Empiric antibiotic prescribing can be supported by guidelines and/or local antibiograms, but these have limitations. We sought to use data from a comprehensive electronic health record to use statistical learning to develop predictive models for individual antibiotics that incorporate patient- and hospital-specific factors. This paper reports on the development and validation of these models with a large retrospective cohort. This was a retrospective cohort study including hospitalized patients with positive urine cultures in the first 48 h of hospitalization at a 1,500-bed tertiary-care hospital over a 4.
KEYWORDS: urinary tract infection, predictive models, antibiotic coverage, antibiotic resistance
ABSTRACT
Empiric antibiotic prescribing can be supported by guidelines and/or local antibiograms, but these have limitations. We sought to use data from a comprehensive electronic health record to use statistical learning to develop predictive models for individual antibiotics that incorporate patient- and hospital-specific factors. This paper reports on the development and validation of these models with a large retrospective cohort. This was a retrospective cohort study including hospitalized patients with positive urine cultures in the first 48 h of hospitalization at a 1,500-bed tertiary-care hospital over a 4.5-year period. All first urine cultures with susceptibilities were included. Statistical learning techniques, including penalized logistic regression, were used to create predictive models for cefazolin, ceftriaxone, ciprofloxacin, cefepime, and piperacillin-tazobactam. These were validated on a held-out cohort. The final data set used for analysis included 6,366 patients. Final model covariates included demographics, comorbidity score, recent antibiotic use, recent antimicrobial resistance, and antibiotic allergies. Models had acceptable to good discrimination in the training data set and acceptable performance in the validation data set, with a point estimate for area under the receiver operating characteristic curve (AUC) that ranged from 0.65 for ceftriaxone to 0.69 for cefazolin. All models had excellent calibration. We used electronic health record data to create predictive models to estimate antibiotic susceptibilities for urinary tract infections in hospitalized patients. Our models had acceptable performance in a held-out validation cohort.
INTRODUCTION
Prescribing antibiotics for a patient before the results of a clinical culture are reported can be complex and challenging for clinicians. They must weigh the severity of the patient’s presentation, patient-specific risk factors for adverse outcomes, the likelihood of antibiotic resistance in the hospital and surrounding community, and the risk to other hospitalized patients and society when a broad-spectrum antibiotic is used. Several tools are available to help clinicians with empiric antibiotic prescribing, including guidelines and antibiograms. However, each of these tools has drawbacks, most notably that they do not provide recommendations personalized to the individual patient. With the rise in adoption of electronic health records (EHR), we have the opportunity to leverage this information to provide clinicians with decision support at the point of care that incorporates critical data about the hospital environment, patient-specific risks, and institutional priorities.
We showed previously how focusing on syndromic antibiograms rather than organism-specific antibiograms may be useful for empiric prescribing (1). However, that work was limited in its application, as it did not incorporate patient-specific risks or recent clinical history. Here, we describe the use of patient-specific data from the EHR to develop a series of models intended to predict the probability of susceptibility to antibiotics commonly used for empiric treatment of community-onset urinary tract infections in a hospital setting. In this work, for each antibiotic model, the infection was determined to be “susceptible” to each antibiotic when all organisms that grew in the urine culture were susceptible to the antibiotic in vitro or, in the case of no reported laboratory data, were determined to be likely susceptible to that antibiotic by expert guidance. These model predictions are intended to provide patient-, location (hospital)-, and time (making use of recent data)-specific guidance for empiric antibiotic prescribing.
RESULTS
The final analysis data set included 6,366 patients after all exclusions (Fig. 1). Table 1 summarizes patient demographic and clinical characteristics, overall and by susceptibility for each of the five antibiotics. Overall, the mean age in the cohort was 61.9 (standard deviation [SD], 17.8) with more female patients (67.1%) than male, and the majority identified as being of white race (72.6%). A small proportion were documented as being admitted from a nursing home (8.1%). The number of records that were dropped due to missing susceptibility data preprocessing ranged from a low of 1 record for cefazolin (0.02%) to a high of 320 records for ceftriaxone (5.0%). The percentages of urine cultures susceptible to the antibiotics were as follows: cefazolin, 48.7% (3,100/6,365); cefepime, 64.6% (4,100/6,359); ceftriaxone, 59.1% (3,575/6,046), ciprofloxacin, 48.1% (3,064/6,364); and piperacillin-tazobactam, 82.0% (5,219/6,363). To evaluate the sensitivity of the rule order used to fill in susceptibility data that were not reported, we randomly shuffled the rules 100 times and compared the filled-in data sets. At most, 3,519 of the 74,885 unreported outcomes (5%) were sensitive to the rule order.
FIG 1.
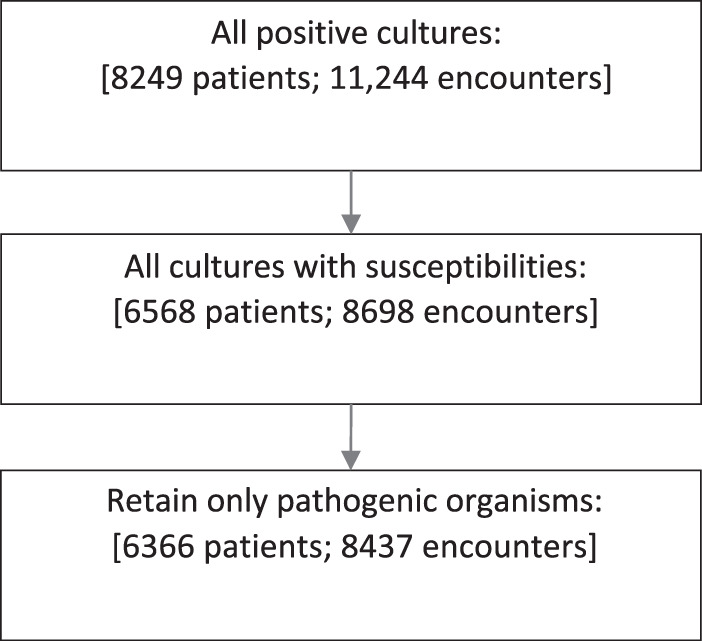
Cohort development.
TABLE 1.
Cohort demographic and clinical characteristics by antibiotic susceptibility
| Characteristic | Valuea
for: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| All (n = 6366) | Cefazolin |
Cefepime |
Ceftriaxone |
Ciprofloxacin |
Piperacillin-tazobactam |
||||||
| S (n = 3,100) | R (n = 3,265) | S (n = 4,110) | R (n = 2,249) | S (n = 3,575) | R (n = 2,471) | S (n = 3,064) | R (n = 3,300) | S (n = 5,219) | R (n = 1,144) | ||
| Age | 61.9 (17.8) | 60.7 (18.8) | 63.0 (16.7) | 61.4 (18.4) | 62.7 (16.6) | 61.1 (18.6) | 62.9 (16.5) | 61.1 (8.7) | 62.6 (16.9) | 62.0 (18) | 61.5 (16.6) |
| Sex | |||||||||||
| Female | 4,272 (67.1) | 2,354 (55.12) | 1,917 (44.88) | 2,942 (68.93) | 1,326 (31.07) | 2,666 (62.95) | 1,569 (37.05) | 2,266 (53.04) | 2,006 (46.96) | 3,585 (83.96) | 685 (16.04) |
| Male | 2,094 (32.9) | 746 (35.63) | 1,348 (64.37) | 1,168 (55.86) | 923 (44.14) | 909 (43.68) | 1,172 (56.32) | 798 (38.15) | 1,294 (61.85) | 1,634 (78.07) | 459 (21.93) |
| Race | |||||||||||
| White | 4,619 (72.6) | 2,135 (46.23) | 2,483 (53.77) | 2,878 (62.38) | 1,736 (37.62) | 2,474 (53.95) | 2,112 (46.05) | 2,136 (46.26) | 2,481 (53.74) | 3,772 (71.68) | 846 (18.32) |
| Black | 1,497 (23.5) | 833 (55.64) | 664 (44.36) | 1,068 (71.44) | 427 (28.56) | 952 (64.24) | 530 (35.76) | 789 (52.71) | 708 (47.29) | 1,251 (83.68) | 244 (16.32) |
| Other | 250 (3.90) | 132 (52.80) | 118 (47.20) | 164 (65.60) | 86 (34.40) | 149 (60.08) | 99 (39.92) | 139 (55.60) | 111 (44.40) | 196 (78.40) | 54 (21.60) |
| History of smoking | |||||||||||
| Yes | 3,688 (57.9) | 1,808 (49.04) | 1,879 (50.96) | 2,378 (64.58) | 1,304 (35.42) | 2,075 (56.57) | 1,593 (43.43) | 1,753 (47.56) | 1,933 (52.44) | 3,020 (71.93) | 666 (18.07) |
| No | 2,678 (42.1) | 1,292 (48.24) | 1,386 (51.76) | 1,732 (64.70) | 945 (35.30) | 1,500 (56.65) | 1,148 (43.35) | 1,311 (48.95) | 1,367 (51.05) | 2,199 (72.14) | 478 (17.86) |
| Admission from: | |||||||||||
| Nursing home | 514 (8.1) | 185 (35.99) | 329 (64.01) | 285 (55.45) | 229 (44.55) | 225 (44.47) | 281 (55.53) | 189 (36.77) | 325 (63.23) | 368 (71.73) | 145 (28.27) |
| Other | 5,852 (91.90) | 2,915 (49.82) | 2,936 (50.18) | 3,825 (65.44) | 2,020 (34.56) | 3,350 (57.66) | 2,460 (42.34) | 2,875 (49.15) | 2,975 (50.85) | 4,851 (82.92) | 999 (17.08) |
Age is given as mean (SD); other values are number (percent) of patients.
Final model covariates, coefficients, areas under the receiver operating characteristic curve (AUCs), and goodness-of-fit (GOF) tests are presented in Table 2. The penalized regression models that included grouping for interaction effects and the penalized models that allowed interaction terms without grouping of main effects returned comparable or less favorable AUCs in the training/test data set compared to those with only main effect terms and were considerably more complex. Final models included only main effect terms. The AUCs of the models in the training cohort ranged from 68.3 (95% confidence interval [CI], 66.8 to 69.8) for cefepime to 71.5 (95% CI, 70.1 to 72.9) for cefazolin. They were generally similar, but lower, in the validation, with a range from 65.0 for cefepime to 69.1 for cefazolin. All final prediction models for each antibiotic had modest to good discrimination with excellent model calibration.
TABLE 2.
Final prediction model covariatesa
| Category | Parameter |
Value for: |
||||
|---|---|---|---|---|---|---|
| Cefazolin (0.63935) | Cefepime (1.17828) | Ceftriaxone (0.94419) | Ciprofloxacin (0.43047) | Piperacillin-tazobactam (2.18690) | ||
| Demographics | Age | 0.00019 | 0.01168 | |||
| Age squared | 0.00010 | 0.00007 | ||||
| Age cubed | −0.00001 | −0.00001 | −0.00001 | −0.00001 | ||
| Sex: maleb | −0.71695 | −0.45621 | −0.70641 | −0.53074 | −0.16150 | |
| History of smokingb | 0.12988 | 0.08713 | ||||
| Race: blackb | 0.30388 | 0.36458 | 0.36887 | 0.23907 | 0.01110 | |
| Race: otherb | 0.29024 | 0.09832 | 0.22255 | 0.28785 | −0.36885 | |
| Medical history | Admission from nursing homeb | −0.29785 | −0.18948 | −0.29092 | −0.10103 | −0.27365 |
| 1 ED visitb ,c | 0.30499 | 0.27674 | 0.18183 | 0.02006 | 0.29449 | |
| ≥2 ED visitsb ,c | 0.01528 | 0.44160 | ||||
| 1 ED to inpatientb ,c | −0.08453 | −0.22877 | −0.08165 | −0.05306 | −0.29808 | |
| ≥2 ED to inpatientb ,c | −0.12733 | −0.16352 | −0.00564 | |||
| 1 inpatient visitb ,c | −0.22318 | −0.20348 | −0.21192 | −0.21073 | ||
| ≥2 inpatient visitsb ,c | 0.00571 | −0.11337 | ||||
| ICU location in first 24 hoursb | 0.07366 | 0.11776 | 0.15119 | |||
| Elixhauser score | −0.01383 | −0.01102 | −0.01155 | −0.00357 | −0.00658 | |
| Antimicrobials in past 30 days | Sulfonamide | −0.25820 | −0.21812 | −0.17295 | −0.05543 | −0.06770 |
| Quinolone | −0.51539 | −0.65805 | −0.53185 | −1.29377 | −0.61818 | |
| Cephalosporin | −0.59423 | −0.25652 | −0.60599 | −0.43066 | ||
| Penicillin | −0.11334 | 0.06402 | −0.14594 | −0.32084 | ||
| Carbapenem | −0.32128 | −0.54253 | −0.44776 | −0.45540 | ||
| Antimicrobials in past 30–90 days | Sulfonamide | −0.19242 | −0.18646 | −0.19487 | −0.14391 | −0.19486 |
| Quinolone | −0.29001 | −0.31754 | −0.32479 | −0.44247 | −0.01208 | |
| Cephalosporin | −0.12215 | −0.11174 | 0.00980 | |||
| Penicillin | −0.08546 | −0.05267 | −0.30380 | |||
| Carbapenem | −0.14987 | −0.30757 | −0.16412 | |||
| Resistant organisms in past 6 months | MDRO | −0.76878 | −0.51511 | −0.80317 | −0.51611 | −0.66253 |
| MRSA | 0.58438 | 0.44875 | 0.63166 | 0.45980 | ||
| VRE | 0.16156 | 0.04077 | −0.00365 | |||
| CephR Klebsiella | −0.85975 | |||||
| ESBL | 0.57457 | 0.22556 | ||||
| MDR Acinetobacter | 0.67298 | 0.46587 | 0.56299 | |||
| Resistant organisms in past 6–12 months | MDRO | −0.97448 | −0.61115 | −1.02161 | −0.10190 | −1.09366 |
| MRSA | 1.13502 | 0.53233 | 1.04338 | 1.22636 | ||
| VRE | 0.17833 | 0.40149 | −0.05931 | 0.52085 | ||
| Antibiotic resistance in past 6 months | Sulfonamide | 0.25044 | 0.37538 | 0.31095 | 0.10899 | |
| Quinolone | −0.31482 | −0.17482 | −0.34883 | −1.49533 | −0.40215 | |
| Cephalosporin | −0.81174 | −0.53988 | −0.56341 | −0.47589 | ||
| Penicillin | 0.35729 | 0.33486 | 0.40173 | 0.38544 | 0.23992 | |
| Carbapenem | −0.05393 | 0.02238 | −0.32193 | 0.07712 | ||
| Documented medication allergy | Penicillin | −0.04274 | −0.05982 | −0.07149 | −0.20527 | |
| Quinolone | −0.56013 | −0.28042 | −0.43616 | |||
| Sulfonamide | −0.06064 | 0.01466 | 0.17357 | |||
| Cephalosporin | −0.14100 | 0.00746 | −0.09309 | −0.11350 | −0.16364 | |
| Glycopeptide | −0.24265 | −0.37125 | −0.27998 | −0.13965 | ||
| Aminoglycoside | 0.25439 | 0.28761 | ||||
| Metronidazole | 0.52051 | 0.32151 | 0.28773 | 0.06804 | ||
| Carbapenem | −1.22837 | |||||
| Final model characteristics | Goodness of fit (P value) | 0.25 | 0.09 | 0.22 | 0.11 | 0.20 |
| AUC (95% CI): training/test | 71.5 (70.1–72.9) | 68.3 (66.8–69.8) | 71.3 (69.9–72.7) | 69.5 (68.1–71.0) | 69.9 (68.7–72.5) | |
| AUC (95% CI): validation | 69.1 (66.3–72.1) | 65.0 (61.8–68.1) | 68.8 (65.9–71.7) | 65.3 (62.3–68.3) | 68.2 (64.1–72.4) | |
Values in parentheses after the drug names are intercepts. Shading indicates covariates included in the model; bold indicates covariates associated with a decrease in the odds of susceptibility to the antibiotic. ED, emergency department; ICU, intensive care unit; VRE, vancomycin-resistant Enterococcus; ESBL, extended-spectrum beta-lactamase; CephR, cephalosporin resistant.
Reference groups: female sex, nonsmoker, white race, no admission from nursing home, no ED visits, no ED visits to inpatient visits, no inpatient visits, non-ICU stay.
In the past 90 days.
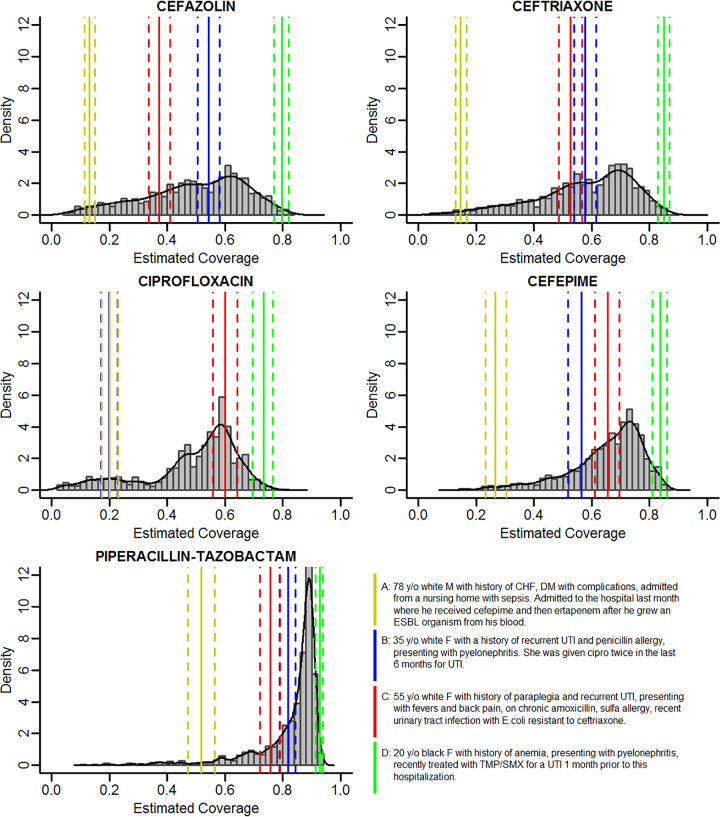
Four patient risk profiles are provided as example results and to facilitate comparisons across antibiotics (Fig. 2). These four profiles were developed by the study team based on common clinical scenarios, without knowledge of the resulting estimated probability. These include an elderly male patient with a recent multidrug-resistant organism; a young female patient with a history of recurrent urinary tract infection (UTI) and penicillin allergy with recent ciprofloxacin use; an older female patient with paraplegia, recurrent UTI, and chronic antibiotic use; and a young, healthy, female patient recently exposed to trimethoprim-sulfamethoxazole. The estimated probabilities of susceptibility for each of these patients are shown in Fig. 2, superimposed on the distribution of estimated probabilities for each patient in the validation cohort.
FIG 2.
Estimated susceptibility (coverage) probability for each antibiotic in the validation set with example cases. y/o, year old; M, male; F, female; CHF, congestive heart failure; DM, diabetes; ESBL, extended-spectrum beta-lactamase; UTI, urinary tract infection; cipro, ciprofloxacin; TMP/SMX, trimethoprim-sulfamethoxazole.
DISCUSSION
We used statistical learning techniques to develop a series of predictive models to estimate the probability of susceptibility for five commonly used antibiotics for UTI. In order to develop these models, we created a robust data preprocessing algorithm which included the use of a novel data completion framework to collect subject matter expertise and a consensus meeting. Our models show acceptable performance in terms of discrimination, with AUCs from 68.3 to 71.5 in the training/test data and lower but comparable AUCs in the validation set (65.0 to 69.1). Although the validation had modest AUC performance, the models show excellent calibration, as measured by GOF, suggesting that our estimates closely agree with the actual outcomes in the group.
In prior research, the inclusion of local resistance data in predictive models has led to more effective empiric treatment algorithms for multiple syndromes, including pneumonia and sepsis (2, 3). Park and colleagues developed a risk-scoring system to predict infection with potentially drug-resistant pathogens in cases of health care-associated pneumonia (HCAP) (4). Compared to the established HCAP criteria for predicting resistant infections, additional stratification by patient-specific risk factors significantly improved diagnostic accuracy (4). Recent works specific to UTI have included prior urine culture results to predict resistance and optimize empirical antimicrobial therapy (5, 6).
Our models were optimized to predict the outcome of antibiotic susceptibility and were not focused on investigating the impact of individual covariates on outcomes. Nevertheless, there are several covariates that deserve discussion. Male sex was associated with lower probability of susceptibility, which has been seen in previous studies (7). Race was a significant predictor in all 5 models. This is likely a marker for factors that are not well captured in EHR data and should not be interpreted as causative. Previous inpatient hospitalizations, antibiotic use, and higher comorbidity scores strongly contributed to a lower probability of susceptibility. A history of multidrug-resistant pathogens (MDRO) is associated with a decreased probability of susceptibility across all the models. Counterintuitively, individual MDRO, such as methicillin-resistant Staphylococcus aureus (MRSA), are associated with an increased probability in susceptibility in some of the models. MDRO is a composite variable that includes individual MDRO such as MRSA. When both the composite (MDRO) and individual (MRSA) variables are included in the model, the effect of one is adjusted for the other, so interpretation becomes difficult. These variables were kept in the model, as they were beneficial in model prediction and calibration, but should not be interpreted alone.
Figure 2 demonstrates the impact of the models on 4 example patients. Patient A was designed as a high-risk patient. He has a history of drug resistance, recent broad-spectrum antibiotic exposure, and recent hospitalization. He has the lowest probability of susceptibility for all antibiotics modeled. Patients B and C were designed as medium-risk patients. For cefazolin, ceftriaxone, and cefepime, their estimated risks are similar and fall between those of our high- and low-risk patients. Interestingly, patient B, who has a penicillin allergy and was recently given ciprofloxacin, has a low estimation of ciprofloxacin susceptibility but high estimation of piperacillin-tazobactam susceptibility. This finding is consistent with our previous work focused on the association of penicillin allergy with ciprofloxacin susceptibility (8). Finally, patient D was designed as a low-risk patient and has a high probability of susceptibility to all antibiotics tested. Other interesting findings from Fig. 2 include the distribution of risk scores. The narrower spectrum antibiotics have a more uniform distribution, whereas for broader-spectrum antibiotics, more patients had a higher estimated probability of susceptibility. Despite cefepime and piperacillin-tazobactam generally having equivalent susceptibility for Gram-negative pathogens, cefepime does not cover Enterococcus, which was our second most common pathogen after Escherichia coli (see Table S4 in the supplemental material).
Our study has several unique strengths. First, we focused on susceptibility to a particular antibiotic of all urinary pathogens, not on modeling risk of a particular organism or a multidrug-resistant organism. This better reflects the real-world decision-making environment of a provider prescribing an empiric antibiotic. Next, we used a reproducible method for filling in unreported susceptibility data using subject matter experts. Our team developed an application which allows subject matter experts to easily express human-understandable rules to quickly and transparently fill in unreported data. These rules can be shared across institutions to replicate our assumptions and methods without requiring sharing of data. Moreover, we utilized a careful model selection process that included an iterative cross-validation step to optimize model selection as well as a final validation step on a held-out cohort.
There are several limitations to this study. Approximately half of the susceptibility data in the final data set were filled in by subject matter experts rather than tested directly in the clinical laboratory. This raises the concern that the filled-in data could be incorrect. However, our data-preprocessing algorithm is similar to the process that most infectious diseases providers perform when they review culture results. They “fill in” the data that are not visible based on their knowledge of the organisms. In addition, we asked our experts to rank the rules as low/medium and high confidence: 80% of cells were filled in with rules marked as high, and only 3% with rules marked as low. We used subject matter experts instead of a simple literature or guideline review, because some decisions depended upon subjective interpretation of the likelihood of antibiotic effectiveness. Organism susceptibility decisions require consideration of a specific infection, including the body site, other susceptibility information on the same culture, and local resistance patterns. Attendings infectious disease physicians make these determinations on a daily basis, and we sought to formalize that decision making. A strength of our approach to data processing is that susceptibility decisions can be made locally and incorporated into data sets before modeling. A second limitation is the retrospective nature of our EHR data set. When extracting data on covariates, we validated these variables with a random subset chart review. However, given the size of our data set and the number of variables, some misclassification is possible. We identified a UTI as one in which the urine culture is positive and the patient has a diagnosis for UTI during that hospitalization. Because of this, we could have included patients that did not have a symptomatic UTI. However, our goal in this work is to improve on the traditional antibiogram, which is based on positive culture rather than disease status. It is reasonable to assume that clinical factors and prior microbiology data will predict antibiotic resistance of organisms that colonize the bladder as well as those that cause symptomatic UTI, so misclassification should not affect our findings. These models do not determine whether the patient should receive antibacterials; rather, they provide information to clinicians as to what antibacterials are likely to cover the organism in the urine. Finally, we are limited to the records on prior antibiotic use, resistance, and hospitalization associated with an encounter at our institution.
These models are the first to be presented from a larger project focused on developing tools to improve empiric antibiotic prescribing for providers and antibiotic stewardship programs. In order for these models to be clinically useful, further knowledge must be incorporated for them to work as clinical decision support tools. We are testing a process which will order antibiotic preferences based on predicted susceptibility, breadth, and adverse side effects (9). In addition, our models could be used to risk-stratify patients who would benefit from costly diagnostic tests (e.g., molecular diagnostic tests [10]).
Conclusion.
In this study, we developed a robust set of models for prediction of antibiotic susceptibility for UTI in hospitalized patients. Our models performed well in a held-out validation cohort. The models make use of innovations to develop reproducible data pathways aimed at better leveraging complex clinical data on infectious diseases.
MATERIALS AND METHODS
Setting and population.
This study was performed at a 1,500-bed academic, tertiary-care medical center located in the midwestern United States. All data were extracted from the information warehouse. The study was approved by the Institutional Review Board at The Ohio State University.
Cohort development.
The cohort included all adult patients who were admitted to an acute-care unit from 1 November 2011 through 1 July 2016, who had a positive urine culture in the first 48 h of admission, and who were discharged with a diagnosis of urinary tract infection. Only the first culture was included. Organisms that were not tested for susceptibility by the clinical laboratory were excluded (3,806 organisms; see Table S1 in the supplemental material), except for organisms for which susceptibility is routinely assumed (e.g., group B Streptococcus is susceptible to penicillin) and so not routinely checked. Based on subject matter expert feedback, organisms were excluded if they were not considered highly pathogenic and had very low colony counts.
Predictors and outcomes.
Variables collected from EHR data included demographics, comorbidities, smoking history, recent antibiotic use and allergies, antibiotic resistance from recent culture data, and recent health care and nursing home exposure (Table 1; also, see Table S2).
Five antibiotics—cefazolin, ceftriaxone, ciprofloxacin, cefepime, and piperacillin-tazobactam—were chosen for investigation based on a formal survey of our antibiotics stewardship team (9). For each of the five antibiotics of interest, a binary outcome of susceptible or resistant was determined. The infection was considered to be susceptible if all organisms that grew in culture were susceptible or assumed to be susceptible to that antibiotic. We did not model trimethoprim-sulfamethoxazole, fosfomycin, or nitrofurantoin, because in our experience these are not common empiric recommendations for patients being admitted to the hospital with UTI.
Data preprocessing.
Patient urine culture data were collected into one file such that each row represented a unique organism that grew during an infection episode. Any susceptibility information that was tested in vitro in the laboratory was included in this data set in the form of the clinical laboratory’s susceptibility interpretation, consisting of R (resistant), S (susceptible), or I (intermediate). All susceptibilities interpreted by the lab as I were considered R for our purposes. However, for each organism, susceptibilities are reported for only a subset of antibiotics based on guidelines and institutional preference, resulting in a sparse data set for cohort susceptibility information. To address unreported susceptibility information, we engaged subject matter experts to create an analysis-ready filled-in data set, using ICARUS, a data completion framework that allows experts to apply rules to the data set in real time (11). Three members of the study team (C. Hebert, M. Lustberg, and P. Pancholi) used ICARUS to fill in unreported data via rules (e.g., “Cefazolin does not cover Enterococcus”), which were then semiautomatically consolidated and reviewed at a consensus meeting. During this meeting, the experts reviewed the rules that were found to have conflicting effects on the data set (i.e., one rule would fill in R and one S for the same cell). Through a round-table discussion, a final rule was agreed upon.
Some rules were dependent on each other, and hence, the order in which they were applied affects the final filled-in data. In our application, the order was assigned by subject matter experts. To test the robustness of rule ordering, we randomly shuffled the rules 100 times and compared the filled-in data sets. Major rules used to fill in the data for relevant antibiotics can be found in Table S3 in the supplemental material.
After data completion rules were applied, the data set was collapsed across infection episodes such that there was one row per episode. If a urine culture had only one organism that resulted, there was no need to collapse, and this record remain unchanged. To collapse rows where a patient had more than one organism grow in culture, we applied the following logic:
-
1.
If any organism was resistant to the antibiotic, the culture was resistant in the final collapsed record.
-
2.
If all organisms were susceptible to an antibiotic, the culture was susceptible in the final collapsed record.
-
3.
If there were remaining missing data and all other organisms in culture were susceptible, the culture had unknown susceptibility and was not included in the modeling.
Statistical methods.
Using the most recent hospitalization for each patient, a multistage statistical learning approach was taken to develop each antibiotic susceptibility model. Model selection and initial performance of multivariable models were evaluated in a large training set, an 80% random sample of the full UTI data set. The remaining 20% was used for model validation. Several functional forms of continuous covariates were included for age (a linear, squared, and cubed term). In all models, age was centered at the mean value of the cohort.
In the training/test set, predictive models for susceptibility of each antibiotic were developed using penalized logistic regression methods incorporating 10-fold cross-validation (CV). All patient characteristics retrieved from the EHR (Table 1; also, see Table S2) were included as potential covariates. Modeling allowed for inclusion of any pairwise interactions, except in cases where these interactions had sparse cells counts (n < 10) and varied functional forms of continuous covariates. Three penalized logistic regression methods were used for model selection and regularization: smoothly clipped absolute deviation (SCAD) (12), minimax concave penalty (MCP) (13), and least absolute shrinkage and selection operator (LASSO) (14). Tenfold CV was repeated 100 times, and the distributions of the model area under the receiver operating characteristic curve (AUC), penalization parameter, and number and frequency of predictors were summarized (15, 16). Once the optimal penalization parameter was chosen across the 100 CV iterations, this penalization was used in each method for model selection. In addition, penalized logistic regression models which considered grouped variables for interaction terms were fitted in each cross-validation fold. In these grouped methods, each predefined group contained two main effects and their interaction (17, 18).
Model calibration was assessed by a modified Hosmer-Lemeshow goodness-of-fit (GOF) test on each of the models (19). Final model selection was based on the model discrimination (AUC), number of predictors, model calibration (GOF), and model AUC variability across the 100 CV iterations. Better models were considered those with higher overall AUC, lower AUC variability, acceptable GOF, and fewer predictors.
In the remaining 20% of data, the best model found at the training/test stage was evaluated. Model prediction accuracy was assessed by the AUC and 95% confidence interval, as estimated from a logistic regression model with each final training/test model risk score as the predictor. Each antibiotic risk score was calculated as the linear combination of the model coefficients from penalized regression models from the training/test stage with the data from the validation cohort.
All analyses were performed using R software and associated packages (20) (https://www.Rproject.org/).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number R01AI116975 and by grant UL1TR001070 from the National Center for Advancing Translational Sciences.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Advancing Translational Sciences or the National Institutes of Health.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Hebert C, Ridgway J, Vekhter B, Brown EC, Weber SG, Robicsek A. 2012. Demonstration of the weighted-incidence syndromic combination antibiogram: an empiric prescribing decision aid. Infect Control Hosp Epidemiol 33:381–388. doi: 10.1086/664768. [DOI] [PubMed] [Google Scholar]
- 2.Becher RD, Hoth JJ, Rebo JJ, Kendall JL, Miller PR. 2012. Locally derived versus guideline-based approach to treatment of hospital-acquired pneumonia in the trauma intensive care unit. Surg Infect (Larchmt) 13:352–359. doi: 10.1089/sur.2011.056. [DOI] [PubMed] [Google Scholar]
- 3.Miano TA, Powell E, Schweickert WD, Morgan S, Binkley S, Sarani B. 2012. Effect of an antibiotic algorithm on the adequacy of empiric antibiotic therapy given by a medical emergency team. J Crit Care 27:45–50. doi: 10.1016/j.jcrc.2011.05.023. [DOI] [PubMed] [Google Scholar]
- 4.Park SC, Kang YA, Park BH, Kim EY, Park MS, Kim YS, Kim SK, Chang J, Jung JY. 2012. Poor prediction of potentially drug-resistant pathogens using current criteria of health care-associated pneumonia. Respir Med 106:1311–1319. doi: 10.1016/j.rmed.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 5.MacFadden DR, Ridgway JP, Robicsek A, Elligsen M, Daneman N. 2014. Predictive utility of prior positive urine cultures. Clin Infect Dis 59:1265–1271. doi: 10.1093/cid/ciu588. [DOI] [PubMed] [Google Scholar]
- 6.Dickstein Y, Geffen Y, Andreassen S, Leibovici L, Paul M. 2016. Predicting antibiotic resistance in urinary tract infection patients with prior urine cultures. Antimicrob Agents Chemother 60:4717–4721. doi: 10.1128/AAC.00202-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lagace-Wiens PR, Simner PJ, Forward KR, Tailor F, Adam HJ, Decorby M, Karlowsky J, Hoban DJ, Zhanel GG, Canadian Antimicrobial Resistance Alliance (CARA) . 2011. Analysis of 3789 in- and outpatient Escherichia coli isolates from across Canada—results of the CANWARD 2007–2009 study. Diagn Microbiol Infect Dis 69:314–319. doi: 10.1016/j.diagmicrobio.2010.10.027. [DOI] [PubMed] [Google Scholar]
- 8.Dewart CM, Gao Y, Rahman P, Mbodj A, Hade EM, Stevenson K, Hebert CL. 2018. Penicillin allergy and association with ciprofloxacin coverage in community-onset urinary tract infection. Infect Control Hosp Epidemiol 39:1127–1128. doi: 10.1017/ice.2018.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patterson ES, Dewart CM, Stevenson K, Mbodj A, Lustberg M, Hade EM, Hebert C. 2018. A mixed methods approach to tailoring evidence-based guidance for antibiotic stewardship to one medical system. Proc Int Symp Hum Factors Ergon Healthc 7:224–231. doi: 10.1177/2327857918071053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tchesnokova V, Avagyan H, Rechkina E, Chan D, Muradova M, Haile HG, Radey M, Weissman S, Riddell K, Scholes D, Johnson JR, Sokurenko EV. 2017. Bacterial clonal diagnostics as a tool for evidence-based empiric antibiotic selection. PLoS One 12:e0174132. doi: 10.1371/journal.pone.0174132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rahman P, Hebert C, Nandi A. 2018. ICARUS: minimizing human effort in iterative data completion. Proceedings VLDB Endowment 11:2263–2276. doi: 10.14778/3275366.3284970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan JQ, Li RZ. 2001. Variable selection via nonconcave penalized likelihood and its oracle properties. J Am Stat Assoc 96:1348–1360. doi: 10.1198/016214501753382273. [DOI] [Google Scholar]
- 13.Zhang CH. 2010. Nearly unbiased variable selection under minimax concave penalty. Ann Statist 38:894–942. doi: 10.1214/09-AOS729. [DOI] [Google Scholar]
- 14.Tibshirani R. 1996. Regression shrinkage and selection via the lasso. J R Stat Soc Series B (Methodol) 58:267–288. doi: 10.1111/j.2517-6161.1996.tb02080.x. [DOI] [Google Scholar]
- 15.Friedman J, Hastie T, Tibshirani R. 2010. Regularization paths for generalized linear models via coordinate descent. J Stat Softw 33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 16.Breheny P, Huang J. 2011. Coordinate descent algorithms for nonconvex penalized regression, with applications to biological feature selection. Ann Appl Stat 5:232–253. doi: 10.1214/10-AOAS388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breheny P, Huang J. 2015. Group descent algorithms for nonconvex penalized linear and logistic regression models with grouped predictors. Stat Comput 25:173–187. doi: 10.1007/s11222-013-9424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan M, Lin Y. 2006. Model selection and estimation in regression with grouped variables. J R Stat Soc B 68:49–67. doi: 10.1111/j.1467-9868.2005.00532.x. [DOI] [Google Scholar]
- 19.Paul P, Pennell ML, Lemeshow S. 2013. Standardizing the power of the Hosmer-Lemeshow goodness of fit test in large data sets. Stat Med 32:67–80. doi: 10.1002/sim.5525. [DOI] [PubMed] [Google Scholar]
- 20.R Core Team. 2016. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.