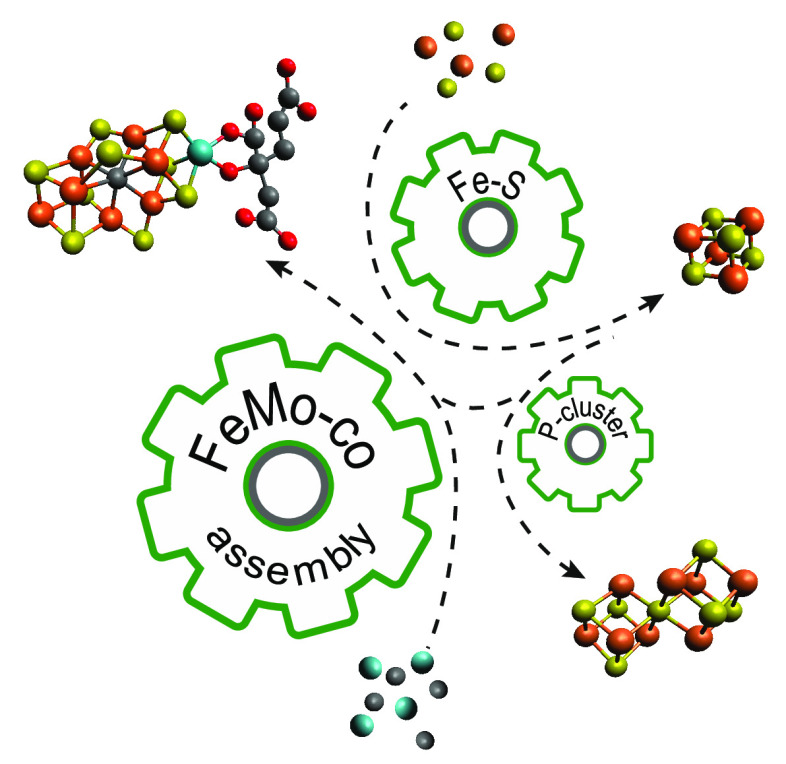
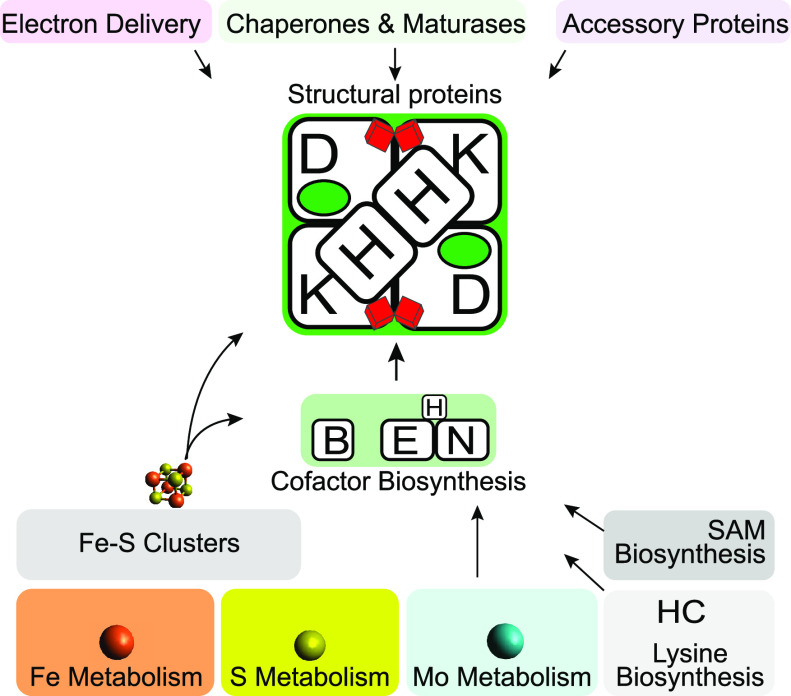
Abstract
Nitrogenase harbors three distinct metal prosthetic groups that are required for its activity. The simplest one is a [4Fe-4S] cluster located at the Fe protein nitrogenase component. The MoFe protein component carries an [8Fe-7S] group called P-cluster and a [7Fe-9S-C-Mo-R-homocitrate] group called FeMo-co. Formation of nitrogenase metalloclusters requires the participation of the structural nitrogenase components and many accessory proteins, and occurs both in situ, for the P-cluster, and in external assembly sites for FeMo-co. The biosynthesis of FeMo-co is performed stepwise and involves molecular scaffolds, metallochaperones, radical chemistry, and novel and unique biosynthetic intermediates. This review provides a critical overview of discoveries on nitrogenase cofactor structure, function, and activity over the last four decades.
1. Introduction
The molybdenum (Mo) nitrogenase enzyme consists of two interacting metalloproteins (the MoFe protein and the Fe protein) that together catalyze the reduction of N2 into NH3 in a reaction that depends on intra- and intermolecular electron transfer and the energy released by ATP hydrolysis.1,2 The MoFe protein (named so because of the presence of Mo and Fe in its active-site metalloclusters, also known as Component I in early work or simply as dinitrogenase) is a dinitrogenase that binds and reduces N2. The Fe protein (named so because of Fe being the sole metal present in its clusters, also known as Component II or dinitrogenase reductase) is a reductase whose function is to provide the MoFe protein with the electrons required for N2 reduction. In addition, the Fe protein plays an essential role in the maturation of the metalloclusters of the MoFe protein. Each electron transfer event from the Fe protein to the MoFe protein involves the proteins to associate and dissociate3 with the following sequence of events: ATP binding to the Fe protein, complex formation with the MoFe protein, electron transfer, ATP hydrolysis at the Fe protein, Pi release, and nitrogenase complex dissociation.4
The Fe protein is a homodimer of the nifH gene product that carries a single [4Fe-4S] cluster bridging the two subunits.5 The MoFe protein is a α2β2 heterodimer of the nifD and nifK gene products that carries in each αβ half a pair of complex metal cofactors, called the P-cluster and the iron–molybdenum cofactor (FeMo-co).6 The three metal clusters are all essential for nitrogenase function and are synthesized by complex biochemical pathways involving the products of many other N2 fixation genes.7,8
As the structural polypeptides of the nitrogenase require metallocluster-prosthetic groups for catalytic activity, two main hypotheses on the mechanisms for the assembly of such clusters have been put forward. In the first mechanism, a cluster is assembled or finalized in situ, on the nitrogenase component. In the second mechanism, a cluster is assembled elsewhere and then inserted into the nitrogenase component. In fact, it is now clear that both mechanisms occur. Namely, the P-clusters are formed via condensation of simpler [Fe–S] cluster precursors already bound at its final location in the MoFe protein,9 while the FeMo-co is assembled and completed outside of the MoFe protein10 by the sequential activities of several biosynthetic proteins prior delivery to the MoFe protein.
Because of its complexity, the biosynthesis of nitrogenase metal clusters has served as model to understand the biosynthesis of simple and complex clusters in other enzymes. The existence of an accessory protein (NifEN) functioning as molecular scaffold for external assembly of a complex metallocluster,11 prior insertion in the real enzyme, was a revolutionary hypothesis derived from nitrogenase research that could later be expanded to other systems such as the hydrogenases.12 By investigating the roles of NifU and NifS in the maturation of nitrogenase components, a new concept of scaffold-controlled [Fe–S] cluster assembly was conceived, giving birth to an entire new research field.13,14 A wealth of high-quality structural and spectroscopic information along with the development of new techniques is owed to nitrogenase researchers. The discovery of FeMo-co15 and the development of an in vitro FeMo-co synthesis assay16 are reviewed here in depth because of the crucial importance to find and investigate proteins and factors required for FeMo-co synthesis. This assay permitted the elucidation of homocitrate as an integral component of FeMo-co;17,18 the isolation of NifB-co, the FeMo-co biosynthetic precursor of unique structure that results from the first committed step in FeMo-co biosynthesis and that connects simpler [Fe–S] clusters into a more complex structure;19,20 the confirmation that NifEN acts as a molecular scaffold for FeMo-co synthesis;21 and finally, the demonstration that FeMo-co can be synthesized in vitro from elemental Fe, S, Mo, and homocitrate using only three purified protein components (NifB, NifH, and NifEN).22
However, our understanding of the nitrogenase metal cofactors formation is still incomplete. Despite decades of investigation, many crucial aspects of nitrogenase maturation are still unclear. Only very recently, new factors affecting maturation of the MoFe protein have been described.23 The role of NifM in processing the Fe protein polypeptide and its effect on Fe protein activity is still unknown, as is the exact mechanism by which NifEN acts as scaffold to transform NifB-co into FeMo-co.24
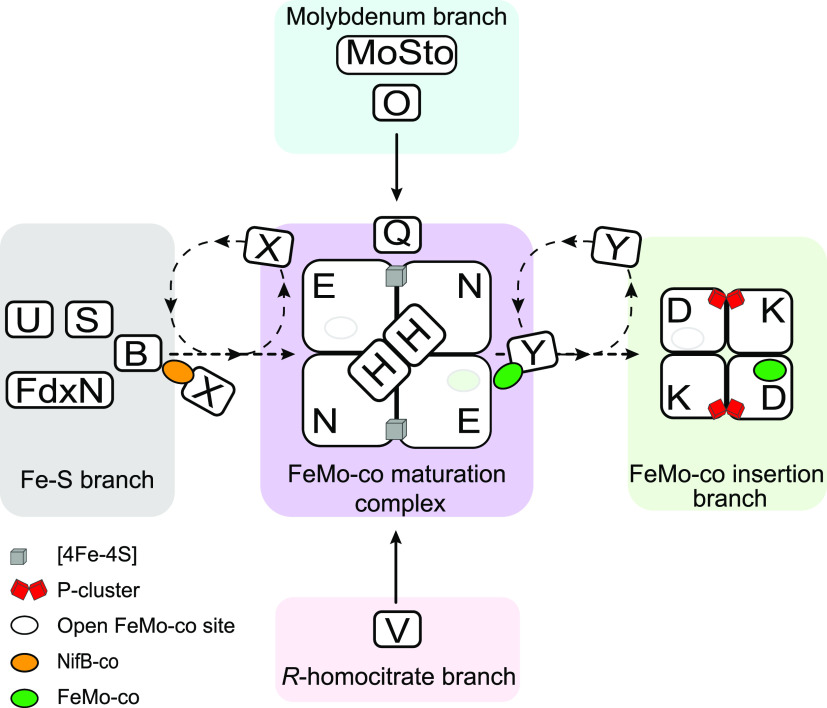
One important conceptual outcome from studying the nitrogenase cofactor biosynthesis is that the proteins involved can be classified into three main groups: (1) proteins acting as molecular scaffolds for the assembly of the inorganic network of atoms (NifU, NifB, NifQ, and NifEN); (2) proteins acting as chaperones both by carrying and protecting metalloclusters in their transit between assembly scaffolds and to their final targets (NifX and NafY) or by maturing the nitrogenase polypeptides (NifM, NafH, NifW, and NifZ); and (3) proteins with enzymatic activities that provide substrates that are used as cofactor parts (NifS, NifV, and NifB).7 In addition, the Fe protein is also essential to P-cluster and FeMo-co biosynthesis, although its role does not appear to fit in any of these three groups.
This review spans research reported from 1966 until 2019. We have tried to include all those original reports that have been essential to our understanding of nitrogenase metallocluster formation. If we have missed important work, we apologize to those authors involved. Also, this review refers to the information obtained from biochemical work on nitrogenase in vitro as well as that obtained from the numerous gene mutants generated. Many of the methods used today were not available for the detailed assessment of many of these mutants at the time they were generated. Initial analyses were often constrained to measuring acetylene reduction activities, the presence and functionality of Fe protein and MoFe protein components, and the ability for diazotrophic growth. Nevertheless, there is much information to be found in these early works. This is especially true for persons about to enter the nitrogenase field. However, it is important to highlight that studying the microbial physiology alone has some limitations, for example, the interpretation of leaky mutant phenotypes due to nonessential functions, gene redundancy, or dispensability under certain growth conditions. Studies based on the use of mutant strains with gene disruptions also present limitations when the ultimate goal is to define the specific function of proteins with several activities, such as the Fe protein, which is involved in P-cluster maturation, FeMo-co synthesis, and electron donation to the MoFe protein for nitrogen reduction, or when there are multiple proteins that are functionally related, such as Fe proteins for the three nitrogenase systems. On the other hand, we must acknowledge that regarding a function solely because it has been proven in vitro can be misleading, as the experimental conditions are often far from the actual situation in vivo. For example, to protect the purified nitrogenase proteins from traces of O2, and to provide electrons to nitrogenase not depending on its physiological donors, assays are normally performed under very artificial conditions in the presence of dithionite (DTH) at low mM concentrations. Therefore, results obtained from the in vitro and in vivo experiments should be compared and interpreted as a whole whenever possible.
Finally, there are some aspects of nitrogenase cofactor formation that are strongly controversial, including the mechanisms of incorporation of Mo into the FeMo-co, the role of accessory proteins in P-cluster maturation, and the confusing nomenclature used by the different groups working in this field. We have tried to be comprehensive albeit critical in treating these aspects and we hope that this review will be helpful to the general reader.
2. Structure of Mo–Nitrogenase Complex
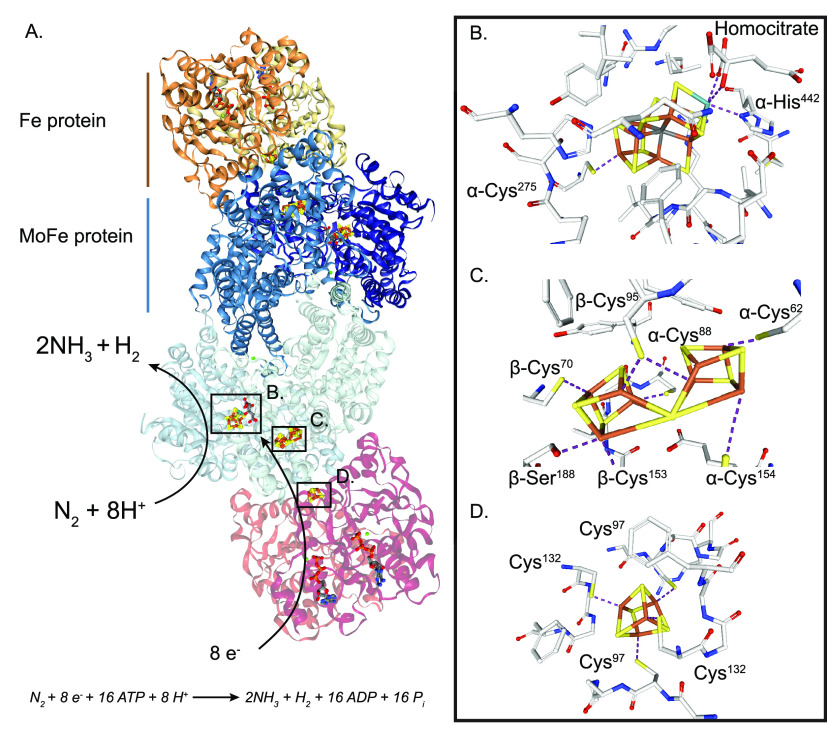
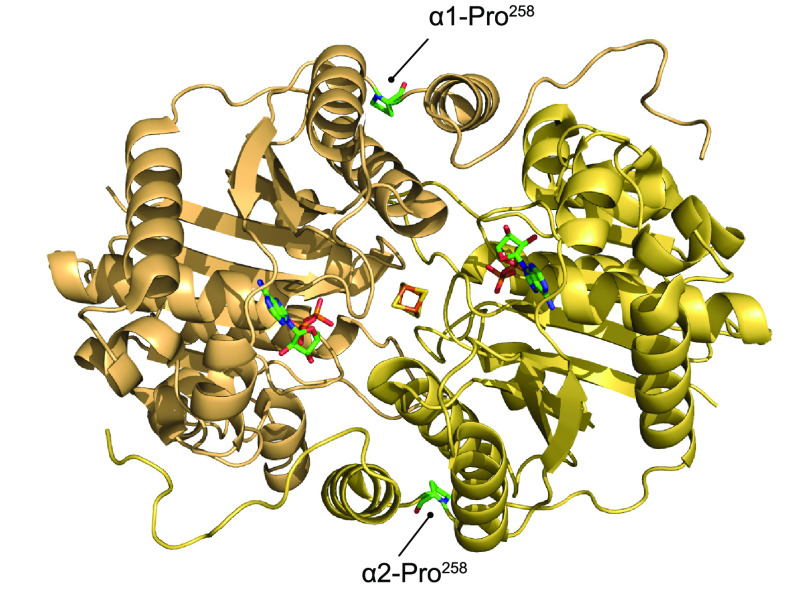
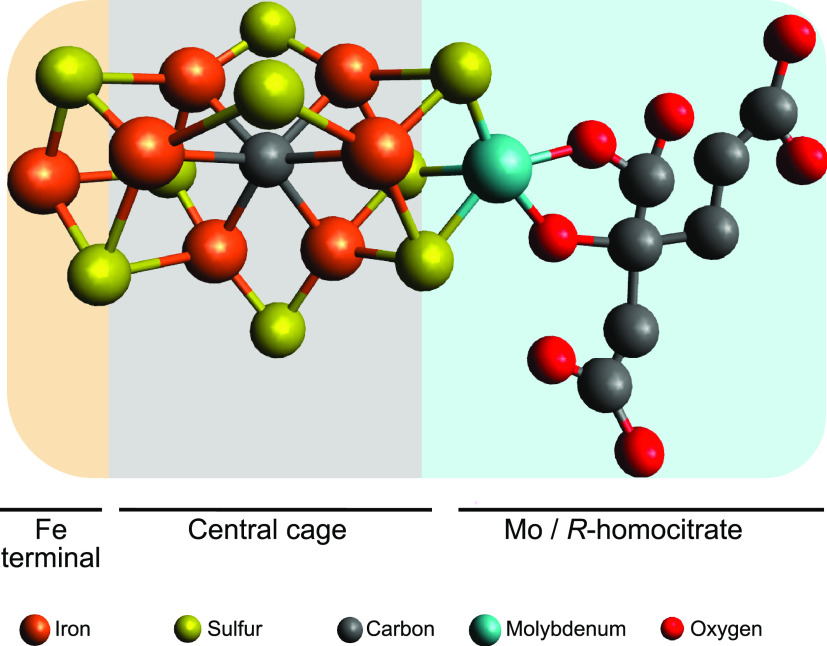
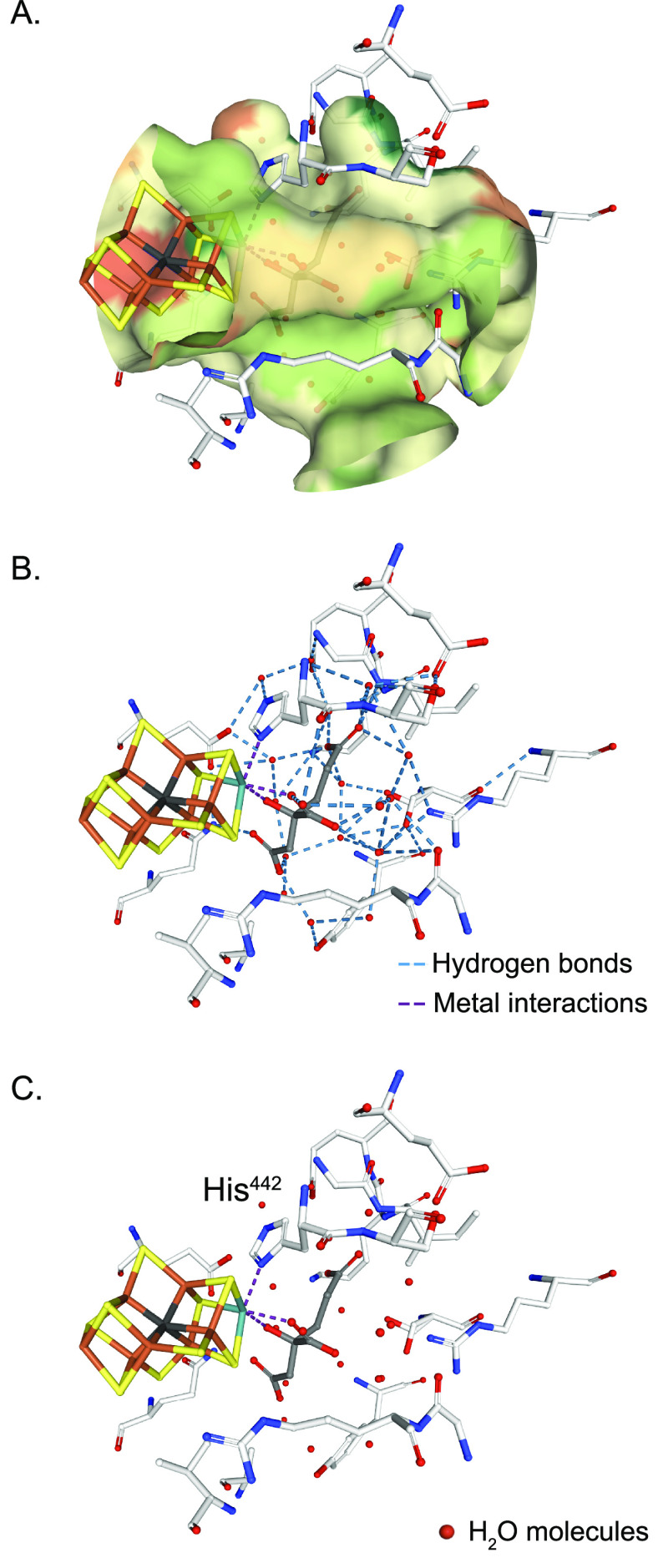
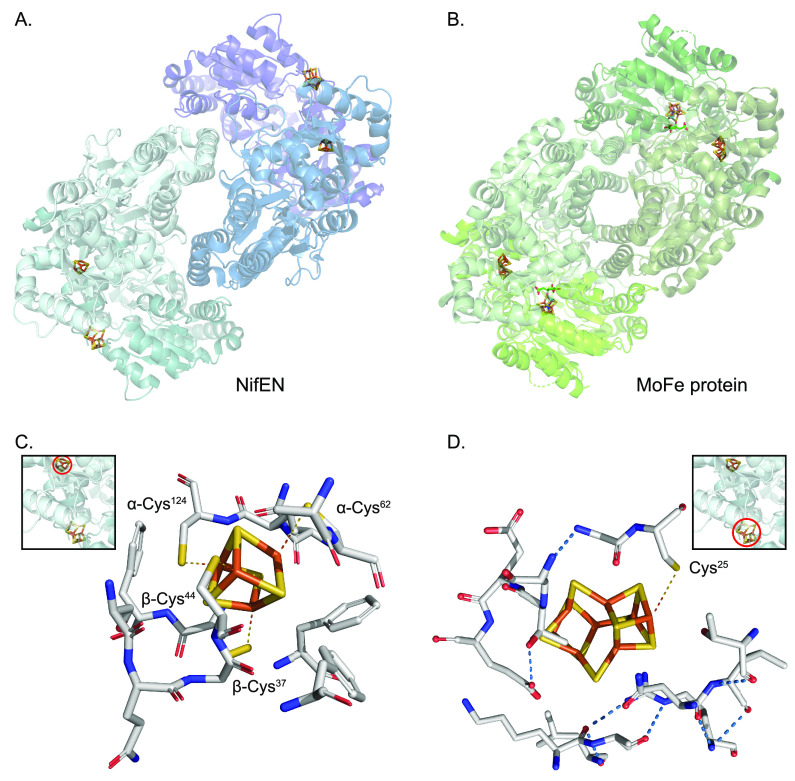
Figure 1 shows the structure of the Mo nitrogenase complex including its component polypeptides, metal prosthetic groups, and bound nucleotides.25 The complex is composed of one central MoFe protein tetramer with α2β2 subunit composition and one Fe protein homodimer bound to each αβ half (Figure 1A). The metal clusters are positioned to facilitate electron transfer from each Fe protein [4Fe-4S] cluster via the surface-located P-cluster to the iron–molybdenum cofactor (FeMo-co) located at the active site buried 10 Å beneath the surface of each α subunit. FeMo-co is composed of an inorganic [7Fe-9S-C-Mo] cluster and a molecule of R-homocitrate exclusively bound to the Mo atom.6,26,27 FeMo-co is ligated by a Cys residue to its terminal Fe and by a His residue to the Mo atom (α-Cys275 and α-His442 in the Azotobacter vinelandii MoFe protein) (Figure 1B). The environment around FeMo-co is mostly hydrophilic, especially at the homocitrate site that is surrounded by a pool of water molecules that participate in a H-bonding network.28
Figure 1.
Structure of Mo nitrogenase. (A) A. vinelandii MoFe protein and Fe protein complex (PDB: 1G21). Top half shows polypeptide secondary structure. Partial transparency has been applied to the polypeptide chains of the bottom half to visualize metal clusters and nucleotides. Ligands and surrounding environment for (B) FeMo-co, (C) P-cluster and (D) [4Fe-4S] cluster are shown. Mo nitrogenase reaction is shown at the bottom. Images created with NGL viewer36 and RCSB PDB.
The P-clusters are [8Fe-7S] groups located at the interface of the α and β subunits within each αβ half of the MoFe protein.6,29 Their positioning is consistent with its function as intermediate carrier of electrons from the Fe protein to FeMo-co.30 The conformation of the P-cluster depends on its oxidation state. The oxidized P-cluster is covalently bound to six Cys residues (Cys62, Cys88, and Cys154 from the α chain, and Cys70, Cys95, and Cys153 from the β chain in A. vinelandii) and to the Ser188 residue from the β chain (Figure 1C). In this conformation, one-half of the P-cluster exists as a standard [4Fe-4S] cubane and the other half as a distorted [4Fe-3S] unit.29 Upon reduction, two Fe atoms from one-half dissociate from residues α-Cys88 and β-Ser188 and bind to a corner S from the other cubane, resulting in two almost regular [4Fe-4S] cubanes sharing one S atom. The residues that coordinate the P-cluster in the A. vinelandii and Klebsiella pneumoniae MoFe proteins have been extensively studied by site-directed mutagenesis. Substitutions of α-Cys62, α-Cys154, and β-Cys70 diminished nitrogenase activity with more than 99%,31−33 while substitutions of the other four coordinating residues resulted in MoFe protein variants that could support some diazotrophic growth.29,31−35
In the Fe protein, the [4Fe-4S] cluster occupies a bridging position at the interface of both subunits and is symmetrically coordinated by two Cys residues from each one (Cys97 and Cys132 in A. vinelandii) (Figure 1D). As a P-loop ATPase, each Fe protein subunit also binds a nucleotide (MgATP or MgADP) in a channel located at the opposite face from the [4Fe-4S] cluster.5
3. Organization of Mo–Nitrogenase Genes and Proposed Functions of Their Products
3.1. Genomic Organization of A. vinelandii Mo–Nitrogenase Genes
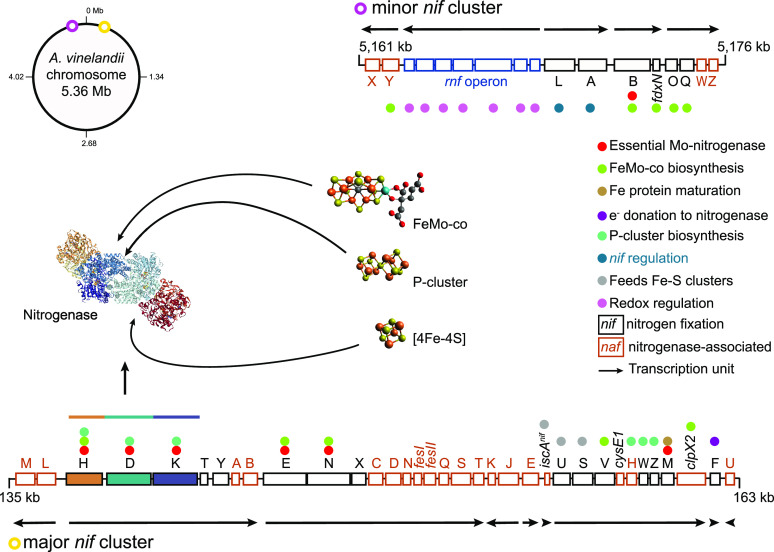
In A. vinelandii, Mo-nitrogenase nitrogen fixation genes (nif) are clustered in two different regions adjacent and equidistant to the replication origin of its single 5.35 Mb chromosome.37 These regions were originally named as major and minor nif gene clusters according to their size.38,39 The major nif cluster is located 135–163 kb downstream of the replication origin. It contains the three structural genes for the MoFe protein (nifD for the α subunit and nifK for the β subunit) and the Fe protein (nifH) and 32 additional genes organized in nine transcriptional units (Figure 2). Genes in this region encode proteins involved in the biosynthesis of MoFe protein cofactors (nifE and nifN), the maturation of Fe protein (nifM) and MoFe protein (nafH, nifW, nifZ) polypeptides, proteins for the biosynthesis of [Fe–S] clusters (nifU, nifS, iscAnif, cysE1), a homocitrate synthase (nifV), proteins involved in electron transfer to nitrogenase (nifF) and in protection against oxidation (feSII), and proteins involved in recycling biosynthetic proteins (clpX2). The region contains well characterized nif genes as well as nitrogenase accessory factors (naf).
Figure 2.
Organization and proposed functions of Mo nitrogenase genes in A. vinelandii. Figure shows the chromosomal location and genetic organization of the major and minor nif gene clusters. Numbers to the left and right of each gene cluster indicate chromosomal location. Transcriptional units are depicted by arrows. Proposed roles in Mo nitrogenase are color coded in the legend. Metal clusters embedded in Mo nitrogenase and a surface structure of the complex are shown.
The minor operon is in the 5.16–5.17 Mb region upstream from the replication origin, and it contains 17 genes in five transcriptional units (Figure 2). Among them is the essential nifB gene, which product catalyzes the first committed step in the biosynthesis of FeMo-co.19,22,40,41 Additional genes in the nifB operon are important for nifB function (fdxN) or for Mo processing for nitrogenase (nifO and nifQ). The products of the nifLA operon form an activator (NifA)/antiactivator (NifL) regulatory pair that controls the expression of nif genes in response to environmental signals such as nitrogen assimilation rates, presence of O2, cellular redox status, and ATP levels.42−44 Divergent from the nifLA operon appear the rnf1 and the nafYX operons. The product of nafY has a dual role in MoFe protein stabilization and in FeMo-co insertion. The rnf genes were first discovered in Rhodobacter capsulatus and encode the components of a membrane protein complex proposed to be involved in electron transport to nitrogenase based on their requirement for nitrogenase activity in vivo but not in vitro.45 Rnf complexes have been shown to catalyze NADH-dependent reduction of ferredoxin (Fd) coupled to an electrochemical gradient.46,47 Initial genetic analysis of the A. vinelandii rnf1 operon, however, did not support a role in electron transport in this bacterium but rather a role in a redox mechanism to control expression and maturation of nitrogenase components.48 However, a recent study showed that the A. vinelandii Δrnf1 phenotype was obscured because its function was compensated by the products of fixABCX, which utilized a bifurcating mechanism to generate low potential electrons for nitrogenase catalysis.49
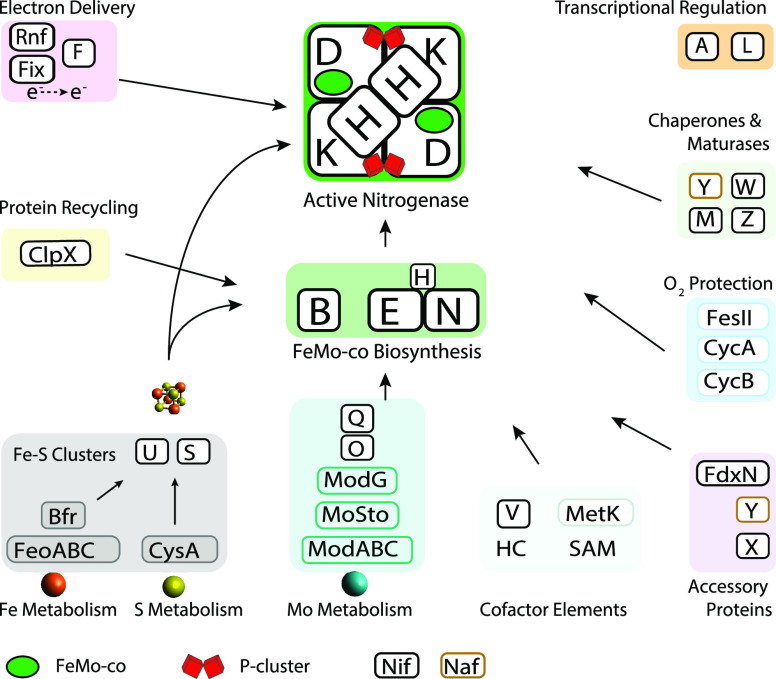
One striking feature of the A. vinelandii nif gene clusters is the large number of genes present in comparison with most other N2 fixing organisms studied to date. Maturation of nitrogenase components from translated structural polypeptides to the metallocluster containing active forms requires more accessory proteins in A. vinelandii than in many other organisms (Figure 3). This could be because A. vinelandii is a strict aerobe while nitrogenase structural and biosynthetic proteins are extremely sensitive to oxidative damage. Although A. vinelandii has mechanisms to protect nitrogenase against O2, such as its very high respiratory activity coupled to high affinity terminal oxidases that deplete intracellular O2,50 it must certainly cope with stressing conditions that might complicate nitrogenase maturation. Finally, the stoichiometry of all these essential and accessory factors must be tightly controlled for efficient N2 fixation. Cellular Nif protein concentrations are exquisitely balanced in A. vinelandii with FeMo-co biosynthetic proteins accumulating 50- to 100-fold less than the structural proteins.51 In this context, mutations of nonregulatory genes were shown to produce dramatic outcomes in nif gene expression and system balance overall.51
Figure 3.
Processes for Mo-nitrogenase biogenesis in the model bacterium A. vinelandii. Proteins involved in each process are shown.
In addition to the nif gene clusters, A. vinelandii carries gene complements required for a V-dependent nitrogenase (vnf genes) and an Fe-only nitrogenase (anf genes). At least five nif genes, nifB, nifU, nifS, nifV, and nifM, have been reported to be required for alternative nitrogenases.40,52,53 The alternative nitrogenases will not be reviewed here.
3.2. Proposed Functions of nif Gene Products
Table 1 summarizes Nif phenotypes, obtained by genetic analysis, and roles proposed for their products. Five out of six genes regarded as the minimum complement for functional Mo-nitrogenase based on genetic and biochemical evidence (nifH, nifD, nifK, nifE and nifN, labeled in Figure 2 with red dots)22 are present in the major nif cluster, whereas the essential nifB gene is located in the minor nif cluster. This minimum gene complement has been used as criterion to identify N2-fixing organisms in protein databases.54 It should be noted that the product of nifM, which is essential for Mo-nitrogenase in A. vinelandii, K. pneumoniae, and other well studied diazotrophs, is not universally essential as the Fe proteins from many N2 fixing organisms are NifM independent.
Table 1. A. vinelandii Genes Involved in Mo-Dependent N2 Fixation and Roles of Their Products.
| genea | Nif phenotypeb | identity/role(s) |
|---|---|---|
| nifH | – | Fe protein. Obligate electron donor to the MoFe protein. Also required for FeMo-co biosynthesis and apo-MoFe maturation. |
| nifD | – | MoFe protein α-subunit. FeMo-co binding subunit. Forms an α2β2 tetramer with NifK. |
| nifK | – | MoFe protein β-subunit. Forms an α2β2 tetramer with NifD. |
| nifT | + | Unknown |
| nifY | + | Interacts with apo-MoFe protein prior to FeMo-co insertion. |
| nafA | + | Unknown function. |
| nifE | – | Scaffold for NifB-co maturation into FeMo-co. Forms an α2β2 tetramer with NifN. |
| nifN | – | Scaffold for NifB-co maturation into FeMo-co. Forms an α2β2 tetramer with NifE. |
| nifX | + | Binds NifB-co and VK-cluster. Interacts with apo-NifEN. Proposed carrier of FeMo-co precursors. |
| nafC | + | Unknown function. |
| feSII | + | Shethna protein II. Nitrogenase protection against O2. |
| nifIscA | + | Fe–S scaffolding Nif protein. Contains a [4Fe-4S] cluster. |
| nifU | – | Supplies [Fe–S] clusters precursors for nitrogenase proteins and cofactors. |
| nifS | ± | Mobilizes S for [Fe–S] cluster synthesis on NifU. |
| nifV | ± | Homocitrate synthase. Involved in FeMo-co synthesis. |
| cysE1 | + | Serine O-acetyltransferase. Participates in cysteine formation. |
| nafH | + | Binds to apo-MoFe protein preceding P-cluster maturation. |
| nifW | ± | Associates with apo-MoFe protein preceding P-cluster maturation. |
| nifZ | ± | Involved in P-cluster maturation in combination with the Fe protein. |
| nifM | – | Essential for the maturation of the Fe protein. Proposed peptidyl-prolyl cis–trans isomerases that would act over the Pro258 residue of NifH. |
| clpx2 | + | Regulates NifEN and NifB protein levels. |
| nifF | + | Flavodoxin. Physiological electron donor to the Fe protein. |
| nifL | + | Sensor protein. Acts as NifA antiactivator. |
| nifA | – | Transcriptional activator. Forms a two-component regulatory system with NifL. |
| nifB | – | Essential for FeMo-co synthesis. Catalyzes NifB-co formation, the first committed step in FeMo-co biosynthesis. |
| fdxN | ± | 2x[4Fe-4S] ferredoxin. Required for NifB in the formation of NifB-co. |
| nifO | + | Required for simultaneous activation of nitrogenase and nitrate reductase. |
| nifQ | – | Processes Mo prior to its incorporation into FeMo-co. |
| nafW | + | Rhodanase-like protein. Also called rhdN. Unknown function. |
| nafZ | + | Glutaredoxin-like protein. Also called grx5nif. Unknown function. |
| rnfAc | ± | Transmembrane protein. Part of the Rnf complex, which is involved in supplying low potential electrons to the nitrogenase. |
| rnfBc | ± | Contains two [Fe–S] cluster binding sites. It is part of the Rnf complex. |
| rnfCc | ± | Contains [4Fe-4S] binding site. Electron transport component of the Rnf complex. |
| rnfDc | ± | Transmembrane protein of the Rnf complex. Includes a flavin binding motive. |
| rnfGc | ± | Last electron acceptor component of the Rnf complex. Contains FMN binding site. |
| rnfEc | ± | Transmembrane protein of the Rnf complex. |
| rnfHc | ± | Soluble protein of the rnf gene cluster. |
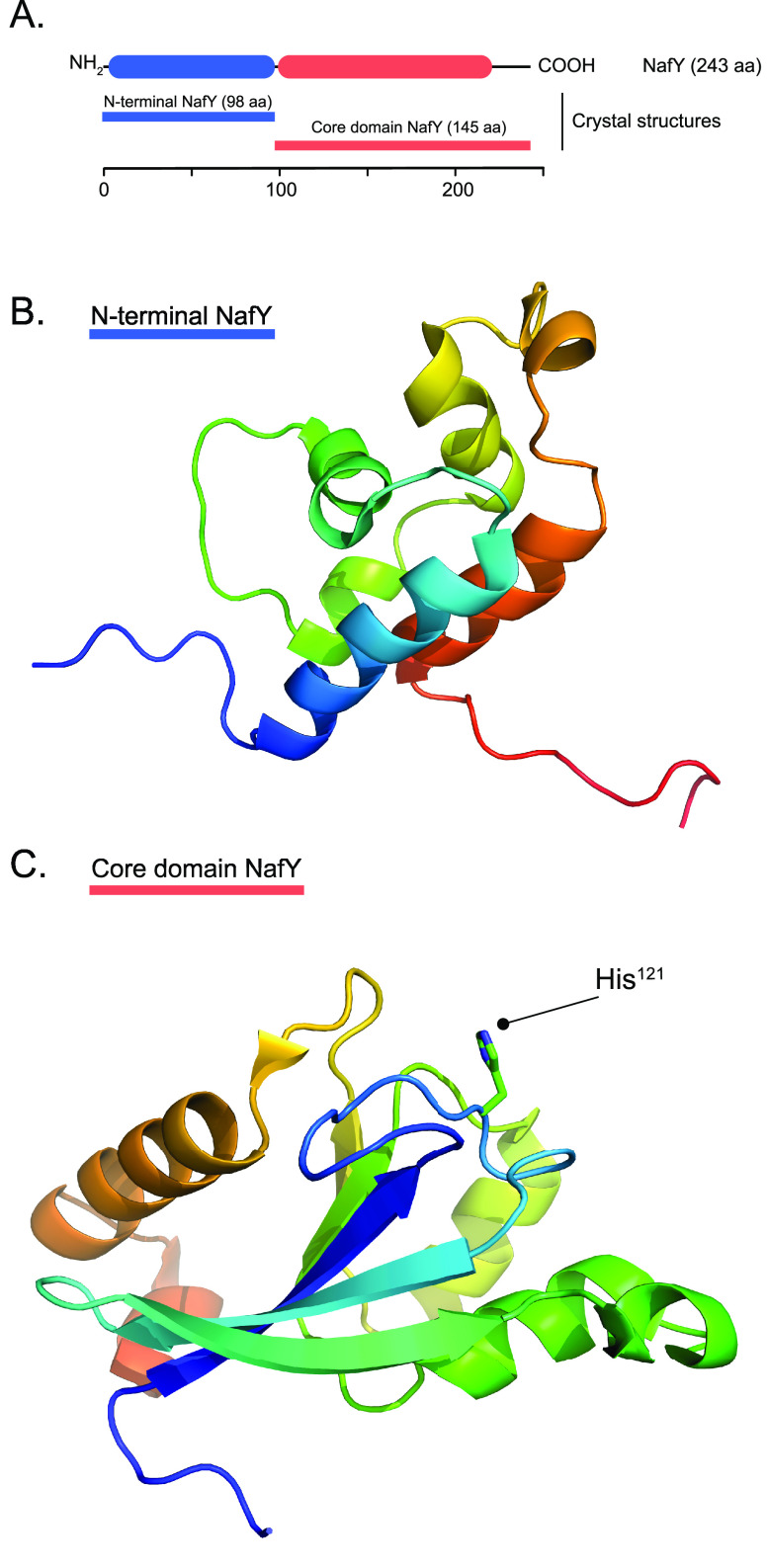
| nafY | + | In A. vinelandii, γ subunit of apo-MoFe protein. Stabilizes apo-MoFe protein prior to FeMo-co insertion. Also binds FeMo-co specifically. |
| mosA | + | Mo storage protein α subunit. Forms an α3β3 hexamer with MosB. |
| mosB | + | Mo storage protein β subunit. |
Minimum set of essential Mo-nitrogenase genes is shown in bold.
+ Diazotrophic growth, ± slow diazotrophic growth, – no diazotrophic growth.
Phenotype of rnf genes was determined by deleting the entire rnf operon.
3.3. Essential and Ancillary Proteins for Mo–Nitrogenase
Biochemical studies have shown that the [4Fe-4S] cluster of the Fe protein can be formed into cluster-less Fe protein in vitro, either by chemical reconstitution using Fe and S under reducing conditions or by transfer of [4Fe-4S] clusters preformed at NifU.55 In addition, the minimum protein and substrate requirements for maturation and activation of the MoFe protein have been established. Simplified, these processes can be divided into three steps: (1) P-cluster formation (at the MoFe protein), (2) FeMo-co synthesis (at the NifB and NifEN proteins), and (3) insertion of FeMo-co into the P-cluster containing MoFe protein. P-cluster formation can be accomplished in vitro by incubating the MoFe protein isolated from a nifH deletion mutant, which contains P-cluster precursors, with Fe protein and ATP under DTH reducing conditions. The P-cluster containing apo-MoFe protein can then be activated by the simple addition of FeMo-co.56 For FeMo-co synthesis, NifB catalysis to render the FeMo-co precursor, NifB-co, can be accomplished in vitro by addition of S-adenosylmethionine (SAM), DTH, Fe, and S,22,41,57,58 or from using SAM together with preformed [4Fe-4S] clusters.59 Conversion of NifB-co into FeMo-co in NifEN only requires the Fe protein, Mo, homocitrate, DTH, and ATP, and de novo synthesized FeMo-co can be directly transferred to the apo-MoFe protein.60,61
Importantly, the complete in vitro FeMo-co synthesis and formation of active MoFe protein could be achieved using the purified protein products of only six nif genes (NifB, NifEN, Fe protein, and MoFe protein containing P-clusters) when supplemented with Fe, S, SAM, Mo, homocitrate, and ATP under reducing conditions.22 This study demonstrated that these six proteins are sufficient for the formation of a functional Mo-nitrogenase in vitro. These six proteins are either structural components (Fe protein and MoFe protein) or involved in P-cluster formation (Fe protein) or FeMo-co synthesis (NifB, NifEN, and the Fe protein). Consistently, genomic studies showed that nearly all organisms containing Mo-nitrogenases carry nifB, nifE, nifN, nifH, nifD, and nifK.54 Interestingly, some diazotrophs contain fusions of these essential genes. For instance, the nifE-N fusion found in the nitrogenase 2 gene cluster of Anabaena variabilis,62 the nifH-E fusion found in some Archaea,54 or the nifN-B fusion found in the genomes of many Clostridia.63,64
It is important to note that many of the proteins used in these assays have been isolated with small epitope-tags, added to facilitate and improve the purification procedures by both shortening the length of the protocol and increasing the purity of the isolated protein. The location, size, and properties of these tags can influence the function of the proteins, for example, by altering their activities or their possibilities to interact to other protein partners. Although functionality of many of these tagged protein variants has been confirmed in vivo, it is important to remember that their function might not be identical to their corresponding native proteins. In addition, much of our understanding of the nitrogenase comes from model organisms that carry many ancillary Nif proteins, for example, K. oxytoca, C. pasteurianum, or A. vinelandii. In this regard, the number of nif related genes is exacerbated in A. vinelandii, probably to cope with its strict aerobic metabolism and because of the existence of not only one, but three, different nitrogenases. Other diazotrophic organisms with distinct lifestyles and metabolic requirements carry fewer auxiliary genes or even different ones.65
3.4. Biosynthesis of Genetically Simpler Mo–Nitrogenases
Some organisms with less complex nitrogenase machineries have been genetically characterized, suggesting that specific gene requirements can be associated with certain lifestyles. For instance, the nine nif-gene cluster (nifBHDKENXhesAnifV) of the Gram-positive, facultative anaerobic rhizobacterium, Paenibacillus sp. WLY78, has been shown to direct the formation of active Mo-nitrogenase when expressed in E. coli.66 In addition to the products of the six essential nif genes and the homocitrate synthase encoding gene nifV, this cluster contains hesA and nifX. The clustering of nifX and nifEN genes often occurs in diazotrophic bacteria,67 which is surprising given the nonessential character of NifX and its lack of any demonstrated catalytic activity over the FeMo-co precursors.24 The presence of hesA in nif gene clusters is much less frequent. HesA is similar to MoeB, a protein that catalyzes the acyladenylation of the MoaD protein prior to S transfer during the biosynthesis of the molybdopterin-based Mo-cofactor, but its exact role in nitrogenase has not yet been elucidated.68
One can envision nitrogenase biogenesis in a diazotroph carrying only the six conserved nif genes (nifHDKBEN) (Figure 4) as a process in which [4Fe-4S] clusters are provided by a housekeeping system (e.g., isc or suf machineries),14,69 homocitrate is provided by a homocitrate synthase (e.g., one that is involved in lysine biosynthesis),70 and in which Mo is incorporated directly into FeMo-co without the need for a Mo concentrating mechanism (e.g., MoSto),71 a Mo partitioning mechanism executed by NifO,72 or the specific Mo delivery by NifQ.73 The NifB protein would be active without a specific FdxN protein or a NifX-like domain.74 In this hypothetical organism, the biosynthetic gene products NifB, NifE, and NifN could form a complex that would interact with some Fe protein to support FeMo-co biosynthesis and to transfer FeMo-co directly to P-cluster containing apo-MoFe protein. Under optimal conditions, carrier proteins such as NifX, NifY, and NafY would not be needed, as their functions would only marginally improve pathway efficiency. In this organism, P-cluster formation could even occur at the same complex by the action of the Fe protein on the P-cluster precursor-containing apo-MoFe protein. No additional factors such as NifM, NifZ, NifW, or NafH would be needed because the biosynthetic rates of Fe protein and MoFe protein would not be as demanding as in A. vinelandii or any other highly efficient diazotroph. One could even imagine a simpler genetic machinery for a Mo-nitrogenase composed of nifHBDK genes and lacking the nifEN genes, as it is the case of the putative diazotroph Roseiflexus sp.54 For such a nitrogenase, FeMo-co biosynthesis would be initiated on NifB and then finished directly on the apo-MoFe protein scaffold. Although this limited system would likely not provide high levels of N2 fixation activity, these could be enough to support diazotrophic growth under certain conditions. If demonstrated, the existence of this genetically simpler nitrogenase could be beneficial to engineer it in a eukaryotic host such as plants.75
Figure 4.
Simplification of genetic requirements for Mo-nitrogenase biogenesis achieved by its integration with housekeeping processes. The essential six gene core is highlighted in green in the middle. Housekeeping processes provide the functions of nitrogenase ancillary proteins.
4. Biosynthesis of Simple [Fe-S] Clusters for Nitrogenase: Roles of NifU and NifS
4.1. Information from nifU and nifS Mutagenesis
A. vinelandii with a disrupted nifU gene did not grow under diazotrophic conditions, indicating that its product is required for the function of Mo nitrogenase.38,39 Corresponding nifS mutant strains showed very slow growth, similarly to what had been observed in K. pneumoniae,44 and therefore, nifS was deemed important but not strictly essential under N2-fixing conditions. Replacing the nif-regulated nifUS gene cluster with a sucrose-regulated copy of nifUS in the A. vinelandii strain DJ1475 showed that addition of sucrose under N2-fixing conditions rescued diazotrophic growth, conclusively proving NifU and NifS requirement for nitrogenase functionality.76 Deletion of isc (iron–sulfur cluster) genes, required for the maturation of [Fe–S] proteins involved in general metabolic processes, showed that while the function of IscU could be rescued by NifU, NifS was not capable of replacing IscS.76,77 Diazotrophic growth of A. vinelandii with deleted nifU could only be partially rescued by iscU overexpression,77 proving that the NifU and NifS proteins are specific and essential for nitrogenase [Fe–S] cluster maturation in vivo.
Fe protein and MoFe protein activities in extracts of A. vinelandii with disrupted nifU or nifS genes were lowered by 95% and 75%, respectively.39 Disruption of both genes simultaneously abolished Fe protein activity and lowered MoFe protein activity by more than 90%. MoFe protein could not be activated in vitro by addition of FeMo-co even if complemented with saturating amounts of Fe protein, indicating that the MoFe protein factored in strains lacking nifU or nifS is nonfunctional and likely does not even contain P-cluster precursors. Similar results were also observed in K. pneumoniae.44 Later studies confirmed that disrupting nifU and nifS genes abolished or severely impaired A. vinelandii growth under diazotrophic conditions requiring each of the three types of nitrogenases.52 This result suggests that NifU and NifS proteins are also essential to the alternative V- and Fe-only nitrogenases and that no additional proteins performing the exact same functions exist for these alternative systems.
4.2. NifS Is a Cysteine Desulfurase Involved in Metallocluster Biosynthesis
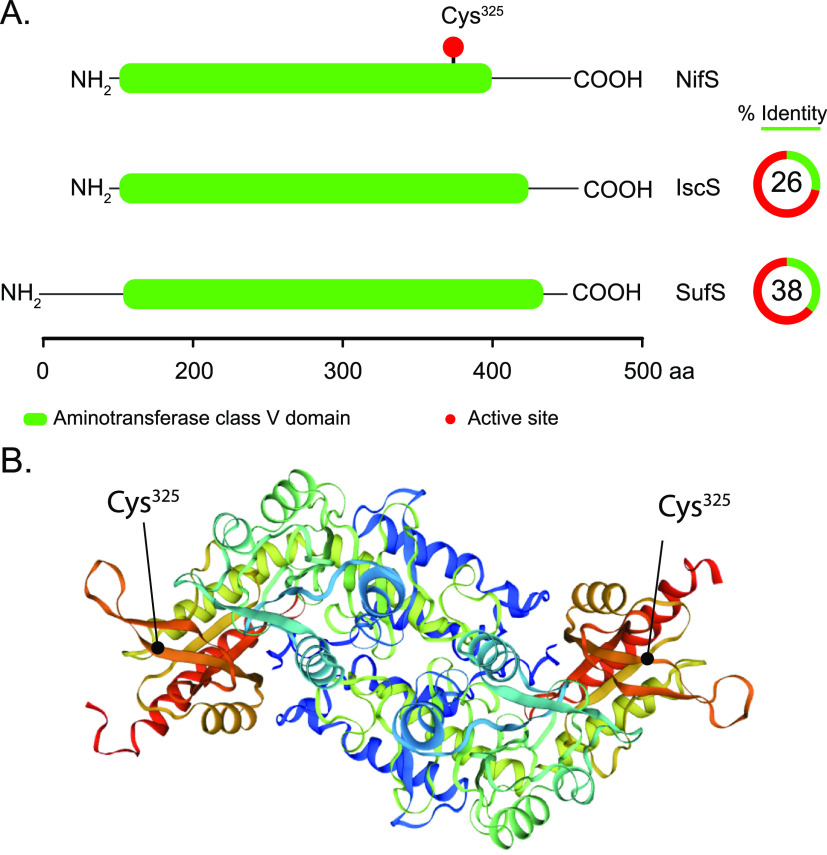
Heterologous expression and purification of A. vinelandii NifS from E. coli produced yellow-colored protein with a molecular weight of about 44 kDa.13 Native molecular weight determination indicated that NifS was a homodimer of about 88 kDa. Extraction of the chromophore suggested a NifS-bound pyridoxal 5-phosphate (PLP) cofactor characteristic of enzymes catalyzing a diverse group of elimination and replacement reactions involving amino acids. Incubation with l-cysteine but none of the other 19 l-amino acids or the d-cysteine isomer altered the protein UV–visible (UV–vis) absorption spectrum. Incubation of NifS with l-cysteine for longer than 10 min formed a cloudy precipitate of elemental sulfur (S).13 The overall reaction catalyzed by NifS under nonreducing conditions showed to be desulfuration of l-cysteine to yield S and l-alanine. Cysteine desulfurases are highly conserved enzymes (Figure 5A). It was later found that A. vinelandii carries a NifS homologue, called IscS, that functions in general housekeeping [Fe–S] cluster biosynthesis.14 It was suggested that NifS generates S destined for nitrogenase [Fe–S] clusters via formation of a persulfide bound at residue Cys325 (Figure 5B).78 NifS was capable of catalyzing the activation of apo-Fe protein (Fe protein with its [4Fe-4S] cluster removed by chelation in vitro) in a reconstitution reaction containing l-cysteine, ferrous (II) iron, dithiothreitol (DTT), and MgATP.79 A modified version of NifS in which the Cys325 residue was replaced by Ala could not activate the apo-Fe protein.
Figure 5.
A. vinelandii NifS cysteine desulfurase. (A) Domain composition and % identity of A. vinelandii NifS (Uniprot C1DH19) compared to IscS (Uniprot C1DH19) and SufS (Uniprot C1DH19). (B) Structure of the NifS homologue protein CsdB showing the surface position of the active-site Cys residue. Image created with NGL viewer36 and RCSB PDB.
4.3. NifU Is a Molecular Scaffold for Assembly of Nitrogenase-Destined [4Fe-4S] Clusters
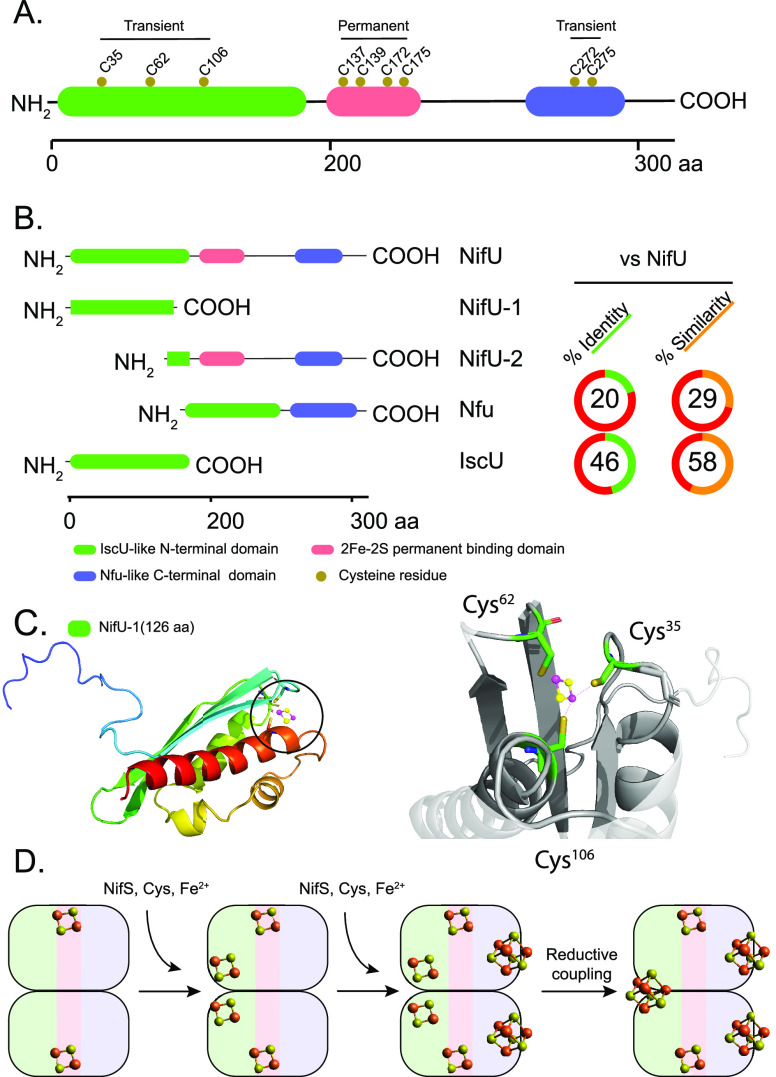
Expression of A. vinelandii NifU in E. coli produced cell pellets with a red color enriched in a 33 kDa protein.80 Size exclusion chromatography indicated that NifU was a homodimer of about 63 kDa, and Fe quantification suggested that NifU contained two Fe atoms per monomer. UV–vis absorption spectra indicated a dithionite (DTH)-reducible [2Fe-2S]2+/1+ cluster in each NifU subunit, which was confirmed by using other spectroscopic techniques such as X-band electron paramagnetic resonance (EPR), variable temperature magnetic circular dichroism (VT-MCD), and resonance Raman.80 Comparison of the primary sequences of A. vinelandii and K. pneumoniae NifU proteins had previously identified nine conserved Cys residues.81 It was observed that NifU redox potential (−254 ± 20 mV) was not low enough to serve as electron donor to the Fe protein. On the other hand, the number and arrangement of conserved Cys residues (being more than needed to coordinate one [2Fe-2S] cluster) and the close phenotypical connection to NifS suggested that NifU could instead be involved in sequestering Fe and S for nitrogenase [Fe–S] cluster formation.80
The [2Fe-2S]2+/1+ clusters attached to as-isolated NifU were resistant to Fe chelation,80 indicating that these clusters are of a “permanent” nature and likely important for NifU activity, but not precursor clusters destined for nitrogenase. On the basis of sequence comparison to other [2Fe-2S] cluster containing proteins, the four Cys residues located at the middle of the NifU primary sequence (Cys137, Cys139, Cys172, Cys175) were proposed to bind the permanent cluster, leaving three Cys residues (Cys35, Cys62, Cys106) in the N-terminal part of the protein, and two Cys residues (Cys272, Cys275) at the C-terminal part, available for transient Fe coordination80 (Figure 6A). Substituting any of the first seven NifU Cys residues to Ala affected diazotrophic growth in A. vinelandii, while Cys272 and Cys275 appeared as not important for NifU functionality.82 Purification of each of these nine Cys-mutated NifU variants from recombinant E. coli confirmed that Cys137, Cys139, Cys172, and Cys175 provided ligands for the permanent [2Fe-2S]2+/1+ cluster. The modular nature of the NifU protein was further confirmed by expressing and purifying two truncated variants: NifU-1, the IscU-like N-terminal fragment of NifU encompassing residues 1–131 with the conserved Cys35, Cys62, and Cys106 residues; and NifU-2 starting at residue 126 with the permanent [2Fe-2S]2+ cluster coordinated by Cys137, Cys139, Cys172, and Cys175 together with C-terminal residues Cys272 and Cys275 (Figure 6B). NfU-2 generated spectral characteristics similar to the full-length protein, confirming that the initial three Cys residues (Cys35, Cys62, Cys106) were not involved in the coordination of the [2Fe-2S]2+/1+ cluster.82 Consistently, site-directed mutagenesis of NifU-1 residues Cys35, Cys62, and Cys106 suggested they were rather involved in transient binding of Fe destined for nitrogenase [Fe–S] cluster formation.
Figure 6.
Modularity of NifU structure and function. (A) A. vinelandii NifU domain architecture and conserved Cys residues (Uniprot C1DH18). Three distinct NifU domains are shown in green, pink, and blue, and their roles in coordinating permanent or transient [Fe–S] clusters are indicated. (B) Alignment of full-length NifU with the NifU-1 and NifU-2 truncated variants and with the homologous proteins IscU (Uniprot C1DE67) and human NfU (Uniprot C1DLW0). Conserved domains are color coded. % Identity and similarity between NifU and NfU or IscU are shown to the right. (C). Structural model of the N-terminal domain of NifU generated with Swiss-Model.87 The protein pocket with ligands to a transient [2Fe-2S] cluster is magnified to the right of the structure. Graphics generated with the PyMOL Molecular Graphics System, Version 2.3.2 Schrödinger, LLC. (D) Proposed model for NifS-mediated assembly of [4Fe-4S] clusters in the N-terminal and C-terminal domains of the NifU.88
Later studies showed that human Nfu, a [Fe–S] cluster scaffold protein with significant sequence similarity to the C-terminal domain of NifU (Figure 6B), could assemble one labile [4Fe-4S] cluster per Nfu dimer in vitro.83 While Cys272 and Cys275 mutations in A. vinelandii did not significantly affect diazotrophic growth,82 they could still provide a second assembly site for transient [Fe–S] clusters and explain why NifU proteins with mutations at the N-terminal Cys35, Cys62, and Cys106 residues supported some growth under N2-fixing conditions. In support of this hypothesis, it was observed that replacing any or all of the three N-terminal Cys35, Cys62, or Cys106 residues with Ala affected diazotrophic growth to the same extent. However, if Cys275 was also changed by mutation, a significantly more severe growth phenotype was observed, indicating that the N-terminal and C-terminal domains of NifU to some extent have independent but additive functions.55
4.4. NifS-Mediated Assembly of Transient [Fe-S] Clusters at NifU
Using catalytic amounts of NifS together with l-Cys, ferric (III) iron, and β-mercaptoethanol (β-ME), the formation of an extremely reductant-sensitive [2Fe-2S] cluster could be observed at the N-terminal part of NifU (one [2Fe-2S] per dimer).84Figure 6C shows a model of the N-terminal half of NifU coordinating this [2Fe-2S] cluster. NifS was subsequently found to also catalyze the formation of one [4Fe-4S]2+ cluster at the IscAnif homodimer, a nif-specific homologue of IscA located immediately upstream of nifU, which could provide an alternative scaffold for the assembly of nitrogenase-destined [Fe–S] clusters.85 Interestingly, the formation of the IscAnif [4Fe-4S]2+ cluster was also time-dependent and formed via an intermediate [2Fe-2S]2+ cluster. Similar results had also been observed using the homologous IscS and IscU proteins of A. vinelandii, where IscU is homologous to the N-terminal part of NifU with the three conserved Cys35, Cys62, and Cys106 residues.14 Cluster assembly in IscU, monitored via anaerobic anion exchange chromatography that allowed for a more detailed time-course study and avoided the formation of iron sulfides, showed that also IscS-mediated cluster assembly proceeded in sequential steps with one [2Fe-2S]2+ per dimer followed by two [2Fe-2S]2+ per dimer and finally one [4Fe-4S]2+ per dimer.86
Finally, NifS-mediated assembly of [4Fe-4S]2+ clusters at NifU was also shown.88 Careful analysis of the NifU cluster assembly using the two truncated NifU variants, NifU-1 and NifU-2, showed that both cluster assembly domains could transfer [4Fe-4S]2+ clusters to, and thereby activate, apo-Fe protein. While one [4Fe-4S]2+ cluster per NifU monomer was rapidly factored at the C-terminal Nfu-type domain (likely without [2Fe-2S] cluster intermediates), [4Fe-4S]2+ assembly at the N-terminal IscU-like domain was slow and progressed via [2Fe-2S]2+ clusters (Figure 6D).88
The reason for NifU having two scaffold sites is not clear. It could reflect the large number of nitrogenase components requiring [Fe–S] clusters for their activity and, therefore, the huge demand for [Fe–S] cluster formation. It could also be that each [Fe–S] cluster scaffold targets distinct proteins. Other explanations could be that the different [Fe–S] scaffolds are involved either in de novo cluster synthesis or in the reassembly of damaged clusters, respectively, or even that they show different activities under distinct metabolic conditions. For example, IscAnif was found capable of accepting [4Fe-4S]2+ clusters from NifU and to activate apo-Fe protein in vitro but also to cycle between forms containing one [2Fe-2S]2+ or one [4Fe-4S]2+ per homodimer in response to O2 exposure and DTT-induced two-electron reductive coupling.89 In this regard, it is interesting to note that, only if cells were cultured under low-O2 conditions, normal levels of NifU were capable of replacing the function of IscU in vivo.77 In this regard, the rescue of IscU ablation by NifU required the N-terminal domain of NifU, while Cys275 at its C-terminal scaffold domain was not important,77 possibly indicating that the function of the C-terminal part of NifU is more specialized.
4.5. NifS and NifU Transfer of [4Fe-4S] Cluster to Fe Protein
As extracts of A. vinelandii with disrupted nifU and nifS genes almost completely lacked Fe protein activity,39 and as functional Fe protein requires a subunit-bridging [4Fe-4S] cluster5 (Figure 1), it seemed reasonable that NifU and NifS were the sources of the Fe protein [4Fe-4S] cluster. To test this hypothesis in vitro, NifU and NifS were expressed in E. coli at equimolar ratios. Purified NifU and NifS proteins were then incubated with ferrous (II) ion, Cys, β-ME, and [4Fe-4S] cluster-free apo-Fe protein prepared by Fe chelator-treatment.55 Under these conditions, about 90% Fe protein activation could be achieved. Specificity of the reaction was shown by the inability of the Cys325-mutated NifS to activate Fe protein. The fact that the initial cluster was assembled at the NifU scaffold became evident from reactions where NifS was first used to build cluster on NifU and then removed before the NifU-mediated apo-Fe protein activation.55 NifU protein where the conserved Cys35, Cys62, and Cys106 residues were replaced by Ala was still capable of activating the apo-Fe protein, albeit at a higher NifU ratio, corroborating the ability of NifU with mutated N-terminal Cys residues to support diazotrophic growth. Additional mutation of the C-terminal Cys275 residue abolished in vitro Fe-protein activation by NifU and confirmed redundancy in the function of the N- and C-terminal NifU [Fe–S] cluster assembly sites. Later work showed that [4Fe-4S]2+ clusters assembled at both the N- and C-terminal domains of NifU could be transferred to the apo-Fe protein.88 Importantly, cluster transfer appeared to be immediate and to not require additional accessory proteins.
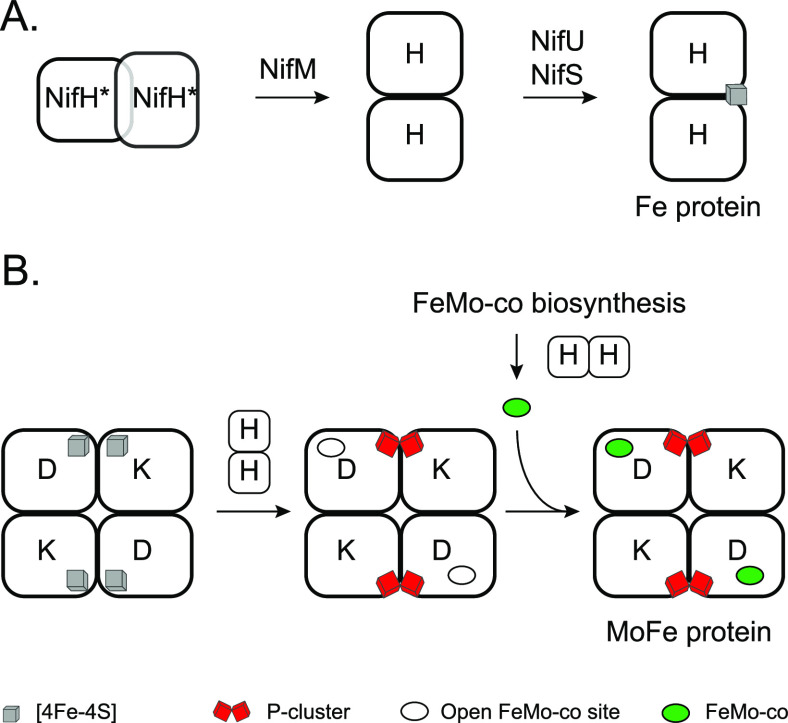
5. Fe Protein Maturation
Fe protein maturation is divided into two processes: the correct folding and formation of the NifH homodimer, and the acquisition of the [4Fe-4S] cluster to generate active Fe protein (Figure 7A). As [4Fe-4S] cluster acquisition has been described in Section 4, here we will focus on NifH folding and the role that NifM plays on it. It is important to note that the Fe protein has at least three roles in the Mo-nitrogenase system: (1) a dinitrogenase reductase role that requires fully mature [4Fe-4S] cluster-containing Fe protein; (2) a role in the formation of the P-clusters in complex with the MoFe protein; and (3) a role in the biosynthesis of FeMo-co in complex with NifEN, a paralog of the MoFe protein (Figure 7B). Although controversial, there is biochemical evidence supporting that [4Fe-4S] cluster-deficient apo-Fe protein functions in P-cluster and FeMo-co biosynthesis.48,90
Figure 7.
Fe protein maturation and the roles of the Fe protein in MoFe protein maturation. (A) Simplified model for the roles of NifM, NifU and NifS in Fe protein maturation. (B) Requirement of Fe protein for MoFe protein maturation. MoFe protein variants from left to right correspond to the ΔnifH apo-MoFe protein, the ΔnifB apo-MoFe protein, and the holo-MoFe protein. Fe protein is shown as NifH homodimer lacking [4Fe-4S] cluster to indicate that apo-Fe protein is competent in P-cluster formation and in FeMo-co biosynthesis.
5.1. Role of NifM
Early work using K. pneumoniae mutants showed that extracts prepared from nifM– strains exhibited negligible activity of Fe protein and about 10% of MoFe protein activity compared to the wild type.44 The leaky phenotype for the MoFe protein could indicate that some P-cluster maturation and FeMo-co synthesis happened in vivo in the nifM mutants. However, it could also be an experimental artifact due to some in vitro MoFe protein maturation by Fe protein added during the acetylene reduction assay. The nifM mutants accumulated low, but significant, amounts of Fe protein polypeptide and normal amounts of MoFe protein polypeptides.44 The NifM dependency of Fe protein was later experimentally tested in a heterologous expression system in Escherichia coli. The Fe protein produced in the absence of NifM was much less stable and accumulated as completely inactive protein.91 It was proposed that NifM would be involved in [4Fe-4S] cluster insertion in the Fe protein. Given the enormous difference in expression levels between NifH and NifM in a N2-fixing bacterium, it was proposed that the action of NifM over NifH would be catalytic.
A study to define K. pneumoniae genes necessary to produce not only Fe protein but also apo-MoFe protein with mature P-cluster but lacking FeMo-co, confirmed that active Fe protein required coexpression of nifH and nifM.92,93 However, inactive Fe protein produced by cells lacking nifM was sufficient to produce significant amount of apo-MoFe protein activatable by FeMo-co, which suggested that P-cluster maturation at the MoFe protein did not strictly require active Fe protein but only the structural NifH polypeptide.93 It should be noted, however, that as the FeMo-co insertion assay was followed by addition of purified K. pneumoniae Fe protein, it is difficult to exclude that some apo-MoFe protein formation (P-cluster maturation) happened in vitro by the exogenously added Fe protein.
The Nif– phenotype of nifM mutants was later confirmed in an A. vinelandii strain with a deleted nifM gene. This strain contained negligible levels of Fe protein activity and 30% MoFe protein activity compared to wild type.39 In contrast to the phenotype in K. pneumoniae,91 NifH polypeptide accumulation appeared not to be affected in the A. vinelandii nifM deleted strain although polypeptide turnover measurements were not carried out to determine protein stability.39 MoFe protein activity was increased 30% by the addition of exogenous FeMo-co, which indicates FeMo-co limitation in the extract and suggests that FeMo-co synthesis might be affected in nifM mutants. However, like in the work with K. pneumoniae proteins discussed above, some P-cluster maturation could have taken place in vitro during the assay. Although these studies presented compelling data for the necessity of NifM to mature the Fe protein, the mode of action for the distinct NifM proteins or the physical effects on their NifH targets appeared different. For instance, while the lack of K. pneumoniae NifM protein affected both Fe protein stability and dimerization neither of those appeared affected in the A. vinelandii nifM mutant strain.
The importance of NifM to produce active Fe protein has also been shown in heterologous expression in yeast. Direct physical interaction between the nifH gene product from Rhizobium meliloti and the nifM product from K. pneumoniae was established using a yeast two-hybrid system.94 It was also shown that coexpression with NifM was required for R. meliloti Fe protein dimer formation and NifH polypeptide stability in yeast.95 Recently, coexpression of the A. vinelandii nifM and nifH genes and targeting of their products to the matrix of mitochondria rendered Fe protein active as electron donor to the MoFe protein.96
5.2. Proposed Function of NifM in Fe Protein Maturation
Analysis of the A. vinelandii NifM sequence identifies a PpiC-type domain significant of peptidyl-prolyl cis–trans isomerases (PPIase),97,98 a group of enzymes that accelerate protein folding by catalyzing the cis–trans isomerization of proline imidic peptide bonds. By examining the consensus sequence of 60 different nifH gene products, seven conserved Pro residues were identified.99 Mutagenesis of the Pro residues in the A. vinelandii Fe protein showed that exchanging Pro258 for Ser converted the Fe protein into a NifM-independent variant and suggested that this residue could act as substrate for NifM PPIase activity. Pro258 is located at the C-terminus of the Fe protein in a stretch that wraps around the other subunit of the homodimer, potentially explaining the importance of NifM on Fe protein function (Figure 8).99 In this regard, the reported leaky phenotype of nifM mutants could be explained by nonspecific isomerization of the target Pro residue at the Fe protein, or by the activity of other isomerase(s) at lower rates, resulting in low amounts of functional Fe protein.
Figure 8.
Location of Pro258 in a top-view structure of the A. vinelandii Fe protein (PDB 1FP6). The two monomers, the central [4Fe-4S] cluster, and the two nucleotides are easily identified. Graphics generated with the PyMOL Molecular Graphics System, Version 2.3.2 Schrödinger, LLC.
In addition to its requirement for the Fe protein of the Mo-nitrogenase, NifM appears to be required for the Fe proteins of the alternative nitrogenases as A. vinelandii nifM mutants did not show diazotrophic growth in Mo-deficient medium.53 In support of the proposed function for NifM, Pro residues corresponding to Pro258 are also present in the A. vinelandii AnfH and VnfH proteins. However, more recent work has shown that NifM was not essential to produce functional FeFe-nitrogenase in an engineered E. coli strain carrying all necessary anf genes.100 It should also be noted that not much recent research has been reported regarding NifM. Examples of open questions are how the Fe protein is matured in organisms lacking nifM, or how an Fe protein with another structural architecture (tetramer) is formed.101 Another interesting topic is what evolutionary benefit NifM-dependence could provide if mutating a certain Pro residue of the Fe protein would render the protein to be NifM-independent. It is therefore probable that the proposed model for the NifM mechanism is not complete and that it requires more attention.
6. Interaction of Maturation Factors with Cofactor Deficient MoFe Protein
The maturation of the MoFe protein is a sequential process driven by the stepwise action of accessory proteins that interact with and convert catalytically inactive NifD and NifK polypeptides into a MoFe protein tetramer equipped with two P-clusters and two FeMo-co molecules. The MoFe protein has a symmetric α2β2 structure,6 and because each αβ subunit pair of the protein has one P-cluster and one FeMo-co, the cluster composition is heterogeneous and different along a very complex process of polypeptide and prosthetic group maturation. One important difference between the acquisition of P-cluster and FeMo-co is that the first one is matured in situ, that is, from precursor clusters that are already bound to the NifDK polypeptides while the latter is assembled elsewhere (see below) and then inserted into a P-cluster containing form of apo-MoFe protein.102
MoFe protein maturation can be rationalized by dividing it into stages defined by the properties of the MoFe protein variants that have been isolated from A. vinelandii strains with mutations in genes essential to P-cluster or FeMo-co biosynthesis (Figure 7B).103 First, apo-MoFe protein lacking both the P-cluster and FeMo-co (but containing P-cluster precursors) is isolated from strains with nifH mutations.104−106 This protein cannot be activated by the simple addition of FeMo-co and requires prior maturation of its P-clusters.107,108 Second is apo-MoFe protein that contains mature P-clusters but lacks FeMo-co and can therefore be activated in vitro by the simple addition of pure FeMo-co.56,109,110 This form is isolated from strains with mutations in nifB, nifE, or nifN specifically disrupting FeMo-co biosynthesis and is typically used in the in vitro FeMo-co synthesis and insertion assays because it is the more stable apo-MoFe protein variant and does not require in vitro P-cluster maturation. Third, mature and functional protein equipped with two pairs of P-clusters and FeMo-co (holo-MoFe protein or simply MoFe protein). As described below, even more complex and heterogeneous apo-MoFe protein populations are found in strains with mutations affecting nonessential gene products involved in MoFe protein maturation.
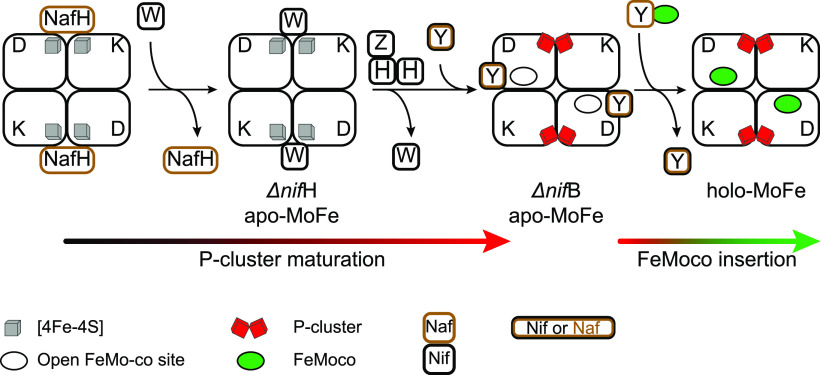
Important to note is that most studies have used MoFe protein variants isolated under conditions that disrupted the labile interactions between immature apo-MoFe protein and its maturation factors. Recent use of milder Strep-tag affinity chromatography (STAC),111 which enables protein purification avoiding metal-affinity resins that can remove labile nitrogenase [Fe–S] clusters (that can be important for nitrogenase protein functions/interactions) and that does not require the use of the organic compound imidazole for protein elution (that can disrupt nitrogenase protein interactions),112−114 has permitted the isolation and reevaluation of complexes of the MoFe protein and these factors.23 The rationale of this work was that proteins specifically trapped at certain stages of apo-MoFe protein maturation should be involved in prior maturation steps. For instance, NifW, NifZ, and NafH copurified with MoFe protein from an A. vinelandii ΔnifH strain that is impaired in P-cluster maturation, suggesting their involvement in P-cluster maturation or in a preceding reaction. By deleting each one of these genes individually, a NafH–NifW–NifZ stepwise sequence of interactions preceding the binding of Fe protein was firmly established (Figure 9).23 This sequence of interactions also reflects their relative positions within the A. vinelandii chromosome (Figure 2). Neither NafH, NifW, nor NifZ was bound to the P-cluster containing, but FeMo-co deficient, apo-MoFe protein isolated from cells with impaired FeMo-co synthesis (ΔnifB or ΔnifE strains).23,114 Instead, two other accessory proteins, NifY and NafY, were found. This observation suggests that NifZ dissociation, the last protein in the above stepwise interaction sequence, is followed by P-cluster maturation, which triggers NafY/NifY binding. NifY binding to apo-MoFe protein was unexpected as no mutant phenotype has yet been shown for a ΔnifY A. vinelandii strain (see Section 15.2).
Figure 9.
Sequential and differential interaction of NafH, NifW and NifZ maturation factors with apo-MoFe protein. From left to right, three steps of MoFe protein maturation related to P-cluster formation and one step for the insertion of FeMo-co are shown. NifZ aids the Fe protein in P-cluster formation. NafH and NifW interaction with the MoFe protein precedes the NifH/NifZ reaction. Formation of the P-clusters releases NifW, changes apo-MoFe protein conformation to make the FeMo-co sites accessible, and promotes the binding of the NifY or NafY factors. FeMo-co insertion releases NifY/NafY and generates holo-MoFe protein.
These accessory proteins could also be used as baits to capture their corresponding apo-MoFe protein isoform by using affinity chromatography. The trapped apo-MoFe proteins, likely representing snapshots at distinct steps of the maturation process, could be characterized. For example, apo-MoFe proteins containing P-cluster precursors at different degree of maturation could be isolated using NifW (see below). Unfortunately, the roles of NifW and that of the recently identified NafH factor remain unknown. In contrast, the population of apo-MoFe protein that was captured by either NafY or NifY was homogeneous with mature P-clusters but no FeMo-co.23 Interestingly, although NafY and NifY share a high degree of primary structure similarity, they did not coexist and were found at distinct apo-MoFe protein molecules, suggesting that they have different roles in the maturation of A. vinelandii MoFe protein.
7. Formation of MoFe Protein P-Clusters
The current model for the maturation of the A. vinelandii apo-MoFe protein up to the stage containing mature P-clusters involves the participation of seven proteins: the Fe protein, NifZ, NifU, NifS, NifM, NifW, and NafH. From this group of proteins, only the Fe protein has been conclusively demonstrated to be essential for P-cluster maturation.23,104,107,108,115−117
7.1. NifU and NifS
The transformation of simpler [Fe–S] precursors into the P-cluster occurs in situ at the MoFe protein. These precursors are believed to be [4Fe-4S] clusters delivered by NifU, although direct involvement of NifU and NifS has not been strictly demonstrated.39,55,79,84 Disruption of nifU or nifS genes in A. vinelandii or K. pneumoniae showed their essentiality for N2 fixation as they almost completely abolished Fe protein activity and seriously impaired MoFe protein activity.38,39,44 It is possible, but not yet shown, that a ΔnifUS strain would produce a MoFe protein devoid of Fe–S clusters. It may seem difficult to separate direct effects of nifU and nifS mutations on the MoFe protein from those derived from the production of [4Fe-4S] cluster-deficient Fe protein. However, it must be noted that two independent reports showed that [4Fe-4S] cluster-deficient Fe protein is able to mature apo-MoFe protein,48,90 strongly suggesting direct requirement for NifU and NifS in MoFe protein maturation. Detailed analysis of apo-MoFe protein isolated from a ΔnifH ΔnifUS mutant strain would clarify this point.
7.2. Fe Protein Is Required for P-Cluster Formation
The requirement of Fe protein for the maturation of MoFe protein was shown in extracts of A. vinelandii nifH mutant strains. These mutants lacked both Fe protein and MoFe protein activity and accumulated apo-MoFe protein with α2β2 tetramer composition that, in contrast to the α2β2γ2 apo-MoFe hexamers present in extracts of nifB or nifE mutant strains (see Section 15.2 for details on the identity of the γ subunit),56,109 could not be activated by the simple addition of FeMo-co. Incubation of nifH mutant extracts with Fe protein and MgATP under DTH reducing conditions was required prior to activation by FeMo-co insertion.107,108 It was proposed that FeMo-co binding sites would be unavailable prior to P-cluster maturation, and, after a conformational change driven by P-cluster formation, they would be exposed and ready for FeMo-co binding. Both events are supported by additional experimental evidence. First, apo-MoFe with mature P-clusters have the FeMo-co-binding α-Cys275 residue exposed to solvent, in contrast to the apo-MoFe protein with immature P-clusters.118 Second, small-angle X-ray scattering (SAXS) suggests that P-cluster formation induces a major conformational change that brings together the α and β subunits.119 Interestingly, it has been recently shown that VnfH was able to replace, to some extent, Fe protein function in apo-MoFe protein maturation in vivo.23
The construction of a new A. vinelandii ΔnifH strain in which the apo-MoFe protein was equipped with a His-tag to facilitate fast isolation of high quality protein enabled more detailed spectroscopy and biochemical analyses.104 The ΔnifH apo-MoFe protein lacked EPR signals from FeMo-co (S = 3/2; g = 4.3, 3.6, and 2.01) and the P-cluster (g = 11.8 in parallel mode) but presented a novel S = 1/2 EPR signal in the g = 2 region proposed to originate from a pair of [4Fe-4S] clusters that would serve as P-cluster precursor. The structure and electronic properties of the metal clusters in ΔnifH apo-MoFe have also been investigated by extended X-ray absorption fine structure (EXAFS)120 and VT-MCD,121 which concur with EPR assignments. Incubation with the Fe protein, DTH, and MgATP caused disappearance of the S = 1/2 signal and appearance of the P-cluster signal.117 On the basis of the dynamics of EPR signal changes, it was suggested that the first P-cluster of each tetramer was rapidly matured, while maturation of the second was slow. As detailed below, this hypothesis has been challenged by new data.116
It was also possible to mature the P-clusters in the absence of Fe protein using the strong reductant Ti(III) citrate.122 This was achieved by briefly incubating ΔnifH apo-MoFe protein with Ti(III) citrate, which produced disappearance of the EPR signal from the P-cluster precursor and subsequent oxidation with indigo disulfonate (IDS) leading to the formation of P-clusters. Involvement of all ferrous [4Fe-4S]0 intermediates for P-cluster formation was proposed. However, given the very reducing conditions of this study (12 mM Ti(III) citrate), the physiological relevance of its results needs to be interpreted with care.
Despite substantial progress in understanding P-cluster maturation, it is important to note that the exact Fe protein role in this process is still unknown. Several site-specific Fe protein mutant variants with deficiencies in MgATP binding or hydrolysis, or in complex formation or electron transfer to the MoFe protein, were investigated for their capacity to drive P-cluster formation.123 The results of these experiments suggested that MgATP binding and interaction with apo-MoFe protein were the only properties essential to P-cluster maturation. Neither MgATP hydrolysis nor the presence of [4Fe-4S] cluster in the Fe protein was essential to this process in vitro.90,123 However, a different study using only purified proteins showed a strict dependence for P-cluster maturation on ATP hydrolysis since site-directed Fe protein mutants defective in ATP hydrolysis, or the use of nonhydrolyzable ATP analogs, prevented P-cluster formation.9 It is unclear whether these opposing results was due to different assay conditions, for example, the genetic background the MoFe protein or the use of purified protein components instead of cell-free extracts.
7.3. NifZ Is Involved in P-Cluster Formation
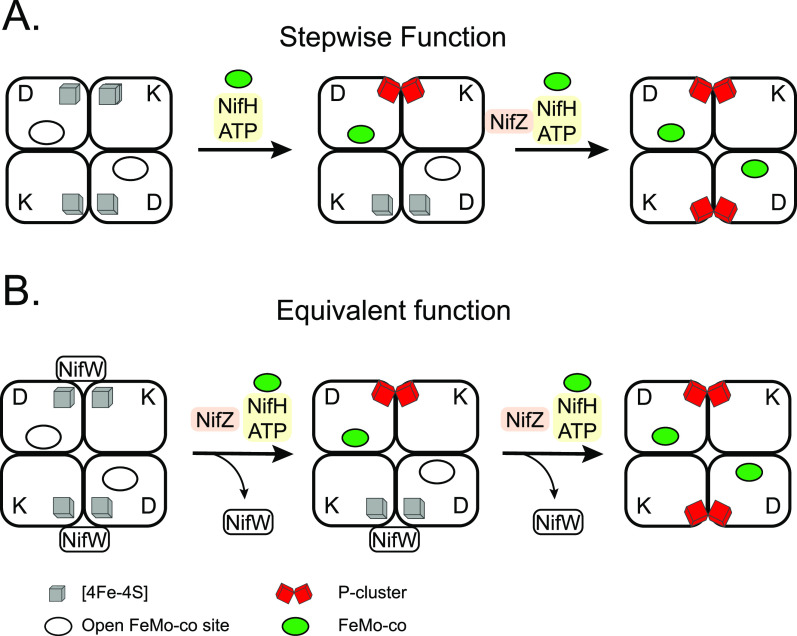
Deletion of nifZ in A. vinelandii had no effect on Fe protein activity but decreased MoFe protein activity by 66%. Addition of FeMo-co to extracts of a ΔnifZ strain did not activate MoFe protein,39 suggesting a role for NifZ in P-cluster formation or in earlier stage. Two different models have been put forward to explain the role of NifZ (Figure 10).
Figure 10.
Models for the participation of NifZ in P-cluster maturation. (A) In the stepwise model, the first P-cluster of each tetramer is matured by the Fe protein alone, while both NifZ and the Fe protein are essential for the maturation of the second P-cluster. (B) In the equivalent function model, NifZ participates in the formation of both P-clusters, but it is not essential for either process. In both models, ATP is required for P-cluster formation. The equivalent model incorporates information about the release of NifW upon P-cluster maturation. Figure adapted from ref (116). Copyright 2019 ASBMB under CC BY 4.0 http://creativecommons.org/licenses/by/4.0/.
7.3.1. Model 1: NifZ Is Only Required for Maturation of Second P-Cluster in Each Apo-MoFe Protein Molecule
His-tagged MoFe protein isolated from an A. vinelandii nifZ mutant exhibited partial activity and mixed EPR signals arising from FeMo-co, P-cluster, and precursors to the P-cluster.115 These results were interpreted as if one αβ half of the MoFe protein was completely mature while the other half was locked at the stage prior to P-cluster formation. Further, apo-MoFe protein from a ΔnifZ ΔnifB double mutant lacked FeMo-co EPR signals (as expected due to deletion of nifB) and maintained EPR signals both from P-cluster precursors and mature P-cluster. The absence of FeMo-co in ΔnifB ΔnifZ apo-MoFe protein facilitated its investigation by VT-MCD, which supported the conclusions obtained from EPR analysis.124 It followed that the in situ assembly of the P-clusters occurred stepwise and was controlled by NifZ, which would be a chaperone that induced a conformational change required for the formation of the P-cluster in the second half of apo-MoFe protein. Consistent with this model, only the concerted action of both the NifZ and the Fe protein produced apo-MoFe protein with both P-clusters matured (Figure 10A).
The order in which NifZ and the Fe protein acted was inferred by breaking up the in vitro P-cluster maturation assay into two phases. First, ΔnifB ΔnifZ apo-MoFe protein was incubated with NifZ alone. Then apo-MoFe protein exposed to NifZ was reisolated and incubated with the Fe protein and MgATP under reducing conditions. Performing the assay in that order, but not in the reverse, generated apo-MoFe protein with mature P-clusters capable of almost full activation by FeMo-co, indicating that the action of NifZ preceded that of the Fe protein.9 The disappearance of the S = 1/2 signal associated with P-cluster precursors during the first 15 min of incubation and the increase of the g = 11.8 EPR signal during the first hour of incubation was interpreted as resulting from time-dependent maturation of the P-cluster.
7.3.2. Model 2: NifZ Is Involved in Maturation of Both P-Clusters
The stepwise Model 1 assumes that in a ΔnifZ strain, the MoFe protein is homogeneous and locked in a state having only one mature P-cluster. However, heterogeneous MoFe protein populations were recently identified using different methodology. Namely, careful analysis of MoFe protein isolated from ΔnifZ strains using STAC methodology that better preserves weak protein interactions and [Fe–S] clusters identified variants that included, at a minimum, MoFe protein with both halves containing only P-cluster precursors, MoFe protein having in one-half only P-cluster precursors and in the other half P-clusters and FeMo-co, and MoFe protein replete with FeMo-co and mature P-cluster.116 In addition, NifW and NafH were found associated with the preparations of purified MoFe protein. The findings of this study, which are described in detail below, revealed a continuous process strongly supporting a model in which NifZ has equivalent function in the maturation of both P-clusters (Figure 10B). It follows that the stepwise model represents only a discrete snapshot rather than the overall process.
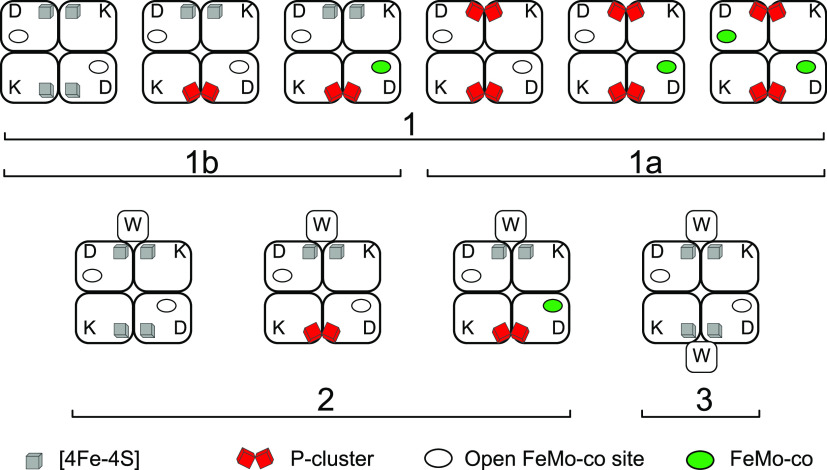
Both his-tagged and strep-tagged MoFe protein variants purified from ΔnifZ strains were heterogeneous as shown by further fractionation by anion exchange chromatography. Attachment of different amounts of the acidic protein NifW to the MoFe protein determined the charge of the complex and hence column retention time (Figure 11). Fraction 1 contained MoFe protein not associated with NifW. Its specific activity was close to 50% of the MoFe protein isolated from wild-type cells, and it showed EPR signals arising from FeMo-co and P-cluster precursors. This fraction contained the MoFe protein forms described in previous work.115 Fraction 2 contained MoFe protein with NifW bound at 1:1 ratio, its specific activity was 25% of wild-type, and it presented EPR signals arising from FeMo-co and P-cluster precursors. The third Fraction contained inactive MoFe protein:NifW complex at 1:2 ratio, showed strong EPR signal arising from P-cluster precursors and very weak FeMo-co signal. Such activities and spectroscopic properties resembled the ΔnifH MoFe protein.104 Fraction 1 was further separated into two subfractions using excess NifW as bait for affinity purification of the MoFe proteins.116 The subfraction bound to the NifW bait showed much lower activity and EPR signals from FeMo-co and P-cluster precursors (Figure 11, fraction 1b). However, the subfraction rejecting the NifW bait was highly enriched in fully mature MoFe protein (Figure 11, fraction 1a), demonstrating that NifZ was not strictly required to mature the second P-cluster, as proposed in Model 1.
Figure 11.
Theoretical apo-MoFe variants and holo-MoFe protein that could accumulate in an A. vinelandii ΔnifZ strain. Chromatography fractions and subfractions in which these proteins were identified are number as 1, 1a, 1b, 2, and 3 (see text). Legend indicates the presence of [4Fe-4S] P-cluster precursors, FeMo-co, and the NifW maturation factor. Figure adapted from ref (116). Copyright 2019 ASBMB under CC BY 4.0 http://creativecommons.org/licenses/by/4.0/.
In Model 2, NifZ aids in the maturation of both P-clusters to the same extent but is not essential to any of them. Consistently, the levels of mature MoFe protein in the ΔnifZ strain slowly increased over time following nitrogenase derepression. In support of this model, it was shown that supplementing ΔnifZ cell extracts only with MgATP and DTH resulted in time-dependent and NifZ-independent P-cluster maturation and MoFe protein activation.116 Surprisingly, this result differs from a previous study showing absolute NifZ dependency of Fe protein driven P-cluster maturation.9 Perhaps some additional factors present in the ΔnifZ cell extracts, for example, NifW, would aid in NifZ-independent maturation.
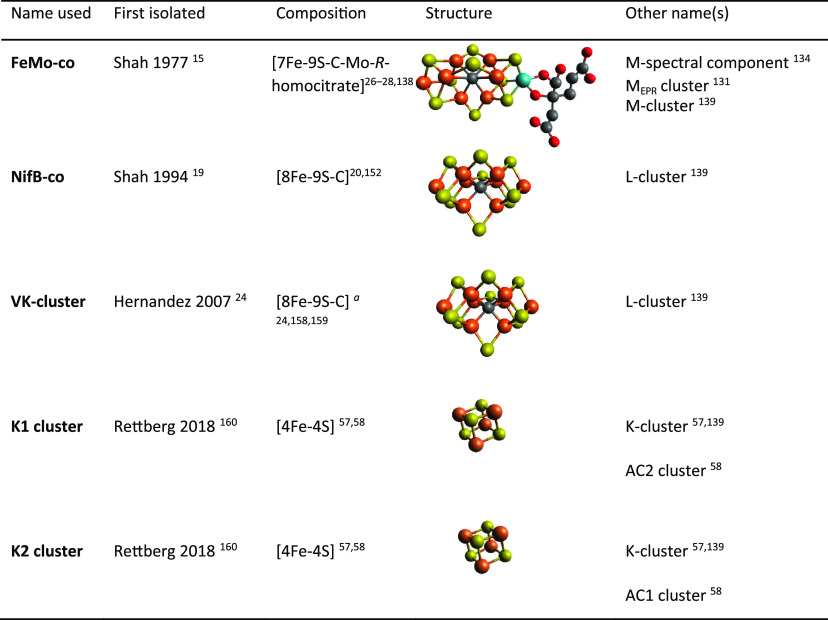
8. FeMo-co: Description of the Cofactor and Methods to Measure Its Biosynthesis
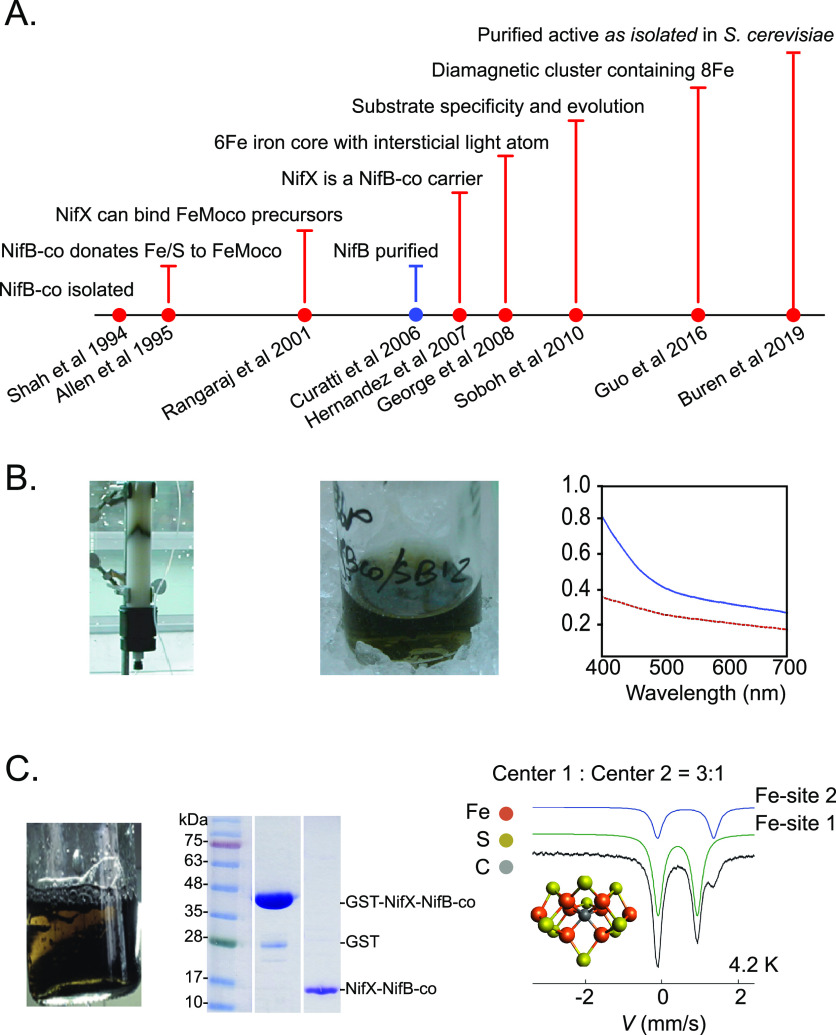
8.1. Discovery and Isolation of FeMo-co
Before its isolation and characterization FeMo-co was reported as an “activating factor” capable of reconstituting inactive nitrogenase produced in A. vinelandii UW45,125,126 a strain carrying the nif-45 mutation later shown to locate to the nifB gene.40 UW45 accumulated active Fe protein but inactive MoFe protein.126,127 Activation of MoFe protein in UW45 extracts was achieved by adding the activating factor extracted from acid-denatured MoFe protein,126 using a method analogous to the extraction of molybdenum cofactor (Mo-co) from xanthine oxidase.128 Acid-treated MoFe protein was completely inactivated after removal of its activating factor. The activating factor could equally be obtained from Clostridium pasteurianum, K. pneumoniae, or Rhodospirillum rubrum MoFe proteins.126 Similar results were obtained with the UN106 strain of K. pneumoniae (carrying the nif-4106 mutation later described to locate to the nifB gene129). UN106 showed expression of active Fe protein but inactive MoFe protein that could be converted into functional protein in vitro by the activating factor.130
The reconstitution of inactive MoFe protein in extracts of UW45 was used as proxy to isolate the activating factor to homogeneity.15 Purified factor contained Fe and Mo at an estimated ratio of 8 to 1 (an accurate initial estimate compared the real 7 to 1 ratio) and was given the name iron–molybdenum cofactor or FeMo-co. Isolated FeMo-co did not reduce acetylene unless incorporated into MoFe protein. Activation by FeMo-co followed a saturation kinetics with almost 100% MoFe protein reconstitution. FeMo-co was extremely oxygen labile, completely losing activity after 1 min exposure to air. It was also unstable in protic solvents such as water, lasting only a few hours in aqueous buffer under anaerobic conditions. A method to isolate FeMo-co into anaerobic N-methyl formamide (NMF) following acid denaturation of MoFe protein was developed to stabilize the cofactor, which kept full activity after 10 days of anaerobic storage in NMF. In addition to the A. vinelandii MoFe protein, FeMo-co was isolated from K. pneumoniae, C. pasteurianum, R. rubrum, and Bacillus polymyxa, and it was determined that all FeMo-cofactors were very similar or identical based on metal content, activity,15 and EPR spectra.131 A modified method to obtain FeMo-co preparations at large scale has also been reported.132
Initially, there was confusion about commonality of molybdenum cofactors from different molybdoenzymes such as nitrogenase, xanthine oxidase, and nitrate reductase. Pienkos et al. used the same NMF-based method to extract FeMo-co from highly purified MoFe protein and Mo-co from xanthine oxidase.133 It was shown that FeMo-co could activate inactive MoFe protein in extracts of A. vinelandii UW45 but not nitrate reductase in extracts of Neurospora crassa Nit1–1, whereas Mo-co could activate nitrate reductase but not the inactive MoFe protein. It was concluded that Mo-co and FeMo-co were different cofactors. Interestingly, it was reported that A. vinelandii synthesized both Mo-co and FeMo-co when cells were fixing N2, an early observation of a peculiar property of A. vinelandii, which can simultaneously synthesize nitrogenase and nitrate reductase (see Section 11.2).
FeMo-co solutions are greenish brown and distinct from simpler Fe–S clusters and exhibit a nondescript spectrum with a steady decrease in absorbance from 400 to 700 nm. Exposure to air destroys the cofactor and decreases absorbance. The EPR spectrum of DTH-reduced FeMo-co corresponds to a S = 3/2 center with g values (gy = 4.6, gx = 3.3, gz = 1.93) and line shape similar to the M spectral component of the MoFe protein,134 although considerably broader due to differences in the ligand environment.131 Incubation of FeMo-co with ligands135 or with the FeMo-co binding protein NafY136 narrow the EPR signal resembling that of FeMo-co in MoFe protein. The EPR signal changes observed in MoFe protein under turnover conditions are attributed to FeMo-co, and additional evidence that FeMo-co constitutes the active site of the enzyme.137
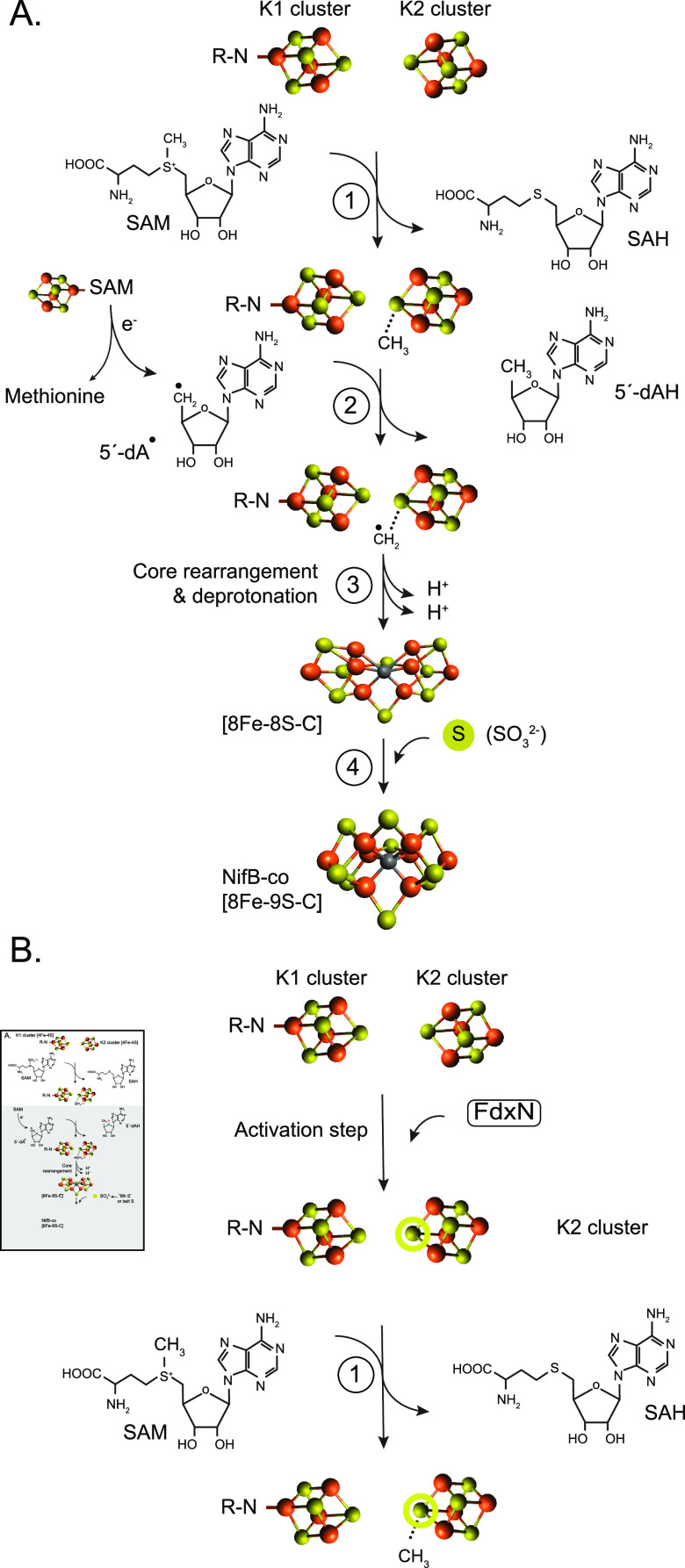
FeMo-co was considered a completely inorganic cofactor until homocitrate was identified as integral part of the cofactor many years later.18 The atomic structure of FeMo-co was finally solved along with that of the MoFe protein by X-ray crystallography.28 FeMo-co was best described as a [4Fe-3S] cluster and a [Mo-3Fe-3S] cluster bridged by a belt of 3 equivalent S atoms. The molecule of (R)-homocitrate was bound the Mo atom by its C-2 carboxyl and hydroxyl groups. A higher-resolution MoFe protein structure identified an electron density at the center of the cofactor, which was proposed to correspond to either N, O, or C.26 This provided a new perspective of the cofactor in which a central atom would be symmetrically coordinated by six Fe atoms forming one 6Fe-9S-X trigonal prism capped by one Fe atom and one end and one Mo atom at the other end. The central atom was finally identified as C by using X-ray emission spectroscopy, by determining the MoFe protein crystal structure at 1 Å resolution, and by radioactive carbon tracing experiments.27,138,139Figure 12 shows the FeMo-co atomic structure.
Figure 12.
Structure of FeMo-co. The structure shows the trigonal prism at the center of the cofactor, the capping Fe and Mo atoms, and the location of R-homocitrate.
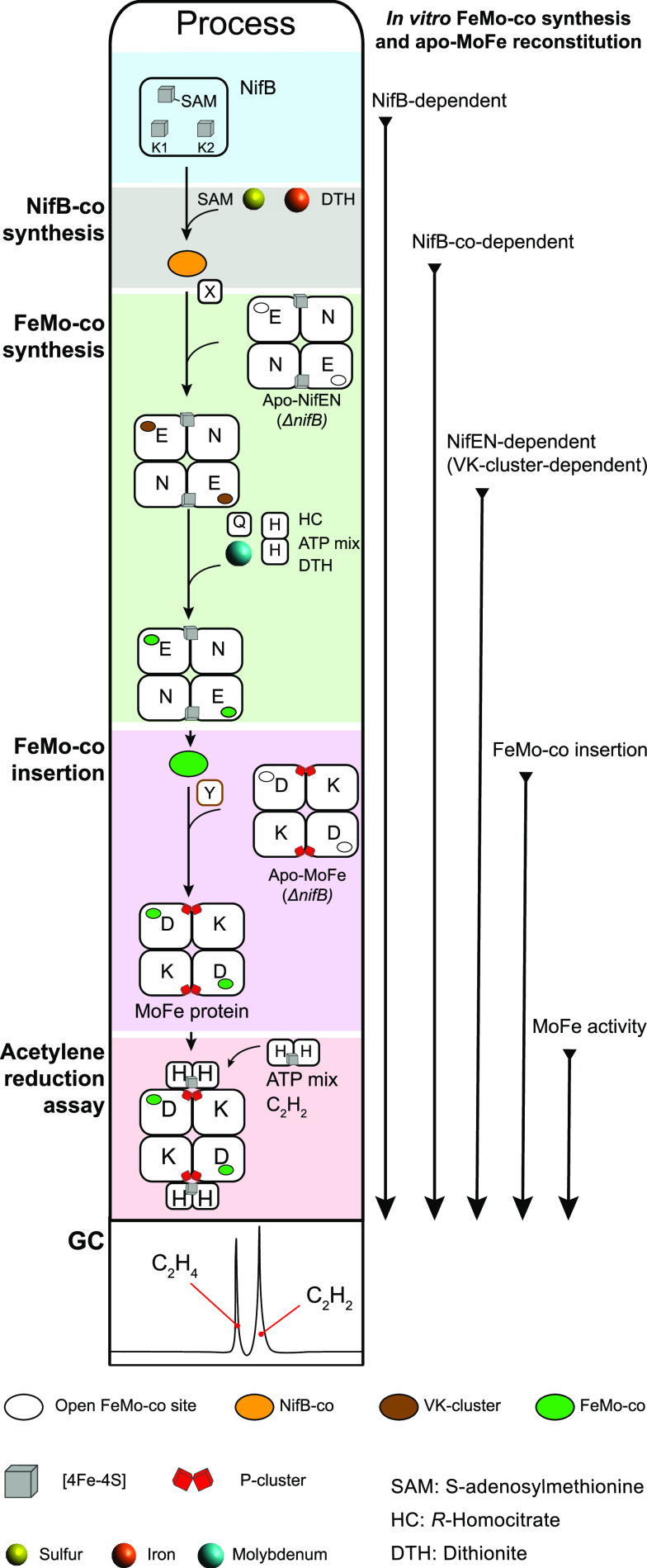
8.2. In Vitro Systems for FeMo-Cofactor Synthesis and Insertion
The first systems for in vitro FeMo-co insertion have been described above and consisted on the activation of inactive MoFe protein in extracts of A. vinelandii UW45 by addition of a mysterious activating factor126 and later by FeMo-co isolated from crystallized MoFe protein.15 Soon followed the first assay for in vitro FeMo-co synthesis, designed to elucidate specific functions of known participant proteins, such as NifB and NifEN, to find novel functions and proteins and, altogether, to understand the FeMo-co biosynthetic pathway.16 This assay was performed by mixing cell extracts of A. vinelandii and K. pneumoniae strains with mutations impairing FeMo-co biosynthesis at independent steps, typically A. vinelandii UW45 (nifB–) and K. pneumoniae UN1100 (nifE–). In vitro synthesized FeMo-co was incorporated into the apo-MoFe protein present in the extracts, and the activity of reconstituted MoFe protein was estimated by the acetylene reduction assay after addition of excess Fe protein and an ATP regenerating mixture.140 Addition of exogenous molybdate, a reductant, and MgATP was required for in vitro activation. Addition of exogenous Fe and S was not required because, as shown later, protein-bound Fe–S cluster FeMo-co precursors were present in the mixtures. Participation of homocitrate in FeMo-co was still unknown, and therefore, this component was not added to the assay. It appears that cell extracts provided enough homocitrate as to detect activity.
The in vitro FeMo-co synthesis and insertion assay has been used to elucidate the importance of proteins in FeMo-co biosynthesis. For instance, the essentiality of NifB16 and NifEN,21 the sufficiency of apo-Fe protein in FeMo-co synthesis,90 and the direct Mo transfer from NifQ to a complex of NifEN with Fe protein73 were demonstrated using this assay. Involvement of nonessential proteins such as NifX,141 NafY,22 and FdxN74 was also elucidated. The assay was also used to identify FeMo-co biosynthetic intermediates, most importantly the so-called V factor or homocitrate17 (see Section 12), an Fe–S cluster precursor to FeMo-co produced by NifB and called NifB-co19 (see Section 10.2) and its derivate cluster bound to NifEN and designated as VK-cluster24 (see Section 13.4) in honor of Vinod Shah, who discovered FeMo-co and NifB-co and developed the in vitro FeMo-co synthesis assay.
Currently, in vitro FeMo synthesis assays (with apo-MoFe protein activation) are typically performed in completely defined reactions using only purified proteins and starting from FeMo-co biosynthetic intermediates that might either be protein-bound or added exogenously. Figure 13 summarizes the different assays for in vitro FeMo-co synthesis and insertion with purified components. All reaction mixtures contain components that are extremely oxygen labile. The FeMo-co insertion assay only requires ΔnifB apo-MoFe protein and exogenous FeMo-co solution in NMF (less than 5% NMF in final reaction mixture). The NifEN-dependent assay requires holo-NifEN (with bound VK-cluster), Fe protein, molybdate, R-homocitrate, MgATP, DTH, and ΔnifB apo-MoFe protein. The NifB-co-dependent assay requires exogenous NifB-co aqueous solution, ΔnifB apo-NifEN (without bound precursor), Fe protein, molybdate, R-homocitrate, MgATP, DTH, and ΔnifB apo-MoFe protein. The NifB-dependent assay requires [Fe–S] cluster-reconstituted NifB, SAM, ΔnifB apo-NifEN (without bound precursor), Fe protein, molybdate, R-homocitrate, MgATP, DTH, and ΔnifB apo-MoFe protein. All assays require activity determination of the in vitro reconstituted MoFe protein, for which excess of complementary Fe protein component (40:1 molar ratio of Fe protein to MoFe protein), a MgATP regenerating mixture, and excess reductant (DTH) are added. Nitrogenase activities determined may include reduction of acetylene into ethylene, reduction of protons into H2, and reduction of N2 into NH3.
Figure 13.
Assays for in vitro FeMo-co synthesis and apo-MoFe protein reconstitution. This schematic diagram shows the most relevant in vitro FeMo-co synthesis assays described in the literature and labeled to the right as NifB-dependent, NifB-co-dependent, NifEN-dependent, FeMo-co insertion, and MoFe protein activity, relating to the starting component (protein or cluster precursor) being tested. The assays can be performed by mixing cell extracts containing the indicated components or by using highly purified proteins, precursors and chemicals. The most complex assay would be the NifB-dependent (requires competent NifB containing its [Fe–S] cluster complement), which includes NifB, SAM, ΔnifB apo-NifEN, Na2MoO4, homocitrate, Fe protein, DTH, ΔnifB apo-MoFe protein, and ATP mix. NifX and NafY are not essential but stimulate FeMo-co synthesis and insertion. Na2MoO4 can be replaced by NifQ.
Not all nif or naf gene products are similarly required for in vitro or in vivo FeMo-co synthesis. The nifD and nifK products are not required for FeMo-co synthesis in vivo(10) or in vitro.142 The products of nifH, nifB, nifE, and nifN are absolutely essential both in vivo and in vitro. The products of nifU and nifS are required in vivo(39,44) but not in vitro as current systems provide either Fe and S or [Fe–S] biosynthetic intermediates to FeMo-co. NifM in vivo requirement is not unambiguously demonstrated, but this protein is certainly not required in vitro as addition of Fe protein waives its function. NifQ is required in vivo only under Mo limitation143 and has been successfully used in the in vitro assay,73 although it is usually replaced by molybdate. NifV is not absolutely essential in vivo,144 and it is usually replaced by R-homocitrate in vitro. NifX and NafY are neither essential in vivo(145) nor in vitro, but they stimulate in vitro FeMo-co synthesis when added at precise stoichiometry with respect to NifEN and MoFe protein.22,113,141
Finally, Curatti et al. showed that FeMo-co synthesis and apo-MoFe protein activation in vitro could be achieved by using only NifB, NifEN, Fe protein, and apo-MoFe protein if FeMo-co components (Fe, S, Mo, and homocitrate) in their appropriate chemical forms, a strong reductant, SAM and MgATP were provided to the assay. This result unambiguously showed that reactions carried out by these six gene products, which had also deemed essential by genetic analysis (Figure 4), were sufficient for FeMo-co synthesis and that the in vitro system mimicked the process in vivo.22
9. Model for FeMo-co Biosynthesis
Our current simplified model for FeMo-co biosynthesis is shown in Figure 14. This model derives from our best understanding of genetic, biochemical and biophysical evidence accumulated over decades of investigations from many research groups. We note that some aspects of this model are not free of controversy, most importantly those related to the incorporation of Mo into the cofactor. All features of this model, including its controversial aspects, will be discussed in detail in following sections.
Figure 14.
Simplified model for FeMo-co biosynthesis centered in the hub formed by NifEN and the Fe protein designated in the figure as FeMo-co maturation complex. The pathways providing the FeMo-co Fe–S cage, molybdate, and homocitrate are shown and include the proteins involved. Roles of NifX and NafY in delivering NifB-co and FeMo-co to NifEN and the MoFe protein, respectively, are indicated.
FeMo-co is synthesized outside of the MoFe protein. The MoFe protein paralog, NifEN, occupies a central role in the FeMo-co biosynthetic pathway, functioning as scaffold for the transformation of a symmetric [8Fe-9S–C] cluster into the final cofactor containing Mo and homocitrate. In this role, NifEN interacts with the Fe protein, NifQ, NifX, and probably with NafY and NifB. Interaction with NifV has not been observed, and NifV does not seem necessary other than to synthesize homocitrate. Figure 14 shows three independent branches converging into NifEN to provide all required Fe and S in a single [Fe–S–C] cluster called NifB-co, Mo in an Fe–S cluster ligand environment, and R-homocitrate, with one output that is FeMo-co. The Fe–S core branch involves NifU, NifS, NifB, FdxN, Fe, cysteine, SAM, and a reductant, roles of which will be discussed in Section 10. The Mo branch involves molybdate, NifQ, NifO, and a reductant, and it will be discussed in Section 11. Homocitrate production will be discussed in Section 12. NifEN biochemical properties and reactions taking place in NifEN, including those promoted by the Fe protein, will be discussed in Sections 13 and 14, respectively. The roles of NifX and NafY will be discussed in Section 15.
A functional categorization of proteins in this pathway has been previously proposed7 that shows proteins acting as molecular scaffolds for the assembly of [Fe–S] clusters (NifU, NifB, NifQ, and NifEN), enzymes acting as donors of FeMo-co components (NifS, NifB, and NifV), and proteins carrying labile metallocluster precursors between scaffolds and finally FeMo-co to the target MoFe protein.
10. Biosynthesis of FeMo-co Fe-S Core: Roles of NifU, NifS, NifB, and FdxN
10.1. NifS and NifU Assembly of Precursor [Fe-S] Clusters for FeMo-co
As mentioned above, A. vinelandii or K. pneumoniae strains with disrupted nifU or nifS genes had reduced nitrogenase activity and no (or slow) growth under N2-fixing conditions. This growth phenotype correlated with lower activities for their corresponding Fe and MoFe proteins in vitro.39,44,146 As MoFe protein could not be activated by the addition of FeMo-co in vitro, P-cluster maturation (required for and preceding the insertion of FeMo-co into MoFe protein) was affected in these cells. The exact consequence from NifU and NifS absence on FeMo-co synthesis was therefore not clear.39 A way to solve this dilemma was by isolating a FeMo-co biosynthetic precursor, synthesized by NifB and called NifB-co (Figure 15), from the cytosolic membranes of the nifN mutant K. pneumoniae strain UN1217, and demonstrating that functional nifU and nifS genes were required for most NifB-co accumulation,147 which suggested that NifU and NifS were the main providers of [Fe–S] clusters precursors to NifB-co (and therefore FeMo-co).
Figure 15.
NifB-co is a diamagnetic [8Fe-9S–C] cluster. (A) Timeline showing the milestones from the discovery of NifB-co to its final characterization together with the first author and year of the reports. (B) NifB-co as isolated from membranes of K. pneumoniae. NifB-co solutions are greenish brown with nondescript visible spectra in the 400–700 nm region. (C) Isolation of an in vivo formed NifX-NifB-co complex and its Mössbauer analysis revealing two spectroscopically different Fe sites at 3:1 ratio. UV–visible spectrum reprinted with permission from ref (19). Copyright 1994 ASBMB. Mössbauer spectra adapted with permission from ref (20). Copyright 2016 John Wiley and Sons.
Replacement of the endogenous nifB gene with an IPTG-controlled gene variant that produced a glutathione S-transferase (GST)-tagged NifB protein (GST-NifB) enabled the purification of NifB from a K. pneumoniae nifN mutant strain (UC17), or from cells with additionally disrupted nifUS genes (UC18). GST-NifB purified from UC17 cells contained about 10 Fe per NifB monomer and did not require addition of Fe, S, and SAM to support in vitro FeMo-co synthesis and nitrogenase activation when combined with extracts from the A. vinelandii nifB– strain UW45.147 On the contrary, GST-NifB isolated from UC18 cells could not support in vitro FeMo-co synthesis to the extent of the corresponding GST-NifB protein produced by UC17 cells, further indicating the NifU and NifS contribution to provide FeMo-co precursors to NifB.
10.2. NifB and NifB-co
10.2.1. Information from nifB Mutagenesis
The importance of the nifB gene product became obvious from mutant strains of N2-fixing bacteria unable to grow using N2 as sole nitrogen source. The history of nifB research is closely connected to the discovery of FeMo-co and the development of the in vitro FeMo-co synthesis assay. From genetic mapping experiments, it was shown that nifB was essential for the production of a so-called “activation factor” in A. vinelandii and K. pneumoniae.40,126,129,130 The activation of MoFe protein in cell extracts of nifB mutant strains upon insertion of purified FeMo-co confirmed that NifB directly affected FeMo-co biosynthesis.15,44 The NifB protein was later identified as a thermosensitive protein of 51.5 kDa in K. pneumoniae(148) that lacked close homologues in nondiazotrophic organisms. In vitro complementation assays using extracts confirmed that the nifB gene product was essential for FeMo-co synthesis and extremely sensitive to O2 exposure.16 This work also predicted that in vitro FeMo-co synthesis would be possible using a completely defined system, a prophecy that was fulfilled 21 years later.22
By using the A. vinelandii nifB mutant strains CA30 and UW45, it was also shown that NifB was essential for N2 fixation under all growth conditions (+Mo, + V, or Mo and V deficient), suggesting that all three dinitrogenase types (and not only the MoFe protein) require NifB for their function40,149 and that the NifB produced factor should be a common precursor to the active-site cofactors of all dinitrogenases.
10.2.2. Identification and Isolation of NifB-co, the Product of NifB Activity
A breakthrough to understand the NifB factor essential to FeMo-co synthesis came from the attempts aiming to purify the NifB protein from K. pneumoniae. Using the nifN mutant strain UN1217,129 a highly elaborate and methodological screening procedure resulted in a three-day protocol where NifB-co was found to elute from UN1217 membranes upon several rounds of membrane washes and freeze–thaw cycles (Figure 15A).19 The eluted NifB-co could then be bound and purified using Sephacryl S-200 and phenyl-Sepharose columns. However, purification of the NifB protein was unsuccessful, and NifB in this way sticks out as its product was isolated before the protein itself.
As predicted, NifB-co was a low molecular weight molecule. Like FeMo-co solutions, NifB-co solutions had a green–brown color and exhibited broad absorbance between 400 and 700 nm without distinctive peaks or shoulders (Figure 15B). In contrast to FeMo-co, NifB-co was EPR silent in its DTH-reduced state. NifB-co did not contain any detectable metals other than Fe, in line with the concept of NifB-co being a common [Fe–S] precursor for the active-site metalloclusters (FeMo-co, FeV-co, and FeFe-co) of all three types of nitrogenases, although this assumption has not yet been experimentally tested in vitro. NifB-co was found to be resistant to repeated freeze–thaw cycles, relatively stable upon heat-treatment but extremely sensitive to O2 exposure (half-life of less than 15 s).19 NifB-co was not detected in wild-type K. pneumoniae membrane preparations indicating that the product is rapidly processed to FeMo-co in vivo. Importantly, when the level of MoFe protein activation was related to the atomic Fe content of NifB-co, activation per nmol Fe was similar to the values obtained using FeMo-co, and suggested that NifB-co served as the [Fe–S] cluster source for FeMo-co synthesis.19 This was later shown biochemically using NifB-co with isotope-labeled Fe or S (Figure 15A).150 Additionally, Fe from NifB-co accumulated at NifEN in vitro when MgATP, Mo, homocitrate, or Fe protein were absent, or when the MoFe protein was first loaded with FeMo-co. Altogether, these results strongly supported a stepwise model for FeMo-co synthesis where the NifB-co is first delivered to NifEN and then further converted into FeMo-co.
10.2.3. Interstitial Atom of FeMo-co Is Present at NifB-co
No NifB-co X-ray crystal structure is yet available. Instead, the composition and structure of NifB-co has been studied using other spectroscopic techniques. In vitro FeMo-co biosynthesis using 55Fe-labeled NifB-co as precursor showed that 55Fe was bound to NifX when MoFe protein was absent, suggesting that NifX could accumulate FeMo-co precursors.151 The function of NifX acting as a promiscuous reservoir for FeMo-co precursors was further confirmed when it was seen that A. vinelandii NifX could accumulate both NifB-co for delivery to NifEN, and a novel FeMo-co precursor, called VK-cluster, that was formed at NifEN (see below).24 The function of NifX and other carrier proteins involved in metallocluster trafficking during nitrogenase maturation will be discussed in Section 15. NifX expressed and purified from E. coli was found to readily accept NifB-co in the absence of other protein components in vitro, indicating a direct and specific interaction. The NifX-NifB-co complex reproduced the absorbance spectrum of NifB-co isolated from K. pneumoniae.24
Taking advantage of the interaction between NifB-co and its intracellular carrier protein NifX, metal-free NifX purified from E. coli was used as acceptor for the NifB-co produced by K. pneumoniae UN1217. K. pneumoniae cultures were grown with either naturally abundant 56Fe, or 57Fe, as source of Fe and used for X-ray absorption spectroscopy (XAS) and EXAFS, or EXAFS together with nuclear resonance vibrational spectroscopy (NRVS) analysis, respectively. Using NafY loaded with FeMo-co as control sample, the oxidation states and the ligand environments of the Fe atoms for both NifB-co/NifX and FeMo-co/NafY were found to be similar.152 No Mo was detected in NifB-co, confirming previous studies.19 EXAFS strongly suggested the presence of a 6Fe-9S-X core in NifB-co, and three working models with six, seven, or eight Fe atoms were constructed based on the structure of FeMo-co. Of the three models, the six Fe model gave the best fit to the experimentally observed data. However, as explained below, Mössbauer provided stronger evidence to support the eight Fe atom model. Importantly, EXAFS data and NVRS analysis provided the first evidence that the single interstitial light atom X (C, N, or O), at the time suggested to be part of FeMo-co,26 was already present at NifB-co (Figure 15A).
Some information regarding NifB-co was also obtained from the generation of a MoFe protein containing NifB-co instead of FeMo-co at its active site, obtained from passing purified FeMo-co deficient A. vinelandii ΔnifB apo-MoFe protein through a Sephacryl column loaded with NifB-co isolated from the K. pneumoniae nifN mutant strain UN1217.153 The chimeric MoFe protein had a brown–green color similar to the corresponding FeMo-co containing protein, and Fe analysis was indicative of two NifB-co molecules per MoFe tetramer. The MoFe protein-bound NifB-co was EPR silent, and no Mo was found even when apo-MoFe protein reconstitution was carried out in the presence of exogenous molybdate and homocitrate. While MoFe protein with bound NifB-co showed significant Fe protein- and MgATP-dependent H+ (and some C2H2) reduction, no N2 to NH3 conversion was observed. Whether NifB-co could be inserted into MoFe protein in vivo or if cellular mechanisms preventing insertion of premature clusters exist, or whether NifB-co-loaded MoFe protein could represent an early ancestor (not capable of FeMo-co biosynthesis) of nitrogenase awaits further investigation.153
10.2.4. NifB-co Is an [8Fe-9S–C] Biosynthetic Precursor to FeMo-co
The NifB product has also been studied when isolated from NifEN in A. vinelandii strains not capable of completing FeMo-co maturation. In this regard, EPR and XAS/EXAFS analyses of the FeMo-co precursor bound to His-tagged NifEN from the A. vinelandii strain DJ1041154 (ΔnifHDKTYnafAB) showed that the cluster could be best modeled as an eight Fe analog of FeMo-co, where the terminal Mo atom was replaced by an Fe atom, and homocitrate was missing.155 However, this FeMo-co precursor originated from NifEN, and modifications to NifB-co at NifEN, even in the absence of Fe protein, have been reported.24 Therefore, the definite answer to the Fe composition of NifB-co came from NifX-bound NifB-co subjected to Mössbauer spectroscopy and density functional theory (DFT) analysis (Figure 15C).20 Two distinct methods were used to prepare 57Fe-enriched NifX/NifB-co complexes. In the first, A. vinelandii NifX was loaded with NifB-co isolated from K. pneumoniae. In the second, GST-tagged NifX was expressed in a K. pneumoniae strain UN1217 where FeMo-co biosynthesis was interrupted at the level of NifEN, resulting in the in vivo accumulation of a GST-NifX/NifB-co complex. In vivo generated NifX/NifB-co contained 7.4 mol Fe per mol NifX, and the Mössbauer analysis provided evidence for the NifB-co being an eight Fe cluster with two distinct types of Fe, where six of the eight Fe atoms have delocalized valences, and two Fe atoms have localized valences.20 The NifB-cofactor is in this regard different from FeMo-co where all Fe have delocalized valences.156 High-field Mössbauer measurements further identified NifB-co as a diamagnetic (S = 0) eight Fe-cluster,20 consistent with previous EPR studies of NifX/NifB-co19,24 and NifB-co/MoFe protein.153
Taken together, the collective experimental data obtained from analysis of the NifB-cofactor points to NifB-co being a diamagnetic cluster, best described either as 4Fe2+-4Fe3+ or a 6Fe2+-2Fe3+ cluster having valence-delocalized states.20 Taking into consideration the presence of an interstitial atom in the Fe–S cage of NifB-co,152 and the high-resolution structures and X-ray emission spectroscopy of the MoFe protein showing that the interstitial light atom of FeMo-co is carbon (C),27,138 it is logical to propose a structure of NifB-co with an [8Fe-9S–C] arrangement. This atomic composition is also supported from NifB mechanistic studies showing that the NifB-cofactor is factored from a SAM-dependent fusion of two [4Fe-4S] clusters with a concomitant insertion of a ninth S (see below)59 and that C derived from SAM can be found at the NifEN-bound precursor157 and be followed to FeMo-co in a NifB-dependent reaction.139
10.2.5. Rosetta Stone of Nitrogenase Metalloclusters
Unfortunately, different research groups use different nomenclature for the FeMo-co biosynthetic intermediates and even for FeMo-co itself. This situation obscures results, precludes direct comparison, and makes literature interpretation, in general, very difficult to the nonexpert. Table 2 shows chemical equivalences and names of FeMo-co [Fe–S]-cluster biosynthetic intermediates found in the literature. In this review, we use the original names given to these [Fe–S]-clusters by the researchers that isolated and characterized them first.
Table 2. Rosetta Stone: Equivalences and Names of FeMo-co Intermediates Found in the Literature.
VK-cluster is electronically distinct from NifB-co.
10.3. NifB Protein and Its Activity
10.3.1. Three Distinct NifB Protein Architectures Exist in Nature
Identification of the nifB gene products from K. pneumoniae, A. vinelandii, Rhizobium, and Bradyrhizobium japonicum in the mid-1980s suggested they were polypeptides with molecular masses of about 50–55 kDa.40,148,161,162 Later work showed that this is not always true, as “truncated” (but functional) NifB homologues of about 35 kDa were found in N2-fixing archaea such as Methanosarcina acetivorans, Methanobacterium thermoautotrophicum, and Methanocaldococcus infernus.57,58,163 While these proteins contain the NifB-conserved N-terminal SAM-radical domain, they lack the C-terminal NifX-like extension seen in the other “more complex” variants such as NifB of K. pneumoniae and A. vinelandii (Figure 16A). Additionally, the archaea NifB variants were monomers in contrast to the K. pneumoniae and A. vinelandii NifB homodimers.64 Analysis of the protein sequences from functional NifB proteins revealed several SAM-domain specific signatures such as the strictly conserved SAM-radical protein AdoMet motif (CxxxCxxC) at the active site located at the N-terminus of the SAM-domain. The three cysteine residues of this hallmark signature are known to coordinate a [4Fe-4S] cluster in SAM-radical proteins, where the forth Fe atom is ligated by the N and O atoms of the amino- and carboxy groups of SAM (Figure 16B,C).164,165 In addition to the AdoMet motif, an ExRP motif, an AGPG motif, a TxTxN motif, and a CxxCRxDAxG motif were identified (Figure 16A). Application of these signature motifs as filter for 390 putative NifB sequences from a diverse selection of organisms (with proven or believed diazotrophic life-style) generated a data set with 289 NifB proteins that could be grouped into three classes according to their protein architecture (Figure 17).64 The most common type of NifB protein contained an N-terminal SAM-radical domain fused to a NifX-like domain, for example, the K. pneumoniae and A. vinelandii NifB proteins. Interestingly, no archaeal NifB was found to contain the NifX-like domain, while almost 75% of the NifB proteins found in bacteria were of the two-domain type. In addition, a few bacteria expressed NifN proteins in the form of translational NifNB fusions.63
Figure 16.
(A) NifB domain architectures and conserved amino acid motifs. The simplest NifB proteins consist of a stand-alone SAM radical motif. More complex NifB proteins include a C-terminal NifX-like, or N-terminal NifN-like domain and C-terminal NifX-like domain. (B) Structure of the NifB homologue MoaA showing the [4Fe-4S]-SAM active site and its ligating Cys residues. Images created with NGL viewer36 and RCSB PDB. No NifB crystal structure is yet available. (C) Structural model, generated with SWISS-MODEL,87 of a 43 amino acid fragment of the A. vinelandii NifB sequence including the AdoMet motif Cx3Cx2C.
Figure 17.
NifB phylogeny and distribution of NifB domain architectures among putative diazotrophs. The tree shows overall distribution and relative frequency of NifB architectures. Figure reprinted from ref (64). Copyright 2017 Arragain, Jiménez-Vicente, Scandurra, Burén, Rubio, and Echavarri-Erasun under CC BY 4.0 http://creativecommons.org/licenses/by/4.0/.
Since NifX is capable of binding NifB-co,24,151 it is reasonable to imagine that the NifX-like domain of NifB could exert protection of the synthesized NifB-co product or facilitate its transfer to subsequent maturation factors. However, this has not been shown experimentally. On the contrary, FeMo-co synthesis and MoFe protein activation assays have shown that the NifX-like domain is not required for efficient transfer of NifB-co in vitro, even when using pure protein components isolated from the two-domain NifB host A. vinelandii.57,58,166 The same result was observed in vivo by complementation of a ΔnifB K. pneumoniae strain with the SAM-domain-only NifB from M. infernus, or with a truncated version of the Chlorobium tepidum NifB lacking the NifX-like domain.64 Although full understanding of these findings requires additional experimental work, they could explain why mutations affecting the C-terminus of NifB generated a leaky phenotype compared to other nifB mutant strains being Nif–.40,129,149
10.3.2. NifB Requires SAM for Its Activity
Biochemical analysis of the NifB protein was for long hampered by the difficulties to isolate it. In a breakthrough study, 12 years after the isolation of the NifB product, NifB-co, a functional NifB protein was purified from A. vinelandii.41 For this, the A. vinelandii nifB gene was replaced with a His-tagged version placed under the control of the nifH promoter. The N-terminal His-tag did not hamper in vivo complementation, and isolation under anaerobic conditions generated a yellow/brown preparation of a NifB homodimer containing O2-sensitive [Fe–S] clusters. As-isolated NifB protein carried an average of 12 Fe atoms per NifB dimer. Addition of Fe and S under reducing conditions increased the Fe content and converted the protein into an active form that together with Fe, S, and SAM could replace isolated NifB-co in the in vitro FeMo-co synthesis assay using A. vinelandii nifB– (UW45) extracts.41 Low activity was obtained when SAM was absent from the reaction or when SAM was replaced by the nonreactive analog S-adenosyl homocysteine (SAH), proving the SAM-dependent nature of the NifB-co formation. It also showed that the IMAC purified A. vinelandii NifB did not accumulate NifB-co at detectable levels although being equipped with a NifX-like domain. In a follow-up work using purified protein components only, the minimal in vitro system for the NifB-dependent FeMo-co synthesis and apo-MoFe protein activation was defined to be composed of a FeMo-co precursor-deficient NifEN (called apo-NifEN) and Fe protein, together with Fe, S, SAM, Mo, homocitrate, and Mg-ATP, in addition to NifB and apo-MoFe protein (Figure 13).22
10.3.3. NifB Contains Accessory [Fe–S] Clusters in Addition to Catalytic [4Fe-4S]-SAM Cluster
Because NifB-co is an [8Fe-9S–C] product, it could be imagined that those eight Fe atoms originated from the fusion of two [4Fe-4S] precursors at NifB. Being a member of the Radical SAM protein superfamily first identified in 2001,167 whose proteins generate radical species by the reductive cleavage of SAM through an unusual [Fe–S] center, a possibility was that NifB contained three [Fe–S] clusters; one SAM-coordinated [Fe–S] cluster required for catalytic activity and two [4Fe-4S] precursor clusters. In fact, [Fe–S] cluster reconstitution of A. vinelandii NifB was shown to increase its Fe content to nine Fe atoms per monomer.41 Similarly, as isolated K. pneumoniae NifB contained 10 Fe atoms per monomer, consistent with the presence of three [4Fe-4S] clusters, it was readily active in the in vitro FeMo-co synthesis assay without the need for [Fe–S] cluster reconstitution.147 Alternative explanations included that the SAM-coordinated [Fe–S] cluster also functioned as a precursor cluster or that not all precursor clusters were directly bound to NifB, although this seemed less likely due to the lack of NifU or other [Fe–S] assembly factors in the minimal in vitro NifB-dependent FeMo-co synthesis reaction described above.
In a further study, Wiig and colleagues created a His-tagged NifN-B fusion, similar to that naturally found in Clostridia,63 for expression in the A. vinelandii DJ1041 (ΔnifHDKTYnafAB) mutant background,154 generating strain YM65A.168 The purified fusion protein was capable of acting as a FeMo-co donor to apo-MoFe-protein after incubation with DTH, Fe protein, MgATP, Mo, and homocitrate. EPR analysis showed that it, in addition to the NifEN bound FeMo-co precursor produced and delivered by NifB, NifEN-B also contained additional [4Fe-4S] cluster(s) whose signal intensity decreased upon addition of SAM.168 Differences in 420 nm absorption between oxidized and reduced protein were interpreted as NifEN-B containing 3.4 mol [4Fe-4S] cluster per mol protein. As each NifB unit could harbor one SAM-coordinated [4Fe-4S] cluster, this study corroborated the presence of additional non-SAM coordinated [4Fe-4S] clusters bound to NifB. These additional cluster(s) were later named as the K-cluster.139 Importantly, incubation with SAM caused a dose-dependent reduction in the EPR signature of the additional [4Fe-4S] clusters,168 confirming the SAM-dependent chemistry of the NifB mediated reaction previously observed by Curatti and colleagues.41
The requirement of Fe protein, Mo, and homocitrate, for NifEN-B-dependent apo-MoFe activation, suggested that the FeMo-co precursor bound to NifEN-B (L-cluster) was likely identical to the much earlier described NifB-co.19 This was also consistent with the first description of the L-cluster as the FeMo-co precursor synthesized by NifB and delivered to NifEN.159 Therefore, and to avoid confusion, when applicable, the L-cluster will be hereinafter described as NifB-co. However, it is important to note that, when investigated in its NifEN-bound form, the L-cluster would correspond to the VK-cluster, a previously described FeMo-co precursor isolated from NifEN24 with the same atomic composition as NifB-co but with different EPR properties (Table 2).
10.3.4. Coordination of NifB-Associated [4Fe-4S] Clusters
Using site-directed mutagenesis, the three [Fe–S] clusters of M. infernus NifB were characterized.58 Ala substitution of Cys residues in the N-terminal AdoMet motif (CxxxCxxC) or in the C-terminal CxxC U-type motif, reduced Fe and S content of NifB, changed the NifB UV–vis absorption spectra (Figure 18A–C) and abolished NifB activity.58 Importantly, the UV–vis spectrum and Fe and S content of a double mutant version eliminating binding of both clusters suggested the presence of a third[4Fe-4S] cluster. This variant lacking two clusters presented an EPR signal arising from a single [4Fe-4S] cluster, while multiple S = 1/2 [Fe–S] clusters were seen in the WT and single cluster mutants. Using the double cluster-deficient variant as a starting point for analysis, simulation of the spectra from the other variants suggested that a total of three [4Fe-4S] clusters were present at reconstituted NifB (Figure 18D), which was supported by their temperature-dependent EPR-spectra. The clusters were denoted as RS for the SAM-binding cluster, and AC1 and AC2 for the auxiliary clusters. An additional cluster was observed in the version where the SAM-cluster ligands were mutated. However, this presumably [2Fe-2S] cluster was only detected in this mutant and was assigned to a degraded cluster (ACx). We note that this review will use the SAM-cluster, K1, and K2 nomenclature instead, with equivalences shown in Table 2.
Figure 18.
Spectroscopic signals from NifB [Fe–S] clusters. (A) SDS gel of purified M. infernus NifB used for spectroscopic analyses. (B) UV–vis spectra of as-isolated and Fe–S cluster reconstituted NifB variants. All spectra were recorded in DTH-reduced samples. (C) Fe content and Fe–S cluster assignment for the NifB variants. (D) Deconvolution of the EPR signal arising from the three distinct [4Fe-4S] clusters of NifB. Recorded data are shown in black and the simulations in blue. The signal from the RS ([4Fe-4S]-SAM cluster) component is shown in red, that of the K1 component in blue, and that of the K2 component in pink. Figure adapted from ref (58). Copyright 2016 ACS.
M. infernus NifB does not contain enough conserved Cys residues for a complete cysteinyl coordination of three [4Fe-4S] clusters. However, its sequence contains two well-conserved His residues as well as several conserved Arg residues that could be involved in cluster coordination.58 Similarly, sequence analysis identified three groups of highly conserved Cys residues in M. acetivorans NifB, each group consisting of three Cys believed to coordinate one [Fe–S] cluster.160 Cys to Ala substitution was employed to generate NifB variants capable of only coordinating a single cluster. Following reconstitution using synthetic [4Fe-4S] clusters,59 the presence of three distinct [4Fe-4S] at NifB was confirmed, and their Cys-coordination was shown,160 in agreement with previously published data for M. infernus NifB.58 In addition, a nitrogenous ligand likely originating from a His residue was found to coordinate the K1 cluster in both studies,58,160 while no N coupling was found to the K2 cluster.58
10.3.5. NifB-co Central Carbide Originates from SAM during NifB Catalysis
The fact that NifB-co already contained a central atom,152 the identification of C as the light interstitial atom of FeMo-co,27,138 and the involvement of SAM for methyl transfer and organic radical chemistry,165 led to the hypothesis that NifB could be responsible for C insertion into NifB-co. This hypothesis was first tested using the NifEN-B protein purified from A. vinelandii.139 First, cleavage of SAM into SAH and 5′-deoxyadenosine (5′-dAH) by NifEN-B was shown, indicating that at least two distinct SAM molecules were involved in the overall reaction (Figure 19A). NifB-dependent carbide insertion into NifB-co was shown using [methyl-14C]-SAM. This 14C signal could subsequently be traced to FeMo-co (M-cluster, see Table 2) by performing in vitro FeMo-co synthesis and insertion into apo-MoFe protein. Replacement of [methyl-14C]-SAM with [carboxyl-14C]-SAM in the reaction mixture did not result in 14C-insertion, proving that the carbide specifically originated from the methyl group of SAM. Further, hydrogen atom abstraction from the methyl group was shown from incubation of NifEN-B with deuterium-labeled [methyl-d3]-SAM. A mixture of deuterium-labeled 5′-dAD and nonlabeled 5′-dAH was detected, where presence of 5′-dAD suggested 5′-dA• mediated H abstraction from the labeled methyl group. Concomitant production of 5′-dAH suggested abortive cleavage of SAM, with 5′-dA• mediating H abstraction from the solvent or from other unlabeled methyl groups generated by NifEN-B.139
Figure 19.
Model of the mechanism of NifB-co formation by NifB. (A) Proposed steps based on biochemical evidence: the K1 cluster presents mixed Cys-His coordination; SAM-dependent methyl group transfer to the K2 cluster; reductive cleavage of a second SAM molecule and formation of a 5′-dA radical at the active site; methyl group H atom abstraction by the 5′-dA radical; oxidation/deprotonation events to remove to H atoms; core rearrangement and insertion of a S atom at the belt of the precursor. (B) Hypothesized role of FdxN in reduction of the K2 cluster to prepare it for methyl group acceptance.
Two mechanisms for the stepwise and SAM-dependent carbide insertion were proposed. First, a methyl group would be transferred to an Fe or S atom of a K cluster, generating SAH as byproduct. Second, a 5′-dA• radical generated by reductive cleavage of another molecule of SAM would abstract a hydrogen atom from the cluster-attached methyl group, generating 5′-dAH as a byproduct. Continued deprotonation (and insertion of a ninth S atom) would result in NifB-co. While both mechanisms require two molecules of SAM for NifB-co formation, they differ in the identity of the target atom for the methyl group transfer (Fe or S) and therefore the involvement of a reductant for the methyl transfer reaction.139
These findings using NifEN-B were later confirmed using “truncated” SAM-domain-only NifB proteins from the methanogenic, N2-fixing organisms M. acetivorans and M. thermoautotrophicum.57 Upon expression in E. coli, these monomeric NifB variants accumulated as soluble proteins permitting more specific NifB cluster analysis, as the influence of signals from NifEN [Fe–S] clusters could be avoided. In vitro [Fe–S] cluster reconstitution suggested the presence of three distinct [4Fe-4S] clusters per NifB.57 The clusters were later characterized in more detail using a related NifB protein from M. infernus.58,64,163 As for A. vinelandii NifEN-B, these methanogenic NifB proteins were capable of cleaving SAM into SAH and 5′-dAH and produced more SAH than 5′-dAH upon addition SAM under reaction conditions.57,58 This could indicate that only a single hydrogen atom is abstracted by a 5′-dA• radical and that the two remaining hydrogen atoms are removed by oxidation/deprotonation events via a SAM-independent mechanism.
Addition of SAM to the NifEN-B or NifB proteins decreased the DTH-reduced S = 1/2 EPR signal, consistent with SAM-dependent conversion of the [4Fe-4S] K-clusters present at NifB into another entity. On the contrary, SAM-treatment converted the otherwise EPR silent IDS-oxidized proteins into proteins with EPR signatures, which could be indicative of proteins with FeMo-co precursor.57 However, this result conflicts with another report showing that NifB-co is a diamagnetic cluster.20 The product of SAM-treated NifB could be transferred to apo-NifEN, where it was converted into a cluster capable of activating apo-MoFe protein. 14C-tracing experiments using [methyl-14C]-SAM also confirmed the previous study that SAM is the source of carbide in NifB-co.139 No methylation event could be detected on the NifB polypeptide as a result of SAM exposure, as has been shown to be the case for other well-characterized Radical SAM enzymes performing similar reactions,169,170 suggesting direct methyl transfer from SAM to the K1 or K2 cluster.
10.3.6. NifB-co Formation Is Initiated by SAM-Dependent Methyl Group Transfer to S at K2 Cluster
Analysis of acid-treated NifEN-B released methanethiol (CH3SH), suggesting that the SAM-derived methyl group was linked to an acid-labile sulfur atom.171 The SAM-derived origin of the methyl group was confirmed using [methyl-d3]-SAM. Using allyl SAM (a SAM analogue with an allyl group instead of a methyl group) in the NifEN-B reaction mixture generated SAH but not 5′-dAH. Acid treatment of this NifEN-B protein released allylthiol. These results indicate that the allyl SAM was not capable of undergoing homolytic cleavage for the generation of the 5′-dA• radical required for hydrogen abstraction, suggesting that the methyl group transfer either precedes or is independent from hydrogen abstraction.
Taking advantage of the possibility to produce NifB proteins with unique and defined clusters, M. acetivorans NifB variants with two clusters (the SAM-cluster together with either the K1 cluster or the K2 cluster) were generated. Only the version equipped with the SAM-cluster and the K2 cluster was capable of SAM-dependent methyl transfer and generation of 5′-dAH. Together these results strongly suggest that the first step of NifB-co formation involves methyl transfer from SAM to an S at the K2 cluster and that the SAM-dependent reactions are initiated by the SAM-cluster but require the presence of the K2-cluster (Figure 19A).160
10.3.7. Insertion of Interstitial C Precedes Incorporation of “Ninth” S into NifB-co
NifB-co appears to contain one more S atom than present in the K1 and K2 clusters. Therefore, an external S donor would be required. Using synthetic [4Fe-4S] clusters instead of adding Fe and S under reducing conditions (or using NifU or NifS), NifB from M. acetivorans could be reconstituted with [4Fe-4S] precursors avoiding attachment of extra S aggregates.59 This method allowed testing of the capacity of distinct S-based products to support K1 and K2 conversion into NifB-co and then into FeMo-co. Only sulfite (SO32–), but not sulfide (S2–) or sulfate (SO42–), generated clusters capable of activating apo-MoFe protein. Addition of 35SO32– to [Fe–S] cluster reconstituted NifB in the absence of SAM produced no significant 35S-labeled NifB or NifB-co, suggesting that incorporation of the S into NifB-co happens after (or simultaneous to) the SAM-dependent carbide insertion. While reconstituted NifB was unable to provide a precursor cluster for FeMo-co synthesis in a reaction mixture containing SAM but not SO32–, SAM-cleavage products were detected, and EPR analysis showed that cluster conversion happened similarly to the mixture containing SO32–, indicative of SAM-dependent carbide insertion. This putative cluster intermediate was tentatively assigned as a [8Fe-8S–C] cluster (Figure 19A).59 Albeit functional in the in vitro FeMo-co synthesis assay, the physiological relevance of SO32– as donor of the ninth S has not yet been investigated, and it remains possible that NifS or another S-donating enzyme could perform this role in vivo.
10.4. Genes Coexpressed with nifB
10.4.1. Mutational Analysis of nifB Gene Cluster
Isolation and sequencing of a 3.8 kb fragment that corrected the NifB– phenotype of the A. vinelandii strains CA30 and UW45 identified four complete open reading frames,40 where the first one encoded a 54 kDa protein homologue to the 51 kDa and 54 kDa nifB gene products of K. pneumoniae and Rhizobium species.161 The Tn5 insertion and the nif-45 mutation of CA30 and UW45, respectively, were both located within this nifB gene. In contrast to the K. pneumoniae nifB gene,129 which was followed by the nifQ gene involved in incorporation of Mo into the FeMo-co of Mo-type nitrogenase (see Section 11.3),143 the A. vinelandii nifB and nifQ genes were separated by fdxN and nifO (originally named as orf2 and orf3, respectively).40,172 While the fdxN gene was found to encode a protein containing Cys residues organized in a pattern characteristic of ferredoxins, the function of the nifO gene product was unknown but showed some similarity to arsenate reductase.173
Expression and phenotypic analysis of the A. vinelandii nifBQ region showed that disruption of fdxN reduced nitrogenase activity to less than half under all nitrogen-fixing conditions (+Mo, + V, -Mo).172 Its effect on all three nitrogenase systems, together with the coordinated expression with nifB, suggested that FdxN could donate electrons for nitrogenase activity or for cofactor biosynthesis. Mutants of fdxN in other N2-fixing organisms had shown similar effects: a Nif– phenotype in R. meliloti and Herbaspirillum seropedicae,174,175 and 50% decrease in nitrogenase activity in Bradyrhizobium japonicum.176 The partial effect from fdxN disruption could be explained by complementation of other ferredoxins. Later studies confirmed the coordinated expression of fdxN with nifB in A. vinelandii.51,177 The expression of the nifB and fdxN genes,172 and their kinetics,51 are also linked in A. vinelandii supporting a concerted action of their products. However, it is intriguing that while in A. vinelandii fdxN is coexpressed with nifB and important for its activity (see below), fdxN is absent from many N2-fixing organisms (e.g., K. pneumoniae).
10.4.2. FdxN Is Important for NifB Activity and NifB-co Production
Analysis of a ΔfdxN A. vinelandii strain showed impaired diazotrophic growth, decreased MoFe protein levels, and slightly increased Fe protein levels.74 The ΔfdxN strain produced fully active Fe protein, while the MoFe protein accumulated as a mixture of functional holo-protein and inactive apo-MoFe protein. Purified ΔfdxN MoFe was less than 50% active, contained 19 Fe and 1 Mo atoms per tetramer, and presented lower FeMo-co-dependent EPR signals intensity, consistent with it containing two P-clusters and one FeMo-co.74 The NafY protein was found to be strongly upregulated in the ΔfdxN strain,51 a proxy indicative of apo-MoFe protein accumulation.109,178
Although a definite role for FdxN is yet to be established, current results identify FdxN as an important but poorly studied component involved in FeMo-co, and more specifically in NifB-co, formation. Cell extracts of a ΔfdxN ΔnifENX double mutant strain showed 20% NifB-co activity compared to extracts containing FdxN.74 An electron donor is required to start the reductive cleavage of SAM leading to 5′-dA• formation. This electron donor could be FdxN. Another possible role for FdxN that would also affect NifB activity could be to poise the K2 cluster to a certain redox potential suitable for SAM-derived methyl group acceptance (Figure 19B). Participation of FdxN in NifB-co formation is also supported by early work showing that fdxN gene disruption in A. vinelandii affected N2 fixation under conditions depending on each of the three nitrogenases,172 which is to be expected as NifB-co is believed to be a common precursor for FeMo-co, FeV-co, and FeFe-co biosynthesis. Finally, a very recent study of NifB expressed in yeast confirmed that FdxN is important for NifB activity and specifically that NifB protein produced in the absence of FdxN appears to be deficient of two of the three [Fe–S] clusters (RS- and K1-clusters).112 The results from this study are particularly interesting not only because a eukaryotic cell enables expression of the components in an environment free from other Nif proteins but also because the NifB proteins were isolated by STAC and did not require [Fe–S] cluster reconstitution for activity.
11. Molybdenum Uptake, Storage and Processing for Nitrogenase: Roles of MoSto, NifQ, and NifO
11.1. Chelation and Mo Uptake from Medium
This section describes the characteristics of molybdenum (Mo) scavenging, uptake, storage, and processing for incorporation into FeMo-co in the model bacterium A. vinelandii (Figure 20A). When genetic or biochemical data were obtained from other organisms, such as K. pneumoniae or R. capsulatus, it will be specifically indicated.
Figure 20.
Mo uptake, storage, and processing for nitrogenase in A. vinelandii. (A) Scheme summarizing current understanding of molybdenum trafficking in A. vinelandii. Molybdate in the medium is chelated by siderophores protochelin and azotochelin. Internalization to the periplasm space is believed to occur via outer membrane porins. Active transport of molybdate (and tungstate) to the cytoplasm is mediated by ABC transporters composed of ModABC polypeptides. Most molybdate in A. vinelandii is stored at MoSto complex in a process energized by ATP hydrolysis. However, in Mo-starved A. vinelandii cells, MoSto can store tungstate forming WSto.186 Mo in MoSto is available to the molybdoenzymes nitrogenase and nitrate reductase. Pathway branching appears to occur before involvement of ModG, a trimer that binds eight molybdates at the interface of its subunits. NifO is also involved in directing Mo toward FeMo-co against the Mo-co branch. NifQ carries a [Mo-3Fe-4S] cluster shown to deliver Mo to the NifEN scaffold protein where it will be incorporated into an Fe–S cluster precursor to generate FeMo-co. The structures of a ModA2B2C2 transporter (PDB: 2ONK), the α3β3 MoSto (PDB: 6GU5) and WSto (PDB: 2OGX), an α3 ModG (PDB 1H9M), α2β2 NifEN (PDB: 3PDI), and the Fe protein (PDB: 1NIP) are shown. Structure images created with NGL viewer36 and RCSB PDB. (B) Molybdenum and tungsten electronic shells and their molybdate and tungstate forms. (C) Common A. vinelandii metalophores. (D) Inside details of the MoSto protein crystal structure shown in panel A including POMs (molybdenum-based polyoxometalates) and ATP sites.
Molybdate (MoO42–) is the predominant form of bioavailable Mo. Despite its high solubility at neutral and basic pH, it is generally the least abundant transition metal with a biological function present in soils.179 Thus, N2-fixing organisms must frequently cope with Mo scarcity. Even when molybdate levels are enough to support N2 fixation (10 nM), the similar tungstate (WO42–) at concentration as low as 100 nM might outcompete molybdate and poison FeMo-co biosynthesis (Figure 20B). In this regard, there is one report of incorporation of W into the active site of R. capsulatus MoFe protein rendering an inactive WFe component.180 In A. vinelandii, tungstate toxicity is lowered by a mechanism in which siderophores discriminate between the two metals. The production of distinct siderophores and their relative abundance are tightly regulated and adapted to the concentrations of molybdate and tungstate in the medium, among other metals, imposing a first barrier against W toxicity (Figure 20C).181 In addition, while the siderophores protochelin and azotochelin chelate both Mo and W, upon entry into the periplasm through porins, the membrane transport systems discriminate against W-catechol acting as second barrier against tungstate. It is possible that additional mechanisms exist that discriminate between tungstate and molybdate before entering in the periplasm, for example, porins of the outer membrane.182,183
Molybdate is transported across the cytoplasmic membrane by high affinity transporters from the ATP-binding cassette (ABC) family composed of the products of the modA, modB, and modC genes184 (Figure 20A). In the periplasm, siderophores present molybdate and tungstate to the molybdate-binding ModA protein.185 Since uptake of free tungstate is as fast as that of free molybdate, it is possible that the relatively fast dissociation of the metal from Mo-catechol compared to W-catechol causes discrimination against tungstate.181 Both metals can be transported to the cytosol through the channel provided by the membrane protein ModB. The ModC component binds and hydrolyzes the MgATP required to energize the active transport. The importance of achieving sufficient intracellular Mo for N2 fixation is highlighted by the presence of three homologous modABC operons in A. vinelandii.37 Mutants in the modA1B1C1 operon, located nearby the minor nif cluster, have been found to be affect 99Mo uptake and the activity of the molybdoenzyme nitrate reductase.184
11.2. Mo Storage and Homeostasis in A. vinelandii
Each MoFe protein contains two Mo atoms. As the MoFe protein concentration in a N2-fixing cell has been shown to be in the range of 50 μM,51 a [Mo] of about 0.1 mM is directly bound to nitrogenase. Considering that A. vinelandii accumulates up to 25-times more Mo than what is needed for maximum N2 fixation rates,187,188 a rather impressive intracellular [Mo] of 2.5 mM can be estimated. This is mainly accomplished by binding of intracellular molybdate to a Mo storage protein (MoSto),187,189 a unique cage-shape heterohexameric α3β3 protein formed by the mosA and mosB gene products. MoSto can accumulate up to 120 Mo or W atoms per hexamer in the form of polyoxometalate clusters (Figure 20D).190,191In vitro experiments have determined that molybdate incorporation into MoSto is dependent on MgATP hydrolysis, while Mo release is an ATP independent process above neutral pH.192 Interestingly, it has been shown that MoSto can serve as Mo donor for FeMo-co synthesis in vitro, although this process is thought to be mediated by NifQ in vivo.71A. vinelandii requires MoSto for storage of Mo but also to control its homeostasis and to properly repress expression of V and Fe-only alternative nitrogenases in conditions of transient Mo scarcity.71 Deletion of the mosAB genes makes A. vinelandii less resilient to Mo starvation, and growth is much more affected by tungstate inhibition.
A. vinelandii can fix N2 and assimilate nitrate simultaneously,193 in contrast to its close relative A. chroococcum.194 As both processes depend on the activity of two distinct molybdoenzymes, the MoFe protein, and the nitrate reductase, simultaneous use of Mo is ensured by a tightly regulated mechanism in which NifO seems to play an essential role.172 The nifO gene is grouped together with nifQ in the same gene cluster as nifB, fdxN, and rhdN (Figure 2).40,195 The stop codon of nifO overlaps with the start codon of nifQ, a type of translational coupling that has been reported to ensure coexpression of two consecutive genes.40,196 However, the expression of the genes within this operon is affected by metals present in the medium in different ways. For instance, under Mo starvation, nifO expression is enhanced, while nifQ expression decreases.172
In the presence of nitrate, nifO mutants have lower N2 fixation rates but higher nitrate reductase activity.40,172 On the other hand, constitutive nifO expression enhanced repression of the nitrate reductase genes.72 This phenotype led to the suggestion that NifO may direct Mo toward FeMo-co synthesis under conditions where nitrate is simultaneously being used.72 Therefore, NifO appears to be important for the ability of A. vinelandii to simultaneously assimilate nitrate and N2, although its exact role is not known. The protein contains a domain that is also found in a subfamily of arsenate reductases, enzymes that catalyze the reduction of arsenate to arsenite using glutathione. NifO also shows similarity to the Azospirillum brasilense DraB, a protein of unknown function encoded in the draTGB operon.197 In R. rubrum, and some other diazotrophs, the DraG-DraT system is responsible for the reversible inactivation of the Fe protein by ADP ribosylation.198
A function for the distribution of Mo between the MoFe protein and nitrate reductase has also been suggested for the product of modG, whose expression is regulated by ModE in response to molybdate.199 The Mo requirements for N2 fixation and nitrate assimilation are shifted in the modEG double mutant, which could not grow at 10 μM molybdate in the medium but exhibited maximum nitrate reductase activity at 10 nM molybdate.184 The role of ModG is not known, except for its atomic structure that shows a protein forming a homotrimer that binds eight atoms of Mo (Figure 20A).200
11.3. Role of NifQ in FeMo-co Biosynthesis
11.3.1. Information from nifQ Mutagenesis
The analysis of K. pneumoniae mutants requiring high concentration of molybdate in the medium for diazotrophic growth led to the identification of genes associated with Mo incorporation into FeMo-co and specifically to nifQ.129,143 Although not impaired in molybdate transport, nifQ mutants were defective in Mo accumulation and had lower levels of Mo in the MoFe protein. Supplementing the growth media with mM levels of molybdate suppressed the nifQ phenotype.143 Similar nifQ phenotypes were later observed in A. vinelandii(40,172) and R. capsulatus.201 In the symbiotic N2-fixing bacterium Sinorhizobium fredii, nifQ deletion did not affect N2 fixation but metal homeostasis and accumulation of certain types of porphyrins that could be involved in metal chelation in the nodules.202
An early observation associated with the study of K. pneumoniae nifQ mutants was that if cysteine was used as source of S in the medium, their molybdate requirements were similar to those of the wild type strain.203 Because both high molybdate and cysteine in the medium suppressed the nifQ phenotype, it was proposed that NifQ functioned in a reaction involving a Mo–S intermediate that could also be generated in absence of NifQ if the levels of Mo and S were high. Finally, because K. pneumonia nifQ mutants were not defective in nitrate reductase activity, the role of NifQ was assumed to be specific for FeMo-co biosynthesis.143
11.3.2. NifQ Carries a Novel [Mo-3Fe-4S] Cluster and Additional MoS2O2 Species
Heterologous expression of NifQ in E. coli appears to be difficult195,204 or produced inactive protein.73 To obtain NifQ protein from A. vinelandii, the nifQ gene was cloned downstream of the strong nifH promoter. The IMAC-purified A. vinelandii NifQ protein was described as a monomer with 3.1 Fe atoms and 0.3 Mo atoms per molecule, in contrast to the Mo-free protein produced in E. coli that instead contained up to 4.6 Fe atoms per protein.73
Three possible reasons have been presented to account for the substoichiometric levels of Mo bound to NifQ. First, Mo binding to NifQ might be labile and easily lost during the purification. Second, as NifQ is normally present at very low levels in nitrogenase-derepressed A. vinelandii, its overexpression could affect the capacity to load it with Mo. Similarly, the lack of Mo found at the E. coli protein could be explained by NifQ accessory proteins being deficient in this heterologous expression system. Third, if NifQ was involved in Mo delivery, any as-isolated NifQ preparation obtained from a wild type strain with ongoing FeMo-co biosynthesis should comprise loaded and unloaded NifQ molecules.
Amino acid sequence comparison of NifQ proteins revealed a conserved Cx4Cx2Cx5C motif at the C-terminus of the protein that could be involved in coordinating a metal cluster. Indeed, EPR spectroscopy showed that NifQ preparations contained redox-responsive [3Fe-4S]+ clusters and [Mo-3Fe-4S]3+ clusters and that incubation with Mo and sulfide increased the signal of the [Mo-3Fe-4S]3+ clusters.73 Consistent with the third possibility mentioned above, this result was interpreted as if each NifQ protein carried either a [3Fe-4S] cluster (Mo unloaded form) or a [Mo-3Fe-4S] cluster (Mo loaded form). It was hypothesized that these two forms were interconvertible by the uptake or release of Mo.173 Additional investigation using Mo and Fe K-edge EXAFS revealed the presence of two different Mo coordination environments in NifQ.205 In one of these environments, the Mo atom was coordinated to Fe (at 2.71 Å), S (at 2.34 Å), and O (at 2.12 Å), which corresponded to the [Mo-3Fe-4S] cluster previously observed in EPR spectroscopy.73 The second Mo environment did not involve an [Fe–S] cluster but rather two O atoms (at 1.73 Å) and two S atoms (at 2.23 Å), similar to Mo in dithiomolybdate (Figure 21). It was determined that each environment accommodated about half of the Mo bound to NifQ. Fe EXAFS also identified the Mo-free [3Fe-4S] cluster consistent with EPR assignments. Both Mo species were shown to be relevant for FeMo-co synthesis as elimination of the [Mo-3Fe-4S] cluster by chelation with α,α′-bipyridyl affected the ability of NifQ to serve as Mo donor, and displacement of the MoS2O2 species by CuCl2 abolished Mo donation by NifQ to a NifEN/Fe protein complex. The fact that both Mo environments in NifQ were relevant for in vitro FeMo-co synthesis was not surprising as conversion of molybdate (Mo6+) into the form of Mo present in FeMo-co (Mo3+) requires both replacement of O-ligands by S ligands and reduction of Mo for insertion into an [Fe–S] cluster.205,206 The [3Fe-4S] cluster of NifQ is the obvious site for reductive incorporation of Mo to yield a [Mo-3Fe-4S] cluster in dynamic equilibrium. The role of the MoS2O2 species is less obvious as it may represent an intermediate poised for incorporation into the [3Fe-4S] or a product released from the [Mo-3Fe-4S] cluster to be donated for FeMo-co synthesis.
Figure 21.
Metal cluster interconversions in NifQ. NifQ preparations carry substoichiometric amounts of [Mo-3Fe-4S-2O] cluster (0.3 mol cluster per mol NifQ) and an independent MoO2S2 species. A [3Fe-4S] cluster species makes up the full cluster complement (0.7 mol cluster per mol NifQ). Redox-gated incorporation of Mo into the [3Fe-4S] cluster has been shown. Molybdenum bound to NifQ is delivered to a NifEN/Fe protein complex for FeMo-co biosynthesis.
12. Discovery of FeMo-co Organic Moiety: The Homocitrate Synthase NifV
For many years after its isolation in 1977, FeMo-co was thought to contain exclusively Mo, Fe, and acid-labile S.15 However, unlike other [Fe–S] clusters, FeMo-co was unusually resistant to Fe chelators and showed net negative charge at neutral pH that was inconsistent with its proposed atomic composition. In addition, EXAFS analysis had suggested that the first coordination sphere of the Mo atom in FeMo-co contained three S atoms and three N or O atoms.207 A putative organic moiety in the cofactor was investigated but sugars, amino acid, or other common moieties could not be detected.208 In an extremely detailed investigation, Hoover and Ludden discovered in 1989 that the organic acid homocitrate was an integral part of FeMo-co.18 The crystal structure of the Mo-nitrogenase corroborated this finding and suggested that the oxygen atoms from the C2 hydroxyl and carboxyl groups were the O ligands to the Mo atom detected by early EXAFS studies (Figure 22).28
Figure 22.
Coordination around homocitrate in the A. vinelandii MoFe protein (PDB: 1M1N). (A) Depiction of the FeMo-co pocket showing solvent accessible tunnels to the cofactor. (B) Network of hydrogen bonds among homocitrate, nearby amino acid residues, and the pool of water molecules inside the pocket. (C) Interactions of the Mo atom with His442 and with the C2 hydroxyl and carboxyl groups of homocitrate. Images created with NGL viewer36 and RCSB PDB.
While homocitrate synthase catalyzes the first and committed step in lysine biosynthesis in many fungi and certain archaea,70 homocitrate for the nitrogenase cofactor is synthesized by the nifV gene product. The elucidation of NifV involvement in FeMo-co synthesis started many years earlier by the analysis of nifV mutant phenotypes. K. pneumoniae nifV mutant strains showed slow diazotrophic growth because they could not reduce N2 effectively.144,209 However, nifV strains reduced H+ and C2H2, which suggested that they were affected in the catalytic properties of nitrogenase. Similar results were shown later for the A. vinelandii nifV mutant strains.39 Hawkes and colleagues showed that the nifV defect lay in FeMo-co and, by doing so, presented direct evidence of FeMo-co being the site for substrate reduction. This experiment consisted in extracting the “activating factor” from MoFe protein of a nifV mutant and inserting it into another apo-MoFe protein. The reconstituted MoFe protein showed the altered substrate reduction characteristic of nifV strains.210
By using the in vitro FeMo-co synthesis assay, the requirement of a low molecular weight factor became evident. This factor was present in the extracts of all A. vinelandii and K. pneumoniae mutant strains tested except for nifV mutants and was therefore designated as the V factor.17 Production of the V factor was repressed by ammonium in the medium consistent with it being a metabolite produced by a Nif protein. Using the in vitro FeMo-co synthesis assay, the V factor was isolated and shown by nuclear magnetic resonance (NMR) spectroscopy and mass spectroscopy to be homocitric acid (R-2-hydroxy-1,2,4-butanetricarboxylic acid).211 Exogenously added homocitrate was then shown to be able to replace the V factor in the assay. Final demonstration that homocitrate was an integral part of FeMo-co in wild type MoFe proteins was obtained by preparing and adding 3H-homocitrate to the in vitro FeMo-co synthesis assay for its incorporation into apo-MoFe protein. Labeled MoFe protein was mixed with a larger amount of unlabeled enzyme and the mixture used to reisolated FeMo-co, which was further tested in independent FeMo-co insertion assays. Co-purification of 3H label with FeMo-co activity was found.18
A correlation between functional nifV gene and the accumulation of homocitrate in nitrogenase derepressed K. pneumoniae cells was shown.212 Moreover, phenotypic reversion of a nifV mutant strain was achieved by addition of homocitrate to the growth medium. The product of the nifV gene was later confirmed to be a homocitrate synthase by expression and purification from recombinant E. coli cells.213 Purified NifV was shown to be enough to catalyze the condensation of acetyl coenzyme A and α-ketoglutarate to form homocitrate.
As mentioned above, nifV mutant strains showed very low levels of N2 reducing activity. It was hypothesized that another organic acid could be partially incorporated into FeMo-co in these mutants. In fact, MoFe protein purified from a K. pneumoniae nifV mutant was shown to contain citrate.214 In this context, it had also been reported that citrate could replace homocitrate in the in vitro FeMo-co synthesis assay, although 100-fold excess of citrate was required to observe appreciable activity.211 Extensive analysis of organic acids that could replace homocitrate in the in vitro FeMo-co synthesis assay has been performed. Incorporation of several analogs into the cofactor resulted in reconstituted MoFe proteins with altered substrate specificities.215 Analysis of these MoFe proteins suggested that the following structural features of homocitrate are essential to obtain catalytically competent FeMo-co: its 1- and 2-carboxyl groups, its hydroxyl group, the R configuration of its chiral center, and a 4–6 carbon chain length with two terminal carboxyl groups.
13. NifEN Scaffold as Central Node in FeMo-co Biosynthesis
13.1. General Introduction to NifEN
The requirement of nifE for N2 fixation was first established in K. pneumoniae.216 From the analysis of a large number of Nif–K. pneumoniae strains by two-dimensional polyacrylamide gels, it was established that nifE and nifN gene products had mutual requirements for stability44 and were sensitive to O2 exposure.217 Absence of either NifE or NifN eliminated MoFe protein activity but not that of the Fe protein. Further in vitro assays suggested their involvement in FeMo-co biosynthesis.44
Taking advantage of the assay to follow FeMo-co biosynthetic activity in vitro,16 the nifE and nifN gene products could be purified from A. vinelandii, which confirmed their importance for FeMo-co synthesis.21 It was shown that NifE and NifN form a complex (hereafter called NifEN) of about 200 kDa. An α2β2 subunit structure similar to that of the MoFe protein was suggested. As it is the case for most other Nif components, purified NifEN was extremely sensitive to O2 exposure with a half-life of 1 min in air. Purified NifEN samples, initially obtained by conventional long procedures, contained 4.6 mol Fe per mol NifEN and presented DTH-responsive UV–vis absorption spectra with a shoulder at around 400 nm in the oxidized protein.21
Because NifE and NifN primary sequences resembled those of NifD and NifK (around 30–35% and 55–60% identity and similarity between NifD and NifE, and around 25–30% and 50–55% identity and similarity between NifK and NifN),67 and shared several conserved Cys residues with NifD and NifK (among which the FeMo-co binding α-Cys275),218 an evolutionary relationship was proposed.11,67 A revolutionary idea, two decades before it was biochemically shown, predicted that NifEN functioned as a scaffold on which FeMo-co was constructed and then delivered to the MoFe protein.11 The NifEN-bound [Fe–S] cluster(s) was suggested to either be a FeMo-co biosynthetic intermediate or a redox-active cluster enabling NifEN to alter the oxidation state of Mo for incorporation into FeMo-co. Later studies have shown that NifEN possesses some redox capacity, being catalytically competent to reduce simpler substrates (although at extremely low rates), for example, acetylene (C2H2) and azide (N3–), but not N2.219
13.2. NifEN Acts as Scaffold for FeMo-co Synthesis
The first biochemical evidence that NifEN has the ability to accumulate FeMo-co biosynthetic intermediates came from differential migration profiles on anoxic native gels.220 With the rationale that eventual FeMo-co precursors bound to NifEN would affect its electrophoretic mobility under native conditions, NifEN from different genetic backgrounds was analyzed. NifEN from extracts of the A. vinelandii ΔnifHDK strain CA12 migrated as a sharp band of higher mobility, compared to the slower and more diffuse NifEN band observed in extracts of the ΔnifB ΔnifDK strain DJ677. During the isolation of NifEN from CA12, its migration changed and converted to that of NifEN from the DJ677 strain, indicating the presence of a labile factor that dissociated during the purification process. On the contrary, addition of increasing amounts of purified NifB-co19 to NifEN from the DJ677 mutant background increased its migration to that observed using CA12 extracts, suggesting that the factor weakly bound to ΔnifHDK NifEN was in fact NifB-co or a derivative of it.220 No change in NifEN migration was observed upon addition of O2-exposed NifB-co, Fe(II) SO42–, or FeMo-co, indicating that the change in mobility specifically reflected the interaction of NifEN with functional NifB-co. It also suggested that NifEN produced in an A. vinelandii ΔnifHDK strain, such as CA12, could accumulate NifB-co in vivo.
Direct biochemical proof of NifEN acting as an intermediate scaffold for NifB-co-dependent FeMo-co biosynthesis came from the appearance of 55Fe label on NifEN after incubation with 55Fe-labeled NifB-co in incomplete in vitro FeMo-co synthesis mixtures, where either one of MgATP, Mo, homocitrate, or the Fe protein (all required for FeMo-co synthesis) was excluded from the mixture, or if apo-MoFe protein was previously saturated with exogenously added FeMo-co.150
13.3. Characterization of NifEN and NifEN-Bound FeMo-co Precursor
Our progressive understanding of the NifEN protein and its mechanism of action have been confounded by incompatible conclusions drawn from studies where different laboratories have employed different purification methods. Interestingly, these methods were often applied to the very same strain, generated at the Dean laboratory to facilitate rapid purification of large amounts of NifEN for accurate characterization.154 As such, much of our knowledge about NifEN is derived from IMAC-assisted purification of an overproduced His-tagged NifEN variant expressed in the A. vinelandii strain DJ1041 (ΔnifHDKTYnafAB) that places nifEN under the control of the strong promoter of nifH (see chromosomal region in Figure 2).154
This purified His-NifEN variant showed an α2β2 subunit conformation with UV–vis absorption spectra characteristic of proteins with [4Fe-4S]2+/1+ clusters. Molar extinction coefficients were consistent with two clusters per NifEN tetramer, which was also consistent with the measured protein-bound Fe content, EPR analysis, and VT-MCD spectra.154 The resonance Raman spectra additionally suggested an all cysteinyl coordination for these clusters, supported by the presence of sufficient Cys residues conserved in NifEN proteins.11 A structural model would locate these [4Fe-4S] clusters at positions equivalent to those of the P-clusters of the MoFe protein. The Fe content of 15 atoms per NifEN tetramer was, however, significantly higher than required for its two identical [4Fe-4S] clusters. Importantly, a NifEN-bound cluster exhibiting an axial EPR signal with a g value of 1.95 in the thionin-oxidized state was identified.221 It was suggested that this EPR signal originated from a trapped FeMo-co precursor since strain DJ1041 had both FeMo-co synthesis and delivery to apo-MoFe protein interrupted by mutations in nifH and nifDK, respectively.
Disruption of the nifB gene in the A. vinelandii strain DJ1041 generated the YM9A and UW243 strains (ΔnifB ΔnifHDKTYnafAB; hereafter designated as ΔnifB NifEN or apo-NifEN for simplicity) at the Ribbe and Ludden laboratories, respectively. These strains enabled the purification of a less colored NifEN protein lacking any bound FeMo-co precursor.24,60 Comparison of the IDS-oxidized ΔnifB NifEN protein to the NifEN isolated from the DJ1041 strain showed that the g = 1.95 EPR signal, previously identified by Goodwin and Dean,221 was missing in the ΔnifB NifEN spectrum.60 Consistent with previous studies,150 it was also shown that the NifEN-bound FeMo-co precursor did not contain homocitrate or Mo.
More information about the chemistry of the NifEN-bound FeMo-co precursor was obtained from a series of related studies that used more advanced methods to analyze high-quality preparations of His-tagged NifEN.158,222−225 First, DJ1041 NifEN protein was compared to the YM9A ΔnifB NifEN using XAS/EXAFS data analysis.158 The NifEN-bound FeMo-co precursor signal was estimated by subtracting the signal originating from the so-called permanent [4Fe-4S] clusters of ΔnifB NifEN. Comparison of the EXAFS data to structural models derived from known high-nuclearity [Fe–S] and mixed metal–sulfur models implied a seven or eight Fe precursor of FeMo-co-like structure, while a six Fe cluster variant was precluded due to the intensity of short-range Fe–Fe scattering.158 The eight Fe model was proposed as the most likely form when comparing the EXAFS fit results with previous biochemical characterization.60 Finally, an [8Fe-9S] FeMo-co precursor was assumed from X-ray structural studies of NifEN, although the electron density was not well resolved (see below).159
13.4. NifEN Readily Converts NifB-co into Next FeMo-co Biosynthetic Intermediate: Isolation of VK-Cluster
FeMo-co biosynthetic intermediates produced by NifB and NifEN have been analyzed using the natural FeMo-co-precursor carrier protein NifX, a tool used to avoid spectroscopic interference from other [Fe–S] clusters present in NifB and NifEN. The NifX protein does neither have permanent [Fe–S] clusters nor binds Fe or other [Fe–S] clusters.24 Using the A. vinelandii NifX produced and purified from E. coli, and NifB-co purified from K. pneumoniae, a NifX/NifB-co complex could be generated in vitro. NifB-co binding produced changes in the circular dichroism (CD) spectrum of NifX, which were used to titrate binding and estimate a Kd for NifB-co of about 1 μM. Transfer of NifB-co from NifX to apo-NifEN could be followed by anoxic native gel electrophoresis. Reciprocal transfer of Fe from the precursor-charged NifEN to NifX was also observed. No stable protein complex of NifEN with NifX was observed, suggesting transient interaction and dynamic exchange of FeMo-co precursors between NifX and NifEN. It was therefore suggested that NifX could function as a reservoir of FeMo-co precursors to buffer their flow, in addition to its role as carrier of NifB-co from NifB to NifEN.24 This hypothesis was consistent with NifX not being essential for FeMo-co synthesis in vitro.22,141 The effect of nifX disruption in vivo was evident during N2 fixation under conditions of Mo or Fe limitation, which are supposed to impair or slow down cofactor biosynthesis.145,226
The NifEN [Fe–S] cluster extracted by NifX (defined as the VK-cluster24) did neither contain Mo nor homocitrate and showed the same requirements for in vitro FeMo-co synthesis as NifB-co and the NifEN-bound FeMo-co precursor (i.e., NifEN, Fe protein, Mo, homocitrate, MgATP, and DTH), indicating that this VK-cluster was either NifB-co or an intermediate cluster formed at NifEN at the expense of NifB-co (Table 2). Importantly, the VK-cluster bound to NifX was EPR active under both oxidized (axial S = 1/2 signal) and reduced (isotropic S = 1/2 signal) conditions, and underwent fully reversible two electron transitions,24 in contrast to the diamagnetic NifX/NifB-co complex,20,24 NifB-co alone,19 or NifB-co bound to the MoFe protein.153 This indicated that NifB-co and the VK-cluster were different entities, or at least metalloclusters with distinct electromagnetic signatures. The EPR signal of the isolated VK-cluster was also different from the S = 1/2 EPR signal observed in IDS-oxidized NifEN, which had been attributed to a FeMo-co precursor60,221 but is reminiscent of a [3Fe-4S]+1 type of cluster. Whether these electronic state differences are physiologically and chemically relevant for FeMo-co maturation requires further investigation.
13.5. Identification of Additional [Mo-3Fe-4S] Cluster at DJ1041 NifEN Protein
As mentioned above, the technique employed to isolate NifEN protein can significantly affect the composition of the protein-bound metal clusters. A study in which the protocol for the isolation of the DJ1041 NifEN protein was modified by using IMAC with Co2+ instead of Zn2+ (distinct metal ions can affect the His-tag binding selectivity), produced NifEN preparations with significantly higher Fe content,61 24 mol Fe per mol NifEN tetramer compared to the previously reported values of 9.5,154 or 16 Fe atoms.60,221,222 Importantly, the NifEN protein also contained Mo at levels of about 0.3–0.4 atoms per tetramer, eliminating the strict requirement for external molybdate addition in a defined in vitro FeMo-co synthesis assays using purified protein components.61 Furthermore, apo-MoFe protein activation in absence of exogenously added molybdate increased in proportion to the amount of heat-denatured NifEN used to release NifEN-bound Mo, strongly indicative of the NifEN protein serving as Mo source. It is unlikely that the Mo was adventitiously bound to NifEN as the buffers used throughout the purification procedure contained only negligible amounts of molybdate, and since addition of an excess tungstate during purification decreased the levels of Mo, but not Fe, in NifEN.61 In addition, apo-NifEN purified from strain UW243 (a ΔnifB derivative of DJ1041) neither contained Mo nor functioned as Mo source after heat denaturation, indicating that Mo binding to NifEN occurs after NifB-co binding or is stabilized by it, in agreement with other observations.227 The study also suggested that the Fe protein was not strictly necessary to move Mo into NifEN. Because the VK-cluster did not contain Mo, it was proposed that NifEN would have a site for transient Mo species prior incorporation into the VK-cluster. This hypothesis was confirmed by XAS/EXAFS measurements that detected a Mo environment in purified NifEN with similar oxidation state but different ligand field to Mo in the MoFe protein. The Mo environment was found to resemble that of a [Mo-3Fe-4S] cubane.228 EXAFS analysis did not elucidate whether the [Mo-3Fe-4S] cluster was a fragment of a larger cluster. As a transient cluster proposed to act as a Mo-donor to the VK-cluster, the [Mo-3Fe-4S] cluster would be extremely labile, which would explain why its presence or absence depends on the procedure used to purify NifEN.
The homocitrate content of purified NifEN was not specifically determined. However, in vitro reaction mixtures lacking molybdate and homocitrate resulted in some apo-MoFe protein activation, suggesting that NifEN could also provide the organic acid.61 Mo and homocitrate provided by NifEN were suboptimal in the in vitro FeMo-co synthesis assay as addition of external molybdate and homocitrate elevated FeMo-co synthesis more than 10-fold.
13.6. NifEN Atomic Structure Determination
The structure of NifEN loaded with VK-cluster has been solved by X-ray crystallography, confirming a tetramer of two αβ dimer subunits similar to the MoFe protein (Figure 23A,B).159 Each permanent [4Fe-4S] cluster locates at the αβ interface and is coordinated by Cys37, Cys62, and Cys124 from the α-subunit and Cys44 from the β-subunit, much like the P-cluster at the MoFe protein (Figure 23C). The VK-cluster (L-cluster, see Table 2) is ligated by Cys25 from the α-subunit in an almost surface exposed position, in contrast to FeMo-co at the MoFe protein, which is buried below the surface.6 Unfortunately, the electron density was not resolved enough to unambiguously determine the structure of the VK-cluster nor involvement of additional amino acid residues in cluster ligation (Figure 23D). The labile [Mo-3Fe-4S] cluster was absent, as expected, given the method used to purify the protein. Using structural information, it was proposed that, upon incorporation of Mo and homocitrate into the VK-cluster, matured FeMo-co would be inserted into an open funnel at NifEN, similarly to the position of FeMo-co at the MoFe protein.159 Subsequent interactions between NifEN and the MoFe protein were hypothesized to induce a conformational shift at NifEN that would then relocate FeMo-co back to the surface for delivery to the apo-MoFe protein.
Figure 23.
Comparison of NifEN and MoFe protein structures. (A) Cartoon depiction of NifEN atomic structure (PDB: 3PDI). Partial transparency has been applied to the polypeptide chains to visualize metal clusters. (B) Cartoon depiction of MoFe protein atomic structure (PDB: 3K1A). (C) [4Fe-4S] cluster structure and coordination by Cys residues in NifEN. (D) VK-cluster structure and coordination to α-Cys25 of NifEN. Note superficial position of the VK-cluster in the protein. Each αβ half of NifEN contains a [4Fe-4S] cluster and a VK-cluster. Graphics generated with the PyMOL Molecular Graphics System, Version 2.3.2 Schrödinger, LLC.
14. Incorporation of Mo and Homocitrate into NifEN-Bound FeMo-co Precursor
14.1. Conversion of VK-Cluster into FeMo-co in NifEN
Advanced spectroscopic studies were used to follow the conversion of VK-cluster into FeMo-co within NifEN. This was achieved by eliminating the terminal FeMo-co acceptor (apo-MoFe protein) from an otherwise complete reaction mixture of a NifEN-dependent in vitro FeMo-co synthesis assay (Figure 13) containing NifEN, Fe protein, Mo, homocitrate, MgATP, and DTH, a process designated as “NifEN activation”.222 After incubation, the “activated NifEN” was reisolated and analyzed. The EPR signal attributed to the IDS-oxidized FeMo-co precursor60 disappeared from NifEN. Activated NifEN contained 1 mol Mo per mol NifEN, and XAS/EXAFS analysis of the Mo environment pointed to the incorporation of Mo into an [Fe–S] cluster. However, the S = 3/2 EPR signal characteristic of FeMo-co was not observed, suggesting that the new cluster had a different spin state from both the VK-cluster and FeMo-co.222 More recently, a NifEN-bound cluster with EPR and XAS/EXAFS spectroscopic properties more similar to FeMo-co was identified when the concentration of DTH in the reaction was increased to 20 mM.224 Activated NifEN was capable of activating apo-MoFe protein in these new reaction mixtures. It was claimed that FeMo-co delivery from activated NifEN involved direct interaction with apo-MoFe protein. However, to our knowledge, no experimental evidence supporting this interaction has been provided. In addition, by using the mild Strep-tag affinity chromatography (STAC) assay, which has been successful in identifying other protein partners transiently interacting with the MoFe protein in various genetic backgrounds,23 no NifEN polypeptides have been detected so far.114
14.2. Fe Protein Is Essential for Maturation of VK-Cluster into FeMo-co
An early assay for NifB-co-dependent in vitro FeMo-co synthesis and insertion into apo-MoFe protein using purified components (including apo-NifEN, NifB-co, NifX, Fe protein, apo-MoFe protein, MgATP, Mo, homocitrate, and DTH) was first developed,141 and later on improved,227 by Ludden and colleagues (Figure 13). Elimination of the Fe protein from the reaction mixtures prevented activation of apo-MoFe protein, showing its requirement for this process. This requirement for the Fe protein was also observed in NifEN-dependent in vitro FeMo-co synthesis assays in which VK-cluster-containing NifEN substituted for NifB-co and apo-NifEN (Figure 13).60,61 In these studies, removal of the Fe protein resulted in negligible apo-MoFe protein reconstitution.
There are, however, contradictory reports about the biochemical requirements for the Fe protein to function in FeMo-co synthesis. Hu and collaborators reported the essentiality of the [4Fe-4S] cluster of the Fe protein.223 However, it is worth noting that there are many reports ruling out an obligate redox role for the Fe protein in FeMo-co biosynthesis, among them reports showing that nifH mutants impaired in electron transfer to the MoFe protein were active in FeMo-co synthesis in vivo,48,125,229 and a report showing that apo-Fe proteins functions in FeMo-co synthesis.90 Similarly, the ability of the Fe protein to hydrolyze MgATP appeared to be required for in vitro FeMo-co synthesis in some studies,60,223 whereas other studies show that Fe protein variants capable of MgATP binding, but not hydrolysis, were competent in FeMo-co synthesis in vivo and in vitro.123,230,231 Perhaps the most detailed analysis of Fe protein requirements to function in the in vitro FeMo-co synthesis assay was provided by Ludden and collaborators and is summarized in Table 3 and Figure 24.123,232 From these studies, a correlation between MgATP binding and ability to support FeMo-co synthesis can be inferred. However, many variants that failed to hydrolyze MgATP were shown active in the in vitro FeMo-co synthesis assay.
Table 3. Functionality of Altered Forms of NifH in in Vitro P-Cluster Maturation and in Vitro FeMo-co Synthesis Assaysc.
e– transfer to the MoFe protein.
Green tick/red cross denote the ability/inability to function in the properties tested; – means not determined.
Table from ref (232), modified with permission. Copyright 2004 Springer Nature.
Figure 24.
Position in the A. vinelandii Fe protein of amino acid residues (from Table 3) mutated to investigate their involvement in Fe protein function. (A) Semi transparent surface and loop representation. (B) Ribbon diagram in the same orientation. (C) Main chain loop representation. Graphics generated with the PyMOL Molecular Graphics System, Version 2.3.2 Schrödinger, LLC. ADP molecules are represented as filled spheres at the bottom part of the protein. The [4Fe-4S] cluster is depicted with stick and balls in the upper part of the protein. Each mutated amino acid residue is assigned a different color.
Finally, while there is compelling evidence that the Fe protein is required for the incorporation of Mo into the VK-cluster, there are disagreements in the exact role played by the Fe protein in this process. While some groups argue that the Fe protein functions as a Mo/homocitrate insertase that delivers Mo covalently bound to homocitrate into NifEN (Figure 25A), others claim that it is NifQ that delivers Mo to NifEN in the presence of the Fe protein (Figure 25B). Previously, we proposed a mechanism, accommodating most published data, in which the Fe protein would dock at the surface of NifEN and induce a conformational change in the complex that results in Mo transfer from the transient NifEN [Mo-3Fe-4S] cluster into the VK-cluster.7 Homocitrate previously bound to NifEN could be incorporated into the VK-cluster in the same reaction. In our opinion, despite a considerable number of experiments during the past decade, no strong new data have been presented that would significantly argue against this model. The next two sections describe in detail the experimental evidence supporting both claims.
Figure 25.
Two models for NifEN metal cluster composition and Mo incorporation into an Fe–S cluster FeMo-co precursor to mature the cofactor within NifEN. Each model shows the protein and metal cluster nomenclature used. EPR signals used as spectroscopic evidence of protein-bound FeMo-co, FeMo-co precursors, and other MoFeS clusters are shown. (A) Model in which the product of NifB, called L-cluster, is delivered by NifB to apo-NifEN via direct interaction to generate the NifENL form. In subsequent step, the Fe protein delivers a Mo-homocitrate complex that replaces one terminal Fe atom in the L-cluster to generate the M-cluster found in a NifENM species. The M-cluster is transferred from NifENM to apo-MoFe protein by direct interaction to generate active MoFe protein. (B) Model in which the product of NifB, called NifB-co, is transferred to apo-NifEN via NifX. Within NifEN, NifB-co is rapidly transformed into the VK-cluster by NifEN. VK-cluster can be extracted from NifEN by NifX. Both NifX/NifB-co and NifX/VK-cluster complexes have been spectroscopically analyzed. In addition to the VK-cluster, NifEN contains a [Mo-3Fe-4S] cluster proposed to be donated by NifQ, which carries a similar cluster. Homocitrate is donated by NifV. Once all FeMo-co precursors are bound to NifEN, the docked Fe protein exerts a conformational change in NifEN that promotes FeMo-co formation. Mature FeMo-co is transferred to apo-MoFe protein via NafY. NifQ inset adapted from ref (73). Copyright 2008 PNAS.
14.3. Does Fe Protein Function as Mo/Homocitrate Insertase?
Ludden and co-workers found Mo attached to NifEN in an in vitro FeMo-co synthesis reaction containing NifEN, Fe protein, NifX, NifB-co, MgATP, DTH, isotope-labeled 99Mo, and homocitrate.227 It is important to mention that this study was performed prior to the generation of strain DJ1041 (that accumulates FeMo-co precursor at NifEN due to its ΔnifHDKTYnafAB genotype), when purified preparations were obtained by tedious and long procedures that rendered precursor-free apo-NifEN. Under these in vitro conditions, removal of either MgATP, Fe protein, or NifB-co from the mixture prevented 99Mo accumulation at NifEN, indicative of Mo insertion into the NifEN-bound precursor being an Fe protein-dependent process requiring prior NifB-co transfer into NifEN. Some 99Mo label was found associated to the Fe protein in the complete reactions, and it was therefore proposed that Fe protein might serve as the entry point for Mo incorporation into NifEN. Interestingly, 99Mo label of the Fe protein was negligible in the absence of the rest of the components, discarding the ability of the Fe protein to bind to Mo by itself.227
Further insight into the Fe protein role was obtained from the analysis of NifEN and Fe protein reisolated from reaction mixtures in the procedure described above as “NifEN activation”.223 Each reisolated NifEN or Fe protein was tested for their ability to support FeMo-co synthesis and apo-MoFe protein activation in reactions containing new NifEN and Fe proteins, but without additional Mo or homocitrate. Not only the activated NifEN but also the Fe protein was found to accumulate Mo (at about 2 mol Mo per mol Fe protein) and to support NifEN-dependent FeMo-co synthesis and apo-MoFe protein activation. Similar to the previous study,227 these results suggested that the Mo and homocitrate required for NifEN-dependent FeMo-co biosynthesis were provided by the Fe protein, which was then assigned a role as Mo/homocitrate insertase.223 Some activity was also observed when the Fe protein was incubated with apo-NifEN in the initial activation reaction, indicating that accumulation of Mo and homocitrate at the Fe protein was not strictly dependent on NifEN containing the VK-cluster. Along with NifEN, MgATP hydrolysis, the [4Fe-4S] cluster containing Fe protein, and DTH were required for Mo incorporation into the Fe protein.223 XAS/EXAFS analysis of reisolated Fe protein showed that its interaction with Mo did not depend on homocitrate. However, a refined study indicated that, to be productive, the incorporation of Mo and homocitrate had to be simultaneous and not sequential.225 A NifEN-bound cluster containing Mo but lacking homocitrate could be generated but was inactive as FeMo-co precursor.
To summarize, these studies were interpreted as the Fe protein functioning as insertase that delivers a Mo/homocitrate complex into NifEN for the maturation of the VK-cluster into FeMo-co (Figure 25A).158,222−225 In our opinion, this interpretation has some limitations. First, if the role of the Fe protein in FeMo-co synthesis was to transfer Mo/homocitrate to NifEN, it should be possible to make this Mo/homocitrate/Fe protein complex in vitro in the absence of NifEN, whereas all results described above showed that NifEN are required for a significant accumulation of Mo (or 99Mo) at the Fe protein. Second, as isolated Fe protein preparations neither contain Mo nor can be used as a Mo donor for in vitro FeMo-co synthesis.61 Third, the isolation of a NifEN protein partially equipped with a [Mo-3Fe-4S] cluster from an A. vinelandii nifH mutant background argues against the Fe protein functioning in Mo delivery to NifEN.228 One explanation for this could be that another protein, for example, VnfH, could partially replace the Fe protein in vivo. In support of this hypothesis, it has been shown that the low amount of FeMo-co produced in vivo in a nifH mutant background is minimized in the nifHvnfH double mutant.23 Fourth, it has been shown that the Fe protein can bind molybdate ions at the position corresponding to the γ phosphate of ATP,5 which would be incompatible with the requirement of Fe protein ATP hydrolysis. It must be noted, however, that this binding occurred in a crystallization solution containing 0.2 M molybdate, which is certainly not physiological and four orders of magnitude above the concentration used in the in vitro FeMo-co synthesis assays (18 μM molybdate). Fifth, in vitro FeMo-co synthesis reactions starting from 55Fe labeled NifB-co also results in 55Fe-labeled Fe protein (with similar requirements as for the 99Mo label). The simplest explanation to account for this multiple labeling is that the Fe protein interacts with FeMo-co or the FeMo-co precursor in a complex with NifEN and that, upon complex dissociation, a part of this label ends up in each protein.
14.4. NifQ Delivers Mo to NifEN for Its Incorporation into FeMo-co Precursor
A major step to understand the various processes converging at NifEN came from the identification of NifQ as direct donor of Mo for FeMo-co synthesis.73 Co2+ IMAC purification of overexpressed and His-tagged NifQ from A. vinelandii identified the protein as a monomer containing a [Mo–Fe–S] cluster with substoichiometric amounts of Mo. EPR spectroscopy suggested a heterogeneous NifQ population with redox responsive [3Fe-4S]+ clusters and [Mo-3Fe-4S]3+ clusters (see Section 11.3). Therefore, NifQ fulfills the requirements expected of a Mo donor.
As isolated NifQ supported in vitro FeMo-co synthesis and apo-MoFe protein activation in a reaction mixture lacking Mo but containing apo-NifEN (isolated from the A. vinelandii strain UW243), Fe protein, NifB-co, homocitrate, MgATP, and DTH.73 In support of previous studies, removal of Fe protein or homocitrate from the reaction mixture prevented or strongly reduced FeMo-co synthesis. Heat-denatured NifQ could not support FeMo-co synthesis, proving the direct requirement of NifQ for the reaction. Mo transfer was not observed when NifQ was incubated with NifEN or the Fe protein separately. However, coincubation of all three protein components resulted in Mo transfer from NifQ to both NifEN and the Fe protein,73 as previously observed from 99Mo labeling and anoxic native gels,227 and indicative of the formation of a transient complex between NifEN and Fe protein that permits or enhances Mo unloading from NifQ. Whether NifQ transfers only Mo or its entire cluster is not known. In this regard, it should be noted that the [Mo-3Fe-4S] cluster of NifQ resembles the transient [Mo-3Fe-4S] cluster found in NifEN preparations. Interestingly, not all NifQ-bound Mo was transferred. In attempts to understand the function of each of the two distinct Mo sites in NifQ (see Section 11.3), individual elimination of each Mo site lowered or even suppressed the ability of NifQ to provide Mo the FeMo-co biosynthesis.205
15. Metallocluster Trafficking in FeMo-co Synthesis: Roles of NifY, NafY, and NifX
Although activation of apo-MoFe protein in vitro is readily accomplished by the simple addition of isolated FeMo-co, this is less likely to happen in vivo due to the oxidative lability of the cofactor and its instability in aqueous solutions.15In vivo, both mature FeMo-co and its precursors appear to be delivered/protected by cluster carrier proteins, that is, NifX and NafY, which are proposed to transfer the clusters from NifB to NifEN and from NifEN to the MoFe protein, respectively. Early analysis of NifY and NifX amino acid sequences from A. vinelandii, K. pneumoniae, and R. capsulatus showed that NifX and the C-terminal 150 amino acids of NifY shared conserved residues.38,233 The NifX sequences of R. capsulatus(233) and A. vinelandii(145) also showed similarity to the C-terminal domain of their respective NifB proteins. The implications from the presence of a NifX-like domain in NifB have been discussed above (Section 10.3). Co-transcription of the nifY gene with the structural nitrogenase genes nifH, nifD, and nifK, and that of nifX with the FeMo-co assembly scaffold genes nifE and nifN in many diazotrophs, including A. vinelandii and K. pneumoniae,38,177,234 suggested that NifY was associated with the MoFe protein, whereas NifX would be associated with the NifEN complex. In this context, it is worth noting that the nifDKY and nifENX regions presumably resulted from a gene duplication event.67
15.1. NifX
Disruption of the nifX gene does not to cause significant effects on N2-fixing capability in a range of diazotrophs tested.38,233,235,236 Although early work in K. pneumoniae suggested that NifX could act as a negative regulator of nif gene expression,236 later work pointed to a more direct role in FeMo-co synthesis as metallocluster carrier capable of NifB-co binding,20,24,151,152 which implies that eventual regulatory effects could be of indirect nature. In contrast to the lack of phenotype reported from earlier in vivo studies, work in vitro showed that NifX stimulated FeMo-co synthesis.141 NifX was proposed to be transiently attached to NifEN during some stage of FeMo-co synthesis, a hypothesis that has been experimentally confirmed.114
The importance of NifX for efficient FeMo-co biosynthesis was later validated in vivo, when A. vinelandii or H. seropedicae was grown under stressing N2-fixing conditions in media presenting either Mo or Fe deficiency.145,226 A more careful study of the components required for in vitro synthesis of FeMo-co showed that although NifX was not strictly essential for the reaction, the protein did stimulate FeMo-co synthesis, and the effect was dose-dependent.22 Higher concentrations of NifX inhibited FeMo-co synthesis and apo-MoFe activation in vitro, indicating a potential competition for FeMo-co precursors or the finalized cofactor between NifX and other proteins (e.g., NifEN and apo-MoFe protein).
VnfX, a NifX homologue expressed together with vnfE and vnfN, was found to be involved in the synthesis of the FeV-cofactor for the vanadium nitrogenase.237 VnfX could bind NifB-co in vitro but also an O2-sensitive homocitrate-free FeV-co precursor containing Fe, S, and V. As for NifX, the vnfX gene was not required for FeV-co synthesis in vivo or for V-dependent diazotrophic growth. That NifX could also act as reservoir for other FeMo-co precursors, similar to VnfX in the V-nitrogenase system,237 which was proposed when A. vinelandii NifX was found to extract the VK-cluster from NifEN.24 One difference of NifX compared to VnfX is that the later was shown to bind a V-equipped precursor,237 whereas no heterometal (i.e., Mo) was found at the NifX-bound precursors.24 However, in vitro FeMo-co synthesis using isotope-labeled 99Mo has shown accumulation of 99Mo in NifX. On the basis of this result, we hypothesize that a precursor containing Fe, S, and Mo could be bound to NifX if isolated from the appropriate mutant background.227
In summary, NifX acts as carrier of NifB-co between NifB and NifEN and as a temporal storage protein for the NifB-co immediately processed by NifEN (in the form of the VK-cluster). An additional population of NifX stable bound to NifEN has been shown by STAC experiments.114 Its function must be explored, but it is easy to imagine a function as stabilizing factor similar to that provided to apo-MoFe protein by NafY. To date, the lack of structural information for NifX limits our understanding for how the protein interacts with NifEN and NifB as well as cofactors.
15.2. NifY and NafY
15.2.1. NifY and Gamma Protein Form Stable Complex with Apo-MoFe Protein Containing Mature P-Clusters
The nifY gene is located together with the structural nitrogenase genes in the A. vinelandii and the K. pneumoniae chromosomes (Figure 2). It was early described as dispensable for growth under normal N2 fixation conditions in A. vinelandii.38 However, the NifY protein was first identified in K. pneumoniae associated to the apo-MoFe protein from the nifB– strain UN106,238 or the nifN– mutant strain UN1217.109 About two molecules of NifY were found attached to each apo-MoFe protein tetramer. Apo-MoFe protein in extracts of a K. pneumoniae a nifY nifB double mutant could not be activated by FeMo-co to the extent of apo-MoFe protein in extracts of a nifB mutant strain. Importantly, upon activation by FeMo-co, NifY dissociated from the reconstituted MoFe protein.109 NifY was then proposed to facilitate FeMo-co activation by maintaining apo-MoFe protein in a conformation competent for cofactor insertion.
A better understanding of the mechanism of FeMo-co insertion was obtained from experiments with apo-MoFe protein produced by the A. vinelandii nifH mutant strain UW97.108 First, FeMo-co activation of this MoFe protein required prior action of the Fe protein and MgATP to convert the ΔnifH apo-MoFe protein into a variant competent for FeMo-co insertion. Second, apo-MoFe protein thus prepared for FeMo-co insertion was found to readily bind a protein designated as the Gamma protein forming a α2β2γ2 complex.108 Gamma was not the product of the A. vinelandii nifY gene, but it appeared to play the same role as NifY in K. pneumoniae. However, the primary sequence of the Gamma protein (later identified as NafY, see below) and the Gamma-encoding gene were unknown at the time. As the crystal structure of the active MoFe protein showed that FeMo-co was buried within the protein,6 it was proposed that Gamma could function by making the FeMo-co binding site more accessible for the cluster or that it could promote interactions with other factors involved in FeMo-co insertion.108 Together with the work in K. pneumoniae, this study supported a stepwise maturation of the MoFe protein where at least one intermediate step involves the association between the apo-MoFe protein and NifY/Gamma.
15.2.2. Gamma Binds FeMo-co
The stepwise model for apo-MoFe protein activation and the function of Gamma in A. vinelandii became more elaborate when a third Gamma population was identified, in addition to the unassociated (free Gamma) and the apo-MoFe protein-bound forms. This third Gamma pool was found to bind to FeMo-co and be able to donate it to the apo-MoFe protein.178 Therefore, Gamma appeared to play two roles: one that stabilizes apo-MoFe protein in a conformation that facilitates FeMo-co insertion, and one that acts as an intermediate FeMo-co carrier/insertase. The latter role was supported by work showing that FeMo-co accumulated in Gamma in A. vinelandii strains lacking nifD or nifK(178) and that Gamma bound FeMo-co synthesized in the in vitro FeMo-co synthesis assay performed in the absence of apo-MoFe protein.142,150,227
15.2.3. nafY Gene of A. vinelandii Codes for Gamma Protein
Interestingly, in A. vinelandii, the nifY gene does not code for the Gamma protein, and no biochemical activity has yet been assigned to it. The identification of nafY (for nitrogenase accessory factor Y) as the Gamma gene was important because a wealth of biochemical information could then be linked to a particular amino acid sequence and a new family of proteins with similar amino acid sequences and functions was revealed.145 The nafY gene is located at the minor nif cluster together with the rnf genes and regulated independently of the nif genes (Figure 2).
NafY showed sequence similarity to NifY, VnfY, NifX, VnfX, and to the C-terminal domain of NifB (Figure 26). In particular, NifY and NafY share about 40% identity, with NafY having a short C-terminal extension dominated by negative amino acids. The roles of NifY, NifX, and VnfX have been discussed above. VnfY appears to be involved in the maturation of V-nitrogenase as an A. vinelandii strain with deleted vnfY has almost no VFe activity and shows defective incorporation of V into the VFe protein.239 The vnfY gene is located downstream vnfK in a location analogous to nifY. In addition, another nafY homologue designated as nafX has recently been found downstream of nafY at the minor nif gene cluster (Figure 2).23 The function of NafX is unknown.
Figure 26.
Core domain conservation among members of NafY family of proteins. NafY and homologues shown are from A. vinelandii. UniProt accession numbers are NafY (C1DMA1), NifY (C1DH00), NifX (C1DH05), VnfX (C1DI38), VnfY (C1DI21), and NifB (C1DMB1). Core domain is shown in orange, SAM-radical domain in green, and the sterile α motif domain in blue.
Deletion of the nafY gene did not cause any significant effect on Mo-dependent diazotrophic growth, in line with other work suggesting that cluster-binding proteins (e.g., NifX and NifY) are nonessential.38,233,235,236 Because of the possible functional overlap between the NafY, NifY, and NifX proteins, A. vinelandii strains with one, two, or three of these genes removed were generated.145 The mutant strains showed only minor growth phenotypes under standard N2-fixing conditions. However, when cultures were grown under Mo-stressing conditions, the strains lacking nafY and nifX genes were significantly affected. No significant effect from nifY deletion was observed. Apo-MoFe protein levels in the ΔnifB ΔnafY strains were severely affected, indicating that NafY function was important for apo-MoFe protein stability. Consistently, the reconstitution of apo-MoFe protein by addition of FeMo-co in the ΔnifB ΔnafY strain was 50% compared to the ΔnifB strain.145
15.2.4. Structure and Function of NafY Protein Domains
Sequence alignments of the A. vinelandii NifX, VnfX, NafY, NifY, and VnfY proteins, and the K. pneumoniae NifY, show that NafY and NifY have N-terminal extensions (Figure 26).240 Functional interaction of full-length A. vinelandii NafY and an N-terminal truncated NafY variant (designated as the “core domain”) with FeMo-co and with the apo-MoFe protein was investigated. While both NafY variants could efficiently bind FeMo-co, binding to apo-MoFe protein was weakened in the truncated one, indicating that FeMo-co binds to the C-terminal core domain of NafY and that the function of the N-terminal extension could be to convey interaction between the NafY and the apo-MoFe protein.240
The structure of full-length NafY could not be resolved because of endogenous proteolytic activity. Isolation of NafY using a different purification protocol, or the use of cleavage-resistant site-directed NafY variants, might be useful for this. To date, the two NafY domains have been independently characterized at the atomic level (Figure 27A). The structure of the core domain was solved by X-ray crystallography and shown to fold as a mixed five-stranded β-sheet flanked by five α-helices in a conformation that resemble proteins from the ribonuclease H superfamily (Figure 27B).240 Such a fold has no structural similarities to the FeMo-co binding pockets at NifEN or the MoFe protein. The N-terminal domain structure was solved by NMR and found to have an all-helical fold similar to that of a putative protein interaction module named as the sterile alpha motif and normally involved in protein–protein or protein-RNA interactions (Figure 27C).241,242 Both NafY domains appear to fold independently of each other and to have very limited interactions in the full-length protein, consistent with their divergent biochemical functions.113
Figure 27.
Atomic structures of NafY N-terminal and core domains. (A) N-terminal and C-terminal (core) domains of NafY. (B) NafY N-terminal domain structure (98 residues) solved by NMR (PDB 2KIC). (C) NafY core domain structure (145 residues) solved by X-ray crystallography (PDB 1P90). The His121 residue essential to FeMo-co binding is labeled. Graphics generated with the PyMOL Molecular Graphics System, Version 2.3.2 Schrödinger, LLC.
M. thermoautotrophicum MTH1175 protein is a homologue of NifX. MTH1175 structure has also been determined by NMR and consists of an α/β topology with a single mixed β-sheet, two flexible loops, and an unstructured C-terminal tail.243 Both MTH1175 and NafY share sequence similarity to the IssA protein from the thermophilic archaeon Pyrococcus furiosus, a protein storing Fe and S in the form of thioferrate. Interestingly, IssA also contained an unstructured C-terminal tail that was suggested to bind to thioferrate.244 If these findings relate to the mechanism by which NifY/NafY/NifX carry [Fe–S] clusters is not clear. Unfortunately, the C-terminal 11 amino acids of NafY were missing from the core domain and therefore not resolved by X-ray crystallography.
Further biochemical characterization established that NafY was capable of binding one molecule of FeMo-co with high affinity, with the NafY His121 being essential for FeMo-co binding.136 His121 is part of an HFG domain located at the surface of NafY240 and is conserved in the K. pneumoniae NifY but not in the A. vinelandii NifY. No FeMo-co binding was detected in the absence of the entire C-terminal core domain. Both unassociated and FeMo-co bound NafY were found to be monomers, in contrast to previous work reporting that cofactor-less Gamma protein was a homodimer.178 NafY was also shown to bind NifB-co, although with much lower affinity than FeMo-co. The preferential binding to one specific metallocluster for members of this family of proteins is thought to be of importance for correct cluster delivery in vivo.113 Similar to the effect of NifX on the in vitro FeMo-co synthesis assay, low levels of NafY increased apo-MoFe protein activation, while higher levels inhibited it,22 potentially explained by competition for FeMo-co between NafY and apo-MoFe protein.
The fact that the N-terminal domain of NafY is responsible for apo-MoFe protein binding was initially revealed by pull-down experiments, and interactions studies in which GST-tagged NafY variants were used as bait to test their affinity for apo-Mofe protein.113 Recently, better understanding of NafY (and NifY) interaction with apo-MoFe protein was obtained from STAC experiments,23,114 in which apo-MoFe protein could be isolated with bound NafY or NifY proteins. These interactions are easily lost during IMAC because of the use of imidazole.113 NafY or NifY binding to apo-MoFe protein was found to protect Cys275, which together with His442 anchors FeMo-co to the MoFe protein. Mutation of either Cys275 or His442 prevented the dissociation of NifY or NafY from the MoFe protein.23 Interestingly, NafY and NifY were never found simultaneously at the apo-MoFe protein, but the specific order in which they bind to immature apo-MoFe protein remains to be elucidated.
In summary, NafY (and the K. pneumoniae NifY) stabilizes the P-cluster containing apo-MoFe protein by protecting the FeMo-co ligand α-Cys275 from oxidation. NafY also acts as carrier of matured FeMo-co from NifEN facilitating FeMo-co insertion into the apo-MoFe protein, although this function is neither essential in vivo nor in vitro. The exact mechanism remains to be determined, but it is possible that NafY increases the rate of FeMo-co transfer or that NafY makes FeMo-co insertion more selective by discriminating against the insertion of a FeMo-co biosynthetic precursor.
16. Concluding Remarks
Research on nitrogenase metal cofactor formation has delivered results, concepts, and methodologies with broad implications in all biology. In particular, the biosynthesis of FeMo-co is a process of enormous complexity involving molecular assembly scaffolds, enzymes to provide cofactor parts, and metallocluster carrier proteins.
NifB is one of the most interesting proteins involved in FeMo-co assembly as it produces the unique [8Fe-9S–C] NifB-co cluster that is used as precursor for the biosynthesis of the active-site cofactors in all nitrogenases. Our understanding of NifB has progressed enormously during the last years. Still, several mechanistic questions need to be addressed; among them are the in vivo source of sulfite proposed to donate the “ninth” S required for NifB-co formation and how the last two H atoms of the K2 cluster-bound methyl group are removed following the first 5′-dA• radical-dependent H abstraction. Also, NifB has one SAM-cluster, but at least two SAM molecules are required for NifB-co formation. It is not clear whether both molecules are presented by the SAM-cluster. Regarding SAM, it is also unclear why NifB generates more SAH than 5′-dAH,57,58 which would be expected to accumulate at a 1:1 ratio if the currently proposed NifB mechanism was entirely correct (Figure 19A). The exact role of FdxN on NifB-co formation is also intriguing and awaits further investigation.
Related to the mechanism and stability of NifB it is also not known whether the protein is capable of multiple turnovers (producing several consecutive NifB-co molecules). The question is relevant as inactivation of the clpX2 gene, a protease that is upregulated during N2-fixing conditions in A. vinelandii, reduces the rate at which NifB is degraded in vivo.245 One function of the ClpX2 protein could be to degrade cluster-less NifB protein. This hypothesis is supported by the fact that disruption of the nifENX genes, whose products are acceptors of NifB-co, made the (presumably cluster-loaded) NifB protein more resistant to degradation.
Many of these open questions could be answered by having a three-dimensional atomic structure of NifB. The structure would also help to understand why, in many N2-fixing organisms, NifB has an additional C-terminal NifX-like domain that is neither strictly required for NifB-co production nor for transfer of NifB-co in vitro and in vivo.
NifQ is another protein producing a unique [Mo-3Fe-4S] cluster destined for FeMo-co biosynthesis.73 As it is the case of NifB, there is no three-dimensional atomic structure of NifQ that would help elucidate how this cluster is formed and how can it serve as Mo donor. One open question about Mo donation in the in vitro FeMo-co synthesis assay is whether the current assays are appropriate to investigate this step of cofactor biosynthesis. The fact that free molybdate, NifQ, the MoSto protein, some NifEN preparations, and Fe protein reisolated from a “NifEN activation” assay can all serve as Mo donors in vitro suggests that the current in vitro FeMo-co synthesis assay might not be properly reproducing the in vivo reaction or that Mo can be incorporated through multiple pathways.
Our understanding of the maturation of MoFe protein and Fe protein polypeptides is still scarce. On one hand, the role of NifM and its essentiality are unclear, as is the phylogenetic distribution of NifM-dependent and NifM-independent Fe proteins. Despite substantial progress in understanding P-cluster maturation, it is important to note that the exact role of the Fe protein in this process is still unknown. The ability of apo-Fe protein to aid in P-cluster formation and FeMo-co biosynthesis makes its role difficult to understand. Finally, the MoFe protein maturation is more complex than initially conceived, at least in the model diazotroph A. vinelandii. Nevertheless, it is expected that novel methods and technologies, such as the powerful and nondisruptive STAC, which has already unveiled and detailed the action of several MoFe protein accessory factors,23,112,114 will provide more precise mapping of the Nif protein complexes. Investigating not only the functions of individual Nif proteins but also their interactions and concert actions under different stimuli and conditions will be important for a global view on nitrogenase biogenesis. It will also be crucial for researchers aiming to transfer the complete nitrogenase molecular machinery into eukaryotes such as plants.75 In this regard, several works addressing methodologies and expression of functional Nif proteins in yeast and plants have recently been published.96,166,246−249 A key study showing in vivo synthesis of NifB-co in yeast mitochondria has also been published,112 a manifest that indicates that we might be closer than ever to reach the longstanding idea of engineering plants capable of utilizing atmospheric N2 as direct nitrogen source.250,251
Acknowledgments
This work has been funded by the Bill & Melinda Gates Foundation Grant No. OPP1143172 and by FEDER / Ministerio de Ciencia, Innovación y Universidades – Agencia Estatal de Investigación / Proyecto 2017-88475-R.
Glossary
Abbreviations
- β-ME
β-mercaptoethanol
- GST
glutathione S-transferase
- IDS
indigo disulfonate
- DTH
dithionite
- NMF
N-methylformamide
- STAC
strep tag affinity chromatography
- IMAC
immobilized metal affinity chromatography
- SAM
S-adenosylmethionine
- SAH
S-adenosylhomocysteine
- EPR
electron paramagnetic resonance
- UV–vis
UV–visible
- NMR
nuclear magnetic resonance
- NRVS
nuclear resonance vibrational spectroscopy
- SAXS
small angle X-ray scattering
- EXAFS
extended X-ray absorption fine structure
- XAS
X-ray absorption spectroscopy
- VT-MCD
variable temperature magnetic circular dichroism
Biographies
Stefan Burén received his M.Sc. degree in Engineering Biology from the Umeå University (Sweden) in 2004 and obtained his Ph.D. degree in Cell and Molecular Biology of Plants at the Umeå Plant Science Centre (UPSC), also at the Umeå University, in 2010. After postdoctoral training at the Spanish National Cancer Research Centre (CNIO), he joined the laboratory of Prof. Luis Rubio at the Centro de Biotecnología y Genómica de Plantas (UPM-INIA) where he currently holds a postdoctoral position. His research is mainly devoted to the functional transfer of nitrogen fixation genes to yeast and plants.
Emilio Jiménez-Vicente completed his Ph.D. degree in Biotechnology and Genetic Resources of Plants and Associated Microorganisms from the Universidad Politécnica de Madrid (UPM) in 2014. Since 2016, he works in the group of Prof. Dennis Dean at Virginia Tech. His current research focuses in the study of key factors involved in the early steps of nitrogenase maturation process.
Carlos Echavarri-Erasun is Assistant Professor at the Departamento de Biotecnología y Biología Vegetal of the Universidad Politécnica de Madrid (UPM). He received his B.S. degree in Biology from Universidad de Navarra, Spain (1996) and his Ph.D. degree in Microbiology from University of Wisconsin-Madison (2005). After postdoctoral training as EU Marie Curie fellow at the CBS-KNAW (Netherlands), he joined Prof. Luis Rubio’s lab at the Centro de Biotecnología y Genómica de Plantas (UPM-INIA). His main research interests are in the biosynthesis of the iron–molybdenum cofactor of nitrogenase and other metalloenzyme systems.
Luis Rubio is Associate Professor at the Departamento de Biotecnología y Biología Vegetal of the Universidad Politécnica de Madrid (UPM), and Deputy Director of the Centro de Biotecnología y Genómica de Plantas (UPM-INIA). He obtained his Ph.D. from the University of Seville in 1999. He worked with Paul Ludden at the University of Wisconsin-Madison (1999–2002) and the University of California-Berkeley (2002–2008). Since 2008, he directs the Biochemistry of Nitrogen Fixation research group of CBGP. His work is devoted to nitrogenase engineering in eukaryotes, the study of nitrogenase iron–molybdenum cofactor biosynthesis, and the optimization of hydrogen production by nitrogenase.
The authors declare no competing financial interest.
References
- Hoffman B. M.; Lukoyanov D.; Yang Z.-Y. Y.; Dean D. R.; Seefeldt L. C. Mechanism of Nitrogen Fixation by Nitrogenase: The Next Stage. Chem. Rev. 2014, 114, 4041–4062. 10.1021/cr400641x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulen W. A.; LeComte J. R. The Nitrogenase System from Azotobacter: Two-Enzyme Requirement for N2 Reduction, ATP-Dependent H2 Evolution, and ATP Hydrolysis. Proc. Natl. Acad. Sci. U. S. A. 1966, 56, 979–986. 10.1073/pnas.56.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hageman R. V.; Burris R. H. Nitrogenase and Nitrogenase Reductase Associate and Dissociate with Each Catalytic Cycle. Proc. Natl. Acad. Sci. U. S. A. 1978, 75, 2699–2702. 10.1073/pnas.75.6.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval S.; Danyal K.; Shaw S.; Lytle A. K.; Dean D. R.; Hoffman B. M.; Antony E.; Seefeldt L. C. Electron Transfer Precedes ATP Hydrolysis during Nitrogenase Catalysis. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 16414–16419. 10.1073/pnas.1311218110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiadis M. M.; Komiya H.; Chakrabarti P.; Woo D.; Kornuc J. J.; Rees D. C. Crystallographic Structure of the Nitrogenase Iron Protein from Azotobacter Vinelandii. Science 1992, 257, 1653–1659. 10.1126/science.1529353. [DOI] [PubMed] [Google Scholar]
- Kim J.; Rees D. C. Crystallographic Structure and Functional Implications of the Nitrogenase Molybdenum-Iron Protein from Azotobacter Vinelandii. Nature 1992, 360, 553–560. 10.1038/360553a0. [DOI] [PubMed] [Google Scholar]
- Rubio L. M.; Ludden P. W. Biosynthesis of the Iron-Molybdenum Cofactor of Nitrogenase. Annu. Rev. Microbiol. 2008, 62, 93–111. 10.1146/annurev.micro.62.081307.162737. [DOI] [PubMed] [Google Scholar]
- Ribbe M. W.; Hu Y.; Hodgson K. O.; Hedman B. Biosynthesis of Nitrogenase Metalloclusters. Chem. Rev. 2014, 114, 4063–4080. 10.1021/cr400463x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y.; Fay A. W.; Lee C. C.; Ribbe M. W. P-Cluster Maturation on Nitrogenase MoFe Protein. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 10424–10429. 10.1073/pnas.0704297104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugalde R. A.; Imperial J.; Shah V. K.; Brill W. J. Biosynthesis of Iron-Molybdenum Cofactor in the Absence of Nitrogenase. J. Bacteriol. 1984, 159, 888–893. 10.1128/JB.159.3.888-893.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigle K. E.; Weiss M. C.; Newton W. E.; Dean D. R. Products of the Iron-Molybdenum Cofactor-Specific Biosynthetic Genes, NifE and NifN, Are Structurally Homologous to the Products of the Nitrogenase Molybdenum-Iron Protein Genes, NifD and NifK. J. Bacteriol. 1987, 169, 1547–1553. 10.1128/JB.169.4.1547-1553.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlynn S. E.; Shepard E. M.; Winslow M. A.; Naumov A. V.; Duschene K. S.; Posewitz M. C.; Broderick W. E.; Broderick J. B.; Peters J. W. HydF as a Scaffold Protein in [FeFe] Hydrogenase H-Cluster Biosynthesis. FEBS Lett. 2008, 582, 2183–2187. 10.1016/j.febslet.2008.04.063. [DOI] [PubMed] [Google Scholar]
- Zheng L.; White R. H.; Cash V. L.; Jack R. F.; Dean D. R. Cysteine Desulfurase Activity Indicates a Role for NIFS in Metallocluster Biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 1993, 90, 2754–2758. 10.1073/pnas.90.7.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L.; Cash V. L.; Flint D. H.; Dean D. R. Assembly of Iron-Sulfur Clusters. Identification of an IscSUA-HscBA-Fdx Gene Cluster from Azotobacter Vinelandii. J. Biol. Chem. 1998, 273, 13264–13272. 10.1074/jbc.273.21.13264. [DOI] [PubMed] [Google Scholar]
- Shah V. K.; Brill W. J. Isolation of an Iron-Molybdenum Cofactor from Nitrogenase. Proc. Natl. Acad. Sci. U. S. A. 1977, 74, 3249–3253. 10.1073/pnas.74.8.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah V. K.; Imperial J.; Ugalde R. A.; Ludden P. W.; Brill W. J. In Vitro Synthesis of the Iron-Molybdenum Cofactor of Nitrogenase. Proc. Natl. Acad. Sci. U. S. A. 1986, 83, 1636–1640. 10.1073/pnas.83.6.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover T. R.; Shah V. K.; Roberts G. P.; Ludden P. W. NifV-Dependent, Low-Molecular-Weight Factor Required for in Vitro Synthesis of Iron-Molybdenum Cofactor of Nitrogenase. J. Bacteriol. 1986, 167, 999–1003. 10.1128/JB.167.3.999-1003.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover T. R.; Imperial J.; Ludden P. W.; Shah V. K. Homocitrate Is a Component of the Iron-Molybdenum Cofactor of Nitrogenase. Biochemistry 1989, 28, 2768–2771. 10.1021/bi00433a004. [DOI] [PubMed] [Google Scholar]
- Shah V. K.; Allen J. R.; Spangler N. J.; Ludden P. W. In Vitro Synthesis of the Iron-Molybdenum Cofactor of Nitrogenase. Purification and Characterization of NifB Cofactor, the Product of NIFB Protein. J. Biol. Chem. 1994, 269, 1154–1158. [PubMed] [Google Scholar]
- Guo Y.; Echavarri-Erasun C.; Demuez M.; Jimenez-Vicente E.; Bominaar E. L.; Rubio L. M. The Nitrogenase FeMo-Cofactor Precursor Formed by NifB Protein: A Diamagnetic Cluster Containing Eight Iron Atoms. Angew. Chem., Int. Ed. 2016, 55, 12764–12767. 10.1002/anie.201606447. [DOI] [PubMed] [Google Scholar]
- Paustian T. D.; Shah V. K.; Roberts G. P. Purification and Characterization of the NifN and NifE Gene Products from Azotobacter Vinelandii Mutant UW45. Proc. Natl. Acad. Sci. U. S. A. 1989, 86, 6082–6086. 10.1073/pnas.86.16.6082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curatti L.; Hernandez J. A.; Igarashi R. Y.; Soboh B.; Zhao D.; Rubio L. M. In Vitro Synthesis of the Iron-Molybdenum Cofactor of Nitrogenase from Iron, Sulfur, Molybdenum, and Homocitrate Using Purified Proteins. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 17626–17631. 10.1073/pnas.0703050104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Vicente E.; Yang Z.-Y.; Ray W. K.; Echavarri-Erasun C.; Cash V. L.; Rubio L. M.; Seefeldt L. C.; Dean D. R. Sequential and Differential Interaction of Assembly Factors during Nitrogenase MoFe Protein Maturation. J. Biol. Chem. 2018, 293, 9812–9823. 10.1074/jbc.RA118.002994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez J. A.; Igarashi R. Y.; Soboh B.; Curatti L.; Dean D. R.; Ludden P. W.; Rubio L. M. NifX and NifEN Exchange NifB Cofactor and the VK-Cluster, a Newly Isolated Intermediate of the Iron-Molybdenum Cofactor Biosynthetic Pathway. Mol. Microbiol. 2007, 63, 177–192. 10.1111/j.1365-2958.2006.05514.x. [DOI] [PubMed] [Google Scholar]
- Schindelin H.; Kisker C.; Schlessman J. L.; Howard J. B.; Rees D. C. Structure of ADP-AIF4 Stabilized Nitrogenase Complex and Its Implications for Signal Transduction. Nature 1997, 387, 370–376. 10.1038/387370a0. [DOI] [PubMed] [Google Scholar]
- Einsle O.; Tezcan F. A.; Andrade S. L. A.; Schmid B.; Yoshida M.; Howard J. B.; Rees D. C. Nitrogenase MoFe-Protein at 1.16 A Resolution: A Central Ligand in the FeMo-Cofactor. Science 2002, 297, 1696–1700. 10.1126/science.1073877. [DOI] [PubMed] [Google Scholar]
- Spatzal T.; Aksoyoglu M. M. M. M.; Zhang L.; Andrade S. L. A.; Schleicher E.; Weber S.; Rees D. C.; Einsle O. Evidence for Interstitial Carbon in Nitrogenase FeMo Cofactor. Science 2011, 334, 940. 10.1126/science.1214025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan M. K.; Kim J.; Rees D. C. The Nitrogenase FeMo-Cofactor and P-Cluster Pair: 2.2 Å Resolution Structures. Science 1993, 260, 792–794. 10.1126/science.8484118. [DOI] [PubMed] [Google Scholar]
- Peters J. W.; Stowell M. H.; Soltis S. M.; Finnegan M. G.; Johnson M. K.; Rees D. C. Redox-Dependent Structural Changes in the Nitrogenase P-Cluster. Biochemistry 1997, 36, 1181–1187. 10.1021/bi9626665. [DOI] [PubMed] [Google Scholar]
- Peters J. W.; Fisher K.; Newton W. E.; Dean D. R. Involvement of the P Cluster in Intramolecular Electron Transfer within the Nitrogenase MoFe Protein. J. Biol. Chem. 1995, 270, 27007–27013. 10.1074/jbc.270.45.27007. [DOI] [PubMed] [Google Scholar]
- Dean D. R.; Setterquist R. A.; Brigle K. E.; Scott D. J.; Laird N. F.; Newton W. E. Evidence That Conserved Residues Cys-62 and Cys-154 within the Azotobacter Vinelandii Nitrogenase MoFe Protein Alpha-Subunit Are Essential for Nitrogenase Activity but Conserved Residues His-83 and Cys-88 Are Not. Mol. Microbiol. 1990, 4, 1505–1512. 10.1111/j.1365-2958.1990.tb02061.x. [DOI] [PubMed] [Google Scholar]
- Kent H. M.; Ioannidis I.; Gormal C.; Smith B. E.; Buck M. Site-Directed Mutagenesis of the Klebsiella Pneumoniae Nitrogenase. Effects of Modifying Conserved Cysteine Residues in the Alpha- and Beta-Subunits. Biochem. J. 1989, 264, 257–264. 10.1042/bj2640257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May H. D.; Dean D. R.; Newton W. E. Altered Nitrogenase MoFe Proteins from Azotobacter Vinelandii. Analysis of MoFe Proteins Having Amino Acid Substitutions for the Conserved Cysteine Residues within the Beta-Subunit. Biochem. J. 1991, 277, 457–464. 10.1042/bj2770457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousafzai F. K.; Buck M.; Smith B. E. Isolation and Characterization of Nitrogenase MoFe Protein from the Mutant Strain PHK17 of Klebsiella Pneumoniae in Which the Two Bridging Cysteine Residues of the P-Clusters Are Replaced by the Non-Coordinating Amino Acid Alanine. Biochem. J. 1996, 318, 111–118. 10.1042/bj3180111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutledge H. L.; Rittle J.; Williamson L. M.; Xu W. A.; Gagnon D. M.; Tezcan F. A. Redox-Dependent Metastability of the Nitrogenase P-Cluster. J. Am. Chem. Soc. 2019, 141, 10091–10098. 10.1021/jacs.9b04555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose A. S.; Bradley A. R.; Valasatava Y.; Duarte J. M.; Prlić A.; Rose P. W. NGL Viewer: Web-Based Molecular Graphics for Large Complexes. Bioinformatics 2018, 34, 3755–3758. 10.1093/bioinformatics/bty419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setubal J. C. J. C.; dos Santos P.; Goldman B. S.; Ertesvag H.; Espin G.; Rubio L. M.; Valla S.; Almeida N. F.; Balasubramanian D.; Cromes L.; et al. Genome Sequence of Azotobacter Vinelandii, an Obligate Aerobe Specialized to Support Diverse Anaerobic Metabolic Processes. J. Bacteriol. 2009, 191, 4534–4545. 10.1128/JB.00504-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson M. R.; Brigle K. E.; Bennett L. T.; Setterquist R. A.; Wilson M. S.; Cash V. L.; Beynon J.; Newton W. E.; Dean D. R. Physical and Genetic Map of the Major Nif Gene Cluster from Azotobacter Vinelandii. J. Bacteriol. 1989, 171, 1017–1027. 10.1128/JB.171.2.1017-1027.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson M. R.; Cash V. L.; Weiss M. C.; Laird N. F.; Newton W. E.; Dean D. R. Biochemical and Genetic Analysis of the NifUSVWZM Cluster from Azotobacter Vinelandii. Mol. Gen. Genet. 1989, 219, 49–57. 10.1007/BF00261156. [DOI] [PubMed] [Google Scholar]
- Joerger R. D.; Bishop P. E. Nucleotide Sequence and Genetic Analysis of the NifB-NifQ Region from Azotobacter Vinelandii. J. Bacteriol. 1988, 170, 1475–1487. 10.1128/JB.170.4.1475-1487.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curatti L.; Ludden P. W.; Rubio L. M. NifB-Dependent in Vitro Synthesis of the Iron-Molybdenum Cofactor of Nitrogenase. Proc. Natl. Acad. Sci. U. S. A. 2006, 103, 5297–5301. 10.1073/pnas.0601115103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Argudo I.; Little R.; Shearer N.; Johnson P.; Dixon R. The NifL-NifA System: A Multidomain Transcriptional Regulatory Complex That Integrates Environmental Signals. J. Bacteriol. 2004, 186, 601–610. 10.1128/JB.186.3.601-610.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R.; Kahn D. Genetic Regulation of Biological Nitrogen Fixation. Nat. Rev. Microbiol. 2004, 2, 621–631. 10.1038/nrmicro954. [DOI] [PubMed] [Google Scholar]
- Roberts G. P.; MacNeil T.; MacNeil D.; Brill W. J. Regulation and Characterization of Protein Products Coded by the Nif (Nitrogen Fixation) Genes of Klebsiella Pneumoniae. J. Bacteriol. 1978, 136, 267–279. 10.1128/JB.136.1.267-279.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmehl M.; Jahn A.; Meyer zu Vilsendorf A.; Hennecke S.; Masepohl B.; Schuppler M.; Marxer M.; Oelze J.; Klipp W. Identification of a New Class of Nitrogen Fixation Genes in Rhodobacter Capsulatus: A Putative Membrane Complex Involved in Electron Transport to Nitrogenase. Mol. Gen. Genet. 1993, 241, 602–615. 10.1007/BF00279903. [DOI] [PubMed] [Google Scholar]
- Poudel S.; Colman D. R.; Fixen K. R.; Ledbetter R. N.; Zheng Y.; Pence N.; Seefeldt L. C.; Peters J. W.; Harwood C. S.; Boyd E. S. Electron Transfer to Nitrogenase in Different Genomic and Metabolic Backgrounds. J. Bacteriol. 2018, 200, e00757-17 10.1128/JB.00757-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biegel E.; Schmidt S.; González J. M.; Müller V. Biochemistry, Evolution and Physiological Function of the Rnf Complex, a Novel Ion-Motive Electron Transport Complex in Prokaryotes. Cell. Mol. Life Sci. 2011, 68, 613–634. 10.1007/s00018-010-0555-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curatti L.; Brown C. S.; Ludden P. W.; Rubio L. M. Genes Required for Rapid Expression of Nitrogenase Activity in Azotobacter Vinelandii. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 6291–6296. 10.1073/pnas.0501216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledbetter R. N.; Garcia Costas A. M.; Lubner C. E.; Mulder D. W.; Tokmina-Lukaszewska M.; Artz J. H.; Patterson A.; Magnuson T. S.; Jay Z. J.; Duan H. D.; et al. The Electron Bifurcating FixABCX Protein Complex from Azotobacter Vinelandii: Generation of Low-Potential Reducing Equivalents for Nitrogenase Catalysis. Biochemistry 2017, 56, 4177–4190. 10.1021/acs.biochem.7b00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole R. K.; Hill S. Respiratory Protection of Nitrogenase Activity in Azotobacter Vinelandii--Roles of the Terminal Oxidases. Biosci. Rep. 1997, 17, 303–317. 10.1023/A:1027336712748. [DOI] [PubMed] [Google Scholar]
- Poza-Carrión C.; Jiménez-Vicente E.; Navarro-Rodríguez M.; Echavarri-Erasun C.; Rubio L. M. Kinetics of Nif Gene Expression in a Nitrogen-Fixing Bacterium. J. Bacteriol. 2014, 196, 595–603. 10.1128/JB.00942-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy C.; Dean D. The NifU, NifS and NifV Gene Products Are Required for Activity of All Three Nitrogenases of Azotobacter Vinelandii. Mol. Gen. Genet. 1992, 231, 494–498. 10.1007/BF00292722. [DOI] [PubMed] [Google Scholar]
- Kennedy C.; Gamal R.; Humphrey R.; Ramos J.; Brigle K.; Dean D. The NifH, NifM and NifN Genes of Azotobacter Vinelandii: Characterisation by Tn5 Mutagenesis and Isolation from PLAFR1 Gene Banks. Mol. Gen. Genet. 1986, 205, 318–325. 10.1007/BF00430445. [DOI] [Google Scholar]
- Dos Santos P. C.; Fang Z.; Mason S. W.; Setubal J. C.; Dixon R. Distribution of Nitrogen Fixation and Nitrogenase-like Sequences amongst Microbial Genomes. BMC Genomics 2012, 13, 162. 10.1186/1471-2164-13-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos P. C.; Smith A. D.; Frazzon J.; Cash V. L.; Johnson M. K.; Dean D. R. Iron-Sulfur Cluster Assembly: NifU-Directed Activation of the Nitrogenase Fe Protein. J. Biol. Chem. 2004, 279, 19705–19711. 10.1074/jbc.M400278200. [DOI] [PubMed] [Google Scholar]
- Paustian T. D.; Shah V. K.; Roberts G. P. Apodinitrogenase: Purification, Association with a 20-Kilodalton Protein, and Activation by the Iron-Molybdenum Cofactor in the Absence of Dinitrogenase Reductase. Biochemistry 1990, 29, 3515–3522. 10.1021/bi00466a014. [DOI] [PubMed] [Google Scholar]
- Fay A. W.; Wiig J. a.; Lee C. C.; Hu Y. Identification and Characterization of Functional Homologs of Nitrogenase Cofactor Biosynthesis Protein NifB from Methanogens. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 14829–14833. 10.1073/pnas.1510409112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcoxen J.; Arragain S.; Scandurra A. A.; Jimenez-Vicente E.; Echavarri-Erasun C.; Pollmann S.; Britt R. D.; Rubio L. M. Electron Paramagnetic Resonance Characterization of Three Iron–Sulfur Clusters Present in the Nitrogenase Cofactor Maturase NifB from Methanocaldococcus Infernus. J. Am. Chem. Soc. 2016, 138, 7468–7471. 10.1021/jacs.6b03329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanifuji K.; Lee C. C.; Sickerman N. S.; Tatsumi K.; Ohki Y.; Hu Y.; Ribbe M. W. Tracing the ‘Ninth Sulfur’ of the Nitrogenase Cofactor via a Semi-Synthetic Approach. Nat. Chem. 2018, 10, 568–572. 10.1038/s41557-018-0029-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y.; Fay A. W.; Ribbe M. W. Identification of a Nitrogenase FeMo Cofactor Precursor on NifEN Complex. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 3236–3241. 10.1073/pnas.0409201102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soboh B.; Igarashi R. Y.; Hernandez J. A.; Rubio L. M. Purification of a NifEN Protein Complex That Contains Bound Molybdenum and a FeMo-Co Precursor from an Azotobacter Vinelandii DeltanifHDK Strain. J. Biol. Chem. 2006, 281, 36701–36709. 10.1074/jbc.M606820200. [DOI] [PubMed] [Google Scholar]
- Thiel T.; Lyons E. M.; Erker J. C.; Ernst A. A Second Nitrogenase in Vegetative Cells of a Heterocyst-Forming Cyanobacterium. Proc. Natl. Acad. Sci. U. S. A. 1995, 92, 9358–9362. 10.1073/pnas.92.20.9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.-S. S.; Toth J.; Kasap M. Nitrogen-Fixation Genes and Nitrogenase Activity in Clostridium Acetobutylicum and Clostridium Beijerinckii. J. Ind. Microbiol. Biotechnol. 2001, 27, 281–286. 10.1038/sj.jim.7000083. [DOI] [PubMed] [Google Scholar]
- Arragain S.; Jimenez-Vicente E.; Scandurra A. A.; Burén S.; Rubio L. M.; Echavarri-Erasun C. Diversity and Functional Analysis of the FeMo-Cofactor Maturase NifB. Front. Plant Sci. 2017, 8, 1947. 10.3389/fpls.2017.01947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd E. S.; Costas A. M. G.; Hamilton T. L.; Mus F.; Peters J. W. Evolution of Molybdenum Nitrogenase during the Transition from Anaerobic to Aerobic Metabolism. J. Bacteriol. 2015, 197, 1690–1699. 10.1128/JB.02611-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.; Zhang L.; Liu Z.; Zhao D.; Liu X.; Zhang B.; Xie J.; Hong Y.; Li P.; Chen S.; et al. A Minimal Nitrogen Fixation Gene Cluster from Paenibacillus Sp. WLY78 Enables Expression of Active Nitrogenase in Escherichia Coli. PLoS Genet. 2013, 9, e1003865 10.1371/journal.pgen.1003865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fani R.; Gallo R.; Lio P. Molecular Evolution of Nitrogen Fixation: The Evolutionary History of the NifD, NifK, NifE, and NifN Genes. J. Mol. Evol. 2000, 51, 1–11. 10.1007/s002390010061. [DOI] [PubMed] [Google Scholar]
- Leimkuhler S.; Wuebbens M. M.; Rajagopalan K. V. Characterization of Escherichia Coli MoeB and Its Involvement in the Activation of Molybdopterin Synthase for the Biosynthesis of the Molybdenum Cofactor. J. Biol. Chem. 2001, 276, 34695–34701. 10.1074/jbc.M102787200. [DOI] [PubMed] [Google Scholar]
- Takahashi Y.; Tokumoto U. A Third Bacterial System for the Assembly of Iron-Sulfur Clusters with Homologs in Archaea and Plastids. J. Biol. Chem. 2002, 277, 28380–28383. 10.1074/jbc.C200365200. [DOI] [PubMed] [Google Scholar]
- Bulfer S. L.; Scott E. M.; Couture J.-F.; Pillus L.; Trievel R. C. Crystal Structure and Functional Analysis of Homocitrate Synthase, an Essential Enzyme in Lysine Biosynthesis. J. Biol. Chem. 2009, 284, 35769–35780. 10.1074/jbc.M109.046821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Rodríguez M.; Buesa J. M.; Rubio L. M. Genetic and Biochemical Analysis of the Azotobacter Vinelandii Molybdenum Storage Protein. Front. Microbiol. 2019, 10, 579. 10.3389/fmicb.2019.00579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez J. C.; Santero E.; Tortolero M. Ammonium Repression of the Nitrite-Nitrate (NasAB) Assimilatory Operon of Azotobacter Vinelandii Is Enhanced in Mutants Expressing the NifO Gene at High Levels. Mol. Gen. Genet. 1997, 255, 172–179. 10.1007/s004380050486. [DOI] [PubMed] [Google Scholar]
- Hernandez J. A.; Curatti L.; Aznar C. P.; Perova Z.; Britt R. D.; Rubio L. M. Metal Trafficking for Nitrogen Fixation: NifQ Donates Molybdenum to NifEN/NifH for the Biosynthesis of the Nitrogenase FeMo-Cofactor. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 11679–11684. 10.1073/pnas.0803576105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Vicente E.; Navarro-Rodríguez M.; Poza-Carrión C.; Rubio L. M. Role of Azotobacter Vinelandii FdxN in FeMo-Co Biosynthesis. FEBS Lett. 2014, 588, 512–516. 10.1016/j.febslet.2013.12.018. [DOI] [PubMed] [Google Scholar]
- Burén S.; Rubio L. M. State of the Art in Eukaryotic Nitrogenase Engineering. FEMS Microbiol. Lett. 2018, 365, fnx274. 10.1093/femsle/fnx274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D. C.; Dos Santos P. C.; Dean D. R. NifU and NifS Are Required for the Maturation of Nitrogenase and Cannot Replace the Function of Isc-Gene Products in Azotobacter Vinelandii. Biochem. Soc. Trans. 2005, 33, 90–93. 10.1042/BST0330090. [DOI] [PubMed] [Google Scholar]
- Dos Santos P. C.; Johnson D. C.; Ragle B. E.; Unciuleac M.-C.; Dean D. R. Controlled Expression of Nif and Isc Iron-Sulfur Protein Maturation Components Reveals Target Specificity and Limited Functional Replacement between the Two Systems. J. Bacteriol. 2007, 189, 2854–2862. 10.1128/JB.01734-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L.; White R. H.; Cash V. L.; Dean D. R. Mechanism for the Desulfurization of L-Cysteine Catalyzed by the NifS Gene Product. Biochemistry 1994, 33, 4714–4720. 10.1021/bi00181a031. [DOI] [PubMed] [Google Scholar]
- Zheng L.; Dean D. R. Catalytic Formation of a Nitrogenase Iron-Sulfur Cluster. J. Biol. Chem. 1994, 269, 18723–18726. [PubMed] [Google Scholar]
- Fu W.; Jack R. F.; Morgan T. V.; Dean D. R.; Johnson M. K. NifU Gene Product from Azotobacter Vinelandii Is a Homodimer That Contains Two Identical [2Fe-2S] Clusters. Biochemistry 1994, 33, 13455–13463. 10.1021/bi00249a034. [DOI] [PubMed] [Google Scholar]
- Beynon J.; Ally A.; Cannon M.; Cannon F.; Jacobson M.; Cash V.; Dean D. Comparative Organization of Nitrogen Fixation-Specific Genes from Azotobacter Vinelandii and Klebsiella Pneumoniae: DNA Sequence of the NifUSV Genes. J. Bacteriol. 1987, 169, 4024–4029. 10.1128/JB.169.9.4024-4029.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agar J. N.; Yuvaniyama P.; Jack R. F.; Cash V. L.; Smith A. D.; Dean D. R.; Johnson M. K. Modular Organization and Identification of a Mononuclear Iron-Binding Site within the NifU Protein. JBIC, J. Biol. Inorg. Chem. 2000, 5, 167–177. 10.1007/s007750050361. [DOI] [PubMed] [Google Scholar]
- Tong W.-H.; Jameson G. N. L.; Huynh B. H.; Rouault T. A. Subcellular Compartmentalization of Human Nfu, an Iron-Sulfur Cluster Scaffold Protein, and Its Ability to Assemble a [4Fe-4S] Cluster. Proc. Natl. Acad. Sci. U. S. A. 2003, 100, 9762–9767. 10.1073/pnas.1732541100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuvaniyama P.; Agar J. N.; Cash V. L.; Johnson M. K.; Dean D. R. NifS-Directed Assembly of a Transient [2Fe-2S] Cluster within the NifU Protein. Proc. Natl. Acad. Sci. U. S. A. 2000, 97, 599–604. 10.1073/pnas.97.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs C.; Agar J. N.; Smith A. D.; Frazzon J.; Dean D. R.; Huynh B. H.; Johnson M. K. IscA, an Alternate Scaffold for Fe–S Cluster Biosynthesis. Biochemistry 2001, 40, 14069–14080. 10.1021/bi015656z. [DOI] [PubMed] [Google Scholar]
- Agar J. N. J. N.; Krebs C.; Frazzon J.; Huynh B. H. B. H.; Dean D. R. D. R.; Johnson M. K. K. M. K. IscU as a Scaffold for Iron-Sulfur Cluster Biosynthesis: Sequential Assembly of [2Fe-2S] and [4Fe-4S] Clusters in IscU. Biochemistry 2000, 39, 7856–7862. 10.1021/bi000931n. [DOI] [PubMed] [Google Scholar]
- Waterhouse A.; Bertoni M.; Bienert S.; Studer G.; Tauriello G.; Gumienny R.; Heer F. T.; De Beer T. A. P.; Rempfer C.; Bordoli L. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296. 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. D.; Jameson G. N. L.; Dos Santos P. C.; Agar J. N.; Naik S.; Krebs C.; Frazzon J.; Dean D. R.; Huynh B. H.; Johnson M. K. NifS-Mediated Assembly of [4Fe-4S] Clusters in the N- and C-Terminal Domains of the NifU Scaffold Protein. Biochemistry 2005, 44, 12955–12969. 10.1021/bi051257i. [DOI] [PubMed] [Google Scholar]
- Mapolelo D. T.; Zhang B.; Naik S. G.; Huynh B. H.; Johnson M. K. Spectroscopic and Functional Characterization of Iron-Sulfur Cluster-Bound Forms of Azotobacter Vinelandii (Nif)IscA. Biochemistry 2012, 51, 8071–8084. 10.1021/bi3006658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangaraj P.; Shah V. K.; Ludden P. W. ApoNifH Functions in Iron-Molybdenum Cofactor Synthesis and Apodinitrogenase Maturation. Proc. Natl. Acad. Sci. U. S. A. 1997, 94, 11250–11255. 10.1073/pnas.94.21.11250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard K. S.; McLean P. A.; Hansen F. B.; Lemley P. V.; Koblan K. S.; Orme-Johnson W. H. Klebsiella Pneumoniae NifM Gene Product Is Required for Stabilization and Activation of Nitrogenase Iron Protein in Escherichia Coli. J. Biol. Chem. 1986, 261, 772–778. [PubMed] [Google Scholar]
- Paul W.; Merrick M. The Roles of the NifW, NifZ and NifM Genes of Klebsiella Pneumoniae in Nitrogenase Biosynthesis. Eur. J. Biochem. 1989, 178, 675–682. 10.1111/j.1432-1033.1989.tb14497.x. [DOI] [PubMed] [Google Scholar]
- Harris G. S.; White T. C.; Flory J. E.; Orme-Johnson W. H. Genes Required for Formation of the ApoMoFe Protein of Klebsiella Pneumoniae Nitrogenase in Escherichia Coli. J. Biol. Chem. 1990, 265, 15909–15919. [PubMed] [Google Scholar]
- Petrova N.; Gigova L.; Venkov P. NifH and NifM Proteins Interact as Demonstrated by the Yeast Two-Hybrid System. Biochem. Biophys. Res. Commun. 2000, 270, 863–867. 10.1006/bbrc.2000.2499. [DOI] [PubMed] [Google Scholar]
- Petrova N.; Gigova L.; Venkov P. Dimerization of Rhizobium Meliloti NifH Protein in Saccharomyces Cerevisiae Cells Requires Simultaneous Expression of NifM Protein. Int. J. Biochem. Cell Biol. 2002, 34, 33–42. 10.1016/S1357-2725(01)00102-9. [DOI] [PubMed] [Google Scholar]
- Lopez-Torrejon G.; Jimenez-Vicente E.; Buesa J. M.; Hernandez J. A.; Verma H. K.; Rubio L. M. Expression of a Functional Oxygen-Labile Nitrogenase Component in the Mitochondrial Matrix of Aerobically Grown Yeast. Nat. Commun. 2016, 7, 11426. 10.1038/ncomms11426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Gebali S.; Mistry J.; Bateman A.; Eddy S. R.; Luciani A.; Potter S. C.; Qureshi M.; Richardson L. J.; Salazar G. A.; Smart A.; et al. The Pfam Protein Families Database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A. L.; Attwood T. K.; Babbitt P. C.; Blum M.; Bork P.; Bridge A.; Brown S. D.; Chang H.-Y.; El-Gebali S.; Fraser M. I.; et al. InterPro in 2019: Improving Coverage, Classification and Access to Protein Sequence Annotations. Nucleic Acids Res. 2019, 47, D351–D360. 10.1093/nar/gky1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavini N.; Tungtur S.; Pulakat L. Peptidyl-Prolyl Cis/Trans Isomerase-Independent Functional NifH Mutant of Azotobacter Vinelandii. J. Bacteriol. 2006, 188, 6020–6025. 10.1128/JB.00379-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.; Xie X.; Wang X.; Dixon R.; Wang Y.-P. Reconstruction and Minimal Gene Requirements for the Alternative Iron-Only Nitrogenase in Escherichia Coli. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, E3718 10.1073/pnas.1411185111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo A. L.; Zinder S. H. Nitrogenase in the Archaebacterium Methanosarcina Barkeri 227. J. Bacteriol. 1990, 172, 6789–6796. 10.1128/JB.172.12.6789-6796.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio L. M.; Ludden P. W. Maturation of Nitrogenase: A Biochemical Puzzle. J. Bacteriol. 2005, 187, 405–414. 10.1128/JB.187.2.405-414.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J. W.; Boyd E. S.; Hamilton T. L.; Rubio L. M.. Biochemistry of Mo Nitrogenase. In Nitrogen Cycling in Bacteria; Moir J. W. B., Ed.; Caister Academic Press: Norfolk, U.K., 2011. [Google Scholar]
- Ribbe M. W. W.; Hu Y.; Guo M.; Schmid B.; Burgess B. K. K. The FeMoco-Deficient MoFe Protein Produced by a NifH Deletion Strain of Azotobacter Vinelandii Shows Unusual P-Cluster Features. J. Biol. Chem. 2002, 277, 23469–23476. 10.1074/jbc.M202061200. [DOI] [PubMed] [Google Scholar]
- Robinson A. C.; Dean D. R.; Burgess B. K. Iron-Molybdenum Cofactor Biosynthesis in Azotobacter Vinelandii Requires the Iron Protein of Nitrogenase. J. Biol. Chem. 1987, 262, 14327–14332. [PubMed] [Google Scholar]
- Robinson A. C.; Burgess B. K.; Dean D. R. Activity, Reconstitution, and Accumulation of Nitrogenase Components in Azotobacter Vinelandii Mutant Strains Containing Defined Deletions within the Nitrogenase Structural Gene Cluster. J. Bacteriol. 1986, 166, 180–186. 10.1128/JB.166.1.180-186.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson A. C.; Chun T. W.; Li J. G.; Burgess B. K. Iron-Molybdenum Cofactor Insertion into the Apo-MoFe Protein of Nitrogenase Involves the Iron Protein-MgATP Complex. J. Biol. Chem. 1989, 264, 10088–10095. [PubMed] [Google Scholar]
- Allen R. M.; Homer M. J.; Chatterjee R.; Ludden P. W.; Roberts G. P.; Shah V. K. Dinitrogenase Reductase- and MgATP-Dependent Maturation of Apodinitrogenase from Azotobacter Vinelandii. J. Biol. Chem. 1993, 268, 23670–23674. [PubMed] [Google Scholar]
- Homer M. J.; Paustian T. D.; Shah V. K.; Roberts G. P. The NifY Product of Klebsiella Pneumoniae Is Associated with Apodinitrogenase and Dissociates upon Activation with the Iron-Molybdenum Cofactor. J. Bacteriol. 1993, 175, 4907–4910. 10.1128/JB.175.15.4907-4910.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen J.; Goodwin P. J.; Lanzilotta W. N.; Seefeldt L. C.; Dean D. R. Catalytic and Biophysical Properties of a Nitrogenase Apo-MoFe Protein Produced by a NifB-Deletion Mutant of Azotobacter Vinelandii. Biochemistry 1998, 37, 12611–12623. 10.1021/bi981165b. [DOI] [PubMed] [Google Scholar]
- Schmidt T. G. M.; Skerra A. The Strep-Tag System for One-Step Purification and High-Affinity Detection or Capturing of Proteins. Nat. Protoc. 2007, 2, 1528–1535. 10.1038/nprot.2007.209. [DOI] [PubMed] [Google Scholar]
- Burén S.; Pratt K.; Jiang X.; Guo Y.; Jimenez-Vicente E.; Echavarri-Erasun C.; Dean D. R.; Saaem I.; Gordon D. B.; Voigt C. A.; et al. Biosynthesis of the Nitrogenase Active-Site Cofactor Precursor NifB-Co in Saccharomyces Cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 2019, 116, 25078–25086. 10.1073/pnas.1904903116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez J. A.; Phillips A. H.; Erbil W. K.; Zhao D.; Demuez M.; Zeymer C.; Pelton J. G.; Wemmer D. E.; Rubio L. M. A Sterile Alpha-Motif Domain in NafY Targets Apo-NifDK for Iron-Molybdenum Cofactor Delivery via a Tethered Domain. J. Biol. Chem. 2011, 286, 6321–6328. 10.1074/jbc.M110.168732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Vicente E.; Martin Del Campo J. S.; Yang Z.-Y.; Cash V. L.; Dean D. R.; Seefeldt L. C. Application of Affinity Purification Methods for Analysis of the Nitrogenase System from Azotobacter Vinelandii. Methods Enzymol. 2018, 613, 231–255. 10.1016/bs.mie.2018.10.007. [DOI] [PubMed] [Google Scholar]
- Hu Y.; Fay A. W.; Dos Santos P. C.; Naderi F.; Ribbe M. W. Characterization of Azotobacter Vinelandii NifZ Deletion Strains: Indication of Stepwise MoFe Protein Assembly. J. Biol. Chem. 2004, 279, 54963–54971. 10.1074/jbc.M408983200. [DOI] [PubMed] [Google Scholar]
- Jimenez-Vicente E.; Yang Z.-Y.; Martin Del Campo J. S.; Cash V. L.; Seefeldt L. C.; Dean D. R. The NifZ Accessory Protein Has an Equivalent Function in Maturation of Both Nitrogenase MoFe Protein P-Clusters. J. Biol. Chem. 2019, 294, 6204–6213. 10.1074/jbc.RA119.007905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. C.; Blank M. A.; Fay A. W.; Yoshizawa J. M.; Hu Y.; Hodgson K. O.; Hedman B.; Ribbe M. W. Stepwise Formation of P-Cluster in Nitrogenase MoFe Protein. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 18474–18478. 10.1073/pnas.0909149106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson J. K.; Paustian T. D.; Shah V. K.; Dean D. R.; Roberts G. P.; Rees D. C.; Howard J. B. Nitrogenase Iron-Molybdenum Cofactor Binding Site: Protein Conformational Changes Associated with Cofactor Binding. Tetrahedron 1997, 53, 11971–11984. 10.1016/S0040-4020(97)00710-2. [DOI] [Google Scholar]
- Corbett M. C.; Hu Y.; Fay A. W.; Tsuruta H.; Ribbe M. W.; Hodgson K. O.; Hedman B. Conformational Differences between Azotobacter Vinelandii Nitrogenase MoFe Proteins as Studied by Small-Angle X-Ray Scattering. Biochemistry 2007, 46, 8066–8074. 10.1021/bi7005064. [DOI] [PubMed] [Google Scholar]
- Corbett M. C.; Hu Y.; Naderi F.; Ribbe M. W.; Hedman B.; Hodgson K. O. Comparison of Iron-Molybdenum Cofactor-Deficient Nitrogenase MoFe Proteins by X-Ray Absorption Spectroscopy: Implications for P-Cluster Biosynthesis. J. Biol. Chem. 2004, 279, 28276–28282. 10.1074/jbc.M403156200. [DOI] [PubMed] [Google Scholar]
- Broach R. B.; Rupnik K.; Hu Y.; Fay A. W.; Cotton M.; Ribbe M. W.; Hales B. J. Variable-Temperature, Variable-Field Magnetic Circular Dichroism Spectroscopic Study of the Metal Clusters in the DeltanifB and DeltanifH Mofe Proteins of Nitrogenase from Azotobacter Vinelandii. Biochemistry 2006, 45, 15039–15048. 10.1021/bi061697p. [DOI] [PubMed] [Google Scholar]
- Rupnik K.; Lee C. C.; Wiig J. A.; Hu Y.; Ribbe M. W.; Hales B. J. Nonenzymatic Synthesis of the P-Cluster in the Nitrogenase MoFe Protein: Evidence of the Involvement of All-Ferrous [Fe4S4] 0 Intermediates. Biochemistry 2014, 53, 1108–1116. 10.1021/bi401699u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangaraj P.; Ryle M. J.; Lanzilotta W. N.; Ludden P. W.; Shah V. K. In Vitro Biosynthesis of Iron-Molybdenum Cofactor and Maturation of the Nif-Encoded Apodinitrogenase. Effect of Substitution for NifH with Site-Specifically Altered Forms of NifH. J. Biol. Chem. 1999, 274, 19778–19784. 10.1074/jbc.274.28.19778. [DOI] [PubMed] [Google Scholar]
- Cotton M. S.; Rupnik K.; Broach R. B.; Hu Y.; Fay A. W.; Ribbe M. W.; Hales B. J. VTVH-MCD Study of the Delta NifB Delta NifZ MoFe Protein from Azotobacter Vinelandii. J. Am. Chem. Soc. 2009, 131, 4558–4559. 10.1021/ja807525m. [DOI] [PubMed] [Google Scholar]
- Shah V. K.; Davis I. C.; Gordon J. K.; Orme-Johnson W. H.; Brill W. J. Nitrogenase. 3. Nitrogenaseless Mutants of Azotobacter Vinelandii: Activities, Cross-Reactions and EPR Spectra. Biochim. Biophys. Acta, Bioenerg. 1973, 292, 246–255. 10.1016/0005-2728(73)90269-7. [DOI] [PubMed] [Google Scholar]
- Nagatani H. H.; Shah V. K.; Brill W. J. Activation of Inactive Nitrogenase by Acid-Treated Component I. J. Bacteriol. 1974, 120, 697–701. 10.1128/JB.120.2.697-701.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop P. E.; Brill W. J. Genetic Analysis of Azotobacter Vinelandii Mutant Strains Unable to Fix Nitrogen. J. Bacteriol. 1977, 130, 954–956. 10.1128/JB.130.2.954-956.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketchum P. A.; Cambier H. Y.; Frazier W. A. 3rd; Madansky C. H.; Nason A. In Vitro Assembly of Neurospora Assimilatory Nitrate Reductase from Protein Subunits of a Neurospora Mutant and the Xanthine Oxidizing or Aldehyde Oxidase Systems of Higher Animals. Proc. Natl. Acad. Sci. U. S. A. 1970, 66, 1016–1023. 10.1073/pnas.66.3.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeil T.; MacNeil D.; Roberts G. P.; Supiano M. A.; Brill W. J. Fine-Structure Mapping and Complementation Analysis of Nif (Nitrogen Fixation) Genes in Klebsiella Pneumoniae. J. Bacteriol. 1978, 136, 253–266. 10.1128/JB.136.1.253-266.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John R. T.; Johnston H. M.; Seidman C.; Garfinkel D.; Gordon J. K.; Shah V. K.; Brill W. J. Biochemistry and Genetics of Klebsiella Pneumoniae Mutant Strains Unable to Fix N2. J. Bacteriol. 1975, 121, 759–765. 10.1128/JB.121.3.759-765.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings J.; Shah V. K.; Chisnell J. R.; Brill W. J.; Zimmermann R.; Münck E.; Orme-Johnson W. H. Novel Metal Cluster in the Iron-Molybdenum Cofactor of Nitrogenase. Spectroscopic Evidence. J. Biol. Chem. 1978, 253, 1001–1004. [PubMed] [Google Scholar]
- Ma L.; Gavini N.; Liu H. I.; Hedman B.; Hodgson K. O.; Burgess B. K. Large Scale Isolation and Characterization of the Molybdenum-Iron Cluster from Nitrogenase. J. Biol. Chem. 1994, 269, 18007–18015. [PubMed] [Google Scholar]
- Pienkos P. T.; Shah V. K.; Brill W. J. Molybdenum Cofactors from Molybdoenzymes and in Vitro Reconstitution of Nitrogenase and Nitrate Reductase. Proc. Natl. Acad. Sci. U. S. A. 1977, 74, 5468–5471. 10.1073/pnas.74.12.5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munck E.; Rhodes H.; Orme-Johnson W. H.; Davis L. C.; Brill W. J.; Shah V. K. Nitrogenase. VIII. Mossbauer and EPR Spectroscopy. The MoFe Protein Component from Azotobacter Vinelandii. Biochim. Biophys. Acta, Protein Struct. 1975, 400, 32–53. 10.1016/0005-2795(75)90124-5. [DOI] [PubMed] [Google Scholar]
- Mascharak P. K.; Smith M. C.; Armstrong W. H.; Burgess B. K.; Holm R. H. Fluorine-19 Chemical Shifts as Structural Probes of Metal-Sulfur Clusters and the Cofactor of Nitrogenase. Proc. Natl. Acad. Sci. U. S. A. 1982, 79, 7056–7060. 10.1073/pnas.79.22.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio L. M.; Singer S. W.; Ludden P. W. Purification and Characterization of NafY (Apodinitrogenase Gamma-Subunit) from Azotobacter Vinelandii. J. Biol. Chem. 2004, 279, 19739–19746. 10.1074/jbc.M400965200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann R.; Munck E.; Brill W. J.; Shah V. K.; Henzl M. T.; Rawlings J.; Orme-Johnson W. H. Nitrogenase X: Mossbauer and EPR Studies on Reversibly Oxidized MoFe Protein from Azotobacter Vinelandii OP. Nature of the Iron Centers. Biochim. Biophys. Acta, Protein Struct. 1978, 537, 185–207. 10.1016/0005-2795(78)90504-4. [DOI] [PubMed] [Google Scholar]
- Lancaster K. M.; Roemelt M.; Ettenhuber P.; Hu Y.; Ribbe M. W.; Neese F.; Bergmann U.; DeBeer S. X-Ray Emission Spectroscopy Evidences a Central Carbon in the Nitrogenase Iron-Molybdenum Cofactor. Science 2011, 334, 974–977. 10.1126/science.1206445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiig J. A.; Hu Y.; Lee C. C.; Ribbe M. W. Radical SAM-Dependent Carbon Insertion into the Nitrogenase M-Cluster. Science 2012, 337, 1672–1675. 10.1126/science.1224603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart W. D.; Fitzgerald G. P.; Burris R. H. In Situ Studies on Nitrogen Fixation with the Acetylene Reduction Technique. Science 1967, 158, 536. 10.1126/science.158.3800.536. [DOI] [PubMed] [Google Scholar]
- Shah V. K.; Rangaraj P.; Chatterjee R.; Allen R. M.; Roll J. T.; Roberts G. P.; Ludden P. W. Requirement of NifX and Other Nif Proteins for in Vitro Biosynthesis of the Iron-Molybdenum Cofactor of Nitrogenase. J. Bacteriol. 1999, 181, 2797–2801. 10.1128/JB.181.9.2797-2801.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen R. M.; Roll J. T.; Rangaraj P.; Shah V. K.; Roberts G. P.; Ludden P. W. Incorporation of Molybdenum into the Iron-Molybdenum Cofactor of Nitrogenase. J. Biol. Chem. 1999, 274, 15869–15874. 10.1074/jbc.274.22.15869. [DOI] [PubMed] [Google Scholar]
- Imperial J.; Ugalde R. A.; Shah V. K.; Brill W. J. Role of the NifQ Gene Product in the Incorporation of Molybdenum into Nitrogenase in Klebsiella Pneumoniae. J. Bacteriol. 1984, 158, 187–194. 10.1128/JB.158.1.187-194.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean P. A.; Smith B. E.; Dixon R. A. Nitrogenase of Klebsiella Pneumoniae NifV Mutants. Biochem. J. 1983, 211, 589–597. 10.1042/bj2110589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio L. M.; Rangaraj P.; Homer M. J.; Roberts G. P.; Ludden P. W. Cloning and Mutational Analysis of the Gamma Gene from Azotobacter Vinelandii Defines a New Family of Proteins Capable of Metallocluster Binding and Protein Stabilization. J. Biol. Chem. 2002, 277, 14299–14305. 10.1074/jbc.M107289200. [DOI] [PubMed] [Google Scholar]
- Kennedy C.; Dean D. The nifU, nifS and nifV gene products are required for activity of all three nitrogenases of Azotobacter vinelandii. Mol. Gen. Genet. 1992, 231, 494–498. 10.1007/BF00292722. [DOI] [PubMed] [Google Scholar]
- Zhao D.; Curatti L.; Rubio L. M. Evidence for NifU and NifS Participation in the Biosynthesis of the Iron-Molybdenum Cofactor of Nitrogenase. J. Biol. Chem. 2007, 282, 37016–37025. 10.1074/jbc.M708097200. [DOI] [PubMed] [Google Scholar]
- Sibold L.; Quiviger B.; Charpin N.; Paquelin A.; Elmerich C. Cloning and Expression of a DNA Fragment Carrying a His NifA Fusion and the NifBQ Operon from a Nif Constitutive Mutant of Klebsiella Pneumoniae. Biochimie 1983, 65, 53–63. 10.1016/S0300-9084(83)80029-7. [DOI] [PubMed] [Google Scholar]
- Joerger R. D.; Premakumar R.; Bishop P. E. Tn5-Induced Mutants of Azotobacter Vinelandii Affected in Nitrogen Fixation under Mo-Deficient and Mo-Sufficient Conditions. J. Bacteriol. 1986, 168, 673–682. 10.1128/JB.168.2.673-682.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen R. M.; Chatterjee R.; Ludden P. W.; Shah V. K. Incorporation of Iron and Sulfur from NifB Cofactor into the Iron- Molybdenum Cofactor of Dinitrogenase. J. Biol. Chem. 1995, 270, 26890–26896. 10.1074/jbc.270.45.26890. [DOI] [PubMed] [Google Scholar]
- Rangaraj P.; Ruttimann-Johnson C.; Shah V. K.; Ludden P. W. Accumulation of 55Fe-Labeled Precursors of the Iron-Molybdenum Cofactor of Nitrogenase on NifH and NifX of Azotobacter Vinelandii. J. Biol. Chem. 2001, 276, 15968–15974. 10.1074/jbc.M100907200. [DOI] [PubMed] [Google Scholar]
- George S. J.; Igarashi R. Y.; Xiao Y.; Hernandez J. A.; Demuez M.; Zhao D.; Yoda Y.; Ludden P. W.; Rubio L. M.; Cramer S. P. Extended X-Ray Absorption Fine Structure and Nuclear Resonance Vibrational Spectroscopy Reveal That NifB-Co, a FeMo-Co Precursor, Comprises a 6Fe Core with an Interstitial Light Atom. J. Am. Chem. Soc. 2008, 130, 5673–5680. 10.1021/ja0755358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soboh B.; Boyd E. S.; Zhao D.; Peters J. W.; Rubio L. M. Substrate Specificity and Evolutionary Implications of a NifDK Enzyme Carrying NifB-Co at Its Active Site. FEBS Lett. 2010, 584, 1487–1492. 10.1016/j.febslet.2010.02.064. [DOI] [PubMed] [Google Scholar]
- Goodwin P. J.; Agar J. N.; Roll J. T.; Roberts G. P.; Johnson M. K.; Dean D. R. The Azotobacter Vinelandii NifEN Complex Contains Two Identical [4Fe- 4S] Clusters. Biochemistry 1998, 37, 10420–10428. 10.1021/bi980435n. [DOI] [PubMed] [Google Scholar]
- Fay A. W.; Blank M. A.; Lee C. C.; Hu Y.; Hodgson K. O.; Hedman B.; Ribbe M. W. Spectroscopic Characterization of the Isolated Iron-Molybdenum Cofactor (FeMoco) Precursor from the Protein NifEN. Angew. Chem., Int. Ed. 2011, 50, 7787–7790. 10.1002/anie.201102724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S. J.; Angove H. C.; Papaefthymiou V.; Burgess B. K.; Münck E. Mössbauer Study of the MoFe Protein of Nitrogenase from Azotobacter Vinelandii Using Selective 57Fe Enrichment of the M-Centers. J. Am. Chem. Soc. 2000, 122, 4926–4936. 10.1021/ja000254k. [DOI] [Google Scholar]
- Lancaster K. M.; Hu Y.; Bergmann U.; Ribbe M. W.; DeBeer S.; Hu Y.; Lancaster K. M.; DeBeer S.; Ribbe M. W. X-Ray Spectroscopic Observation of an Interstitial Carbide in NifEN-Bound FeMoco Precursor. J. Am. Chem. Soc. 2013, 135, 610–612. 10.1021/ja309254g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett M. C.; Hu Y.; Fay A. W.; Ribbe M. W.; Hedman B.; Hodgson K. O. Structural Insights into a Protein-Bound Iron-Molybdenum Cofactor Precursor. Proc. Natl. Acad. Sci. U. S. A. 2006, 103, 1238–1243. 10.1073/pnas.0507853103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser J. T.; Hu Y.; Wiig J. A.; Rees D. C.; Ribbe M. W. Structure of Precursor-Bound NifEN: A Nitrogenase FeMo Cofactor Maturase/Insertase. Science 2011, 331, 91–94. 10.1126/science.1196954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rettberg L. A.; Wilcoxen J.; Lee C. C.; Stiebritz M. T.; Tanifuji K.; Britt R. D.; Hu Y. Probing the Coordination and Function of Fe4S4 Modules in Nitrogenase Assembly Protein NifB. Nat. Commun. 2018, 9, 2824. 10.1038/s41467-018-05272-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buikema W. J.; Klingensmith J. A.; Gibbons S. L.; Ausubel F. M. Conservation of Structure and Location of Rhizobium Meliloti and Klebsiella Pneumoniae NifB Genes. J. Bacteriol. 1987, 169, 1120–1126. 10.1128/JB.169.3.1120-1126.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noti J. D.; Folkerts O.; Turken A. N.; Szalay A. A. Organization and Characterization of Genes Essential for Symbiotic Nitrogen Fixation from Bradyrhizobium Japonicum I110. J. Bacteriol. 1986, 167, 774–783. 10.1128/JB.167.3.774-783.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echavarri-Erasun C.; Arragain S.; Scandurra A. A.; Rubio L. M.. Expression and Purification of NifB Proteins from Aerobic and Anaerobic Sources. In Metalloproteins: Methods and Protocols; Fontecilla-Camps J. C., Nicolet Y., Eds.; Humana Press: Totowa, NJ, 2014; pp 19–31. [DOI] [PubMed] [Google Scholar]
- Kozbial P. Z.; Mushegian A. R. Natural History of S-Adenosylmethionine-Binding Proteins. BMC Struct. Biol. 2005, 5, 19. 10.1186/1472-6807-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick J. B.; Duffus B. R.; Duschene K. S.; Shepard E. M. Radical S - Adenosylmethionine Enzymes. Chem. Rev. 2014, 114, 4229–4317. 10.1021/cr4004709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burén S.; Jiang X.; Lopez-Torrejon G.; Echavarri-Erasun C.; Rubio L. M. Purification and In Vitro Activity of Mitochondria Targeted Nitrogenase Cofactor Maturase NifB. Front. Plant Sci. 2017, 8, 1567. 10.3389/fpls.2017.01567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofia H. J.; Chen G.; Hetzler B. G.; Reyes-Spindola J. F.; Miller N. E. Radical SAM, a Novel Protein Superfamily Linking Unresolved Steps in Familiar Biosynthetic Pathways with Radical Mechanisms: Functional Characterization Using New Analysis and Information Visualization Methods. Nucleic Acids Res. 2001, 29, 1097–1106. 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiig J. A.; Hu Y.; Ribbe M. W. NifEN-B Complex of Azotobacter Vinelandii Is Fully Functional in Nitrogenase FeMo Cofactor Assembly. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 8623–8627. 10.1073/pnas.1102773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boal A. K.; Grove T. L.; McLaughlin M. I.; Yennawar N. H.; Booker S. J.; Rosenzweig A. C. Structural Basis for Methyl Transfer by a Radical SAM Enzyme. Science 2011, 332, 1089–1092. 10.1126/science.1205358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove T. L.; Benner J. S.; Radle M. I.; Ahlum J. H.; Landgraf B. J.; Krebs C.; Booker S. J. A Radically Different Mechanism for S-Adenosylmethionine-Dependent Methyltransferases. Science 2011, 332, 604–607. 10.1126/science.1200877. [DOI] [PubMed] [Google Scholar]
- Wiig J. A.; Hu Y.; Ribbe M. W. Refining the Pathway of Carbide Insertion into the Nitrogenase M-Cluster. Nat. Commun. 2015, 6, 8034. 10.1038/ncomms9034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Quinones F.; Bosch R.; Imperial J. Expression of the NifBfdxNnifOQ Region of Azotobacter Vinelandii and Its Role in Nitrogenase Activity. J. Bacteriol. 1993, 175, 2926–2935. 10.1128/JB.175.10.2926-2935.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez J. A.; George S. J.; Rubio L. M. Molybdenum Trafficking for Nitrogen Fixation. Biochemistry 2009, 48, 9711–9721. 10.1021/bi901217p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klipp W.; Reilander H.; Schluter A.; Krey R.; Puhler A. The Rhizobium Meliloti FdxN Gene Encoding a Ferredoxin-like Protein Is Necessary for Nitrogen Fixation and Is Cotranscribed with NifA and NifB. Mol. Gen. Genet. 1989, 216, 293–302. 10.1007/BF00334368. [DOI] [PubMed] [Google Scholar]
- Souza A. L. F.; Invitti A. L.; Rego F. G. M.; Monteiro R. A.; Klassen G.; Souza E. M.; Chubatsu L. S.; Pedrosa F. O.; Rigo L. U. The Involvement of the Nif-Associated Ferredoxin-like Genes FdxA and FdxN of Herbaspirillum Seropedicae in Nitrogen Fixation. J. Microbiol. 2010, 48, 77–83. 10.1007/s12275-009-0077-y. [DOI] [PubMed] [Google Scholar]
- Ebeling S.; Noti J. D.; Hennecke H. Identification of a New Bradyrhizobium Japonicum Gene (FrxA) Encoding a Ferredoxinlike Protein. J. Bacteriol. 1988, 170, 1999–2001. 10.1128/JB.170.4.1999-2001.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton T. L.; Ludwig M.; Dixon R.; Boyd E. S.; Dos Santos P. C.; Setubal J. C.; Bryant D. A.; Dean D. R.; Peters J. W. Transcriptional Profiling of Nitrogen Fixation in Azotobacter Vinelandii. J. Bacteriol. 2011, 193, 4477–4486. 10.1128/JB.05099-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homer M. J.; Dean D. R.; Roberts G. P. Characterization of the Gamma Protein and Its Involvement in the Metallocluster Assembly and Maturation of Dinitrogenase from Azotobacter Vinelandii. J. Biol. Chem. 1995, 270, 24745–24752. 10.1074/jbc.270.42.24745. [DOI] [PubMed] [Google Scholar]
- Pope M. T.; Still E. R.; Williams R. J. P.. A Comparison between the Chemistry and Biochemistry of Molybdenum and Related Elements. In Molybdenum and Molybdenum-Containing Enzymes; Coughlan M. P., Ed.; Pergamon Press: Oxford, U.K., 1980; pp 1–40. [Google Scholar]
- Siemann S.; Schneider K.; Oley M.; Müller A. Characterization of a Tungsten-Substituted Nitrogenase Isolated from Rhodobacter Capsulatus. Biochemistry 2003, 42, 3846–3857. 10.1021/bi0270790. [DOI] [PubMed] [Google Scholar]
- Wichard T.; Bellenger J. P.; Loison A.; Kraepiel A. M. Catechol Siderophores Control Tungsten Uptake and Toxicity in the Nitrogen-Fixing Bacterium Azotobacter Vinelandii. Environ. Sci. Technol. 2008, 42, 2408–2413. 10.1021/es702651f. [DOI] [PubMed] [Google Scholar]
- Page W. J.; von Tigerstrom M. Iron- and Molybdenum-Repressible Outer Membrane Proteins in Competent Azotobacter Vinelandii. J. Bacteriol. 1982, 151, 237–242. 10.1128/JB.151.1.237-242.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellenger J. P.; Wichard T.; Kustka A. B.; Kraepiel A. M. L. Uptake of Molybdenum and Vanadium by a Nitrogen-Fixing Soil Bacterium Using Siderophores. Nat. Geosci. 2008, 1, 243. 10.1038/ngeo161. [DOI] [Google Scholar]
- Mouncey N. J.; Mitchenall L. A.; Pau R. N. Mutational Analysis of Genes of the Mod Locus Involved in Molybdenum Transport, Homeostasis, and Processing in Azotobacter Vinelandii. J. Bacteriol. 1995, 177, 5294–5302. 10.1128/JB.177.18.5294-5302.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraepiel A. M.; Bellenger J. P.; Wichard T.; Morel F. M. Multiple Roles of Siderophores in Free-Living Nitrogen-Fixing Bacteria. BioMetals 2009, 22, 573–581. 10.1007/s10534-009-9222-7. [DOI] [PubMed] [Google Scholar]
- Schemberg J.; Schneider K.; Fenske D.; Muller A. Azotobacter Vinelandii Metal Storage Protein: “Classical” Inorganic Chemistry Involved in Mo/W Uptake and Release Processes. ChemBioChem 2008, 9, 595–602. 10.1002/cbic.200700446. [DOI] [PubMed] [Google Scholar]
- Pienkos P. T.; Brill W. J. Molybdenum Accumulation and Storage in Klebsiella Pneumoniae and Azotobacter Vinelandii. J. Bacteriol. 1981, 145, 743–751. 10.1128/JB.145.2.743-751.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah V. K.; Ugalde R. A.; Imperial J.; Brill W. J. Molybdenum in Nitrogenase. Annu. Rev. Biochem. 1984, 53, 231–257. 10.1146/annurev.bi.53.070184.001311. [DOI] [PubMed] [Google Scholar]
- Fenske D.; Gnida M.; Schneider K.; Meyer-Klaucke W.; Schemberg J.; Henschel V.; Meyer A.-K. K.; Knöchel A.; Müller A. A New Type of Metalloprotein: The Mo Storage Protein from Azotobacter Vinelandii Contains a Polynuclear Molybdenum-Oxide Cluster. ChemBioChem 2005, 6, 405–413. 10.1002/cbic.200400263. [DOI] [PubMed] [Google Scholar]
- Kowalewski B.; Poppe J.; Demmer U.; Warkentin E.; Dierks T.; Ermler U.; Schneider K. Nature’s Polyoxometalate Chemistry: X-Ray Structure of the Mo Storage Protein Loaded with Discrete Polynuclear Mo-O Clusters. J. Am. Chem. Soc. 2012, 134, 9768–9774. 10.1021/ja303084n. [DOI] [PubMed] [Google Scholar]
- Schemberg J.; Schneider K.; Demmer U.; Warkentin E.; Muller A.; Ermler U. Towards Biological Supramolecular Chemistry: A Variety of Pocket-Templated, Individual Metal Oxide Cluster Nucleations in the Cavity of a Mo/w-Storage Protein. Angew. Chem., Int. Ed. 2007, 46, 2408–2413. 10.1002/anie.200604858. [DOI] [PubMed] [Google Scholar]
- Poppe J.; Brunle S.; Hail R.; Wiesemann K.; Schneider K.; Ermler U. The Molybdenum Storage Protein: A Soluble ATP Hydrolysis-Dependent Molybdate Pump. FEBS J. 2018, 285, 4602–4616. 10.1111/febs.14684. [DOI] [PubMed] [Google Scholar]
- Ramos F.; Blanco G.; Gutierrez J. C.; Luque F.; Tortolero M. Identification of an Operon Involved in the Assimilatory Nitrate- Reducing System of Azotobacter Vinelandii. Mol. Microbiol. 1993, 8, 1145–53. 10.1111/j.1365-2958.1993.tb01659.x. [DOI] [PubMed] [Google Scholar]
- Drozd J. W.; Tubb R. S.; Postgate J. R. A Chemostat Study of the Effect of Fixed Nitrogen Sources on Nitrogen Fixation, Membranes and Free Amino Acids in Azotobacter Chroococcum. J. Gen. Microbiol. 1972, 73, 221–232. 10.1099/00221287-73-2-221. [DOI] [PubMed] [Google Scholar]
- Bosch R.; Rodríguez-Quiñones F.; Imperial J. Identification of Gene Products from the Azotobacter Vinelandii NifBfdxNnifOQ Operon. FEMS Microbiol. Lett. 1997, 157, 19–25. 10.1111/j.1574-6968.1997.tb12747.x. [DOI] [PubMed] [Google Scholar]
- Oppenheim D. S.; Yanofsky C. Translational Coupling during Expression of the Tryptophan Operon of Escherichia Coli. Genetics 1980, 95, 785–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Burris R. H.; Roberts G. P. Cloning, Sequencing, Mutagenesis, and Functional Characterization of DraT and DraG Genes from Azospirillum Brasilense. J. Bacteriol. 1992, 174, 3364–3369. 10.1128/JB.174.10.3364-3369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J. H.; Nielsen G. M.; Lies D. P.; Burris R. H.; Roberts G. P.; Ludden P. W. Mutations in the DraT and DraG Genes of Rhodospirillum Rubrum Result in Loss of Regulation of Nitrogenase by Reversible ADP-Ribosylation. J. Bacteriol. 1991, 173, 6903–6909. 10.1128/JB.173.21.6903-6909.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouncey N. J.; Mitchenall L. A.; Pau R. N. The ModE Gene Product Mediates Molybdenum-Dependent Expression of Genes for the High-Affinity Molybdate Transporter and ModG in Azotobacter Vinelandii. Microbiology 1996, 142, 1997–2004. 10.1099/13500872-142-8-1997. [DOI] [PubMed] [Google Scholar]
- Delarbre L.; Stevenson C. E. M.; White D. J.; Mitchenall L. A.; Pau R. N.; Lawson D. M. Two Crystal Structures of the Cytoplasmic Molybdate-Binding Protein ModG Suggest a Novel Cooperative Binding Mechanism and Provide Insights into Ligand-Binding Specificity. J. Mol. Biol. 2001, 308, 1063–1079. 10.1006/jmbi.2001.4636. [DOI] [PubMed] [Google Scholar]
- Moreno-Vivian C.; Hennecke S.; Puhler A.; Klipp W. Open Reading Frame 5 (ORF5), Encoding a Ferredoxinlike Protein, and NifQ Are Cotranscribed with NifE, NifN, NifX, and ORF4 in Rhodobacter Capsulatus. J. Bacteriol. 1989, 171, 2591–2598. 10.1128/JB.171.5.2591-2598.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad M. M.; Michalet S.; Fossou R.; Putnik-Delic M.; Crevecoeur M.; Meyer J.; de Malezieux C.; Hopfgartner G.; Maksimovic I.; Perret X. Loss of NifQ Leads to Accumulation of Porphyrins and Altered Metal-Homeostasis in Nitrogen-Fixing Symbioses. Mol. Plant-Microbe Interact. 2019, 32, 208–216. 10.1094/MPMI-07-18-0188-R. [DOI] [PubMed] [Google Scholar]
- Ugalde R. A.; Imperial J.; Shah V. K.; Brill W. J. Biosynthesis of the Iron-Molybdenum Cofactor and the Molybdenum Cofactor in Klebsiella Pneumoniae: Effect of Sulfur Source. J. Bacteriol. 1985, 164, 1081–1087. 10.1128/JB.164.3.1081-1087.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddavattam D.; Singh M.; Klingmüller W. Structure of the NifQ Gene from Enterobacter Agglomerans 333 and Its Overexpression in Escherichia Coli. Mol. Gen. Genet. 1993, 239, 435–440. 10.1007/BF00276942. [DOI] [PubMed] [Google Scholar]
- George S. J.; Hernandez J. A.; Jimenez-Vicente E.; Echavarri-Erasun C.; Rubio L. M. EXAFS Reveals Two Mo Environments in the Nitrogenase Iron-Molybdenum Cofactor Biosynthetic Protein NifQ. Chem. Commun. 2016, 52, 11811–11814. 10.1039/C6CC06370E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spatzal T.; Schlesier J.; Burger E.-M.; Sippel D.; Zhang L.; Andrade S. L. A.; Rees D. C.; Einsle O. Nitrogenase FeMoco Investigated by Spatially Resolved Anomalous Dispersion Refinement. Nat. Commun. 2016, 7, 10902. 10.1038/ncomms10902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer S. P.; Gillum W. O.; Hodgson K. O.; Mortenson L. E.; Stiefel E. I.; Chisnell J. R.; Brill W. J.; Shah V. K. The Molybdenum Site of Nitrogenase. 2. A Comparative Study of Mo-Fe Proteins and the Iron-Molybdenum Cofactor by x-Ray Absorption Spectroscopy. J. Am. Chem. Soc. 1978, 100, 3814–3819. 10.1021/ja00480a027. [DOI] [Google Scholar]
- Burgess B. K. The Iron-Molybdenum Cofactor of Nitrogenase. Chem. Rev. 1990, 90, 1377–1406. 10.1021/cr00106a002. [DOI] [Google Scholar]
- McLean P. A.; Dixon R. A. Requirement of NifV Gene for Production of Wild-Type Nitrogenase Enzyme in Klebsiella Pneumoniae. Nature 1981, 292, 655–656. 10.1038/292655a0. [DOI] [PubMed] [Google Scholar]
- Hawkes T. R.; McLean P. A.; Smith B. E. Nitrogenase from NifV Mutants of Klebsiella Pneumoniae Contains an Altered Form of the Iron-Molybdenum Cofactor. Biochem. J. 1984, 217, 317–321. 10.1042/bj2170317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover T. R.; Robertson A. D.; Cerny R. L.; Hayes R. N.; Imperial J.; Shah V. K.; Ludden P. W. Identification of the V Factor Needed for Synthesis of the Iron-Molybdenum Cofactor of Nitrogenase as Homocitrate. Nature 1987, 329, 855–857. 10.1038/329855a0. [DOI] [PubMed] [Google Scholar]
- Hoover T. R.; Imperial J.; Ludden P. W.; Shah V. K. Homocitrate Cures the NifV- Phenotype in Klebsiella Pneumoniae. J. Bacteriol. 1988, 170, 1978–1979. 10.1128/JB.170.4.1978-1979.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L.; White R. H.; Dean D. R. Purification of the Azotobacter Vinelandii NifV-Encoded Homocitrate Synthase. J. Bacteriol. 1997, 179, 5963–5966. 10.1128/JB.179.18.5963-5966.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J.; Madden M.; Shah V. K.; Burris R. H. Citrate Substitutes for Homocitrate in Nitrogenase of a NifV Mutant of Klebsiella Pneumoniae. Biochemistry 1990, 29, 8577–8581. 10.1021/bi00489a011. [DOI] [PubMed] [Google Scholar]
- Imperial J.; Hoover T. R.; Madden M. S.; Ludden P. W.; Shah V. K. Substrate Reduction Properties of Dinitrogenase Activated in Vitro Are Dependent upon the Presence of Homocitrate or Its Analogues during Iron-Molybdenum Cofactor Synthesis. Biochemistry 1989, 28, 7796–7799. 10.1021/bi00445a040. [DOI] [PubMed] [Google Scholar]
- Dixon R.; Kennedy C.; Kondorosi A.; Krishnapillai V.; Merrick M. Complementation Analysis of Klebsiella Pneumoniae Mutants Defective in Nitrogen Fixation. Mol. Gen. Genet. 1977, 157, 189–198. 10.1007/BF00267397. [DOI] [PubMed] [Google Scholar]
- Roberts G. P.; Brill W. J. Gene-Product Relationships of the Nif Regulon of Klebsiella Pneumoniae. J. Bacteriol. 1980, 144, 210–216. 10.1128/JB.144.1.210-216.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean D. R.; Brigle K. E. Azotobacter Vinelandii NifD- and NifE-Encoded Polypeptides Share Structural Homology. Proc. Natl. Acad. Sci. U. S. A. 1985, 82, 5720–5723. 10.1073/pnas.82.17.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y.; Yoshizawa J. M.; Fay A. W.; Lee C. C.; Wiig J. A.; Ribbe M. W. Catalytic Activities of NifEN: Implications for Nitrogenase Evolution and Mechanism. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 16962–16966. 10.1073/pnas.0907872106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll J. T.; Shah V. K.; Dean D. R.; Roberts G. P. Characteristics of NIFNE in Azotobacter Vinelandii Strains. Implications for the Synthesis of the Iron-Molybdenum Cofactor of Dinitrogenase. J. Biol. Chem. 1995, 270, 4432–4437. 10.1074/jbc.270.9.4432. [DOI] [PubMed] [Google Scholar]
- Goodwin P. J. Biosynthesis of the Nitrogenase FeMo-Cofactor from Azotobacter Vinelandii: Involvement of the NifEN Complex, NifX and the Fe Protein. Thesis Diss. Virginia Technol. 1999, 1–210. [Google Scholar]
- Hu Y.; Corbett M. C.; Fay A. W.; Webber J. A.; Hodgson K. O.; Hedman B.; Ribbe M. W. FeMo Cofactor Maturation on NifEN. Proc. Natl. Acad. Sci. U. S. A. 2006, 103, 17119–17124. 10.1073/pnas.0602647103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y.; Corbett M. C.; Fay A. W.; Webber J. A.; Hodgson K. O.; Hedman B.; Ribbe M. W. Nitrogenase Fe Protein: A Molybdate/Homocitrate Insertase. Proc. Natl. Acad. Sci. U. S. A. 2006, 103, 17125–17130. 10.1073/pnas.0602651103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa J. M.; Blank M. A.; Fay A. W.; Lee C. C.; Wiig J. A.; Hu Y.; Hodgson K. O.; Hedman B.; Ribbe M. W. Optimization of FeMoco Maturation on NifEN. J. Am. Chem. Soc. 2009, 131, 9321–9325. 10.1021/ja9035225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay A. W.; Blank M. A.; Yoshizawa J. M.; Lee C. C.; Wiig J. A.; Hu Y.; Hodgson K. O.; Hedman B.; Ribbe M. W. Formation of a Homocitrate-Free Iron-Molybdenum Cluster on NifEN: Implications for the Role of Homocitrate in Nitrogenase Assembly. Dalton Trans. 2010, 39, 3124–3130. 10.1039/c000264j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klassen G.; de Oliveira Pedrosa F.; de Souza E. M.; Yates M. G.; Rigo L. U. Nitrogenase Activity of Herbaspirillum Seropedicae Grown under Low Iron Levels Requires the Products of NifXorf1 Genes. FEMS Microbiol. Lett. 2003, 224, 255–259. 10.1016/S0378-1097(03)00453-1. [DOI] [PubMed] [Google Scholar]
- Rangaraj P.; Ludden P. W. Accumulation of 99Mo-Containing Iron-Molybdenum Cofactor Precursors of Nitrogenase on NifNE, NifH, and NifX of Azotobacter Vinelandii. J. Biol. Chem. 2002, 277, 40106–40111. 10.1074/jbc.M204581200. [DOI] [PubMed] [Google Scholar]
- George S. J.; Igarashi R. Y.; Piamonteze C.; Soboh B.; Cramer S. P.; Rubio L. M. Identification of a Mo-Fe-S Cluster on NifEN by Mo K-Edge Extended X-Ray Absorption Fine Structure. J. Am. Chem. Soc. 2007, 129, 3060–3061. 10.1021/ja0663428. [DOI] [PubMed] [Google Scholar]
- Wolle D.; Kim C.; Dean D.; Howard J. B. Ionic Interactions in the Nitrogenase Complex. Properties of Fe-Protein Containing Substitutions for Arg-100. J. Biol. Chem. 1992, 267, 3667–3673. [PubMed] [Google Scholar]
- Wolle D.; Dean D. R.; Howard J. B. Nucleotide-Iron-Sulfur Cluster Signal Transduction in the Nitrogenase Iron-Protein: The Role of Asp125. Science 1992, 258, 992–995. 10.1126/science.1359643. [DOI] [PubMed] [Google Scholar]
- Gavini N.; Burgess B. K. FeMo Cofactor Synthesis by a NifH Mutant with Altered MgATP Reactivity. J. Biol. Chem. 1992, 267, 21179–21186. [PubMed] [Google Scholar]
- Ludden P. W.; Rangaraj P.; Rubio L. M.. Biosynthesis of the Iron-Molybdenum and Iron-Vanadium Cofactors of the Nif- and Vnf- Encoded Nitrogenases. In Catalyests for Nitrogen Fixation: Nitrogenases, Relevant Chemical Models and Commercial Processes; Smith B. E., Richards R. L., Newton W. E., Eds.; Kluwer Academic Publishers: The Netherlands, 2004; pp 219–254. [Google Scholar]
- Moreno-Vivian C.; Schmehl M.; Masepohl B.; Arnold W.; Klipp W. DNA Sequence and Genetic Analysis of the Rhodobacter Capsulatus NifENX Gene Region: Homology between NifX and NifB Suggests Involvement of NifX in Processing of the Iron-Molybdenum Cofactor. Mol. Gen. Genet. 1989, 216, 353–363. 10.1007/BF00334376. [DOI] [PubMed] [Google Scholar]
- Simon H. M.; Gosink M. M.; Roberts G. P. Importance of Cis Determinants and Nitrogenase Activity in Regulated Stability of the Klebsiella Pneumoniae Nitrogenase Structural Gene MRNA. J. Bacteriol. 1999, 181, 3751–3760. 10.1128/JB.181.12.3751-3760.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klassen G.; Pedrosa F. O.; Souza E. M.; Yates M. G.; Rigo L. U. Sequencing and Functional Analysis of the NifENXorf1orf2 Gene Cluster of Herbaspirillum Seropedicae. FEMS Microbiol. Lett. 1999, 181, 165–170. 10.1111/j.1574-6968.1999.tb08840.x. [DOI] [PubMed] [Google Scholar]
- Gosink M. M.; Franklin N. M.; Roberts G. P. The Product of the Klebsiella Pneumoniae NifX Gene Is a Negative Regulator of the Nitrogen Fixation (Nif) Regulon. J. Bacteriol. 1990, 172, 1441–1447. 10.1128/JB.172.3.1441-1447.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruttimann-Johnson C.; Staples C. R.; Rangaraj P.; Shah V. K.; Ludden P. W. A Vanadium and Iron Cluster Accumulates on VnfX during Iron-Vanadium-Cofactor Synthesis for the Vanadium Nitrogenase in Azotobacter Vinelandii. J. Biol. Chem. 1999, 274, 18087–18092. 10.1074/jbc.274.25.18087. [DOI] [PubMed] [Google Scholar]
- White T. C.; Harris G. S.; Orme-Johnson W. H. Electrophoretic Studies on the Assembly of the Nitrogenase Molybdenum-Iron Protein from the Klebsiella Pneumoniae NifD and NifK Gene Products. J. Biol. Chem. 1992, 267, 24007–24016. [PubMed] [Google Scholar]
- Ruttimann-Johnson C.; Rubio L. M.; Dean D. R.; Ludden P. W. VnfY Is Required for Full Activity of the Vanadium-Containing Dinitrogenase in Azotobacter Vinelandii. J. Bacteriol. 2003, 185, 2383–2386. 10.1128/JB.185.7.2383-2386.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer D. H.; Rubio L. M.; Thoden J. B.; Holden H. M.; Ludden P. W.; Rayment I. The Three-Dimensional Structure of the Core Domain of Naf Y from Azotobacter Vinelandii Determined at 1.8-A Resolution. J. Biol. Chem. 2003, 278, 32150–32156. 10.1074/jbc.M304264200. [DOI] [PubMed] [Google Scholar]
- Schultz J.; Ponting C. P.; Hofmann K.; Bork P. SAM as a Protein Interaction Domain Involved in Developmental Regulation. Protein Sci. 1997, 6, 249–253. 10.1002/pro.5560060128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C. A.; Bowie J. U. SAM Domains: Uniform Structure. Trends Biochem. Sci. 2003, 28, 625–628. 10.1016/j.tibs.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Cort J. R.; Yee A.; Edwards A. M.; Arrowsmith C. H.; Kennedy M. A. NMR Structure Determination and Structure-Based Functional Characterization of Conserved Hypothetical Protein MTH1175 from Methanobacterium Thermoautotrophicum. J. Struct. Funct. Genomics 2000, 1, 15–25. 10.1023/A:1011348803324. [DOI] [PubMed] [Google Scholar]
- Vaccaro B. J.; Clarkson S. M.; Holden J. F.; Lee D.-W.; Wu C.-H.; Poole Ii F. L.; Cotelesage J. J. H.; Hackett M. J.; Mohebbi S.; Sun J.; et al. Biological Iron-Sulfur Storage in a Thioferrate-Protein Nanoparticle. Nat. Commun. 2017, 8, 16110. 10.1038/ncomms16110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Noël G.; Curatti L.; Hernandez J. A.; Rubio L. M. NifB and NifEN Protein Levels Are Regulated by ClpX2 under Nitrogen Fixation Conditions in Azotobacter Vinelandii. Mol. Microbiol. 2011, 79, 1182–1193. 10.1111/j.1365-2958.2011.07540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burén S.; Young E. M.; Sweeny E. A.; Lopez-Torrejon G.; Veldhuizen M.; Voigt C. A.; Rubio L. M. Formation of Nitrogenase NifDK Tetramers in the Mitochondria of Saccharomyces Cerevisiae. ACS Synth. Biol. 2017, 6, 1043–1055. 10.1021/acssynbio.6b00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.; Xie X.; Xiang N.; Tian Z.-X.; Dixon R.; Wang Y.-P. Polyprotein Strategy for Stoichiometric Assembly of Nitrogen Fixation Components for Synthetic Biology. Proc. Natl. Acad. Sci. U. S. A. 2018, 115, E8509–E8517. 10.1073/pnas.1804992115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen R. S.; Tilbrook K.; Warden A. C.; Campbell P. C.; Rolland V.; Singh S. P.; Wood C. C. Expression of 16 Nitrogenase Proteins within the Plant Mitochondrial Matrix. Front. Plant Sci. 2017, 8, 287. 10.3389/fpls.2017.00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.; Xie X.; Yang M.; Dixon R.; Wang Y.-P. Modular Electron-Transport Chains from Eukaryotic Organelles Function to Support Nitrogenase Activity. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, E2460–E2465. 10.1073/pnas.1620058114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good A. Toward Nitrogen-Fixing Plants. Science 2018, 359, 869–870. 10.1126/science.aas8737. [DOI] [PubMed] [Google Scholar]
- Hardy R. W.; Havelka U. D. Nitrogen Fixation Research: A Key to World Food?. Science 1975, 188, 633–643. 10.1126/science.188.4188.633. [DOI] [PubMed] [Google Scholar]