Abstract
Plastic marine debris (PMD) affects spatial scales of life from microbes to whales. However, understanding interactions between plastic and microbes in the “Plastisphere”—the thin layer of life on the surface of PMD—has been technology‐limited. Research into microbe–microbe and microbe–substrate interactions requires knowledge of community phylogenetic composition but also tools to visualize spatial distributions of intact microbial biofilm communities. We developed a CLASI‐FISH (combinatorial labelling and spectral imaging – fluorescence in situ hybridization) method using confocal microscopy to study Plastisphere communities. We created a probe set consisting of three existing phylogenetic probes (targeting all Bacteria, Alpha‐, and Gammaproteobacteria) and four newly designed probes (targeting Bacteroidetes, Vibrionaceae, Rhodobacteraceae and Alteromonadaceae) labelled with a total of seven fluorophores and validated this probe set using pure cultures. Our nested probe set strategy increases confidence in taxonomic identification because targets are confirmed with two or more probes, reducing false positives. We simultaneously identified and visualized these taxa and their spatial distribution within the microbial biofilms on polyethylene samples in colonization time series experiments in coastal environments from three different biogeographical regions. Comparing the relative abundance of 16S rRNA gene amplicon sequencing data with cell‐count abundance data retrieved from the microscope images of the same samples showed a good agreement in bacterial composition. Microbial communities were heterogeneous, with direct spatial relationships between bacteria, cyanobacteria and eukaryotes such as diatoms but also micro‐metazoa. Our research provides a valuable resource to investigate biofilm development, succession and associations between specific microscopic taxa at micrometre scales.
Keywords: biofilm, CLASI‐FISH, confocal microscopy, marine plastic, succession
1. INTRODUCTION
Plastic marine debris (PMD) is receiving increased attention because it is a visible and steadily increasing pollutant in the ocean (Jambeck et al., 2015; Rochman et al., 2013). Although the impacts of PMD on marine animals have been extensively documented, interactions between PMD and microscopic life and especially microbes are poorly understood. Upon entering the aquatic environment, a layer of organic and inorganic substances quickly coats the plastic surface followed by a rapid colonization of bacterial and eukaryotic (micro)organisms within hours depending on location and season (Foulon et al., 2016). After a few days to several weeks, dense biofilms form on plastics and these can have multiple effects on the fate of PMD (Rummel, Jahnke, Gorokhova, Kühnel, & Schmitt‐Jansen, 2017). Colonization or biofouling can change the buoyancy of floating plastic causing neutral and/or negative buoyancy and settling deeper into the water column (Kooi, Van Nes, Scheffer, & Koelmans, 2017; Ye & Andrady, 1991). Biofilms can also change the physical and chemical properties of plastic and influence further colonization by metazoan larvae (Chung et al., 2010; Hadfield, 2010). Microbes and biofilms can promote degradation of PMD (Chauhan, Agrawal, Deshmukh, Roy, & Priyadarshini, 2018; Kumari, Chaudhary, & Jha, 2019; Montazer, Habibi Najafi, & Levin, 2018; Shah, Hasan, Hameed, & Ahmed, 2008; Zettler, Mincer, & Amaral‐Zettler, 2013), but they can also hinder abiotic ageing by shielding PMD from ultraviolet (UV) radiation in the surface ocean, arguably the most important and rapid abiotic process of plastic breakdown in the environment (Song et al., 2017). Moreover, biofilms can make plastic smell and taste like food to animals such as birds and fish that rely on chemoreception for food selection (Savoca, Tyson, McGill, & Slager, 2017; Savoca, Wohlfeil, Ebeler, & Nevitt, 2016).
Recent awareness of PMD has kindled interest in biofilms that form on this new anthropogenic substrate. Despite media depictions of large intact plastic litter that forms massive floating “islands” in the ocean's interior, most PMD is in the microplastic (<5‐mm) size range (Eriksen et al., 2014) and thus a habitat for primarily microbial and other microscopic life. Despite the demonstrated ubiquity of biofilms on PMD in marine environments, we know very little about the composition and spatial distribution of “Plastisphere”—the thin layer of life on the surface of PMD—members or their geographical and temporal variations. Some studies use scanning electron microscopy (SEM) to visualize microbial communities (Carson, Nerheim, Carroll, & Eriksen, 2013; Masó, Fortuño, De Juan, & Demestre, 2016; Sieburth & Pratt, 1975) and others use molecular microbial profiling to determine community composition (De Tender et al., 2017; Jiang, Zhao, Zhu, & Li, 2018; Oberbeckmann, Osborn, & Duhaime, 2016). SEM provides good spatial resolution but poor taxonomic resolution, while sequencing data reveal the taxonomic composition of the community but not the spatial distribution and interaction between different taxa. One of the properties that biofilms share across different environments is that they are three‐dimensional structures: this imparts an added challenge in studying them in their most intact state because most molecular methods fail to capture this spatial arrangement of life (Flemming & Wuertz, 2019). Understanding the micrometre‐scale distribution of specific groups of microbes and potential associations and interactions has so far not been possible. Whole cell fluorescence in situ hybridization (FISH) methods hold promise to combine taxonomic and spatial resolution (Amann & Fuchs, 2008; Hasegawa et al., 2010), but the extremely heterogeneous communities typically observed in the Plastisphere compared to the surrounding seawater present a challenge in applying these methods to microbial biofilms on PMD (Zettler et al., 2013).
To overcome these barriers, we applied a suite of existing and novel group‐specific probes (phylum to class‐level probes) in a nested approach to target the most commonly occurring members of the Plastisphere. We used an innovative epifluorescence microscopy technique called combinatorial labelling and spectral imaging – fluorescence in situ hybridization (CLASI‐FISH) (Mark Welch, Rossetti, Rieken, Dewhirst, & Borisy, 2016; Valm, Mark Welch, & Borisy, 2012; Valm, Oldenbourg, & Borisy, 2016). This method uses fluorophore‐labelled oligonucleotides to identify microbial taxa, and enables simultaneous use of as many as 16 fluorophores (Valm et al., 2016) whose spectral signatures are distinguished using fluorescence spectral imaging. Whereas Valm et al. (2012), Valm et al. (2016) employed probes in binary combinations to discriminate large numbers of distinct microbial types, here we employed nested probe sets with domain‐, phylum‐, class‐ and family‐level probes so that each taxon was characterized by a spectral signature consisting of one, two or three fluorophores. This nested labelling permitted us to differentiate seven major microbial groups simultaneously in one image. With this technique we showed evidence for inter‐ and intradomain interactions in the Plastisphere from a variety of marine systems that inform our understanding of this new and expanding habitat.
2. MATERIALS AND METHODS
2.1. In situ incubation setup and sampling
Figure 1a–f illustrates our experimental design and workflow. Five‐millimetre‐wide strips of polyethylene (PE) were submerged in the coastal ocean surface waters of three locations: the North Atlantic Ocean (Vineyard Sound; 41.5°N, 70.7°W; from July 10 to August 14, 2013), the tropical Atlantic Ocean (True Blue Bay; 12.0°N, 61.8°W from May 23 to June 6, 2013) and the Wadden Sea (North Sea, Texel, the Netherlands; 53.0°N, 4.8°E; April 18 to May 3, 2018). We used post‐consumer PE plastic from 1‐gallon milk jugs (Vineyard Sound and True Blue Bay) or heavy‐duty plastic bags (Wadden Sea). The plastic was cleaned and then sterilized using 70% ethanol immediately before being deployed in the seawater, and was suspended ~1 m below the surface. For long‐term conservation of the biofilm samples for CLASI‐FISH and SEM, samples were fixed by submerging the plastic in 4% (w/v) paraformaldehyde in 0.2 M phosphate buffered saline (PBS) for 3 hr at 4°C immediately after removing the samples from seawater, then transfered and stored in PBS/ethanol (50:50, v/v) at −20°C. The samples we used in this study were stored for up to 5 years before imaging and still possessed an intact biofilm suitable for CLASI‐FISH.
Figure 1.
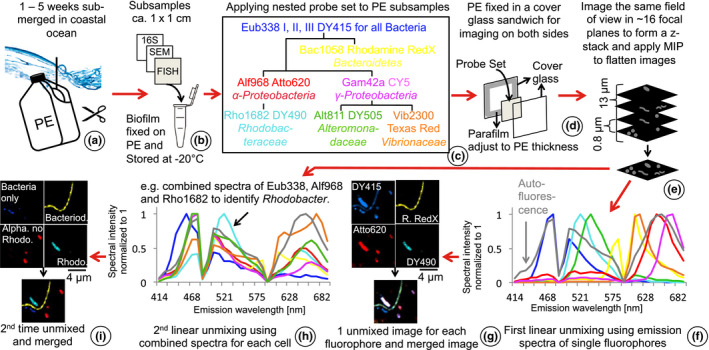
Schematic set‐up of our experiment and workflow. (a) Polyethylene (PE) was submerged in surface seawater (~1‐m depth) for up to 5 weeks. (b) Triplicates were sampled each week for 16S rRNA amplicon sequencing, scanning electron microscopy (SEM) and CLASI (combinatorial laser and spectral imaging) – FISH (fluorescence in situ hybridization). (c) A nested probe set of nine different probes was developed based on the most abundant taxa on plastic marine debris (PMD) identified by amplicon sequencing (Amaral‐Zettler et al., 2015; De Tender et al., 2017). Each bacterial cell on PE hybridized with up to three different fluorescent probes. (d) To image both sides of PE pieces, we sandwiched PE pieces between two glass cover slips and imaged 10 different fields of view per side. (e) The uneven surface of the PE made it impossible to focus the entire biofilm in one field of view, so we performed z‐stack and maximum intensity projections (MIP) to bring the biofilm into one focal plane. (f) We further processed images by applying linear unmixing using emission spectra of each fluorophore. This resulted in separate images for each of the seven fluorophores and autofluorescence from phytoplankton and other autofluorescent cells. (g) Merging these false‐coloured images caused a colour mix that hampered clear identification of different taxa because overlaying three colours per cell resulted in whitish cells. Combined emission spectra of up to three different fluorophores were retrieved from images from the first linear unmixing process. (h) The second linear unmixing was applied to the same unprocessed images used for the first linear unmixing. (i) The second unmixing produced images showing single colours also for double‐ and triple‐labelled cells. Colour‐codes in the first and second linear unmixing plots refer to the different fluorophores and taxa shown in the nested probe set in the tree diagram, respectively
For this “proof of concept” adaptation of the CLASI‐FISH approach applied to Plastisphere samples, we employed eight samples: four from the North Atlantic submerged for 1, 2, 3 and 5 weeks, two from the tropical Atlantic submerged for 1 and 2 weeks, and two from the North Sea submerged for 1 and 2 weeks. To compare amplicon data relative abundances with those from CLASI‐FISH results, we used existing published datasets (selected from the Amaral‐Zettler et al., 2015 study—available from the VAMPS platform) from the North Atlantic samples that were collected from the same piece of plastic at the same time as the CLASI‐FISH samples. Information from amplicon sequencing enabled the design of a CLASI‐FISH probe set to investigate the spatial distribution of the most abundant taxa in the biofilm and to explore microbe–microbe interactions. Additionally, subsamples were imaged using SEM to investigate the morphology of the Plastisphere biofilm and the plastic surface. To our knowledge, this is the first report of the complementary use of SEM and CLASI‐FISH and their quantitative comparison to amplicon sequencing results from the same samples.
2.2. Design and validation of probes and probe set
To visualize the bacterial community in our samples with both breadth and specificity, we developed a probe set with a nested design in which each taxon is targeted by at least two probes. To target the entire domain Bacteria we used the probes Eub338‐I, Eub338‐II and Eub338‐III (Amann et al., 1990; Daims, Brühl, Amann, Schleifer, & Wagner, 1999) labelled with the same fluorophore: we hereafter refer to this mixture as a single probe, Eub338. In addition, we employed six probes each labelled with a distinct fluorophore, targeting the phylum Bacteroidetes, the classes Alphaproteobacteria and Gammaproteobacteria (Manz, Amann, Ludwig, Wagner, & Schleifer, 1992; Neef, 1997), the alphaproteobacterial family Rhodobacteraceae, and the gammaproteobacterial families Alteromonadaceae and Vibrionaceae. The entire probe set thus contained seven oligonucleotide probes labelled with a total of seven different fluorophores, and was capable of distinguishing seven distinct bacterial types (Figure 1c): Rhodobacteraceae; Alphaproteobacteria that were not Rhodobacteraceae; Alteromonadaceae; Vibrionaceae; Gammaproteobacteria that were not Alteromonadaceae or Vibrionaceae; Bacteroidetes; and Bacteria that were not members of any of these other groups. This nested probe set that included probes targeting the phylum, class and family levels improved the likelihood of correct taxonomic identification due to double and triple labelling of single cells (e.g., cells identified as Rhodobacteraceae had to be labelled with the probes Rho1682, Alf968 and Eub338; Figure 1c). Cyanobacteria and diatoms were also distinguishable by their morphology and autofluorescence.
Of the seven probes, four were newly designed targeting the phylum Bacteroidetes (Bac1058) and the families Rhodobacteraceae (Rho1682), Alteromonadaceae (Alt811) and Vibrionaceae (Vib2300) (Table 1). We used arb software (Ludwig et al., 2004) with SILVA arb database release 132 (Quast et al., 2012) for probe design. For in silico prediction of probe hybridization efficiency we used mathfish (Yilmaz, Parnerkar, & Noguera, 2011) (Table S1) and for in silico prediction of probe specificity we used mathfish and probecheck (Loy et al. 2008). To validate probe specificities, we applied individual probes and the seven‐probe set to pure cultures of the target taxa, hybridized and imaged under the same conditions as plastic samples (Figure 2; Tables S2 and S3).
Table 1.
Probes and attached fluorophores used in this study and their properties
| Probe | Specificity | Sequence 5′3′ | Target site | Fluorophore labelled (dual) | References | Tested isolates/true positives | Tested isolates/true negatives |
|---|---|---|---|---|---|---|---|
| Eub338 | Bacteria | GCTGCCTCCCGTAGGAGT | 16S | DY415 | Amann et al. (1990) | 55/55 | |
| Eub338‐II | Planctomycetales | GCAGCCACCCGTAGGTGT | 16S | DY415 | Daims et al. (1999) | ||
| Eub338‐III | Verrucomicrobiales | GCTGCCACCCGTAGGTGT | 16S | DY415 | Daims et al. (1999) | ||
| Alf968 | Alphaproteobacteria | GGTAAGGTTCTGCGCGTT | 16S | Atto620 | Neef (1997) | 11/12 | 25/25 |
| Gam42a | Gammaproteobacteria | GCCTTCCCACATCGTTT | 23S | Cy5 | Manz et al. (1992) | 19/20 | 23/23 |
| Bac1058 | Bacteroidetes | TGAATGGCTGCTTCCAAGCCAACA | 23S | Rhodamine RedX | This paper | 3/3 | 17/17 |
| Rho1682 | Rhodobacteraceae | CCTTCTCGCGAACTTACGGAGG | 23S | DY490 | This paper | 5/5 | 15/15 |
| Vib2300 | Vibrionaceae | TAACCTCACGATGTCCAACCGTG | 23S | Texas RedX | This paper | 3/3 | 9/11 |
| Alt811 | Alteromonadaceae | ACAGCTAGTAGACAGCGTTTACG | 16S | DY505 | This paper | 5/6 | 14/14 |
| Fla891 | Flavobacteriaceae | AGTTTGTCAGGAATTGGTAGGCG | 23S | DY490 | This paper | 0/3 | 1/1 |
| Act1894 | Actinobacteria | AGTTACCACCGCCGTTTACTGG | 23S | DY490 | This paper | 1/3 | |
| Vib1759 | Vibrionaceae | AGCCACCTGGTATCTGCGACT | 23S | Texas RedX | This paper | 5/5 | 9/19 |
| Rho420 | Rhodobacteraceae | TCAGTAAGGAGTACTTAGCCTTCG | 23S | DY490 | This paper | 1/2 |
Probes were tested on 55 different cultured isolates to show true positive and true negative hybridizations. Columns 7 and 8 (true positives or true negatives): the first number is the number of cultures that show results as expected, while the second number is the number of cultures tested in total for the specific probe.
Figure 2.
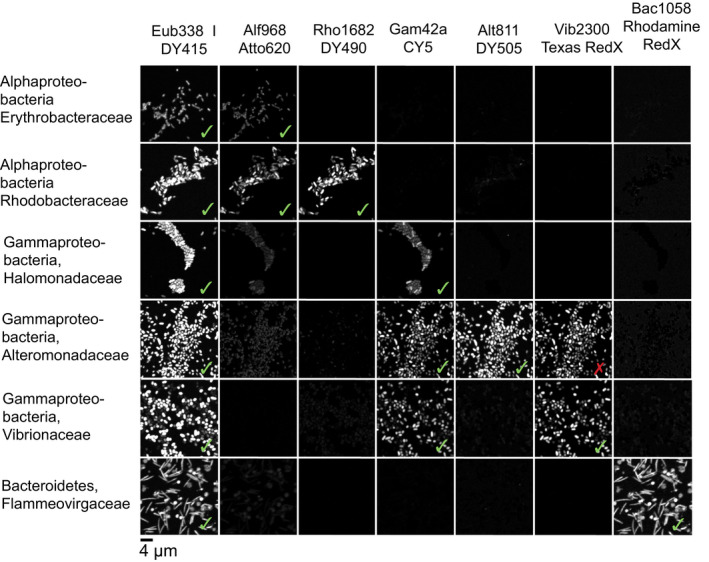
A set of seven probes was validated simultaneously to different pure marine cultures. Up to three different cultured species of the same taxonomic group were tested to ensure specific as well as broad specificity of the probe set. Imaging and linear unmixing were applied under the same conditions as the samples. The probe set hybridized as expected (checkmarks) showing cells labelled specifically with up to three different probes (compare within rows), except for Vib2300 (cross). Although in silico testing of Vib2300 did not show any mismatches, Vib2300 hybridized with some species of the family Alteromonadaceae when tested with pure cultures. Despite this cross‐hybridization, we were able to differentiate members of the Alteromonadaceae from the Vibrionaceae because Alt811 is specific to the former
2.3. Plastisphere CLASI‐FISH
The CLASI‐FISH methodology was developed to visualize microbial spatial organization in biofilms prepared as whole mount spreads or as semithin sections (Mark Welch, Hasegawa, McNulty, Gordon, & Borisy, 2017; Mark Welch et al., 2016). Here we adapted and customized this protocol to visualize biofilms on the relatively thick, hard and uneven surfaces of PMD (see overview in Figure 1). Key modifications included mounting the sample in a sandwich between two coverslips so that both sides of the same piece could be imaged and compensating for the uneven surface by acquiring images as z‐stacks followed by a maximum intensity projection to computationally flatten the image into a single plane. Plastic samples (5 mm × 5 mm) were dehydrated through an ethanol series (70%, 85%, 100%; for 15 min each), air‐dried and incubated in 250 µl of hybridization solution (900 mM NaCl, 20 mM Tris pH 7.5, 0.01% SDS, 20% formamide and each probe at a final concentration of 2 µM) at 46°C for 3 hr. Plastic pieces were then transferred into wash buffer (215 mM NaCl, 20 mM Tris pH 7.5, 5 mM EDTA) at 48°C for 15 min to remove unbound probe. Samples were subsequently dipped in ice‐cold water, drained, air‐dried, and mounted in ProLong Gold Antifade Solution (ThermoFisher) within a frame constructed of Parafilm (Beemis Co.) of the same thickness as the plastic samples. This sample spacer was embedded between two glass coverslips each 0.17 mm thick to permit imaging of both sides of the samples (Figure 1d). Samples were stored in the dark overnight before imaging with a Zeiss LSM 780 confocal microscope (Carl Zeiss) with a 40 × 1.4 N.A. Plan‐Apochromat oil immersion objective.
The pinhole in the light path of a confocal microscope eliminates most of the fluorescence from out‐of‐focus portions of the sample and permits high‐resolution imaging of three‐dimensional structure by acquiring a z‐stack, that is successive images of the same field of view at different focal planes. For imaging the thin biofilms formed in the early stages of colonization of plastic, we selected segments of plastic that were as flat as possible. Nonetheless, there was a slight curvature of the plastic, such that only a portion of the approximately 200 × 200 × 1‐μm field of view was occupied by biofilm in focus at any given focal plane. To compensate for this curvature we acquired z‐stacks of up to 17 µm in thickness and then applied a maximum intensity projection to each z‐stack. This projection selects the brightest plane of the stack, which is generally the most in‐focus plane, at each pixel position and results in a flat image in which cells across the entire field of view are in focus. Ten fields of view per sample side were imaged in this way to acquire spectral images using simultaneous excitation with the 405‐, 488‐ and 594‐nm laser lines with laser intensities of 5%, 10% and 4%, respectively, and laser gain of 760.
Flattened spectral images were processed by linear unmixing with Zeiss zen software. Reference spectra were measured from experimental images or from cultured cells hybridized and imaged under the same conditions used for acquiring experimental data. The linear unmixing algorithm is then used to express the recorded fluorescence spectrum from each pixel in an image as a weighted sum of the reference spectra.
Linear unmixing with reference spectra corresponding to individual fluorophores was performed as previously described (Mark Welch et al., 2017, 2016; Valm et al., 2012) and the resulting false‐coloured images were superimposed to identify cells labelled with multiple fluorophores from the nested probe set (Figure 1f,g). In these unmixed images, cells appeared in one, two or three images depending on the number of probes targeting them in our nested probe set. To simplify the separation of each taxon we repeated the unmixing of the raw data using reference spectra corresponding to the spectral signatures of each distinct cell type, composed of combinations of one, two or three fluorophores (Figure 1h,i). These combined reference spectra were measured from cells in the experimental images of plastisphere biofilms that exhibited a specific combination of fluorophores (Figure 1h). In the resulting unmixed images, cells of different taxonomic levels were clearly distinguishable; for example, Rhodobacteraceae were clearly distinguishable from Alphaproteobacteria that were not Rhodobacteraceae (Figure 1i). Images unmixed with combined reference spectra were subjected to image segmentation as described by Mark Welch et al. (2017) and automated cell counting using fiji (Schindelin et al., 2012).
Brightly autofluorescent objects can interfere with the imaging of fluorescent probes. Phytoplankton cells and protists containing fluorescent pigments were present in our samples and visible in the unmixed images. However, these autofluorescent cells generally did not prevent the imaging of nearby cells, for several reasons. The emission spectra of autofluorescent cells overlapped the emission spectra of the fluorophores but had a different shape (Figure 1f) and thus could be included as a reference spectrum in the linear unmixing and unmixed into a separate channel. Probe‐conferred fluorescence from the fluorophores used in this study was bright enough to be visible adjacent to, and even on top of, phytoplankton cells, in part because of the dual labelling of our probes on both their 3′ and 5′ ends with the same fluorophore. Confocal imaging, by removing out‐of‐focus light, improves the signal‐to‐noise ratio in the plane of focus and thus improves the differentiation of labelled cells from adjacent autofluorescent objects. Collectively these factors made it possible to carry out CLASI‐FISH even in the presence of phytoplankton and other autofluorescent cells.
3. RESULTS
3.1. Identification of abundant taxa to target with CLASI‐FISH
To select an appropriate phylogenetic probe set to visualize the most abundant taxa in our 1‐ to 5‐week‐old samples, we first examined the bacterial taxon composition in our PE subsamples (16S rRNA amplicon sequencing data, Figure 1b) and in published amplicon sequencing data from PE from diverse marine regions (all data published in Amaral‐Zettler et al., 2015 and analysed taxonomically in a comparative meta‐analysis presented in De Tender et al., 2017). By comparing sequence data from our early‐stage biofilms with biofilms on PE samples randomly sampled in the Atlantic and Pacific Ocean and in the North Sea (De Tender et al., 2017), we were able to determine the most commonly occurring taxa in PMD across biogeographically distant habitats. The retention time of the samples from Amaral Zettler et al. (2015) in the ocean and with it the age of the biofilms are unknown and are most likely a mix of young and matured biofilms. The higher‐level taxonomic compositions (e.g., phylum, class, family) of our experimental early‐stage biofilms and of the field‐collected biofilms of unknown ages were the same when the two or three most abundant taxa were compared. Proteobacteria, Cyanobacteria and Bacteroidetes were the most abundant phyla in all Plastisphere biofilms independent of age. Cyanobacteria could be identified due to their distinct morphology and inherent autofluorescence, so Proteobacteria and Bacteroidetes were the primary targets of our probe design. Within the Proteobacteria, Alpha‐ and Gammaproteobacteria were the most abundant classes and Rhodobacteraceae and Alteromonadaceae were the most common families within these classes, respectively, when the biofilms of mixed aged were considered (De Tender et al., 2017). Alteromonadaceae were less abundant on our incubation experiments but still an important group in the Plastisphere community in general. In our experiments, up to 90% of the relative sequence abundances were assigned to the Bacteroidetes and two Proteobacteria groups with Rhodobacteraceae as the most abundant family; therefore, we designed our probes against these most commonly occurring taxa. We also designed a probe targeting the gammaproteobacterial class Vibrionaceae due to the importance of vibrios as potentially pathogenic colonizers of marine surfaces.
3.2. Design and validation of new oligonucleotide probes
To design new probes targeting the phylum Bacteroidetes and families Rhodobacteraceae, Altermonadaceae and Vibrionaceae, we used arb software to mark sequences in the SILVA database for each of the four target groups. We then used the “probe design” function of arb to select potential probe sites and assess the fraction of sequences within the target group that exactly matched the potential probe, as well as the number of exactly matching sequences outside the target group (Table S1). Where possible we selected probes that were an exact match to at least 80% of the target sequences and had two or more mismatches to nontarget sequences from marine organisms. We then used mathfish to calculate the predicted free energy of the hybridization reaction (ΔG overall) and the predicted hybridization efficiency of each potential probe to target and nontarget sequences. We selected candidate probes with a ΔG overall of less than − 14 kcal/mol and a predicted hybridization efficiency not lower than 99% in a hybridization solution containing 20% formamide. Where necessary, we lengthened probes to bring their predicted hybridization into these ranges. The final probes we selected were 22–24 nucleotides in length. An overview of the expected probe coverage, specificity and predicted hybridization thermodynamics is given in Table S1.
We tested the probes against a panel of 55 different pure cultures of marine bacteria, 27 of which were isolated from PMD, to validate probe effectiveness against the target taxon and lack of cross‐reactivity against nontarget taxa (Table S2). Table 1 summarizes the tested cultures and the hybridization success rate. We then combined the six most effective oligonucleotide probes plus Eub338, each labelled with a distinct fluorophore, into a probe set which we tested simultaneously against a panel of six pure cultures to verify effectiveness and specificity of the probe set as a whole (Figure 2). All probes in our probe set except for Vib2300 reacted specifically only with their target taxon as expected (Figure 2; Tables 1, Tables S2 and S3). Eub338, which targets all bacteria, hybridized with all tested cultures. Alf968, which targets Alphaproteobacteria, showed false negative results in only one culture out of 37 (Tables 1 and Table S2). Similarly, Gam42a for Gammaproteobacteria and labelled with the fluorophores Cy5 or DY505 showed one false negative result out of 43 (Tables 1 and Table S2). However, we encountered results consistent with nonspecific binding of the fluorophore Atto647N attached to Gam42a on cultured isolates of Alphaproteobacteria. We observed strong fluorescence on this nontarget taxon when the Gam42a probe was labelled with Atto647N but not when it was labelled with Cy5 or DY505. Hughes, Rawle, and Boxer (2014) showed that Atto647N maleimide strongly associated with membranes, resulting in nonspecific binding and fluorescence that can lead to false positives. Rho1682, which targets the family clade Rhodobacteraceae, behaved as expected in all 20 tested cultures. The same was true for the probe Bac1058 targeting Bacteroidetes with 20 tested cultures. Alt811 for Alteromonadaeceae was tested in 20 cultures and showed one false‐negative result (Table S2).
Vib2300 showed hybridization not only with Vibrionaceae but also with some members of the families Alteromonadaceae and Pseudoalteromonadaceae when tested in pure cultures. Estimation with arb of the specificity and coverage range of Vib2300, based on the presence of two or more mismatches, did not predict these false positives (Table S1). However, mathfish showed hybridization efficiency with sequences from Alteromonadaceae and Pseudoalteromonadaceae of 43% using a 20% formamide concentration, suggesting the possibility of false positive hybridization depending on the precise target sequence and conditions of hybridization. We included Vib2300 in our probe set and identified cells hybridized with both probes Vib2300 and Alt811 as members of the family Alteromondaceae, and cells hybridized only with Vib2300 as members of Vibrionaceae. Although amplicon data assigned Vibrionaceae as relatively low‐abundance targets in our samples, this taxon is of particular interest as it is one of the early colonizers on PMD and it has been detected on PMD in the open Atlantic Ocean (Zettler et al., 2013) and North and Baltic Seas (Kirstein et al., 2016). Given that some members of the family Vibrionaceae can be pathogenic to humans and other animals and can occur in high relative abundance on some microplastics, it is important to investigate their occurrence and distribution in the Plastisphere.
In summary, we observed only in three of 55 tested isolates a lack of hybridization with the target taxon and we did not observe false positive binding of our probes in any of the 55 tested cultures except for Vib2300. Additionally, the nested nature of the probe set increased the likelihood of detecting any cross‐hybridization. Thus, our nested probe set was specific enough for a clear identification of the desired taxa and minimized false positives.
We designed two additional probes to detect the family Flavobacteriaceae and the class Actinobacteria. In silico analysis predicted high hybridization efficiency for both probes and no mismatches to the target taxa. However, we tested the probes on taxon‐specific cultured isolates and observed no signal or weak signals (Table S2) and thus excluded these probes from further testing. We can only speculate on why the two probes were ineffective: inaccessibility due to tertiary structure, protein occlusion, cell wall thickness or low ribosome content of the targeted cells. Additionally, we designed another probe for Vibrionaceae (Vib1749) that hybridized unexpectedly with nontarget taxa such as Alteromonadaceae, Halomonadaceae, Rhodobacteracea and Flavobacteriaceae. Thus, we rejected this probe. Another probe targeting Rhodobacteraceae (Rho420) was less specific and covered fewer groups in the Rhodobacteraceae clade than Rho1682 (Tables 1, Tables S1 and S2), so we employed Rho1682 for further experiments. Our experience demonstrates the importance of probe selection, design and thorough testing for successful application of this technique to other biofilms.
3.3. Spatial distribution of bacteria and phytoplankton on plastic surfaces
We aimed to analyse the spatial organization and succession of early biofilms on PE surfaces immersed in surface seawater in the western North Atlantic Ocean, Tropical Atlantic Ocean (metadata and sequencing details of samples in Amaral‐Zettler et al., 2015) and Wadden Sea. For each sample we imaged 10 fields of view from each of the two sides of the piece of plastic. We prepared images for visual inspection and also carried out semi‐automated cell counts using fiji; representative images are shown in Figures 3, 4, 5 and quantification of cells is shown in Figures 6 and 7.
Figure 3.
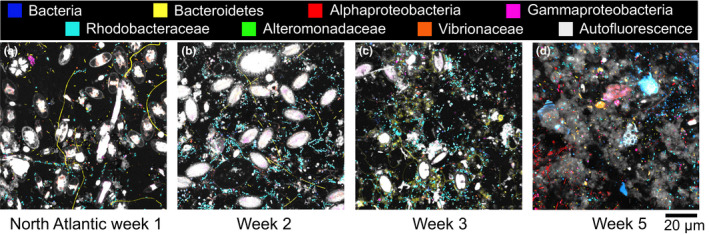
Succession of the biofilm community on polyethylene (PE) after 1–5 weeks in the surface (~1 m) of the North Atlantic Ocean (Vineyard Sound), Woods Hole, MA, USA. The black background is the surface of the plastic pieces. Ellipitical white structures are diatom (phytoplankton) cells attached to the PE surface, and are visible due to their autofluorecent pigments such as chlorophyll a. Variously coloured smaller cells are bacteria hybridized with phylogenetic probes targeting different levels of taxonomic resolution. Greyish and coloured material in the week 5 sample are unidentified organic material (perhaps biofilm EPS) with captured autofluorescent pigments making these structures visible
Figure 4.
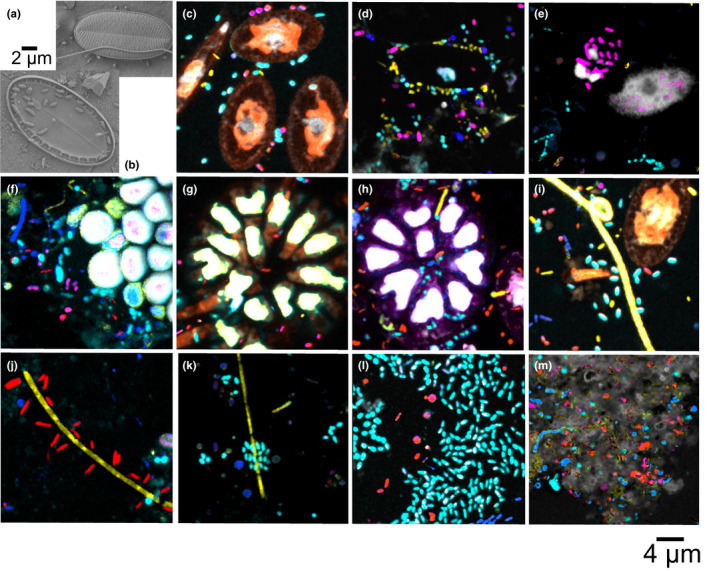
(a,b) Scanning electron micrographs and (c–m) CLASI‐FISH images. (a, c) Diatom surrounded by bacteria cells after 1 week in the surface seawater of the North Atlantic Ocean (Vineyard Sound). (b, d) Lower diatom frustules attached to PE after 2–3 weeks of incubation. (b, d, e) Frustules were surrounded and invaded by different bacterial taxa such as Bacteroidetes (yellow), Rhodobacteraceae (cyan) and Gammaproteobacteria (magenta). Colour code is the same as shown in Figure 3. (f) Bryozoan‐like structure surrounded by bacteria after 1 week in the surface seawater of the Tropical Atlantic Ocean (Grenada). (g, h) Bryozoan‐like structure and (i) filamentous Cyanobacteria surrounded by bacteria after 1 week in the surface seawater of the North Atlantic Ocean. (j, k) Filamentous Bacteroidetes surrounded by Alphaproteobacteria and Rhodobacteraceae after 1 week in the tropical Atlantic Ocean. (l) Clusters of Rhodobacteraceae surrounded by Alphaproteobacteria and other bacteria after 1 week in the Tropical Atlantic Ocean. (m) Diverse bacteria embedded in a fluorescent EPS‐like structure after 5 weeks in the North Atlantic Ocean
Figure 5.
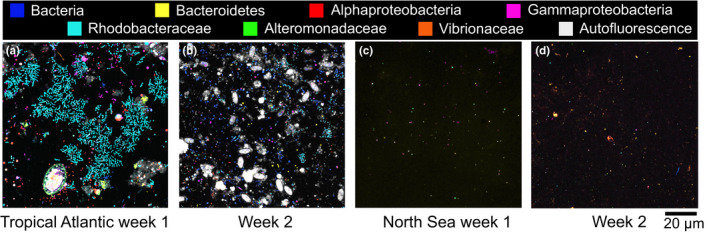
Succession of the biofilm community on polyethylene (PE) after 1–2 weeks in the Tropical Atlantic Ocean (True Blue Bay), Grenada, West Indies (a, b) and in the Wadden Sea, Texel, The Netherlands, EU (c, d). The black background is the surface of the plastic pieces. Ellipitical white structures are phytoplankton cells attached to the PE surface, and are visible due to their autofluorecent pigments such as chlorophyll a. Variously coloured smaller cells are bacteria hybridzed with phylogenetic probes targeting different levels of taxonomic resolution as indicated in the legend. Greyish material in the week 2 sample of the Tropical Atlantic Ocean (b) are unidentified organic material (perhaps biofilm EPS) with captured autofluorescent pigments making these structures visible
Figure 6.
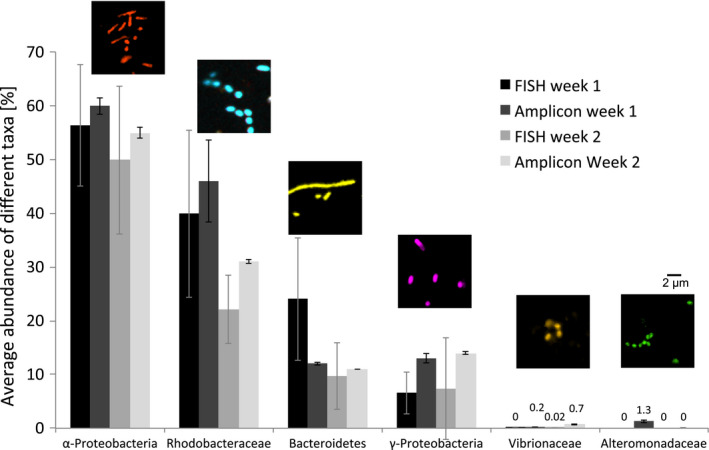
Relative average abundance of different taxa ascertained from cell counting of micrographs (mean of 20 images per sample ± SD) from CLASI‐FISH experiments and from 16S rRNA amplicon sequencing data of the same samples. Each week we sampled three replicates for amplicon sequencing but only one sample for CLASI‐FISH (error bars indicate the variations within the biofilm of each sample: for sequencing, variation between three replicate pieces of plastic; for imaging, variation between 20 fields of view from a single piece of plastic). Epifluourescence microscope images above bars show examples of taxon‐specific cells. Filamentous Bacteroidetes were segmented into individual cells with fiji software before counting. Colour coding of the cells is as in Figure 3. Note that Rhodobacteraceae are included within the Alphaprotobaceria and Vibrionaceae and Alteromonadaceae are included within the Gammaprotobacteria
Figure 7.
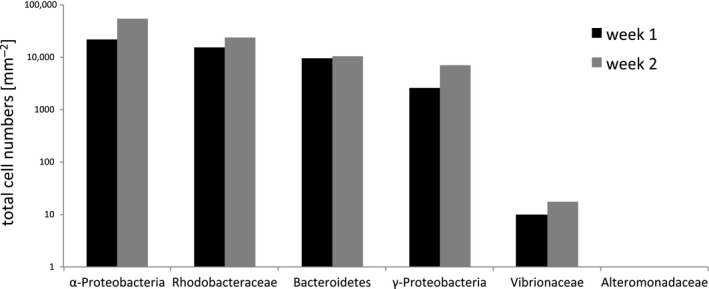
Total cell numbers obtained by CLASI‐FISH on polyethylene submerged for 1 or 2 weeks in surface seawater (1 m) of the North Atlantic Ocean (Vineyard Sound), Woods Hole, MA, USA
Figure 3 shows representative examples of biofilm succession on PE immersed in the coastal North Atlantic Ocean over 5 weeks. The biofilm grew thicker and denser with bacterial abundance increasing from 40,000 cells/mm2 in week 1 to 107,000 cells/mm2 by week 2, followed by a slower increase in bacterial density thereafter. In addition, cellular debris and extracellular polymeric substances (EPS) appeared in week 3 with broad variations in coverage density dependent on the field of view of the imaged plastic surface (grey autofluorescence in Figure 3c,d). At all times, bacteria and phytoplankton were distributed unevenly on the plastic surfaces with some cluster formation of Rhodobacteraceae and filamentous organization of Bacteroidetes. Diatoms, Cyanobacteria and the colonial micro‐animals such as bryozoans colonized plastic within the first week and covered a significant amount of the PE surface. We observed fewer diatoms after the first week, perhaps due to competition, grazing or viral/bacterial infections. We commonly observed the lower diatom frustules (hypothecae) attached to the plastic in SEM images (Figure 4b) after week 2. Diverse bacterial communities often surrounded and appeared to have colonized the hypotheca (Figure 4a‐e). Either bacteria contributed to the death of diatoms or they might be attracted by organic nutrient release during cell lysis. In the 5‐week sample few diatoms were observed. After the first week, 52% of bacteria cells that surrounded diatoms belonged to the family clade Rhodobacteraceae and 20% to the phylum Bacteroidetes. An additional 14% and 10% were Alpha‐ and Gammaproteobacteria, respectively. After 2 weeks, the bacterial community around diatoms changed in composition with a decline in Rhodobactereaceae and Bacteroidetes abundance to 34% and 15%, respectively. Gammaproteobacteria abundance around diatoms increased to 16% while cell numbers of Alphaproteobacteria remained stable. The proportion of bacterial cells that we could not identify taxonomically increased from 3% to 21%, indicating that additional taxa were recruited that settled around diatoms.
The bacterial community composition overall changed dramatically over time with week 1 surfaces dominated by Rhodobacteraceae (average 40%) and filamentous Bacteroidetes (25%) (Figure 6), followed by an increase of Alphaproteobacteria (other than Rhodobacteraceae) and Gammaproteobacteria (other than Alteromonadaceae and Vibrionaceae) until week 5. Almost no Bacteroidetes were observed after 5 weeks while Rhodobacteraceae remained abundant (Figure 3). Bacterial groups of unknown taxonomic affiliation (only hybridized with the Eub338 probe set) increased in abundance over the 5 weeks. We detected only a few individual cells of Alteromonadaceae and Vibrionaceae attached to our PE samples during the entire study in agreement with low abundances of these taxa according to amplicon sequencing data of the same samples (Figure 6). We compared the relative abundance of all examined bacterial groups using amplicon sequencing data and the cell count data from CLASI‐FISH in the first 2 weeks. Both methods showed similar proportions of representative taxa (Figure 6), demonstrating that both approaches provided comparable quantitative results for biofilm composition. Correlation analyses of cell counts versus. amplicon abundances for the six taxonomic groups in Figure 6 showed that the relationship was highly significant (Figure S1). The advantage of CLASI‐FISH over amplicon sequencing is that cell number and size can be determined directly and this provides further information about biofilm density and biomass. Total cell numbers imaged on the 1‐week and 2‐week North Atlantic samples are presented in Figure 7.
We know that in some cases plastics in marine environments are selective for specific microbial communities (Kirstein, Wichels, Krohne, & Gerdts, 2018; Oberbeckmann, Kreikemeyer, & Labrenz, 2018; Zettler et al., 2013) and that there are regional and seasonal differences in the development and composition of biofilms (Amaral‐Zettler et al., 2015; Oberbeckmann, Osborn, & Duhaime, 2016). To investigate this variation, we also examined biofilm succession on pieces of PE from the Tropical Atlantic Ocean (Grenada) during the same summer as our North Atlantic samples (summer 2013), as well as samples incubated in the Wadden Sea off the island of Texel, the Netherlands, in spring 2018.
In the Tropical Atlantic Ocean, phytoplankton such as diatoms and cyanobacteria covered far less of the PE surfaces than in the North Atlantic after 1 week and only increased slightly in abundance in the second week (Figure 5a,b). However, the bacterial community was more abundant and more complex in the Tropics compared to the North Atlantic Ocean after 1 week with large clusters of Rhodobacteraceae dominating the community (Figure 5a). Bacteroidetes were also common and mainly organized in filaments. More Alpha‐ and Gammaproteobacteria that were not Rhodobacteraceae, Alteromonadaceae or Vibrionaceae, as well as more cells only labelled with Eub338, appeared in the first week compared to the North Atlantic Ocean. This suggests a more diverse bacterial community with additional or different taxa in the biofilm compared to the North Atlantic samples. Additionally, autofluorescent organic debris and EPS covered much of the PE surface after only 2 weeks (Figure 5b), in contrast to the North Atlantic samples where the abiotic material (i.e., EPS) was not common until week 3.
In contrast to the relatively rapid colonization of plastic in the Atlantic Ocean samples, the Wadden Sea spring PE samples were still free of a coherent biofilm and organic debris even after 2 weeks (Figure 5c,d). Only individual bacterial cells, mainly Gammaproteobacteria and Alteromonadaceae, were attached to the PE at the end of week 1. After 2 weeks, a thin autofluorescent film covered parts of the PE pieces and bacterial abundance increased slightly (Figure 5d).
4. DISCUSSION
4.1. Application of CLASI‐FISH to marine biofilm communities
CLASI‐FISH has been successfully applied to oral and gut microbiomes (Mark Welch et al., 2017, 2016), but this is the first report of its application to a complex microbiome from the marine environment. The challenges associated with applying CLASI‐FISH to the Plastisphere in the marine environment are several‐fold: (a) the Plastisphere microbiome is highly diverse and variable in nature; (b) Plastisphere microbiomes show biogeographical, environmental and seasonal variability; and (c) many members of the microbiota on floating PMD derive their energy from photosynthesis and possess inherent autofluorescence (e.g., Cyanobacteria and diatoms). Here we present a strategy to overcome these barriers and successfully characterize biofilm succession on PMD and other marine substrates. Surprisingly, amplification methods such as CARD‐FISH (Bobrow, Harris, Shaughnessy, & Litt, 1989) or hybridization chain reaction (Choi, Beck, & Pierce, 2014), which are often required for FISH applied to environmental water‐column samples, were not necessary to enhance the brightness of the fluorescent signals in our experiments. One possible explanation for this was that we imaged early colonizers, which may have had a relatively high ribosome content in their cells due to high growth rates while colonizing new substrate. Bacterial cells that hybridized with our dual‐labelled probes were bright enough for a clear phylogenetic identification even close to or on top of autofluorescent cells (Figure 4c). In addition, the relatively high signal to noise ratio obtained using a laser scanning confocal microscope may have facilitated the imaging of these marine bacteria using direct‐labelled, unamplified fluorescent probes.
CLASI‐FISH was successfully used to investigate the diversity and spatial organization of the microbial community in the Plastisphere due to the simultaneous use of multiple probes. With our nested probe set we could determine the taxonomy of up to 90% of the bacteria in our samples, at least for the higher taxonomic ranks such as class and family. New combinations of different probes or the design of additional probes will advance the field of biofilm investigation within but also beyond the Plastisphere to other marine environments, substrates and microbiomes. Probe and fluorophore variations can be adjusted to expand the number of bacteria that can be identified taxonomically for the determination of a broader or finer variation of taxonomic levels from phylum to species. Another major advantage of CLASI‐FISH is that it provides spatial and taxonomic information about early‐stage biofilm formation that may have low biomass that presents challenges for amplicon sequencing.
CLASI‐FISH can be adapted to target different taxa as broadly or finely as desired, as long as the taxa to be targeted have distinct ribosomal RNA targets. The method can also be adapted to other substrates and environments. However, each new ecosystem probably requires some new probe/fluorophore combination development and testing. Newly designed probes must be tested for effectiveness as well as specificity, because carrying out probe design using thermodynamics‐based mathematical modelling of hybridization can improve the likelihood of success but does not increase it to 100% (Yilmaz & Noguera, 2004; Yilmaz et al., 2011). Each new combination of a probe set has to be tested for hybridization efficiency, possible mismatches, sufficient brightness and specificity of the fluorophore emission spectrum for clear discrimination. Another consideration for the CLASI‐FISH application is that, in its present form, the preparation and imaging of samples including analysis of images represents about 1 week of work per sample. The benefit of CLASI‐FISH is that it allows us to answer questions that we have been unable to address with other approaches. Its application may become more streamlined in the future through automated analysis methods.
4.2. The “Plastisphere” meets the “Phycosphere”
Perhaps the most common interactions we encountered in our CLASI‐FISH samples were between diatoms and bacteria in the Atlantic incubations. Our SEM images from this study (Figure 4a,b) showed diatoms and bacteria in close association on the surface of plastic and other marine substrates, but SEM alone was not able to determine which groups of bacteria were present. CLASI‐FISH, by contrast, enabled us to identify Bacteroidetes, Rhodobacteraceae and Gammaproteobacteria in direct contact with diatoms. Most of our knowledge of bacteria–diatom interactions comes from cultures, where Proteobacteria and Bacteroidetes are most frequently represented (Amin, Parker, & Armbrust, 2012). Interactions between bacteria and diatoms can take on many forms including parasitism, competition and synergism.
We also observed a heterogeneous bacterial community around phytoplankton and bryozoan structures (Figure 4c–i). One interpretation of this spatial distribution is that these interdomain associations are based on the attraction of bacteria to exuded nutrients by phytoplankton (the “Phycosphere”) and other organisms. An alternative explanation is that the distribution of bacteria near eukaryotes is based on hydrodynamic effects due to the occurrence of elevated shapes on the surface layer. In other words: Would bacteria collect around any object in general or specifically around phytoplankton cells? Schäfer, Abbas, Witte, and Muyzer (2002) and Guannel, Horner‐Devine, and Rocap (2011) examined diatom–bacteria associations using laboratory isolates. They found Alphaproteobacteria, especially different clades of the family Rhodobacteraceae with Roseobacter as the most prevalent group, as well as Bacteroidetes and Gammaproteobacteria in association with all tested diatoms. We observed the same taxa in the vicinity of diatoms in our Atlantic Ocean samples (Figure 4c–e). Diatoms can actively release organic exudates forming transparent exopolymer particles (TEPs) or passively add to the carbon pool via cell lysis (Fukao, Kimoto, & Kotani, 2010; Lebeau & Robert, 2003). TEPs attract certain types of bacteria and often become colonized by bacteria by serving as a nutrient for heterotrophs, and bacteria release EPS in the presence of phytoplankton to initiate attachment (Amin et al., 2012). Synergistic interactions can occur when phytoplankton release nutrients, such as dissolved organic matter (DOM) and/or organic nitrogen that is consumed by heterotrophic bacteria, and in return, bacteria provide vitamins, and biologically available forms of nitrogen or iron for algae. However, the interaction can also be parasitic or even fatal when heterotrophic bacteria release lethal molecules or attach and consume diatoms (Amin et al., 2012; Armbrust, 2009) as may be occurring in Figure 4(e). The arrangements of bacteria and algae that we observed on our PE samples suggest an active interaction of bacteria and phytoplankton.
4.3. Microbe–metazoan interactions
Bryant et al. (2016) also examined the microbial community on PMD samples in the Pacific gyre using SEM and metagenomic sequencing techniques. They observed similar groups of dominant organisms attached to plastic as we did, including micro‐animals such as Bryozoa (Figure 4f–h), and microbes including Cyanobacteria (Figure 4i), Alphaproteobacteria and Bacteroidetes. Furthermore, they observed that plastics were a net autotrophic hot spot in the oligotrophic open ocean and that microbial assemblages on PMD were distinct from free‐living microbes in the surrounding seawater. Additionally, Reisser et al. (2014) observed diatoms as the most diverse group of plastic colonizers together with Bryozoans, Cyanobacteria and other bacteria as important members of the Plastisphere. Bryozoans are very common hitchhikers on flotsam (marine debris) in the ocean and have been recognized as important colonizers on plastic for decades (Winston, 1982).
4.4. Bacteria–bacteria interactions
The most abundant and dominant taxon in our North and Tropical Atlantic samples was the family Rhodobacteraceae, which was arranged in large cell clusters (Figures 4l and 5a) either due to fast cell division or recruitment from the water column. Dang, Li, Chen, & Huang, (2008) described Rhodobacteraceae especially the clade Roseobacter as a ubiquitous dominant early colonizer on substrates in Atlantic and Pacific water. Roseobacter are able to grow quickly even at low nutrient concentrations. They also produce antibiotics to outcompete other bacterial taxa and thus modulate the biofilm composition. Furthermore, Rhodobacterales produce EPS, converting the low nutrient status of the biofilm to a high‐nutrient status (Dang et al., 2008). We observed EPS‐like material after 2–3 weeks on our tropical and North Atlantic Ocean samples with a diverse bacterial community embedded in this material (Figures 3d, 4m and 5b). In addition to being a nutrient source, EPS can stabilize and shape the spatial structure of microcolonies and biofilms (Lawrence, Korber, Hoyle, Costerton, & Caldwell, 1991; Tolker‐Nielsen & Molin, 2000; Wolfaardt, Lawrence, Robarts, & Caldwell, 1998). EPS‐producing bacteria might have an advantage over bacteria without EPS production when their descendants embedded in EPS become elevated into the nutrient‐ and oxygen‐rich top layer of the biofilm (Jayathilake et al., 2017; Kim, Racimo, Schluter, Levy, & Foster, 2014). However, we found a heterogeneous bacterial community in the EPS material, suggesting no distinct advantages of specific taxa. We observed diverse associations between many bacterial taxa such as between Alphaproteobacteria and Bacteroidetes (Figure 4j), Rhodobacteraceae and Bacteroidetes (Figure 4k), Rhodobacteracaee and Cyanobacteria (Figure 4i), etc. Bacteria living in communities are in competition with each other for nutrients, space and oxygen. However, flexible metabolic strategies and a heterogeneous distribution of metabolic specialization are common in multispecies communities, showing that metabolic syntrophism is a viable strategy (reviewed by Stubbendieck, Vargas‐Bautista, & Straight, 2016). Thus, bacteria in multispecies communities have access to a wider range of nutrients such as vitamins, trace metals and amino acids than single species (Ponomarova & Patil, 2015). Due to competition in bacterial communities, there will be selection against species with redundant and cost‐intensive metabolic functions and selection for syntrophy to save energy such as for growth and division (Morris, Lenski, & Zinser, 2012). Thus different species that occur in close proximity benefit from nutrient exchange with neighbours that possess complementary metabolic functions and demands. This might be a reason for the multispecies conglomerates we frequently observed on our plastic samples from the Atlantic Ocean. However, we cannot know the exact nature of multispecies interactions that occur on the plastic surface based on our image data alone. Additional methods need to be applied in combination with CLASI‐FISH to understand metabolic pathways, interactions, competitions and synergisms in the Plastisphere.
5. CONCLUSIONS
Visualizing microbial communities using CLASI‐FISH is a powerful and novel tool to investigate different aspects of biofilms: (a) diverse microbes in close proximity reveal potential interactions for further investigation; (b) early colonizers can be detected when sequencing methods are complicated due to low biomass; and (c) imaging allows for identification of biofilm membership, structure and succession. Future applications of CLASI‐FISH can help to visualize specific groups of microbes to see how they are interacting with the substrate surface, including possible polymer hydrolysers contributing to PMD degradation.
Author Contributions
CS and LAA‐Z conceived the project. CS, JLMW, LAA‐Z, and ERZ designed the research. CS, AMK and ERZ performed research. LAA‐Z and JLMW contributed reagents and analytical tools. CS, JLMW, AMK, ERZ and LAA‐Z analyzed data. CS, JLMW, ERZ and LAA‐Z wrote the paper. All authors read and approved the final draft of the manuscript.
Supporting information
ACKNOWLEDGMENTS
C.S. gratefully acknowledges support from the “Deutsche Forschungsgemeinschaft” (DFG) for her “Forschungsstipendium” (SCHL 2141/2‐1), as well as resource support from NOAA grant NA17NOS9990024 awarded to L.A.Z. and funding from the American Chemistry Council awarded to L.A.Z. and E.Z. J.M.W. was supported by the U.S. National Science Foundation under Grant No. 1650141. The Woods Hole Partnership Education Program supported A.M.K.'s internship on the project. We acknowledge Clare Morrall, Jody Daniel and Julia Brunet from St. George's University for Grenada samples, Keven Dooley for Woods Hole samples, and Fons de Vogel for Wadden Sea samples. We acknowledge Tracy Mincer for sharing some of his culture collection for probe testing. We thank Gary Borisy and Blair Rossetti for helpful discussions and Tabita Ramírez‐Puebla and Louis Kerr for help with microscopy and image analysis.
Schlundt C, Mark Welch JL, Knochel AM, Zettler ER, Amaral‐Zettler LA. Spatial structure in the “Plastisphere”: Molecular resources for imaging microscopic communities on plastic marine debris. Mol Ecol Resour. 2019;20:620–634. 10.1111/1755-0998.13119
DATA AVAILABILITY STATEMENT
Amplicon sequence data are available on the vamps database (https://vamps2.mbl.edu/visuals/visuals_index) as described by De Tender et al. (2017). Raw images files are available from the corresponding author upon request.
REFERENCES
- Amann, R. I. , Binder, B. J. , Olson, R. J. , Chisholm, S. W. , Devereux, R. , & Stahl, D. A. (1990). Combination of 16S rRNA‐targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Applied and Environmental Microbiology, 56(6), 1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann, R. , & Fuchs, B. M. (2008). Single‐cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nature Reviews Microbiology, 6(5), 339–348. 10.1038/nrmicro1888 [DOI] [PubMed] [Google Scholar]
- Amaral‐Zettler, L. A. , Zettler, E. R. , Slikas, B. , Boyd, G. D. , Melvin, D. W. , Morrall, C. E. , … Mincer, T. J. (2015). The biogeography of the Plastisphere: Implications for policy. Frontiers in Ecology and the Environment, 13(10), 541–546. 10.1890/150017 [DOI] [Google Scholar]
- Amin, S. A. , Parker, M. S. , & Armbrust, E. V. (2012). Interactions between diatoms and bacteria. Microbiology and Molecular Biology Reviews, 76(3), 667–684. 10.1128/MMBR.00007-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbrust, E. V. (2009). The life of diatoms in the world’s oceans. Nature, 459(7244), 185–192. 10.1038/nature08057 [DOI] [PubMed] [Google Scholar]
- Bobrow, M. N. , Harris, T. D. , Shaughnessy, K. J. , & Litt, G. J. (1989). Catalyzed reporter deposition, a novel method of signal amplification application to immunoassays. Journal of Immunological Methods, 125(1–2), 279–285. 10.1016/0022-1759(89)90104-X [DOI] [PubMed] [Google Scholar]
- Bryant, J. A. , Clemente, T. M. , Viviani, D. A. , Fong, A. A. , Thomas, K. A. , Kemp, P. , … DeLong, E. F. (2016). Diversity and activity of communities inhabiting plastic debris in the North Pacific Gyre. mSystems, 1(3), e00024‐16 10.1128/mSystems.00024-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson, H. S. , Nerheim, M. S. , Carroll, K. A. , & Eriksen, M. (2013). The plastic‐associated microorganisms of the North Pacific Gyre. Marine Pollution Bulletin, 75(1–2), 126–132. 10.1016/j.marpolbul.2013.07.054 [DOI] [PubMed] [Google Scholar]
- Chauhan, D. , Agrawal, G. , Deshmukh, S. , Roy, S. S. , & Priyadarshini, R. (2018). Biofilm formation by Exiguobacterium sp. DR11 and DR14 alter polystyrene surface properties and initiate biodegradation. RSC Advances, 8(66), 37590–37599. 10.1039/C8RA06448B [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, H. M. T. , Beck, V. A. , & Pierce, N. A. (2014). Next‐generation in situ hybridization chain reaction: Higher gain, lower cost, greater durability. ACS Nano, 5, 4284–4294. 10.1021/nn405717p [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, H. C. , Lee, O. O. , Huang, Y.‐L. , Mok, S. Y. , Kolter, R. , & Qian, P.‐Y. (2010). Bacterial community succession and chemical profiles of subtidal biofilms in relation to larval settlement of the polychaete Hydroides elegans . The ISME Journal, 4(6), 817–828. 10.1038/ismej.2009.157 [DOI] [PubMed] [Google Scholar]
- Daims, H. , Brühl, A. , Amann, R. , Schleifer, K.‐H. , & Wagner, M. (1999). The domain‐specific probe EUB338 is insufficient for the detection of all Bacteria: Development and evaluation of a more comprehensive probe set. Systematic and Applied Microbiology, 22(3), 434–444. 10.1016/S0723-2020(99)80053-8 [DOI] [PubMed] [Google Scholar]
- Dang, H. , Li, T. , Chen, M. , & Huang, G. (2008). Cross‐ocean distribution of rhodobacterales bacteria as primary surface colonizers in temperate coastal marine waters. Applied and Environmental Microbiology, 74(1), 52–60. 10.1128/AEM.01400-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Tender, C. , Schlundt, C. , Devriese, L. I. , Mincer, T. J. , Zettler, E. R. , & Amaral‐Zettler, L. A. (2017). A review of microscopy and comparative molecular‐based methods to characterize “Plastisphere” communities. Analytical Methods, 9(14), 2132–2143. 10.1039/C7AY00260B [DOI] [Google Scholar]
- Eriksen, M. , Lebreton, L. C. M. , Carson, H. S. , Thiel, M. , Moore, C. J. , Borerro, J. C. , … Reisser, J. (2014). Plastic pollution in the World’s oceans: More than 5 trillion plastic pieces weighing over 250,000 tons afloat at sea. PLoS ONE, 9(12), e111913 10.1371/journal.pone.0111913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming, H.‐C. , & Wuertz, S. (2019). Bacteria and Archaea on Earth and their abundance in biofilms. Nature Reviews Microbiology, 17(4), 247–260. 10.1038/s41579-019-0158-9 [DOI] [PubMed] [Google Scholar]
- Foulon, V. , Le Roux, F. , Lambert, C. , Huvet, A. , Soudant, P. , & Paul‐Pont, I. (2016). Colonization of polystyrene microparticles by Vibrio crassostreae: Light and electron microscopic investigation. Environmental Science & Technology, 50, 10988–10996. 10.1021/acs.est.6b02720 [DOI] [PubMed] [Google Scholar]
- Fukao, T. , Kimoto, K. , & Kotani, Y. (2010). Production of transparent exopolymer particles by four diatom species. Fisheries Science, 76(5), 755–760. 10.1007/s12562-010-0265-z [DOI] [Google Scholar]
- Guannel, M. L. , Horner‐Devine, M. C. , & Rocap, G. (2011). Bacterial community composition differs with species and toxigenicity of the diatom Pseudo‐nitzschia . Aquatic Microbial Ecology, 64(2), 117–133. 10.3354/ame01513 [DOI] [Google Scholar]
- Hadfield, M. G. (2010). Biofilms and marine invertebrate larvae: What bacteria produce that larvae use to choose settlement sites. Annual Review of Marine Science, 3(1), 453–470. 10.1146/annurev-marine-120709-142753 [DOI] [PubMed] [Google Scholar]
- Hasegawa, Y. , Welch, J. L. M. , Valm, A. M. , Rieken, C. W. , Sogin, M. L. , & Borisy, G. G. (2010). Imaging marine bacteria with unique 16S rRNA V6 sequences by fluorescence in situ hybridization and spectral analysis. Geomicrobiology Journal, 27(3), 251–260. 10.1080/01490450903456806 [DOI] [Google Scholar]
- Hughes, L. D. , Rawle, R. J. , & Boxer, S. G. (2014). Choose your label wisely: Water‐soluble fluorophores often interact with lipid bilayers. PLoS ONE, 9(2), e87649 10.1371/journal.pone.0087649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambeck, J. R. , Geyer, R. , Wilcox, C. , Siegler, T. R. , Perryman, M. , Andrady, A. , … Law, K. L. . (2015). Plastic waste inputs from land into the ocean. Science, 347(6223), 768–771. 10.1126/science.1260352 [DOI] [PubMed] [Google Scholar]
- Jayathilake, P. G. , Jana, S. , Rushton, S. , Swailes, D. , Bridgens, B. , Curtis, T. , & Chen, J. (2017). Extracellular polymeric substance production and aggregated bacteria colonization influence the competition of microbes in biofilms. Frontiers in Microbiology., 8, 1865 10.3389/fmicb.2017.01865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, P. , Zhao, S. , Zhu, L. , & Li, D. (2018). Microplastic‐associated bacterial assemblages in the intertidal zone of the Yangtze Estuary. Science of the Total Environment, 624, 48–54. 10.1016/J.SCITOTENV.2017.12.105 [DOI] [PubMed] [Google Scholar]
- Kim, W. , Racimo, F. , Schluter, J. , Levy, S. B. , & Foster, K. R. (2014). Importance of positioning for microbial evolution. Proceedings of the National Academy of Sciences, 111(16), E1639–E1647. 10.1073/pnas.1323632111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirstein, I. V. , Kirmizi, S. , Wichels, A. , Garin‐Fernandez, A. , Erler, R. , Löder, M. , & Gerdts, G. (2016). Dangerous hitchhikers? Evidence for potentially pathogenic Vibrio spp. on microplastic particles. Marine Environmental Research, 120, 1–8. 10.1016/j.marenvres.2016.07.004 [DOI] [PubMed] [Google Scholar]
- Kirstein, I. V. , Wichels, A. , Krohne, G. , & Gerdts, G. (2018). Mature biofilm communities on synthetic polymers in seawater ‐ specific or general? Marine Environmental Research, 142, 147–154. 10.1016/J.MARENVRES.2018.09.028 [DOI] [PubMed] [Google Scholar]
- Kooi, M. , Van Nes, E. H. , Scheffer, M. , & Koelmans, A. A. (2017). Ups and downs in the ocean: Effects of biofouling on vertical transport of microplastics. Environmental Science and Technology, 51(14), 7963–7971. 10.1021/acs.est.6b04702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari, A. , Chaudhary, D. R. , & Jha, B. (2019). Destabilization of polyethylene and polyvinylchloride structure by marine bacterial strain. Environmental Science and Pollution Research, 26(2), 1507–1516. 10.1007/s11356-018-3465-1 [DOI] [PubMed] [Google Scholar]
- Lawrence, J. R. , Korber, D. R. , Hoyle, B. D. , Costerton, J. W. , & Caldwell, D. E. (1991). Optical sectioning of microbial biofilms. Journal of Bacteriology, 173(20), 6558–6567. 10.1128/jb.173.20.6558-6567.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeau, T. , & Robert, J.‐M. (2003). Diatom cultivation and biotechnologically relevant products. Part I: Cultivation at various scales. Applied Microbiology and Biotechnology, 60(6), 612–623. [DOI] [PubMed] [Google Scholar]
- Loy, A. , Arnold, R. , Tischler, P. , Rattei, T. , Wagner, M. , & Horn, M. (2008). probeCheck–a central resource for evaluating oligonucleotide probe coverage and specificity. Environmental Microbiology, 10(10), 2894–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig, W. , Strunk, O. , Westram, R. , Richter, L. , Meier, H. , Yadhukumar, A. , … Schleifer, K. H. (2004). ARB: A software environment for sequence data. Nucleic Acids Research, 32(4), 1363–1371. 10.1093/nar/gkh293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manz, W. , Amann, R. , Ludwig, W. , Wagner, M. , & Schleifer, K.‐H. (1992). Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: Problems and solutions. Systematic and Applied Microbiology, 15(4), 593–600. 10.1016/S0723-2020(11)80121-9 [DOI] [Google Scholar]
- Mark Welch, J. L. , Hasegawa, Y. , McNulty, N. P. , Gordon, J. I. , & Borisy, G. G. (2017). Spatial organization of a model 15‐member human gut microbiota established in gnotobiotic mice. Proceedings of the National Academy of Sciences, 114(43), E9105–E9114. 10.1073/pnas.1711596114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark Welch, J. L. , Rossetti, B. J. , Rieken, C. W. , Dewhirst, F. E. , & Borisy, G. G. (2016). Biogeography of a human oral microbiome at the micron scale. Proceedings of the National Academy of Sciences, 113(6), E791–E800. 10.1073/pnas.1522149113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masó, M. , Fortuño, J. M. , De Juan, S. , & Demestre, M. (2016). Microfouling communities from pelagic and benthic marine plastic debris sampled across Mediterranean coastal waters. Scientia Marina, 80(S1), 117–127. 10.3989/scimar.04281.10A [DOI] [Google Scholar]
- Montazer, Z. , Habibi Najafi, M. B. , & Levin, D. B. (2018). Microbial degradation of low‐density polyethylene and synthesis of polyhydroxyalkanoate polymers. Canadian Journal of Microbiology, 65(3), 224–234. 10.1139/cjm-2018-0335 [DOI] [PubMed] [Google Scholar]
- Morris, J. J. , Lenski, R. E. , & Zinser, E. R. (2012). The Black Queen Hypothesis: Evolution of dependencies through adaptive gene loss. mBio, 3(2), e00036–e112. 10.1128/mBio.00036-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef, A. (1997). Anwendung der in situ‐Einzelzell‐identifizierung von bakterien zur populationsanalyse in komplexen mikrobiellen Biozonosen. Ph. D. Thesis, Technische Universitaet Muenchen, http://ci.nii.ac.jp/naid/10030869592/en/
- Oberbeckmann, S. , Kreikemeyer, B. , & Labrenz, M. (2018). Environmental factors support the formation of specific bacterial assemblages on microplastics. Frontiers in Microbiology, 8, 2709 https://www.frontiersin.org/article/10.3389/fmicb.2017.02709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberbeckmann, S. , Osborn, A. M. , & Duhaime, M. B. (2016). Microbes on a bottle: Substrate, season and geography influence community composition of microbes colonizing marine plastic debris. PLoS ONE, 11(8), 1–24. 10.1371/journal.pone.0159289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarova, O. , & Patil, K. R. (2015). Metabolic interactions in microbial communities: Untangling the Gordian knot. Current Opinion in Microbiology, 27, 37–44. 10.1016/j.mib.2015.06.014 [DOI] [PubMed] [Google Scholar]
- Quast, C. , Pruesse, E. , Yilmaz, P. , Gerken, J. , Schweer, T. , Yarza, P. , … Glöckner, F. O. (2012). The SILVA ribosomal RNA gene database project: Improved data processing and web‐based tools. Nucleic Acids Research, 41(D1), D590–D596. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisser, J. , Shaw, J. , Hallegraeff, G. , Proietti, M. , Barnes, D. K. A. , Thums, M. , … Pattiaratchi, C. (2014). Millimeter‐sized marine plastics: A new pelagic habitat for microorganisms and invertebrates. PLoS ONE, 9(6), e100289 10.1371/journal.pone.0100289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochman, C. M. , Browne, M. A. , Halpern, B. S. , Hentschel, B. T. , Hoh, E. , Karapanagioti, H. K. , … Thompson, R. C. (2013). Classify plastic waste as hazardous. Nature, 494, 169 10.1038/494169a [DOI] [PubMed] [Google Scholar]
- Rummel, C. D. , Jahnke, A. , Gorokhova, E. , Kühnel, D. , & Schmitt‐Jansen, M. (2017). Impacts of biofilm formation on the fate and potential effects of microplastic in the aquatic environment. Environmental Science and Technology Letters, 4(7), 258–267. 10.1021/acs.estlett.7b00164 [DOI] [Google Scholar]
- Savoca, M. S. , Tyson, C. W. , McGill, M. , & Slager, C. J. (2017). Odours from marine plastic debris induce food search behaviours in a forage fish. Proceedings of the Royal Society B: Biological Sciences, 284(1860), 20171000 10.1098/rspb.2017.1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savoca, M. S. , Wohlfeil, M. E. , Ebeler, S. E. , & Nevitt, G. A. (2016). Marine plastic debris emits a keystone infochemical for olfactory foraging seabirds. Science Advances, 2(11), e1600395 10.1126/sciadv.1600395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer, H. , Abbas, B. , Witte, H. , & Muyzer, G. (2002). Genetic diversity of ‘satellite’ bacteria present in cultures of marine diatoms. FEMS Microbiology Ecology, 42(1), 25–35. 10.1111/j.1574-6941.2002.tb00992.x [DOI] [PubMed] [Google Scholar]
- Schindelin, J. , Arganda‐Carreras, I. , Frise, E. , Kaynig, V. , Longair, M. , Pietzsch, T. , … Cardona, A. (2012). Fiji: An open‐source platform for biological‐image analysis. Nature Methods, 9(7), 676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, A. A. , Hasan, F. , Hameed, A. , & Ahmed, S. (2008). Biological degradation of plastics: A comprehensive review. Biotechnology Advances, 26(3), 246–265. 10.1016/J.BIOTECHADV.2007.12.005 [DOI] [PubMed] [Google Scholar]
- Sieburth, J. M. , & Pratt, H. L. (1975). Microbial seascapes: A pictorial essay on marine microorganisms and their environments. Baltimore: University Park Press. [Google Scholar]
- Song, Y. K. , Hong, S. H. , Jang, M. , Han, G. M. , Jung, S. W. , & Shim, W. J. (2017). Combined effects of UV exposure duration and mechanical abrasion on microplastic fragmentation by polymer type. Environmental Science & Technology, 51(8), 4368–4376. 10.1021/acs.est.6b06155 [DOI] [PubMed] [Google Scholar]
- Stubbendieck, R. M. , Vargas‐Bautista, C. , & Straight, P. D. (2016). Bacterial communities: Interactions to scale. Frontiers in Microbiology, 7, 1234 https://www.frontiersin.org/article/10.3389/fmicb.2016.01234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolker‐Nielsen, T. , & Molin, S. (2000). Spatial organization of microbial biofilm communities. Microbial Ecology, 40(2), 75–84. 10.1007/s002480000057 [DOI] [PubMed] [Google Scholar]
- Valm, A. M. , Mark Welch, J. L. , & Borisy, G. G. (2012). CLASI‐FISH: Principles of combinatorial labeling and spectral imaging. Systematic and Applied Microbiology, 35(8), 496–502. 10.1016/j.syapm.2012.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valm, A. M. , Oldenbourg, R. , & Borisy, G. G. (2016). Multiplexed spectral imaging of 120 different fluorescent labels. PLoS ONE, 11(7), e0158495 10.1371/journal.pone.0158495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston, J. E. (1982). Drift plastic—An expanding niche for a marine invertebrate? Marine Pollution Bulletin, 13(10), 348–351. 10.1016/0025-326X(82)90038-8 [DOI] [Google Scholar]
- Wolfaardt, G. M. , Lawrence, J. R. , Robarts, R. D. , & Caldwell, D. E. (1998). In situ characterization of biofilm exopolymers involved in the accumulation of chlorinated organics. Microbial Ecology, 35(3–4), 213–223. 10.1007/s002489900077 [DOI] [PubMed] [Google Scholar]
- Ye, S. , & Andrady, A. L. (1991). Fouling of floating plastic debris under Biscayne Bay exposure conditions. Marine Pollution Bulletin, 22(12), 608–613. 10.1016/0025-326X(91)90249-R [DOI] [Google Scholar]
- Yilmaz, L. S. , & Noguera, D. R. (2004). Mechanistic approach to the problem of hybridization efficiency in fluorescent in situ hybridization. Applied and Environmental Microbiology, 70(12), 7126–7139. 10.1128/AEM.70.12.7126-7139.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz, L. S. , Parnerkar, S. , & Noguera, D. R. (2011). mathfish, a web tool that uses thermodynamics‐based mathematical models for in silico evaluation of oligonucleotide probes for fluorescence in situ hybridization. Applied and Environmental Microbiology, 77(3), 1118–1122. 10.1128/AEM.01733-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zettler, E. R. , Mincer, T. J. , & Amaral‐Zettler, L. A. (2013). Life in the “Plastisphere”: Microbial communities on plastic marine debris. Environmental Science and Technology, 47(13), 7137–7146. 10.1021/es401288x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Amplicon sequence data are available on the vamps database (https://vamps2.mbl.edu/visuals/visuals_index) as described by De Tender et al. (2017). Raw images files are available from the corresponding author upon request.


