Abstract
Colistin is considered as a last resort treatment for infections caused by multidrug-resistant Enterobacteriaceae. Plasmid-mediated mobile colistin resistance (mcr) genes contributed to the global spread of colistin resistance. This is the first report of plasmid-mediated colistin resistance mcr-8 gene from a clinical Klebsiella pneumoniae K9 isolate recovered from Lebanon. The isolate was characterized phenotypically and genotypically through both short and long read whole-genome sequencing, plasmid typing and conjugation assays. k9 belonged to sequence type 15 and harbored 31 antimicrobial resistance genes. The mcr-8.1 variant was carried on a novel ~ 300 kb multireplicon plasmid having IncFIA, IncR and IncHI1B. The plasmid was conjugative and carried a plethora of antimicrobial resistance determinants. The introduction of novel mcr variants in Lebanon poses an alarming health concern. Surveillance and screening for colistin resistant Enterobacteriaceae and mcr in livestock, animal farms, imported meat and poultry is highly recommended along with monitoring antibiotic use.
Keywords: Colistin, mcr-8, Hybrid assembly, Plasmid, Klebsiella pneumoniae, IncFIA, IncR, IncHI1B
Introduction
Colistin is the last-line treatment option for infections caused by carbapenem resistant Enterobacteriaceae such as carbapenem-resistant Klebsiella pneumoniae (CRKP) [1]. The first plasmid mediated colistin resistance gene, mcr-1, was identified in 2015 in Enterobacteriaceae, mainly Escherichia coli and K. pneumoniae, collected from animals and humans in China [2]. To date, nine different plasmid-mediated mcr variants have been described, designated as mcr-1 -9, isolated from humans, animals and the environment [3].
mcr-8 located on an IncFII-type conjugative plasmid was first described in 2018 in China in K. pneumoniae collected from both animals and humans [4]. Shortly after, it was detected in Laos [5] and Algeria [6].
The first report of a colistin resistant K. pneumoniae in Lebanon was in 2017 [7]. mcr-1-positive E. coli from a human clinical isolate in Lebanon has been recently reported [8]. On the other hand, mcr-1 was widely detected in Gram-negative bacilli isolated from poultry [9], swine farms [9] and water sources in Syrian refugee camps [10]. This is accordingly, the first report of the complete sequence of a novel multireplicon conjugative plasmid carrying colistin resistance mcr-8.1 gene variant in an ST-15 K. pneumoniae isolate recovered from Lebanon.
Materials and methods
Bacterial isolate
The isolate was recovered from the urine sample of a 50-years old female patient on the 28th of August 2018 at the Clinical Microbiology Laboratory of the American University of Beirut Medical Centre (AUBMC), Lebanon. The identification of the isolate was performed using Matrix-Assisted Laser Desorption/Ionization Time of Flight (MALDI-TOF) system (Bruker Daltonik, GmbH, Bremen, Germany). The isolate was identified as Klebsiella pneumoniae and designated as k9.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed using the disk diffusion assay on Mueller-Hinton agar and included a panel of 24 antibiotics belonging to 16 different classes (Table S1). The obtained data was interpreted according to the guidelines of the clinical & laboratory standards institute (CLSI) [11]. The minimal inhibitory concentrations (MICs) of ertapenem, imipenem and meropenem were determined using the E-test methodology (AB BIODISK, Solna, Sweden) according to the manufacturer’s guidelines. MIC of colistin in k9 and in the colistin resistant E. coli transconjugant was further determined by using the broth microdilution method according to the recommendations from the European Committee on Antimicrobial Susceptibility Testing (EUCAST) [12].
DNA extraction
DNA extraction was performed from fresh bacterial colonies using the Nucleospin® Tissue kit (Macherey-Nagel, Germany) according to the manufacturer’s instructions.
mcr-8 PCR
The presence of mcr-8 was confirmed through PCR using MCR-8F 5′-AACCGCCAGAGCACAGAATT-3′ and MCR-8R 5′-TTCCCCCAGCGATTCTCCAT-3’primer pair as previously described [4].
Plasmid replicon typing
Plasmid characterization was performed using the DIATHEVA plasmid based replicon typing (PBRT) kit (Diatheva, Fano, Italy) through a polymerase chain reaction (PCR) based replicon typing method consisting of eight multiplex PCR assays for the amplification of 25 replicons: A/C, B/O, FIA, FIB, FIB-M, FIC, FII, FIIK, FIIS, HI1, HI2, HIB-M, I1, I2, K, L/M, N, P, R, T, U, W, X1, X2, and Y found in the family Enterobacteriaceae. Positive controls were included for all reactions. All PCR reactions were performed according to the manufacturer’s instructions.
Conjugation assay
Conjugation was performed with an azide-resistant E. coli J53 strain. Selection of transconjuguants was done on Uriselect agar (Bio-Rad, France) supplemented with 100 mg/L sodium azide and 4 μg/ml colistin. E. coli transconjugants were typed by PBRT to detect plasmid replicons.
Whole-genome sequencing
Genomic libraries were constructed using the Nextera XT DNA library preparation kit with dual indexing (Illumina). The libraries were sequenced on an Illumina MiSeq with 250 bp × 2 read length. Genome assembly was performed de novo using Spades Genome Assembler Version 3.6.0 [13]. Quality control check was performed using FastQC version 0.11.5 [14].
The mcr8-carrying plasmid was sequenced using PacBio long-read sequencing technology on the Sequel platform (Pacific Biosciences, CA, USA). Library preparation was performed according to the manufacturer’s instructions for microbial multiplexing. G-tubes (Covaris, USA) were used for DNA shearing, and no size selection was performed.
Genome analysis
The assembled genomes were annotated using the RAST online server (http://rast.nmpdr.org) [15]. MLST 2.0 [16], ResFinder 3.0 [17], PointFinder 3.1.0 [18] and Plasmid Finder 2.1 [19] available on the Centre for Genomic Epidemiology website (www.genomicepidemiology.org) were used to determine the ST, presence of resistance genes and plasmid content, respectively. The sequences encoding for plasmids were then extracted and aligned to references obtained from NCBI as previously described [20]. The ISfinder database (https://www-is.biotoul.fr) [21] was used to identify insertion sequences. In silico capsular typing of the wzi gene was performed using Kaptive (www.kaptive.holtlab.net) [22]. The presence of putative virulence factors was screened using the VF scheme available on http://bigsdb.pasteur.fr. PlasmidsSpades was used to assemble the plasmid sequences [23]. Plasmid sequences were extracted and aligned with corresponding reference strains using BioNumerics software version 7.6.1 (Applied Maths, St-Martens-Latem, Belgium). Blast Ring Image Generator (BRIG) version 0.95 was used to visualize the plasmid comparisons [24].
The presence of known mutations conferring colistin resistance on the pmrA, pmrB, pmrC, pmrD, phoP, phoQ, mgrB, crrA and crrB genes were investigated in silico using both nucleic and amino acid alignments compared to the wild-type sequence of K. pneumoniae MGH 78578 (GenBank accession no. CP000647). SNAP [25] (http://www.hiv.lanl.gov) was used to calculate synonymous and non-synonymous substitution rates based on the codon-aligned nucleotide sequences of pmrA and pmrB against the K. pneumoniae MGH 78578 references.
Results
Genome content
K9 genome size was 5,656,813 bp with a GC content of 56.9%. The best match for the capsular locus was KL107 (Blastn identity: 89.60%) with wzc 931 and wzi 178 allelic types. Four Inc. groups were detected in k9 using PlasmidFinder [19] including IncFIA (HI1) (Accession no. AF250878), IncFIB (pKPHS1) (Accession no. CP003223), IncHI1B (Accession no. JN420336) and IncR (Accession no. DQ449578). The presence of the four replicons was also confirmed by PBRT. k9 also harboured virulence genes encoding for iron uptake (kfuA and kfuC) and fimbriae (mrkB-D, mrkH-J).
k9 was compared to all other mcr-8.1 positive isolates recovered so far (Table 1). These included pKP91 (Accession no. MG736312) [4], pKP95 (Accession no. VDEL010000) [6], R. ornithinolytica QDRO2 (Accession no. MK097469.1) [26] KP91, KP95 and QDRO2 were obtained from swine fecal material in China [4], a clinical oral cavity pus sample in Algeria [6], and chicken cloaca in China [26], respectively (Table 1). k9 harboured significantly more resistant determinants (n = 31) than other mcr-8.1 positive isolates with 16, 18 and 23 antimicrobial resistance genes (AMRs) detected in isolates KP95, KP91 and QDRO2, respectively. The latter isolates represented different STs and were recovered from both humans and animal sources [4, 6, 26].
Table 1.
Characteristics of all reported mcr-8.1carrying isolates. COL: colistin; MIC: minimal inhibitory concentration; CC: clonal complex
| # | Isolate | Source | City, Country | COL MIC (mg/L) | Antibiotic Resistance Genes | ST | CC | Reference |
|---|---|---|---|---|---|---|---|---|
| 1 | K. pneumoniae k9 | Urine (Human) | Beirut, Lebanon | 10 | mcr-8.1, blaDHA-1, blaSHV-12, blaSHV-13, blaSHV-31, blaSHV-86, blaSHV-129, blaSHV-155, blaSHV-172, aac(6′)-Ib-cr, aadA1, aadA16, aadA2b, aph(3′)-Ia, aac(6′)-Ib-cr, oqxA, oqxB, qnrB2, qnrB4, qnrB52, sul1, sul3, fosA, mph(A), mph(E), msr(E), tetD, dfrA27, cmlA1, floR, arr-3 | 15 | 23 | This study |
| 2 | K. pneumoniae KP91 | Swine feces (Animal) | Shandong, China | 16 | mcr-8.1, blaCTX-M-14, blaSHV-1, strA, strB, armA, aph(4)-Ia, mph(E), msr(E), oqxA,qnrB4, sul1, sul2, sul3, tet(A), tet(B), tet(34), dfrA12 | 42 | [4] | |
| 3 | K. pneumoniae KP95 | Oral cavity pus (Human) | Sétif, Algeria | 8 | mcr-8.1, blaOXA-48, blaCTX-M-15, blaSHV-94,blaOXA-1, blaTEM-1B, aph(3′)-la, aac(6′)-lb-cr, aph(3″)-lb, aph(6)-ld, oqxB,oqxA, fosA, catB3, sul2, dfrA14 | 336 | [6] | |
| 4 | R. ornithinolytica QDRO2 | Chicken cloaca (Animal) | Shandong, China | 8 | mcr-8.1, blaTEM-1B, blaOXA-1, blaSHV-73, aac(3)-IVa, aph(4)-Ia, aadA2, fosA, mph(A), mph(E), cat, floR, cml, qnrS4, oqxAB, nrB52, sul1, sul2, sul3, tet(A), tet(34), tet(O), tet(B) | [26] | ||
Antimicrobial susceptibility testing
k9 had a colistin MIC of 10 μg/mL (Table S1). It was resistant to 20 of the tested antibiotics, showed intermediate resistance to cefepime, and remained susceptible to meropenem, imipenem, ertapenem, and fosfomycin and was accordingly classified as being extensively drug resistant (XDR) [27].
Resistance genes
Studying antibiotic resistance mechanisms revealed the presence of 31 different resistance genes on the plasmid including resistance to: β-lactams (blaDHA-1, blaSHV-12, blaSHV-13, blaSHV-31, blaSHV-86, blaSHV-129, blaSHV-155, and blaSHV-172), aminoglycosides (aac(6′)-Ib-cr, aadA1, aadA16, aadA2b and aph(3′)-Ia), quinolones (aac(6′)-Ib-cr, oqxA, oqxB, qnrB2, qnrB4, and qnrB52), sulphonamides (sul1 and sul3), fosfomycin (fosA), macrolides (mph(A),mph(E),msr(E)), tetracycline (tetD), trimethoprim (dfrA27), phenicol (cmlA1, floR), rifampicin (arr-3) and colistin (mcr-8).
The presence of known mutations conferring colistin resistance were also investigated. pmrD, phoP, phoQ and mgrB showed 100% identity when compared to the wild-type reference strain K. pneumoniae MGH 78578, while crrA and crrB were not detected. pmrA and pmrB however, showed 40.1 and 49.1% similarity, respectively, to the wild-type reference strain. The rates of synonymous and non-synonymous substitutions were further investigated. SNAP (http://www.hiv.lanl.gov) revealed a dN/dS ratio of 1.131 for pmrA and 1.181 for pmrB indicating a positive selection for non-synonymous mutations (Figure S1).
Mutations in ompK, an outer membrane porin, were investigated using a codon-aligned nucleotide sequences of an intact ompK35 (Accession no. AJ011501), ompK36 sequence (Accession no. Z33506.1) and ompK37 (Accession no. AJ011502.1). We detected a premature stop codon in ompK35 (Y76*), and 22 different amino acids in ompK36 and two in ompK37. A premature stop codon was also detected in ramR (K194*); a positive regulator of the AcrAB efflux system, compared to an intact ramR reference genes (Accession no. KY465996.1).
mcr-8 genetic environment
Blast analysis revealed that the mcr variant was mcr-8.1 showing 100% identity to mcr-8.1 carried by K. pneumoniae plasmid pKP91 (Accession no. MG736312), a ~ 90-kb IncFII-type plasmid obtained from swine fecal material in China [4] and in Raoutella ornithinolytica QDRO2 (Accession no. MK097469.1) [26]
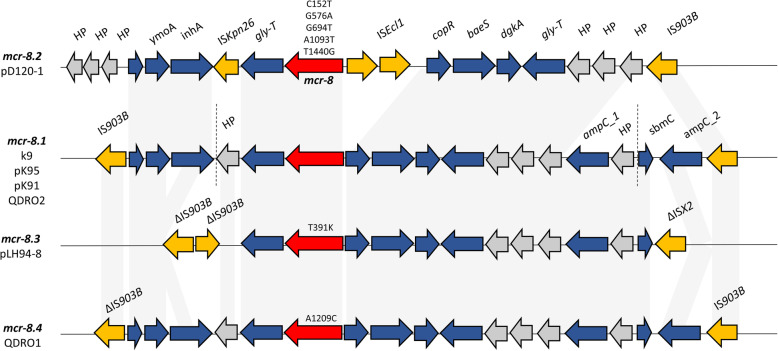
The genetic environment of mcr-8.1 recovered from k9 was further compared with other mcr-8 variants. The amino acid sequences and the genetic environment of mcr-8.1 was most similar to mcr-8.3 and mcr-8.4 then to mcr-8.2, the latter being recovered from K. quasipneumoniae in China [28]. mcr-8.2 also harboured more mutations compared to mcr-8.1 than the other variants (Fig. 1).
Fig. 1.
Schematic representation of the genetic environments of various mcr-8 variants. mcr-8 is shown in red. Amino acid substitutions compared to mcr-8.1 are shown above each arrow. Hypothetical proteins (HP) are shown in grey and insertion sequences in yellow. Dotted vertical lines indicate the contig boundaries carrying mcr-8.1 in k9
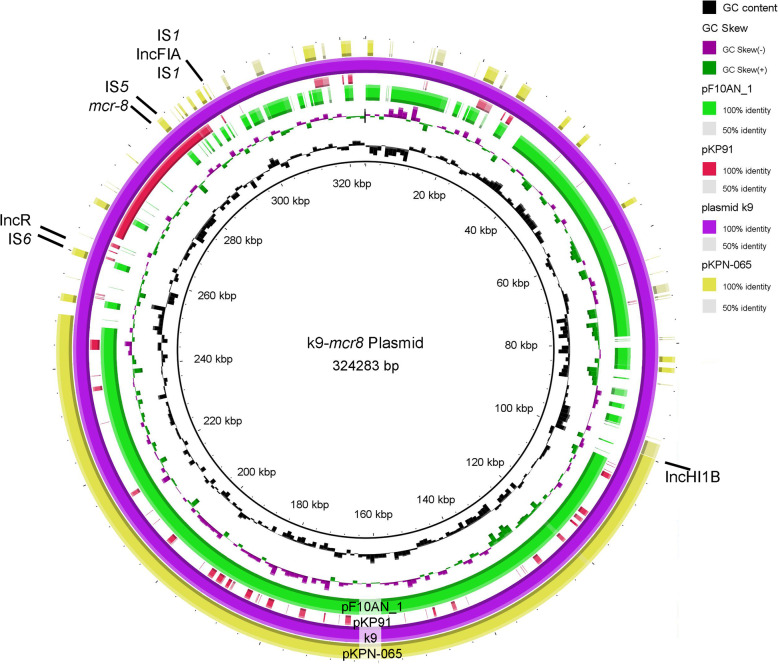
mcr-8.1 was located on a 324,283 bp multireplicon conjugative plasmid pk9 showing highest nucleotide similarity of 99–100% to plasmids: pF10AN_1 (305,552 bp; query coverage = 79.46%; Accession no. CP026154.1) and pKPN-065 (170,926 bp; query coverage = 90.16%; Accession no. CP015026.1) isolated from China and USA [29], respectively. The multireplicon plasmid harboured IncR and IncFIA rep genes also found in pKP91 and IncHI1B rep gene also found in pKPN-065. IncFIA was bracketed by two IS1 insertion sequences while IS6 was present downstream of IncR. Parts of pk9 plasmid carrying various AMR determinants also aligned to pR50–74 (Accession no. CP040362.1) collected from a rabbit fecal sample in China in 2017, pYDC676 collected from patients in the USA in 2014 (Accession no. KT225462.1) [30] and pAR_0158 collected in 2017 (Accession no. CP021699) (Fig. 2).
Fig. 2.
Circular graphical map of mcr-8.1 plasmid in k9. The multireplicon plasmid in k9 was compared to pF10AN_1, AR_0158 plasmid, pKP91, pKPN-065, pR50–74, pYDC676 and pKP91 plasmids. Circles indicate, from inside to outwards: GC skew; GC content; Blast hits to various plasmids used as references. Rep genes and mcr-8 locations are indicated on the figure. Inc. groups are shown in bold
A plethora of resistance genes were detected on pk9. An AMR cassette was located between 14,200 bp and 18,300 bp encoding sul1, aadA16, dfrA27, arr-3 and aac(6′)-Ib-cr5. It closest identity (99.93% identity, query coverage: 86%) to Enterobacter hormaechei plasmid pM206-NDM1 collected in Japan (Accession no. AP018830.1) [31]. Another similar cassette was located between 301,000 bp and 310,200 bp encoding sul1, qnr, sul1, aadA16 and aac(6′)-Ib-cr5 showing 100% identity and 98% coverage to K. pneumoniae Kp_29407 (Accession no. LR736030.1). Other resistance determinants present on the pk9 included tetA, qnrB4, blaDHA-1, apha1b, sul3, aadA1–3, cmlA, floR and ampC. Various sulfonamide resistance genes were identified including four copies of sul1 in addition to sul3 and bcr (bicyclomycin resistance protein).
Discussion
The plasmid-mediated spread of mcr gene variants represents a major health concern worldwide. In this study, we identified a multireplicon plasmid carrying the mcr-8.1 gene variant in a K. pneumoniae clinical isolate recovered from a urine sample in Lebanon. This represents the first report of mcr-8 from Lebanon. mcr-8.1 showed 100% gene sequence similarity to mcr-8.1 encoded by pKP91 plasmid originating from a K. pneumoniae KP91 collected in 2018 from swine fecal material in China [4]. The detection of colistin resistance determinants in both humans and animals directly supports the One Health perspectives that focus on the spread of drug resistance between the environment, humans and animals [31, 32]. Since 2016, nine different plasmid-mediated mcr variants have been described (mcr-1 to mcr-9) from human, animal and environmental sources [2–4], showing the prevalence of the mcr gene in the wildlife and particularly in surface water samples [33].
mcr-8.1 was located on a 324,283 bp multireplicon plasmid showing highest nucleotide similarity (99–100%) to plasmids pF10AN_1 (Accession no. CP026154.1) and pKPN-065 (Accession no. CP015026.1) isolated from China and USA [31], respectively. The multireplicon plasmid harboured IncR and IncFIA rep genes also found in pKP91 [4] and IncHI1B rep gene also found in pKPN-065. Recently, mcr-8.2 was detected on a large, hybrid plasmid containing IncQ, IncR, and IncFII replicons [34] confirming the notion that hybrid multi-replicon plasmids are emerging as vehicles for mcr gene spread [34].
Colistin resistant bacteria and the mcr gene family could be transmitted via the food chain, and so its prudent use in both human and veterinary medicine is of paramount importance [35]. Polymyxins were classified as the “Highest Priority Critically Important Antimicrobials” by the World Health Organization (WHO) because of the increasing usage of colistin to treat serious infections in humans in many parts of the world [35]. Further surveillance and screening for mcr-8 in livestock and animal farms in Lebanon as well as in imported meat and poultry is recommended to track the environmental influences contributing to the development and dissemination of resistance determinants propagating on large mobile genetic vehicles.
Supplementary information
Additional file 1: Table S1. Antimicrobial susceptibility and MIC results of K. pneumoniae k9. Antimicrobial susceptibility was performed using the disk diffusion technique; MICs of ertapenem, imipenem and meropenem were determined using the E-test methodology; MIC of colistin was determined using the broth microdilution method. Antibiotics were divided by their drug categories. GM: gentamycin 10μg; IP: imipenem; MEM: meropenem 10μg; IPM: imipenem 10μg; PM: cefepime; CTX: cefotaxime 30μg; CXM: cefuroxime 30μg; FEP: cefepime 30μg; CZD: ceftazidime 10μg; CZN: cefazolin 30μg; SCF: cefoperazone/sulbactam 75/30μg; FOX: cefoxitin 30μg; NOR: norfloxacin 5μg; CIP: ciprofloxacin 5μg; FAD: fusidic acid 10μg; TGC: tigecycline 15μg; CMN: clindamycin 2μg; ERY: erythromycin 15μg; ATM: aztreonam 30μg; LZD: linezolid 30μg; AMX: amoxicilin 25μg; PIR: pipercacillin 100μg; AMP: amipicillin 10μg; TZP: pipercacillin/tazobactam 100/10μg; FOS: fosfomycin 200μg; TE: tetracycline 30μg; TS: trimethoprim/sulfamethoxazole; CO: colistin; Dark blue: resistant, light blue: intermediate resistance, white: susceptible; MICs are in μg/mL; NA: not available.
Additional file 2: Figure S1. SNAP plots with potential synonymous and non-synonymous substitutions in pmrA (A) and pmrB (B). Alignment was performed against the query sequence of K. pneumoniae MGH 78578 wild-type chromosomal genes.
Abbreviations
- CRKP
Carbapenem resistant Klebsiella pneumoniae
- MALDI-TOF
Matrix-Assisted Laser Desorption/Ionization Time of Flight
- AUBMC
American University of Beirut Medical Centre
- CLSI
Clinical & laboratory standards institute
- MIC
Minimal inhibitory concentration
- EUCAST
European Committee on Antimicrobial Susceptibility Testing
- PBRT
Plasmid based replicon typing
- PCR
Polymerase chain reaction
- BRIG
Blast Ring Image Generator
- AMR
Antimicrobial Resistance
- XDR
Extensively drug resistant
- WHO
World Health Organization
Authors’ contributions
TS: Conceptualization, Data curation, Formal analysis, Writing-original draft, Investigation, Validation. BP: Conceptualization, Data curation, Formal analysis, Writing -original draft, Investigation, Validation. IB: Methodology, Resources, Validation, Funding acquisition. GA: Methodology, Resources, Validation. JH: Methodology, Resources, Validation, Funding acquisition. ST: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Writing - review and editing. The author(s) read and approved the final manuscript.
Funding
This work was supported by Lebanese American University Strategic Research Review Committee Grant (SRRC-R-2019-38) and by the research project grant 17-29239A provided by Czech Health Research Council, by the Charles University Research Fund PROGRES (project number Q39), by the National Sustainability Program I (NPU I) Nr. LO1503 and by the project Nr. CZ.02.1.01/0.0/0.0/16_019/0000787 “Fighting Infectious Diseases” provided by the Ministry of Education Youth and Sports of the Czech Republic.
Availability of data and materials
This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession WTCT00000000. The version described in this paper is version WTCT01000000. The pk9 plasmid sequence was submitted to NCBI on 17/02/2020 under the submission number SUB6977004.
Ethics approval
Ethical approval was not required as the isolate was collected and stored as part of routine clinical care.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Tamara Salloum, Email: tamara.salloum@lau.edu.
Balig Panossian, Email: balig.panossian@lau.edu.
Ibrahim Bitar, Email: ibrahimbitar5@gmail.com.
Jaroslav Hrabak, Email: jaroslav.hrabak@lfp.cuni.cz.
George F. Araj, Email: garaj@aub.edu.lb
Sima Tokajian, Email: stokajian@lau.edu.lb.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13756-020-00759-w.
References
- 1.Moghnieh R, Araj GF, Awad L, Daoud Z, Mokhbat JE, Jisr T, et al. A compilation of antimicrobial susceptibility data from a network of 13 Lebanese hospitals reflecting the national situation during 2015-2016. Antimicrob Resist Infect Control. 2019;8:41. doi: 10.1186/s13756-019-0487-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 3.Carroll LM, Gaballa A, Guldimann C, Sullivan G, Henderson LO, Wiedmann M. Identification of novel mobilized Colistin resistance gene mcr-9 in a multidrug-resistant, Colistin-Susceptible Salmonella enterica Serotype Typhimurium Isolate. mBio. 2019;10:e00853–e00819. doi: 10.1128/mBio.00853-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X, Wang Y, Zhou Y, Li J, Yin W, Wang S, et al. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg Microbes Infect. 2018;7:122. doi: 10.1038/s41426-018-0124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hadjadj L, Baron SA, Olaitan AO, Morand S, Rolain JM. Co-occurrence of variants of mcr-3 and mcr-8 genes in a Klebsiella pneumoniae isolate from Laos. Front Microbiol. 2019;10:2720. doi: 10.3389/fmicb.2019.02720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nabti LZ, Sahli F, Ngaiganam EP, Radji N, Mezaghcha W, Lupande-Mwenebitu D, et al. Development of real-time PCR assay allowed describing the first clinical Klebsiella pneumoniae isolate harboring plasmid-mediated colistin resistance mcr-8 gene in Algeria. J Glob Antimicrob Resist. 2020;20:266–271. doi: 10.1016/j.jgar.2019.08.018. [DOI] [PubMed] [Google Scholar]
- 7.Okdah L, Leangapichart T, Hadjadj L, Olaitan AO, Al-Bayssari C, Rizk R, et al. First report of colistin-resistant Klebsiella pneumoniae clinical isolates in Lebanon. J Glob Antimicrob Resist. 2019;9:15. doi: 10.1016/j.jgar.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 8.Al-Mir H, Osman M, Azar N, Madec JY, Hamze M, Haenni M. Emergence of clinical mcr-1-positive Escherichia coli in Lebanon. J Glob Antimicrob Resist. 2019;19:83. doi: 10.1016/j.jgar.2019.08.019. [DOI] [PubMed] [Google Scholar]
- 9.Dandachi I, Fayad E, El-Bazzal B, Daoud Z, Rolain JM. Prevalence of extended-Spectrum Beta-lactamase-producing gram-negative bacilli and emergence of mcr-1 Colistin resistance gene in Lebanese swine farms. Microb Drug Resist. 2018;25:233–240. doi: 10.1089/mdr.2018.0110. [DOI] [PubMed] [Google Scholar]
- 10.Hmede Z, Sulaiman AA, Jaafar H, Kassem II. Emergence of plasmid-borne colistin resistance gene mcr-1 in multidrug-resistant Escherichia coli isolated from irrigation water in Lebanon. Int J Antimicrob Agents. 2019;54:102–104. doi: 10.1016/j.ijantimicag.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 11.Clinical and Laboratory Standards Institute . Performance Standards for Antimicrobial Susceptibility Testing: 28th Edition M100. Wayne: CLSI; 2018. [Google Scholar]
- 12.The European Committee on Antimicrobial Susceptibility Testing . Breakpoint tables for interpretation of MICs and zone diameters. Version 10.0. 2020. [Google Scholar]
- 13.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010. [Google Scholar]
- 15.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9(1):75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol. 2012;50(4):1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemoth. 2012;67(11):2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zankari E, Allesøe R, Joensen KG, Cavaco LM, Lund O, Aarestrup FM. PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J Antimicrob Chemoth. 2017;72(10):2764–2768. doi: 10.1093/jac/dkx217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carattoli A, Zankari E, García-Fernández A, Larsen MV, Lund O, Villa L, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58(7):3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arabaghian H, Salloum T, Alousi S, Panossian B, Araj GF, Tokajian S. Molecular characterization of Carbapenem resistant Klebsiella pneumoniae and Klebsiella quasipneumoniae isolated from Lebanon. Sci Rep. 2019;9(1):1–2. doi: 10.1038/s41598-018-36554-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siguier P, Varani A, Perochon J, Chandler M. Exploring bacterial insertion sequences with ISfinder: objectives, uses, and future developments: In: Bigot Y. (eds) Mobile Genetic Elements. Methods in Molecular Biology (Methods and Protocols), vol 859. Humana Press; 2012. p. 91–103. 10.1007/978-1-61779-603-6_5. [DOI] [PubMed]
- 22.Wick RR, Heinz E, Holt KE, Wyres KL. Kaptive web: user-friendly capsule and lipopolysaccharide serotype prediction for Klebsiella genomes. J Clin Microbiol. 2018;56(6):e00197–e00118. doi: 10.1128/JCM.00197-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Antipov D, Hartwick N, Shen M, Raiko M, Lapidus A, Pevzner P. plasmidSPAdes: assembling plasmids from whole genome sequencing data. bioRxiv. 2016;1:048942. doi: 10.1093/bioinformatics/btw493. [DOI] [PubMed] [Google Scholar]
- 24.Alikhan NF, Petty NK, Zakour NL, Beatson SA. BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12(1):402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korber B. HIV signature and sequence variation analysis. Computational analysis of HIV molecular sequences, chapter 4, pages 55–72. Allen G. Rodrigo and Gerald H. learn, eds. Dordrecht: Kluwer Academic Publishers; 2000. [Google Scholar]
- 26.Wang X, Wang Y, Zhou Y, Wang Z, Wang Y, Zhang S, et al. Emergence of colistin resistance gene mcr-8 and its variant in Raoultella ornithinolytica. Front Microbiol. 2019;10:228. doi: 10.3389/fmicb.2019.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 28.Yang X, Liu L, Wang Z, Bai L, Li R. Emergence of mcr-8.2-bearing Klebsiella quasipneumoniae of animal origin. J Antimicrob Chemoth. 2019;74(9):2814–2817. doi: 10.1093/jac/dkz213. [DOI] [PubMed] [Google Scholar]
- 29.Hardiman CA, Weingarten RA, Conlan S, Khil P, Dekker JP, Mathers AJ, et al. Horizontal transfer of carbapenemase-encoding plasmids and comparison with hospital epidemiology data. Antimicrob Agents Chemother. 2016;60(8):4910–4919. doi: 10.1128/AAC.00014-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo Q, Spychala CN, McElheny CL, Doi Y. Comparative analysis of an IncR plasmid carrying armA, blaDHA-1 and qnrB4 from Klebsiella pneumoniae ST37 isolates. J Antimicrob Chemoth. 2016;71(4):882–886. doi: 10.1093/jac/dkv444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sugawara Y, Akeda Y, Hagiya H, Sakamoto N, Takeuchi D, Shanmugakani RK, et al. Spreading patterns of NDM-producing Enterobacteriaceae in clinical and environmental settings in Yangon, Myanmar. Antimicrob Agents Chemother. 2019;63(3):e01924–e01918. doi: 10.1128/AAC.01924-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tokajian S, Moghnieh R, Salloum T, Arabaghian H, Alousi S, Moussa J, et al. Extended-spectrum β-lactamase-producing Escherichia coli in wastewaters and refugee camp in Lebanon. Future Microbiol. 2018;13(1):81–95. doi: 10.2217/fmb-2017-0093. [DOI] [PubMed] [Google Scholar]
- 33.Collignon PJ, McEwen SA. One health—its importance in helping to better control antimicrobial resistance. Trop Med Inf Dis. 2019;4:22. doi: 10.3390/tropicalmed4010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma K, Feng Y, Liu L, Yao Z, Zong Z. A cluster of colistin-and carbapenem-resistant Klebsiella pneumoniae carrying blaNDM-1 and mcr-8.2. J Infect Dis. 2019;221(Supplement_2):S237–S242. doi: 10.1093/infdis/jiz519. [DOI] [PubMed] [Google Scholar]
- 35.WHO Advisory Group on Integrated Surveillance of Antimicrobial Resistance (AGISAR). Critically important antimicrobials for human medicine, 6th revision. Geneva: World Health Organization; 2019. Licence: CC BY-NC-SA 3.0 IGO. Document link: https://apps.who.int/iris/bitstream/handle/10665/312266/9789241515528-eng.pdf?ua=1.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Antimicrobial susceptibility and MIC results of K. pneumoniae k9. Antimicrobial susceptibility was performed using the disk diffusion technique; MICs of ertapenem, imipenem and meropenem were determined using the E-test methodology; MIC of colistin was determined using the broth microdilution method. Antibiotics were divided by their drug categories. GM: gentamycin 10μg; IP: imipenem; MEM: meropenem 10μg; IPM: imipenem 10μg; PM: cefepime; CTX: cefotaxime 30μg; CXM: cefuroxime 30μg; FEP: cefepime 30μg; CZD: ceftazidime 10μg; CZN: cefazolin 30μg; SCF: cefoperazone/sulbactam 75/30μg; FOX: cefoxitin 30μg; NOR: norfloxacin 5μg; CIP: ciprofloxacin 5μg; FAD: fusidic acid 10μg; TGC: tigecycline 15μg; CMN: clindamycin 2μg; ERY: erythromycin 15μg; ATM: aztreonam 30μg; LZD: linezolid 30μg; AMX: amoxicilin 25μg; PIR: pipercacillin 100μg; AMP: amipicillin 10μg; TZP: pipercacillin/tazobactam 100/10μg; FOS: fosfomycin 200μg; TE: tetracycline 30μg; TS: trimethoprim/sulfamethoxazole; CO: colistin; Dark blue: resistant, light blue: intermediate resistance, white: susceptible; MICs are in μg/mL; NA: not available.
Additional file 2: Figure S1. SNAP plots with potential synonymous and non-synonymous substitutions in pmrA (A) and pmrB (B). Alignment was performed against the query sequence of K. pneumoniae MGH 78578 wild-type chromosomal genes.
Data Availability Statement
This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession WTCT00000000. The version described in this paper is version WTCT01000000. The pk9 plasmid sequence was submitted to NCBI on 17/02/2020 under the submission number SUB6977004.