Clostridium (Clostridioides) difficile causes toxin-mediated diarrhea and pseudomembranous colitis, primarily among hospital inpatients. Outbreaks of C. difficile infection (CDI) have been caused by strains with acquired antimicrobial resistance, particularly fluoroquinolone resistance, including C. difficile ribotype (RT) 027 in North America and Europe and RT 017, the most common strain in Asia. Despite being the most common cause of hospital-acquired infection in high-income countries, and frequent misuse of antimicrobials in Asia, little is known about CDI in the Asia-Pacific region.
KEYWORDS: Clostridium difficile, epidemiology
ABSTRACT
Clostridium (Clostridioides) difficile causes toxin-mediated diarrhea and pseudomembranous colitis, primarily among hospital inpatients. Outbreaks of C. difficile infection (CDI) have been caused by strains with acquired antimicrobial resistance, particularly fluoroquinolone resistance, including C. difficile ribotype (RT) 027 in North America and Europe and RT 017, the most common strain in Asia. Despite being the most common cause of hospital-acquired infection in high-income countries, and frequent misuse of antimicrobials in Asia, little is known about CDI in the Asia-Pacific region. We aimed to determine the antimicrobial susceptibility profiles of a collection of C. difficile isolates from the region. C. difficile isolates (n = 414) from a 2014 study of 13 Asia-Pacific countries were tested for susceptibility to moxifloxacin, amoxicillin-clavulanate, erythromycin, clindamycin, rifaximin, metronidazole, vancomycin, and fidaxomicin according to the Clinical and Laboratory Standards Institute’s agar dilution method. All isolates were susceptible to metronidazole, vancomycin, amoxicillin-clavulanate, and fidaxomicin. Moxifloxacin resistance was detected in all countries except Australia, all RT 369 and QX 239 strains, and 92.7% of RT 018 and 70.6% of RT 017 strains. All C. difficile RT 012, 369, and QX 239 strains were also resistant to erythromycin and clindamycin. Rifaximin resistance was common in RT 017 strains only (63.2%) and was not detected in Australian, Japanese, or Singaporean isolates. In conclusion, antimicrobial susceptibility of C. difficile varied by strain type and by country. Multiresistance was common in emerging RTs 369 and QX 239 and the most common strain in Asia, RT 017. Ongoing surveillance is clearly warranted.
INTRODUCTION
Clostridium (Clostridioides) difficile, the most common cause of health care-associated infections in high-income countries (1), imposes a heavy burden on national health care systems (2, 3). C. difficile opportunistically infects the gut, causing toxin-mediated diarrhea when the commensal microflora is perturbed, most often due to antimicrobial use. The capability of C. difficile to produce spores that can withstand many disinfectants allows it to survive in health care facility environments, often infecting older patients with a range of comorbidities, and recurrent infections are common due to slow recovery of the gut microflora following antimicrobial treatment. Infection with C. difficile can lead to life-threatening complications, including pseudomembranous colitis, septic shock, and toxic megacolon, and C. difficile-attributable mortality rates generally range from 4 to 7% (2, 4).
C. difficile is intrinsically resistant to β-lactam antibiotics (5), and many C. difficile strains have acquired resistance to a range of other antimicrobials, including the macrolide clindamycin and fluoroquinolones (6). Acquiring new resistance capabilities enables C. difficile strains to emerge and spread, both locally and internationally. For instance, acquired fluoroquinolone resistance via a Thr82Ile mutation in DNA gyrase subunit A was one of the main driving factors in the global dissemination of the epidemic C. difficile strain ribotype (RT) 027 (7), which caused some of the most significant outbreaks of C. difficile infection (CDI) to date, particularly in North America and Europe (7). Similarly, C. difficile RT 017, the predominant C. difficile strain circulating in Asia (8) and a common strain worldwide (9–11), is frequently resistant to clindamycin and fluoroquinolones, a feature that has most likely contributed to its global prominence (12).
In many Asian countries, antimicrobial consumption rates in both humans and animals are among the highest in the world (13, 14), and antimicrobial usage is frequently inappropriate due to unrestricted availability without prescriptions (13). Antimicrobial resistance is escalating among many pathogens, including Staphylococcus aureus, Escherichia coli, Streptococcus pneumoniae, and Neisseria gonorrhoeae in Asia, particularly in Southeast Asia, but often is poorly monitored (15).
Despite the substantial inappropriate use of antimicrobials in Asia and documented antimicrobial resistance of other pathogens, the epidemiology of CDI is largely underinvestigated due to poor awareness and inadequate testing practices in many Asian hospitals (8). The pooled prevalence of CDI among all patients with diarrhea in Asia was calculated at 14.8%, with an estimated incidence of 5.3 cases/10,000 patient days (16). Overall, the burden of CDI appears to be lower in Asian countries than in other regions, with rare occurrences of pseudomembranous colitis (PMC) and toxic megacolon and lower rates of recurrent CDI (17). As mentioned above, the most common strain in Asia, RT 017, frequently is reported as resistant to clindamycin and fluoroquinolones (18). In addition to RT 017, other common C. difficile strains circulating frequently in Asia are often reported as resistant to these agents. These include RTs 018, 002, and 369 (19–21).
A possible explanation for the less severe burden of CDI in Asia is the documented high prevalence in China and Southeast Asia (22–25) of nontoxigenic C. difficile strains, which are incapable of causing CDI. Elsewhere in Asia, the prevalence of nontoxigenic strains is also likely to be high; however, they are not detected and/or confirmed as nontoxigenic unless culture and PCR for toxin genes are performed, and many laboratories lack anaerobic culture facilities. Even if nontoxigenic strains are detected, they may not be reported in publications since they do not cause disease; however, they can also carry antimicrobial resistance genes and thus could contribute to horizontal transfer of these genes to toxigenic strains (26).
This study aimed to evaluate the antimicrobial susceptibility profiles of a collection of C. difficile isolates from a prospective study performed in 2014 in 13 Asia-Pacific countries (17).
RESULTS
All isolates were susceptible to metronidazole (MIC50 = 0.25 mg/liter), vancomycin (MIC50 = 1 mg/liter), fidaxomicin (MIC50 = 0.125 mg/liter), and amoxicillin-clavulanate (MIC50 = 0.5 mg/liter [Table 1]). Resistance to clindamycin was most common (80.7%; MIC50 > 32 mg/liter) overall, followed by erythromycin (55.3%; MIC50 > 256 mg/liter) and moxifloxacin (44.4%; MIC50 = 2 mg/liter), while resistance to rifaximin was least common (15.5%; MIC50 = 0.03 mg/liter [Table 1]).
TABLE 1.
MIC data for eight antimicrobials against C. difficile isolates from the Asia-Pacific region, by ribotype
| Ribotype | Antimicrobiala |
No. (%) resistant |
MIC (mg/liter) |
Geometric mean | ||
|---|---|---|---|---|---|---|
| Range | 50% | 90% | ||||
| All (n = 414) | FDX | 0 | 0.004 to 0.5 | 0.125 | 0.25 | 0.10 |
| VAN | 0 | 0.25 to 4b | 1 | 2 | 1.44 | |
| MTZ | 0 | 0.015 to 2 | 0.25 | 0.25 | 0.22 | |
| RFX | 64 (15.5) | 0.0005 to >32 | 0.03 | >32 | 0.07 | |
| CLI | 334 (80.7) | 0.015 to >32 | >32 | >32 | 12.87 | |
| ERY | 229 (55.3) | 0.03 to >256 | >256 | >256 | 23.75 | |
| AUG | 0 | 0.03 to 8 | 0.5 | 1 | 0.66 | |
| MXF | 184 (44.4) | 0.5 to >32 | 2 | 32 | 6.17 | |
| RT 017 (n = 68) | FDX | 0 | 0.004 to 0.25 | 0.125 | 0.25 | 0.11 |
| VAN | 0 | 0.5 to 2 | 1 | 2 | 1.06 | |
| MTZ | 0 | 0.015 to 2 | 0.25 | 0.25 | 0.19 | |
| RFX | 46 (67.7) | 0.008 to >32 | >32 | >32 | 3.47 | |
| CLI | 64 (94.1) | 0.06 to >32 | >32 | >32 | 21.93 | |
| ERY | 59 (86.8) | 0.5 to >256 | >256 | >256 | 116.78 | |
| AUG | 0 | 0.25 to 2 | 1 | 1 | 0.80 | |
| MXF | 48 (70.6) | 2 to >32 | 32 | 32 | 14.16 | |
| RT 014/020 (n = 45) | FDX | 0 | 0.03 to 0.5 | 0.125 | 0.25 | 0.13 |
| VAN | 0 | 0.5 to 2 | 2 | 2 | 1.47 | |
| MTZ | 0 | 0.06 to 0.5 | 0.25 | 0.25 | 0.23 | |
| RFX | 0 | 0.015 to 0.03 | 0.03 | 0.03 | 0.03 | |
| CLI | 30 (66.7) | 0.015 to 8 | 8 | 8 | 4.45 | |
| ERY | 2 (4.4) | 0.03 to >256 | 2 | 2 | 1.64 | |
| AUG | 0 | 0.25 to 8 | 0.5 | 1 | 0.64 | |
| MXF | 4 (8.9) | 0.5 to 32 | 2 | 4 | 2.48 | |
| RT 018 (n = 41) | FDX | 0 | 0.03 to 0.5 | 0.06 | 0.125 | 0.08 |
| VAN | 0 | 1 to 4b | 2 | 2 | 1.80 | |
| MTZ | 0 | 0.125 to 0.5 | 0.25 | 0.25 | 0.23 | |
| RFX | 3 (7.3) | 0.015 to >32 | 0.015 | 2 | 0.03 | |
| CLI | 39 (95.1) | 1 to >32 | >32 | >32 | 25.99 | |
| ERY | 38 (92.7) | 1 to >256 | >256 | >256 | 174.85 | |
| AUG | 0 | 0.25 to 1 | 1 | 1 | 0.74 | |
| MXF | 38 (92.7) | 2 to >32 | 32 | 32 | 24.68 | |
| RT 002 (n = 38) | FDX | 0 | 0.015 to 0.25 | 0.125 | 0.25 | 0.09 |
| VAN | 0 | 1 to 4b | 2 | 2 | 1.49 | |
| MTZ | 0 | 0.06 to 0.5 | 0.25 | 0.25 | 0.22 | |
| RFX | 1 (2.6) | 0.015 to >32 | 0.03 | 0.03 | 0.03 | |
| CLI | 26 (68.4) | 0.5 to >32 | 8 | >32 | 9.43 | |
| ERY | 15 (39.5) | 0.25 to >256 | 2 | >256 | 10.14 | |
| AUG | 0 | 0.25 to 1 | 1 | 1 | 0.73 | |
| MXF | 18 (47.4) | 2 to >32 | 4 | >32 | 7.30 | |
| RT 012 (n = 20) | FDX | 0 | 0.03 to 0.125 | 0.125 | 0.125 | 0.09 |
| VAN | 0 | 1 to 4b | 2 | 2 | 2.00 | |
| MTZ | 0 | 0.125 to 0.25 | 0.25 | 0.25 | 0.24 | |
| RFX | 0 | 0.015 to 0.125 | 0.015 | 0.03 | 0.02 | |
| CLI | 20 (100.0) | 8 to >32 | >32 | >32 | 28.84 | |
| ERY | 20 (100.0) | 256 to >256 | >256 | >256 | 256.00 | |
| AUG | 0 | 0.5 to 1 | 0.5 | 1 | 0.62 | |
| MXF | 5 (25.0) | 1 to 16 | 2 | 16 | 3.03 | |
| RT 369 (n = 17) | FDX | 0 | 0.03 to 0.25 | 0.125 | 0.25 | 0.10 |
| VAN | 0 | 1 | 1 | 1 | 1.00 | |
| MTZ | 0 | 0.125 to 0.25 | 0.25 | 0.25 | 0.21 | |
| RFX | 0 | 0.015 to 0.25 | 0.03 | 0.03 | 0.02 | |
| CLI | 17 (100.0) | >32 | >32 | >32 | 32.00 | |
| ERY | 17 (100.0) | >256 | >256 | >256 | 1.00 | |
| AUG | 0 | 0.5 to 2 | 1 | 1 | 0.82 | |
| MXF | 17 (100.0) | 8 to >32 | 16 | >32 | 18.83 | |
| QX 239 (n = 15) | FDX | 0 | 0.015 to 0.06 | 0.03 | 0.06 | 0.04 |
| VAN | 0 | 0.5 to 4b | 1 | 4 | 1.45 | |
| MTZ | 0 | 0.06 to 0.25 | 0.25 | 0.25 | 0.18 | |
| RFX | 0 | 0.008 to 0.03 | 0.015 | 0.03 | 0.02 | |
| CLI | 15 (100.0) | 32 to >32 | >32 | >32 | 32.00 | |
| ERY | 15 (100.0) | >256 | >256 | >256 | 256.00 | |
| AUG | 0 | 0.5 to 2 | 1 | 1 | 0.87 | |
| MXF | 15 (100.0) | 16 to >32 | 32 | 32 | 30.55 | |
| QX 032 (n = 15) | FDX | 0 | 0.03 to 0.25 | 0.125 | 0.25 | 0.09 |
| VAN | 0 | 0.25 to 2 | 1 | 2 | 0.95 | |
| MTZ | 0 | 0.125 to 0.25 | 0.25 | 0.25 | 0.22 | |
| RFX | 0 | 0.0005 to 0.03 | 0.03 | 0.03 | 0.02 | |
| CLI | 11 (73.3) | 0.03 to >32 | >32 | >32 | 13.26 | |
| ERY | 11 (73.3) | 0.03 to >256 | >256 | >256 | 48.37 | |
| AUG | 0 | 0.03 to 1 | 0.5 | 1 | 0.48 | |
| MXF | 0 | 2 to 4 | 2 | 2 | 2.00 | |
| RT 001 (n = 13) | FDX | 0 | 0.03 to 0.25 | 0.03 | 0.06 | 0.04 |
| VAN | 0 | 1 to 4b | 2 | 4 | 1.90 | |
| MTZ | 0 | 0.125 to 0.25 | 0.25 | 0.25 | 0.24 | |
| RFX | 0 | 0.015 to 0.03 | 0.03 | 0.03 | 0.02 | |
| CLI | 7 (53.9) | 0.5 to >32 | >32 | >32 | 11.02 | |
| ERY | 7 (53.9) | 1 to >256 | >256 | >256 | 20.89 | |
| AUG | 0 | 0.25 to 1 | 0.5 | 0.5 | 0.43 | |
| MXF | 5 (38.5) | 1 to >32 | 2 | 32 | 4.95 | |
| RT 106 (n = 12) | FDX | 0 | 0.03 to 0.5 | 0.25 | 0.5 | 0.15 |
| VAN | 0 | 1 to 2 | 1 | 2 | 1.33 | |
| MTZ | 0 | 0.125 to 0.25 | 0.25 | 0.25 | 0.24 | |
| RFX | 0 | 0.03 | 0.03 | 0.03 | 0.03 | |
| CLI | 8 (66.7) | 4 to >32 | 8 | >32 | 8.00 | |
| ERY | 3 (25.0) | 1 to >256 | 1 | >256 | 4.76 | |
| AUG | 0 | 0.5 to 1 | 0.5 | 1 | 0.57 | |
| MXF | 2 (16.7) | 2 to 32 | 2 | 32 | 3.36 | |
| RT 046 (n = 11) | FDX | 0 | 0.03 to 0.25 | 0.125 | 0.125 | 0.16 |
| VAN | 0 | 1 to 4b | 2 | 4 | 1.41 | |
| MTZ | 0 | 0.125 to 0.5 | 0.25 | 0.5 | 0.25 | |
| RFX | 0 | 0.015 to 0.125 | 0.015 | 0.03 | 0.03 | |
| CLI | 11 (100.0) | 8 to >32 | >32 | >32 | 8.98 | |
| ERY | 10 (90.9) | 1 to >256 | >256 | >256 | 7.55 | |
| AUG | 0 | 0.5 to 1 | 1 | 1 | 0.63 | |
| MXF | 4 (36.4) | 1 to 32 | 2 | 16 | 3.17 | |
| Others (n = 119) | FDX | 0 | 0.015 to 0.25 | 0.125 | 0.25 | 0.10 |
| VAN | 0 | 0.5 to 4b | 2 | 4 | 1.49 | |
| MTZ | 0 | 0.125 to 0.5 | 0.25 | 0.25 | 0.23 | |
| RFX | 13 (10.9) | 0.015 to >32 | 0.03 | >32 | 0.05 | |
| CLI | 86 (72.3) | 0.5 to >32 | 8 | >32 | 8.11 | |
| ERY | 32 (26.9) | 0.25 to >256 | 2 | >256 | 5.56 | |
| AUG | 0 | 0.25 to 2 | 0.5 | 1 | 0.57 | |
| MXF | 28 (23.5) | 0.5 to >32 | 2 | 32 | 3.28 | |
Abbreviations: FDX, fidaxomicin; VAN, vancomycin; MTZ, metronidazole; RFX, rifaximin; CLI, clindamycin; ERY, erythromycin; AUG, amoxicillin-clavulanate; MXF, moxifloxacin.
Isolates with MICs of ≥2 mg/liter by agar dilution were further tested for vancomycin susceptibility by Etest and confirmed as susceptible. Values reported here are for agar dilution only.
Resistance rates were highest among C. difficile RTs 017, 018, 369, and QX 239. RT 017 isolates had high rates of resistance to clindamycin (94.1%; MIC50 > 32 mg/liter), erythromycin (86.8%; MIC50 > 256 mg/liter), and moxifloxacin (70.6%; MIC50 = 32 mg/liter), and 67.7% were resistant to rifaximin (MIC50 > 32 mg/liter). RT 018 isolates were almost all resistant to erythromycin (92.7%; MIC50 > 256 mg/liter) and moxifloxacin (92.7%; MIC50 = 32 mg/liter) and clindamycin (95.1%; MIC50 > 32 mg/liter), and 7.3% were resistant to rifaximin (MIC50 = 0.015 mg/liter [Table 1]). C. difficile RT 369 and QX 239 isolates were all resistant to clindamycin, erythromycin, and moxifloxacin (MIC50 > 32 mg/liter, MIC50 > 256 mg/liter, and MIC50 = 16 mg/liter, respectively, for RT 369, and MIC50 > 32 mg/liter, MIC50 > 256 mg/liter, and MIC50 = 32 mg/liter, respectively, for QX 239) (Table 1). Intermediate vancomycin resistance (MIC = 4 mg/liter) was found in 27 isolates.
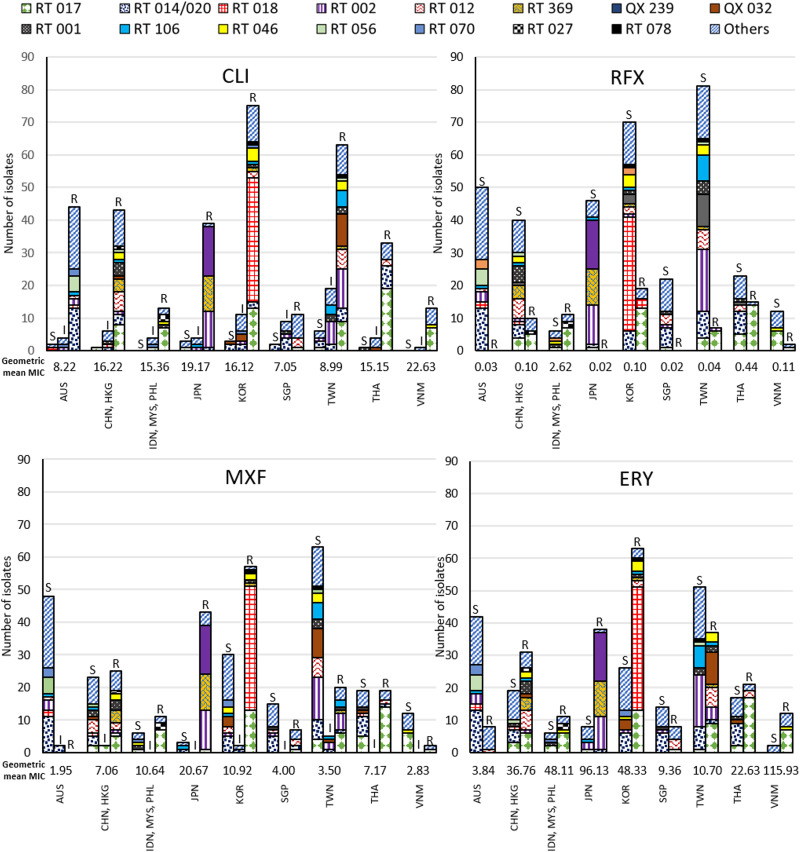
Summaries of country- and RT-specific resistance for clindamycin, rifaximin, moxifloxacin, and erythromycin are shown in Fig. 1. Clindamycin resistance was found in the majority of isolates across all countries. Moxifloxacin resistance was most frequently found in Japan (93.5%), Indonesia (85.7%), Republic of Korea (64.0%), China (52.3%), Thailand (50.0%), Philippines (44.4%), Hong Kong (33.3%), Singapore (31.8%), and Taiwan (22.7%), in RT QX 239 (100.0%), 369 (100.0%), 018 (92.7%), 017 (70.6%), 002 (47.4%), 001 (38.5%), and 046 (36.4%) isolates (Fig. 1 and Table 1). No moxifloxacin resistance was detected in isolates from Australia. Rifaximin resistance was mainly found in RT 017 (67.7%) and some RT 018 (7.3%), 002 (2.6%), and 027 (100.0%) isolates and was not detected in isolates from Australia, Japan, or Singapore (Fig. 1 and Table 1). Erythromycin resistance was found in isolates from all study countries and in the majority of isolates from Vietnam (85.7%), Japan (82.6%), Indonesia (71.4%), Republic of Korea (70.8%), China (63.6%), Philippines (55.6%), Thailand (55.3%), and Hong Kong (50.0%) (Fig. 1).
FIG 1.
Numbers of susceptible (S), intermediate (I), and resistant (R) isolates and geometric mean MIC by ribotype and country or region for clindamycin (CLI), rifaximin (RFX), moxifloxacin (MXF), and erythromycin (ERY). AUS, Australia; CHN, China; HKG, Hong Kong; IDN, Indonesia; MYS, Malaysia; PHL, Philippines; JPN, Japan; KOR, Republic of Korea; SGP, Singapore; TWN, Taiwan; THA, Thailand; VNM, Vietnam.
Mean cumulative resistance scores varied for study countries. The overall mean score was 4.23. Australia (2.58) and Singapore (2.66) had the lowest scores, followed by Hong Kong (3.67) and Taiwan (3.27). The highest scores were found in Indonesia and Malaysia (6.71 and 8.00, respectively [see Fig. S1 in the supplemental material]).
DISCUSSION
The C. difficile isolate collection tested in this study was diverse, comprising a broad array of 79 different RTs representing the most common RTs circulating in Asia-Pacific countries. Various susceptibilities to the agents tested were found across different RTs and between countries. Overall, rates of resistance to clindamycin were highest (80.7% of all isolates), followed by erythromycin (55.3%) and moxifloxacin (44.4%), and a minority of strains was resistant to rifaximin (15.5% [Table 1]).
Some of the most common C. difficile strains in the study isolate collection showed resistance to three or more agents. All RT 369 and QX 239 isolates, and 92.7% of RT 018 isolates, were resistant to ≥3 agents (clindamycin, moxifloxacin, and erythromycin [Table 1]). For RT 017, the most common strain in the collection, 66.1% of isolates were resistant to three agents, and 61.8% were resistant to four agents (clindamycin, erythromycin, moxifloxacin, and rifaximin). Multiresistance to these agents likely has contributed to these strains’ predominance in the Asia-Pacific region.
Rifaximin resistance varied by country, was not detected in Australia, Singapore, or Japan (Fig. 1), and was primarily found in RT 017 isolates (Table 1). Rifampin, a derivative of rifaximin, is one of the most commonly used antituberculosis agents worldwide and is typically used as long-term therapy, placing a substantial selective pressure for developing resistance. High prevalence of C. difficile RT 017 has been reported previously for tuberculosis patients in South Africa (11). The high prevalence of tuberculosis in Southeast Asian countries (27) may contribute to rifaximin resistance and may even contribute to the predominance of RT 017 in particular in these countries. Rapid emergence of rifaximin and rifampin resistance following treatment with rifaximin has been demonstrated many times for S. aureus and has also been shown in patients infected with C. difficile (28–30). Tuberculosis is rare in Australia, Singapore, and Japan, where rifaximin resistance was not detected in the current study, and supports the theory that rifaximin resistance in C. difficile may have emerged in regions where tuberculosis is more prevalent and rifaximin and rifampin are used more frequently for treatment of tuberculosis. In fact, Japan only introduced rifaximin in 2016 (31).
Infection with QX 239 was previously significantly associated with outcome of recurrent CDI in this study (17). With PCR ribotyping, QX 239 gives a banding pattern similar to that of RT 018, differing by one band (17), and corresponds to smz’ in other reports from Japan (H. Kato, personal communication). It was isolated from one site only in the current study, and all QX 239 isolates were resistant to clindamycin, moxifloxacin, and erythromycin, which likely contributed to their association with recurrent infection. These isolates also demonstrated reduced susceptibility to vancomycin by agar dilution (MIC90 = 4 mg/liter [Table 1]); however, by Etest they had MICs of <2 mg/liter.
Meta-analysis suggests that MICs for both vancomycin and metronidazole are increasing (32), which is a cause of major concern since these are the two primary agents used to treat CDI cases. New antimicrobial agents for treatment of CDI have narrow-spectrum activity, targeting C. difficile while conserving the gut microflora, thus reducing the risk of recurrent infection. Fidaxomicin, a macrocyclic antibiotic, is one such narrow-spectrum agent and is already available in most Asia-Pacific countries; however, it is mainly used in cases of recurrent CDI due to its high cost. All isolates in this collection were susceptible to fidaxomicin, with MIC50 values of 0.125 mg/liter and a geometric mean MIC of 0.10 mg/liter (Table 1). Another novel, small-molecule antimicrobial agent, ridinilazole, shows similar highly potent narrow-spectrum activity for C. difficile (33) and is currently in phase III clinical trials (34).
Cumulative resistance scores varied across countries included in the study (Fig. S1). While scores were high for the Southeast Asian countries of Malaysia (8.00), Vietnam (4.21), Philippines (4.11), and Indonesia (3.85), few isolates were collected for these countries, which means that their scores could be overestimated. However, there was a general trend correlating higher cumulative resistance score with decreasing gross domestic product (GDP) per capita (Fig. S2), with higher-income countries such as Singapore and Australia showing lower cumulative resistance scores than lower-income countries like Thailand and China. This reflects findings reported by Collignon et al., who showed an inverse correlation between aggregate antimicrobial resistance and GDP per capita (35). The same study also found positive correlations of higher aggregate antimicrobial resistance with higher temperatures and poorer infrastructure, features which generally apply to Southeast Asian countries which lie close to the equator.
There were some limitations to the study, the primary limitation being the low numbers of isolates collected from some study countries, particularly Indonesia, Malaysia, Vietnam, and Philippines, as mentioned before (17). This was due to poor recruitment numbers in these countries, mainly due to late study commencement after delays in receiving ethics approvals to conduct the study. The study was also performed in India, but due to government restrictions isolates could not be collected and sent overseas, so we were unable to investigate the molecular epidemiology and antimicrobial susceptibility profiles for C. difficile in India in this instance. Furthermore, the diagnostic assay for CDI varied across sites and countries, which may have led to some inconsistencies in identification of CDI cases for recruitment. Notwithstanding these limitations, we collected a significant number of C. difficile isolates, allowing a broad comparison of the molecular epidemiology and antimicrobial susceptibility profiles for the 12 study countries.
In conclusion, the susceptibility of C. difficile to various antimicrobial agents varied highly by strain type and by country across the Asia-Pacific region. C. difficile RTs 369 and QX 239 showed high MICs, and all were resistant to clindamycin, moxifloxacin, and erythromycin. Other common strains in Asia, including the predominant strain RT 017, were resistant to many antimicrobials, which likely facilitated their proliferation in the Asia-Pacific region. Elevated MICs, including possible reduced susceptibility to vancomycin, were recorded for several C. difficile strains. Ongoing surveillance of C. difficile and its resistance profiles is clearly warranted in the Asia-Pacific region.
MATERIALS AND METHODS
Study isolate collection.
We recently published a study of C. difficile in the Asia-Pacific region (17). Briefly, 600 patients with CDI diagnosed by toxin enzyme immunoassay (EIA), tcdB PCR, toxigenic culture or cell culture cytotoxicity neutralization assay provided consent to participate in a prospective observational study of CDI, which was conducted at 40 hospital sites in Australia, China, Hong Kong, India, Indonesia, Japan, Malaysia, Philippines, Singapore, Republic of Korea, Taiwan, Thailand, and Vietnam from March 2014 to January 2015. Diarrheal stool samples were collected from all participants, sent to a central processing laboratory (LSI Medience, Tokyo, Japan, and/or PathWest Laboratory Medicine, Perth, Australia), and cultured for C. difficile. As previously described, PCR ribotyping was performed on all recovered isolates (n = 414 [Table 2]) (17). Isolates from India were not included in the collection due to government restriction. Seventy-nine ribotypes were represented in the isolate collection.
TABLE 2.
Study isolate collection by ribotype and country or regiona
| Ribotype | No. of isolates (%) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AUS | CHN, HKG | IDN, MYS, PHL | JPN | KOR | SGP | TWN | THA | VNM | Total | |
| RT 017 | 0 | 9 (18.0) | 8 (47.1) | 1 (2.2) | 13 (14.6) | 1 (4.5) | 10 (11.4) | 19 (50.0) | 7 (50.0) | 68 (16.4) |
| RT 014/020 | 13 (26.0) | 4 (8.0) | 0 | 1 (2.2) | 6 (6.7) | 6 (2.7) | 8 (9.1) | 7 (18.4) | 0 | 45 (10.9) |
| RT 018 | 2 (4.0) | 1 (2.0) | 0 | 0 | 38 (42.7) | 0 | 0 | 0 | 0 | 41 (9.9) |
| RT 002 | 3 (6.0) | 1 (2.0) | 0 | 12 (26.1) | 1 (1.1) | 1 (4.5) | 20 (22.7) | 0 | 0 | 38 (9.2) |
| RT 012 | 1 (2.0) | 6 (12.0) | 0 | 0 | 2 (2.2) | 3 (13.6) | 6 (6.8) | 2 (5.3) | 0 | 20 (4.8) |
| RT 369 | 0 | 4 (8.0) | 0 | 11 (23.9) | 1 (1.1) | 0 | 1 (1.1) | 0 | 0 | 17 (4.1) |
| QX 239 | 0 | 0 | 0 | 15 (32.6) | 0 | 0 | 0 | 0 | 0 | 15 (3.6) |
| QX 032 | 0 | 1 (2.0) | 0 | 0 | 3 (3.4) | 0 | 10 (11.4) | 1 (2.6) | 0 | 15 (3.6) |
| RT 001 | 0 | 5 (10.0) | 1 (5.9) | 0 | 1 (1.1) | 1 (4.5) | 4 (4.5) | 1 (2.6) | 0 | 13 (3.1) |
| RT 106 | 1 (2.0) | 1 (2.0) | 0 | 1 (2.2) | 1 (1.1) | 0 | 8 (9.1) | 0 | 0 | 12 (2.9) |
| RT 046 | 0 | 2 (4.0) | 1 (5.9) | 0 | 4 (4.5) | 0 | 3 (3.4) | 0 | 1 (7.1) | 11 (2.7) |
| RT 056 | 5 (10.0) | 1 (2.0) | 0 | 0 | 0 | 0 | 1 (1.1) | 0 | 0 | 7 (1.7) |
| RT 070 | 3 (6.0) | 0 | 1 (5.9) | 0 | 2 (2.2) | 0 | 0 | 0 | 0 | 6 (1.4) |
| RT 027 | 0 | 1 (2.0) | 2 (11.8) | 0 | 0 | 0 | 0 | 0 | 0 | 3 (0.7) |
| RT 078 | 0 | 0 | 0 | 0 | 1 (1.1) | 0 | 1 (1.1) | 0 | 0 | 2 (0.5) |
| Other | 22 (44.0) | 14 (28.0) | 4 (23.5) | 5 (10.9) | 16 (18.0) | 10 (45.5) | 16 (18.1) | 8 (21.1) | 6 (42.9) | 101 (24.4) |
| Total | 50 (100.0) | 50 (100.0) | 17 (100.0) | 46 (100.0) | 89 (100.0) | 22 (100.0) | 88 (100.0) | 38 (100.0) | 14 (100.0) | 414 (100.0) |
Abbreviations: AUS, Australia; CHN, China; HKG, Hong Kong; IDN, Indonesia; MYS, Malaysia; PHL, Philippines; JPN, Japan; KOR, Republic of Korea; SGP, Singapore; TWN, Taiwan; THA, Thailand; VNM, Vietnam.
Antimicrobial susceptibility testing.
Antimicrobial susceptibility testing was performed using the agar dilution method according to the guidelines of the Clinical and Laboratory Standards Institute (36). All culture was performed at 35°C in an anaerobic chamber (A3; Don Whitley Scientific Ltd., Shipley, West Yorkshire, United Kingdom) in an atmosphere containing 80% nitrogen, 10% hydrogen, and 10% carbon dioxide at 75% relative humidity. A 0.5 McFarland suspension was prepared in prereduced saline (0.85%) from colonies of 48-h blood agar cultures of test C. difficile and control strains (Bacteroides fragilis ATCC 25285, Bacteroides thetaiotaomicron ATCC 29741, Eubacterium lentum ATCC 43055, and C. difficile ATCC 700057).
A 52-pin inoculum replicator was used to apply approximately 1 to 2 μl of each inoculum onto each test plate (brucella agar supplemented with hemin [5 μg/ml], vitamin K1 [1 μg/ml], and laked sheep blood [5%, vol/vol], incorporated with various concentrations of antimicrobial agents). Test antimicrobial agents were fidaxomicin, vancomycin, metronidazole, rifaximin, clindamycin, erythromycin, amoxicillin-clavulanate, and moxifloxacin. MICs were recorded following 48 h of anaerobic incubation of test plates, and resistance was determined according to recommended clinical breakpoints or epidemiological cutoffs (Table 3). For isolates with MICs of ≥2 mg/liter for vancomycin, Etests (bioMérieux, Marcy l’Etoile, France) were performed to confirm vancomycin susceptibility.
TABLE 3.
Study breakpoints or epidemiological cutoffs for susceptibility, intermediate status, or full resistance to test antimicrobial agents
Resistance rates, MIC50s, MIC90s, and geometric mean MICs were calculated. Cumulative resistance scores were calculated as described by Freeman et al. (37), where isolates were assigned scores determined by their result of susceptible (score of 0), intermediate (score of 1), or fully resistant (score of 2) to each antimicrobial. Scores were summed for all antimicrobials for each isolate and then grouped by country, and mean cumulative resistance scores were calculated for each country.
Ethics approval.
Ethics approval to conduct the observational CDI study was obtained from relevant human research ethics committees at each individual study site.
Supplementary Material
ACKNOWLEDGMENTS
CDAP Study Group members in Australia include the following: Michael Leung, PathWest Laboratory Medicine WA, Nedlands, Western Australia; David McGechie, PathWest Laboratory Medicine WA, Fremantle, Western Australia; and Alison Keed, Royal Perth Hospital, Perth, Western Australia.
CDAP Study Group members in China include the following: Haihui Huang, Huasahan Hospital, Fu Dan University, Shanghai; Fei Liu, Shanghai East Hospital, Shanghai; Yao-Zong Yuan, Ruijin Hospital, Shanghai Jiaotong University School of Medicine, Shanghai; Kaichun Wu, Fourth Military Medical University, Xijing Hospital, Xi’an, Xian Shi; Zhihua Ran, Shanghai Renji Hospital, Shanghai; Yunsong Yu, Sir Run Run Shaw Hospital, Zhejiang University College of Medicine, Hangzhou, Zhejiang; Jinghang Xu, Peking University First Hospital, Beijing; and Ye Chen, Southern Medical University Nanfang Hospital, Guangzhou.
CDAP Study Group members in Hong Kong include the following: Owen Tak Yin Tsang, Princess Margaret Hospital, Hong Kong; Sunny Hei Wong, Prince of Wales Hospital, Shatin; and Ivan Fan Ngai Hung, Queen Mary Hospital, Pok Fu Lam.
CDAP Study Group members in India include the following: Srinivasa Madaiah, Mysore Medical College & Research Institute, Mysore, Karnataka; Nagarjuna Yarlagadda, KIMS Hospitals, Hyderabad, Andhra Pradesh; Phillip Abraham, P.D. Hinduja National Hospital and Medical Research Centre, Mumbai, Maharashtra; and Pravin Gare, Chopda Medicare & Research Centre Pvt. Ltd., Magnum Heart Institute, Nashik.
CDAP Study Group members in Indonesia include the following: Muhammad Hussein Gasem, Rumah Sakit Umum Pusat Dr. Kariadi, Semarang.
CDAP Study Group members in Japan include the following: Shinya Kusachi, Toho University Medical Center, Ohashi Hospital, Meguro-ku, Tokyo, and Makoto Nagashima, Toho University Medical Center, Sakura Hospital, Sakura, Chiba.
CDAP Study Group members in Republic of Korea include the following: Soo Jung Park, Yonsei University Severance Hospital, Seoul, and Sungmin Kiem, Inje University Haeundae Paik Hospital, Busan.
CDAP Study Group members in Malaysia include the following: Christopher K. C. Lee, Hospital Sungai Buloh, Sungai Buloh, Selangor; Jayaram Menon, Clinical Research Centre (CRC), Queen Elizabeth Hospital, Kota Kinabalu, Sabah; and Ting Soo Chow, Hospital Pulau Pinang, Pulau Pinang.
CDAP Study Group members in Philippines include the following: Myrna Mendoza, National Kidney and Transplant Institute, Quezon City; Randy Mercado, St. Luke’s Medical Center, Quezon City; Marilyn Arguillas, Davao Doctors Hospital, Davao City; and Raul Destura, The Medical City, Pasig City.
CDAP Study Group members in Singapore include the following: David Ong Eng Hui, National University Hospital, Singapore; Ang Tiing Leong, Changi General Hospital, Singapore; and Ling Khoon Lin, Singapore General Hospital, Singapore.
CDAP Study Group members in Taiwan include the following: Yi-Hui Wu, E-Da Hospital, Kaohsiung; Po-Ren Hsueh, National Taiwan University Hospital, Taipei; Yuarn-Jang Lee, Taipei Medical University Hospital, Taipei; Jen-Hsien Wang, China Medical University Hospital, Taichung; Yao-Shen Chen, Veterans General Hospital- Kaohsiung, Kaohsiung; and Wen-Chien Ko, National Cheng Kung University Hospital, Tainan.
CDAP Study Group members in Thailand include the following: Chomsri Kositchaiwat, Ramathibodi Hospital, Bangkok, Krung Thep Maha Nakhon; Varocha Mahachai, King Chulalongkorn Memorial Hospital, Bangkok, Krung Thep Maha Nakhon; and Naichaya Chamroonkul, Songklanagarind Hospital, Songkla.
CDAP Study Group members in Vietnam include the following: Nguyen Van Kinh, National Hospital of Tropical Diseases, Ha Noi; Le Thanh Hai, National Pediatric Hospital, Ha Noi; and Hoang Le Phuc, Pediatric Hospital No 1, Ho Chi Minh.
The observational study of C. difficile infection in the Asia-Pacific was funded by Otsuka Pharmaceutical Co., Ltd. Deirdre A. Collins is a recipient of an Early Career Fellowship from the National Health & Medical Research Council.
Kentaro Ouchi is an employee of Otsuka Pharmaceutical Co., Ltd.
Kazuhiro Tateda, Yoshikazu Ishii, Thomas V. Riley, and Deirdre A. Collins developed the study concept and design. Kentaro Ouchi supervised and coordinated patient enrollment and sample collection. Kyung Mok Sohn and the CDAP Study Group recruited patients. Deirdre A. Collins, Yuan Wu, Tanya Lew, and Papanin Putsathit performed laboratory assays and analyzed the results. Deirdre A. Collins interpreted the data and wrote the first draft of the manuscript. All authors critically reviewed and edited the manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Miller BA, Chen LF, Sexton DJ, Anderson DJ. 2011. Comparison of the burdens of hospital-onset, healthcare facility-associated Clostridium difficile infection and of healthcare-associated infection due to methicillin-resistant Staphylococcus aureus in community hospitals. Infect Control Hosp Epidemiol 32:387–390. doi: 10.1086/659156. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2019. Antibiotic resistance threats in the United States, 2019. US Department of Health and Human Services, Atlanta, GA. [Google Scholar]
- 3.Chen Y, Glass K, Liu B, Korda RJ, Riley TV, Kirk MD. 2017. Burden of Clostridium difficile infection: associated hospitalization in a cohort of middle-aged and older adults. Am J Infect Control 45:508–511. doi: 10.1016/j.ajic.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 4.Bauer MP, ECDIS Study Group, Notermans DW, van Benthem BH, Brazier JS, Wilcox MH, Rupnik M, Monnet DL, van Dissel JT, Kuijper EJ, Group ES. 2011. Clostridium difficile infection in Europe: a hospital-based survey. Lancet 377:63–73. doi: 10.1016/S0140-6736(10)61266-4. [DOI] [PubMed] [Google Scholar]
- 5.Toth M, Stewart NK, Smith C, Vakulenko SB. 2018. Intrinsic class D beta-lactamases of Clostridium difficile. mBio 9:e01803-18. doi: 10.1128/mBio.01803-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peng Z, Jin D, Kim HB, Stratton CW, Wu B, Tang YW, Sun X. 2017. Update on antimicrobial resistance in Clostridium difficile: resistance mechanisms and antimicrobial susceptibility testing. J Clin Microbiol 55:1998–2008. doi: 10.1128/JCM.02250-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He M, Miyajima F, Roberts P, Ellison L, Pickard DJ, Martin MJ, Connor TR, Harris SR, Fairley D, Bamford KB, D’Arc S, Brazier J, Brown D, Coia JE, Douce G, Gerding D, Kim HJ, Koh TH, Kato H, Senoh M, Louie T, Michell S, Butt E, Peacock SJ, Brown NM, Riley T, Songer G, Wilcox M, Pirmohamed M, Kuijper E, Hawkey P, Wren BW, Dougan G, Parkhill J, Lawley TD. 2013. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet 45:109–113. doi: 10.1038/ng.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins DA, Hawkey PM, Riley TV. 2013. Epidemiology of Clostridium difficile infection in Asia. Antimicrob Resist Infect Control 2:21. doi: 10.1186/2047-2994-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tickler IA, Obradovich AE, Goering RV, Fang FC, Tenover FC, Consortium H. 2019. Changes in molecular epidemiology and antimicrobial resistance profiles of Clostridioides (Clostridium) difficile strains in the United States between 2011 and 2017. Anaerobe 60:102050. doi: 10.1016/j.anaerobe.2019.06.003. [DOI] [PubMed] [Google Scholar]
- 10.Freeman J, Vernon J, Pilling S, Morris K, Nicholson S, Shearman S, Longshaw C, Wilcox MH, Pan-European Longitudinal Surveillance of Antibiotic Resistance among Prevalent Clostridium difficile Ribotypes Study Group . 2018. The ClosER study: results from a three-year pan-European longitudinal surveillance of antibiotic resistance among prevalent Clostridium difficile ribotypes, 2011–2014. Clin Microbiol Infect 24:724–731. doi: 10.1016/j.cmi.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 11.Kullin B, Wojno J, Abratt V, Reid SJ. 2017. Toxin A-negative toxin B-positive ribotype 017 Clostridium difficile is the dominant strain type in patients with diarrhoea attending tuberculosis hospitals in Cape Town, South Africa. Eur J Clin Microbiol Infect Dis 36:163–175. doi: 10.1007/s10096-016-2790-x. [DOI] [PubMed] [Google Scholar]
- 12.Imwattana K, Knight DR, Kullin B, Collins DA, Putsathit P, Kiratisin P, Riley TV. 2019. Clostridium difficile ribotype 017—characterization, evolution and epidemiology of the dominant strain in Asia. Emerg Microbes Infect 8:796–807. doi: 10.1080/22221751.2019.1621670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Boeckel TP, Gandra S, Ashok A, Caudron Q, Grenfell BT, Levin SA, Laxminarayan R. 2014. Global antibiotic consumption 2000 to 2010: an analysis of national pharmaceutical sales data. Lancet Infect Dis 14:742–750. doi: 10.1016/S1473-3099(14)70780-7. [DOI] [PubMed] [Google Scholar]
- 14.Van Boeckel TP, Brower C, Gilbert M, Grenfell BT, Levin SA, Robinson TP, Teillant A, Laxminarayan R. 2015. Global trends in antimicrobial use in food animals. Proc Natl Acad Sci U S A 112:5649–5654. doi: 10.1073/pnas.1503141112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zellweger RM, Carrique-Mas J, Limmathurotsakul D, Day NPJ, Thwaites GE, Baker S, Southeast Asia Antimicrobial Resistance Network . 2017. A current perspective on antimicrobial resistance in Southeast Asia. J Antimicrob Chemother 72:2963–2972. doi: 10.1093/jac/dkx260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borren NZ, Ghadermarzi S, Hutfless S, Ananthakrishnan AN. 2017. The emergence of Clostridium difficile infection in Asia: a systematic review and meta-analysis of incidence and impact. PLoS One 12:e0176797. doi: 10.1371/journal.pone.0176797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collins DA, Sohn KM, Wu Y, Ouchi K, Ishii Y, Elliott B, Riley TV, Tateda K, Clostridioides difficile Asia-Pacific Study Group . 2020. Clostridioides difficile infection in the Asia-Pacific region. Emerg Microbes Infect 9:42–52. doi: 10.1080/22221751.2019.1702480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imwattana K, Knight DR, Kullin B, Collins DA, Putsathit P, Kiratisin P, Riley TV. 2020. Antimicrobial resistance in Clostridium difficile ribotype 017. Expert Rev Anti Infect Ther 18:17–25. doi: 10.1080/14787210.2020.1701436. [DOI] [PubMed] [Google Scholar]
- 19.Wang B, Peng W, Zhang P, Su J. 2018. The characteristics of Clostridium difficile ST81, a new PCR ribotype of toxin A− B+ strain with high-level fluoroquinolones resistance and higher sporulation ability than ST37/PCR ribotype 017. FEMS Microbiol Lett 365:fny168. doi: 10.1093/femsle/fny168. [DOI] [PubMed] [Google Scholar]
- 20.Kuwata Y, Tanimoto S, Sawabe E, Shima M, Takahashi Y, Ushizawa H, Fujie T, Koike R, Tojo N, Kubota T, Saito R. 2015. Molecular epidemiology and antimicrobial susceptibility of Clostridium difficile isolated from a university teaching hospital in Japan. Eur J Clin Microbiol Infect Dis 34:763–772. doi: 10.1007/s10096-014-2290-9. [DOI] [PubMed] [Google Scholar]
- 21.Cheng VC, Yam WC, Lam OT, Tsang JL, Tse EY, Siu GK, Chan JF, Tse H, To KK, Tai JW, Ho PL, Yuen KY. 2011. Clostridium difficile isolates with increased sporulation: emergence of PCR ribotype 002 in Hong Kong. Eur J Clin Microbiol Infect Dis 30:1371–1381. doi: 10.1007/s10096-011-1231-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collins DA, Gasem MH, Habibie TH, Arinton IG, Hendriyanto P, Hartana AP, Riley TV. 2017. Prevalence and molecular epidemiology of Clostridium difficile infection in Indonesia. New Microbes New Infect 18:34–37. doi: 10.1016/j.nmni.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riley TV, Collins DA, Karunakaran R, Kahar MA, Adnan A, Hassan SA, Zainul NH, Rustam FRM, Wahab ZA, Ramli R, Lee YY, Hassan H. 2018. High prevalence of toxigenic and nontoxigenic Clostridium difficile strains in Malaysia. J Clin Microbiol 56:e00170-18. doi: 10.1128/JCM.00170-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Putsathit P, Maneerattanaporn M, Piewngam P, Kiratisin P, Riley TV. 2017. Prevalence and molecular epidemiology of Clostridium difficile infection in Thailand. New Microbes New Infect 15:27–32. doi: 10.1016/j.nmni.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng JW, Xiao M, Kudinha T, Xu ZP, Sun LY, Hou X, Zhang L, Fan X, Kong F, Xu YC. 2015. The role of glutamate dehydrogenase (GDH) testing assay in the diagnosis of Clostridium difficile infections: a high sensitive screening test and an essential step in the proposed laboratory diagnosis workflow for developing countries like China. PLoS One 10:e0144604. doi: 10.1371/journal.pone.0144604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerding DN, Sambol SP, Johnson S. 2018. Non-toxigenic Clostridioides (formerly Clostridium) difficile for prevention of C. difficile infection: from bench to bedside back to bench and back to bedside. Front Microbiol 9:1700. doi: 10.3389/fmicb.2018.01700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.WHO. 2019. Global tuberculosis report 2019. WHO, Geneva, Switzerland. [Google Scholar]
- 28.Carman RJ, Boone JH, Grover H, Wickham KN, Chen L. 2012. In vivo selection of rifamycin-resistant Clostridium difficile during rifaximin therapy. Antimicrob Agents Chemother 56:6019–6020. doi: 10.1128/AAC.00974-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Padilla E, Oms L, Espejo E, Gómez L, Pagespetit L, Boada N, Bella F, Pérez J. 2018. Rifampin resistance in staphylococci after rifaximin intake for surgical prophylaxis in elective colorectal surgery. Antimicrob Agents Chemother 62:e01353-18. doi: 10.1128/AAC.01353-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang JY, Kim SE, Kim TH, Woo SY, Ryu MS, Joo YH, Lee KE, Lee J, Lee KH, Moon CM, Jung HK, Shim KN, Jung SA. 2017. Emergence of rifampin-resistant staphylococci after rifaximin administration in cirrhotic patients. PLoS One 12:e0186120. doi: 10.1371/journal.pone.0186120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishida S, Hamada K, Nishino N, Fukushima D, Koyanagi R, Horikawa Y, Shiwa Y, Saitoh S. 2019. Efficacy of long-term rifaximin treatment for hepatic encephalopathy in the Japanese. World J Hepatol 11:531–541. doi: 10.4254/wjh.v11.i6.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saha S, Kapoor S, Tariq R, Schuetz AN, Tosh PK, Pardi DS, Khanna S. 2019. Increasing antibiotic resistance in Clostridioides difficile: a systematic review and meta-analysis. Anaerobe 58:35–46. doi: 10.1016/j.anaerobe.2019.102072. [DOI] [PubMed] [Google Scholar]
- 33.Baines SD, Crowther GS, Freeman J, Todhunter S, Vickers R, Wilcox MH. 2015. SMT19969 as a treatment for Clostridium difficile infection: an assessment of antimicrobial activity using conventional susceptibility testing and an in vitro gut model. J Antimicrob Chemother 70:182–189. doi: 10.1093/jac/dku324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carlson TJ, Endres BT, Basseres E, Gonzales-Luna AJ, Garey KW. 2019. Ridinilazole for the treatment of Clostridioides difficile infection. Expert Opin Invest Drugs 28:303–310. doi: 10.1080/13543784.2019.1582640. [DOI] [PubMed] [Google Scholar]
- 35.Collignon P, Beggs JJ, Walsh TR, Gandra S, Laxminarayan R. 2018. Anthropological and socioeconomic factors contributing to global antimicrobial resistance: a univariate and multivariable analysis. Lancet Planet Health 2:e398–e405. doi: 10.1016/S2542-5196(18)30186-4. [DOI] [PubMed] [Google Scholar]
- 36.Clinical and Laboratory Standards Institute. 2007. Methods for antimicrobial susceptibility testing of anaerobic bacteria; approved standard M11-A7, 7th ed, vol 27 CLSI, Wayne, PA. [Google Scholar]
- 37.Freeman J, Vernon J, Morris K, Nicholson S, Todhunter S, Longshaw C, Wilcox MH, Pan-European Longitudinal Surveillance of Antibiotic Resistance among Prevalent Clostridium difficile Ribotypes’ Study Group . 2015. Pan-European longitudinal surveillance of antibiotic resistance among prevalent Clostridium difficile ribotypes. Clin Microbiol Infect 21:248.e9–248.e16. doi: 10.1016/j.cmi.2014.09.017. [DOI] [PubMed] [Google Scholar]
- 38.European Committee on Antimicrobial Susceptibility Testing. 2019. Breakpoint tables for interpretation of MICs and zone diameters, version 9.0. http://wwweucastorg.
- 39.O’Connor JR, Galang MA, Sambol SP, Hecht DW, Vedantam G, Gerding DN, Johnson S. 2008. Rifampin and rifaximin resistance in clinical isolates of Clostridium difficile. Antimicrob Agents Chemother 52:2813–2817. doi: 10.1128/AAC.00342-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clinical and Laboratory Standards Institute. 2019. Performance standards for antimicrobial susceptibility testing: seventh informational supplement M100-S29. CLSI, Wayne, PA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.