Ebola virus (EBOV) is among the most devastating pathogens causing fatal hemorrhagic fever in humans. The epidemics from 2013 to 2016 resulted in more than 11,000 deaths, and another outbreak is currently ongoing. Since there is no FDA-approved drug so far to fight EBOV infection, there is an urgent need to focus on drug discovery. Considering the tight correlation between the high EBOV virulence and its ability to suppress the type I interferon (IFN-I) system, identifying molecules targeting viral protein VP24, one of the main virulence determinants blocking the IFN response, is a promising novel anti-EBOV therapy approach.
KEYWORDS: Ebola virus, EBOV, VP24, IFN signaling, drug development, luciferase assay, flavonoids, quercetin
ABSTRACT
Ebola virus (EBOV) is among the most devastating pathogens causing fatal hemorrhagic fever in humans. The epidemics from 2013 to 2016 resulted in more than 11,000 deaths, and another outbreak is currently ongoing. Since there is no FDA-approved drug so far to fight EBOV infection, there is an urgent need to focus on drug discovery. Considering the tight correlation between the high EBOV virulence and its ability to suppress the type I interferon (IFN-I) system, identifying molecules targeting viral protein VP24, one of the main virulence determinants blocking the IFN response, is a promising novel anti-EBOV therapy approach. Hence, in the effort to find novel EBOV inhibitors, a screening of a small set of flavonoids was performed; it showed that quercetin and wogonin can suppress the VP24 effect on IFN-I signaling inhibition. The mechanism of action of the most active compound, quercetin, showing a half-maximal inhibitory concentration (IC50) of 7.4 μM, was characterized to significantly restore the IFN-I signaling cascade, blocked by VP24, by directly interfering with the VP24 binding to karyopherin-α and thus restoring P-STAT1 nuclear transport and IFN gene transcription. Quercetin significantly blocked viral infection, specifically targeting EBOV VP24 anti-IFN-I function. Overall, quercetin is the first identified inhibitor of the EBOV VP24 anti-IFN function, representing a molecule interacting with a viral binding site that is very promising for further drug development aiming to block EBOV infection at the early steps.
INTRODUCTION
Within the Filoviridae family, the genus Ebolavirus consists of six species, Zaire ebolavirus, Sudan ebolavirus, Reston ebolavirus, Bundibugyo ebolavirus, Taï Forest ebolavirus, and Bombali ebolavirus. The prototype Ebola virus (EBOV) belongs to the species Zaire ebolavirus, which is considered the most virulent species, resulting in up to 90% mortality. No FDA-approved drugs are available for Ebola virus disease (EVD) treatment. However, great efforts have been made in the development of EBOV therapeutics (1). In December 2019, the European Medicines Agency approved the first EBOV vaccine, Ervebo (2). Moreover, the Pamoja Tulinde Maisha (PALM) trial, in Congo, demonstrated the efficacy of two monoclonal antibody therapies, MAb114 and REGN-EB3, in reducing the case fatality rate of EVD (3).
The pathogenesis of EBOV disease involves the dysregulation of both the innate and adaptive immune systems, allowing the virus to replicate, leading to the development of hematological manifestations, such as lymphopenia, thrombocytopenia, hemorrhage, and, finally, death. In particular, the high virulence is tightly correlated with the suppression of the interferon (IFN) response, which represents the first line of defense during a viral infection (4, 5).
Together with EBOV VP35, the multifunctional protein VP24 is one of the main determinants of virulence by virtue of its inhibition of the IFN signaling cascade (4, 5). VP24 exerts its action by binding the NPI-1 subfamily of karyopherin-α proteins (KPNα1, KPNα5, and KPNα6), importins involved in the nuclear transport of phosphorylated STAT1 (P-STAT1) protein, thus blocking the transcription of IFN-stimulated genes (ISGs) (6–8). Recently, the crystal structure of the VP24/KPNα5 complex has been solved (PDB code 4U2X) (9), showing that the KPNα5-VP24 binding region interface includes KPNα5 armadillo (ARM) repeats 7 to 10 and three VP24 clusters: cluster 1 (N130, T131, N135, R137, T138, and R140), cluster 2 (Q184, N185, and H186), and cluster 3 (L201, E203, P204, D205, and S207). Additional amino acids are required for binding: L115, L121, D124, W125, T128, and T129 (9). Mutations of VP24 residues involved in the suppression of the IFN cascade were shown to be responsible for the acquisition of high virulence in animal models (5, 10), suggesting VP24 as a validated target for drug development. Hence, identifying a therapy targeting the IFN-I-inhibitory functions of EBOV represents a promising approach to an early control of the infection (11–14). A number of in silico studies reported molecules potentially able to bind different EBOV targets, including VP24 (15, 16), while two recent studies reported that a macrocyclic peptide and a nucleic acid aptamer could antagonize the binding of VP24 and KPNα in biochemical assays (17, 18). However, no small molecule has ever been reported to inhibit VP24 anti-IFN-I functions in cells and subsequent EBOV replication. Flavonoids have been described for their antiviral activity among the other biological properties (19), and a recent in silico study reported that few of them are able to form stable complexes with VP24 (16). Hence, using a recently developed drug screening assay (20), a small number of flavonoids were screened for their effects against VP24; it was observed that some of them were able to significantly inhibit VP24 anti-IFN-I activity, leading to the restoration of the IFN signaling cascade. In particular, quercetin was shown to inhibit EBOV VP24 interaction with KPNα in vitro and to selectively inhibit EBOV infection.
RESULTS
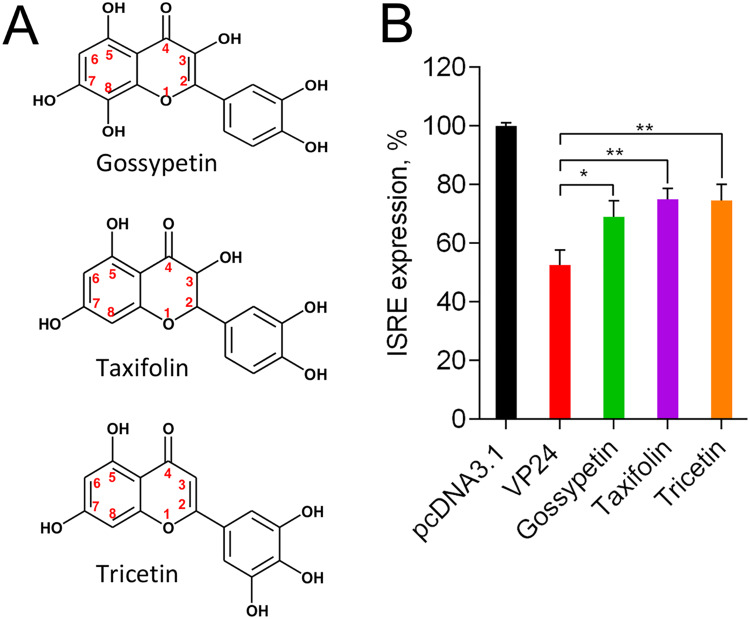
Gossypetin, taxifolin, and tricetin suppress VP24 IFN-inhibitory function.
In a recent in silico docking study, the flavonoids gossypetin, taxifolin, and tricetin (Fig. 1A) were predicted to be potentially able to impair VP24 ability to disrupt the JAK/STAT signaling pathway (16). Hence, we wanted to verify whether these flavonoid derivatives were effectively able to affect VP24 anti-IFN-I function in a dual-luciferase reporter cell-based assay (20). In this assay, HEK293T cells were cotransfected with pISRE-luc, pRL-TK, and a plasmid expressing EBOV VP24. Stimulation with IFN-α resulted in 100% interferon-sensitive response element (ISRE) expression in mock-transfected cells, while cells cotransfected with VP24 showed an inhibition of the IFN-I response. The treatment of VP24-cotransfected cells with 10 μM gossypetin, taxifolin, and tricetin resulted in the partial restoration of the ISRE expression, demonstrating that all three compounds were able to block the VP24 inhibitory effect on IFN signaling cascade in a cell-based assay, confirming in silico studies (Fig. 1B).
FIG 1.
Inhibition of VP24 by gossypetin, taxifolin, and tricetin. (A) Chemical structures of gossypetin, taxifolin, and tricetin. (B) HEK293T cells were transfected with pISRE-luc, Renilla luciferase-thymidine kinase DNA (RL-TK), and pcDNA3.1 or VP24 expression plasmid. Twenty-four hours after transfection, cells were stimulated with IFN-α, and gossypetin, taxifolin, and tricetin were added. Then luciferase activity was measured. Results are shown as percent ISRE expression in VP24-transfected cells over empty vector control. Firefly luciferase activity was normalized to the Renilla luciferase internal control. Data represent the means ± SDs from at least three independent experiments. Asterisks indicate significant difference (two-tailed unpaired Student’s t test, n = 3). *, P < 0.05; **, P < 0.01.
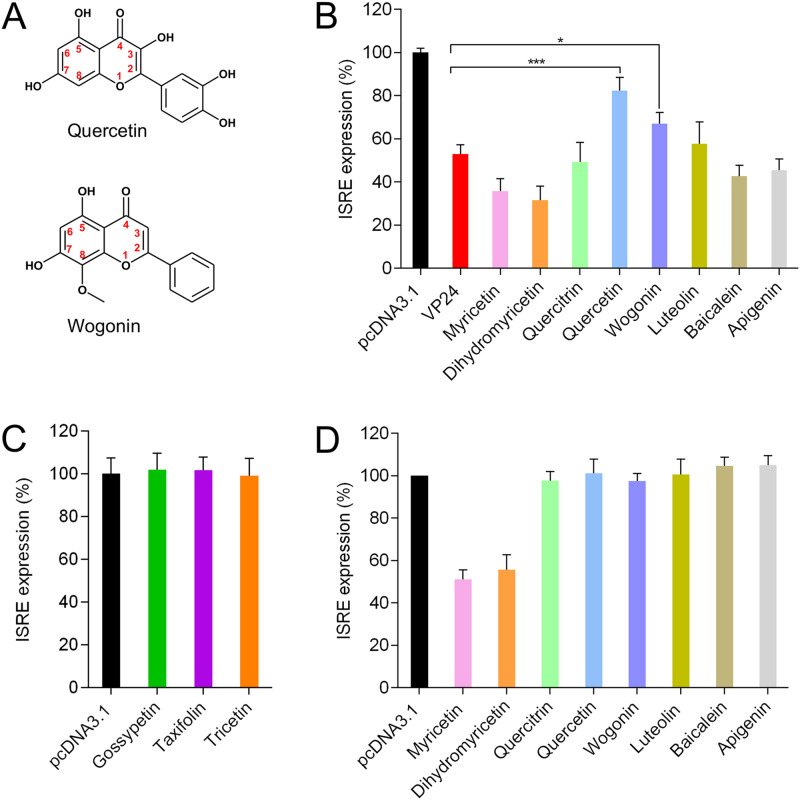
Quercetin and wogonin are active compounds against VP24.
With the aim to identify more active compounds, we performed the screening of a library of eight other flavonoids that share a structure similar to those of gossypetin, taxifolin, and tricetin. Utilization of a dual-luciferase reporter assay showed that two out of eight tested flavonoids, namely, quercetin and wogonin (Fig. 2A), significantly suppressed the anti-IFN function of VP24, restoring IFN stimulation (Fig. 2B). In particular, at a concentration of 10 μM, quercetin was able to restore ISRE expression from ∼50% up to ∼80%, in VP24-cotransfected cells, while wogonin restored the pathway up to ∼70% (Fig. 2B). Overall, among all tested flavonoids, quercetin was the most effective in restoring IFN stimulation (Fig. 2B).
FIG 2.
Effects of screened flavonoids on EBOV VP24 inhibition of IFN signaling. (A) Chemical structures of quercetin and wogonin. (B) HEK293T cells were transfected with pISRE-luc, RL-TK, and pcDNA3.1 or EBOV VP24 expression plasmid. After 24 h, cells were treated with IFN-α and compounds. After 8 h of stimulation, luciferase activity was measured. Results are shown as percent ISRE expression in VP24-transfected cells over pcDNA3.1 control. (C and D) After 24 h of cotransfection with pISRE-luc, RL-TK, and pcDNA3.1, HEK293T cells were stimulated with IFN-α and compounds. Results are shown as percent ISRE expression in compound-treated cells over untreated control. Firefly luciferase activity was normalized to the Renilla luciferase internal control. Data represent the means ± SDs from at least three independent experiments. Asterisks indicate significant difference (two-tailed unpaired Student’s t test, n = 3). *, P < 0.05; ***, P < 0.005.
It has been demonstrated that some flavonoids, including the tested compounds quercetin, luteolin, and apigenin, are able to induce interferon signaling (21, 22). Hence, in order to verify whether the tested flavonoids, at the indicated concentration, acted by targeting specifically VP24 or by enhancing the IFN response employing some VP24-independent mechanisms, we tested them for the ability to boost the IFN protein cascade (Fig. 2C and D). The results demonstrated that none of the screened compounds enhanced ISRE expression in HEK293T cells, confirming that active compounds could possibly specifically interact with VP24. Of note, myricetin and dihydromyricetin reduced ISRE activation (Fig. 2D). In fact, myricetin has been reported to inhibit STAT1 phosphorylation (23), and this can explain the inhibition of ISRE transcription. While no direct data are available for dihydromyricetin, and considering its structural diversity from myricetin, further studies are needed to understand its mechanism of action.
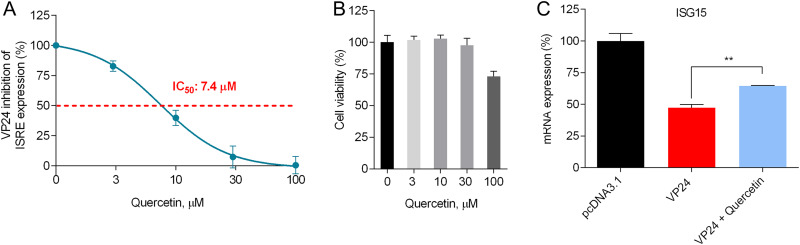
Quercetin dose dependently blocks VP24 inhibition of ISRE expression and affects ISG15 mRNA transcription.
Next, we wanted to better evaluate the effect of the most active compound, quercetin, on the VP24 blockade of IFN signaling by performing a dose-response curve in the presence of the same amount of VP24 plasmid. Of note, quercetin exhibited a dose-dependent reversion of the VP24 inhibition of ISRE IFN-dependent stimulation, with a half-maximal inhibitory concentration (IC50) of 7.4 μM (Fig. 3A). Quercetin cytotoxicity in HEK293T cells was also assessed; the results showed 100% cell viability at concentrations between 3 μM and 30 μM and a half-maximal cytotoxic concentration (CC50) of >100 μM (Fig. 3B). Hence, quercetin showed a selectivity index (SI; CC50/IC50) of >13. We then wanted to examine the effect of quercetin on the modulation of IFN signaling by VP24 using a different endpoint, i.e., by assessing its efficacy on ISG15 gene expression. The analysis confirmed that VP24 was effectively able to inhibit ISG15 mRNA transcription in IFN-stimulated cells compared to cells transfected with an empty vector (Fig. 3C). These results also confirmed that quercetin significantly restored IFN-induced ISG15 gene expression downregulated by VP24, confirming that the compound effectively blocked VP24 function (Fig. 3C).
FIG 3.
Quercetin dose dependently inhibits VP24 IFN-inhibitory function. (A) HEK293T cells were cotransfected with pISRE-luc, RL-TK, and VP24 plasmid. Twenty-four hours after transfection, cells were stimulated with IFN-α and quercetin was added to VP24-cotransfected cells. After 8 h, cells were lysed and luciferase was measured. Results are shown as percent VP24 inhibitory activity on ISRE expression using as a positive control of inhibition (100%) cells not treated with quercetin. Firefly luciferase activity was normalized to the Renilla luciferase internal control. (B) HEK293T cells were treated with quercetin and cytotoxicity was measured. Results are shown as percentage of cells treated with quercetin over untreated cells. (C) HEK293T cells transfected with pcDNA3.1 or VP24 plasmid. After 24 h, cells were stimulated with IFN-α and treated or not with quercetin (30 μM). After 24 h, total RNA was extracted, and RT-qPCR was performed. Results are shown as percent ISG15 mRNA expression in VP24-cotransfected cells (treated or not with quercetin) over empty vector control. Data represent the means ± SDs from three independent experiments. **, P < 0.01, two-tailed unpaired Student’s t test (n = 3).
Quercetin blocks VP24 inhibition of P-STAT1 nuclear transport.
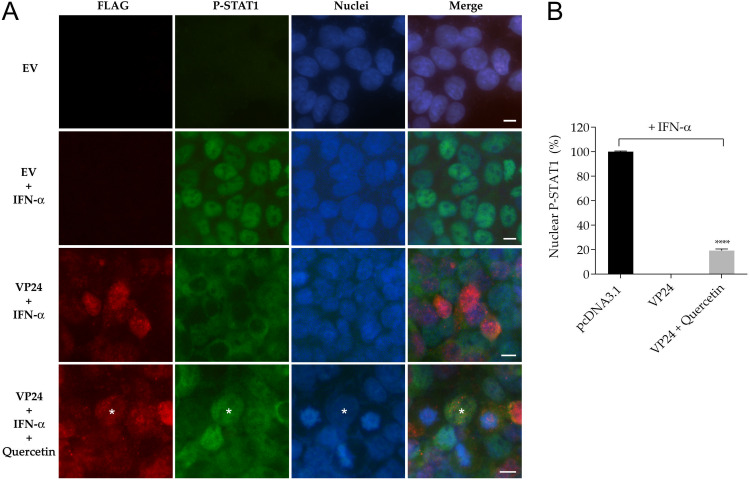
It is well known that VP24 inhibits the translocation of P-STAT1 to the nucleus (6–8). With the aim of verifying whether quercetin could increase the P-STAT1 nuclear transport, we performed an immunofluorescence assay by stimulating cells with a large amount of IFN-α (150 ng/ml) for 30 min, with or without quercetin. As expected, in control cells transfected with an empty vector, 100% of IFN-stimulated cells were positive for nuclear P-STAT1, indicating the activation of the IFN signaling cascade after IFN-α stimulation (Fig. 4A and B). In contrast, in IFN-stimulated cells expressing VP24 after transfection, P-STAT1 nuclear transport was inhibited, as no P-STAT1 was detectable in the nuclei of these cells (Fig. 4A). In this system, the presence of quercetin in IFN-stimulated VP24 transfected cells led to a significant increase (20%) of cells positive for nuclear P-STAT1 (Fig. 4A and B). Of note, this also showed that quercetin was able to rapidly block VP24 anti-IFN function.
FIG 4.
Effect of quercetin on VP24 inhibition of P-STAT1 nuclear translocation. (A) Rows 1 and 2, immunofluorescence of HEK293T cells cotransfected with empty vector (EV) not stimulated (row 1) or stimulated (row 2) with IFN-α for 30 min. Rows 3 and 4, immunofluorescence of HEK293T cells cotransfected with FLAG-tagged VP24 plasmid stimulated with IFN-α and not treated (row 3) or treated (row 4) with quercetin (30 μM). Asterisk indicates P-STAT1 in the nucleus of a VP24 cell treated with quercetin. All cells were stained for FLAG and P-STAT1. Scale bar, 10 μm. (B) Percentage of nuclear P-STAT1 within transfected cells expressing VP24. Data represent the means ± SDs from three independent experiments. ****, P < 0.0001, two-tailed unpaired Student’s t test (n = 3).
In silico and in vitro characterization of quercetin binding mode on VP24-KPNα complex.
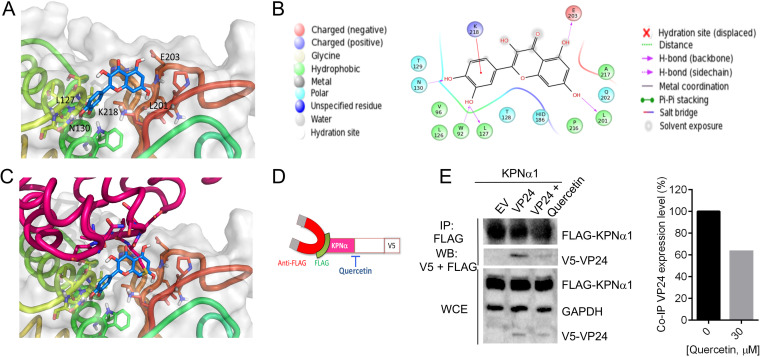
We performed in silico molecular docking experiments to evaluate the putative binding mode of quercetin on VP24. Among the available VP24 crystal structures, the one with PDB code 4M0Q (24) currently has the best resolution (1.92 Å); therefore, it was selected for docking experiments performed by means of Glide-XP (25). Then the best docking complexes were subjected to a postdocking procedure based on energy minimization, applying molecular mechanics generalized Born/surface area (MM-GBSA) method and continuum solvation models (26). The most stable binding mode was analyzed, and the results showed that quercetin occupies a central position in the reported contact area between VP24 and KPNα5 (Fig. 5A). The complex VP24-quercetin was stabilized by hydrogen bonds with the VP24 key residues N130, L201, and E203, while additional residues are involved in anchoring the compound, i.e., L127 and W92 by hydrogen bonds and K218 by cation-π interaction (Fig. 5B). Such in silico analysis allowed us to propose that the presence of quercetin in such a VP24 pocket would prevent KPNα5 ARMs 7 to 10 from moving close to VP24, ultimately inhibiting VP24 binding to KPNα5 (Fig. 5C).
FIG 5.
(A) Three-dimensional representation of the putative binding mode obtained by blind docking of quercetin into VP24. (B) Relative 2D representation of the complexes stabilizing interactions. (C) Alignment of quercetin-VP24 complex obtained by docking experiment with VP24-KPNα5 reported in the model with PDB code 4U2X (9). (D) Schematic representation of KPNα-VP24 co-IP experiment in the presence of quercetin. (E) HEK293T cells were cotransfected with a V5-tagged form of EBOV VP24 and FLAG-tagged KPNα1. Empty vector was used as a negative control. At 24 h posttransfection, cells were lysed and co-IP was performed using anti-FLAG magnetic beads (IP:FLAG). Immunoprecipitated proteins were detected by Western blotting with antibodies to V5 (WB:V5) and FLAG (WB:FLAG). The presence of transfected proteins in whole-cell extracts (WCE) was verified by detection with the corresponding antibodies. Band quantification was performed.
To demonstrate that quercetin was effectively able to interfere with the binding between VP24 and KPNα, coimmunoprecipitation (co-IP) assays were performed. Cells were cotransfected with V5-tagged VP24 and FLAG-tagged KPNα1, and 1 day posttransfection, after stimulation with IFN-α and treatment with quercetin, lysates were precipitated with anti-FLAG magnetic beads and immunocomplexes were analyzed by Western blotting (Fig. 5D and E). As expected, FLAG-KPNα1 was coprecipitated with V5-VP24. When VP24-cotransfected cells were treated with quercetin, there was a significant reduction in VP24 coimmunoprecipitated with KPNα1 (Fig. 5E). This result indicates that quercetin is indeed able to interfere with the binding between VP24 and KPNα1, confirming the proposed in silico model.
Antiviral activity of quercetin against EBOV replication.
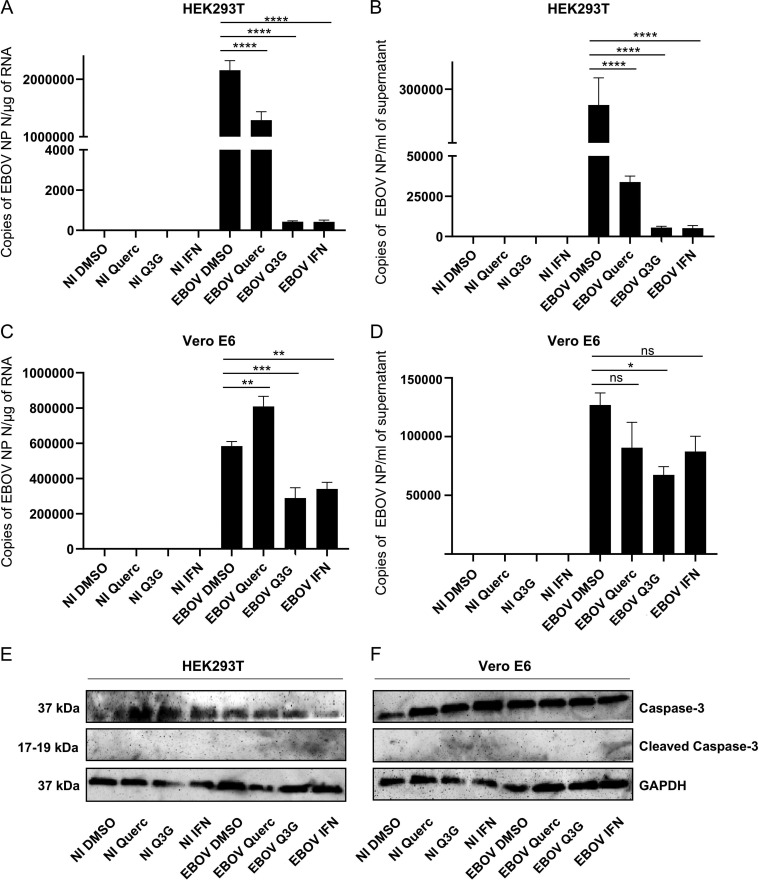
It has been recently demonstrated that quercetin 3-β-O-d-glucoside (Q3G), a glucosylated form of quercetin, has antiviral activity against EBOV both in vitro and in vivo (27). In order to test if quercetin was also able to inhibit EBOV replication, wild-type EBOV Makona-infected cells were treated with quercetin, Q3G, and IFN-α as a control for inhibition. Quercetin significantly inhibited viral replication in HEK293T cells (Fig. 6A), while it was not able to inhibit EBOV in Vero E6 cells (Fig. 6C). EBOV yield in supernatants collected from HEK293T cells treated with the compounds was also significantly reduced (Fig. 6B), while the antiviral effect of quercetin in supernatant from Vero cells was not significant (Fig. 6D). Indeed, as expected, the lack of effect of quercetin on EBOV replication in Vero cells confirmed the proposed mode of action, since Vero E6 cells cannot produce type I IFNs; thus, the JAK-STAT cascade, in total absence of exogenous IFN, is not activated and, hence, VP24 cannot exert its interferon-inhibitory function (28). In contrast to quercetin, Q3G was able to block EBOV replication in both cell lines (Fig. 6). It has been previously demonstrated that Q3G targets the viral entry process (27). The fact that Q3G is more active in HEK293T cells than in Vero cells might suggest a dual mechanism of action: (i) impairing the IFN antagonism of VP24 and (ii) a mechanism independent of that, as proposed by Qiu et al. (27), blocking the level of virus entry.
FIG 6.
Quercetin inhibits EBOV replication in HEK293T cells. HEK293T (A) and Vero E6 (C) cells were treated with DMSO, quercetin (30 μM), Q3G (10 μM), or IFN-α (1,000 IU/ml), infected or not (NI) with EBOV Makona (MOI = 0.1), and cultured for 72 h. Then cells were harvested and analyzed by RT-qPCR for EBOV NP expression in cell lysates. Supernatants from HEK293T (B) and Vero E6 (D) cell cultures were harvested and analyzed by RT-qPCR for EBOV NP expression in supernatants. Results are presented as means ± SEs. Statistical significances between DMSO-treated EBOV-infected cells and quercetin-, Q3G-, and IFN-α-treated EBOV-infected cells were analyzed using one-way ANOVA followed by Tukey’s multiple-comparison test (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant). Lysates from HEK293T (E) and Vero E6 (F) cells were analyzed by Western blotting for caspase-3 and cleaved caspase-3 expression.
To exclude that the decrease in viral titers was attributable to cell toxicity, we measured quercetin and Q3G cytotoxicity, observing that in both HEK293T and VeroE6 cells their CC50 values were >100 μM (data not shown). Furthermore, we performed Western blotting to examine the level of caspase-3 cleavage as a marker of apoptosis (29). We used antibodies recognizing the caspase-3 active cleaved form and its inactive precursor, and no relevant cleaved caspase-3 was detectable in any of the tested samples, despite the expression of large amounts of its inactive precursor protein (Fig. 6E and F). Given the absence of apoptosis, we confirmed that the reduction in viral replication was due only to compound treatment.
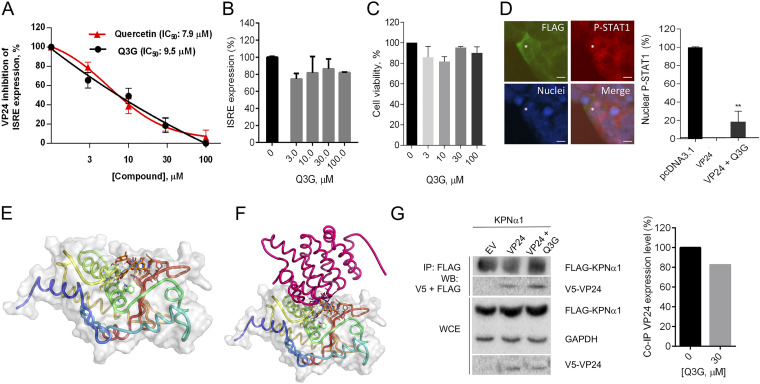
Finally, to confirm this hypothesis, we investigated if Q3G, like the nonglucosylated form, was able to restore the ISRE expression in HEK293T cells. VP24-transfected cells were stimulated with IFN-α and treated with increasing concentrations of Q3G. We observed that Q3G dose dependently inhibited VP24 IFN-inhibitory function, with an IC50 of 9.5 μM (Fig. 7A), showing no alternative VP24-independent mechanism on ISRE stimulation (Fig. 7B) and no cytotoxicity even at high concentrations (Fig. 7C). Similarly to quercetin, Q3G was able to restore the P-STAT1 nuclear transport in VP24-cotransfected cells, resulting in a significant proportion (17%) of nuclear P-STAT1-positive cells (Fig. 7D). Given the similar IC50s for quercetin and Q3G, we hypothesized that the presence of the glucoside did not alter the binding of the molecule with VP24. Indeed, performing the alignment of quercetin-VP24 and Q3G-VP24 complexes, obtained by docking analysis with VP24, we confirmed that the glucoside moiety did not alter the position of the Q3G, compared to quercetin, on VP24 (Fig. 7E and F). In addition, co-IP of VP24 and KPNα1 in cells treated with Q3G showed that the compound affected the binding between VP24 and the importin, resulting in a 20% reduction of coimmunoprecipitated VP24, compared to the untreated control (Fig. 7G). These results suggest that Q3G is slightly less efficient than the nonglucosylated form.
FIG 7.
(A) VP24-transfected HEK293T cells were stimulated with IFN-α and treated with quercetin and Q3G. After 8 h, luciferase was read. Results are shown as percent VP24 inhibitory activity on ISRE expression using as a positive control of inhibition (100%) untreated cells. Data represent the means ± SDs from three independent experiments. (B) HEK293T cells transfected with pISRE-luc, RL-TK, and pcDNA3.1 were treated with IFN-α and Q3G. Results are shown as percent ISRE expression in Q3G-treated cells over the untreated control. (C) HEK293T cells were treated with Q3G and cytotoxicity was measured. Results are shown as percent cell viability in Q3G-treated cells over the untreated cells (100%). (D) Immunofluorescence of HEK293T cells cotransfected with VP24 and treated with IFN-α and Q3G. Asterisks indicate P-STAT1 in the nucleus of a VP24-contransfected cell treated with Q3G. Signal of P-STAT1 was detected and percent cells positive for nuclear P-STAT1 was calculated. **, P < 0.01, two-tailed unpaired Student’s t test (n = 3). Scale bar, 10 μm. (E) Three-dimensional representation of superimposed putative binding modes obtained by blind docking experiments of Q3G (in orange) and quercetin (in blue) into VP24. (F) Alignment of quercetin-VP24 and Q3G-VP24 complexes obtained by docking experiments with VP24 and KPNα5 reported for the model with PDB code 4U2X (9). (G) Co-IP of V5-VP24 with KPNα1 in the presence or absence of Q3G. Band quantification was performed.
DISCUSSION
Among the diversified antiviral strategies developed to counteract the different steps of the viral life cycle (30–34), interfering with the initial phases of infection, such as the viral evasion of the IFN system, is an attractive therapeutic approach against several types of viruses (35–38). Since EBOV inhibition of the IFN response massively contributes to viral pathogenesis (4, 5), restoring the IFN system could represent a promising strategy for EBOV control (13). Two viral proteins have been demonstrated to suppress interferon responses, VP35 and VP24 (13, 39). Different studies have shown that it is possible to block VP35 IFN-inhibitory function with natural compounds (11, 12) and also antibodies (14), thus restoring the IFN production cascade. Recently, Jasenosky et al. demonstrated that the known FDA-approved small molecule nitazoxanide was able to significantly suppress EBOV replication in human cells through enhancing RIG-I pathway sensing, thus counteracting EBOV VP35’s ability to prevent triggering of the host antiviral response (40).
Although several mutational studies confirmed VP24 as a validated therapeutic target (15, 16, 41–44), until now, only two molecules have been reported to block the binding between VP24 and KPNα that is essential for the anti-IFN function of VP24 (17, 18). First, using the RaPID system, which allows the selection of high-affinity binders from an mRNA library of nonstandard peptides, Song et al. were able to identify a macrocyclic peptide binder for VP24, namely, eVpeD2. This represents the first molecule that has been shown to inhibit the VP24-KPNα5 protein-protein interaction, with an IC50 value of 9 μM (17). Second, natural and artificial nucleic acid aptamers with high affinity for VP24 were generated using the capillary electrophoresis-systematic evolution of ligands by exponential enrichment (CE-SELEX). Sharing a common binding site with KPNα1, they compete for the binding with VP24 (Kd [dissociation constant] ≈ 0.1 nM) (18). Even though they have been shown to be active at micromolar and subnanomolar ranges in biochemical assays, to the best of our knowledge, they have not been reported to be tested in cell-based assays.
The identification of compounds able to block VP24 function in human cells was previously limited due to the difficulty of developing a suitable cellular screening system. We first developed a cellular model able to evaluate the inhibition of IFN signaling by VP24 (20) and used this system to test anti-VP24 agents. Unlike biochemical assays, cell-based assays can be used to monitor specific viral proteins and pathways, such as VP24 and ISRE expression, providing the means to test for potent viral inhibitors intracellularly and thus giving extra information about their effect in the more complex cellular context, improving the early phase of the drug discovery process.
Since previous in silico studies suggested the potential anti-EBOV activity of flavonoid derivatives, we performed a screening of different flavonoids in a cell-based assay. It has been shown that flavonoids possess a wide range of biological properties, such as antioxidant, hepatoprotective, antibacterial, anti-inflammatory (19, 45), and antiviral activities against different viruses, such as Japanese encephalitis virus (46), dengue virus type 2 (DENV-2) (47), influenza virus (48, 49), and human immunodeficiency virus (50, 51). Different modes of action have been proposed, such as the inhibition of viral polymerase and capsid proteins by flavonoids (52).
In the present study, we aimed to characterize the effect of flavonoids on IFN inhibition mediated by VP24 and observed that quercetin and wogonin were able to inhibit VP24 significantly, restoring ISRE expression, with quercetin being the most active compound. Quercetin is a molecule largely present in the human diet, being part of many fruits and vegetables. It has been reported that an average of up to 1 g of quercetin is taken in per day, representing 60% to 75% of overall polyphenol ingestion (53). Among its wide range of biological activities, it has been demonstrated to be active on different viruses, including hepatitis C virus (HCV) (54), Mayaro virus (55), influenza A virus (IAV) (56), Chikungunya virus (CHIKV) (57), and Epstein-Barr virus (EBV) (58). In particular, quercetin has been shown to inhibit HCV through binding and inactivating the viral NS3 protease (59). Zandi et al. proposed that quercetin prevents DENV-2 replication by inhibiting viral RNA polymerase (47), and a recent paper by Granato et al. suggested that quercetin may have potential to be used to counteract EBV-driven lymphomagenesis, since it is able to counteract EBV-driven immortalization of B cells and lymphoblastoid cell line (LCL) outgrowth, interrupting the cross talk between interleukin 6 (IL-6) and STAT3 and promoting autophagy (60). Moreover, a quercetin derivative, Q3G, has been demonstrated to inhibit the early steps of EBOV entry (27). Our results illustrate a novel antiviral mechanism adopted by quercetin and its derivative Q3G. In fact, we observed that quercetin inhibits EBOV VP24, leading to a partial restoration of the ISRE expression, ISG15 mRNA transcription, and P-STAT1 nuclear transport in the presence of the viral protein. Docking studies suggest the putative quercetin binding mode at the interface between VP24 and KPNα5. The binding of a small molecule in this surface could prevent KPNα binding. This mechanism was confirmed by the ability of Quercetin to affect the binding of coimmunoprecipitated KPNα and VP24.
We also tested the effect of quercetin on EBOV replication and demonstrated that the compound was able to significantly suppress viral replication. This effect was similar to the previously reported effect of flavonoid derivative Q3G (27). In contrast to that of Q3G, however, the effect of quercetin was not due to an inhibition of the entry process, since quercetin inhibited EBOV only in IFN-competent HEK293T cells, being ineffective in IFN-incompetent Vero cells. This result supports the hypothesis that quercetin affects specifically EBOV evasion of the IFN pathway. In contrast, Q3G inhibited EBOV replication in both cell lines, and its more potent effect on HEK293T cells than on Vero E6 cells possibly suggests a bimodal mechanism. In fact, when Q3G was tested in our cell-based assay, we observed that the glucosylated molecule was able to inhibit EBOV VP24 by reverting both the IFN-α-induced stimulation and the P-STAT1 nuclear translocation blocked by the viral protein. Molecular docking experiments confirmed that Q3G localized in the same VP24 surface as quercetin, suggesting that Q3G also affects the binding between VP24 and KPNα but with lower efficiency. In addition, present data suggest that the glucoside moiety is probably required for the inhibition of viral entry, since this mechanism was not observed for the aglycone form.
In conclusion, we identified a small molecule, already used as a nutritional supplement, for which toxicity and pharmacokinetics are well known (53), whose metabolism is well studied in animal models and that is a promising anti-EBOV agent. In fact, it has been previously demonstrated that only 2 h after oral administration of quercetin (50 and 100 mg/kg of body weight), the total quercetin plasma concentrations in mice, rats, and gerbils reached concentrations between 6 and 12 μM (61). The molecule could rapidly exert its anti-EBOV activity either in plasma (mainly for the indirect effect of its metabolite, Q3G, as previously reported by Qiu et al.) (27) or in tissues where Q3G is again converted into its aglycone form.
Quercetin is the first compound that specifically inhibits EBOV VP24 IFN-inhibitory function, restoring the IFN signaling cascade and leading to the block of viral infection. Given its good tolerability, these findings open original perspectives in the field of EBOV therapeutics development.
MATERIALS AND METHODS
Cells and reagents.
HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (Gibco) and 1% penicillin-streptomycin (Sigma). pISRE-luc and pcDNA3.1-FLAG-VP24 were kindly gifted by Ian Goodfellow (University of Cambridge, UK) and Marco Sgarbanti (Italian National Institute of Health, Italy), respectively. pcDNA3-V5-VP24 and FLAG-tagged KPNα1 were provided by St. Patrick Reid (University of Nebraska Medical Center, NE). pRL-TK plasmid, T-Pro P-Fect transfection reagent, and human recombinant IFN-α were purchased from Promega, T-Pro Biotechnology, and PeproTech, respectively. Gossypetin, taxifolin, and tricetin were from Extrasynthese, while Q3G, myricetin, dihydromyricetin, luteolin, apigenin, quercitrin, quercetin, wogonin, baicalein, anti-FLAG M2 antibodies, and anti-FLAG magnetic beads were from Sigma-Aldrich. Antibodies against P-STAT1, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), anti-caspase-3, anti-cleaved caspase-3, and horseradish peroxidase (HRP)-linked IgG were purchased from Cell Signaling. Alexa Fluor 488 goat anti-mouse IgG, Alexa Fluor 594 goat anti-rabbit IgG, and Pierce ECL Western blotting substrate were from Thermo Fisher Scientific.
Virus.
Zaïre EBOV (Makona strain) (62) was prepared by infecting Vero E6 cells and titrated on Vero E6 cells as well by plaque assay, using crystal violet in the INSERM Jean Mérieux biosafety level 4 (BSL4) laboratory in Lyon, France. 293T and Vero E6 cells were cultured in DMEM GlutaMAX (Life Technologies) supplemented with 10% heat-inactivated fetal bovine serum (HI-FBS; Eurobio), 1% HEPES, 1% nonessential amino acids, 1% sodium pyruvate, and 2% penicillin-streptomycin mix (all from Life Technologies). For treatments and infections, cells were plated in 12-well plates at 2.5 × 105 per well and then were treated with dimethyl sulfoxide (DMSO; Sigma-Aldrich), quercetin (30 μM; Sigma-Aldrich), Q3G (10 μM; Sigma-Aldrich), or IFN-α (1,000 IU/ml; PBL Assay Science) for 1 h at 37°C before being infected or not with EBOV Makona at a multiplicity of infection (MOI) of 0.1 PFU/cell for 1 h at 37°C. Then virus-containing media were removed, and cells were washed with 1× phosphate-buffered saline (PBS). Furthermore, fresh DMEM containing drugs associated with each condition described in the legend of Fig. 6 (HEK293T or Vero E6 cells, infected or not with Makona) was added to cells that were incubated for 72 h at 37°C. Finally, infected cell lysates and supernatants were collected for further analysis, according to validated BSL4 procedures.
VP24 IFN inhibition assay.
EBOV VP24 inhibition of the ISRE-dependent promoter reporter plasmid (pISRE-luc) after IFN-α stimulation has been previously described (20). Briefly, HEK293T cells were cotransfected, using T-pro P-Fect transfection reagent with pISRE-luc (60 ng/well), a constitutively expressed Renilla luciferase reporter plasmid (RL-TK) (10 ng/well), and a FLAG-tagged wild-type VP24 plasmid (30 ng/well) from the Zaire ebolavirus isolate H.sapiens-wt/GIN/2014/Gueckedou-C05, Sierra Leone/guinea strain. Renilla luciferase was used to normalize transfection efficiencies. Twenty-four hours posttransfection, cells were mock or IFN-α stimulated (1 ng/ml) and treated or not with compounds (3 μM, 10 μM, 30 μM, and 100 μM) for 8 h. Then luciferase reporter activities were analyzed using cell lysates and luciferase counts were determined using a Victor3 luminometer.
IC50 calculation.
To determine the IC50 of quercetin, we used the log agonist concentration versus response, variable slope algorithm in GraphPad Prism software where Y = bottom + (top − bottom)/(1 + 10logIC50). IC50 values were calculated based on three independent experiments performed in triplicate.
Cell viability assays.
HEK293T and Vero E6 cells were seeded in 96-well plates. After 24 h, cells were treated with quercetin and Q3G and incubated for 8 h or 72 h at 37°C in 5% CO2. Then PrestoBlue cell viability reagent (Thermo Fisher Scientific) was added to each well. The plate was incubated for 1 h at 37°C in 5% CO2, and fluorescence was read in the Victor3 luminometer.
After 72 h of infection, HEK293T and Vero E6 cells treated and not treated were harvested, washed three times with PBS, and lysed in radioimmunoprecipitation assay (RIPA) lysis buffer (Thermo Scientific) supplemented with 4× Laemmli buffer (Life Technologies) and 10× reducing agent buffer (Invitrogen). Then Western blot analysis was performed using anti-caspase-3 (Cell Signaling) and anti-cleaved caspase-3 (Cell Signaling) antibodies. GAPDH antibody (EMD Millipore) was used as an internal control.
Immunofluorescence.
For immunostaining, HEK293T cells, grown in 6-well plates on glass slides, were cotransfected with Lipofectamine 2000 transfection reagent (Invitrogen) with pcDNA3.1 (2.5 μg/well) or EBOV VP24 plasmid (2.5 μg/well). After 24 h of transfection, cells were treated with IFN-α (150 μg/well) for 30 min and then fixed with 4% paraformaldehyde in PBS for 15 min at room temperature. Permeabilization was performed with ice-cold 100% methanol for 10 min, cells were washed with PBS, and permeabilization was blocked in blocking buffer (PBS containing 0.2% Triton X-100, 10% normal goat serum, and 3% bovine serum albumin) for 1 h at room temperature. Cells were first incubated with mouse anti-FLAG (Sigma-Aldrich; 1:500) and rabbit anti-P-STAT1 (Cell Signaling; 1:400) for 1 h at room temperature and then with goat anti-mouse and anti-rabbit secondary antisera conjugated with Alexa Fluor 488 and Alexa Fluor 594 (Invitrogen; 1:500), for 1 h at room temperature. After incubation with the nuclear dye Hoechst 33258 (Sigma-Aldrich), slides with stained cells were mounted on histology standard slides with glycerol mounting medium with 1,4-diazabicyclo[2.2.2]octane (DABCO) as an antifading reagent and observed with an Olympus BX61 microscope equipped with epifluorescence illumination, and digital images were captured with a Leica DF 450C camera. The cultures were examined at a magnification of ×40. Cells were counted using the cell counter plugin image analysis program ImageJ.
RNA extraction and quantitative real-time PCR.
Total RNA was extracted from transfected cells with TRIzol reagent (Invitrogen). RNA was then reverse transcribed and amplified using a Luna universal one-step quantitative real-time PCR (RT-qPCR) kit (New England BioLabs). RT-qPCR experiments were performed in triplicate. mRNA expression levels were normalized to the level of GAPDH. Results are expressed as percentage of mRNA transcript levels of treated cells versus untreated cells. Regarding BSL4 procedures, total RNAs from cells and supernatants were inactivated with RLT and AVL reagents (Qiagen), respectively, before being extracted using the Nucleospin RNA kit (Macherey-Nagel). Reverse transcription was realized using an iScript cDNA kit (Bio-Rad), and qPCR was realized using a Platinum SYBR green qPCR SuperMix-UDG kit with ROX (Invitrogen). Data were treated using StepOne software, and calculations were done using the threshold cycle (2ΔΔCT) method and normalized with the GAPDH housekeeping gene. Primers used for qPCR were EBOV Makona NP forward (GCT CCT TTC GCC CGA CTT TTG AA) and EBOV Makona NP reverse (CTG TGG CGA CTC CGA GTG CAA).
Coimmunoprecipitation experiment.
HEK293T cells were transfected with empty or expression plasmids for V5-tagged VP24 and FLAG-tagged KPNα1 and lysed in PBS containing 1.5% Triton X-100, 1 mM Na3VO4, 1 mM dithiothreitol (DTT), and 1× cOmplete protease inhibitor cocktail (Roche). Lysates were incubated with anti-FLAG M2 magnetic beads overnight at 4°C. Precipitated proteins were eluted by boiling with SDS sample loading buffer. Whole-cell lysates and immunoprecipitated samples were analyzed by Western blotting. Membranes were probed with anti-FLAG M2 (Sigma-Aldrich; 1 μg/ml), anti-V5 (Thermo Fisher Scientific; 1:200), and anti-GAPDH (Cell Signaling; 1:1,000). Images were captured with the Chemidoc MP imaging system (Bio-Rad). Band quantification was performed using Image Lab software. Coimmunoprecipitated V5-VP24 was normalized to the level of immunoprecipitated FLAG-KPNα1. V5-VP24 and KPNα bands in WCL were normalized to the level of GAPDH.
Molecular docking.
(i) Ligand preparation. The two-dimensional (2D) coordinate structure-data files (SDF) of quercetin and Q3G ligands were downloaded from the PubChem compound repository. Then they were prepared with Maestro GUI and subject to conformational analysis by means of MacroModel program version 9.2 (63). Merck molecular force field (MMFF) (64) was applied as a force field, and it was considered the water implicit generalized born/surface area (GB/SA) solvation model (26). Therefore, the compounds’ geometry was energy minimized using the Polak-Ribier conjugate gradient (PRCG) method, 5,000 iterations, and a convergence criterion of 0.05 kcal/(mol Å). The compounds’ global minimum conformations were considered for the docking studies.
(ii) Docking protein preparation. The coordinates for VP24 were taken from the RCSB Protein Data Bank (65), considering the model with PDB code 4M0Q (24). The protein was prepared using the Protein Preparation Wizard in Maestro (Schrödinger, New York, NY, 2018). Original water molecules were removed. The bond orders, hydrogen atoms, and formal charges were added in the structure. After preparation, the structure was refined in order to optimize the hydrogen bond network using the OPLS_2005 force field (66).
(iii) Docking experiments. Docking inner and outer grids were defined around the refined structure by calculating the centroid of clustered residues interacting with KPNα5 (9). The coordinates for the x, y, and z axes are −2.06, −21.53, and −12.16. The inner box side was increased to 15 Å, while the outer was left at 20 Å. The box resulted in a cube of 35 Å for side in order to include the whole protein. The extra precision (XP) docking algorithm was applied for scoring theoretical poses (25). The other settings were left as default.
(iv) Postdocking. In order to better take into account the induced fit phenomenon occurring at the ligand binding domain, the most energy-favored generated complexes were fully optimized with 10,000 steps of the PRCG minimization method considering OPLS_2005 force field and GB/SA implicit water. The optimization process was performed up to the derivative convergence criterion equal to 0.1 kJ/(mol·Å).
Statistical analysis and graphic visualization.
Statistical analysis and graphic visualization were performed using GraphPad Prism software 6.01 (GraphPad Software, Inc.), PyMOL (the PyMOL molecular graphics system version 1.7; Schrödinger), and Maestro ligand interaction visualization (Schrödinger).
ACKNOWLEDGMENTS
The present study was supported by RAS LR 07/2007 grant no. CRP-78711/F72I15000900002.
E.F. and E.T. conceived and designed the experiments and wrote the paper; E.F., M.I., and S.D. performed the experiments; and E.F., M.I., A.C., M.Q., E.M., and B.H. analyzed data. All authors revised the manuscript and read and approved the final version.
We thank Claire Dumont for her technical support.
We declare no conflict of interest.
REFERENCES
- 1.Fanunza E, Frau A, Corona A, Tramontano E. 2018. Antiviral agents against Ebola virus infection: repositioning old drugs and finding novel small molecules. Annu Rep Med Chem 51:135–173. doi: 10.1016/bs.armc.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henao-Restrepo AM, Longini IM, Egger M, Dean NE, Edmunds WJ, Camacho A, Carroll MW, Doumbia M, Draguez B, Duraffour S, Enwere G, Grais R, Gunther S, Hossmann S, Kondé MK, Kone S, Kuisma E, Levine MM, Mandal S, Norheim G, Riveros X, Soumah A, Trelle S, Vicari AS, Watson CH, Kéïta S, Kieny MP, Røttingen JA. 2015. Efficacy and effectiveness of an rVSV-vectored vaccine expressing Ebola surface glycoprotein: interim results from the Guinea ring vaccination cluster-randomised trial. Lancet 386:857–866. doi: 10.1016/S0140-6736(15)61117-5. [DOI] [PubMed] [Google Scholar]
- 3.Mulangu S, Dodd LE, Davey RT, Mbaya OT, Proschan M, Mukadi D, Manzo ML, Nzolo D, Oloma AT, Ibanda A, Ali R, Coulibaly S, Levine AC, Grais R, Diaz J, Clifford Lane H, Muyembe-Tamfum JJ, Sivahera B, Camara M, Kojan R, Walker R, Dighero KB, Cao H, Mukumbayi P, Mbala-Kingebeni P, Ahuka S, Albert S, Bonnett T, Crozier I, Duvenhage M, Proffitt C, Teitelbaum M, Moench T, Aboulhab J, Barrett K, Cahill K, Cone K, Eckes R, Hensley L, Herpin B, Higgs E, Ledgerwood J, Pierson J, Smolskis M, Sow Y, Tierney J, Sivapalasingam S, Holman W, Gettinger N, Vallée D, Nordwall J. 2019. A randomized, controlled trial of Ebola virus disease therapeutics. N Engl J Med 382:2293–2303. doi: 10.1056/NEJMoa1910993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prins KC, Delpeut S, Leung DW, Reynard O, Volchkova VA, Reid SP, Ramanan P, Cárdenas WB, Amarasinghe GK, Volchkov VE, Basler CF. 2010. Mutations abrogating VP35 interaction with double-stranded RNA render Ebola virus avirulent in guinea pigs. J Virol 84:3004–3015. doi: 10.1128/JVI.02459-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ebihara H, Takada A, Kobasa D, Jones S, Neumann G, Theriault S, Bray M, Feldmann H, Kawaoka Y. 2006. Molecular determinants of Ebola virus virulence in mice. PLoS Pathog 2:e73. doi: 10.1371/journal.ppat.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reid SP, Leung LW, Hartman AL, Martinez O, Shaw ML, Carbonnelle C, Volchkov VE, Nichol ST, Basler CD. 2006. Ebola virus VP24 binds karyopherin α1 and blocks STAT1 nuclear accumulation. J Virol 80:5156–5167. doi: 10.1128/JVI.02349-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mateo M, Reid SP, Leung LW, Basler CF, Volchkov VE. 2010. Ebolavirus VP24 binding to karyopherins is required for inhibition of interferon signaling. J Virol 84:1169–1175. doi: 10.1128/JVI.01372-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reid SP, Valmas C, Martinez O, Mauricio Sanchez F, Basler CF. 2007. Ebola virus VP24 proteins inhibit the interaction of NPI-1 subfamily karyopherin α proteins with activated STAT1. J Virol 81:13469–13477. doi: 10.1128/JVI.01097-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu W, Edwards MR, Borek DM, Feagins AR, Mittal A, Alinger JB, Berry KN, Yen B, Hamilton J, Brett TJ, Pappu RV, Leung DW, Basler CF, Amarasinghe GK. 2014. Ebola virus VP24 targets a unique NLS binding site on karyopherin alpha 5 to selectively compete with nuclear import of phosphorylated STAT1. Cell Host Microbe 16:187–200. doi: 10.1016/j.chom.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Volchkov VE, Chepurnov AA, Volchkova VA, Ternovoj VA, Klenk H. 2000. Molecular characterization of guinea pig-adapted variants of Ebola virus. Virology 277:147–155. doi: 10.1006/viro.2000.0572. [DOI] [PubMed] [Google Scholar]
- 11.Di Petrillo A, Fais A, Pintus F, Santos-Buelga C, González-Paramás AM, Piras V, Orrù G, Mameli A, Tramontano E, Frau A. 2017. Broad-range potential of Asphodelus microcarpus leaves extract for drug development. BMC Microbiol 17:159. doi: 10.1186/s12866-017-1068-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daino GL, Frau A, Sanna C, Rigano D, Distinto S, Madau V, Esposito F, Fanunza E, Bianco G, Taglialatela-Scafati O, Zinzula L, Maccioni E, Corona A, Tramontano E. 2018. Identification of myricetin as an Ebola virus VP35-double-stranded RNA interaction inhibitor through a novel fluorescence-based assay. Biochemistry 57:6367–6378. doi: 10.1021/acs.biochem.8b00892. [DOI] [PubMed] [Google Scholar]
- 13.Fanunza E, Frau A, Corona A, Tramontano E. 2019. Insights into Ebola virus VP35 and VP24 interferon inhibitory functions and their initial exploitation as drug targets. Infect Disord Drug Targets 19:362–374. doi: 10.2174/1871526519666181123145540. [DOI] [PubMed] [Google Scholar]
- 14.Flego M, Frau A, Accardi L, Mallano A, Ascione A, Gellini M, Fanunza E, Vella S, Di Bonito P, Tramontano E. 2019. Intracellular human antibody fragments recognizing the VP35 protein of Zaire Ebola filovirus inhibit the protein activity. BMC Biotechnol 19:64. doi: 10.1186/s12896-019-0554-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plesko S, Volk H, Luksic M, Podlipnik C. 2015. In silico study of plant polyphenols’ interactions with VP24-Ebola virus membrane-associated protein. Acta Chim Slov 62:555–564. [PubMed] [Google Scholar]
- 16.Raj U, Varadwaj PK. 2016. Flavonoids as multi-target inhibitors for proteins associated with Ebola virus: in silico discovery using virtual screening and molecular docking studies. Interdiscip Sci Comput Life Sci 8:132–141. doi: 10.1007/s12539-015-0109-8. [DOI] [PubMed] [Google Scholar]
- 17.Song X, Lu L, Passioura T, Suga H. 2017. Macrocyclic peptide inhibitors for the protein–protein interaction of Zaire Ebola virus protein 24 and karyopherin alpha 5. Org Biomol Chem 15:5155–5160. doi: 10.1039/c7ob00012j. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka K, Kasahara Y, Miyamoto Y, Takumi O, Kasai T, Onodera K, Kuwahara M, Oka M, Yoneda Y, Obika S. 2018. Development of oligonucleotide-based antagonists of Ebola virus protein 24 inhibiting its interaction with karyopherin alpha 1. Org Biomol Chem 16:4456–4463. doi: 10.1039/c8ob00706c. [DOI] [PubMed] [Google Scholar]
- 19.Kumar S, Pandey AK. 2013. Chemistry and biological activities of flavonoids: an overview. Sci World J 2013:1–16. doi: 10.1155/2013/162750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fanunza E, Frau A, Sgarbanti M, Orsatti R, Corona A, Tramontano E. 2018. Development and validation of a novel dual luciferase reporter gene assay to quantify Ebola virus VP24 inhibition of IFN signaling. Viruses 10:98. doi: 10.3390/v10020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tai Z, Lin Y, He Y, Huang J, Guo J, Yang L, Zhang G, Wang F. 2014. Luteolin sensitizes the antiproliferative effect of interferon α/β by activation of Janus kinase/signal transducer and activator of transcription pathway signaling through protein kinase A-mediated inhibition of protein tyrosine phosphatase SHP-2 in cancer cells. Cell Signal 26:619–628. doi: 10.1016/j.cellsig.2013.11.039. [DOI] [PubMed] [Google Scholar]
- 22.Igbe I, Shen X-F, Jiao W, Qiang Z, Deng T, Li S, Liu W-L, Liu H-W, Zhang G-L, Wang F. 2017. Dietary quercetin potentiates the antiproliferative effect of interferon-α in hepatocellular carcinoma cells through activation of JAK/STAT pathway signaling by inhibition of SHP2 phosphatase. Oncotarget 8:113734–113748. doi: 10.18632/oncotarget.22556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scarabelli TM, Mariotto S, Abdel-Azeim S, Shoji K, Darra E, Stephanou A, Chen-Scarabelli C, Marechal JD, Knight R, Ciampa A, Saravolatz L, de Prati AC, Yuan Z, Cavalieri E, Menegazzi M, Latchman D, Pizza C, Perahia D, Suzuki H. 2009. Targeting STAT1 by myricetin and delphinidin provides efficient protection of the heart from ischemia/reperfusion-induced injury. FEBS Lett 583:531–541. doi: 10.1016/j.febslet.2008.12.037. [DOI] [PubMed] [Google Scholar]
- 24.Edwards MR, Johnson B, Mire CE, Xu W, Shabman RS, Speller LN, Leung DW, Geisbert TW, Amarasinghe GK, Basler CF. 2014. The Marburg virus VP24 protein interacts with Keap1 to activate the cytoprotective antioxidant response pathway. Cell Rep 6:1017–1025. doi: 10.1016/j.celrep.2014.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT. 2006. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem 49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 26.Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, Lee M, Lee T, Duan Y, Wang W, Donini O, Cieplak P, Srinivasan J, Case DA, Cheatham TE. 2000. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res 33:889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 27.Qiu X, Kroeker A, He S, Kozak R, Audet J, Mbikay M, Chrétien M. 2016. Prophylactic efficacy of quercetin 3-β-O-D-glucoside against Ebola virus infection. Antimicrob Agents Chemother 60:5182–5188. doi: 10.1128/AAC.00307-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diaz MO, Ziemin S, Le Beau MM, Pitha P, Smith SD, Chilcote RR, Rowley JD. 1988. Homozygous deletion of the α- and β1-interferon genes in human leukemia and derived cell lines. Proc Natl Acad Sci U S A 85:5259–5263. doi: 10.1073/pnas.85.14.5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olejnik J, Nelson EV. 2017. Analyzing apoptosis induction and evasion in Ebola virus-infected cells. Methods Mol Biol 1628:227–241. doi: 10.1007/978-1-4939-7116-9_18. [DOI] [PubMed] [Google Scholar]
- 30.Gastaminza P, Whitten-Bauer C, Chisari FV. 2010. Unbiased probing of the entire hepatitis C virus life cycle identifies clinical compounds that target multiple aspects of the infection. Proc Natl Acad Sci U S A 107:291–296. doi: 10.1073/pnas.0912966107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheahan TP, Sims AC, Graham RL, Menachery VD, Gralinski LE, Case JB, Leist SR, Pyrc K, Feng JY, Trantcheva I, Bannister R, Park Y, Babusis D, Clarke MO, MacKman RL, Spahn JE, Palmiotti CA, Siegel D, Ray AS, Cihlar T, Jordan R, Denison MR, Baric RS. 2017. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med 9:eaal3653. doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abner E, Stoszko M, Zeng L, Chen H-C, Izquierdo-Bouldstridge A, Konuma T, Zorita E, Fanunza E, Zhang Q, Mahmoudi T, Zhou M-M, Filion GJ, Jordan A. 2018. A new quinoline BRD4 inhibitor targets a distinct latent HIV-1 reservoir for reactivation from other “shock” drugs. J Virol 92:e02056-17. doi: 10.1128/JVI.02056-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van De Klundert MAA, Zaaijer HL, Kootstra NA. 2016. Identification of FDA-approved drugs that target hepatitis B virus transcription. J Viral Hepat 23:191–201. doi: 10.1111/jvh.12479. [DOI] [PubMed] [Google Scholar]
- 34.Esposito F, Tramontano E. 2014. Past and future. Current drugs targeting HIV-1 integrase and reverse transcriptase-associated ribonuclease H activity: single and dual active site inhibitors. Antivir Chem Chemother 23:129–144. doi: 10.3851/IMP2690. [DOI] [PubMed] [Google Scholar]
- 35.Khiar S, Lucas-Hourani M, Nisole S, Smith N, Helynck O, Bourgine M, Ruffié C, Herbeuval JP, Munier-Lehmann H, Tangy F, Vidalain PO. 2017. Identification of a small molecule that primes the type i interferon response to cytosolic DNA. Sci Rep 7:2561. doi: 10.1038/s41598-017-02776-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones M, Cunningham ME, Wing P, DeSilva S, Challa R, Sheri A, Padmanabhan S, Iyer RP, Korba BE, Afdhal N, Foster GR. 2017. SB 9200, a novel agonist of innate immunity, shows potent antiviral activity against resistant HCV variants. J Med Virol 89:1620–1628. doi: 10.1002/jmv.24809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zinzula L, Tramontano E. 2013. Strategies of highly pathogenic RNA viruses to block dsRNA detection by RIG-I-like receptors: hide, mask, hit. Antiviral Res 100:615–635. doi: 10.1016/j.antiviral.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pizzolla A, Smith JM, Brooks AG, Reading PC. 2017. Pattern recognition receptor immunomodulation of innate immunity as a strategy to limit the impact of influenza virus. J Leukoc Biol 101:851–861. doi: 10.1189/jlb.4MR0716-290R. [DOI] [PubMed] [Google Scholar]
- 39.Di Palma F, Daino GL, Ramaswamy VK, Corona A, Frau A, Fanunza E, Vargiu AV, Tramontano E, Ruggerone P. 2019. Relevance of Ebola virus VP35 homo-dimerization on the type I interferon cascade inhibition. Antivir Chem Chemother 27:2040206619889220. doi: 10.1177/2040206619889220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jasenosky LD, Cadena C, Mire CE, Borisevich V, Haridas V, Ranjbar S, Nambu A, Bavari S, Soloveva V, Sadukhan S, Cassell GH, Geisbert TW, Hur S, Goldfeld AE. 2019. The FDA-approved oral drug nitazoxanide amplifies host antiviral responses and inhibits Ebola virus. iScience 19:1279–1290. doi: 10.1016/j.isci.2019.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao Z, Martin C, Fan R, Bourne PE, Xie L. 2016. Drug repurposing to target Ebola virus replication and virulence using structural systems pharmacology. BMC Bioinformatics 17:90. doi: 10.1186/s12859-016-0941-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Darapaneni V. 2014. Virion protein 24 of Ebola virus as a potential drug target. Am J Curr Microbiol 3:14–22. [Google Scholar]
- 43.Setlur AS, Naik SY, Skariyachan S. 2017. Herbal lead as ideal bioactive compounds against probable drug targets of Ebola virus in comparison with known chemical analogue: a computational drug discovery perspective. Interdiscip Sci 9:254–277. doi: 10.1007/s12539-016-0149-8. [DOI] [PubMed] [Google Scholar]
- 44.Pinto AK, Williams GD, Szretter KJ, White JP, Proença-Módena JL, Liu G, Olejnik J, Brien JD, Ebihara H, Mühlberger E, Amarasinghe G, Diamond MS, Boon A. 2015. Human and murine IFIT1 proteins do not restrict infection of negative-sense RNA viruses of the Orthomyxoviridae, Bunyaviridae, and Filoviridae families. J Virol 89:9465–9476. doi: 10.1128/JVI.00996-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Umesh CV, Jamsheer AM, Prasad MA. 2018. The role of flavonoids in drug discovery—review on potential applications. Res J Life Sci Bioinformatics Pharm Chem Sci 4:70–77. [Google Scholar]
- 46.Johari J, Kianmehr A, Mustafa MR, Abubakar S, Zandi K. 2012. Antiviral activity of baicalein and quercetin against the Japanese encephalitis virus. Int J Mol Sci 13:16020–16045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zandi K, Teoh BT, Sam SS, Wong PF, Mustafa M, Abubakar S. 2011. Antiviral activity of four types of bioflavonoid against dengue virus type-2. Virol J 8:560. doi: 10.1186/1743-422X-8-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu AL, Wang H, Di Lee SM, Wang YT, Du GH. 2008. Structure-activity relationship of flavonoids as influenza virus neuraminidase inhibitors and their in vitro anti-viral activities. Bioorg Med Chem 16:7141–7147. doi: 10.1016/j.bmc.2008.06.049. [DOI] [PubMed] [Google Scholar]
- 49.Bang S, Li W, Ha TKQ, Lee C, Oh WK, Shim SH. 2018. Anti-influenza effect of the major flavonoids from Salvia plebeia R.Br. via inhibition of influenza H1N1 virus neuraminidase. Nat Prod Res 32:1224–1228. doi: 10.1080/14786419.2017.1326042. [DOI] [PubMed] [Google Scholar]
- 50.Esposito F, Sanna C, Del Vecchio C, Cannas V, Venditti A, Corona A, Bianco A, Serrilli AM, Guarcini L, Parolin C, Ballero M, Tramontano E. 2013. Hypericum hircinum L. components as new single-molecule inhibitors of both HIV-1 reverse transcriptase-associated DNA polymerase and ribonuclease H activities. Pathog Dis 68:116–124. doi: 10.1111/2049-632X.12051. [DOI] [PubMed] [Google Scholar]
- 51.Sanna C, Rigano D, Corona A, Piano D, Formisano C, Farci D, Franzini G, Ballero M, Chianese G, Tramontano E, Taglialatela-Scafati O, Esposito F. 2018. Dual HIV-1 reverse transcriptase and integrase inhibitors from Limonium morisianum Arrigoni, an endemic species of Sardinia (Italy). Nat Prod Res 33:1768–1803. [DOI] [PubMed] [Google Scholar]
- 52.Zakaryan H, Arabyan E, Oo A, Zandi K. 2017. Flavonoids: promising natural compounds against viral infections. Arch Virol 162:2539–2551. doi: 10.1007/s00705-017-3417-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.D’Andrea G. 2015. Quercetin: a flavonol with multifaceted therapeutic applications? Fitoterapia 106:256–271. doi: 10.1016/j.fitote.2015.09.018. [DOI] [PubMed] [Google Scholar]
- 54.Khachatoorian R, Arumugaswami V, Raychaudhuri S, Yeh GK, Maloney EM, Wang J, Dasgupta A, French SW. 2012. Divergent antiviral effects of bioflavonoids on the hepatitis C virus life cycle. Virology 433:346–355. doi: 10.1016/j.virol.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dos Santos AE, Kuster RM, Yamamoto KA, Salles TS, Campos R, De Meneses MDF, Soares MR, Ferreira D. 2014. Quercetin and quercetin 3-O-glycosides from Bauhinia longifolia (Bong.) Steud. show anti-Mayaro virus activity. Parasit Vectors 7:130. doi: 10.1186/1756-3305-7-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim Y, Narayanan S, Chang KO. 2010. Inhibition of influenza virus replication by plant-derived isoquercetin. Antiviral Res 88:227–235. doi: 10.1016/j.antiviral.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 57.Lani R, Hassandarvish P, Chiam CW, Moghaddam E, Jang J, Chu H, Rausalu K, Merits A, Higgs S, Vanlandingham D, Bakar SA, Zandi K, Chu JJH, Rausalu K, Merits A, Higgs S, Vanlandingham D, Abu Bakar S, Zandi K. 2015. Antiviral activity of silymarin against chikungunya virus. Sci Rep 5:11421. doi: 10.1038/srep11421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee M, Son M, Ryu E, Kang H, Shin YS, Kim JG, Kang BW, Cho H. 2015. Quercetin-induced apoptosis prevents EBV infection. Oncotarget 6:12603–12624. doi: 10.18632/oncotarget.3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bachmetov L, Gal-Tanamy M, Shapira A, Vorobeychik M, Giterman-Galam T, Sathiyamoorthy P, Golan-Goldhirsh A, Benhar I, Tur-Kaspa R, Zemel R. 2012. Suppression of hepatitis C virus by the flavonoid quercetin is mediated by inhibition of NS3 protease activity. J Viral Hepat 19:e81–e88. doi: 10.1111/j.1365-2893.2011.01507.x. [DOI] [PubMed] [Google Scholar]
- 60.Granato M, Gilardini Montani MS, Zompetta C, Santarelli R, Gonnella R, Romeo MA, D’Orazi G, Faggioni A, Cirone M. 2019. Quercetin interrupts the positive feedback loop between STAT3 and IL-6, promotes autophagy, and reduces ROS, preventing EBV-driven B cell immortalization. Biomolecules 9:482. doi: 10.3390/biom9090482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yeh SL, Lin YC, Lin YL, Li CC, Chuang CH. 2016. Comparing the metabolism of quercetin in rats, mice and gerbils. Eur J Nutr 55:413–422. doi: 10.1007/s00394-015-0862-9. [DOI] [PubMed] [Google Scholar]
- 62.Baize S, Pannetier D, Oestereich L, Rieger T, Koivogui L, Magassouba N, Soropogui B, Sow MS, Keïta S, De Clerck H, Tiffany A, Dominguez G, Loua M, Traoré A, Kolié M, Malano ER, Heleze E, Bocquin A, Mély S, Raoul H, Caro V, Cadar D, Gabriel M, Pahlmann M, Tappe D, Schmidt-Chanasit J, Impouma B, Diallo AK, Formenty P, Van Herp M, Günther S. 2014. Emergence of Zaire Ebola virus disease in Guinea. N Engl J Med 371:1418–1425. doi: 10.1056/NEJMoa1404505. [DOI] [PubMed] [Google Scholar]
- 63.Mohamadi F, Richards NGJ, Guida WC, Liskamp R, Lipton M, Caufield C, Chang G, Hendrickson T, Still WC. 1990. Macromodel—an integrated software system for modeling organic and bioorganic molecules using molecular mechanics. J Comput Chem 11:440–467. doi: 10.1002/jcc.540110405. [DOI] [Google Scholar]
- 64.Halgren TA. 1996. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J Comput Chem 17:490–519. doi:. [DOI] [Google Scholar]
- 65.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. 2000. The protein data bank. Nucleic Acids Res 28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL. 2001. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J Phys Chem B 105:6474–6487. doi: 10.1021/jp003919d. [DOI] [Google Scholar]