Abstract
Ontamalimab (SHP647) is a fully human, immunoglobulin G2, antihuman mucosal addressin cell adhesion molecule‐1 (MAdCAM‐1) monoclonal antibody being developed for the treatment of ulcerative colitis (UC) and Crohn's disease (CD). A population pharmacokinetic/pharmacodynamic (PK/PD) analysis was conducted using clinical phase 2 study data to evaluate the PK and PD of ontamalimab following subcutaneous administrations of 7.5, 22.5, 75, and 225 mg every 4 weeks in patients with moderate to severe UC or CD. A total of 440 patients with UC (n = 249; 56.6%) or CD (n = 191; 43.4%) were included in the analysis. A 2‐compartment model with parallel linear and nonlinear elimination adequately characterized concentration‐time profiles of ontamalimab. The apparent clearance and volume of distribution were 0.0127 L/h (0.305 L/day) and 6.53 L, respectively. Apparent clearance and volume of distribution were mainly dependent on baseline albumin and body weight, respectively. No differences in the PK properties of ontamalimab were observed between patients with UC or CD. The presence of antidrug antibodies did not impact the PK of ontamalimab. Nonlinear elimination occurred at very low concentrations and was unlikely to contribute to the elimination half‐life under steady‐state conditions. A linear PK/PD model described the relationship between ontamalimab and free MAdCAM‐1. Minimum concentrations of ontamalimab at steady state following 75 mg every 4 weeks were associated with >95% suppression of circulating free MAdCAM‐1. The PK/PD properties characterized support phase 3 testing in UC and CD.
Keywords: Crohn's disease, ulcerative colitis, pharmacokinetics, pharmacodynamics, ontamalimab, MAdCAM‐1
Mucosal addressin cell adhesion molecule‐1 (MAdCAM‐1) is an immunoglobulin superfamily adhesion molecule for lymphocytes that is expressed by mucosal venules and helps to direct lymphocyte traffic into Peyer patches and the intestinal lamina propria. 1 The α4β7 integrin is the recognized ligand for MAdCAM‐1, and expression of this ligand on populations of CD4+ and CD8+ T cells, as well as on subsets of B cells, distinguishes them as unique gut‐homing lymphocytes. MAdCAM‐1 plays a role in gut immune surveillance and appears to facilitate excessive lymphocyte infiltration under conditions of chronic gastrointestinal inflammation. 2
Principal sites of MAdCAM‐1 expression on normal tissue include the intestines, pancreas, stomach, esophagus, spleen, and, to a lesser extent, the lung, liver, and bladder. MAdCAM‐1 is not expressed in the central nervous system. In patients with Crohn's disease (CD), sites of active inflammation have been reported to have increased expression of MAdCAM‐1, suggesting a connection between inflammation and the receptor. 2 , 3
Ontamalimab (SHP647) is an anti‐MAdCAM‐1 monoclonal antibody that prevents the binding of α4β7 + lymphocytes to MAdCAM‐1‐expressing sites in the high endothelial venules of the gastrointestinal tract with high affinity and selectivity. Ontamalimab does not bind to intercellular adhesion molecule‐1 or vascular cell adhesion molecule‐1 and is therefore not expected to affect lymphocyte homing or surveillance in the central nervous system. 4 Ontamalimab is being developed for the treatment of CD and ulcerative colitis (UC).
Methods
The institutional review board/independent ethics committee at each of the participating investigational centers and the sponsor prospectively approved the final protocols and any amendments and informed consent forms used in these studies before use. The investigator, or a person designated by the investigator, obtained written informed consent from each participant or their legal representative before any study‐specific activity was performed. The investigator ensured that each study participant or his/her legal representative was fully informed about the nature and objectives of the study, and possible risks associated with participation. Studies were conducted in accordance with legal and regulatory requirements, as well as the general principles set forth in the International Ethical Guidelines for Biomedical Research Involving Human Subjects (Council for International Organizations of Medical Sciences, 2002), Guidelines for Good Clinical Practice (International Conference on Harmonisation, 1996), and the Declaration of Helsinki (World Medical Association, 1996 and 2008). In addition, the studies were conducted in accordance with protocols, the International Conference on Harmonisation guideline on Good Clinical Practice, and applicable local regulatory requirements and laws.
Description of Clinical Studies and Populations
A phase 2, parallel, dose‐ranging, randomized, double‐blind, placebo‐controlled clinical trial in patients with moderate to severe CD who had a history of treatment failure, or intolerance to immunosuppressants and/or anti–tumor necrosis factor (TNF) agents was performed (A7281006, ClinicalTrials.gov identifier: NCT01276509; OPERA). 5 Patients eligible for this study must have met the entry criteria (details of which have been published previously 5 ), which specified a high‐sensitivity C‐reactive protein (CRP) concentration greater than the upper limit of normal (3.0 mg/L). In this study, 3 subcutaneous (SC) dose levels (22.5, 75, and 225 mg) of ontamalimab were compared with placebo (in a 1:1:1:1 ratio). Patients were randomly assigned to 1 of 4 treatment arms for the 12‐week induction period and stratified by the status of anti‐TNF experience or immunosuppressant intolerance/failure and concomitant immunosuppressant therapy. The primary efficacy end point was the proportion of patients with a decrease in Crohn's Disease Activity Index score of at least 70 points from baseline to week 8 or week 12. 5 A minimum of 240 randomized patients (60 per treatment group) were required to evaluate the primary end point.
A phase 2b, parallel, randomized, double‐blind, placebo‐controlled, dose‐ranging study in patients with moderate to severe UC was performed (A7281009, ClinicalTrials.gov identifier: NCT01620255; TURANDOT). 6 Four dose levels (7.5, 22.5, 75, and 225 mg) of ontamalimab SC were compared with placebo (in a 1:1:1:1:1 ratio). Approximately 300 patients were randomly assigned to 1 of 5 treatment arms for the 12‐week treatment period and stratified by prior anti‐TNF status. The primary efficacy end point was the proportion of patients in clinical remission at week 12, as defined by a total Mayo score of 2 points or lower, with no individual subscore exceeding 1 point and a rectal bleed subscore of 0 or 1. 6
In both studies, patients received ontamalimab or placebo at baseline (day 1), week 4 (day 28), and week 8 (day 56). Blood samples were collected (3 mL, to provide a minimum of 1.5 mL of serum) and assayed for ontamalimab using a validated enzyme‐linked immunosorbent assay. Pharmacokinetic (PK) sampling was performed in all patients on days 1 (predose), 14, 28 (predose), 42, 56 (predose), and 70. Additional PK sampling was performed in a subset of patients with CD (n = 60) in study A7281006 on days 3 ± 3, 7 ± 3, 10 ± 3, 17 ± 3, 35 ± 3, and 63 ± 3.
Blood samples (5 mL, to provide a minimum of 2.5 mL of serum) were collected in both studies at baseline and week 12 for the analysis of soluble MAdCAM‐1 using a validated assay (based on a hybrid of immunocapture and nano‐liquid chromatography–tandem mass spectrometry). 7
A total of 440 patients were included in the PK and PK/pharmacodynamic (PD) analyses, based on the PK populations of the trials, which included all patients who received at least 1 dose of investigational product and had data for at least 1 PK concentration. Baseline characteristics and demographics were summarized using descriptive statistics overall and stratified by disease type (UC or CD).
Population PK Analysis
Multiple PK compartmental models were evaluated (ie, 1‐ and 2‐compartment) and a model discrimination was performed based on the NONMEM minimum objective function value (OFV) (−2 times the log likelihood function) and by looking at pertinent graphical representations of goodness of fit (eg, fitted and observed concentrations vs time, weighted residuals vs time). 8 Models including parallel linear clearance (CLLIN/F) and nonlinear clearance (CLNLIN/F) mechanisms were considered. 9 The total clearance (CLT/F) for average steady‐state concentrations (Cave,ss) of ontamalimab was derived as follows:
where Vmax/F = apparent maximum velocity, Km = Michaelis–Menten constant, and Cave,ss = concentration predicted with the population PK model.
An allometric model accounting for the effect of body weight on clearance and volume parameters was included in the base model (estimated exponent). In a second step, graphical display of the correlation between PK parameters (random effects) and covariates (intrinsic and extrinsic) was evaluated. The following intrinsic covariates were explored: age, sex, race, disease (CD or UC), markers of inflammation (albumin and CRP), and markers of liver function (bilirubin, aspartate aminotransferase, and alanine aminotransferase). In addition, the potential impact of antidrug antibodies (ADAs) as a time‐varying covariate was assessed on the clearance of ontamalimab. For missing ADA, the last observation carried forward was used. A formal covariate analysis was performed using a full model approach. Briefly, all covariates of interest were tested in a univariate manner in a first step and all statistically significant covariates were included in the full model (Δ OFV of 6.63, P < .01 for 1 degree of freedom). A bootstrap resampling analysis was performed to assess the robustness of parameters estimated with the full model. Full‐model parameter estimates and 95% percentile intervals were obtained via nonparametric bootstrapping. A total of 500 bootstrap replicates were performed. The bias of each parameter was calculated by the percentage change between median value derived from the bootstrap and the full model PK estimate. If applicable, the full model was reduced by excluding covariates that were not statistically significant based on nonparametric 95% confidence interval (CI) estimates. 10
Forest plots were used to present the effects of covariates and associated 95%CI on exposure parameters. Changes in exposure parameters were computed relative to their reference categories.
Visual predictive checks (VPCs) were performed (uncorrected and prediction corrected) to allow visual comparison of model‐predicted vs observed values from the original data set.
Concentration‐time profiles were simulated for every‐4‐week (Q4W) dosing regimens (7.5, 22.5, 75, 150, and 225 mg) and the following parameters were derived at week 12: average concentration (Cave), maximum concentration (Cmax), and minimum concentration (Cmin). In addition, steady‐state average concentration (Cave,ss), maximum concentration (Cmax,ss), and minimum concentration (Cmin,ss) were derived.
Individual values of Cave,ss were used for the calculation of total clearance [CLT/F = CLLIN/F + Vmax/F / (Cave,ss + Km)]. The half‐life associated with the alpha (t½α) and beta (t½β) phases were derived using clearance (CLT/F and apparent distributional clearance [CLd/F]) and volume parameters (apparent volume of the central compartment [Vc/F] and of the peripheral compartment [Vp/F]) for typical Cave,ss values. The terminal elimination half‐life is referred to as an apparent half‐life because it was derived assuming a fixed value of Cave,ss.
Population PK/PD Analysis
Exploratory analyses were performed to assess the relationship between concentrations of ontamalimab and free MAdCAM‐1 levels. The relationship between observed ontamalimab concentrations and free MAdCAM‐1 (log‐transformed data) was assessed using a maximum inhibitory model (Imax) as described below:
where E( c ) is the effect for any concentrations of ontamalimab, E0 is the baseline, Imax is the maximum inhibition, C is the concentration of ontamalimab, IC50 is the concentration associated with 50% of the maximum effect, and H is the Hill factor (also referred to as gamma, a parameter used to describe sigmoidicity).
In addition, a linear model linking Ln‐transformed concentrations of ontamalimab and Ln‐transformed concentrations of free MAdCAM‐1 was used as per the following equation:
where EMAdCAM‐1 is the resulting effect of ontamalimab on free MAdCAM‐1 levels, E0 is the baseline, Slope is the linear relationship between Ln‐transformed MAdCAM‐1 and the Ln‐transformed exposure parameter of ontamalimab, and ontamalimab is the concentration of ontamalimab in nanograms per milliliter.
Graphical display of the correlation between PD parameters (random effects) and selected covariates (CRP, fecal calprotectin, and colonoscopy score) was performed. Covariates were tested and potentially included in the full PK/PD using a similar approach as described for PK. Goodness of fit and VPC were derived using a similar method as described for PK.
Bioanalytical Assays for Ontamalimab and MAdCAM‐1
Serum concentrations of ontamalimab were assessed at QPS (Newark, Delaware) using a validated sandwich enzyme‐linked immunosorbent assay, which used Nunc MaxiSorp plates coated with RA‐659. The plate was then washed and blocked. Samples (including standards and quality control samples) were diluted by a minimum required dilution of 10‐fold with a buffer solution and incubated on the plate for 60 minutes. The plate was washed and then incubated with biotin‐mouse antihuman immunoglobulin G2. After 60 minutes, the plate was washed and then incubated with horseradish peroxidase–conjugated streptavidin for 30 minutes. 3,3′,5,5′‐tetramethylbenzidine was used as the substrate for horseradish peroxidase. The reaction was stopped with 1M hydrochloric acid. The color intensity is directly proportional to the quantity of ontamalimab. The calibration range was 10 to 200 ng/mL with a dilution factor of up to 1:5000. The lower limit of quantification was 10 ng/mL. The assay accuracy (relative error) and precision (coefficient of variation for the mean) were within acceptance criteria (±20%) for a majority of quality control samples. Based on quality control samples (10, 30, 90, 145, and 200 ng/mL), interbatch precision ranged from 2.6% to 4.7%, and the interbatch accuracy ranged from −12.0% to −7.0%.
Serum concentrations of soluble MAdCAM‐1 were assessed at Q2 Solutions (Ithaca, New York) using a hybrid of immunocapture and nano‐liquid chromatography–tandem mass spectrometry. Free soluble MAdCAM‐1 was isolated from human serum by immunoprecipitation with biotinylated ontamalimab acting as the capture agent. The internal standard for MAdCAM‐1 (AQUA Peptide 1) was added to the samples, and then trypsin digestion and liquid chromatography–tandem mass spectrometry were carried out for separation and detection, respectively. 7 The immunocapture conditions of the assay were optimized to provide good recovery of endogenous soluble MAdCAM‐1 levels and minimal disruption of the preexisting drug/target equilibrium in the samples using low concentrations of biotinylated ontamalimab and a short incubation time. 7 The following ion transition m/z was monitored for MAdCAM‐1 and the internal standard: 738.9→890.4 and 743.8→900.4, respectively. Overall precision and accuracy were acceptable at ≤15.2% and ≤16.2%, respectively. 7 Calibration standard responses for free soluble MAdCAM‐1 were linear over the range of 0.5 to 512 pmol/L in serum.
Software
Population PK and PK/PD analyses were performed using NONMEM (version 7.3) with the GNU Fortran 95 compiler with the first‐order conditional estimation and the INTERACTION option. Preparation, exploration, and visualization of data sets were carried out using R (version 3.4.1) and Microsoft Office Excel 2016.
Results
Baseline Characteristics
Of the 440 patients included in the population PK analysis, 191 (43.4%) had CD and 249 (56.6%) had UC. Overall, 225 patients (51.1%) were female and 215 (48.9%) were male. The majority of patients were white (86.1%); Asian (including Japanese) patients accounted for 10.0% of the population. Descriptive statistics of continuous baseline characteristics are presented in Table 1. Patients with CD and UC had similar mean ages (36.0 and 40.4 years, respectively), body weights (70.6 and 72.5 kg, respectively), and albumin values (39.6 and 38.3 g/L). Patients with CD had mean bilirubin levels approximately 50% lower than patients with UC (0.288 and 0.419 mg/dL, respectively). Patients with CD had mean CRP concentrations 2.8‐fold higher than patients with UC (2.82 and 1.02 mg/dL, respectively). Patients with CD and UC had similar mean free MAdCAM‐1 (259 and 282 pmol/L, respectively) and fecal calprotectin (2730 and 2850 µg/g, respectively) levels. Descriptive statistics for additional baseline characteristics in patients with CD and UC are presented in Table S1.
Table 1.
Descriptive Statistics of Baseline Characteristics
| Characteristic | Patients With CD (n = 191) | Patients With UC (n = 249) | Overall (n = 440) | |
|---|---|---|---|---|
| Age, y | Mean (SD) | 36.0 (11.5) | 40.4 (13.2) | 38.5 (12.7) |
| Median (min‐max) | 35.0 (19.0‐68.0) | 39.0 (18.0‐65.0) | 37.0 (18.0‐68.0) | |
| 95%CI | 19.8‐60.3 | 19.2‐63.0 | 19.0‐62.0 | |
| Body weight, kg | Mean (SD) | 70.6 (20.2) | 72.5 (16.8) | 71.7 (18.4) |
| Median (min‐max) | 67.3 (35.6‐155) | 70.0 (40.5‐140) | 68.8 (35.6‐155) | |
| 95%CI | 44.9‐121 | 47.6‐113 | 45.0‐116 | |
| Albumin, g/L | Mean (SD) | 39.6 (4.35) | 38.3 (4.34) | 38.9 (4.39) |
| Median (min–max) | 40.0 (25.0‐49.0) | 39.0 (23.0‐47.0) | 39.0 (23.0‐49.0) | |
| 95%CI | 30.5‐46.3 | 28.2‐46.8 | 29.0‐47.0 | |
| CRP, mg/dL | Mean (SD) | 2.82 (3.29) | 1.02 (1.68) | 1.80 (2.66) |
| Median (min‐max) | 1.79 (0.0330‐18.0) | 0.407 (0.0100‐17.0) | 0.837 (0.0100‐18.0) | |
| 95%CI | 0.154‐11.6 | 0.0130‐4.76 | 0.0299‐8.83 | |
|
Free MAdCAM‐1 at baseline, pmol/L |
Mean (SD) | 259 (216) | 282 (220) | 273 (218) |
| Median (min‐max) | 212 (1.58‐819) | 234 (1.24‐874) | 226 (1.24‐874) | |
| 95%CI | 66.6‐701 | 97.3‐777 | 77.2‐757 | |
| Missing, n (%) | 39 (20.4) | 8 (3.2) | 47 (10.7) | |
| Fecal calprotectin, µg/g | Mean (SD) | 2730 (3560) | 2850 (3360) | 2800 (3440) |
| Median (min‐max) | 1580 (22.8‐31 600) | 1950 (28.5‐29 600) | 1810 (22.8‐31 600) | |
| 95%CI | 113‐10 100 | 86.3‐8990 | 106‐9970 | |
| Missing, n (%) | 19 (9.9) | 25 (10.0) | 44 (10.0) |
ADA, antidrug antibody; CD, Crohn's disease; CI, confidence interval; CRP, C‐reactive protein; MAdCAM‐1, mucosal addressin cell adhesion molecule‐1; SD, standard deviation; UC, ulcerative colitis.
Safety and Antidrug Antibody
Ontamalimab has been generally well tolerated in patients with UC and CD with no evidence of a safety signal or increased incidence of adverse events at higher doses. 5 , 6 , 11 Furthermore, no cases of progressive multifocal leukoencephalopathy have been observed in the current clinical studies. 5 , 6 , 11 At week 12, the 7.5‐, 22.5‐, 75‐, and 225‐mg dose levels were associated with 6 (10.9%), 5 (3.8%), 10 (7.8%), and 5 (4.0%) samples associated with a positive ADA status (Table S2). For ADA assessment, a confirmatory cut‐point value of 12.5% inhibition with ontamalimab was established based on a 1% false‐positive rate for inhibition of 30 individual CD serum samples; and a confirmatory cut‐point value of 16.7% inhibition with ontamalimab was established based on a 1% false‐positive rate for inhibition of 30 individual UC serum samples, which suggests relatively low false‐positive rates for both disease populations. None of the ADA‐positive patients with CD developed neutralizing ADAs, and only 7 ADA‐positive UC patients developed neutralizing ADAs. Given the small number of neutralizing ADA‐positive samples, there was no discernible impact on absolute lymphocyte count, safety, or efficacy.
Analyses of Observed PK and PD Data
Individual concentration‐time profiles of ontamalimab and MAdCAM‐1 by dose levels in patients with CD and UC are presented in Figure 1. Concentration–time profiles of ontamalimab after the first, second, and third doses were associated with an important variability across dose levels. Very low concentrations of ontamalimab were observed following Q4W dosing of 7.5 mg. Concentration‐time profiles of ontamalimab for Q4W dosing of 22.5 mg were associated with an apparent faster decline, which may suggest the presence of nonlinear elimination (target mediated) at low concentrations. Concentration‐time profiles of ontamalimab for Q4W dosing of 75 and 225 mg were associated with a marked increase in ontamalimab concentrations and lower variability. For ontamalimab, 9.4% (202/2138) of samples had concentrations below the limit of quantitation. These samples were set as “missing” for the population PK analysis. Concentrations of free MAdCAM‐1 decreased in a dose‐dependent manner. All samples assayed for MAdCAM‐1 presented a concentration above the lower limit of quantitation.
Figure 1.
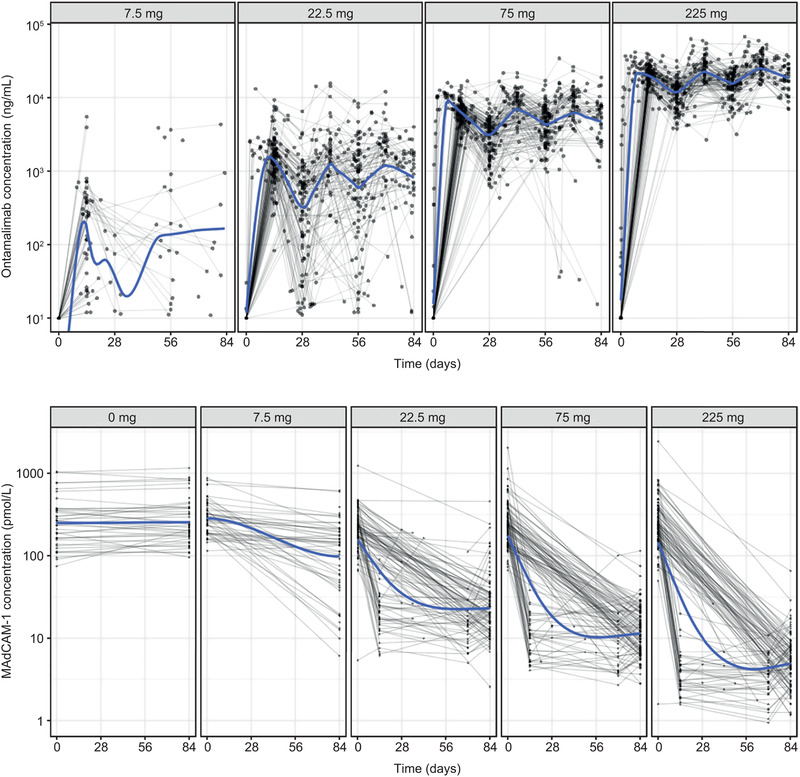
Concentration‐time profiles of ontamalimab (top panel) and free MAdCAM‐1 (bottom panel) in patients with CD and UC. CD, Crohn's disease; MAdCAM‐1, mucosal addressin cell adhesion molecule‐1; UC, ulcerative colitis.
Population PK Analysis
A 2‐compartment model with parallel linear and nonlinear elimination, allometric function on clearance and volume parameters, and a first‐order rate constant of absorption and absorption lag time resulted in the lowest objective function and acceptable goodness of fit (OFV = 30592.327; Tables S3 and S4 and Figure S1). Based on exploratory covariate plots (Figures S2–S11), the following covariates on apparent clearance (CL/F) were first tested in a univariate manner: disease status (CD or UC), albumin, CRP, sex, and time‐varying ADA. In addition, disease status (CD or UC) was tested on Vc/F. No covariate analysis was performed on absorption rate constant owing to the high shrinkage most likely due to the limited samples available during the absorption phase. Time‐varying ADA and sex on CL/F were not statistically significant (Table S5). In addition, disease status (CD or UC) on CL/F and Vc/F were not statistically significant. All other covariates were retained in the full model, and a bootstrap resampling analysis was performed to derive median and 95%CIs (Table S6). The effects of albumin and CRP were retained in the final model. Median values of covariates were all within 20% of those derived in the original analysis.
The performance of the final population PK model of ontamalimab, including covariates, is presented in Figure 2. Observed concentrations of ontamalimab vs both individual‐predicted and population‐predicted values fell along the line of identity in all studies. A bias was observed for concentrations <100 ng/mL (on the log‐scale), which represented approximately 6% of the total number of available samples. Conditional weighted residual values were homogeneously distributed around 0. Results of the VPC are presented in Figure 3. The VPC illustrates that the observed data fall largely within the simulated range, with the 5th, 50th, and 95th percentiles of the observed data in close agreement with the corresponding 95% prediction intervals of the simulations. Only 25 observed concentrations were below the 95%CI of the model‐predicted 5th percentile after the first dose (up to week 4) suggesting that the overprediction of low concentrations is likely to be negligible. Additional VPCs by dose level (and prediction corrected) confirmed that the model adequately described the observed data across the dose range study (Figures S12 to S16). Typical population PK parameters of ontamalimab derived with the final model are presented in Table 2. Population estimates of CL/F and Vc/F were 0.0127 L/h (0.305 L/day) and 6.53 L, respectively. The rate of absorption following SC administration was 0.0187/h, corresponding to an absorption half‐life of 37 hours. Population estimates of Vmax/F and Km were 5.87 µg/h and 19.0 ng/mL, respectively. Based on a bootstrap resampling analysis, the median (95%CIs) Vmax/F and Km were 5.91 µg/h (3.68‐9.43) and 19.0 ng/mL (12.9‐23.2), respectively. Overall, the above results suggest that Vmax and Km parameters were estimated with a sufficient level of confidence. The residual variability of predicted ontamalimab concentrations was low based on the additive and proportional error model. The PK of ontamalimab was characterized using parallel CLLIN/F and CLNLIN/F mechanisms. At high concentrations, the CLT/F is governed by CLLIN/F (since Vmax/F / [C + Km] will trend to zero). The CLT/F is governed by CLNLIN at low concentrations (since Vmax/F / [C + Km] will increase) as presented (Figures S17 and S18). The mean apparent half‐life of ontamalimab associated with average steady‐state concentrations of the 22.5‐, 75‐, and 225‐mg dose levels was 12.3, 16.5, and 18.6 days, respectively.
Figure 2.
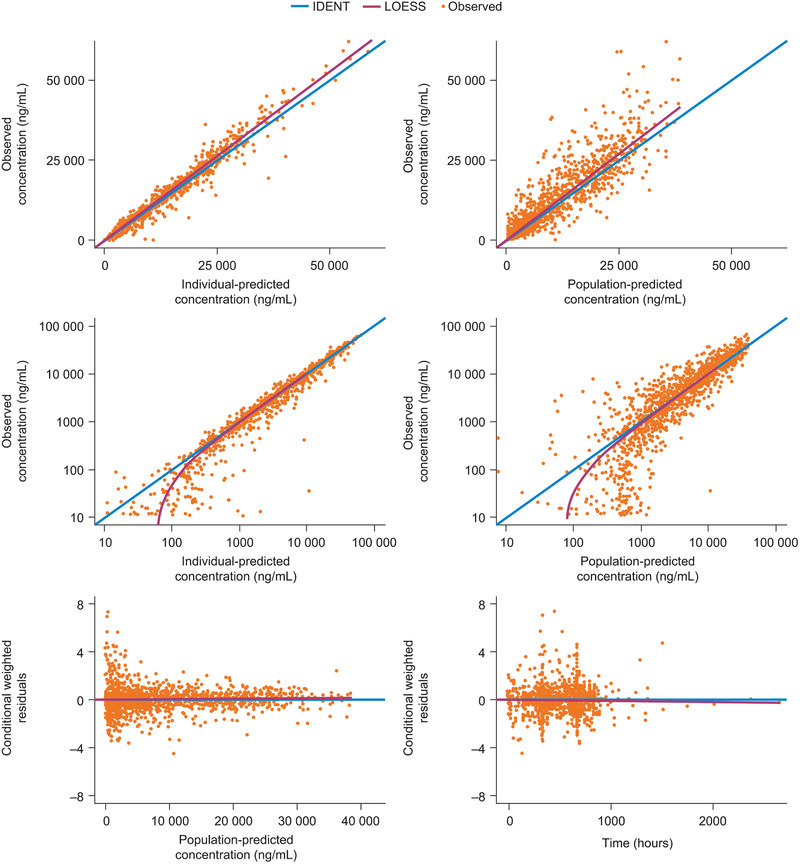
Goodness of fit of final population PK model of ontamalimab. IDENT, line of identity; LOESS, locally weighted scatterplot smoothing; PK, pharmacokinetic.
Figure 3.
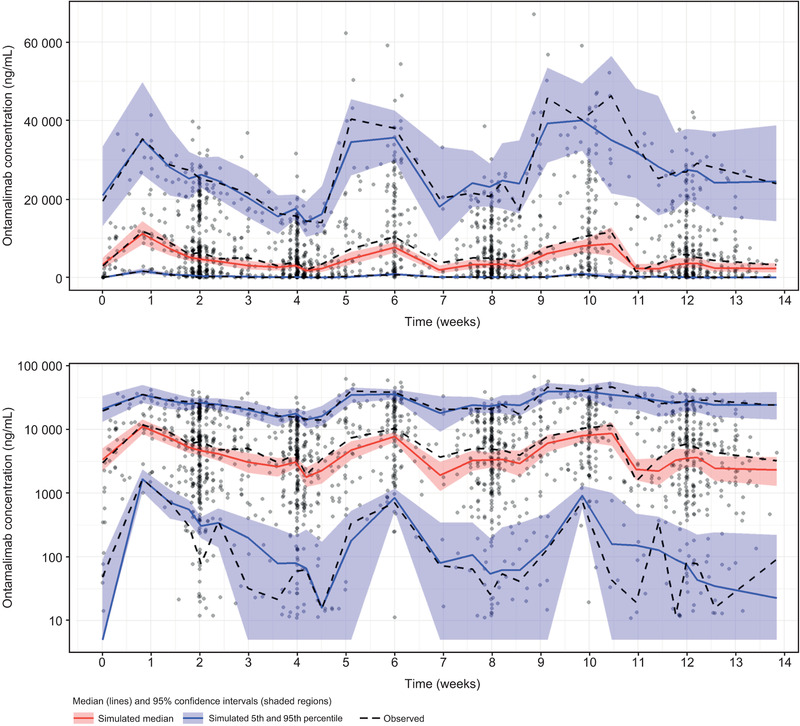
VPC of final population PK model of ontamalimab on linear (top panel) and semilog (bottom panel) scales. PK, pharmacokinetic; VPC, visual predictive check.
Table 2.
Population PK Parameters of Ontamalimab
| Parameter | Estimate (RSE%) | BSV% (RSE%) | Shrinkage (%) |
|---|---|---|---|
| Lag, h | 2.61 (16.5) | NA | NA |
| Ka, h−1 | 0.0187 (10.8) | 61.8 (19.6) | 72.1 |
| CL/F, L/h |
0.0127 (3.73) × (WT/70)0.0034 × (CRP/0.837)0.147 × (ALB/39)−0.889 |
54.6 (7.15) | 10.4 |
| Vc/F, L |
6.53 (2.84) × (WT/70)0.635 |
41.0 (10.6) | 25.0 |
| CLd/F, L/h |
0.000345 (16.3) × (WT/70)0.0034 |
NA | NA |
| Vp/F, L |
0.0216 (16.5) × (WT/70)0.635 |
NA | NA |
| Vmax/F, µg/h |
5.87 (20.8) × (WT/70) 1 .89 |
NA | NA |
| Km, ng/mL | 19.0 (38.3) | NA | NA |
| Error model | |||
| Additive, ng/mL | 166 (34.4) | NA | NA |
| Proportional | 19.6% (10.2) | NA | NA |
ALB, albumin in grams per liter; BSV, between‐subject variability; CL/F, apparent clearance; CLd/F, apparent distributional clearance between the central compartment and the peripheral compartment; CRP, C‐reactive protein; Ka, absorption rate constant; Km, Michaelis‐Menten constant; Lag, absorption lag time; NA, not applicable; RSE, relative standard error; Vc/F, apparent volume of the central compartment; Vmax/F, apparent maximum velocity for the nonlinear elimination; Vp/F, apparent volume of the peripheral compartment; WT, body weight.
The reference subject was a 70‐kg patient with UC or CD, with albumin and CRP levels of 39 g/L and 0.837 mg/dL, respectively.
The condition number is 16 010 (ie, ratio of larger over smaller eigenvalues).
The impact of covariates on the Cave of ontamalimab at week 12 for the 75‐mg Q4W regimen are presented in Figure 4. The 95%CI lower value of albumin (29 g/L) was associated with a 23% reduction in Cave, while the 95%CI upper value of CRP (8.83 mg/dL) was associated with a 24% reduction in Cave. The upper 95%CI of body weight (116 kg) was associated with a 13% decrease in the Cave of ontamalimab. The impact of covariates on the Cmax of ontamalimab at week 12 for the 75‐mg Q4W regimen was also derived (Figure S19). Descriptive statistics of the PK parameters of ontamalimab in subpopulations of interest are presented in Tables S7 through S19.
Figure 4.
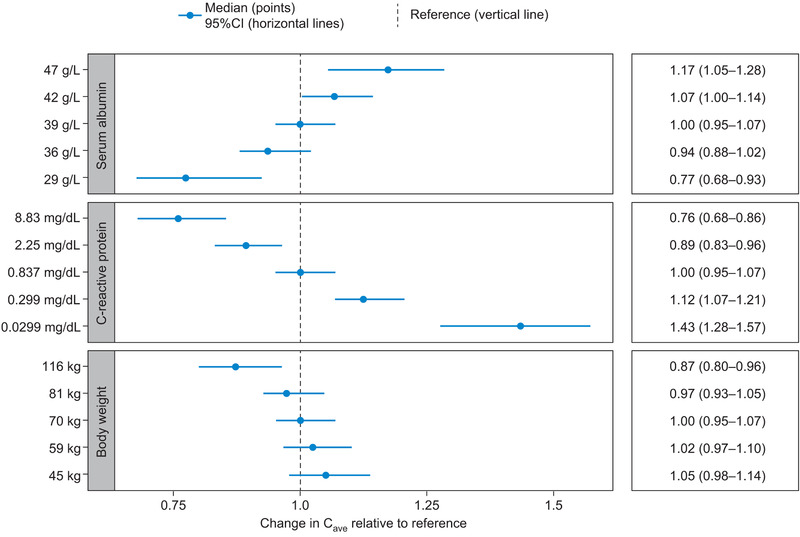
Impact of covariates on average concentrations of ontamalimab at week 12 for the 75‐mg Q4W regimen. Note: The reference subject was a 70‐kg patient with UC or CD, with albumin and CRP levels of 39 g/L and 0.837 mg/dL, respectively. CD, Crohn's disease; CI, confidence interval; CRP, C‐reactive protein; Q4W, every 4 weeks; UC, ulcerative colitis.
Population PK/PD Analysis of Free MAdCAM‐1
An Imax resulted in biased predictions, especially for low values of free MAdCAM‐1 (Table S20). An Imax with a Hill factor <1 better predicted the free MAdCAM‐1 profile, in which an initial rapid drop was followed by a slightly slower decline from baseline. The half maximal effective concentration was not robustly estimated and was very close to the limit of quantitation of ontamalimab. The goodness of fit of the PK/PD model is presented in Figure S20. Observed concentrations of MAdCAM‐1 vs both individual‐predicted and population‐predicted values fell along the line of identity in all studies. Conditional weighted residual values were homogeneously distributed around 0. Scatter plots and box plots were used to display relationships between random effects of PD parameters vs continuous or categorical covariates (Figure S21). The covariates CRP, fecal calprotectin, and colonoscopy score were first tested in a univariate manner on the PD parameters E0 and Slope prior to development of the full model. None of the covariates were statistically significant (Table S21). Results of the VPC are presented in Figure 5. The VPC illustrates that the observed data fall largely within the simulated range, with the 5th, 50th, and 95th percentiles of the observed data in close agreement with the corresponding prediction intervals of the simulations. The 95th percentiles of observed free MAdCAM‐1 value should be interpreted with caution because a few very high values skewed the estimate. VPC by dose levels confirmed the robustness of the model across dose levels (refer to Figures S22 through S25). The overall and dose‐specific VPCs confirmed the predictive performance of the PK/PD model for free MAdCAM‐1 concentrations. Typical parameters derived with the final PK/PD model are presented in Table 3. Based on the PK/PD model, the concentrations of ontamalimab associated with a 50%, 90%, and 95% reduction of MAdCAM‐1 were 51.0, 455, and 2836 ng/mL, respectively. The E0 and Slope estimated (on log scale) were 5.48 and −0.375, respectively. Bootstrap estimates were consistent with those derived from the original analysis (Tables S22 and S23). The predicted effect of ontamalimab at week 12 for each dose level on the reduction of free MAdCAM‐1 is presented in Table 3. Based on geometric mean values of Cmin of ontamalimab at week 12, hypothetical (simulated) doses of 75, 150, and 225 mg would be expected to result in more than 95% suppression of MAdCAM‐1 levels. A similar suppression of free MAdCAM‐1 was observed for steady exposure of ontamalimab (Table S24).
Figure 5.
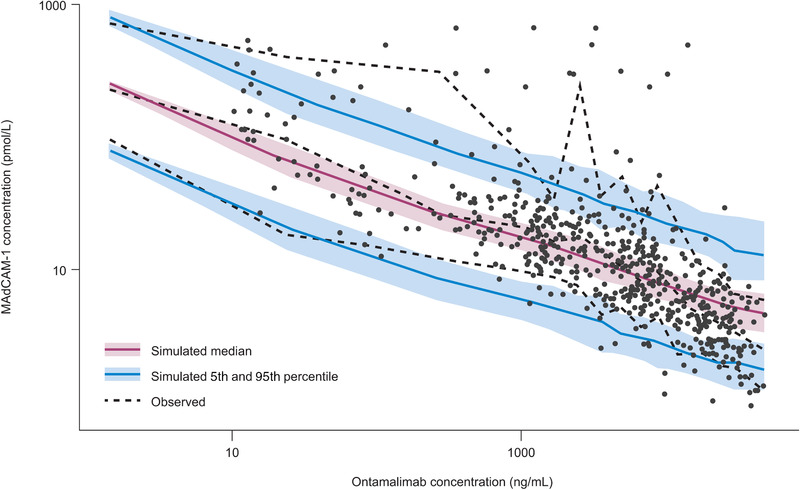
VPC of final population PK/PD model of free MAdCAM‐1. MAdCAM‐1, mucosal addressin cell adhesion molecule‐1; PD, pharmacodynamic; PK, pharmacokinetic; VPC, visual predictive check.
Table 3.
PK/PD Relationship: Percentage Reduction of MAdCAM‐1 at Week 12
| PK Parameter | Dose, mg | Geometric Mean Ontamalimab, ng/mL | Predicted Free MAdCAM‐1, pmol/L | Percentage Change From Baseline of Free MAdCAM‐1, % |
|---|---|---|---|---|
| Cave | 0 | 0 | 240 | 0.0 |
| 7.5 | 461 | 24.1 | −90.0 | |
| 22.5 | 1930 | 14.1 | −94.1 | |
| 75 | 8160 | 8.21 | −96.6 | |
| 150 | 11 500 | 7.22 | −97.0 | |
| 225 | 27 700 | 5.19 | −97.8 | |
| Cmax | 0 | 0 | 240 | 0.0 |
| 7.5 | 986 | 18.1 | −92.5 | |
| 22.5 | 3300 | 11.5 | −95.2 | |
| 75 | 12 000 | 7.11 | −97.0 | |
| 150 | 17 400 | 6.18 | −97.4 | |
| 225 | 38 100 | 4.61 | −98.1 | |
| Cmin | 0 | 0 | 240 | 0.0 |
| 7.5 | 2.13 | 157 | −34.8 | |
| 22.5 | 304 | 28.1 | −88.3 | |
| 75 | 3670 | 11.1 | −95.4 | |
| 150 | 5190 | 9.73 | −95.9 | |
| 225 | 14 600 | 6.60 | −97.2 |
Cave, average concentration; Cmax, maximum (peak) concentration; Cmin, minimum concentration; MAdCAM‐1, mucosal addressin cell adhesion molecule‐1; PK, pharmacokinetic.
Discussion
The interaction of the α4β7 integrin and the MAdCAM‐1 endothelial cell receptor is believed to contribute to chronic inflammation in bowel diseases. The interference of lymphocyte homing by preventing the binding of α4β7 integrin to the MAdCAM‐1 receptor is a well‐established mechanism of action, as demonstrated by the efficacy of natalizumab and vedolizumab in patients with CD. 9 Ontamalimab presents a novel mechanism of action by preventing leukocyte migration secondary to a specific targeting of MAdCAM‐1, while vedolizumab acts by targeting the α4β7 integrin and natalizumab acts by targeting both the α4β1 and α4β7 integrins in tandem. In terms of migration, MAdCAM‐1 is selectively expressed on mucosal endothelial cells, driving memory T‐cell recirculation through mucosal tissues. Since ontamalimab does not bind to the α4β1 integrin and does not affect lymphocyte homing or surveillance in the CNS, minimizing off‐target effects such as progressive multifocal leukoencephalopathy is expected, while continuing to inhibit leukocyte migration into gut mucosa. 12 , 13 Similar to vedolizumab, 14 no cases of progressive multifocal leukoencephalopathy have been observed following repeated administration of ontamalimab in the current clinical studies. 5 , 6 , 11
Population estimates of CL/F and Vc/F for ontamalimab were 0.0127 L/h (0.305 L/day) and 6.53 L, respectively. The mean apparent half‐life of ontamalimab associated with average steady‐state concentrations for the 22.5‐, 75‐, and 225‐mg dose levels was 12.3, 16.5, and 18.6 days, respectively. The rate of absorption following SC administration was slow, corresponding to an absorption half‐life of 37 hours. Complete drug absorption is therefore expected to occur within 8 days of SC dosing of ontamalimab. The half‐life of ontamalimab at the 22.5‐ and 225‐mg dose levels was consistent with those reported for vedolizumab in patients with UC and CD following intravenous administration of 2 to 10 mg/kg (mean values ranging from 15.1 to 22.0 days), as well as for other monoclonal antibodies reported in the literature. 15 , 16
Baseline albumin had the most important impact on the CL/F of ontamalimab and the magnitude of effect (estimated exponent of −0.889) was consistent with that reported for vedolizumab (estimated exponent of −1.18) in patients with CD or UC. 17 The estimated t½β of ontamalimab for a 225‐mg dose in patients with typical baseline albumin values of 29 and 47 g/L was 11.5 and 17.6 days, respectively. These results are consistent with those reported for vedolizumab, suggesting that extreme albumin levels may have an impact on the CL/F in patients with CD or UC. 17 The impact of extreme values of albumin, CRP, and body weight on exposure parameters of ontamalimab was assessed. Overall, changes in exposure ontamalimab were not deemed clinically relevant in light of the PK/PD relationship of MAdCAM‐1. Based on the above information, no changes in dose or frequency of administration of ontamalimab are recommended for patients with extreme values of albumin, CRP, or body weight.
The total volume of distribution (Vss) of ontamalimab (6.6 L) was consistent with plasma volume in humans, and that previously reported for vedolizumab (4.84 L) in patients with UC or CD, as well as for other monoclonal antibodies. 16 , 17
Population estimates of Vmax/F and Km were 5.87 µg/h and 19.0 ng/mL, respectively. The estimated Km was very low, suggesting that nonlinear elimination (target mediated) occurred at low concentrations and is less likely to contribute to the elimination half‐life at therapeutic concentrations. The Michaelis–Menten approximation of the target‐mediated drug disposition model assumes quasi steady state and rapid binding/dissociation between the target (free MAdCAM‐1) and ontamalimab. A wide range of concentrations is typically required to estimate Vmax/F and Km accurately, and simultaneously fitting high‐dose and low‐concentration data is recommended. 18 , 19 The current data set provided a wide range of doses (7.5‐225 mg) with ontamalimab concentrations ranging from 10.2 to 67 100 ng/mL. A bootstrap resampling analysis was preformed to derive median and nonparametric CIs and ultimately assess the uncertainty of Vmax/F and Km estimates. The median (95%CIs) Vmax/F and Km were 5.85 µg/h (3.68‐9.76) and 18.5 ng/mL (13.5‐23.2), respectively, suggesting that the parameters were estimated with a sufficient level of confidence.
A low number of patients (<5%) presented with ADAs at baseline. Patients with a positive ADA status at baseline remained positive over the course of treatment. Patients with a negative ADA status at baseline did not become ADA positive over the course of treatment.
The PK of ontamalimab was similar in patients with UC and CD, and the variability in CL/F was mainly explained by differences in baseline albumin levels. Repeated administrations of 75‐, 150‐, and 225‐mg Q4W ontamalimab would be expected to result in more than 95% suppression of free MAdCAM‐1 levels after 12 weeks of dosing. It should be noted that doses of 75 mg and 225 mg have been tested in clinical trials of ontamalimab, 5 , 6 whereas 150 mg is a hypothetical dose. Suppression of MAdCAM‐1 presents a novel targeted approach for the inhibition of trafficking of α4β7 + lymphocytes in the high endothelial venules of the gastrointestinal tract. The effect of MAdCAM‐1 suppression on clinical end points in patients with CD or UC remains to be determined following long‐term administration of ontamalimab.
Conclusions
Nonlinear elimination of ontamalimab occurred at very low concentrations and is unlikely to contribute to the elimination half‐life at therapeutic concentrations. The primary drivers of PK variability appeared to be baseline levels of CRP and albumin in patients with CD and UC. Repeated SC administration of ontamalimab markedly and sustainably suppresses circulating MAdCAM‐1 levels in patients with UC and CD. The PK/PD properties observed in this study support phase 3 testing of ontamalimab.
Conflicts of Interest
Y.W. and P.M. are employees of Shire, a member of the Takeda group of companies. J.F.M., J.L., and N.K. are paid consultants of Certara.
Funding
This analysis was funded by Shire Human Genetic Therapies Inc., a member of the Takeda group of companies. Editorial support was provided by PharmaGenesis London, with funding provided by Shire International GmbH, a member of the Takeda group of companies.
Data Sharing
The data sets, including the redacted study protocol, redacted statistical analysis plan, and individual participants’ data supporting the results reported in this article, will be made available within 12 months from initial request, to researchers who provide a methodologically sound proposal. The data will be provided after their deidentification, in compliance with applicable privacy laws, data protection, and requirements for consent and anonymization.
Supporting information
Supplement Information
Nastya Kassir and Jean‐Francois Marier are Fellows of the American College of Clinical Pharmacology (FCP)
References
- 1. Shyjan AM, Bertagnolli M, Kenney CJ, Briskin MJ. Human mucosal addressin cell adhesion molecule‐1 (MAdCAM‐1) demonstrates structural and functional similarities to the alpha 4 beta 7‐integrin binding domains of murine MAdCAM‐1, but extreme divergence of mucin‐like sequences. J Immunol. 1996;156(8):2851‐2857. [PubMed] [Google Scholar]
- 2. Steffen BJ, Breier G, Butcher EC, Schulz M, Engelhardt B. ICAM‐1, VCAM‐1, and MAdCAM‐1 are expressed on choroid plexus epithelium but not endothelium and mediate binding of lymphocytes in vitro. Am J Pathol. 1996;148(6):1819‐1838. [PMC free article] [PubMed] [Google Scholar]
- 3. Ghosh S, Goldin E, Gordon FH, et al. Natalizumab for active Crohn's disease. N Engl J Med. 2003;348(1):24‐32. [DOI] [PubMed] [Google Scholar]
- 4. Reinisch W, Hung K, Hassan‐Zahraee M, Cataldi F. Targeting endothelial ligands: ICAM‐1/alicaforsen, MAdCAM‐1. J Crohns Colitis. 2018;12(suppl_2):S669‐S677. [DOI] [PubMed] [Google Scholar]
- 5. Sandborn WJ, Lee SD, Tarabar D, et al. Phase II evaluation of anti‐MAdCAM antibody PF‐00547659 in the treatment of Crohn's disease: report of the OPERA study. Gut. 2018;67(10):1824‐1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vermeire S, Sandborn WJ, Danese S, et al. Anti‐MAdCAM antibody (PF‐00547659) for ulcerative colitis (TURANDOT): a phase 2, randomised, double‐blind, placebo‐controlled trial. Lancet. 2017;390(10090):135‐144. [DOI] [PubMed] [Google Scholar]
- 7. Fernandez Ocana M, Zhang JY, Jones BR, Martin SW, Goetsch M, Neubert H. Poster presentation. P066 Validation of assay for detection of free soluble mucosal addressin cell adhesion molecule‐1 (MAdCAM‐1) in human serum and cerebrospinal fluid Presented at 14th Congress of ECCO, March 6‐9, 2019, Copenhagen, Denmark. [Google Scholar]
- 8. Nguyen TH, Mouksassi MS, Holford N, et al. Model evaluation of continuous data pharmacometric models: metrics and graphics. CPT Pharmacometrics Syst Pharmacol. 2017;6(2):87‐109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Feagan BG, Greenberg GR, Wild G, et al. Treatment of active Crohn's disease with MLN0002, a humanized antibody to the alpha4beta7 integrin. Clin Gastroenterol Hepatol. 2008;6(12):1370‐1377. [DOI] [PubMed] [Google Scholar]
- 10. Harrell FE, Jr. Multivariable modeling strategies. In: Regression Modeling Strategies. Switzerland: Springer International Publishing; 2015:chap 4. [Google Scholar]
- 11. D'Haens G, Vermeire S, Vogelsang H, et al. Effect of PF‐00547659 on central nervous system immune surveillance and circulating beta7+ T cells in Crohn's disease: report of the TOSCA study. J Crohns Colitis. 2018;12(2):188‐196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Major EO. Progressive multifocal leukoencephalopathy in patients on immunomodulatory therapies. Annu Rev Med. 2010;61:35‐47. [DOI] [PubMed] [Google Scholar]
- 13. Warnke C, Menge T, Hartung HP, et al. Natalizumab and progressive multifocal leukoencephalopathy: what are the causal factors and can it be avoided? Arch Neurol. 2010;67(8):923‐930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Card T, Xu J, Liang H, Bhayat F. What is the risk of progressive multifocal leukoencephalopathy in patients with ulcerative colitis or Crohn's disease treated with vedolizumab? Inflamm Bowel Dis. 2018;24:953‐959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rosario M, Dirks NL, Milch C, et al. A review of the clinical pharmacokinetics, pharmacodynamics, and immunogenicity of vedolizumab. Clin Pharmacokinet. 2017;56(11):1287‐1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dostalek M, Gardner I, Gurbaxani BM, Rose RH, Chetty M. Pharmacokinetics, pharmacodynamics and physiologically‐based pharmacokinetic modelling of monoclonal antibodies. Clin Pharmacokinet. 2013;52(2):83‐124. [DOI] [PubMed] [Google Scholar]
- 17. Rosario M, Dirks NL, Gastonguay MR, et al. Population pharmacokinetics‐pharmacodynamics of vedolizumab in patients with ulcerative colitis and Crohn's disease. Aliment Pharmacol Ther. 2015;42(2):188‐202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gibiansky L, Gibiansky E, Kakkar T, Ma P. Approximations of the target‐mediated drug disposition model and identifiability of model parameters. J Pharmacokinet Pharmacodyn. 2008;35(5):573‐591. [DOI] [PubMed] [Google Scholar]
- 19. Gabrielsson J, Weiner D. Pharmacokinetic & Pharmacodynamic Data Analysis: Concepts and Applications. 5th ed Stockholm, Sweden: Swedish Pharmaceutical Society; 2016:131. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement Information


