert R, Whichard JM. 2011. Characterization of blaCMY-encoding plasmids among Salmonella isolated in the United States in 2007. Foodborne Pathog Dis 8:1289–1294. doi: 10.1089/fpd.2011.0944. [DOI] [PubMed] [Google Scholar]
- 27.Poppe C, Martin LC, Gyles CL, Reid-Smith R, Boerlin P, McEwen SA, Prescott JF, Forward KR. 2005. Acquisition of resistance to extended-spectrum cephalosporins by Salmonella enterica subsp. enterica serovar Newport and Escherichia coli in the turkey poult intestinal tract. Appl Environ Microbiol 71:1184–1192. doi: 10.1128/AEM.71.3.1184-1192.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rozwandowicz M, Brouwer MS, Zomer AL, Bossers A, Harders F, Mevius DJ, Wagenaar JA, Hordijk J. 2017. Plasmids of distinct IncK lineages show compatible phenotypes. Antimicrob Agents Chemother 61:e01954-16. doi: 10.1128/AAC.01954-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamruzzaman M, Iredell J. 2019. A ParDE-family toxin antitoxin system in major resistance plasmids of Enterobacteriaceae confers antibiotic and heat tolerance. Sci Rep 9:9872. doi: 10.1038/s41598-019-46318-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Midonet C, Barre FX. December 2014. Xer site-specific recombination: promoting vertical and horizontal transmission of genetic information. Microbiol Spectr 2:. doi: 10.1128/microbiolspec.MDNA3-0056-2014. [DOI] [PubMed] [Google Scholar]
- 31.Mo SS, Slettemeas JS, Berg ES, Norstrom M, Sunde M. 2016. Plasmid and host strain characteristics of Escherichia coli resistant to extended-spectrum cephalosporins in the Norwegian broiler production. PLoS One 11:e0154019. doi: 10.1371/journal.pone.0154019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borjesson S, Jernberg C, Brolund A, Edquist P, Finn M, Landen A, Olsson-Liljequist B, Tegmark Wisell K, Bengtsson B, Englund S. 2013. Characterization of plasmid-mediated AmpC-producing E. coli from Swedish broilers and association with human clinical isolates. Clin Microbiol Infect 19:E309–E311. doi: 10.1111/1469-0691.12192. [DOI] [PubMed] [Google Scholar]
- 33.Paivarinta M, Latvio S, Fredriksson-Ahomaa M, Heikinheimo A. 2020. Whole genome sequence analysis of antimicrobial resistance genes, multilocus sequence types and plasmid sequences in ESBL/AmpC Escherichia coli isolated from broiler caecum and meat. Int J Food Microbiol 315:108361. doi: 10.1016/j.ijfoodmicro.2019.108361. [DOI] [PubMed] [Google Scholar]
- 34.Seiffert SN, Carattoli A, Schwendener S, Collaud A, Endimiani A, Perreten V. 2017. Plasmids carrying blaCMY-2/4 in Escherichia coli from poultry, poultry meat, and humans belong to a novel IncK subgroup designated IncK2. Front Microbiol 8:407. doi: 10.3389/fmicb.2017.00407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rozwandowicz M, Brouwer MSM, Fischer J, Wagenaar JA, Gonzalez-Zorn B, Guerra B, Mevius DJ, Hordijk J. 2018. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J Antimicrob Chemother 73:1121–1137. doi: 10.1093/jac/dkx488. [DOI] [PubMed] [Google Scholar]
- 36.Call DR, Singer RS, Meng D, Broschat SL, Orfe LH, Anderson JM, Herndon DR, Kappmeyer LS, Daniels JB, Besser TE. 2010. blaCMY-2-positive IncA/C plasmids from Escherichia coli and Salmonella enterica are a distinct component of a larger lineage of plasmids. Antimicrob Agents Chemother 54:590–596. doi: 10.1128/AAC.00055-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fernandez-Alarcon C, Singer RS, Johnson TJ. 2011. Comparative genomics of multidrug resistance-encoding IncA/C plasmids from commensal and pathogenic Escherichia coli from multiple animal sources. PLoS One 6:e23415. doi: 10.1371/journal.pone.0023415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo YF, Zhang WH, Ren SQ, Yang L, Lu DH, Zeng ZL, Liu YH, Jiang HX. 2014. IncA/C plasmid-mediated spread of CMY-2 in multidrug-resistant Escherichia coli from food animals in China. PLoS One 9:e96738. doi: 10.1371/journal.pone.0096738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ambrose SJ, Harmer CJ, Hall RM. 2018. Evolution and typing of IncC plasmids contributing to antibiotic resistance in Gram-negative bacteria. Plasmid 99:40–55. doi: 10.1016/j.plasmid.2018.08.001. [DOI] [PubMed] [Google Scholar]
- 40.Harmer CJ, Hall RM. 2014. pRMH760, a precursor of A/C(2) plasmids carrying blaCMY and blaNDM genes. Microb Drug Resist 20:416–423. doi: 10.1089/mdr.2014.0012. [DOI] [PubMed] [Google Scholar]
- 41.Li R, Lin D, Chen K, Wong MH, Chen S. 2015. First detection of AmpC beta-lactamase bla(CMY-2) on a conjugative IncA/C plasmid in a Vibrio parahaemolyticus isolate of food origin. Antimicrob Agents Chemother 59:4106–4111. doi: 10.1128/AAC.05008-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McIntosh D, Cunningham M, Ji B, Fekete FA, Parry EM, Clark SE, Zalinger ZB, Gilg IC, Danner GR, Johnson KA, Beattie M, Ritchie R. 2008. Transferable, multiple antibiotic and mercury resistance in Atlantic Canadian isolates of Aeromonas salmonicida subsp. salmonicida is associated with carriage of an IncA/C plasmid similar to the Salmonella enterica plasmid pSN254. J Antimicrob Chemother 61:1221–1228. doi: 10.1093/jac/dkn123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang R, Lou J, Li J. 2019. A mobile restriction modification system consisting of methylases on the IncA/C plasmid. Mob DNA 10:26. doi: 10.1186/s13100-019-0168-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.San Millan A, MacLean RC. September 2017. Fitness costs of plasmids: a limit to plasmid transmission. Microbiol Spectr 5 . doi: 10.1128/microbiolspec.MTBP-0016-2017. [DOI] [PubMed] [Google Scholar]
- 45.Agerso Y, Jensen JD, Hasman H, Pedersen K. 2014. Spread of extended spectrum cephalosporinase-producing Escherichia coli clones and plasmids from parent animals to broilers and to broiler meat in a production without use of cephalosporins. Foodborne Pathog Dis 11:740–746. doi: 10.1089/fpd.2014.1742. [DOI] [PubMed] [Google Scholar]
- 46.Dame-Korevaar A, Fischer EAJ, Stegeman A, Mevius D, van Essen-Zandbergen A, Velkers F, van der Goot J. 2017. Dynamics of CMY-2 producing E. coli in a broiler parent flock. Vet Microbiol 203:211–214. doi: 10.1016/j.vetmic.2017.03.024. [DOI] [PubMed] [Google Scholar]
- 47.National Veterinary Assay Laboratory. 2009. A report on the Japanese veterinary antimicrobial resistance monitoring system 2000 to 2007. JVARM report:51 https://www.maff.go.jp/nval/english/pdf/jvarm2000_2007_final_201005.pdf.
- 48.National Veterinary Assay Laboratory. 2013. A report on the Japanese veterinary antimicrobial resistance monitoring system 2008 to 2011. JVARM report:31 https://www.maff.go.jp/nval/tyosa_kenkyu/taiseiki/pdf/jvarm2008_2011.pdf.
- 49.National Veterinary Assay Laboratory. 2016. A report on the Japanese Veterinary Antimicrobial Resistance Monitoring System 2012 to 2013. JVARM report:32 https://www.maff.go.jp/nval/english/pdf/jvarm_report_2012_2013.pdf.
- 50.National Veterinary Assay Laboratory. 2018. A report on the Japanese Veterinary Antimicrobial Resistance Monitoring System 2014 to 2015. JVARM report:38 https://www.maff.go.jp/nval/english/pdf/jvarm_report_2014_2015.pdf.
- 51.Barton BM, Harding GP, Zuccarelli AJ. 1995. A general method for detecting and sizing large plasmids. Anal Biochem 226:235–240. doi: 10.1006/abio.1995.1220. [DOI] [PubMed] [Google Scholar]
- 52.Koort JM, Lukinmaa S, Rantala M, Unkila E, Siitonen A. 2002. Technical improvement to prevent DNA degradation of enteric pathogens in pulsed-field gel electrophoresis. J Clin Microbiol 40:3497–3498. doi: 10.1128/jcm.40.9.3497-3498.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Coil D, Jospin G, Darling AE. 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 31:587–589. doi: 10.1093/bioinformatics/btu661. [DOI] [PubMed] [Google Scholar]
- 54.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 56.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koboldt DC, Chen K, Wylie T, Larson DE, McLellan MD, Mardis ER, Weinstock GM, Wilson RK, Ding L. 2009. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25:2283–2285. doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, Wishart DS. 2016. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res 44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol 5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, Parkhill J, Harris SR. 2015. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamashita A, Sekizuka T, Kuroda M. 2014. Characterization of antimicrobial resistance dissemination across plasmid communities classified by network analysis. Pathogens 3:356–376. doi: 10.3390/pathogens3020356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Akiba M, Sekizuka T, Yamashita A, Kuroda M, Fujii Y, Murata M, Lee K, Joshua DI, Balakrishna K, Bairy I, Subramanian K, Krishnan P, Munuswamy N, Sinha RK, Iwata T, Kusumoto M, Guruge KS. 2016. Distribution and relationships of antimicrobial resistance determinants among extended-spectrum-cephalosporin-resistant or carbapenem-resistant Escherichia coli isolates from rivers and sewage treatment plants in India. Antimicrob Agents Chemother 60:2972–2980. doi: 10.1128/AAC.01950-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Warnes GR, Bolker B, Bonebakker L, Gentleman R, Liaw W, Lumley T, Maechler M, Magnusson A, Moeller S, Schwartz M, Venables B. 2015. Gplots: various R programming tools for plotting data. R package version 2.17.0. http://CRAN.R-project.org/package=gplots.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw short-read sequence data were deposited in the DNA Data Bank of Japan (BioSample IDs, SAMD00179258 to SAMD00179339; DRA accession no. DRA008702 and DRA008648). The assembled draft sequences of plasmids were deposited in the DDBJ/EMBL/GenBank database (accession no. LC501464 to LC501701).