Ceftazidime-avibactam and cefiderocol are two of the latest generation β-lactam agents that possess expanded activity against highly drug-resistant bacteria, including carbapenem-resistant Enterobacterales. Here, we show that structural changes in AmpC β-lactamases can confer reduced susceptibility to both agents. A multidrug-resistant Enterobacter cloacae clinical strain (Ent385) was found to be resistant to ceftazidime-avibactam and cefiderocol without prior exposure to either agent.
KEYWORDS: ceftazidime, avibactam, cefiderocol, beta-lactamase
ABSTRACT
Ceftazidime-avibactam and cefiderocol are two of the latest generation β-lactam agents that possess expanded activity against highly drug-resistant bacteria, including carbapenem-resistant Enterobacterales. Here, we show that structural changes in AmpC β-lactamases can confer reduced susceptibility to both agents. A multidrug-resistant Enterobacter cloacae clinical strain (Ent385) was found to be resistant to ceftazidime-avibactam and cefiderocol without prior exposure to either agent. The AmpC β-lactamase of Ent385 (AmpCEnt385) contained an alanine-proline deletion at positions 294 and 295 (A294_P295del) in the R2 loop. AmpCEnt385 conferred reduced susceptibility to ceftazidime-avibactam and cefiderocol when cloned into Escherichia coli TOP10. Purified AmpCEnt385 showed increased hydrolysis of ceftazidime and cefiderocol compared to AmpCEnt385Rev, in which the deletion was reverted. Comparisons of crystal structures of AmpCEnt385 and AmpCP99, the canonical AmpC of E. cloacae complex, revealed that the two-residue deletion in AmpCEnt385 induced drastic structural changes of the H-9 and H-10 helices and the R2 loop, which accounted for the increased hydrolysis of ceftazidime and cefiderocol. The potential for a single mutation in ampC to confer reduced susceptibility to both ceftazidime-avibactam and cefiderocol requires close monitoring.
INTRODUCTION
Antimicrobial resistance in Gram-negative bacteria represents one of the top public health priorities that is expected to worsen further in the absence of interventions to mitigate it. Carbapenem resistance in Enterobacter spp. is increasingly recognized as a clinical challenge against which few existing agents remain active. Resistance is typically due to a combination of mechanisms—derepressed production of the intrinsic AmpC β-lactamase, reduced outer membrane permeability, and augmented efflux—with or without the production of carbapenemase (1). The recent introduction of novel β-lactamase inhibitors, in particular avibactam, has significantly improved treatment of infections caused by carbapenem-resistant Enterobacterales, including Enterobacter spp. (2). Avibactam is a potent diazabicyclooctane inhibitor of class A and class C β-lactamases (Fig. 1a), the latter representing the AmpC group of enzymes. Resistance to ceftazidime-avibactam has emerged in about 10% of patients treated with this agent for infections caused by Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae; this resistance is in many cases due to amino acid substitutions in the omega loop of KPC (3). However, the frequency and clinical relevance of ceftazidime-avibactam resistance in Enterobacter spp. are still unclear.
FIG 1.
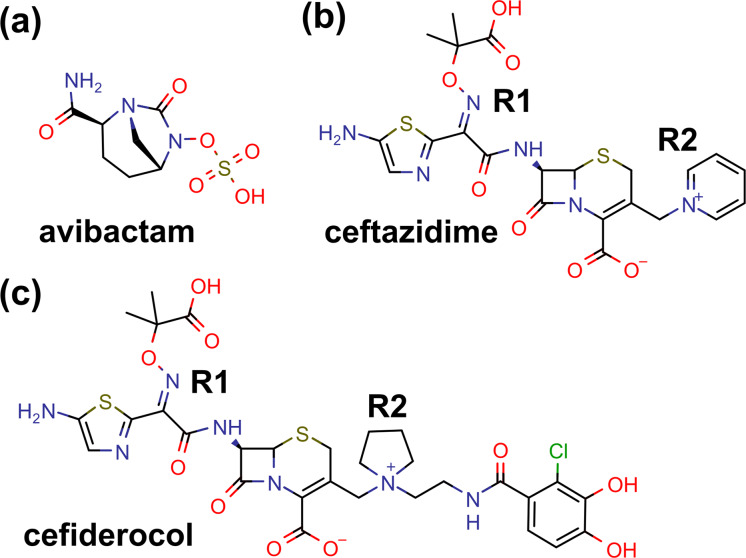
Chemical structures of avibactam (a), ceftazidime (b), and cefiderocol (c). The R1 and R2 side chains of cephalosporin are labeled. This figure was prepared by using MarvinSketch 19.20 (2019; ChemAxon).
Cefiderocol is a novel catechol-substituted siderophore cephalosporin that has been approved for the treatment of complicated urinary tract infection in patients who have limited or no alternative treatment options (Fig. 1c) (4). Cefiderocol is actively transported across the Gram-negative outer membrane into the periplasmic space due to the halogenated catechol moiety on the C-3 side chain that chelates ferric ion and binds primarily to PBP3 (5). It is stable against all classes of β-lactamases, including metallo-β-lactamases, owing to the presence of the cyclic quaternary ammonium moiety on the same side chain. Thanks to these unique structural modifications, cefiderocol demonstrates potent activity across Gram-negative bacterial species, including carbapenem-resistant Enterobacter spp. In a surveillance of more than 9,000 contemporary clinical strains from North America and Europe, the MICs of cefiderocol were ≤4 μg/ml (the susceptibility breakpoint endorsed by the Clinical and Laboratory Standards Institute [CLSI]) for 99.9% of Enterobacterales clinical strains, 99.9% of P. aeruginosa strains, 97.6% of A. baumannii strains, 100% of S. maltophilia strains, and 93.8% of Burkholderia cepacia strains (6). More recently, the U.S. Food and Drug Administration has approved a susceptibility breakpoint of 2 μg/ml for Enterobacterales. For the 214 E. cloacae isolates tested in this surveillance study, the cefiderocol MIC90 was 1 μg/ml, with MICs ranging between 0.008 and 128 μg/ml. The mechanisms underlying resistance in isolates with high MICs have not been elucidated.
Here, we report the genetic, biochemical, and structural characterization of a variant AmpC identified in an E. cloacae clinical strain resistant to both ceftazidime-avibactam and cefiderocol.
RESULTS AND DISCUSSION
E. cloacae strain with reduced susceptibility to meropenem, ceftazidime-avibactam, and cefiderocol.
E. cloacae Ent385 was isolated from a patient who had undergone small-bowel transplant and presented to a hospital in Pennsylvania for fever, vomiting, abdominal pain, and distention. The patient received courses of meropenem and piperacillin-tazobactam, but not cephalosporins. On hospital day 30, the patient developed ventilator-associated pneumonia that was empirically treated with meropenem. A diagnostic bronchoalveolar lavage culture grew carbapenem-resistant E. cloacae (Ent385) with meropenem and ceftazidime-avibactam MICs of 16 and 8 μg/ml, respectively. Given the concerning susceptibility pattern, the strain was subjected to MIC testing of cefiderocol. The cefiderocol MIC of E. cloacae Ent385 was >16 μg/ml, which was unusually high and interpreted as resistant according to the provisional susceptibility breakpoint provided by the Clinical and Laboratory Standards Institute (CLSI; ≤4 μg/ml). By whole-genome sequencing, the strain belonged to sequence type (ST) 456 and carried blaDHA-1, blaOXA-1, and blaTEM-1 acquired β-lactamase genes, in addition to intrinsic ampC. DHA-1 is an acquired AmpC β-lactamase that is nearly identical to the intrinsic AmpC of Morganella morganii (7). The ceftazidime-avibactam MIC90 against clinical isolates that produce DHA-1 is 0.5 μg/ml (8). OXA-1 and TEM-1 are an oxacillinase and a penicillinase, respectively, and confer resistance to penicillins. While no carbapenemase gene was identified, ompC and ompF possessed nonsynonymous mutations that corresponded to loss of the start codon (Met→Arg) and an early stop codon at position 66 (Ser→Stop), respectively. Functional loss of these two major outer membrane protein genes, coupled with production of DHA-1, likely accounted for meropenem resistance of E. cloacae Ent385 (9, 10). These mutations may have resulted from the patient’s previous exposure to meropenem, but none of the prior cultures grew E. cloacae; thus, we could not examine this possibility.
AmpC R2 loop deletion as the source of reduced ceftazidime-avibactam and cefiderocol susceptibility in E. cloacae Ent385.
The whole-genome sequencing of E. cloacae Ent385 identified an alanine-proline deletion at positions 294 and 295 (A294_P295del) and a leucine-to-valine substitution at position 296 (L296V) in the ampC gene. Various insertions, deletions, and amino acid substitutions in the H-10 helix or the R2 loop of ampC in various Enterobacterales and P. aeruginosa clinical strains have previously been associated with extension of the spectrum of hydrolysis to include cefepime, which is usually resistant to hydrolysis (11–17). We therefore hypothesized that A294_P295del observed in ampC might be the source of reduced susceptibility to cefiderocol and possibly also ceftazidime-avibactam.
To exclude the involvement of novel β-lactamases that had not been previously characterized or not catalogued in ResFinder, we conducted functional genome cloning of E. cloacae Ent385 using ampicillin resistance as the marker for β-lactamase production. As a result, several E. coli TOP10 transformants harboring recombinant pBCSK(–) plasmids were obtained. They all showed reduced susceptibility to ceftazidime-avibactam and cefiderocol compared to empty vector control. The recombinant plasmid with the shortest inserted fragment of ∼2.3 kb (pAmpCEnt385) containing part of ampR and the entire ampC was used for subsequent experiments.
Ceftazidime-avibactam and cefiderocol MICs of E. coli TOP10(pAmpCEnt385) were both 2 μg/ml. The corresponding MICs of E. coli TOP10(pAmpCEnt385Rev), in which A294_P295del was reverted to wild type by site-directed mutagenesis, were both 0.5 μg/ml (Table 1). These results confirmed the contribution of this R2 loop deletion to the ceftazidime-avibactam and cefiderocol-resistant phenotype in E. cloacae Ent385. The higher cefiderocol MIC of the parental E. cloacae strain may be further augmented by increased expression of pAmpCEnt385.
TABLE 1.
β-Lactam MICs against clinical and laboratory-generated strains with or without variant AmpC
| Strain | AmpC variant | MIC (μg/ml)a
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CAZ | CZA | FDC | FEP | MEM | IPM | MVB | I-R | ATM | ATA | ||
| E. cloacae Ent385 | A294_P295del | >512 | 8 | >16 | 128 | 16 | 16 | 16 | 4 | >128 | 8 |
| E. coli TOP10(pAmpCEnt385) | A294_P295del | 256 | 2 | 2 | 4 | ≤0.06 | 0.5 | 0.03 | 0.25 | 16 | 0.25 |
| E. coli TOP10(pAmpCEnt385Rev) | Wild type | 128 | 0.5 | 0.5 | 0.25 | ≤0.06 | 0.5 | 0.03 | 0.25 | 16 | 0.25 |
| E. coli TOP10(pBCSK(–)) | Vector control | 0.5 | ≤0.25 | ≤0.03 | ≤0.25 | ≤0.06 | 0.25 | 0.03 | 0.25 | 0.25 | ≤0.125 |
CAZ, ceftazidime; CZA, ceftazidime-avibactam; FDC, cefiderocol; FEP, cefepime; MEM, meropenem; IPM, imipenem; MVB, meropenem-vaborbactam; I-R, imipenem-relebactam; ATM, aztreonam; ATA, aztreonam-avibactam.
AmpCEnt385 shows augmented hydrolysis of ceftazidime and cefiderocol.
To further define the altered kinetic properties imparted by A294_P295del, AmpCEnt385 and AmpCEnt385Rev were purified and subjected to measurements of the steady-state kinetic parameters kcat and Km for nitrocefin, cephalothin, ceftazidime, and cefiderocol (Table 2). Compared to AmpCEnt385Rev, AmpCEnt385 showed substantial (∼5-fold) reduction in kcat values for nitrocefin and cephalothin, while the Km values were within a 2-fold difference. This resulted in overall lower hydrolytic efficiency of AmpCEnt385 for these substrates. For ceftazidime, however, kcat was significantly higher with AmpCEnt385 in comparison with AmpCEnt385Rev, yielding an ∼1,000-fold higher hydrolytic efficiency. For cefiderocol, the difference was with the affinity, with AmpCEnt385 showing 10-fold lower Km, whereas the kcat values were comparable. Overall for cefiderocol, this resulted in an ∼20-fold higher hydrolytic efficiency with AmpCEnt385 compared to AmpCEnt385Rev. The kinetic studies indicated that AmpCEnt385 shows a broad substrate specificity, including cefiderocol, and superior hydrolysis of ceftazidime, whereas its catalytic property against more classic cephalosporin substrates is slightly inferior to the native AmpC. The 50% inhibitory concentrations (IC50s) of avibactam, measured using nitrocefin as the reporter substrate, were 0.04 ± 0.04 μM and 0.08 ± 0.02 μM, respectively, for AmpCEnt385 and AmpCEnt385Rev, demonstrating that the potencies of inhibition by avibactam are almost identical. Thus, we speculate that the enhanced hydrolytic efficiency of ceftazidime primarily contributes to the reduced ceftazidime-avibactam susceptibility exhibited by AmpCEnt385.
TABLE 2.
Kinetic parameters of AmpCEnt385 and AmpCEnt385Rev
| Substrate | AmpCEnt385 |
AmpCEnt385Rev |
(kcat/Km)Ent385/(kcat/Km)Ent385Rev | ||||
|---|---|---|---|---|---|---|---|
| kcat (s−1) | Km (μM) | kcat/Km (μM−1 s−1) | kcat (s−1) | Km (μM) | kcat/Km (μM−1 s−1) | ||
| Nitrocefin | 18.4 ± 1.8 | 11.0 ± 4.7 | 1.67 | 92.6 ± 8.1 | 21.1 ± 3.6 | 4.39 | 0.38 |
| Cephalothin | 73.1 ± 9.9 | 0.34 ± 0.12 | 215 | 432 ± 150 | 0.17 ± 0.10 | 2,540 | 0.85 × 10−1 |
| Ceftazidime | 25.8 ± 3.8 | 49.0 ± 25.2 | 0.53 | (0.30 ± 0.07) × 10–1 | 80.0 ± 29.6 | 0.38 × 10−3 | 1,394.7 |
| Cefiderocol | 0.14 ± 0.03 | 49.8 ± 8.6 | 0.28 × 10−2 | 0.08 ± 0.02 | 533 ± 267 | 0.15 × 10−3 | 18.6 |
The overall structure of AmpCEnt385 indicates a local structural change of the R2 loop.
To obtain further structural insights on AmpCEnt385 showing superior catalytic properties against ceftazidime and cefiderocol, we determined crystal structures of the AmpCEnt385 free form and its complexes with ceftazidime or avibactam (referred to as the AmpCEnt385-CAZ complex and the AmpCEnt385-AVI complex, respectively). The data collection and refinement statistics are summarized in Table 3. The AmpCEnt385 crystal belongs to the space group P21, and there are two AmpCEnt385 monomers contained in a crystallographic asymmetric unit. The qualities of the electron density maps were better for chain A in common to the AmpCEnt385 structures, and almost all residues except for the C-terminal Q259 residue could be assigned in the chain A structures. On the other hand, several disordered regions were observed in chain B of the AmpCEnt385 structures, and we could not model the following structures: residues of P140 to A143 (P140–A143) and N289–P295 for the free form; residues of T113–N128, P140–A151, and D279–P295 for the AmpCEnt385-CAZ complex; and residues of T111–Q120, Q139–A151, and V280–P295 for the AmpCEnt385-AVI complex. Thus, we describe the following results and discussion of the AmpCEnt385 structures based on the chain A structures.
TABLE 3.
Data collection and structure refinement statisticsa
| Data set | Free form | AVI complex | CAZ complex |
|---|---|---|---|
| Data collection | |||
| Source | Photon Factory BL-17A | Photon Factory BL-17A | Photon Factory BL-17A |
| Wavelength (Å) | 0.9800 | 0.9800 | 0.9800 |
| Space group | P21 | P21 | P21 |
| Unit cell parameters | |||
| Length (Å) | a = 50.6, b = 75.2, c = 103.4 | a = 50.4, b = 73.7, c = 102.1 | a = 50.4, b = 74.1, c = 102.3 |
| Angle (°) | β = 95.1 | β = 94.5 | β = 94.8 |
| Resolution range (Å) | 46.9–1.40 (1.48–1.40) | 46.5–1.60 (1.70–1.60) | 46.7–1.65 (1.75–1.65) |
| No. of observed reflections | 1,039,223 (169,454) | 668,610 (109,167) | 630,466 (100,564) |
| No. of unique reflections | 149,918 (23,824) | 96,869 (15,241) | 89,346 (14,132) |
| Multiplicity | 6.9 (7.1) | 6.9 (7.2) | 7.1 (7.1) |
| Completeness (%) | 98.7 (97.2) | 98.4 (96.3) | 98.1 (96.6) |
| Rmerge (%)b | 4.9 (66.0) | 5.9 (66.6) | 5.7 (77.3) |
| ⟨I/σ (I)⟩ | 18.66 (2.59) | 16.86 (2.45) | 18.84 (2.35) |
| Refinement | |||
| Resolution (Å) | 46.9–1.40 (1.42–1.40) | 37.6–1.60 (1.62–1.60) | 43.7–1.65 (1.67–1.65) |
| No. of reflections used | 149,881 (4,852) | 96,856 (2,991) | 89,321 (2,904) |
| Rwork (%)c | 14.7 (23.1) | 16.7 (23.6) | 16.3 (29.9) |
| Rfree (%)d | 16.8 (25.5) | 18.9 (23.3) | 19.4 (36.8) |
| No. of nonhydrogen atoms | 6,613 | 6,059 | 6,003 |
| Protein | 5,811 | 5,342 | 5,281 |
| Ligands | 110 | 113 | 122 |
| Solvent | 696 | 604 | 600 |
| RMSD from ideality | |||
| Bond length (Å) | 0.004 | 0.005 | 0.009 |
| Bond angle (°) | 0.763 | 0.775 | 0.928 |
| Avg B-factor | 24.8 | 29.5 | 29.4 |
| Protein | 22.8 | 27.9 | 27.8 |
| Ligands | 54.6 | 56.1 | 48.8 |
| Solvent | 37.4 | 39.0 | 39.8 |
| Ramachandran plot | |||
| Favored region (%) | 98.85 | 98.80 | 98.63 |
| Allowed region (%) | 1.15 | 1.20 | 1.37 |
| Outlier region (%) | 0.00 | 0.00 | 0.00 |
| Clashscore | 2.12 | 2.22 | 1.40 |
| PDB ID | 6LC7 | 6LC8 | 6LC9 |
Values in parentheses denote the highest-resolution shell.
Rmerge = 100 × Σhkl Σi |Ii(hkl) − ⟨I(hkl)⟩|/Σhkl Σi Ii(hkl), where ⟨I(hkl)⟩ is the mean value of I(hkl).
Rwork = 100 × Σhkl | |Fo| − |Fc| |/Σhkl |Fo|, where Fo and Fc the observed and calculated structure factors, respectively.
Rfree is calculated as for Rwork, but for the test set comprising 5% reflections not used in refinement.
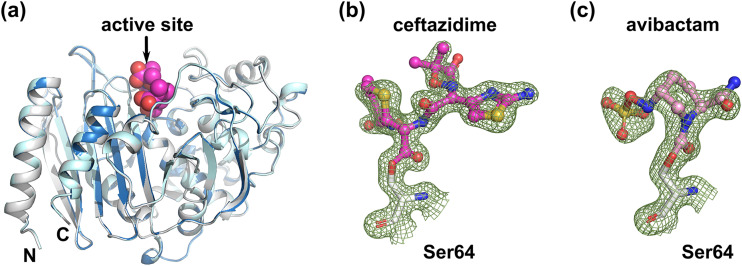
According to the previous structural studies, the overall fold of class C β-lactamase consists of an α-helical domain and a mixed α/β domain, and the active site is located between the two domains (18). AmpCEnt385 also has a typical fold of class C β-lactamase (Fig. 2a). The overall structure of the AmpCEnt385 free form is quite similar to those of the AmpCEnt385–CAZ complex and the AmpCEnt385–AVI complex, with 0.16 Å and 0.18 Å of the root mean square deviation (RMSD) values for the corresponding Cα positions for 356 residues, respectively (Fig. 2a). These results suggest that AmpCEnt385 does not require an overall structural change for substrate recognition. Electron density maps corresponding to the molecules of ceftazidime and avibactam were observed in the active site of each AmpCEnt385-drug complex structure. These electron density maps were continuously extended to the catalytic residue S64 of AmpCEnt385, indicating that Oγ atom of S64 is covalently bound to C-8 atom of ceftazidime or the C-7 atom of avibactam, and these structures displayed acyl-enzyme intermediates of the AmpCEnt385-drug complex structures (Fig. 2b and c). The R1 side chain of cefiderocol is identical to that of ceftazidime (Fig. 1b and c). The R2 side chain of cefiderocol is a bulkier pyrrolidine group with the catechol moiety, while that of ceftazidime is a pyridinium group. However, these R2 side chain are liberated during the hydrolysis reaction (19). Thus, we hypothesize that the final AmpCEnt385 acyl-complex structure with cefiderocol is identical to the AmpCEnt385-CAZ complex structure.
FIG 2.
Overall structures of AmpCEnt385. (a) The AmpCEnt385 structures are shown as cartoon representations and colored white for the AmpCEnt385 free form, blue for the AmpCEnt385-CAZ complex, and cyan for the AmpCEnt385-AVI complex. To indicate the active site, the ceftazidime molecule is shown as CPK (space-filling model) representation colored magenta. (b and c) 2mFo-DFc maps are shown as green mesh contoured 1.5σ. The molecules of ceftazidime and avibactam are shown as ball-and-stick representations colored magenta and pink, respectively.
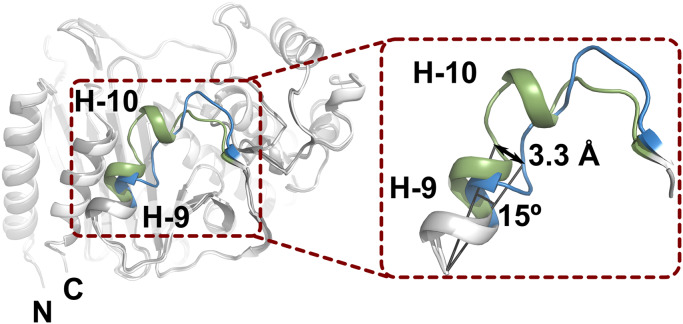
To clarify the structural changes of AmpCEnt385 due to A294_P295del, the crystal structure of AmpCEnt385 was superimposed on the crystal structure of AmpC P99 isolated from Enterobacter hormaechei (referred to as AmpCP99, PDB ID 5XHR) (20), having 84% sequence identity over 359 residues. As a result, a structural difference between AmpCEnt385 and AmpCP99 was observed in V283–V294 residues of AmpCEnt385, and the RMSD value for the corresponding Cα positions of this region is over at least 1 Å, while the overall RMSD value for the corresponding Cα positions for 351 residues is 0.65 Å. In detail, relative to the AmpCP99 structure, H-9 helix of AmpCEnt385 is slightly slanted (Fig. 3). The Cα positions of S287 located at the C-terminal side of H-9 helix shift 3.3 Å, and the Cα position of S287 of AmpCEnt385 and that of AmpCP99 subtends approximately 15° from the Cα position of D279 of AmpCEnt385. Slanted H-9 helix is directly connected with the R2 loop, and the H-10 helix disappears in the AmpCEnt385 structure.
FIG 3.
Structure comparison of AmpCEnt385 and AmpCP99. The structures of AmpCEnt385 and AmpCP99 are shown as cartoon representations and colored white and gray, respectively. To clarify the difference, the colors of the V283–V294 residue structures in AmpCEnt385 and the corresponding residues in AmpCP99 appear in blue and green, respectively.
Structural changes of AmpCEnt385 account for its altered kinetic properties.
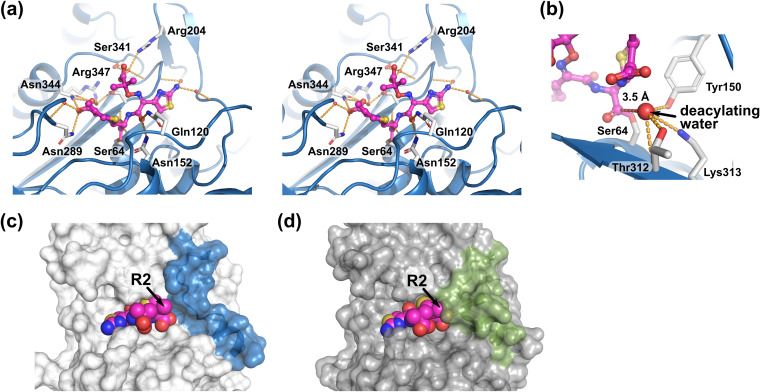
The active site of the AmpCEnt385-CAZ complex is shown in Fig. 4. A total of 12 hydrogen bonds were observed in the AmpCEnt385-CAZ complex, and AmpCEnt385 recognizes ceftazidime by a tight hydrogen bond network in a manner that is similar to ceftazidime recognition by the native AmpC isolated from E. coli (referred to as AmpCEc, having 73% sequence identity over 356 residues with AmpCEnt385, PDB ID 1IEL; Fig. 4a) (21). The carbonyl oxygen atom of the ester bond between AmpCEnt385 and ceftazidime forms hydrogen bonds to main chain nitrogen atoms of S64 and S316. The C4 carboxyl group of the dihydrothiazine ring of ceftazidime forms hydrogen bonds to side chains of N289 and N344 and two water molecules. The amide group at the R1 side chain of ceftazidime forms hydrogen bonds to side chains of Q120 and N152. The carboxyl group and the aminothiazole ring at the R1 side chain of ceftazidime form hydrogen bonds to one and two water molecules, respectively. The deacylating water molecule, which is predicted by the AmpCEc structure, was observed in the AmpCEnt385–CAZ complex structure, and its position and interaction with AmpCEnt385 were similar to those of AmpCEc (Fig. 4b). In the native AmpCEc-CAZ complex structure, the position of the R2 side chain (R2 site) is surrounded by the R2 loop, including the H-10 helix (Fig. 4d).
FIG 4.
Ceftazidime recognition by AmpCEnt385. (a) Stereo view of the ceftazidime binding. The ceftazidime molecule as shown in ball-and-stick representations colored magenta. Hydrogen bonds are shown as orange dashed lines. Residues and the water molecules participating the hydrogen bond network are shown as white stick and red spheres, respectively. (b) The deacylating water molecule is shown as a large red sphere. Red dashed line indicates the distance between C-8 atom of ceftazidime and the deacylating water molecule. (c and d) Comparison of the substrate-binding sites of the AmpCEnt385-CAZ complex and the AmpCEc-CAZ complex. Transparent molecular surfaces of AmpCEnt385 (c) and AmpCEc (d) are colored white and gray, respectively. To clarify the difference, the colors of the V283–V294 residue structures, including R2 loop in AmpCEnt385, and corresponding residues in AmpCP99 appear in blue and green, respectively. Arrowheads indicate the R2 side chain positions of ceftazidime.
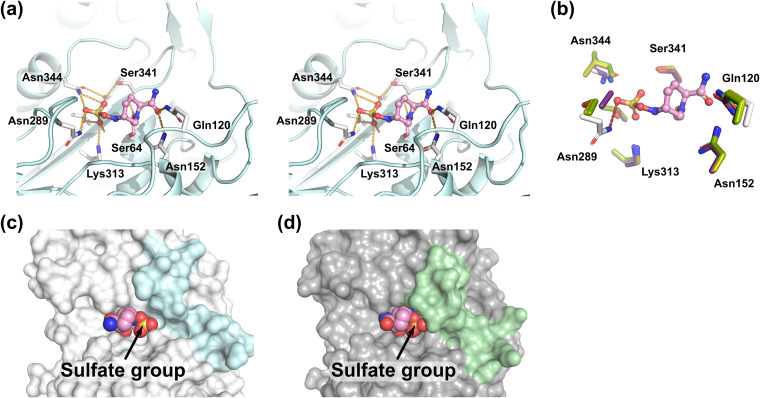
Figure 5 shows the active site of the AmpCEnt385-AVI complex. AmpCEnt385 recognizes avibactam through several hydrogen bonds that also recognize ceftazidime. The carbonyl oxygen atom of the ester bond between S64 of AmpCEnt385 and avibactam forms hydrogen bonds to main chain nitrogen atoms of S64 and S316. The carboxamide group of piperidine ring of avibactam forms hydrogen bonds to side chains of Q120 and N152. The sulfate group of avibactam forms hydrogen bonds to side chains of N289, K313, T314, and N344, main chain oxygen atom of T314 and one water molecule. To date, the following four crystal structures of the class C β-lactamase complex with avibactam have been reported: PDC-1 (PDB ID 4OOY and 4HEF) (22, 23), FOX-4 (PDB ID 5ZA2) (24), and TRU-1 (PDB ID 6FM7) (25). Comparison of the AmpCEnt385-AVI complex structure with the other structures of class C β-lactamase complex with avibactam indicates that the hydrogen bond between N289 of AmpCEnt385 and the sulfate group of avibactam is an interaction unique to the AmpCEnt385-AVI complex, while the rest of the hydrogen bond network involved in the recognition of avibactam is similar (Fig. 5b). This unique interaction could be attributed to the translocation of N289 due to A294_P295del. In addition, the AmpCEnt385-AVI complex structure shows that the sulfate group of avibactam bound to AmpCEnt385 at the R2 site is exposed due to the disappearance of H-10 helix (Fig. 5c and d).
FIG 5.
Avibactam recognition by AmpCEnt385. (a) Stereo view of the avibactam binding. The avibactam molecule is shown as ball-and-stick representations colored pink. Hydrogen bonds are shown as orange dashed lines. Residues and the water molecules participating the hydrogen bond network are shown as white stick and red spheres, respectively. (b) Structure comparisons of the AmpCEnt385-AVI complex to the other class C β-lactamase complex with avibactam. Colors are white for AmpCE385, purple for PDC-1 (PDB ID 4OOY), green for FOX-4 (PDB ID 5ZA2), and yellow for TRU-1 (PDB ID 6FM7). Red dashed line indicates a unique hydrogen bond between N289 of AmpCEnt385 and the sulfate group of avibactam. Residues of AmpCEnt385 are labeled. (c and d) Comparison of the substrate-binding sites of the AmpCEnt385-AVI complex and the PDC-1–AVI complex (PDB ID 4OOY). Transparent molecular surfaces of AmpCEnt385 (c) and PDC-1 (d) are colored in white and gray, respectively. To clarify the difference, the colors of the V283–V294 residue structures, including the R2 loop in AmpCEnt385, and corresponding residues in PDC-1 appear in cyan and pale green, respectively. Arrowheads indicate the sulfate group of avibactam.
These structural studies of AmpCEnt385 revealed that A294_P295del induces local structural change from the H-9 helix to the R2 loop and the disappearance of the H-10 helix (Fig. 3), while the hydrogen bond network between AmpCEnt385 and ceftazidime or avibactam and the key residues of AmpCEnt385 involved in the reaction are largely conserved compared to that of the native AmpC (Fig. 4 and 5). The results indicated that the structural change in AmpCEnt385 enables extension of the substrate-binding site to the bulk solvent region and recognition of cephalosporins having bulkier R2 side chains such as cefiderocol, thus allowing for relevant levels of hydrolysis to occur (Fig. 4c). Cefiderocol will escape recognition by the native AmpC due to the steric hindrance between the bulkier R2 side chain of cefiderocol and the R2 site of the native AmpC, which conceals this binding site with the H-10 helix (Fig. 1c and 4d). These observations are consistent with the results of the kinetic studies showing ∼10-fold lower Km of AmpCEnt385 for cefiderocol compared to AmpCEnt385Rev (Table 2). With cefiderocol, the substrate-binding to the active site may be essential for the occurrence of relevant levels of hydrolysis. In addition, the kinetic parameters show that ceftazidime is a better substrate for AmpCEnt385 (Table 2). Ceftazidime has a smaller R2 side chain than that of cefiderocol (Fig. 1b and c), and it is well known that the native AmpC modestly hydrolyzes ceftazidime, indicating that its binding to the native AmpC is sufficient for relevant levels of ceftazidime hydrolysis to occur. We speculate that expansion of the substrate-binding site of AmpCEnt385 also increases the rates of both the substrate binding and the product leaving of ceftazidime, resulting in improved turnover of ceftazidime, whereas the substrate affinity is largely maintained.
Genetic changes (deletions, insertions, and amino acid substitutions) in the R2 site of AmpC have historically been associated with extension of the AmpC substrate specificity to cefepime in several species, including E. cloacae, Klebsiella aerogenes, Serratia marcescens, Citrobacter freundii, E. coli and Pseudomonas aeruginosa (11–18). Crystal structures of AmpC, including these genetic changes in the R2 site, were reported on AmpCD (containing three amino acid deletions at positions 286 to 288, PDB ID 2ZJ9) (26) and AmpC BER (containing two alanine residues insertions at positions 294 and 295, PDB ID 5JOC) (27), and these structures show similar local structural changes at the R2 site observed in the AmpCEnt385 structure. Although it is not known whether these previously reported variant AmpCs also confer reduced susceptibility to ceftazidime-avibactam and/or cefiderocol, this suggests that similar variants may arise in species other than E. cloacae as reported here.
Conclusion.
There continues to be a need for new therapeutics to treat highly drug-resistant Gram-negative bacterial infections. Ceftazidime-avibactam has been successfully used in the treatment of carbapenem-resistant Enterobacterales infection in regions where KPC is the predominant carbapenemase type. Cefiderocol, a novel siderophore cephalosporin, possesses broad-spectrum activity against Gram-negative pathogens. Based on microbiological surveillance studies, cefiderocol maintains potency across Gram-negative bacterial species of clinical relevance, regardless of the β-lactamase background (6, 28, 29). From a clinical standpoint, cefiderocol is notably active against most carbapenem-resistant Enterobacterales strains, including those producing KPC, NDM, and OXA-48-group carbapenemases. While isolates resistant to cefiderocol have been rarely identified in surveillance studies, the underlying mechanisms have not been reported. Here, we demonstrated that a two amino acid deletion in the R2 loop of AmpC β-lactamases can confer reduced susceptibility to ceftazidime-avibactam and cefiderocol, which can be attributed to the drastic structural changes of the H-9 and H-10 helices and the R2 loop induced by the deletion and consequent increased hydrolysis of ceftazidime and cefiderocol. The findings provide new insights into how multidrug-resistant Gram-negative bacteria might evolve their β-lactamases to survive selective pressure from the latest β-lactam agents that are engineered to combat them.
MATERIALS AND METHODS
Strains and plasmids.
E. cloacae Ent385 was a clinical strain isolated from the respiratory tract of a hospitalized patient. The isolate was initially identified as E. cloacae complex by using a matrix-assisted laser desorption ionization--time of flight approach and later confirmed to belong to E. cloacae species as described below. E. coli TOP10 and cloning vector pBCSK(–) were used for initial cloning of the ceftazidime-avibactam and cefiderocol resistance determinant. E. coli BL21(DE3) and expression vector pET30b were used for purification of AmpCEnt385 and AmpCEnt385Rev (revertant of the R2 loop deletion in AmpCEnt385).
Whole-genome sequencing.
The genome of strain Ent385 was sequenced by Illumina NextSeq using a paired-end protocol. The reads were assembled into contigs by CLC Genomics Workbench 11. The species was confirmed with the average nucleotide identity method, and the sequence type was determined in silico (https://pubmlst.org/ecloacae/). Antimicrobial resistance genes were identified by ResFinder version 3.2 (https://cge.cbs.dtu.dk/services/ResFinder/).
Cloning and sequencing of AmpCEnt385.
The cefiderocol resistance determinant was sought by shotgun cloning of the E. cloacae Ent385 genome. In brief, genomic DNA was extracted, purified. and partially digested with Sau3AI, which was then ligated with BamHI-digested pBCSK(–). E. coli TOP10 was transformed with the ligated products, and transformants were selected by growth on lysogenic broth agar containing 50 μg/ml of ampicillin. The recombinant plasmids were sequenced on both strands by Sanger sequencing.
Susceptibility testing.
MICs of E. cloacae Ent385 and E. coli producing AmpCEnt385 were obtained by standard broth microdilution methods. For cefiderocol, iron-depleted cation-adjusted Mueller-Hinton broth plates provided by Shionogi, Inc., through International Health Management Associates (Schaumburg, IL) were used to test susceptibility (30).
Generation of AmpCEnt385Rev.
AmpCEnt385Rev was generated by site-directed mutagenesis according to the manufacturer’s protocol of the Q5 site-directed mutagenesis kit (New England Biolabs, Ipswich, MA). Briefly, engineered forward (5′-CGCCGTTGCCGGTCGCAGAAGTGA-3′) and reverse (CCAGTGCGACCTTATTGTCG) primers including the six deleted nucleotides were used to amplify AmpCEnt385. The PCR system contained 10 ng template, 2× Q5 Hot Start High-Fidelity Master Mix, and 0.5 μM concentrations of each primer in a total volume of 25 μl. The PCR began with an initial denaturation at 98°C (30 s), followed by 25 cycles at 98°C for 10 s, 60°C for 30 s, and 72°C for 1 min 45 s and then an extension step at 72°C for 7 min. The product (ampCEnt385Rev) was sequenced to confirm addition of the six nucleotides. ampCEnt385 and ampCEnt385Rev were subcloned into pET30b using NdeI and EcoRI and transferred into the E. coli BL21(DE3) strain.
Purification of AmpCEnt385 and AmpCEnt385Rev for kinetic measurements.
Lysogenic broth plus kanamycin (50 μg/ml) was inoculated with E. coli BL21(DE3) harboring the pET30b-AmpCEnt385 or pET30b-AmpCEnt385Rev recombinant plasmids and grown to an optical density at 600 nm of 0.5 to 0.7. A final concentration of 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added, and the culture was incubated for an additional 2 h. The cells were harvested by centrifugation; the pellet was then resuspended in 20 mM Tris-HCl (pH 7.0) buffer, sonicated, and centrifuged at 8,000 rpm for 10 min at 4°C to remove cell debris. The supernatants, containing the active enzymes, were ultracentrifuged at 100,000 × g and filtered through a 0.45 μm Durapore membrane filter (Millipore, Billerica, MA). The crude extract was loaded onto a HiTrap SP HP (GE Healthcare) previously equilibrated with 20 mM Tris-HCl (pH 7.0). The AmpC enzymes were then eluted with a NaCl gradient at a flow rate of 5 ml/min. Fractions containing active enzyme were pooled, concentrated, and dialyzed into PBS (pH 7.2) using an Amicon Ultra spin column with a cut off size of 10 kDa (Millipore) to yield salt-free enzymes approximately 10 mg in quantity and >95% in purity as determined by SDS-PAGE.
Kinetic measurements.
The steady-state kinetic parameters were determined in phosphate-buffered saline (PBS; pH 7.2) with a constant amount of enzyme and various concentrations of the substrates using a UV spectrophotometer (V-750; JASCO, Easton, MD). Substrates tested included nitrocefin, cephalothin, ceftazidime, and cefiderocol. For avibactam, the concentration of avibactam which gave a 50% reduction in the hydrolysis of nitrocefin (IC50) was measured after 5 min of preincubation of the enzyme with the inhibitor at 25°C (room temperature) and nitrocefin as the substrate at 100 μM. Three independent measurements were performed for each enzyme and substrate or inhibitor combination.
Purification of AmpCEnt385 for the X-ray crystallography.
The gene encoding residues 21 to 379 of AmpCEnt385 without the signal peptide was cloned into the expression plasmid vector pET30b between the restriction enzyme sites of NdeI and EcoRI. Purification was conducted in the same manner as the proteins for kinetic measurements. In addition, further purification was performed with a HiTrap Blue HP column (GE Healthcare, Chicago, IL) chromatography, and the proteins were eluted with a linear gradient of 0 to 2 M NaCl in 20 mM Tris-HCl (pH 7.0). The purity of AmpCEnt385 was evaluated by SDS-PAGE analysis, and the β-lactamase activity was confirmed as described previously (31). The active fractions were collected and concentrated using a Vivaspin Turbo 4 centrifugal concentrator (MWCO 10,000; Sartorius, Gottingen, Germany), and the buffer was exchanged to 20 mM Tris-HCl (pH 7.0) by several rounds of dilution and concentration. The purified active AmpCEnt385 solution was finally concentrated to 50 mg/ml and stored in 20 μl aliquots at –80°C until used in crystallization experiments.
Crystallization.
Initial screening of AmpCEnt385 was performed with the commercial crystal screening kits Index, Crystal Screen, and Crystal Screen 2 (Hampton Research, Aliso Viejo, CA) using the hanging-drop vapor diffusion method by mixing 1.0 μl of the AmpCEnt385 solution (20 mg/ml) and 1.0 μl of the reservoir solution at 20°C. The cluster crystals of AmpCEnt385 appeared in no. 23 crystallization condition of Crystal Screen 2 kit within 2 months. Single crystals of AmpCEnt385 suitable for the X-ray crystallography were obtained by using the multiple rounds of streak-seeding methods with aforementioned cluster crystals, and the droplets were prepared by mixing 1.5 μl of the AmpCEnt385 solution (20 mg/ml) and 1.5 μl of the reservoir solution containing 1.8 M ammonium sulfate, 0.1 M MES (pH 5.5), and 5% 1,4-dioxane and were preequilibrated for 1 week at 4°C. Crystals of the AmpCEnt385-CAZ complex and the AmpCEnt385-AVI complex were obtained by soaking the AmpCEnt385 crystals in a solution containing 10 mM ceftazidime or 10 mM avibactam, 1.8 M ammonium sulfate, 0.1 M MES (pH 5.5), and 5% 1,4-dioxane at 4°C for 3 h.
Data collection, structure determination, and refinement.
Prior to the X-ray experiments, the AmpCEnt385 crystals were transferred into a cryoprotectant solution composed of reservoir solution containing 30% glycerol and then flash frozen in liquid nitrogen. Synchrotron experiments were carried out using Photon Factory BL-17A (High Energy Accelerator Research Organization, Tsukuba, Japan). Diffraction data sets were collected at –173°C using an EIGER X16M detector, and data sets were processed and scaled using XDS (32). The initial phase of the AmpCEnt385 structure was determined by the molecular replacement method using Molrep (33) from the CCP4 program suite (34), with the coordinate (PDB ID 5XHR) serving as the search model (20). Manual model rebuilding was performed with COOT (35). Structure refinements were performed with phenix.refine from the PHENIX package (36). The AmpCEnt385 structure was refined with the anisotropic atomic displacement parameters. The structures of the AmpCEnt385-CAZ complex and the AmpCEnt385-AVI complex were refined with the atomic displacement parameters using the translation, liberation, and screw (TLS) method, and TLS groups were determined by using phenix.find_tls_groups. The stereochemical quality of the final structures were evaluated by MolProbity (37). All molecular graphics were prepared using PyMOL v2.3.2 (Schrödinger, New York, NY).
Data availability.
The contigs of the E. cloacae Ent385 genome have been submitted to the NCBI under GenBank accession number WNXG00000000. The atomic coordinates of AmpCEnt385 have been submitted to the Protein Data Bank (PDB) under PDB accession numbers 6LC7 for the free form, 6LC8 for the complex with avibactam, and 6LC9 for the complex with ceftazidime.
ACKNOWLEDGMENTS
We thank Masahiro Suzuki for help in confirming the species of AmpCP99-producing Enterobacter cloacae complex strain. Synchrotron experiments were performed with the approval of the Photon Factory Program Advisory Committee (proposal 2017G554). We are grateful to the beamline staff for their support of our synchrotron experiments and to Shionogi, Inc., for the provision of cefiderocol.
This study was supported by grant R21AI151362 from the National Institutes of Health. R.K.S. was supported by NIH grants K08AI114883 and R03AI144636. Y.D. was supported by NIH grants R01AI104895 and R21AI135522.
REFERENCES
- 1.Szabó D, Silveira F, Hujer AM, Bonomo RA, Hujer KM, Marsh JW, Bethel CR, Doi Y, Deeley K, Paterson DL. 2006. Outer membrane protein changes and efflux pump expression together may confer resistance to ertapenem in Enterobacter cloacae. Antimicrob Agents Chemother 50:2833–2835. doi: 10.1128/AAC.01591-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Duin D, Lok JJ, Earley M, Cober E, Richter SS, Perez F, Salata RA, Kalayjian RC, Watkins RR, Doi Y, Kaye KS, Fowler VG, Paterson DL, Bonomo RA, Evans S, Antibacterial Resistance Leadership Group . 2018. Colistin versus ceftazidime-avibactam in the treatment of infections due to carbapenem-resistant Enterobacteriaceae. Clin Infect Dis 66:163–171. doi: 10.1093/cid/cix783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shields RK, Nguyen MH, Chen L, Press EG, Kreiswirth BN, Clancy CJ. 2018. Pneumonia and renal replacement therapy are risk factors for ceftazidime-avibactam treatment failures and resistance among patients with carbapenem-resistant Enterobacteriaceae infections. Antimicrob Agents Chemother 62:e02497-17. doi: 10.1128/AAC.02497-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Food and Drug Administration. 2019. FDA news release. U.S. FDA, Bethesda, MD: https://www.fda.gov/news-events/press-announcements/fda-approves-new-antibacterial-drug-treat-complicated-urinary-tract-infections-part-ongoing-efforts. Accessed 19 November 2019. [Google Scholar]
- 5.Aoki T, Yoshizawa H, Yamawaki K, Yokoo K, Sato J, Hisakawa S, Hasegawa Y, Kusano H, Sano M, Sugimoto H, Nishitani Y, Sato T, Tsuji M, Nakamura R, Nishikawa T, Yamano Y. 2018. Cefiderocol (S-649266), A new siderophore cephalosporin exhibiting potent activities against Pseudomonas aeruginosa and other gram-negative pathogens including multidrug-resistant bacteria: structure activity relationship. Eur J Med Chem 155:847–868. doi: 10.1016/j.ejmech.2018.06.014. [DOI] [PubMed] [Google Scholar]
- 6.Karlowsky JA, Hackel MA, Tsuji M, Yamano Y, Echols R, Sahm DF. 2019. In vitro activity of cefiderocol, a siderophore cephalosporin, against Gram-negative bacilli isolated by clinical laboratories in North America and Europe in 2015–2016: SIDERO-WT-2015. Int J Antimicrob Agents 53:456–466. doi: 10.1016/j.ijantimicag.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 7.Barnaud G, Arlet G, Verdet C, Gaillot O, Lagrange PH, Philippon A. 1998. Salmonella enteritidis: AmpC plasmid-mediated inducible β-lactamase (DHA-1) with an ampR gene from Morganella morganii. Antimicrob Agents Chemother 42:2352–2358. doi: 10.1128/AAC.42.9.2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karlowsky JA, Biedenbach DJ, Kazmierczak KM, Stone GG, Sahm DF. 2016. Activity of ceftazidime-avibactam against extended-spectrum- and AmpC β-lactamase-producing Enterobacteriaceae collected in the INFORM Global Surveillance Study from 2012 to 2014. Antimicrob Agents Chemother 60:2849–2857. doi: 10.1128/AAC.02286-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen YG, Zhang Y, Yu YS, Qu TT, Wei ZQ, Shen P, Li LJ. 2008. In vivo development of carbapenem resistance in clinical isolates of Enterobacter aerogenes producing multiple β-lactamases. Int J Antimicrob Agents 32:302–307. doi: 10.1016/j.ijantimicag.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 10.Lee K, Yong D, Choi YS, Yum JH, Kim JM, Woodford N, Livermore DM, Chong Y. 2007. Reduced imipenem susceptibility in Klebsiella pneumoniae clinical isolates with plasmid-mediated CMY-2 and DHA-1 β-lactamases co-mediated by porin loss. Int J Antimicrob Agents 29:201–206. doi: 10.1016/j.ijantimicag.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 11.Barnaud G, Labia R, Raskine L, Sanson-Le Pors MJ, Philippon A, Arlet G. 2001. Extension of resistance to cefepime and cefpirome associated with a 6-amino-acid deletion in the H-10 helix of the cephalosporinase of an Enterobacter cloacae clinical isolate. FEMS Microbiol Lett 195:185–190. doi: 10.1111/j.1574-6968.2001.tb10519.x. [DOI] [PubMed] [Google Scholar]
- 12.Mammeri H, Poirel L, Bemer P, Drugeon H, Nordmann P. 2004. Resistance to cefepime and cefpirome due to a 4-amino-acid deletion in the chromosome-encoded AmpC β-lactamase of a Serratia marcescens clinical isolate. Antimicrob Agents Chemother 48:716–720. doi: 10.1128/aac.48.3.716-720.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doi Y, Wachino J-i, Ishiguro M, Kurokawa H, Yamane K, Shibata N, Shibayama K, Yokoyama K, Kato H, Yagi T, Arakawa Y. 2004. Inhibitor-sensitive AmpC β-lactamase variant produced by an Escherichia coli clinical isolate resistant to oxyiminocephalosporins and cephamycins. Antimicrob Agents Chemother 48:2652–2658. doi: 10.1128/AAC.48.7.2652-2658.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mammeri H, Poirel L, Nordmann P. 2007. Extension of the hydrolysis spectrum of AmpC β-lactamase of Escherichia coli due to amino acid insertion in the H-10 helix. J Antimicrob Chemother 60:490–494. doi: 10.1093/jac/dkm227. [DOI] [PubMed] [Google Scholar]
- 15.Ahmed AM, Shimamoto T. 2008. Emergence of a cefepime- and cefpirome-resistant Citrobacter freundii clinical isolate harbouring a novel chromosomally encoded AmpC β-lactamase, CMY-37. Int J Antimicrob Agents 32:256–261. doi: 10.1016/j.ijantimicag.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 16.Rodríguez-Martínez JM, Fernández-Echauri P, Fernández-Cuenca F, Diaz de Alba P, Briales A, Pascual A. 2012. Genetic characterization of an extended-spectrum AmpC cephalosporinase with hydrolysing activity against fourth-generation cephalosporins in a clinical isolate of Enterobacter aerogenes selected in vivo. J Antimicrob Chemother 67:64–68. doi: 10.1093/jac/dkr423. [DOI] [PubMed] [Google Scholar]
- 17.Berrazeg M, Jeannot K, Ntsogo Enguéné VY, Broutin I, Loeffert S, Fournier D, Plésiat P. 2015. Mutations in β-lactamase AmpC increase resistance of Pseudomonas aeruginosa isolates to antipseudomonal cephalosporins. Antimicrob Agents Chemother 59:6248–6255. doi: 10.1128/AAC.00825-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Powers RA. 2016. Structural and functional aspects of extended-spectrum AmpC cephalosporinases. Curr Drug Targets 17:1051–1060. doi: 10.2174/1573399811666150615144707. [DOI] [PubMed] [Google Scholar]
- 19.Perez-Inestrosa E, Suau R, Montanez MI, Rodriguez R, Mayorga C, Torres MJ, Blanca M. 2005. Cephalosporin chemical reactivity and its immunological implications. Curr Opin Allergy Clin Immunol 5:323–330. doi: 10.1097/01.all.0000173788.73401.69. [DOI] [PubMed] [Google Scholar]
- 20.Pan X, He Y, Chen T, Chan KF, Zhao Y. 2017. Modified penicillin molecule with carbapenem-like stereochemistry specifically inhibits class C β-lactamases. Antimicrob Agents Chemother 61:e01288-17. doi: 10.1128/AAC.01288-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Powers RA, Caselli E, Focia PJ, Prati F, Shoichet BK. 2001. Structures of ceftazidime and its transition-state analogue in complex with AmpC β-lactamase: implications for resistance mutations and inhibitor design. Biochemistry 40:9207–9214. doi: 10.1021/bi0109358. [DOI] [PubMed] [Google Scholar]
- 22.Lahiri SD, Johnstone MR, Ross PL, McLaughlin RE, Olivier NB, Alm RA. 2014. Avibactam and class C β-lactamases: mechanism of inhibition, conservation of the binding pocket, and implications for resistance. Antimicrob Agents Chemother 58:5704–5713. doi: 10.1128/AAC.03057-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lahiri SD, Mangani S, Durand-Reville T, Benvenuti M, De Luca F, Sanyal G, Docquier JD. 2013. Structural insight into potent broad-spectrum inhibition with reversible recyclization mechanism: avibactam in complex with CTX-M-15 and Pseudomonas aeruginosa AmpC β-lactamases. Antimicrob Agents Chemother 57:2496–2505. doi: 10.1128/AAC.02247-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nukaga M, Papp-Wallace KM, Hoshino T, Lefurgy ST, Bethel CR, Barnes MD, Zeiser ET, Johnson JK, Bonomo RA. 2018. Probing the mechanism of inactivation of the FOX-4 cephamycinase by avibactam. Antimicrob Agents Chemother 62:e02371-17. doi: 10.1128/AAC.02371-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pozzi C, Di Pisa F, De Luca F, Benvenuti M, Docquier JD, Mangani S. 2018. Atomic-resolution structure of a class C β-lactamase and its complex with avibactam. ChemMedChem 13:1437–1446. doi: 10.1002/cmdc.201800213. [DOI] [PubMed] [Google Scholar]
- 26.Yamaguchi Y, Sato G, Yamagata Y, Doi Y, Wachino J-i, Arakawa Y, Matsuda K, Kurosaki H. 2009. Structure of AmpC β-lactamase (AmpCD) from an Escherichia coli clinical isolate with a tripeptide deletion (Gly286-Ser287-Asp288) in the H10 helix. Acta Crystallogr Sect F Struct Biol Cryst Commun 65:540–543. doi: 10.1107/S1744309109014249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Na JH, Cha SS. 2016. Structural basis for the extended substrate spectrum of AmpC BER and structure-guided discovery of the inhibition activity of citrate against the class C β-lactamases AmpC BER and CMY-10. Acta Crystallogr D Struct Biol 72:976–985. doi: 10.1107/S2059798316011311. [DOI] [PubMed] [Google Scholar]
- 28.Jacobs MR, Abdelhamed AM, Good CE, Rhoads DD, Hujer KM, Hujer AM, Domitrovic TN, Rudin SD, Richter SS, van Duin D, Kreiswirth BN, Greco C, Fouts DE, Bonomo RA. 2018. ARGONAUT-I: activity of cefiderocol (S-649266), a siderophore cephalosporin, against Gram-negative bacteria, including carbapenem-resistant nonfermenters and Enterobacteriaceae with defined extended-spectrum β-lactamases and carbapenemases. Antimicrob Agents Chemother 63:e01801-18. doi: 10.1128/AAC.01801-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kazmierczak KM, Tsuji M, Wise MG, Hackel M, Yamano Y, Echols R, Sahm DF. 2019. In vitro activity of cefiderocol, a siderophore cephalosporin, against a recent collection of clinically relevant carbapenem-nonsusceptible Gram-negative bacilli, including serine carbapenemase- and metallo-β-lactamase-producing isolates (SIDERO-WT-2014 Study). Int J Antimicrob Agents 53:177. doi: 10.1016/j.ijantimicag.2018.10.007.-] [DOI] [PubMed] [Google Scholar]
- 30.Hackel MA, Tsuji M, Yamano Y, Echols R, Karlowsky JA, Sahm DF. 2019. Reproducibility of broth microdilution MICs for the novel siderophore cephalosporin, cefiderocol, determined using iron-depleted cation-adjusted Mueller-Hinton broth. Diagn Microbiol Infect Dis 94:321–325. doi: 10.1016/j.diagmicrobio.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 31.Sawai T, Takahashi I, Yamagishi S. 1978. Iodometric assay method for β-lactamase with various β-lactam antibiotics as the substrates. Antimicrob Agents Chemother 13:910–913. doi: 10.1128/aac.13.6.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kabsch W. 2010. XDS. Acta Crystallogr D Biol Crystallogr 66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vagin A, Teplyakov A. 2010. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr 66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 34.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen VB, Arendall WB III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The contigs of the E. cloacae Ent385 genome have been submitted to the NCBI under GenBank accession number WNXG00000000. The atomic coordinates of AmpCEnt385 have been submitted to the Protein Data Bank (PDB) under PDB accession numbers 6LC7 for the free form, 6LC8 for the complex with avibactam, and 6LC9 for the complex with ceftazidime.