Abstract
Presented here is a protocol to study pharmacodynamics, stem cell potential, and cancer differentiation in prostate epithelial organoids. Prostate organoids are androgen responsive, three-dimensional (3D) cultures grown in a defined medium that resembles the prostatic epithelium. Prostate organoids can be established from wild-type and genetically engineered mouse models, benign human tissue, and advanced prostate cancer. Importantly, patient derived organoids closely resemble tumors in genetics and in vivo tumor biology. Moreover, organoids can be genetically manipulated using CRISPR/Cas9 and shRNA systems. These controlled genetics make the organoid culture attractive as a platform for rapidly testing the effects of genotypes and mutational profiles on pharmacological responses. However, experimental protocols must be specifically adapted to the 3D nature of organoid cultures to obtain reproducible results. Described here are detailed protocols for performing seeding assays to determine organoid formation capacity. Subsequently, this report shows how to perform drug treatments and analyze pharmacological response via viability measurements, protein isolation, and RNA isolation. Finally, the protocol describes how to prepare organoids for xenografting and subsequent in vivo growth assays using subcutaneous grafting. These protocols yield highly reproducible data and are widely applicable to 3D culture systems.
Keywords: Cancer Research, Issue 152, prostate organoids, prostate cancer, second generation anti-androgens, drug resistance, primary cell culture, prostate model systems
Introduction
Drug resistance is one of the major clinical problems in cancer treatment. Metastatic prostate cancer (PCa) treatment is primarily directed at the androgen-signaling axis. Next-generation anti-androgen therapies (e.g., enzalutamide and abiraterone) have showed great clinical success, but virtually all PCa eventually progresses towards an androgen-independent state, or castration resistant prostate cancer (CRPC).
Recent genomic and transcriptomic profiling of CRPC revealed there are three general mechanisms of resistance in prostate cancer: 1) activating mutations resulting in the restoration of androgen receptor (AR) signaling1; 2) activation of bypass signaling, as exemplified in a pre-clinical model for next-generation anti-androgen therapy resistance in which activation of the glucocorticoid receptor (GR) can compensate for loss of AR signaling2; and 3) the recently identified process of lineage plasticity, in which tumor cells acquire resistance by switching lineages from a cell type dependent on the drug target to another cell type that is not dependent on this (which, in PCa, is represented as AR-negative and/or neuroendocrine disease [NEPC])3,4. However, the molecular mechanisms that cause drug resistance are not understood. Moreover, acquired anti-androgen resistance may lead to therapeutic vulnerabilities that can be exploited. Therefore, it is essential to evaluate drug responses in model systems that mimic patient phenotypes and genotypes.
Prostate organoids are organotypic cultures grown in a 3D protein matrix with a defined medium. Importantly, prostate organoids can be established from benign and cancerous tissue of murine or human origin, and they retain phenotypic and genotypic features found in vivo5,6. Importantly, both anti-androgen sensitive PCa and CRPC cells are represented in the current compendium of organoids. Moreover, prostate organoids are easily genetically manipulated using CRISPR/Cas9 and shRNA5. Thus, prostate organoids are a suitable model system for testing drug responses and elucidating resistance mechanisms. Here, a detailed protocol is described to perform drug testing and analyze pharmacological responses using prostate organoids.
Protocol
All work described in this protocol has been performed with previously established murine organoids and patient-derived organoids. All animal work was performed in compliance with the guidelines of Research Animal Resource Center of Memorial Sloan Kettering Cancer Center (IACUC: 06-07-012). All patient-derived tissues were collected in compliance with rules and regulations of Memorial Sloan Kettering Cancer Center (IRB: 12001).
1. Medium and buffer preparation
Thaw basement membrane matrix (e.g., Matrigel) at 4 °C overnight before starting the experiment. Keep it on ice during use.
Place culture plates at 37 °C for 24 h prior to experiments. This will help the basement membrane matrix dome (hereafter referred to as matrix dome) to polymerize. Plating organoids is described in step 2.1.2.
Prepare the organoid medium according to the established protocol5.
Prepare the organoid medium without the addition of epidermal growth factor (EGF; see Table of Materials for components). EGF suppresses the AR transcriptional output and confers anti-androgen resistance5.
-
Prepare Dulbecco’s Modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS).
NOTE: This media is used to inhibit the enzymatic digestion with trypsin replacement in sections 2–4.
Prepare drug solutions according to the manufacturer’s protocol. For enzalutamide/mdv3100 (hereafter referred to as second generation antiandrogen), prepare a stock solution of 100 μM in dimethyl sulfoxide (DMSO). Stock can be stored at −20 °C for up to 6 months and does not have to be made fresh.
| Reagent | Source | Catalog Number | Comments |
|---|---|---|---|
| A83–01 | Tocris | 2939 | Organoid medium component: Final concentration 200 nM |
| ADMEM/F12 | Gibco/Life technologies | 12634028 | Organoid medium component |
| B27 | Gibco/Life technologies | 17504–044 | Organoid medium component |
| Cell culture plates | Fisher | 657185 | |
| Cell Titer Glo | Promega | G7571 | |
| DHT | Sigma-Aldrich | D-073 | Organoid medium component: Final Concentration 1 nM |
| DMSO | Fisher | BP231–100 | |
| EGF | Peprotech | 315–09 | Organoid medium component: Final concentration 50 ng/ml for mouse, 5 ng/nl for Human |
| FGF10 | Peprotech | 100–26 | Human specific organoid medium component: Final concentration 10 ng/ml |
| FGF2 | Peprotech | 100–18B | Human specific organoid medium component: Final concentration 5 ng/ml |
| Glutamax | Gibco/Life technologies | 35050079 | Organoid medium component |
| HEPES | MADE IN-HOUSE | N/A | Organoid medium component: Final concentration 10 mM |
| Matrigel (Growthfactor reduced & Phenol Red free) | Corning | CB-40230C | Organoid medium component |
| N-Acetylcysteine | Sigma-Aldrich | A9165 | Organoid medium component: Final concentration 1.25 mM |
| Nicotinamide | Sigma-Aldrich | N0636 | Human specific organoid medium component: Final concentration 10 mM |
| NOGGIN | Peprotech or stable transfected 293t cells with Noggin construct (Karthaus et al. 2014) | 120–10C | Organoid medium component: Final Concentration 10% conditioned medium or 100 ng/ml |
| Penicillin/Streptavidin | Gemini Bio-Products | 400–109 | Organoid medium component |
| Phospatase inhibitors | Merck Millipore | 524629 | |
| Prostaglandin E2 | Tocris | 3632464 | |
| Protease Inhibitors | Merck Millipore | 539131 | |
| R-SPONDIN | Peprotech or stable transfected 293t cells with R-Spondin1 construct (Karthaus et al. 2014) | 120–38 | Organoid medium component: Final Concentration 10% conditioned medium or 500 ng/ml |
| RIPA buffer | Merck | 20–188 | |
| RNA-easy minikit | Qiagen | 74104 | |
| SB202190 | Sigma-Aldrich | 152121-30-7 | Human specific organoid medium component: Final concentration 10 μM |
| TryplE | ThermoFisher | 12605036 | |
| Y-27632 | Selleckchem | S1049 | Organoid medium component: Final Concentration 10 μM |
2. Isolation, enzymatic digestion, and establishment of organoids
- Isolate prostate organoids from mouse or human tissue according to the previously established protocols5,7. A brief description is provided below.
- Mince and enzymatically digest prostate tissue to produce a single cell suspension. In this experiment, 1 mL of 5 mg/mL collagenase type II in ADMEM/F12 was used for the digestion of 50 mg of prostate tissue.
- Allow the domes to solidify and add media onto the tops of the domes so that they are completely covered.
Grow organoids to the desired quantity for downstream applications. Cell number can be determined by standard counting methods. See the application-specific section of the protocol for additional details on density.
Using a P1000 pipette, draw up the medium and pipette up and down to disrupt. When basement membrane matrix is fully disrupted, transfer the suspension to a 15 mL conical tube. Do not place more than 10 domes per single 15 mL conical tube. Centrifuge at 300 × g for 5 min.
Draw off the supernatant and wash the cell pellet with 5 mL of PBS. Centrifuge at 300 × g for 5 min.
Draw off the supernatant and resuspend the pellet in 4 mL of trypsin replacement. Digest for 5–10 min with shaking at 37 °C. Add an equal volume of organoid medium + 10% FBS to inhibit the trypsin replacement. Centrifuge at 300 × g for 5 min.
Draw off the supernatant and resuspend in 1 mL of PBS.
-
Filter the suspension with a 40 μm filter to ensure a single cell suspension. Quantify the cell number using a hemocytometer or equivalent counting device.
NOTE: If obtaining viable single cells is difficult, use flow sorting to obtain a single cell solution.
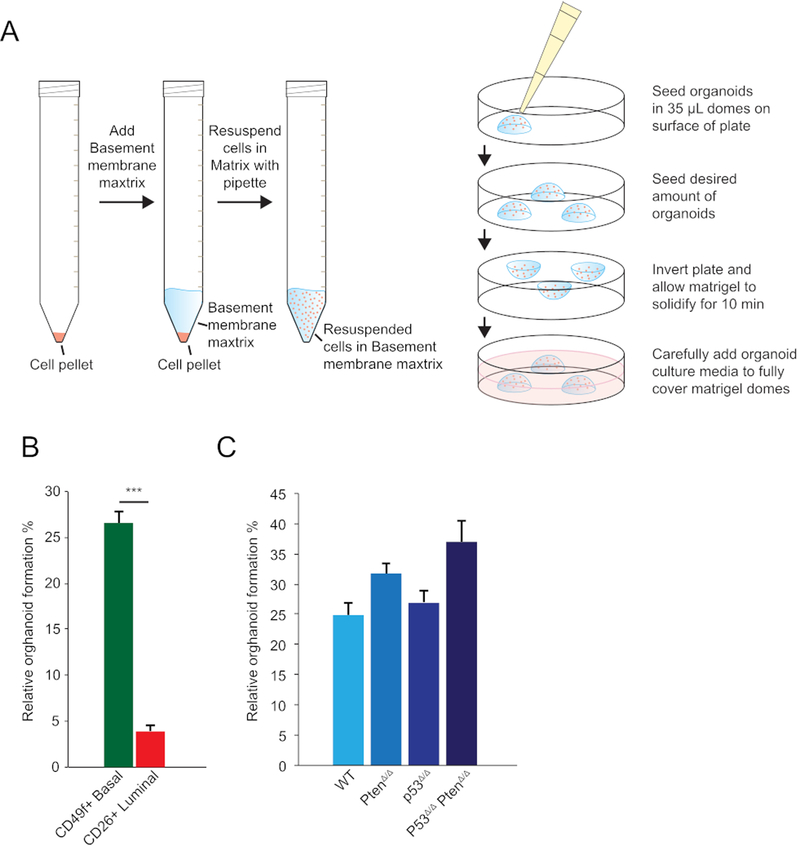
Figure 1: Measuring organoid formation rates of human and mouse prostate cells.
(A) Schematic overview of cell resuspension in basement membrane matrix (left) and organoid seeding in matrix domes (right). (B) Relative organoid formation of human (CD49f+)-derived basal and (CD26+)-derived luminal cells (%, y-axis; mean ± SD) in the presence of 1 nM DHT. A total of 200 cells were seeded and the number of organoids was quantified 7 days post-seeding (n = 3, ***p < 0.01, t-test). (C) Relative organoid formation of murine WT, PtenΔ/Δ, P53Δ/Δ, and P53Δ/Δ PtenΔ/Δ organoids (%, y-axis; mean ± SD) in the presence of 1 nM DHT. A total of 200 cells were seeded and the number of organoids was quantified 7 days post-seeding (n = 3). p53 and Pten loss was mediated by the gRNA’s targeting of the p53 and/or Pten locus in organoids expressing Cas9 constitutively under a Rosa26 promoter.
3. Assessing organoid formation capacity
NOTE: To determine the percentage of cells that can generate an organoid, a seeding assay can be performed as a proxy for the stem/progenitor potential. The organoid formation capacity is also important for defining a cell seeding number for the viability assays.
Dilute the cell suspension obtained in step 2.7 to 100 cells per 10 μL of the suspension using organoid medium containing 10 μM Rho kinase inhibitor Y-27632.
Transfer 1,100 cells (110 μL of suspension) to a new conical tube.
-
Add 285 μL of basement membrane matrix and resuspend the cells. This will result in a ~70% matrix concentration.
NOTE: Dilution of the basement membrane matrix during seeding greatly reduces the variation in dome size, caused by the viscosity of the protein matrix.
Seed cells in 35 μL of matrix domes in a pre-warmed 24 well plate, resulting in 200 cells/well. Plate 3–5 replicates per sample (also see plating method in Figure 1A and step 2.1.2).
To ensure that the cells remain within the matrix dome, flip the plate and place it in a cell incubator to solidify the basement membrane matrix.
After 10 min, remove the plate from the incubator and add medium containing the Rho kinase inhibitor.
Refresh the medium every 2 days. After 7 days, quantify the number of organoids. Keep the Rho kinase inhibitor in media throughout the experiment.
-
Count the number of organoids established per dome and calculate the percent of organoids formed out of the total number of cells plated (200 cells).
NOTE: Organoid establishment ratios vary from 3%−60% depending on the cell type and genotype.
4. Determining pharmacological responses of organoids
-
Continuing from step 2.7, seed 1,000–10,000 cells in a matrix dome. Use the organoid formation efficiency and growth speed as a proxy for determining the final cell number.
NOTE: Recommended cell numbers are provided in Table 1. Use three to five replicates per condition per analysis. Use a final concentration of 70% basement membrane matrix to reduce pipetting errors induced by the viscosity.
Seed 35 μL of matrix domes in a 24 well plate and let the domes solidify as done in section 3 (also see plating method in Figure 1A and step 2.1.2). Add medium containing the Rho kinase inhibitor and drug of choice. This method can be applied to all drugs, but in this protocol, a second-generation anti-androgen is used at 10 μM for an example. To determine half maximal inhibitory concentration (IC50), perform a log10 incremental, and as a control, use the vehicle in which the drug was dissolved.
-
Refresh the medium every two or three days and analyze the organoids on day 7 to determine the pharmacological response of the drug. The timepoints may vary among the choice of experiment and drug.
NOTE: Organoids do not have to be trypsinized to perform these assays.
To keep organoids intact, using a P1000 pipette, draw up the medium and pipette up and down to disrupt the basement membrane matrix.
When the basement membrane matrix is fully disrupted, transfer the suspension to a 15 mL conical tube. Do not transfer more than 10 matrix domes per 15 mL conical tube.
Centrifuge at 300 × g for 5 min. Draw off the supernatant and wash with 5 mL of PBS.
Resuspend organoids in 1 mL of PBS and disrupt the organoids using trituration and a glass Pasteur pipette.
Quantify the number of organoid fragments. Seed 5 replicates containing 100 organoid fragments as described in step 2.1.2.
Perform the cell viability assay as described below in section 7.
Table 1: Cell seeding numbers used to assess pharmacological response based on organoid formation capacity.
Table by column (left to right) includes organoid formation capacity ranges and the corresponding number of cells to seed per matrix dome.
| Organoid seeding efficiency (%) | Cell-seeding number (per dome) |
|---|---|
| 1–10% | 100000 |
| 10–20% | 5000 |
| 20–60% | 2500 |
| 60–100% | 1000 |
5. RNA isolation from organoids
NOTE: Commercially available column-based methods yield good quantity and quality of RNA. To ensure good quantity RNA, use a minimum of one dome per sample; however, using three domes is recommended, which can be seeded in a single well of a 12 well plate.
Add ß-mercaptoethanol (1%) to the glutathione lysis buffer in the RNA isolation kit.
Draw off the medium from the basement membrane domes containing organoids and add 750 μL of this buffer. Pipette up and down using a P1000 pipette. Check that all the basement membrane matrix has been dissolved.
Add 750 μL of 70% ethanol and mix by pipetting. Subsequently transfer 700 μL of the mixture to the column, centrifuge at 12,000 × g for 1 min, and repeat with the remainder of the lysate.
Perform washes and on-column DNase treatment according to manufacturer’s instructions. Elute RNA in 30–50 μL of RNAse-free water.
Measure the concentrations using a fluorometer at OD = 260 nm and 280 nm and store at −80 °C or continue with downstream applications.
6. Protein isolation from organoids
NOTE: For protein isolation, prepare standard RIPA buffer containing phosphatase and protease inhibitors (Table of Materials). Using at least three domes is recommended, which can be seeded in a single 12 well.
Using a P1000 pipette, draw up the medium from the cell with the basement membrane domes containing organoids and pipette up and down to disrupt the basement membrane matrix.
When fully disrupted, transfer the suspension to a 15 mL conical tube. Centrifuge at 300 × g for 5 min.
Draw off the supernatant and wash with 5 mL of ice-cold PBS. Centrifuge at 300 × g for 5 min.
Draw off the supernatant and resuspend the pellet in 4 mL of trypsin replacement. Digest for 5–10 min while shaking at 37 °C.
-
Add an equal volume of organoid medium + 10% FCS to inhibit the trypsin replacement. Centrifuge at 300 × g for 5 min.
NOTE: Post-centrifugation, no basement membrane matrix should be visible in the pellet.
Draw off the supernatant and wash with 5 mL of ice-cold PBS. Draw off the supernatant and resuspend the cell pellet in 300 μL of lysis buffer using a P1000 pipet, then transfer to a 1.5 mL microcentrifuge tube.
Incubate on ice for 10 min and subsequently sonicate 2x for 30 s each at cooled water with a temperature of 4 °C. Place the tube back on ice and perform protein quantification using standard methods.
Denature the protein by adding sodium dodecyl sulfate (SDS) containing loading dye and boil for 5 min at 95 °C. Store lysates at −80 °C or continue with downstream applications.
7. Cell viability assay with organoids
NOTE: Cell viability can be assessed using the commercially available cell viability assay kit and a luminometer. Prepare buffers according to the manufacturer’s instructions. Five replicates per condition is recommended: one replicate consisting of one 35 μL basement membrane matrix dome in one well of a 24 well plate.
Draw off the medium of the organoid culture, being careful to leave the matrix domes intact.
Add 65 μL of PBS and pipette up and down to disrupt the matrix dome.
Add 100 μL of the cell viability assay kit buffer and resuspend by pipetting.
Incubate at room temperature (RT) for 10 min with shaking.
Transfer 100 μL of mixture to a non-translucent plate suitable for the luminometer and perform reading according to the manufacturer’s instructions for the cell viability assay kit.
8. Preparation of organoids for xenografting
NOTE: Organoids are also amenable for subcutaneous grafting in both immune compromised animals, as well as, isogenic mice. To ensure injected organoids are distinguishable in vivo, label organoids with a constitutively expressing fluorophore5. It is recommended to perform a pilot experiment for grafting using 5 × 105 cells to 2 × 106 cells per injection, with increments of 5 × 106 cells, as grafting efficiency varies between organoid lines.
-
Using a P1000 pipette, draw up the medium and pipette up and down to disrupt the basement membrane matrix. When fully disrupted, transfer the suspension to a 15 mL conical tube. Centrifuge at 300 × g for 5 min.
NOTE: Do not transfer more than 10 matrix domes per 15 mL conical tube.
Draw off the supernatant and wash with 5 mL of PBS. Centrifuge at 300 × g for 5 min.
Draw off the supernatant and resuspend the pellet in 4 mL of trypsin replacement. Digest for 5–10 min while shaking at 37 °C.
Add equal volume of organoid medium + 10% FBS to inhibit trypsin replacement. Centrifuge at 300 × g for 5 min.
Draw off the supernatant and resuspend in 1 mL of PBS. Filter the suspension with a 40 μm filter to ensure a single cell suspension. Quantify cells using standard methods.
Spin down and resuspend the cells in PBS + Rho inhibitor to a concentration of 2 × 106 cells per 100 μL (see Table 2 for cell concentrations and absolute cell number needed for varying concentrations). Use an equal volume of basement membrane matrix to generate a 1:1 suspension. Place the suspension on ice.
Inject cells according to standard protocols and monitor xenograft growth using standard methods8.
Table 2: Recommended cell numbers for organoid xenografting experiments.
Columns from left to right include absolute total cell number for 10 injections, total volume of PBS + Y-27632 for 10 injections, basement membrane matrix volume, and cell number per injection.
| Total Cell number | PBS0+ Y-27632 | Matrigel volume | cell concentration / Injection |
|---|---|---|---|
| 5 × 106 | 500 μl | 500 μl | 5 × 105 |
| 10 × 106 | 500 μl | 500 μl | 1 × 106 |
| 15 × 106 | 500 μl | 500 μl | 1.5 × 106 |
| 20 × 106 | 500 μl | 500 μl | 2 × 106 |
Representative Results
Seeding efficiency
Organoid formation capacity is determined by phenotype and genotype. Wild-type (WT) prostate basal cells showed superior organoid formation capacity (30%−40%) compared to luminal cells (3%) (Figure 1A). After organoid establishment, the formation capacity increased drastically. Typically, 25%−30% of cells derived from a WT organoid can form a new organoid (Figure 1B). CRISPR/Cas9-mediated loss of Pten (PtenΔ/Δ) or p53 (p53Δ/Δ) resulted in a minor increase in organoid formation capacity. Loss of both p53 and Pten further increased formation capacity (Figure 1B).
Pharmacological response
Based on seeding efficiency, seeding of 1,000–10,000 cells in 35 μL of basement membrane matrix dome in a 24 well plate was performed. Recommended cell seeding numbers based on organoid formation efficiency is provided in Table 1. However, organoid proliferation speeds can differ greatly depending on genotype. Additional changes to the cell seeding number can be made based on proliferation.
In Figure 2, the effects of anti-androgenic molecules on growth were tested in murine organoids with different genotypes. A total of 2,500 cells were seeded from murine organoids with a WT genotype, p53 loss, Pten loss, or dual p53 and Pten loss. p53 and Pten loss was initiated by lentiviral introduction of a gRNA targeting the p53 and/or Pten locus in organoids constitutively expressing Cas9 under the control of the Rosa26 promoter with a C57/Bl6 genetic background9.
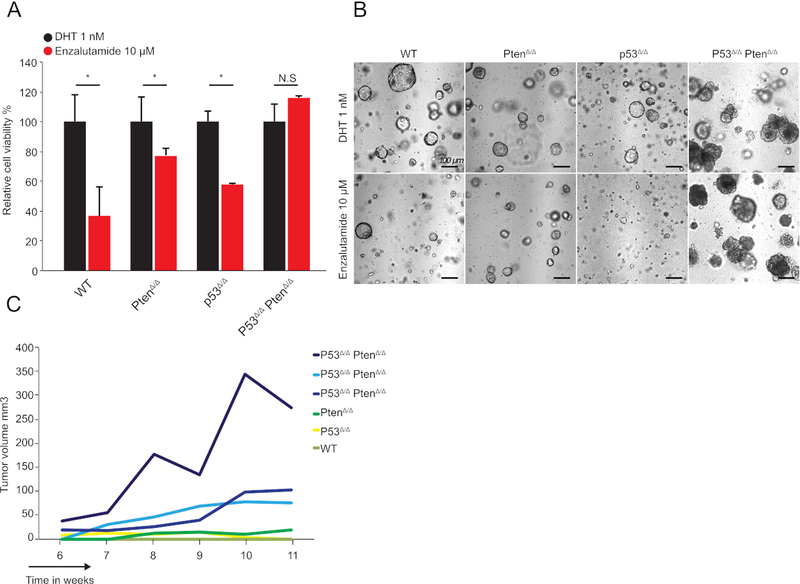
Figure 2: Assessing the pharmacological response of organoids derived from genetically engineered mice.
(A) Relative cell proliferation of murine WT, PtenΔ/Δ, P53Δ/Δ, and P53Δ/Δ PtenΔ/Δ organoids (y-axis, mean ± SD, measured by cell viability assay kit) of 2,500 cells 7 days post-establishment of organoids; (n = 3, *p < 0.05, t-test) with 1 nM DHT or 10 μM of second-generation antiandrogen as indicated. (B) Representative brightfield images of WT, PtenΔ/Δ, P53Δ/Δ, and P53Δ/Δ PtenΔ/Δ organoid cultures treated with 1 nM DHT or 10 μM second-generation anti-androgen as indicated. (C) Representative growth curve of WT, PtenΔ/Δ, P53Δ/Δ, and P53Δ/Δ PtenΔ/Δ organoids injected subcutaneously in the flank of isogenic C57/Bl6 mice. Only P53Δ/Δ PtenΔ/Δ organoids showed growth. A total of 1 × 106 cells were injected. Three independent curves of P53Δ/Δ PtenΔ/Δ organoids are shown to show heterogeneity in growth speed.
Loss of p53 did not cause resistance to the anti-androgenic molecules. Loss of Pten increased resistance to anti-androgenic compound, as shown previously10. Dual loss of p53 and Pten, however, resulted in complete resistance to the second-generation anti-androgen (Figure 2A). AR inhibition also altered organoid phenotypes. In control Cas9+/+ organoids, as well as P53Δ/Δ-deleted and PtenΔ/Δ organoids, a decrease in organoid lumen size was observed (Figure 2B). p53Δ/Δ PtenΔ/Δ organoids were phenotypically unaffected (Figure 2B). In line with these results, when 1 × 106 cells were grafted subcutaneously in the flank, only p53Δ/Δ PtenΔ/Δ organoids grew (Figure 2C). Overall, these results demonstrate that p53Δ/Δ PtenΔ/Δ co-deletion results in resistance to the second-generation anti-androgen in murine organoids.
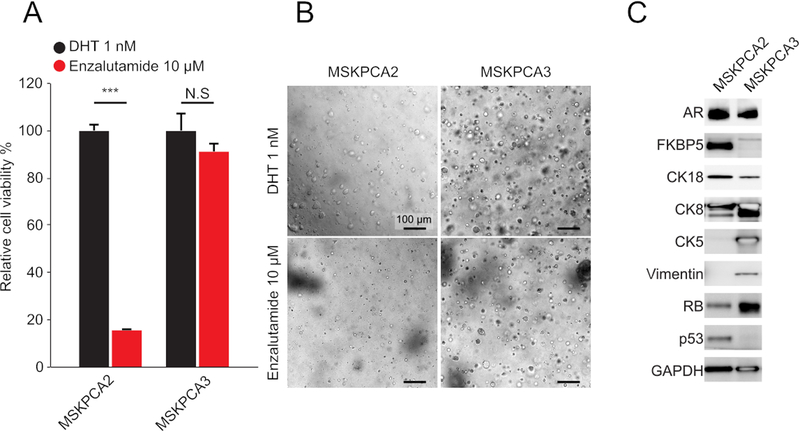
Patient-derived PCa organoids are heterogeneous in phenotype and genotype11,12; therefore, responses to drugs can differ greatly between human PCa organoid lines. In Figure 3, the anti-androgenic molecules response of two distinct human PCa organoids, MSKPCA2 and MSKPCA3 are shown. Proliferation of MSKPCA2 organoids was strongly inhibited by anti-androgenic molecules, whereas MSKPCA3 organoids remained unaffected (Figure 3A,B). MSKCPCA2 organoids expressed high levels of AR and the AR-target FKBP5, and they expressed hallmark luminal proteins such as CK8 and CK18. In contrast, MSKPCA3 organoids also expressed basal (CK5) and mesenchymal (Vimentin) markers and showed no expression of FKBP5. These results suggest that these organoids model a non-luminal androgen-independent phenotype.
Figure 3: Assessing the pharmacological response of organoids derived human prostate cancer biopsies.
(A) Relative cell proliferation of patient-derived MSKPCA2 and MSKPCA2 organoids (y-axis, mean ± SD, measured by cell viability assay kit) of 5,000 cells 7 days post-establishment of organoids (n = 4, *p < 0.05, t-test) with 1 nM DHT or 10 μM second-generation anti-androgen as indicated. (B) Representative brightfield images of MSKPCA2 and MSKPCA3 organoids cultures treated with 1 nM DHT or 10 μM second-generation anti-androgen as indicated. (C) Western blot analysis of AR, FKBP5 (AR-target gene), CK8 and CK18 (luminal markers), CK5 (basal marker), and Vimentin (mesenchymal marker) in MSKPCA2 and MSKPCA3 organoids. GAPDH was used as a loading control.
Discussion
Understanding the molecular mechanisms underlying anti-androgen resistance and discovering potential therapeutic vulnerabilities requires testing of pharmacological responses in model systems mimicking prostate cancer. Described here is a detailed protocol for the reliable analysis of pharmacological responses in patient-derived and genetically engineered prostate organoids and preparation of these organoid samples for downstream applications.
There are two critical steps in this protocol. The first is determining the seeding efficiency and growth rate of organoids. Organoid growth speed varies greatly. This is dependent on species, as murine derived organoids grow about two-fold faster than human-derived organoids. Apart from species, growth speed is dependent on genotype and phenotype. However, when seeding efficiency and growth speed are determined, this protocol can be adapted to all prostate organoid types.
The second critical step is working with the protein-matrix-based 3D culture to prepare for subsequent downstream applications. The introduction of seeding variation by viscosity of the basement membrane matrix during plating can be avoided by using diluted (70%) basement membrane matrix, as described. Properly breaking up the polymerized matrix without excessively disturbing the organoids is also described in detail. This protocol enables disruption of the matrix without introducing variation in the organoid readout, which can be adapted for the screening of drug libraries for different genetic backgrounds in PCa. Moreover, by performing CRISPR/Cas9- or shRNA-based expression interference, genes conferring drug resistance can be queried.
A point of consideration is that the medium composition of prostate organoid culture can influence the pharmacological response. For example, EGF, a component of both murine and human prostate organoid culture, greatly reduces sensitivity to anti-androgen. Hence, EGF is omitted in this protocol from the medium, and sensitivity to anti-androgen is restored. It is advised to determine if any organoid ingredients influence sensitivity for the drug being tested. This holds especially true for the complex human prostate organoid culture medium, which (apart from EGF, Noggin, R-spondin1, DHT, and A83–001 [the composition of murine organoid medium]) contains fibroblast growth factor 10 (FGF10), FGF2, prostaglandin E2, nicotinamide, and the p38i inhibitor SB202190.
The organoid medium composition favors the growth of benign prostate epithelium over cancer tissue, thus no primary hormone sensitive PCa organoid lines have been established. Currently, all human PCa organoids are derived from patients with advanced metastatic anti-androgen resistant PCa; hence, most of these lines are anti-androgen resistant and suitable for identifying new treatments. As proof-of-concept, delta-like 3 (DLL3) has been identified as a therapeutic target using patient-derived NEPC organoids that are targetable with rovalpituzumab tesirine13. This method is suitable for these types of experiments and is also suitable for prostate organoids from normal benign tissue, primary prostate cancer, and CTCs.
One shortcoming of prostate organoid culture is the absence of a cellular niche. Thus, contributions to drug resistance by non-tumor cells cannot be studied using the current platform. However, co-cultures of colorectal cancer and lung cancer organoids with autologous T-cells have recently been established, enabling studies of interactions between the tumor and immune system14. Other co-culture systems may be established to further study non-cell-autonomous interactions.
In conclusion, this report provides a detailed protocol for the reproducible assessment of pharmacological responses in prostate organoids and subsequent downstream applications. Importantly, this protocol is broadly applicable and can be used for organoid cultures of other organs, including the colon15, small intestine16, stomach17,18, liver19, pancreas20, kidney21, and mammary gland22.
Acknowledgments
K.P. is supported by NIH 1F32CA236126-01. C.L.S. is supported by HHMI; CA193837; CA092629; CA224079; CA155169; CA008748; and Starr Cancer Consortium. W.R.K. is supported by Dutch Cancer Foundation/KWF Buit 2015-7545 and Prostate Cancer Foundation PCF 17YOUN10.
Footnotes
Video Link
The video component of this article can be found at https://www.jove.com/video/60346/
Disclosures
C.L.S serves on the board of directors of Novartis; is a cofounder of ORIC Pharmaceuticals and coinventor of enzalutamide and apalutamide; is a science advisor to Agios, Beigene, Blueprint, Column Group, Foghorn, Housey Pharma, Nextech, KSQ, Petra, and PMV; and is a cofounder of Seragon, purchased by Genentech/Roche in 2014. W.R.K. is a coinventor and patent holder of organoid technology.
References
- 1.Robinson D et al. Integrative Clinical Genomics of Advanced Prostate Cancer. Cell. 162 (2), 454 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Arora VK et al. Glucocorticoid Receptor Confers Resistance to Antiandrogens by Bypassing Androgen Receptor Blockade. Cell. 155 (6), 1309–1322 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ku SY et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science. 355 (6320), 78–83 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mu P et al. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science. 355 (6320), 84–88 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karthaus WR et al. Identification of Multipotent Luminal Progenitor Cells in Human Prostate Organoid Cultures. Cell. 159 (1), 163–175 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao D et al. Organoid Cultures Derived from Patients with Advanced Prostate Cancer. Cell. 159 (1), 176–187 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drost J et al. Organoid culture systems for prostate epithelial and cancer tissue. Nature Protocols. 11 (2), 347–358 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bose R et al. ERF mutations reveal a balance of ETS factors controlling prostate oncogenesis. Nature. 546 (7660), 671–675 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Platt RJ et al. CRISPR-Cas9 Knockin Mice for Genome Editing and Cancer Modeling. Cell. 159 (2), 440–455 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carver BS et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 19 (5), 575–586 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Puca L et al. Patient derived organoids to model rare prostate cancer phenotypes. Nature Communications. 9 (1), 2404 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao D et al. Organoid Cultures Derived from Patients with Advanced Prostate Cancer. Cell. 159 (1), 176–187 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puca L et al. Delta-like protein 3 expression and therapeutic targeting in neuroendocrine prostate cancer. Science Translational Medicine. 11 (484), eaav0891 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dijkstra KK et al. Generation of Tumor-Reactive T Cells by Co-culture of Peripheral Blood Lymphocytes and Tumor Organoids. Cell. 174 (6), 1586–1598.e12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van de Wetering M et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 161 (4), 933–945 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato T et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 459 (7244), 262–265 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Barker N et al. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell stem cell. 6 (1), 25–36 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Bartfeld S et al. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology. 148 (1), 126–136.e6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huch M et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell. 160 (1–2), 299–312 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boj SF et al. Organoid models of human and mouse ductal pancreatic cancer. Cell. 160 (1–2), 324–338 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schutgens F et al. Tubuloids derived from human adult kidney and urine for personalized disease modeling. Nature Biotechnology. 37 (3), 303–313 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Sachs N et al. A Living Biobank of Breast Cancer Organoids Captures Disease Heterogeneity. Cell. 1–25 (2017). [DOI] [PubMed] [Google Scholar]