Each year, 5% to 20% of the population of the United States becomes infected with influenza A virus. Combination therapy with two or more antiviral agents has been considered a potential treatment option for influenza virus infection. However, the clinical results derived from combination treatment with two or more antiviral drugs have been variable. We examined the effectiveness of cotreatment with two distinct classes of anti-influenza drugs, i.e., neuraminidase (NA) inhibitor, laninamivir, and interferon lambda 1 (IFN-λ1), against the emergence of drug-resistant virus variants in vitro.
KEYWORDS: influenza, interferons, laninamivir
ABSTRACT
Each year, 5% to 20% of the population of the United States becomes infected with influenza A virus. Combination therapy with two or more antiviral agents has been considered a potential treatment option for influenza virus infection. However, the clinical results derived from combination treatment with two or more antiviral drugs have been variable. We examined the effectiveness of cotreatment with two distinct classes of anti-influenza drugs, i.e., neuraminidase (NA) inhibitor, laninamivir, and interferon lambda 1 (IFN-λ1), against the emergence of drug-resistant virus variants in vitro. We serially passaged pandemic A/California/04/09 [A(H1N1)pdm09] influenza virus in a human lung epithelial cell line (Calu-3) in the presence or absence of increasing concentrations of laninamivir or laninamivir plus IFN-λ1. Surprisingly, laninamivir used in combination with IFN-λ1 promoted the emergence of the E119G NA mutation five passages earlier than laninamivir alone (passage 2 versus passage 7, respectively). Acquisition of this mutation resulted in significantly reduced sensitivity to the NA inhibitors laninamivir (∼284-fold) and zanamivir (∼1,024-fold) and decreased NA enzyme catalytic activity (∼5-fold) compared to the parental virus. Moreover, the E119G NA mutation emerged together with concomitant hemagglutinin (HA) mutations (T197A and D222G), which were selected more rapidly by combination treatment with laninamivir plus IFN-λ1 (passages 2 and 3, respectively) than by laninamivir alone (passage 10). Our results show that treatment with laninamivir alone or in combination with IFN-λ1 can lead to the emergence of drug-resistant influenza virus variants. The addition of IFN-λ1 in combination with laninamivir may promote acquisition of drug resistance more rapidly than treatment with laninamivir alone.
INTRODUCTION
Influenza virus infection affects millions of people around the world, with mortality estimates of 290,000 to 645,000 deaths per year (1). Influenza virus has been the cause of four pandemics in the past century, including the 1918 Spanish flu and the most recent 2009 H1N1 swine flu [A(H1N1)pdm09]. Neuraminidase inhibitors (NAIs) such as oseltamivir, peramivir, zanamivir, and laninamivir, as well as the recently approved polymerase inhibitor baloxavir marboxil, are the front line of defense in the treatment of influenza virus infection. However, NAI-resistant influenza variants that emerge either naturally or as a result of selective drug pressure have been observed (2–6).
Amino acid mutations that confer resistance to NAIs are often found within the neuraminidase (NA) catalytic site and/or framework region, and changes in these areas may prevent NAIs from effective binding (2). One such mutation, Q136K (N1 numbering here and throughout), was first seen between 2006 and 2008 when 2.3% of isolates circulating in Australia and Southeast Asia acquired this zanamivir-, peramivir-, and laninamivir-resistant mutation (2, 3). In addition, the H275Y mutation is a predominant NA change that confers resistance to oseltamivir and peramivir (2). This mutation emerged rapidly during 2007 to 2009 when the naturally occurring H275Y A(H1N1) variants gained additional permissive mutations that allowed their spread without affecting viral fitness (4, 7). The E119D NA substitution, which resulted in resistance to all four NAIs, has been observed clinically postzanamivir treatment (5, 6). Other N1 NA mutations, including E119A/G/K/V, R152K, I223K/R/V, S247N, and N295S, have also been shown to confer resistance to NAIs in vitro and in vivo (8–11). Thus, the emergence of new drug-resistant influenza virus variants is always a concern.
Laninamivir is a long-lasting anti-influenza NAI, which is inhaled as a prodrug and converted to its active form within the respiratory tract (12). Unlike the other NAIs, laninamivir requires only one dose and remains at a concentration above its influenza 50% inhibitory concentration (IC50) in the lungs and blood plasma for at least 5 days (12). It has been shown that laninamivir remains effective against A(H1N1)pdm09 viruses containing the H275Y NA mutation (13). Despite rapid emergence of drug resistance in cell culture models (8), no viruses displayed resistance to laninamivir over six seasons of clinical observation in Japan (14).
NAIs are effective at preventing influenza virus spread, but the human body also has natural antiviral defense mechanisms, including the ability to express interferon (IFN) proteins with antiviral activity. Virus-induced IFNs are a group of innate cytokines that are produced by many cell types in response to viral infection. IFNs activate a cascade of innate immune responses, including induction of IFN-stimulated genes (ISGs), that control viral infection through a variety of effector functions (15). There have been multiple in vitro and in vivo studies evaluating the anti-influenza potential of type I and type III IFNs (16–23). A recent study with Stat1-deficient mice, which are unable to respond to type I and type II IFNs, showed that these mice displayed a 10-fold lower 50% median lethal dose compared to the wild-type mice following infection with the A/Puerto Rico/8/34 (H1N1) strain (16). Pretreatment with IFN-α/β also resulted in significantly reduced A(H1N1) and A(H5N1) viral replication in mice, demonstrating its potential antiviral protection in the early stages of influenza virus infection (17, 18). In a guinea pig model, human recombinant IFN alpha (IFN-α), administered intranasally, significantly reduced lung and nasal wash titers of a reconstructed 1918 pandemic H1N1 virus as well as a contemporary H5N1 strain (19). Additionally, type III IFN-λs were found to exert variable degrees of antiviral activity in vivo without emergence of resistance against influenza A and B viruses (20, 21), especially in mouse lungs (22). Moreover, IFN-λ1 treatment was shown to be beneficial in the murine upper airway, where sustained induction of ISGs correlated with a significant reduction of viral transmission (23).
In view of the limitations posed by treating influenza with a single drug, combination antiviral drug therapy has been considered an alternative means to reduce development of drug resistance and decrease viral spread. However, variable results have been obtained based on the combinations used so far. While combined use of rimantadine and nebulized zanamivir led to a faster resolution of viral shedding in patients with lower respiratory tract influenza infection (24), the combination of oseltamivir and inhaled zanamivir was less effective than oseltamivir alone in controlling viral infection and reducing disease severity (25). It was hypothesized that there may be a negative interaction at the NA catalytic binding site when antiviral drugs with similar modes of action are used. Therefore, combination therapy with drugs having different modes of action may be more effective in mitigating the emergence of drug resistance, as demonstrated previously when amantadine was combined with oseltamivir in vitro (26). Multiple in vitro and in vivo studies have also evaluated the triple combination of amantadine, oseltamivir, and ribavirin (27–30). A phase II trial of triple-combination therapy indicated that this may be a more effective treatment option because there was a significant decrease in viral shedding, even though no clinical benefits were observed (30). Additionally, our previous evaluation of the combination of oseltamivir and recombinant IFN-β or IFN-λ1 in vitro indicated that the use of an NAI plus IFN may provide an effective influenza treatment (31).
So far, no studies have assessed the effect of combination therapy with laninamivir and type III interferon (IFN-λ1) against A(H1N1)pdm09 influenza virus in vitro. Therefore, we examined whether combined treatment with these two classes of anti-influenza drugs can alter the emergence of drug-resistant variants. We used the human airway epithelial cell line, Calu-3, to serially passage A/California/04/09 influenza virus in the presence of increasing concentrations of laninamivir, laninamivir plus IFN-λ1, or media alone, to generate virus variants that displayed various degrees of NAI resistance. The Calu-3 cell line functionally recapitulates human airway epithelium and provides an ideal in vitro model for predicting the emergence of NA inhibitor-resistant variants and evaluating the mechanism of action of laninamivir−IFN-λ1 interactions against influenza infection.
RESULTS
Susceptibility of A(H1N1)pdm09 virus to laninamivir and IFN-λ1.
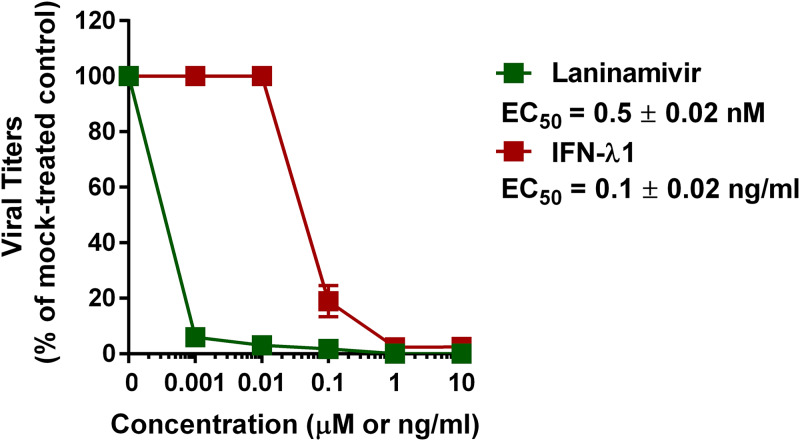
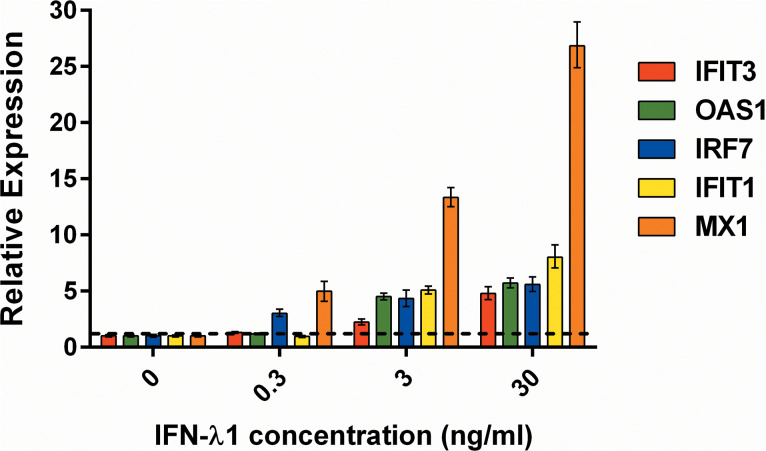
We determined the susceptibility of wild-type (WT) A(H1N1)pdm09 virus to treatment with laninamivir or IFN-λ1 using the virus yield reduction assay in Calu-3 cells (Fig. 1). The 50% effective concentration (EC50) values for laninamivir and IFN-λ1 were 0.5 ± 0.02 nM and 0.1 ± 0.02 ng/ml, respectively, which is consistent with our previous findings (32). We next measured the induction of several ISGs in Calu-3 cells after treatment with IFN-λ1 at a starting concentration of 0.3 ng/ml, which is equal to three times (3×) the EC50 value. ISG expression levels were measured by quantitative real-time PCR (qPCR). As shown in Fig. 2, an approximate 4-fold increase in IRF7 and MX1 gene expression levels was observed in the cells after treatment with 0.3 ng/ml IFN-λ1 compared to the untreated control cells. Therefore, 0.3 ng/ml IFN-λ1 was chosen for use in our antiviral drug combination experiments.
FIG 1.
Antiviral effect of laninamivir (μM) and IFN-λ1 (ng/ml) against the WT virus in Calu-3 cells as measured by the virus yield reduction assay. Mean EC50 values and standard deviations are calculated from two independent experiments.
FIG 2.
Effect of treatment with IFN-λ1 (0.3, 3, and 30 ng/ml) on induction of individual ISGs (i.e., IFIT3, OAS1, IRF7, IFIT1, and MX1) in Calu-3 cells. Expression level values higher than the threshold line are statistically significant (P < 0.05 compared with the results for the mock-treated cells; one-way ANOVA).
Generation of A(H1N1)pdm09 virus variants by serial passaging in Calu-3 cells.
To evaluate the effects of single and combination treatment with laninamivir and IFN-λ1 on viral growth and plaque morphology, WT virus was serially passaged 10 times in the presence of increasing concentrations of laninamivir (0.01 μM to 50 μM) or laninamivir plus a fixed concentration (0.3 ng/ml) of IFN-λ1. To evaluate the effect of serial passaging of the virus without any selective pressure, we passaged the WT virus in Calu-3 in the presence of media alone (no drug). Viral titers and plaque size were measured by plaque assay in Madin-Darby canine kidney (MDCK) cells at each passage (P).
Viruses passaged in the presence of single or combination therapy produced average viral titers of 5.6 ± 0.9 or 6.0 ± 0.8 log10 PFU/ml, respectively (Fig. 3A). The WT virus passaged without selective pressure produced average viral titers of 7.1 ± 1.3 log10PFU/ml. Virus passaged in the presence of laninamivir demonstrated mixed plaque morphology at P3 and P4 with both large plaques of 1.2 ± 0.2 mm and small plaques of 0.2 ± 0.1 mm (Fig. 3B). The WT virus passaged in media alone exhibited significantly increased plaque sizes throughout the 10 passages, ranging from the largest at P8 (1.4 ± 0.3 mm) to the smallest at P1 (0.5 ± 0.2 mm; P < 0.01) (Fig. 3B). At P5, a mixed population was observed by plaque morphology that included two distinct sizes of WT plaques: 1.5 ± 0.3 mm and 0.5 ± 0.2 mm. In contrast, no mixed population was observed in virus that was passaged in the presence of laninamivir plus IFN-λ1 (plaque size ≈ 0.2 ± 0.03) (Fig. 3B).
FIG 3.
Comparison of viral titers (A) and plaque sizes (B) in the presence of increasing concentrations of laninamivir (WT+LAN), laninamivir plus IFN-λ1 (WT+LAN+IFN-λ1), or in the absence of selective pressure (WT with media) over 10 passages in Calu-3 cells. *, P < 0.05; °, P < 0.01 compared to WT by one-way ANOVA; M, mixed population present.
Six virus variants were selected during the passaging protocol, and they were designated as follows: LAN (P5), LAN (P7), and LAN (P10) for viruses passaged in the presence of laninamivir, LAN+IFN (P5) and LAN+IFN (P10) for viruses passaged in the presence of laninamivir plus IFN-λ1, and P10 for virus passaged in the presence of media alone. We next determined the growth kinetics of the WT virus and our selected variants at 24, 48, and 72 h postinfection (hpi) in Calu-3 cells (Fig. 4). The LAN (P7), LAN (P10), LAN+IFN (P5), and LAN+IFN (P10) viruses exhibited significantly increased viral replication titers compared to the WT virus at 24 and 48 hpi (∼3.2-fold increase; P < 0.01). LAN+IFN (P10) also showed significantly increased viral titers at 72 hpi (1.2-fold increase; P < 0.05). The control P10 virus demonstrated significantly higher viral titers than the WT virus at each time point (∼2.3-fold increase; P < 0.01).
FIG 4.
Replication of the WT, LAN (P5), LAN (P7), LAN (P10), LAN+IFN (P5), LAN+IFN (P10), and P10 variants in Calu-3 cells. *, P < 0.05; °, P < 0.01 compared to WT at corresponding time point by one-way ANOVA.
Sequence analysis of selected A(H1N1)pdm09 variants.
We next sequenced the complete genomes of our selected variants (Table 1). The D127E HA1 mutation (H1 hemagglutinin [HA] numbering here and throughout) was found in all viruses sequenced. While T197A and D222G were present as mixed populations in LAN (P10) and LAN+IFN (P5), D222G was present as a pure population in the LAN+IFN (P10) variant (Fig. S1 in the supplemental material). The E119G NA mutation was observed as a mixed population in LAN (P7), and it became a pure population in LAN (P10). Notably, the E119G mutation was found as a pure population in LAN+IFN (P5) and remained as such in LAN+IFN (P10) variant (Fig. S1). Interestingly, the G155E HA1 and Y56C NA mutations were found in the P10 variant only (Table 1). To analyze the speed of emergence of the HA and NA mutations in the variants which were under treatment, we performed additional sequence analysis of the P1 to P4 passages (Fig. S2). We observed that the LAN+IFN combination selective pressure promoted the emergence of T197A HA1 and E119G NA as early as P2, indicating that the latter mutation arose at least five passages sooner than it was observed in the virus passaged in the presence of laninamivir alone [i.e., LAN (P7); Fig. S1]. The D127E HA1 mutation was first seen at P2 after treatment with laninamivir alone or in combination with IFN-λ1 (Fig. S2).
TABLE 1.
Amino acid changes identified in selected A(H1N1)pdm09 variants
| Virus | Passage no. | Laninamivir (μM) | IFN-λ1 (ng/ml) | HA1a,b | NP | NAb,c | M1 | NS1 |
|---|---|---|---|---|---|---|---|---|
| LAN (P5) | 5 | 1.3 | D127E | |||||
| LAN (P7) | 7 | 5.1 | D127E | E119G | ||||
| LAN (P10) | 10 | 50.0 | D127E, T197A, D222G | L108P | E119G | A149G | ||
| LAN+IFN (P5) | 5 | 1.3 | 0.3 | D127E, T197A, D222G | E119G | |||
| LAN+IFN (P10) | 10 | 50.0 | 0.3 | D127E, T197A, D222G | E119G | A227S | ||
| P10 | 10 | D127E, G155E | Y56C |
We next analyzed the frequency of HA and NA amino acid mutations present in a pure population in our variants in ∼35,000 A(H1N1)pdm09 virus sequences (Fig. S3; https://www.fludb.org). The D127E and G155E HA1 mutations were found at the highest frequency of 1.4% in 2012 and the lowest frequency of ∼0.1% in 2018. The D222G HA1 mutant fluctuated in frequency from 2.6% in 2010 to ∼0.1% in 2018. The E119G NA mutation was observed sporadically (∼0.4% in 2014), and the Y56C NA variant was only seen in one virus in 2009 (Fig. S3).
Effect of HA changes on HA receptor affinity.
To evaluate the effect of the D127E and G155E HA1 mutations observed as a pure population on HA binding properties, we examined the affinity of the WT, LAN (P7), and P10 variants to four biotinylated α2,6-sialylglycopolymers (6-Su-6′SLN, YDS, 6′SLN, and 6′SL) using a direct binding assay (Fig. 5) (33, 34). Affinity for α2,3 glycans was not measured because the affinity of the WT virus for these receptors was minimal (35). LAN (P7) containing the D127E mutation exhibited a significant decrease in affinity (2.8-fold) to 6′SL compared to WT (P < 0.01). In contrast, P10 with the double HA1 mutation, D127E and G155E, demonstrated a statistically significant decrease in binding affinity to 6Su-6′SLN, YDS, and 6′SL glycans compared to the WT virus (3.4-fold; P < 0.01) (Fig. 5).
FIG 5.
HA receptor affinity of the WT, LAN (P7), and P10 variants. °, in color, P < 0.01, compared to WT by one-way ANOVA. *, in black, P < 0.05; °, in black, P < 0.01, compared to LAN (P7) by one-way ANOVA.
Effect of NA mutations on susceptibility to NAIs and enzymatic activity.
We next examined the effect of the NA mutations found in our selected variants on susceptibility to three NAIs, oseltamivir carboxylate, zanamivir, and laninamivir, using an enzyme-based NA inhibition assay (Table 2). LAN (P7) carrying the E119G NA mutation as a mixed population exhibited reduced inhibition by laninamivir (11.5-fold increase in IC50) compared to WT. The LAN (P10), LAN+IFN (P5), and LAN+IFN (P10) variants containing the E119G mutation demonstrated highly reduced inhibition by zanamivir (∼1,024-fold increase in IC50) and laninamivir (∼284-fold increase in IC50) compared to WT, but all three of these viral variants remained susceptible to oseltamivir carboxylate. The P10 variant remained susceptible to all NAIs tested despite the presence of the Y56C NA mutation (Table 2).
TABLE 2.
Sensitivity of selected A(H1N1)pdm09 variants to NAIsa
| Virus | Passage no. | Oseltamivir carboxylate |
Zanamivir |
Laninamivir |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Laninamivir (μM) | IFN-λ1 (ng/ml) | IC50 (nM) | Fold change (phenotype) | IC50 (nM) | Fold change (phenotype) | IC50 (nM) | Fold change (phenotype) | ||
| WT | 1.7 ± 0.5 | 1.1 ± 0.1 | 0.2 ± 0.1 | ||||||
| LAN (P5) | 5 | 1.3 | 1.9 ± 0.3 | 1.1 (S) | 1.1 ± 0.1 | 1.0 (S) | 0.3 ± 0.1 | 1.5 (S) | |
| LAN (P7) | 7 | 5.1 | 2.4 ± 0.8 | 1.4 (S) | 9.3 ± 1.4 | 8.5 (S) | 2.3 ± 0.7 | 11.5 (RI) | |
| LAN (P10) | 10 | 50.0 | 2.9 ± 0.2 | 1.6 (S) | 1,158.5 ± 218.5 | 1,053.2 (HRI) | 67.8 ± 2.1 | 339.0 (HRI) | |
| LAN+IFN (P5) | 5 | 1.3 | 0.3 | 5.7 ± 0.1 | 3.4 (S) | 934.6 ± 45.5 | 849.6 (HRI) | 50.3 ± 2.6 | 251.5 (HRI) |
| LAN+IFN (P10) | 10 | 50.0 | 0.3 | 2.5 ± 0.4 | 1.5 (S) | 1,287.5 ± 99.7 | 1,170.5 (HRI) | 52.1 ± 4.0 | 260.5 (HRI) |
| P10 | 10 | 1.2 ± 0.1 | 0.7 (S) | 1.9 ± 0.1 | 1.7 (S) | 0.2 ± 0.1 | 1.0 (S) | ||
Viral sensitivities were determined by NA inhibition assay. The values were fit to nonlinear regression curves with the variable slope model to determine the IC50, using GraphPad Prism. Values are means ± standard deviation from two or more independent experiments. IC50-fold changes were calculated relative to the WT virus. Phenotypes are described as follows: S, susceptible (<10-fold increase in IC50 compared to WT); RI, reduced inhibition (10- to 100-fold increase in IC50 compared to WT); HRI, highly reduced inhibition (>100-fold increase in IC50 compared to WT).
To evaluate the effect of the E119G NA mutation on NA enzymatic activity, a fluorescence-based assay based on 2′-(4-methylumbelliferyl)α-d-N-acetylneuraminic acid sodium salt hydrate (MUNANA) catalyzation was used to measure changes in the relative enzymatic activity (Vmax), Michaelis-Menten constant (Km), and catalytic efficiency (kcat/Km) of mutant NA compared to WT NA (Table 3). We observed a significant decrease in enzymatic activity of LAN+IFN (P10) carrying the E119G mutation (Vmax ratio relative to WT = 0.2; P < 0.05). Moreover, LAN+IFN (P10) demonstrated a significantly reduced MUNANA affinity (37.6-fold) and catalytic efficiency (7.0-fold) compared to the WT virus.
TABLE 3.
NA enzyme kinetics properties of purified influenza A(H1N1)pdm09 viruses
| Virus | Vmax (μM/min) | Vmax ratioa | Km (μM)b | Kcat/Km (μM min)−1c |
|---|---|---|---|---|
| WT | 5.2 ± 0.7 | 1.0 | 203.7 ± 71.4 | 7.7 ± 2.8 |
| LAN+IFN (P10) | 1.1 ± 0.8c | 0.2c | 7,666.0 ± 843.2c | 0.1 ± 0.04c |
The Vmax was calculated using nonlinear regression of the curve according to the Michaelis-Menten equation, and then the ratio of the respective viruses’ NA Vmax to the Vmax of the wild-type CA virus was determined.
The Km represents the Michaelis-Menten constant at which the reaction rate is half of Vmax. The enzyme kinetic data were fit to the Michaelis-Menten equation using GraphPad Prism, version 7.0. Values are the means ± 95% confidence interval from 3 independent determinations.
P < 0.05 compared with the value for WT by unpaired t test.
Drug-drug interaction of laninamivir and IFN-λ1.
Due to the more rapid emergence of drug-resistant NA mutations when the WT virus was passaged in the presence of laninamivir plus IFN-λ1 versus laninamivir alone (P2 versus P7), we examined potential drug-drug interactions between laninamivir and IFN-λ1 at concentrations similar to those used for serial passaging in Calu-3 cells. We observed drug antagonism (−2.04%) when 0.13 μM laninamivir was combined with 0.3 ng/ml IFN-λ1 (Fig. 6). These drug concentrations were equivalent to those used at P1.
FIG 6.
Three-dimensional plot showing the interaction of laninamivir and IFN-λ1. Antagonism was calculated at a 99.9% confidence level.
DISCUSSION
NAIs provide the front line of defense in the treatment of influenza virus infection. However, drug-resistant mutants can arise either naturally or as a result of drug pressure imposed by the clinical use of NAIs. Therefore, it is important to identify additional novel therapeutic options in case drug-resistant virus variants arise. Combination therapy with two or more antiviral agents has been considered a potential option for influenza treatment. However, the clinical results derived from combination treatment with two or more antiviral drugs have been variable. In this study, we evaluated the effectiveness of laninamivir combined with IFN-λ1 against the emergence of A(H1N1)pdm09 drug-resistant variants.
We observed that laninamivir used as a single drug or in combination with IFN-λ1 promoted emergence of the E119G NA mutation, which correlated with significantly reduced susceptibility to laninamivir and zanamivir (i.e., 283.7- and 1,024.4-fold decreases, respectively) in Calu-3 cells. These results are consistent with similar findings reported by Samson et al., which showed that the E119G NA mutation was associated with significantly decreased sensitivity of the A/Quebec/144147/2009 [A(H1N1)pdm09] influenza virus to laninamivir and zanamivir (8). The amino acid residue at position 119 is the key NA residue in the influenza virus that determines susceptibility to laninamivir and zanamivir. Both laninamivir and zanamivir contain a 4-guanidino group, which forms hydrogen bonds with the viral NA glutamic acid at this position. Therefore, amino acid changes at residue 119 can lead to reduced NA binding by both NAIs (36). Moreover, the E119G NA mutation was also shown to confer resistance to peramivir because of the guanidino group in this drug (8). In contrast to the three NAIs discussed above (i.e., laninamivir, zanamivir, and peramivir), oseltamivir contains an acetamide group instead of guanidine, which explains the high sensitivity of our selected variants containing the E119G mutation to this NAI.
Amino acid changes at residue 119, i.e., E119A/G/K/V, were shown previously to reduce NA activity and decrease viral replication in vitro (37). A(H1N1)pdm09 virus with the E119G mutation demonstrated significantly reduced NA activity and decreased viral fitness and transmissibility in ferrets (38). The E119G NA mutation evolved alone or in combination with H275Y in an immunocompromised infant who was infected with A(H1N1)pdm09 virus and then treated with oseltamivir and zanamivir. This double E119G/H275Y change was associated with highly reduced inhibition by four different NAIs (39). Analysis of the E119G mutation frequency indicated that the emergence of this NA substitution is a rare event (e.g., only 0.4% was observed in 2014). However, if this amino acid change emerges alone or in conjunction with H275Y in circulating viruses, such an influenza strain could display resistance to either three or all four of the NAIs that are currently available for treatment of influenza virus infection (10, 11).
Three HA mutations, D127E, T197A, and D222G, were observed in all variants passaged in the presence of laninamivir alone or laninamivir plus IFN-λ1. Interestingly, the emergence of the D127E HA1 mutation preceded acquisition of the E119G NA mutation in the presence of laninamivir. This mutation appeared five passages earlier than E119G under laninamivir selective pressure and was conserved throughout the drug-mediated selection process. The T197A and D222G HA1 substitutions were selected more rapidly by the combination treatment (P2 and P3, respectively) than by the single-drug pressure (P10). The D127E HA1 mutation correlated with a significant decrease in 6′SL binding affinity of the WT virus and has been shown to contribute to more efficient and prolonged viral replication of A/California/04/09 [A(H1N1)pdm09] virus in the lungs of experimentally infected mice (40). The D222G mutation, associated with the HA receptor binding site and mapped to the antigenic site Ca (41), conferred enhanced virulence to A(H1N1)pdm09 virus via increased binding affinity to α2,3 sialyl receptors while maintaining α2,6 specificity (42). Moreover, the D222G HA1 mutation was associated with higher viral titers in Calu-3 cells (43) and more prolonged hospitalization of patients (44). This mutation was also associated with a higher frequency of severe and fatal cases (44). Taking into account the balance between the functions of the HA and NA glycoproteins (45), we surmise that the mechanism of in vitro laninamivir resistance involves compensating HA mutation(s) that modify virus binding to its cognate cell surface receptors. These changes would result in increased efficiency of viral release from infected cells with less dependence on the reduced NA function.
To monitor the emergence of amino acid changes associated with adaptation of the WT virus to growth in Calu-3 cells, we passaged the parental virus in parallel in the absence of any selective pressure by antivirals. Two mutations in the HA1 protein (D127E and G155E) and one mutation in the NA protein (Y56C) were associated with adaptation of the parental A(H1N1)pdm09 virus to growth in respiratory epithelial cells. We showed previously that the G155E mutation located within the Sa antigenic site is necessary for the enhanced growth of the A/California/04/09 [A(H1N1)pdm09] virus both in vitro in Calu-3 cells and in vivo in mice (43, 46). Further characterization of the receptor specificity of the P10 variant revealed that the G155E HA1 substitution was associated with a significant decrease in binding to α2,6 glycans, i.e., 6Su-6′SLN, YDS, and 6′SL. It is likely that the observed HA1 changes (D127E and G155E) in the P10 variant contributed to the increased ability of the parental virus to infect and spread in mammalian respiratory epithelium.
It was shown previously by others that certain ISGs can enhance virus infectivity (47, 48). The ISG, LY6E, is a member of the LY6-uPAR family that was found to promote HIV-1 virus entry in human cells (47). Moreover, Mar et al. demonstrated that LY6E is also able to promote A/WSN/33 (H1N1) influenza virus entry and uncoating in human airway epithelial cells (48). Based on these findings, one can speculate that combination antiviral drug therapy could trigger proviral ISGs and/or host factors that might explain the antagonistic interaction between laninamivir and IFN-λ1 that we observed in this study. Additional studies will be necessary to identify and measure the expression of proviral ISGs that might be induced by treatment with laninamivir alone or laninamivir in combination with IFN-λ1. Such studies might help to determine if the induction of one or more proviral ISGs accounts for the more rapid emergence of HA and NA mutations after treatment with laninamivir plus IFN-λ1 versus laninamivir alone.
In summary, our results show that resistance to NAIs can be induced by extended exposure to laninamivir alone or laninamivir in combination with IFN-λ1. Development of antiviral drug resistance in Calu-3 cells correlated with acquisition of the E119G NA mutation and was associated with decreased NAI sensitivity and enzyme catalytic activity. This NA mutation emerged together with concomitant HA mutations that modify virus receptor-binding affinity. These observations highlight a critical interrelationship between the HA and NA proteins. The identification of HA/NA mutation patterns that confer resistance to NAIs may help to predict clinical cases of antiviral drug resistance. Therefore, our finding that combined treatment with laninamivir plus IFN-λ1 induced stronger HA and NA mutation pressure than laninamivir alone indicates that this combination may be less effective than using either agent alone in vivo. Caution should be exercised when considering the possibility of combined therapy with this NAI together with recombinant IFN in the treatment of influenza virus infection.
MATERIALS AND METHODS
Cells, viruses, and compounds.
The MDCK and Calu-3 cell lines were obtained from the American Type Culture Collection (Manassas, VA, USA) and were maintained as described previously (31, 49).
Human influenza A/California/04/09 [A(H1N1)pdm09] virus was kindly provided by Robert G. Webster (St. Jude Children’s Research Hospital, Memphis, TN, USA). Stock virus was prepared by one passage in the allantoic cavities of 10-day-old embryonated chicken eggs for 48 h at 37°C, and aliquots were stored at −70°C until use. Stock viruses of six selected variants [i.e., LAN (P5), LAN (P7), LAN (P10), LAN+IFN (P7), LAN+IFN (P10), and P10] were prepared in Calu-3 cells. All experimental work was performed in a biosafety level 2 laboratory approved for use of these strains by the U.S. Department of Agriculture and the U.S. Centers for Disease Control and Prevention.
Human recombinant IFN-λ1 protein was obtained from R&D Systems, Inc. (Minneapolis, MN, USA). Oseltamivir carboxylate was provided by Roche Diagnostics GmbH (Mannheim, Germany). Zanamivir was obtained from Sigma-Aldrich (St. Louis, MO, USA). Laninamivir was provided by Daiichi Sankyo Co., Ltd. (Tokyo, Japan).
Infectivity of A(H1N1)pdm09 influenza viruses.
The infectivity of WT and selected variants was determined by at least three or four independent plaque assays (50). Briefly, confluent cultures of MDCK cells were incubated at 37°C for 1 h with 10-fold serial dilutions of each virus prepared in infection medium (i.e., minimal essential medium [MEM] containing 4% bovine serum albumin [BSA], sodium bicarbonate, 100 U of penicillin per ml, and 1 μg/ml l-[tosylamido-2-phenyl]ethylchloromethylketone [TPCK]-treated trypsin). The cells were then washed and overlaid with MEM containing 0.3% BSA, 0.9% Bacto agar, and 1 μg/ml TPCK-treated trypsin. After 3 days of incubation at 37°C, the cells were stained with 0.1% crystal violet in 10% formaldehyde solution. The number of PFU per milliliter and plaque size of any 10 plaques were determined using a Finescale magnifying comparator.
Virus yield reduction assay.
The extracellular virus yield reduction assay was performed as described previously in 24-well plates containing confluent Calu-3 cells (26). The concentrations of IFN-λ1 ranged from 0.01 to 10 ng/ml (the 50% cytotoxic concentration for IFN-λ1 is >1,000 ng/ml), and cells were pretreated with IFN-λ1 overnight (31). The concentrations of laninamivir ranged from 0.001 to 10 μM, and cells were pretreated with laninamivir for 2 h. After pretreatment, the cells were infected with influenza virus (100 μl/well) at a multiplicity of infection (MOI) of 0.01 PFU/cell for 1 h and then incubated in a drug-containing medium for 72 h at 37°C. Virus yields were determined as the number of PFU/ml in MDCK cells. The drug concentration that caused a 50% decrease in the PFU titer in comparison to control wells without drug was defined as the EC50. The results of two independent experiments, each consisting of two replicates, were averaged.
Virus yield reduction assay was also performed for synergy/antagonism determination for laninamivir combined with IFN-λ1 in Calu-3 cells. Briefly, cells were pretreated overnight with 0, 0.03, 0.3, and 3 ng/ml concentrations of IFN-λ1. Laninamivir was added for 2 h pretreatment at concentrations of 0, 0.13, 1.3, and 10.3 μM. Cells were then infected with WT at an MOI of 0.01 PFU/cell for 1 h, followed by 3-day incubation with media containing the combination of laninamivir and IFN-λ1. Virus yields were determined by plaque assay in MDCK cells. Values are the mean of two independent experiments, each consisting of two replicates.
Measurement of ISG expression.
Quantification of changes in ISG gene expression was carried out by qPCR measurement of individual ISGs, IFIT3, OAS1, IRF7, IFIT1, MX1, IRF3, and CXCL10. Total cellular RNA was isolated from Calu-3 cells stimulated with 0, 0.3, 3, and 30 ng/ml IFN-λ1 for 24 h using RNeasy minikit (Qiagen, Germantown, MD, USA). The RNA samples were then treated with DNase, and 1.6 μg of each purified RNA sample was then reverse transcribed to cDNA with Quantiscript reverse transcriptase (Qiagen). The cDNAs were mixed with RT2 SYBR green qPCR mastermix (Qiagen), and qPCR analyses were performed using the ViiA 7 instrument (Applied Biosystems, Waltham, MA, USA). Graphing and statistical analysis of the qPCR results were performed using Prism 7.0 (GraphPad Software, La Jolla, CA, USA). Values are the means of three independent determinations. No changes in IRF3 or CXCL10 gene expression were observed in Calu-3 cells after treatment with 0, 0.3, 3, or 30 ng/ml IFN-λ1 for 24 h.
Viral replication kinetics.
To determine multistep growth curves, Calu-3 cells were infected with the A(H1N1)pdm09 viruses at an MOI of 1 PFU/cell. After incubation for 1 h, the cells were washed and overlaid with MEM containing 0.3% BSA and 1 μg/ml TPCK-treated trypsin. The supernatants were collected at 24, 48, and 72 hpi and stored at −80°C until titration. The results of at least two or three independent experiments were averaged.
Virus sequence analysis.
Viral RNAs were isolated from virus-containing cell culture fluid after passages in Calu-3 cells. Samples were reverse transcribed and analyzed by PCR using universal primers specific for influenza gene segments as described previously (51). Sequencing was performed by the Research Central Facility for Biotechnology Resources at the U.S. Food and Drug Administration, Silver Spring, MD. DNA sequences were completed and edited by using a Lasergene sequence analysis software package (DNASTAR, Madison, WI, USA).
Receptor-binding assay.
The affinity for biotinylated 6′-glycans was measured in a direct binding assay as described previously (33–35). Briefly, plates were precoated with each virus at 4°C for 16 h, followed by washing with 0.05% Tween 20 in phosphate-buffered saline (PBS). After the addition of biotinylated sialylglycopolymer in PBS supplemented with 0.02% Tween 20, 0.02% BSA, and 3 mM oseltamivir carboxylate, plates were incubated at 4°C for 1 h. Plates were then washed with cold PBS-Tween 20 and incubated with streptavidin-peroxidase (Sigma-Aldrich) at 4°C for 1 h. After washing, tetramethylbenzidine (TMB) substrate solution (KPL, Gaithersburg, MD, USA) was added, and the reaction was stopped with TMB stop solution (KPL). Optical density was determined at 450 nm with a Synergy 2 multimode microplate reader (BioTek Instruments, Winooski, VT, USA). The association constant (Ka; 1/μM sialic acid) values were determined by fitting the data to the one site-total binding equation by using nonlinear regression in Prism 7.0 software (GraphPad Software). The reported data represent the mean of at least four individual and independent experiments for each virus.
Virus purification.
Allantoic fluid was clarified by low-speed centrifugation. The WT and LAN+IFN (P10) viruses were pelleted and then purified through 27% and 49% (wt/vol) sucrose cushions. Virus-containing bands were pelleted and stored in PBS at −80°C until use. NA concentrations were determined by optical densitometry of the SDS-PAGE gel images, and total protein content was determined by bicinchoninic acid protein assay (Pierce Biotechnology, Rockford, IL, USA).
NA enzyme inhibition assay.
Influenza viruses were standardized to equivalent NA activity and incubated for 30 min at 37°C with NAIs at concentrations of 0.0001 to 5 μM and with MUNANA (Sigma-Aldrich) as a substrate for 1 h at 37°C. The reaction was then terminated by adding 14 mM NaOH, and fluorescence was quantified in a Synergy 2 multimode microplate reader. The IC50 concentration of each NAI was determined by plotting the dose-response curve of inhibition of NA activity as a function of the compound concentration. Values represent the mean of at least two independent experiments, each consisting of two or three replicates.
NA enzyme activity.
The NA activity of influenza viruses was measured by a fluorescence-based assay using the fluorogenic substrate MUNANA (Sigma-Aldrich), based on the method of Potier et al. (52) as described previously (32). Briefly, all viruses were standardized to an equivalent NA protein content of 0.2 ng/μl as determined by protein gel electrophoresis using purified and concentrated viruses. This virus dilution was selected as a dilution that converted ≤15% MUNANA substrate to product during the reaction time to meet the requirements for steady-state kinetic analysis (53). Virus dilutions were prepared in enzyme buffer (32.5 mM 2-[N-morpholino]ethanesulfonic acid, 4 mM calcium chloride, pH 6.5) and added (100 μl/well) in duplicate to a flat-bottom 96-well opaque black plate (Corning, Tewksbury, MA, USA). After preincubation for 20 to 30 min at 37°C, the MUNANA substrate at various concentrations (separately preincubated for 20 to 30 min at 37°C) was added to all wells (50 μl/well). Immediately after adding the MUNANA substrate, the plate was transferred to a 37°C prewarmed Synergy 2 multimode microplate reader, and fluorescence was measured every 60 s for 60 min at 37°C, using excitation and emission wavelengths of 360 nm and 460 nm, respectively. Time course data from each concentration of the MUNANA substrate were examined for linearity by linear regression analysis. The kinetic parameters Michaelis-Menten constant (Km), maximum velocity of substrate conversion (Vmax), and catalytic efficiency (kcat/Km) of the NAs were calculated by fitting the data to the appropriate Michaelis-Menten equations by using nonlinear regression in Prism 7.0 software (GraphPad Software). Values represent the means of at least two to four individual and independent determinations.
Statistical analysis and synergy determinations.
ISG expression values, virus yield, plaque size, binding to sialyl receptors, IC50 values, and NA enzyme kinetic parameters (Km, Vmax, and kcat/Km) of the WT and selected variants were compared by unpaired t test or analysis of variance (ANOVA). Probability values ≤0.05 indicate statistically significant differences.
The combination data were analyzed with MacSynergy II software (54). Theoretical additive interactions were calculated from dose-response curves for each drug used individually. This calculated additive surface was then subtracted from the experimentally determined dose-response surface to give regions of nonadditive interactions. The confidence intervals around the experimental dose-response surface were used to evaluate the data statistically, and the volume of peaks was calculated and used to quantify the volume of synergy (or antagonism) produced. The guidelines for volumes of synergy determinations expressed as unit2% (unit × unit × percent) at a 99.9% confidence level were as follows: 0 to 25, insignificant; 25 to 50, minor but significant; 50 to 100, moderate; >100, strong synergy or antagonism. Synergy plots were generated at the 99.9% confidence limit.
Data availability.
Sequences have been deposited in GenBank under accession numbers MT423008 to MT423014.
Supplementary Material
ACKNOWLEDGMENTS
Simone E. Adams was supported in part by an appointment to the Research Participation Program in the Office of Biotechnology Products, Center for Drug Evaluation and Research at the U.S. Food and Drug Administration (FDA) administered by the Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement between the FDA and the U.S. Department of Energy.
We are grateful to Robert G. Webster (St. Jude Children’s Research Hospital, Memphis, TN) for providing A/California/04/09 (H1N1) virus.
We declare that we have no competing interests. This article reflects the views of the authors and should not be construed to represent FDA’s views or policies.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Juliano AD, Roguski KM, Chang HH, Muscatello DJ, Palekar R, Tempia S, Cohen C, Gran JM, Schanzer D, Cowling BJ, Wu P, Kyncl J, Ang LW, Park M, Redlberger-Fritz M, Yu H, Espenhain L, Krishnan A, Emukule G, van Asten L, da Silva SP, Aungkulanon S, Buchholz U, Widdowson MA, Bresee JS, Global Seasonal Influenza-associated Mortality Collaborator Network . 2018. Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet 391:1285–1300. doi: 10.1016/S0140-6736(17)33293-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hussain M, Galvin HD, Haw TY, Nutsford AN, Husain M. 2017. Drug resistance in influenza A virus: the epidemiology and management. Infect Drug Resist 10:121–134. doi: 10.2147/IDR.S105473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hurt AC, Holien JK, Parker M, Kelso A, Barr IG. 2009. Zanamivir-resistant influenza viruses with a novel neuraminidase mutation. J Virol 83:10366–10373. doi: 10.1128/JVI.01200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2009. Update: influenza activity—United States, September 28, 2008–April 4, 2009, and composition of the 2009–10 influenza vaccine. Morb Mortal Wkly Rep 58:369–374. [PubMed] [Google Scholar]
- 5.L'Huillier AG, Abed Y, Petty TJ, Cordey S, Thomas Y, Bouhy X, Schibler M, Simon A, Chalandon Y, van Delden C, Zdobnov E, Boquete-Suter P, Boivin G, Kaiser L. 2015. E119D neuraminidase mutation conferring pan-resistance to neuraminidase inhibitors in an A(H1N1)pdm09 isolate from a stem-cell transplant recipient. J Infect Dis 212:1726–1734. doi: 10.1093/infdis/jiv288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yates PJ, Raimonde DS, Zhao HH, Man CY, Steel HM, Mehta N, Peppercorn AF. 2016. Phenotypic and genotypic analysis of influenza viruses isolated from adult subjects during a phase II study of intravenous zanamivir in hospitalised subjects. Antiviral Res 134:144–152. doi: 10.1016/j.antiviral.2016.08.023. [DOI] [PubMed] [Google Scholar]
- 7.Bloom JD, Gong LI, Baltimore D. 2010. Permissive secondary mutations enable the evolution of influenza oseltamivir resistance. Science 328:1272–1275. doi: 10.1126/science.1187816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samson M, Abed Y, Desrochers FM, Hamilton S, Luttick A, Tucker SP, Pryor MJ, Boivin G. 2014. Characterization of drug-resistant influenza virus A(H1N1) and A(H3N2) variants selected in vitro with laninamivir. Antimicrob Agents Chemother 58:5220–5228. doi: 10.1128/AAC.03313-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tu V, Abed Y, Fage C, Baz M, Boivin G. 2018. Impact of R152K and R368K neuraminidase catalytic substitutions on in vitro properties and virulence of recombinant A(H1N1)pdm09 viruses. Antiviral Res 154:110–115. doi: 10.1016/j.antiviral.2018.04.009. [DOI] [PubMed] [Google Scholar]
- 10.Lackenby A, Besselaar TG, Daniels RS, Fry A, Gregory V, Gubareva LV, Huang W, Hurt AC, Leang SK, Lee RTC, Lo J, Lollis L, Maurer-Stroh S, Odagiri T, Pereyaslov D, Takashita E, Wang D, Zhang W, Meijer A. 2018. Global update on the susceptibility of human influenza viruses to neuraminidase inhibitors and status of novel antivirals, 2016–2017. Antiviral Res 157:38–46. doi: 10.1016/j.antiviral.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen HT, Fry AM, Gubareva LV. 2012. Neuraminidase inhibitor resistance in influenza viruses and laboratory testing methods. Antivir Ther 17:159–173. doi: 10.3851/IMP2067. [DOI] [PubMed] [Google Scholar]
- 12.Ikematsu H, Kawai N. 2011. Laninamivir octanoate: a new long-acting neuraminidase inhibitor for the treatment of influenza. Expert Rev Anti Infect Ther 9:851–857. doi: 10.1586/eri.11.112. [DOI] [PubMed] [Google Scholar]
- 13.Takashita E, Fujisaki S, Shirakura M, Nakamura K, Kishida N, Kuwahara T, Shimazu Y, Shimomura T, Watanabe S, Odagiri T, The Influenza Virus Surveillance Group of Japan . 2016. Influenza A(H1N1)pdm09 virus exhibiting enhanced cross-resistance to oseltamivir and peramivir due to a dual H275Y/G147R substitution, Japan, March 2016. Euro Surveill 21:30258. doi: 10.2807/1560-7917.ES.2016.21.24.30258. [DOI] [PubMed] [Google Scholar]
- 14.Chong Y, Matsumoto S, Kang D, Ikematsu H. 2018. Consecutive influenza surveillance of neuraminidase mutations and neuraminidase inhibitor resistance in Japan. Influenza Other Respir Viruses 13:115–122. doi: 10.1111/irv.12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneider WM, Chevillotte MD, Rice CM. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durbin JE, Fernandez-Sesma A, Lee C-K, Rao TD, Frey AB, Moran TM, Vukmanovic S, García-Sastre A, Levy DE. 2000. Type I IFN modulated innate and specific antiviral immunity. J Immunol 164:4220–4228. doi: 10.4049/jimmunol.164.8.4220. [DOI] [PubMed] [Google Scholar]
- 17.Beilharz MW, Cummins JM, Bennett AL. 2007. Protection from lethal influenza virus challenge by oral type I interferon. Biochem Biophys Res Commun 355:740–744. doi: 10.1016/j.bbrc.2007.02.019. [DOI] [PubMed] [Google Scholar]
- 18.Szretter KJ, Gangappa S, Belser JA, Zeng H, Chen H, Matsuoka Y, Sambhara S, Swayne DE, Tumpey TM, Katz JM. 2009. Early control of H5N1 influenza virus replication by the type I interferon response in mice. J Virol 83:5825–5834. doi: 10.1128/JVI.02144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Hoeven N, Belser JA, Szretter KJ, Zeng H, Staeheli P, Swayne DE, Katz JM, Tumpey TM. 2009. Pathogenesis of 1918 pandemic and H5N1 influenza virus infections in a guinea pig model: antiviral potential of exogenous alpha interferon to reduce virus shedding. J Virol 83:2851–2861. doi: 10.1128/JVI.02174-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mordstein M, Kochs G, Dumoutier L, Renauld J-C, Paludan SR, Klucher K, Staeheli P. 2008. Interferon-λ contributes to innate immunity of mice against influenza A virus but not against hepatotropic viruses. PLoS Pathog 4:e1000151. doi: 10.1371/journal.ppat.1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mordstein M, Neugebauer E, Ditt V, Jessen B, Rieger T, Falcone V, Sorgeloos F, Ehl S, Mayer D, Kochs G, Schwemmle M, Gunther S, Drosten C, Michiels T, Staeheli P. 2010. Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J Virol 84:5670–5677. doi: 10.1128/JVI.00272-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sommereyns C, Paul S, Staeheli P, Michiels T. 2008. IFN-lambda (IFN-λ) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog 4:e1000017. doi: 10.1371/journal.ppat.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klinkhammer J, Schnepf D, Ye L, Schwaderlapp M, Gad HH, Hartmann R, Garcin D, Mahlakõiv T, Staeheli P. 2018. IFN-λ1 prevents influenza virus spread from the upper airways to the lungs and limits virus transmission. Elife 7:e33354. doi: 10.7554/eLife.33354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ison MG, Gnann JW, Nagy-Agren S, Treannor J, Paya C, Steigbigel R, Elliott M, Weiss HL, Hayden FG, NIAID Collaborative Antiviral Study Group . 2003. Safety and efficacy of nebulized zanamivir in hospitalized patients with serious influenza. Antivir Ther 8:183–190. [PubMed] [Google Scholar]
- 25.Duval X, van der Werf S, Blanchon T, Mosnier A, Bouscambert-Duchamp M, Tibi A, Enouf V, Charlois-Ou C, Vincent C, Andreoletti L, Tubach F, Lina B, Mentré F, Leport C, The Bivir Study Group . 2010. Efficacy of oseltamivir-zanamivir combination compared to each monotherapy for seasonal influenza: a randomized placebo-controlled trial. PLoS Med 7:e1000362. doi: 10.1371/journal.pmed.1000362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ilyushina NA, Bovin NV, Webster RG, Govorkova EA. 2006. Combination chemotherapy, a potential strategy for reducing the emergence of drug-resistant influenza A variants. Antiviral Res 70:121–131. doi: 10.1016/j.antiviral.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 27.Nguyen JT, Hoopes JD, Le MH, Smee DF, Patick AK, Faix DJ, Blair PJ, de Jong MD, Prichard MN, Went GT. 2010. Triple combination of amantadine, ribavirin, and oseltamivir is highly active and synergistic against drug resistant influenza virus strains in vitro. PLoS One 5:e9332. doi: 10.1371/journal.pone.0009332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen JT, Smee DF, Barnard DL, Julander JG, Gross M, de Jong MD, Went GT. 2012. Efficacy of combined therapy with amantadine, oseltamivir, and ribavirin in vivo against susceptible and amantadine-resistant influenza A viruses. PLoS One 7:e31006. doi: 10.1371/journal.pone.0031006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seo S, Englund JA, Nguyen JT, Pukrittayakamee S, Lindegardh N, Tarning J, Tambyah PA, Renaud C, Went GT, de Jong MD, Boeckh MJ. 2013. Combination therapy with amantadine, oseltamivir and ribavirin for influenza A infection: safety and pharmacokinetics. Antivir Ther 18:377–386. doi: 10.3851/IMP2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beigel JH, Bao Y, Beeler J, Manosuthi W, Slandzicki A, Dar SM, Panuto J, Beasley RL, Perez-Patrigeon S, Suwanpimolkul G, Losso MH, McClure N, Bozzolo DR, Myers C, Holley HP Jr, Hoopes J, Lane HC, Hughes MD, Davey RT, IRC003 Study Team . 2017. Oseltamivir, amantadine, and ribavirin combination antiviral therapy versus oseltamivir monotherapy for the treatment of influenza: a multicentre, double-blind, randomised phase 2 trial. Lancet Infect Dis 17:1255–1265. doi: 10.1016/S1473-3099(17)30476-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ilyushina NA, Donnelly RP. 2014. In vitro anti-influenza A activity of interferon (IFN)-λ1 combined with IFN-β or oseltamivir carboxylate. Antiviral Res 111:112–120. doi: 10.1016/j.antiviral.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 32.Adams SE, Lee N, Lugovtsev VY, Kan A, Donnelly RP, Ilyushina NA. 2019. Effect of influenza H1N1 neuraminidase V116A and I117V mutations on NA activity and sensitivity to NA inhibitors. Antiviral Res 169:104539. doi: 10.1016/j.antiviral.2019.104539. [DOI] [PubMed] [Google Scholar]
- 33.Ilyushina NA, Lugovtsev VY, Samsonova AP, Sheikh FG, Bovin NV, Donnelly RP. 2017. Generation and characterization of interferon-lambda 1-resistant H1N1 influenza A viruses. PLoS One 12:e0181999. doi: 10.1371/journal.pone.0181999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X, Ilyushina NA, Lugovtsev VY, Bovin NV, Couzens LK, Gao J, Donnelly RP, Eichelberger MC, Wan H. 2017. Amino acids in hemagglutinin antigenic site B determine antigenic and receptor binding differences between A(H3N2)v and ancestral seasonal H3N2 influenza viruses. J Virol 91:e015120-16. doi: 10.1128/JVI.01512-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee N, Khalenkov AM, Lugovtsev VY, Ireland DD, Samsonova AP, Bovin NV, Donnelly RP, Ilyushina NA. 2018. The use of plant lectins to regulate H1N1 influenza A virus receptor binding activity. PLoS One 13:e0195525. doi: 10.1371/journal.pone.0195525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vavricka CJ, Li Q, Wu Y, Qi J, Wang M, Liu Y, Gao F, Liu J, Feng E, He J, Wang J, Liu H, Jiang H, Gao GF. 2011. Structural and functional analysis of laninamivir and its octanoate prodrug reveals group specific mechanisms for influenza NA inhibition. PLoS Pathog 7:e1002249. doi: 10.1371/journal.ppat.1002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pizzorno A, Bouhy X, Abed Y, Boivin G. 2011. Generation and characterization of recombinant pandemic influenza A(H1N1) viruses resistant to neuraminidase inhibitors. J Infect Dis 203:25–31. doi: 10.1093/infdis/jiq010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pizzorno A, Abed Y, Rhéaume C, Bouhy X, Boivin G. 2013. Evaluation of recombinant 2009 pandemic influenza A (H1N1) viruses harboring zanamivir resistance mutations in mice and ferrets. Antimicrob Agents Chemother 57:1784–1789. doi: 10.1128/AAC.02269-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamura D, DeBiasi RL, Okomo-Adhiambo M, Mishin VP, Campbell AP, Loechelt B, Wiedermann BL, Fry AM, Gubareva LV. 2015. Emergence of multidrug-resistant influenza A(H1N1)pdm09 virus variants in an immunocompromised child treated with oseltamivir and zanamivir. J Infect Dis 212:1209–1213. doi: 10.1093/infdis/jiv245. [DOI] [PubMed] [Google Scholar]
- 40.Sakabe S, Ozawa M, Takano R, Iwastuki-Horimoto K, Kawaoka Y. 2011. Mutations in PA, NP, and HA of a pandemic (H1N1) 2009 influenza virus contribute to its adaptation to mice. Virus Res 158:124–129. doi: 10.1016/j.virusres.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu R, Ekiert DC, Krause JC, Hai R, Crowe JE Jr, Wilson IA. 2010. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science 328:357–360. doi: 10.1126/science.1186430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chutinimitkul S, Herfst S, Steel J, Lowen AC, Ye J, van Riel D, Schrauwen EJ, Bestebroer TM, Koel B, Burke DF, Sutherland-Cash KH, Whittleston CS, Russell CA, Wales DJ, Smith DJ, Jonges M, Meijer A, Koopmans M, Rimmelzwaan GF, Kuiken T, Osterhaus AD, García-Sastre A, Perez DR, Fouchier RA. 2010. Virulence-associated substitution D222G in the hemagglutinin of 2009 pandemic influenza A(H1N1) virus affects receptor binding. J Virol 84:11802–11813. doi: 10.1128/JVI.01136-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ilyushina NA, Komatsu TE, Ince WL, Donaldson EF, Lee N, O’Rear JJ, Donnelly RP. 2019. Influenza A virus hemagglutinin mutations associated with use of neuraminidase inhibitors correlate with decreased inhibition by anti-influenza antibodies. Virology J 16:149. doi: 10.1186/s12985-019-1258-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chan PK, Lee N, Joynt GM, Choi KW, Cheung JL, Yeung AC, Lam P, Wong R, Leung BW, So H, Lam WY, Hui DC. 2011. Clinical and virological course of infection with haemagglutinin D222G mutant strain of 2009 pandemic influenza A (H1N1) virus. J Clin Virol 50:320–324. doi: 10.1016/j.jcv.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 45.Wagner R, Matrosovich MN, Klenk H-D. 2002. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev Med Virol 12:159–166. doi: 10.1002/rmv.352. [DOI] [PubMed] [Google Scholar]
- 46.Ilyushina NA, Khalenkov AM, Seiler JP, Forrest HL, Bovin NV, Marjuki H, Barman S, Webster RG, Webby RJ. 2010. Adaptation of pandemic H1N1 influenza viruses in mice. J Virol 84:8607–8616. doi: 10.1128/JVI.00159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu J, Liang C, Liu SL. 2017. Interferon-inducible LY6E protein promotes HIV-1 infection. J Biol Chem 292:4674–4685. doi: 10.1074/jbc.M116.755819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mar KB, Rinkenberger NR, Boys IN, Eitson JL, McDougal MB, Richardson RB, Schoggins JW. 2018. LY6E mediates an evolutionarily conserved enhancement of virus infection by targeting a late entry step. Nat Commun 9:3603. doi: 10.1038/s41467-018-06000-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci U S A 97:6108–6113. doi: 10.1073/pnas.100133697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hayden FG, Cote KM, Douglas RG. 1980. Plaque inhibition assay for drug susceptibility testing of influenza viruses. Antimicrob Agents Chemother 17:865–870. doi: 10.1128/aac.17.5.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol 146:2275–2289. doi: 10.1007/s007050170002. [DOI] [PubMed] [Google Scholar]
- 52.Potier M, Mameli L, Belisle M, Dallaire L, Melancon SB. 1979. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-alpha-D-N-acetylneuraminate) substrate. Anal Biochem 94:287–296. doi: 10.1016/0003-2697(79)90362-2. [DOI] [PubMed] [Google Scholar]
- 53.Marathe BM, Leveque V, Klump K, Webster RG, Govorkova EA. 2013. Determination of neuraminidase kinetic constants using whole influenza virus preparations and correction for spectroscopic interference by a fluorogenic substrate. PLoS One 8:e71401. doi: 10.1371/journal.pone.0071401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prichard MN, Shipman C Jr.. 1990. A three-dimensional model to analyze drug-drug combinations. Antiviral Res 14:181–206. doi: 10.1016/0166-3542(90)90001-N. [DOI] [PubMed] [Google Scholar]
- 55.Burke DF, Smith DJ. 2014. A recommended numbering scheme for influenza A HA subtypes. PLoS One 9:e112302. doi: 10.1371/journal.pone.0112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Colman PM. 1994. Influenza virus neuraminidase: structure, antibodies, and inhibitors. Protein Sci 3:1687–1696. doi: 10.1002/pro.5560031007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequences have been deposited in GenBank under accession numbers MT423008 to MT423014.