Abstract
Norovirus constitutes the most frequently identified infectious cause of disease outbreaks associated with untreated recreational water. When investigating outbreaks related to surface water, a One Health approach is insightful. Historically, there has been a focus on potential contamination of recreational water by bird droppings and a recent publication demonstrating human noroviruses in bird faeces suggested this should be investigated in future water‐related norovirus outbreaks. Here, we describe a One Health approach investigating a norovirus outbreak in a natural playground. On social media, a large amount of waterfowl were reported to defecate near these playground premises leading to speculations about their potential involvement. Surface water, as well as human and bird faecal specimens, was tested for human noroviruses. Norovirus was found to be the most likely cause of the outbreak but there was no evidence for transmission via waterfowl. Cases had become known on social media prior to notification to the public health service underscoring the potential of online media as an early warning system. In view of known risk factors, advice was given for future outbreak investigations and natural playground design.
Keywords: birds, faeces, norovirus, One Health, social media, water
Impacts.
A norovirus outbreak in a natural playground was investigated via a One Health approach testing human faecal, water and bird faecal specimens. Incidental human introduction of norovirus was the most likely cause of the outbreak. Geese were reported to defecate near the outbreak site, leading to speculations about their potential involvement, but there was no evidence for transmission via waterfowl.
Monitoring of public social media revealed that cases had become known online prior to notification to the Public Health Service: this underscores the potential of online media as an early warning system. Additional insights concerning, potential, sources of contamination and outbreak magnitude were also obtained.
Rapid sampling and storage of a variety of environmental specimens is useful so that these can later be tested, not only for typical indicator bacteria, but also for viruses.
1. INTRODUCTION
In July 2018, the Public Health Service (PHS) received several notifications via a national reporting system for water‐quality and water‐related health issues from persons who had become sick following water contact in the vicinity of a natural playground. Reports mostly concerned children who, during the previous weekend, had had water contact in a recreational lake as well as a nearby natural playground which consists of shallow playground water. The outbreak reached the press and fuelled discussion on local social media in which an unusually large number of geese defecating near the playground were discussed among social media users. Outbreak and symptom characteristics, consisting of an acute‐onset and relatively swift disappearance of symptoms of diarrhoea and vomiting, were suggestive of an outbreak with norovirus (NoV) which constitutes the most frequently identified infectious cause of outbreaks associated with untreated recreational water (de Graaf, Beek, & Koopmans, 2016; Graciaa et al., 2018; Matthews et al., 2012; Rockx et al., 2002).
In the current investigation, a multidisciplinary One Health approach taking into account human, animal and environmental factors was applied to determine the source of the outbreak investigating the role of humans, surface water and waterfowl (World Health Organization, 2017).
2. MATERIALS AND METHODS
2.1. Outbreak description
Patient details could be obtained directly because patients had submitted personal notifications to a national reporting system for water‐quality and water‐related health issues (Meldpunt Water). The PHS was notified of the outbreak on July 4. A timetable demonstrating key events is depicted in Table 1.
Table 1.
Date of key events
| Date | Key action |
|---|---|
| 30 June 2018 | Outbreak onset (date first cases) |
| 4 July 2018 |
Online enquiry started following outbreak discussion on Facebook‐platform Notification to Public Health Service |
| 5 July 2018 |
Inspection playground Collection water specimens Fixing water pump playground |
| 6 July 2018 | Collection human stool specimens |
| 12 July 2018 | Collection bird droppings |
To get a more precise overview of outbreak details and proactively screen for further signs of unrest, the PHS monitored public social media via individual queries. On July 4, the day of the notification, a local health‐related Facebook‐platform posted results of an online request that had started the same day to estimate the total number of sick persons. In this online request, people had been asked to report “the number of persons within their household that had become ill following water exposure in the area” and “to actively tag other ill persons.” People were also asked to actively “report the age of all ill persons” (moderator Facebook‐platform, personal communication).
2.2. Patient faeces
N = 7 patients from three families who had submitted personal notifications to the national online reporting system were contacted on July 6 and asked to send in stool specimens for NoV‐testing using previously published methods (Sukhrie et al., 2011; van Beek et al., 2017). For specimens that were positive in RT‐PCR, a fragment of approximately ~1,000 bp overlapping open reading frames 1 and 2 (ORF1 and ORF2) was sequenced, enabling genotyping of both the polymerase and capsid genes using the NoroNet typing tool (Kroneman et al., 2011). For the phylogenetic analyses, norovirus GI.2 capsid sequences were downloaded from ncbi, and not more than two identical sequences per year and location were included (n = 121). Maximum likelihood trees of the capsid gene (260 bp) were inferred by phyml 3.0 software using the general time‐reversible nucleotide substitution model. The trees were visualized in figtree v1.4.3.
2.3. Water
Following the first disease notifications, the municipality placed warning signs in the swimming area and ordered rapid routine testing for indicator bacteria of faecal contamination (Escherichia coli, enterococci) and cyanobacteria of the swimming water of the recreational lake. During the summer months, water testing of designated large natural swimming areas, including this specific lake, is routinely performed on a biweekly basis in the Netherlands according to the European Bathing Water Directive (EU BWD) (Schets et al., 2018; The European Parliament & the Council of the European Union, 2006). Rapid testing for faecal indicator bacteria and cyanobacteria of the playground water, which unlike the lake is not included in routine screenings, was initiated as well.
In close collaboration with local governing bodies, the PHS performed a location visit to the playground on July 5 and sampled n = 6 playground water specimens (1 ½ litres each) at four different locations which were transported to the laboratory under cold conditions for NoV‐testing. Water specimens were concentrated for viral content using polyethylene glycol precipitation (PEG) as previously described (Lewis & Metcalf, 1988; Sima et al., 2011). In brief, 10 ml of 50% PEG 6000 (Sigma‐Aldrich) was added to 40 ml of water sample and incubated overnight at 4°C. Subsequently, specimens were centrifuged at 13,500 g for 1.5 hr, from which the pellets were re‐suspended in glycine buffer followed by chloroform‐butanol (50%/50% by v/v) treatment. Total nucleic acid from each concentrated water specimen was extracted using High Pure RNA Isolation kit (Roche Diagnostics) and subjected to RT‐PCR for NoV‐detection.
2.4. Waterfowl faeces
In light of multiple reports, notably on social media where sustained speculations about their potential role in disease transmission had arisen, of a continuous unusually large amounts of geese defecating near the playground water, a proactive second location visit was performed on July 12. During this visit, n = 6 faecal specimens were collected from sites directly adjacent to or in the playground water and transported to the laboratory under cold conditions for NoV‐testing using previously published methods (Sukhrie et al., 2011; van Beek et al., 2017). Briefly, a faeces suspension was prepared with 100 mg or 200 μl faecal specimen in 800 μl PBS, and after centrifugation (17,000 g), 200 μl was used for RNA isolation with the High Pure RNA isolation kit (Roche Diagnostics) and subjected to RT‐PCR.
3. RESULTS
3.1. Outbreak description
The PHS received n = 21 case notifications via the national online reporting system. Reported symptoms consisted of vomiting, diarrhoea, headache and fever. Symptom onset was acute, within 0–3 days. Symptoms disappeared within 3 days. Secondary cases were also reported, consistent with an infectious aetiology. The first day of disease onset was June 30.
The online query of the local health‐related Facebook‐platform yielded a total of at least n = 100 sick persons, almost exclusively children (Figure 1, personal communication moderator Facebook‐platform, published with permission).
Figure 1.
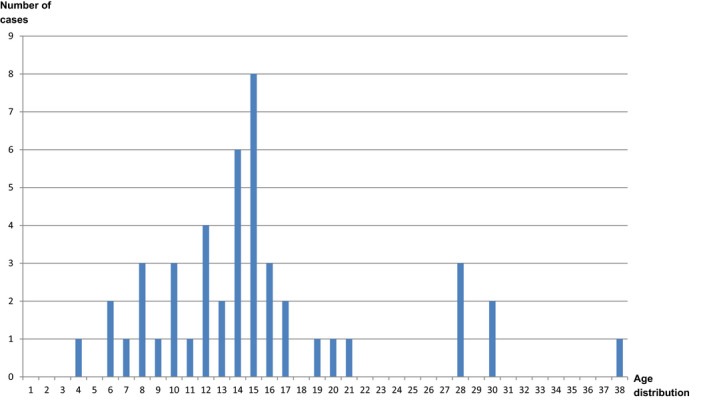
Number and age distribution of cases notified via an online social media enquiry. X‐axis: age distribution; Y‐axis: number of cases (total n = 101, n = 55 age unknown) (personal communication moderator Facebook‐platform, published with permission) [Colour figure can be viewed at wileyonlinelibrary.com]
Patients specifically mentioned visits to the natural playground which they suspected to be the likely source. Patient interviewing by the PHS revealed that there had been no food stands at the location, nor were there other indications of food as a common outbreak source.
The location visit on July 5 revealed the playground area schematically depicted in Figure 2. Briefly, a solar‐energy‐driven water pump distributes water from a nearby river (A) into a natural playground area (B) consisting of two metal water dispensers out of which water flows into a shallow natural playground basin, where mud water is formed, and then into the adjacent lake (C). There were waste bins and, at a nearby beach, public toilets. There were no food trucks. The location visit demonstrated that the water pump designed to refresh the playground basin with fresh river water had broken down. Because of this, a situation with non‐refreshed shallow playground water had existed for an unknown duration. The pump was repaired directly after the outbreak, on July 5, and water was refreshed.
Figure 2.

Schematic depiction of natural playground area. Water from the nearby river (A) is pumped into a natural playground area (B) from where it flows into the adjacent lake (C). Waterfowl (insert) reside near the playground area. Map: © OpenStreetMap contributors https://www.openstreetmap.org/copyright [Colour figure can be viewed at wileyonlinelibrary.com]
3.2. Patient faeces
n = 4 patients out of two different families returned stool specimens. 4/4 specimens were positive for NoV GI. One stool specimen, additionally, tested positive for enterovirus and one for adenovirus genotype non‐40/41. All specimens were sequenced to determine the infecting GI genotype, and 4/4 specimens contained NoV genotype GI.P2‐GI.2 as determined by the NoroNet typing tool and phylogenetic analysis (Figure 3). Norovirus GI‐related outbreaks are less frequent compared to GII outbreaks, but GI.P2‐G1.2 has previously been detected in outbreaks in the Netherlands (van Beek et al., 2018).
Figure 3.

Phylogenetic tree demonstrating norovirus interrelatedness. Maximum‐likelihood trees of ORF 2 were inferred by phyml 3.0 software using the general time reversible nucleotide substitution model. Scale bars indicate nucleotide substitutions per site. The sequences of two outbreak samples are depicted in red [Colour figure can be viewed at wileyonlinelibrary.com]
3.3. Water
Test values for Escherichia coli, enterococci and cyanobacteria on water specimens from the recreational lake as well as from the refreshed playground water specimens were below cut‐off values set by the EU BWD (The European Parliament & the Council of the European Union, 2006). 6/6 playground water specimens tested negative for NoVs GI and GII.
3.4. Waterfowl faeces
During the second location visit, fuelled by reports of large numbers of defecating geese, significant quantities of thin herbivoral faecal specimens, most likely geese faeces, were spotted near the playground water. 6/6 tested faecal specimens were negative for NoVs GI and GII.
4. DISCUSSION
This outbreak investigation illustrates a number of important considerations.
Norovirus, incidentally introduced by a child, or multiple children, recreating in the playground water, was the most likely cause of the outbreak. Preventive measures, including installing a water pump, public toilets and waste bins, were taken when designing the playground, and it is difficult to assess the exact role of a defunct pump, as norovirus is highly infectious in small quantities. However, nearly all risk factors for increased faecal contamination were present in this outbreak setting, that is shallow, poorly circulating water, frequented by young children and animals (Graciaa et al., 2018).
In accordance with the 10‐steps of outbreak investigation, the outbreak was structurally investigated and followed a One Health approach (Prikazsky, V. (Ed.)).
Monitoring of public social media proved of added value in the outbreak investigation. Interestingly, cases had already become known via social media, enabling an online investigation to be started, before notification to the PHS (Table 1). Although exact reliability can be an issue, previous publications have demonstrated the value of online monitoring strategies as early warning systems (Ginsberg et al., 2009; Liu et al., 2017; van de Belt et al., 2018).
From a One Health perspective, an outbreak related to surface water warrants a multidisciplinary approach investigating human, environmental and animal factors (Graciaa et al., 2018; Summa, Henttonen, & Maunula, 2018). As surface water is not routinely tested for NoVs, water specimens were only included for NoV‐testing following notifications of disease when the playground water had already been refreshed. In future outbreak settings, it is advisable to obtain water specimens directly following disease outbreaks near recreational water. These can be stored under cold conditions and tested at a later moment. The same holds true for other environmental specimens, such as subsurface sand specimens, which can be positive for NoV while the water is negative (Schets et al., 2018).
In light of a recent publication on detection of human norovirus (HuNoV) in bird faeces in non‐outbreak settings, it is of importance that, in our case, bird faecal specimens underwent testing for HuNoV‐testing as part of the outbreak investigation and were found to be negative (Summa et al., 2018). There has been a lot of historical attention on the disease‐transmitting potential of bird faeces with a focus on bacteria and parasites (Benskin, Wilson, Jones, & Hartley, 2009; Elmberg, Berg, Lerner, Waldenstrom, & Hessel, 2017; Gorham & Lee, 2016; Graczyk, Majewska, & Schwab, 2008; Meerburg, Koene, & Kleijn, 2011). In this respect, however, a recent review found evidence suggesting HuNoV‐transmission could in principle rather be directed from humans or the environment towards animals (Villabruna, Koopmans, & de Graaf, 2019).
Overall, our findings may serve to stress the fact that routine water testing for faecal bacteria and harmful algal blooms are not always a good indicator of viral‐contamination which is in line with previous publications (Rose, Mullinax, Singh, Yates, & Gerba, 1987; Sinclair, Jones, & Gerba, 2009). It is advisable to collect a variety of environmental specimens, such as water specimens, subsurface sand specimens and animal stool, as soon as possible following an outbreak. Non‐outbreak‐related routine collection can increase knowledge on the background presence and relevance of norovirus.
In an era in which all sorts of natural recreation are gaining popularity, it is prudent that clear guidelines are developed describing minimal requirements for recreational areas. In this case, the playground was well‐designed and thoroughly constructed but a pump that was crucial for water refreshment went defunct. In general, though, measures should incorporate minimal hygiene facilities, such as a sufficient amount of garbage containers, simple shower, toilet, and diaper‐changing facilities and a system avoiding poorly circulating shallow water. The public should be advised to avoid recreational activities in natural water when sick.
Ultimately, in accordance with the One Health approach described here, it should also be realized that these leisure areas are situated in the natural habitat of animals, keeping them in mind in our designs and interventions.
CONFLICT OF INTEREST
None.
Sips GJ, Dirven MJG, Donkervoort JT, et al. Norovirus outbreak in a natural playground: A One Health approach. Zoonoses Public Health. 2020;67:453–459. 10.1111/zph.12689
Funding information
This study was financially supported by the EU H2020 grant COMPARE (643476).
REFERENCES
- Benskin, C. M. , Wilson, K. , Jones, K. , & Hartley, I. R. (2009). Bacterial pathogens in wild birds: A review of the frequency and effects of infection. Biological Reviews of the Cambridge Philosophical Society, 84(3), 349–373. 10.1111/j.1469-185X.2008.00076.x [DOI] [PubMed] [Google Scholar]
- de Graaf, M. , van Beek, J. , & Koopmans, M. P. (2016). Human norovirus transmission and evolution in a changing world. Nature Reviews Microbiology, 14(7), 421–433. 10.1038/nrmicro.2016.48 [DOI] [PubMed] [Google Scholar]
- Elmberg, J. , Berg, C. , Lerner, H. , Waldenstrom, J. , & Hessel, R. (2017). Potential disease transmission from wild geese and swans to livestock, poultry and humans: A review of the scientific literature from a One Health perspective. Infection Ecology and Epidemiology, 7(1), 1300450 10.1080/20008686.2017.1300450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg, J. , Mohebbi, M. H. , Patel, R. S. , Brammer, L. , Smolinski, M. S. , & Brilliant, L. (2009). Detecting influenza epidemics using search engine query data. Nature, 457(7232), 1012–1014. 10.1038/nature07634 [DOI] [PubMed] [Google Scholar]
- Gorham, T. J. , & Lee, J. (2016). Pathogen loading from Canada geese faeces in freshwater: Potential risks to human health through recreational water exposure. Zoonoses Public Health, 63(3), 177–190. 10.1111/zph.12227 [DOI] [PubMed] [Google Scholar]
- Graciaa, D. S. , Cope, J. R. , Roberts, V. A. , Cikesh, B. L. , Kahler, A. M. , Vigar, M. , … Hlavsa, M. C. (2018). Outbreaks associated with untreated recreational water – United States, 2000–2014. MMWR. Morbidity and Mortality Weekly Report, 67(25), 701–706. 10.15585/mmwr.mm6725a1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graczyk, T. K. , Majewska, A. C. , & Schwab, K. J. (2008). The role of birds in dissemination of human waterborne enteropathogens. Trends in Parasitology, 24(2), 55–59. 10.1016/j.pt.2007.10.007 [DOI] [PubMed] [Google Scholar]
- Kroneman, A. , Vennema, H. , Deforche, K. , Avoort, H. , Peñaranda, S. , Oberste, M. S. , … Koopmans, M. (2011). An automated genotyping tool for enteroviruses and noroviruses. Journal of Clinical Virology, 51(2), 121–125. 10.1016/j.jcv.2011.03.006 [DOI] [PubMed] [Google Scholar]
- Lewis, G. D. , & Metcalf, T. G. (1988). Polyethylene glycol precipitation for recovery of pathogenic viruses, including hepatitis A virus and human rotavirus, from oyster, water, and sediment samples. Applied and Environment Microbiology, 54(8), 1983–1988. 10.1128/AEM.54.8.1983-1988.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, K. , Huang, S. , Miao, Z.‐P. , Chen, B. , Jiang, T. , Cai, G. , … Jiang, J. (2017). Identifying potential norovirus epidemics in China via internet surveillance. Journal of Medical Internet Research, 19(8), e282 10.2196/jmir.7855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, J. E. , Dickey, B. W. , Miller, R. D. , Felzer, J. R. , Dawson, B. P. , Lee, A. S. , … Leon, J. S. (2012). The epidemiology of published norovirus outbreaks: A review of risk factors associated with attack rate and genogroup. Epidemiology and Infection, 140(7), 1161–1172. 10.1017/S0950268812000234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerburg, B. G. , Koene, M. G. , & Kleijn, D. (2011). Escherichia coli concentrations in feces of geese, coots, and gulls residing on recreational water in the Netherlands. Vector‐Borne and Zoonotic Diseases, 11(6), 601–603. 10.1089/vbz.2010.0218 [DOI] [PubMed] [Google Scholar]
- Meldpunt Water . Retrieved from https://www.meldpuntwater.nl/
- Prikazsky, V. (Ed.). Field Epidemiology Manual. Outbreak Investigations: 10 steps, 10 pitfalls. Retrieved from https://wiki.ecdc.europa.eu/fem/w/wiki/outbreak-investigations-10-steps-10-pitfalls [Google Scholar]
- Rockx, B. , De Wit, M. , Vennema, H. , Vinje, J. , De Bruin, E. , Van Duynhoven, Y. , & Koopmans, M. (2002). Natural history of human calicivirus infection: A prospective cohort study. Clinical Infectious Diseases, 35(3), 246–253. 10.1086/341408 [DOI] [PubMed] [Google Scholar]
- Rose, J. B. , Mullinax, R. L. , Singh, S. N. , Yates, M. V. , & Gerba, C. P. (1987). Occurrence of rotaviruses and enteroviruses in recreational waters of Oak Creek, Arizona. Water Research, 21(11), 1375–1381. 10.1016/0043-1354(87)90012-1 [DOI] [Google Scholar]
- Schets, F. M. , van den Berg, H. , Vennema, H. , Pelgrim, M. T. M. , Colle, C. , Rutjes, S. A. , & Lodder, W. J. (2018). Norovirus outbreak associated with swimming in a recreational lake not influenced by external human fecal sources in the Netherlands, August 2012. International Journal of Environmental Research and Public Health, 15(11), 2550 10.3390/ijerph15112550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sima, L. C. , Schaeffer, J. , Le Saux, J. C. , Parnaudeau, S. , Elimelech, M. , & Le Guyader, F. S. (2011). Calicivirus removal in a membrane bioreactor wastewater treatment plant. Applied and Environment Microbiology, 77(15), 5170–5177. 10.1128/AEM.00583-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair, R. G. , Jones, E. L. , & Gerba, C. P. (2009). Viruses in recreational water‐borne disease outbreaks: A review. Journal of Applied Microbiology, 107(6), 1769–1780. 10.1111/j.1365-2672.2009.04367.x [DOI] [PubMed] [Google Scholar]
- Sukhrie, F. H. , Beersma, M. F. , Wong, A. , van der Veer, B. , Vennema, H. , Bogerman, J. , & Koopmans, M. (2011). Using molecular epidemiology to trace transmission of nosocomial norovirus infection. Journal of Clinical Microbiology, 49(2), 602–606. 10.1128/JCM.01443-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summa, M. , Henttonen, H. , & Maunula, L. (2018). Human noroviruses in the faeces of wild birds and rodents‐new potential transmission routes. Zoonoses and Public Health, 65(5), 512–518. 10.1111/zph.12461 [DOI] [PubMed] [Google Scholar]
- The European Parliament and the Council of the European Union (2006). Directive 2006/7/EC of the European Parliament and of the Council of 15 February 2006 concerning the management of bathing water quality and repealing Directive 76/160/EEC. Official Journal of the European Union, L64, 37–51. [Google Scholar]
- van Beek, J. , de Graaf, M. , Al‐Hello, H. , Allen, D. J. , Ambert‐Balay, K. , Botteldoorn, N. , … Koopmans, M. P. G. (2018). Molecular surveillance of norovirus, 2005–16: An epidemiological analysis of data collected from the NoroNet network. The Lancet Infectious Diseases, 18(5), 545–553. 10.1016/S1473-3099(18)30059-8 [DOI] [PubMed] [Google Scholar]
- van Beek, J. , van der Eijk, A. A. , Fraaij, P. L. , Caliskan, K. , Cransberg, K. , Dalinghaus, M. , … Koopmans, M. P. (2017). Chronic norovirus infection among solid organ recipients in a tertiary care hospital, the Netherlands, 2006–2014. Clinical Microbiology and Infection, 23(4), 265.e9–265.e13. 10.1016/j.cmi.2016.12.010 [DOI] [PubMed] [Google Scholar]
- van de Belt, T. H. , van Stockum, P. T. , Engelen, L. J. L. P. G. , Lancee, J. , Schrijver, R. , Rodríguez‐Baño, J. , … Voss, A. (2018). Social media posts and online search behaviour as early‐warning system for MRSA outbreaks. Antimicrobial Resistance and Infection Control, 7, 69 10.1186/s13756-018-0359-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villabruna, N. , Koopmans, M. P. G. , & de Graaf, M. (2019). Animals as reservoir for human norovirus. Viruses, 11(5), 478 10.3390/v11050478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2017). One Health. Retrieved from https://www.who.int/features/qa/one-health/en/ [Google Scholar]


