Abstract
The function of membrane-bound proteins often depends on their interactions with the lipid bilayer. Bulk absorption-based linear dichroism has been historically used to investigate molecular orientations in the phospholipid bilayer but cannot resolve the actual distribution of molecules embedded in the membrane and is often limited by a poor signal-to-noise ratio. Here, we present single-molecule orientation determination by fluorescence-detected linear dichroism visualization in Nanodisc grids or SOLVING, to determine the molecular orientation of molecules assembled into nanoscale lipid bilayers. We provide a proof-of-concept by using SOLVING to quantitate the orientation distribution of two commonly used fluorescent dyes, DiO and BODIPY, in 10 nm Nanodiscs. Besides confirming the mean orientation determined by bulk absorption measurement, SOLVING provides the actual distribution of orientations and promises to provide key molecular insights into the topology and interactions of multiprotein complexes, such as those observed in intracellular signal transduction.
Graphical Abstract
Nanodisc technology1-4 empowers quantitative investigation of the function of membrane-bound proteins and protein–lipid interactions, which have been shown to be responsible for orienting and stabilizing the proteins so that they can carry out their function in many processes ranging from blood coagulation5,6 to cancer signaling.7 Nanodiscs involve the self-assembly of phospholipids into a discoidal bilayer, stabilized by two encircling amphipathic membrane scaffold proteins. Membrane proteins can be incorporated into the bilayer in their native functional state for structural and biophysical studies. The monodisperse and homogeneous Nanodisc represents a well-organized lipid bilayer, wherein the lipid composition can be precisely controlled. As a result, Nanodiscs have also successfully been used as a membrane surface with precisely controlled composition for quantitating the interactions of peripheral membrane proteins with the bilayer.8, 9
The first step toward understanding macromolecule–lipid interactions is to define orientations in the lipid bilayer. We have previously used absorption-based linear dichroism to determine the orientation of a Nanodisc-embedded heme protein.10,11 This ensemble approach measures an average orientation but does not resolve the distribution of molecular orientation in Nanodiscs. To address this issue, we introduce single-molecule orientation determination by fluorescence-detected linear dichroism visualization in Nanodisc grids or SOLVING, to resolve the distribution of molecular orientation at the single-molecule level within individual Nanodiscs. This work builds upon previous work that used polarization-modulated, single-molecule detection to resolve molecular orientation within a crystal,12 immobilized on a solid substrate,13-15 or linked to a motor protein, such as myosin.16-18 Single-molecule measurements of membrane-bound proteins on lipid bilayers19,20 or Nanodiscs21,22 have also been reported, but these studies do not extract the molecular orientation of the studied entity. We also emphasize that orientation in this work refers to the angle a molecule makes with the lab z-axis rather than how the N- and C-termini of a protein extend either into the cytosol or extracellular environment in a cell membrane.
■ EXPERIMENTAL SECTION
Construction of TAMRA, DiO, and BODIPY-Labeled Nanodiscs.
Nanodiscs were prepared following standard protocols.23 MSP1D1 was used in all Nanodisc preps. His-tags were removed from the MSPs by tobacco etch virus (TEV) protease cleavage at 35 °C for 4 h in a water bath while agitating, followed by Ni-NTA affinity chromatography to isolate the processed MSP. To construct TAMRA-labeled MSP, MSP1D1(−), which lacks an N-terminal histidine tag, was labeled with an amine-reactive TAMRA dye (5- (and-6)-carboxytetramethylrhodamine, succinimidyl ester, AnaSpec). The dye was dissolved in DMSO at 30 mM and added in 10× molar excess to the MSP after reducing sulfhydryls with 4× molar excess tris(2-carboxyethyl)phosphine (TCEP) for 10 min. The reaction continued for 4 h while stirring and protected from light at room temperature and then allowed to continue at 4 °C overnight. Labeled MSP was purified from free dye and unlabeled protein via gel filtration on a G-25 column. Labeling efficiency was determined using the absorbances at 280 and 556 nm along with a correction factor.8 DiO (3,3′-dioctadecyloxacarbocyanine perchlorate), BODIPY FL C12 (4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-dodecanoic acid), and BODIPY FL DHPE (N-(4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-propionyl)-1,2-dihexadecanoyl-sn-glycero-3-phosphoethanol-amine, triethylammonium salt) were incorporated directly into Nanodiscs during self-assembly. Each dye was added in a 1:1 ratio of dye: Nanodisc. Concentrations of the dyes in the resulting Nanodiscs were measured on a Cary300 UV/vis spectrophotometer.
Imaging Sample Preparation.
Nanodisc imaging was carried out on flat 24 mm × 40 mm glass coverslips purchased from VWR (Cat: 48393230). Glass coverslips were first washed using 1% Alconox and rinsed under running Milli-Q water. The washed coverslips were stored in a coverslip rack immersed in Milli-Q water. Coverslips were then submerged in acetone and sonicated for 30 min. After the coverslip containing rack was rinsed in molecular grade water, the rack was then submerged in 3 M KOH solution, sonicated for 30 min and left undisturbed overnight. After rinsing the coverslips in fresh molecular grade water, the coverslips were submerged in fresh methanol and sonicated for 5 min. The rack with coverslips was finally transferred to a beaker containing fresh molecular grade water and stored until further use. Before imaging, a clean coverslip was air-dried and briefly flamed to remove any adsorbed residual contaminants. A PDMS chamber was carefully assembled on the coverslip in a laminar airflow cabinet. Ten microliters of 10 pM nanodiscs (estimated from the dye concentration) was loaded directly into the PDMS wells. All dilutions were made in the imaging buffer (20 mM HEPES, 150 mM NaCl, and 1 mM MgCl2, pH 7.3).
Single-Molecule Fluorescence Detection.
Objective-based total internal reflection fluorescence microscopy (TIRFM) was used for single-particle imaging. A continuous wavelength (488 or 561 nm, Spectral physics) laser was used as the light source. An inverted microscope (IX73, Olympus) equipped with a 100× oil immersion objective (Olympus, PlanApo, 100×, N.A. 1.49, oil immersion) was used. The laser beam was then expanded, collimated to about 35 mm, and directed into the microscope by a lens (400 mm focal length, Thorlabs LA1725A). Polarization of the incident light was continuously tuned by a half-wave plate mounted on a rotational stage (PI Instruments model DT-31), which sends a TTL pulse to trigger EMCCD acquisition. The incident light was directed through the objective via an exciter (FF01-482/563-25) and a dual-band dichroic filter (Di01-R488/561–25 × 36). The luminescence photons from individual Nanodiscs were collected by the same objective, passing an emitter (FF01-523/610-25) and captured by an electron multiplying charge coupled device (EMCCD) camera (iXon U797, Andor Technology). Individual fluorescently labeled Nanodiscs were detected as diffraction-limited spots. Imaging buffer with no Nanodiscs was used as negative controls.
Data Analysis of Single-Molecule Fluorescence Trajectories.
Videos of single-molecule fluorescence were processed using softwares maintained by the Zhang laboratory. The individual particle trajectories were obtained by selecting only features above an intensity cutoff provided by the user based on the observed signal. After video processing, trajectories were selected for further analysis by a combination of automatic filtering and manual inspection. Single-particle trajectories were analyzed using in-house MATLAB software. Semiautomated filtering of automatically selected particle trajectories used change-point analysis to identify bleaching events.24,25
The trajectories after filtering and manual selection were Fourier transformed to obtain the one-sided frequency spectrum of the signal. For a half-wave plate rotating at frequency ω, the polarization is modulated at a frequency of 4ω (a full p- to s- back to p- cycle occurs when the wave plate rotates 90°). Thus, the peak nearest this frequency yields the amplitude of the signal component due to the rotating wave plate.
The signal generated by the rotating half-wave plate is a sinusoid whose maxima and minima correspond to either p- or s-polarized light. To determine which peak corresponds to which polarization, we assumed that DiO absorbs s-polarized light more strongly than p-polarized light based on its geometry in the bilayer (hydrophobic alkyl chains are assumed to partition into the lipids, while the transition moment lies in the plane of the bilayer). These assumptions are supported by the literature26 and our observation. Since the camera exposure time and the DC-motor frequency are the same in all videos, and the TTL pulse triggers data collection at the same position of the half-wave plate each time, this allows us to discriminate which peak, maximal or minimal, corresponds to p- and s-polarization for any sample. See the Supporting Information for calculation of the dichroic ratio.
■ RESULTS AND DISCUSSION
Scheme of SOLVING.
The overall scheme of SOLVING consists of a polarization-tunable, objective-based total internal reflection fluorescence microscopy (TIRFM), with fluorescently labeled Nanodiscs to be deposited on a flat, thin glass coverslip. Data acquisition is triggered by a TTL pulse generated from the rotating half-wave plate optical mount so that the absolute light polarization can be precisely mapped to the time points of the fluorescence trajectory (Figure 1a). To calculate the dichroic ratio and orientation angle of single molecules, we modified previous work on TIRF-based absorption-based linear dichroism methodologies.27 Our configuration uses a rotating half-wave plate to modulate the incident polarization at a frequency 4ω (where ω is the rotational frequency of the motor driving the half-wave plate). Fourier analysis of the observed emission intensity allows us to recover the amplitude of the component of the signal oscillating at 4ω. This scheme allows for the determination of the sample absorbance under two orthogonal polarizations, TM and TE, also commonly referred to as p- and s-polarized light, relative to the surface normal (Figure S1). We did not detect any significant difference in the reflectivity of s- and p-polarized light in our setup (Figure S2).
Figure 1.
Scheme of instrumentation and theoretical framework. (a) Scheme of polarization-tunable, single-molecule TIRFM. The molecule is represented as a dipole pointing with an orientation angle θ away from the z-axis. (b) The relationship between the orientation angle θ and the dichroic ratio ρ, which is defined as the fluorescence intensity ratio between TE and TM excitation. The internal incident angle was estimated to be 80°.
The orientation angle θ can be derived from the dichroic ratio ρ, the ratio between fluorescence intensity from TE and TM excitation, given the amplitude of each evanescent field component. The measured dichroic ratio is expressed in terms of the evanescent electric field vectors as (see SI for derivation)27
where F is the fluorescence intensity, QY is the quantum yield of the fluorophore, and θ is the orientation angle. The relationship between the orientation angle and the dichroic ratio is shown in Figure 1b. The internal reflection angle was estimated to be 80° based on the numerical aperture of the objective (1.49) and the refractive index of the immersion oil (1.515).
Single-Molecule Detection of Fluorescently Labeled Nanodiscs.
To validate the feasibility of observing single Nanodiscs, we first prepared TAMRA-labeled Nanodiscs using a unique sulfhydryl labeling site provided by an aspartate to cysteine mutation at position 73 of the membrane scaffold protein (MSP) belt. The concentration of the dye–Nanodisc conjugate was adjusted so that approximately 200 diffraction-limited spots were observed in a 80 μm × 80 μm field of view (Figure 2a). The sample was then excited by a 561 nm laser using a 100× oil immersion TIRF objective. A total of 1000 time-stamped fluorescence images were acquired with an exposure time of 60 ms. Individual trajectories were reconstructed using a MATLAB script. Photobleaching of background-subtracted fluorescence trajectories was then confirmed by visual inspection (Figure 2b and 2c). The histogram of average intensity before photobleaching was presented in Figure 2d, which a primary peak at 8 Kcts/s and larger shoulder peaks around 15 Kcts/s, indicative of Nanodiscs labeled with two fluorescent MSP proteins. On the basis of the bulk absorption result, we estimated a 70% labeling efficiency of the encircling membrane scaffold protein (MSP) used to form the two belts around the Nanodisc. Considering that each Nanodisc contains two MSPs, we determined that the average number of TAMRA molecules on each Nanodisc is 1.4. Assuming a Poisson distribution for the labeling efficiency where u is the mean labeling efficiency, and x is the actual labeling efficiency, we expect that the ratio between the number of single-step and double-step photobleaching trajectories should be . Using single-molecule fluorescence measurements, we obtained a ratio of 1.45 (Figure 2e), consistent with the predicted value. This result serves as the quality control of our protocol to prepare and detect single Nanodiscs on the glass substrate.
Figure 2.
(a) Representative image of TAMRA-labeled Nanodiscs (left) and imaging buffer (right). Samples were excited using 561 nm laser with 9.14 mW power and an exposure time of 60 ms. (b) Time-stamped montage of TAMRA-labeled Nanodiscs. (c) Representative background-subtracted trajectories of TAMRA-labeled Nanodiscs showing single (top) and double-step (bottom) photobleaching. (d) The intensity distribution of individual TAMRA-labeled Nanodiscs (N = 250). (e) Distribution of bleaching steps in the individual traces. Percentage of occurrence based on single, double and multiple bleaching step events (N = 1374). Scale bar: 5 μm.
Characterizing the Molecular Orientation of Single Fluorophores in a Nanodisc.
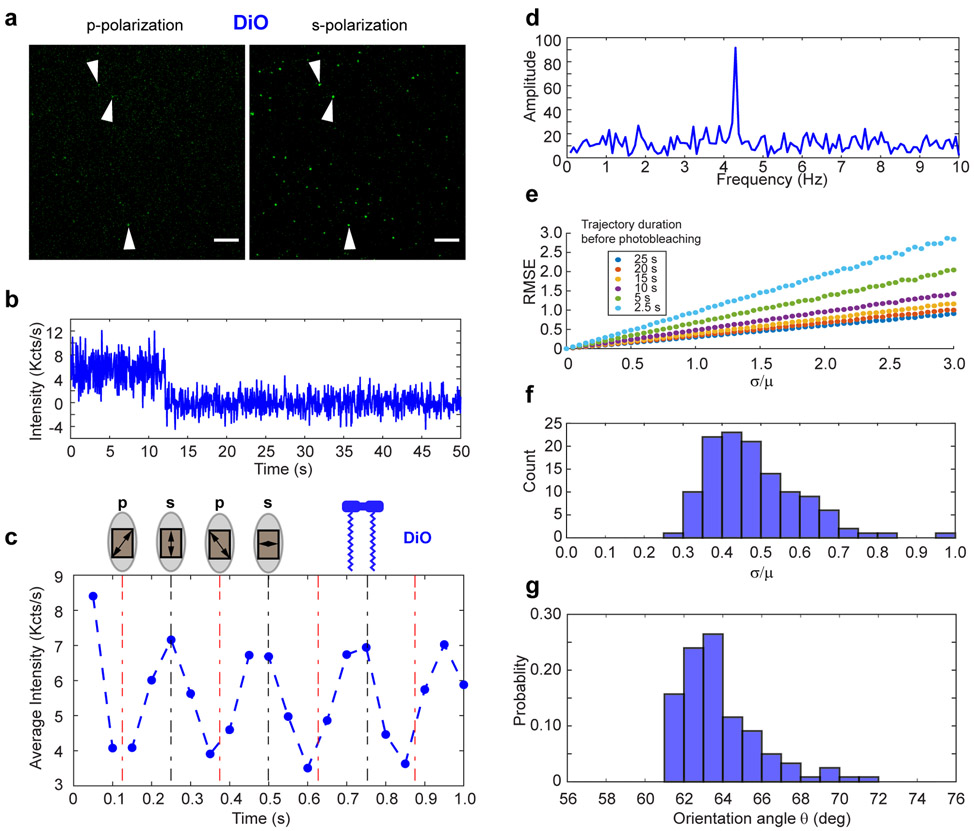
We prepared Nanodiscs labeled with DiO, whose alkyl tails should be anchored in the bilayer. We controlled the stoichiometry between Nanodiscs and DiO so that, on average, there was one DiO molecule in a single Nanodisc. DiO molecules were excited by a 488 nm laser beam, which was modulated by the zero-order quartz half-wave plate (HWP) cut for 488 nm mounted on a motorized rotation stage. A DC voltage of 4.4 V was applied to drive the motor and HWP to rotate at ~1 Hz, which resulted in an approximately 4 Hz frequency for a p-s-p transition on the emission signal. A total of 1000 time-stamped fluorescence images were acquired with an exposure time of 50 ms.
Figure 3a shows DiO Nanodiscs imaged under p- and s-polarized light within the same field of view. Spots at identical positions display significantly higher intensity under s-polarized light excitation. On average the fluorescence undergoes photobleaching within 10–15 s (Figure 3b). Figure 3c shows average fluorescence intensities from 100 particles over time with the positions of the maxima and minima and their corresponding positions of the fast axis of the wave plate. Inspection of averaged trajectories of several DiO-Nanodisc particles indicates that the first valley observed corresponds to p-polarization incident on the sample since the emission intensity of DiO-Nanodisc particles is a minimum here. The Fourier-transform of fluorescence trajectory (Figure 3d) successfully recovered the frequency component at 4.2 Hz. Since the chemically similar dye DiI is known to orient itself by burying its alkyl chains into the phospholipid bilayer,26 s-polarized light is expected undergo stronger absorption than p-polarized light (for a discussion of the relationship between the position of the wave plate and the polarization incident on the sample in our instrument, see the Figure S1). We verified that DiO indeed absorbs s-polarized light more strongly (Figure S3).
Figure 3.
(a) A snapshot showing DiO incorporated into DMPC Nanodiscs illuminated with p- (left) and s- (right) polarized light. Images were acquired using a 488 nm laser at 1.44 mW power and exposure of 50 ms. Scale bar: 5 μm. (b) The background-subtracted intensity trajectory of a single DiO-labeled Nanodisc. (c) DiO fluorescence trajectory under continuous polarization modulation averaged over 100 DiO-labeled Nanodiscs. The maxima occur when s-polarization is incident on the molecules, indicating that DiO is oriented along or near the plane of the bilayer surface. (d) DiO Fluorescence in the frequency domain. The same molecule as in panel b plotted as a one-sided spectrum using the fluorescence signal up to the bleaching event. The peak occurs at ~4 times the frequency of the motor. (e) Simulated root mean square error (RMSE) between the recovered and measured orientation angle of a single molecule for different trajectory durations. The recovered angle was computed by corrupting the real, continuously modulated intensity with added Gaussian noise of varying magnitude (σ refers to the standard deviation of the Gaussian noise; μ is the average intensity of the simulated fluorescence signal). Each data point was averaged over 5000 simulations with resampled Gaussian noise. (f) Noise statistics for DiO-Nanodiscs. A histogram showing the ratio of the standard deviation to the mean of all DiO-Nanodisc molecules. The predominance of values near 0.5 indicates that the RMSE between our measured angles and the true angles rarely exceeds 0.5°. (g) The probability distribution function of the orientation angle of DiO in Nanodiscs recovered by SOLVING (N = 121).
The acquired data is fluorescence intensity as a function of time as modulated by the rotating half-wave plate. To validate our analysis and identify optimal instrumental settings for imaging, simulations were performed using different levels of Gaussian noise imposed on a sinusoidal signal. The random noise was chosen to model the total uncertainty on our detector. The amplitude and offset of the real signal were chosen to reflect observed data during previous experiments. Analysis of simulated signals with various levels of noise relative to the mean signal intensity is shown in Figure 3e. Fourier analysis of even relatively short signals (as short as 2.5 s) with a 50 ms exposure time leads to a systematic error of no more than 1 degree between the recovered and true orientation angle (Figure 3f), assuming the magnitude of the noise equals the mean signal intensity. This feature is important since the photobleaching of many particles often occurs within the first few seconds of acquisition. Thus, our analysis allows us to maximize the number of useful fluorescent spots in a given image stack.
The distribution of orientation angles from SOLVING is shown as a histogram in Figure 3g. We measured the average molecular orientation of the absorption transition dipole to be 61.9° ± 2.1° (Mean ± SD, where SD is the sample standard deviation and depicts the distribution of the orientation angle). The average orientation angle is in excellent agreement with the published value of the chemically similar DiI in erythrocyte ghosts26 and our absorbance-based measurements of DiO in Nanodiscs using an ensemble-averaged slab waveguide setup. For three technical replicates with 500 nM of purified DiO-labeled DMPC Nanodiscs, absorbance-based measurement gave the orientation angle as 61.0° ± 0.7° (Mean ± SD, SD represents the standard deviation of three technical replicates instead of orientation angle distribution). In contrast to the absorbance method, SOLVING allows us to recover the shape of the distribution of orientation angles, which is invisible to ensemble methods.
Determination of the Molecular Orientation of Fluorescent Lipid Molecules.
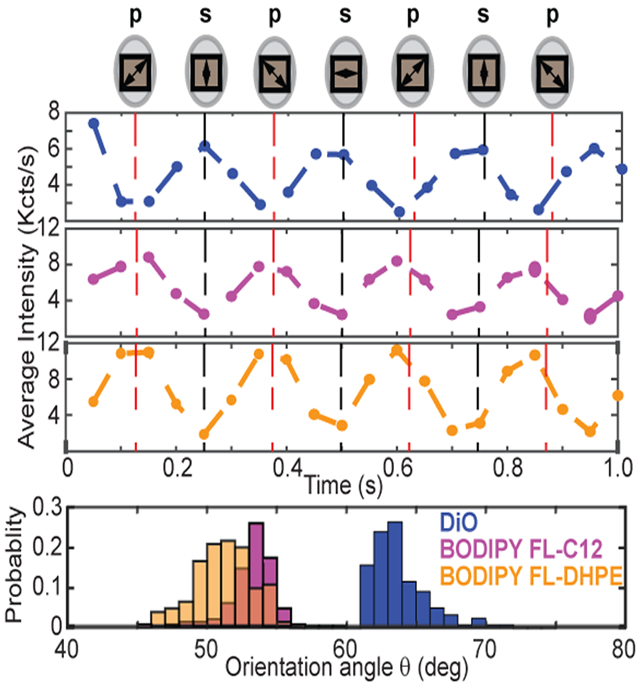
To determine if SOLVING is effective in resolving distinct molecular orientation in the lipid membrane, we also imaged two fluorescently labeled lipid molecules incorporated into Nanodiscs during self-assembly: BODIPY FL C12 and BODIPY FL DHPE. The C12 label is attached at the end of an alkyl chain, while the DHPE label is attached to a modified ethanolamine group on a phospholipid. BODIPY is expected to be parallel to the lipid tails (as opposed to parallel to the bilayer surface like DiO), but their orientation has not been directly studied in Nanodiscs at the single-molecule level. A quick examination of BODIPY-labeled Nanodiscs revealed an opposite trend from DIO-labeled ones—the fluorescence intensity is significantly stronger under p-polarized light excitation (Figure 4a). Typical photobleaching time remains at approximately 10 s (Figure 4b). We aligned 100 traces of the C12 and DHPE samples with the position of the wave plate (Figure 4c). The data unambiguously confirm that BODIPY-labeled lipids absorb p-polarized light more than s-polarized light, indicating that these absorbers are parallel to the lipid tails. Similar to the observation of DiO, Fourier transform of the trajectory recovered the frequency component at 4.2 Hz (Figure 4d). As an additional check on our method, we computed the ratio of the standard deviation to the mean signal intensity for DiO (Figure 3f) and BODIPY (Figure 4e) and found that the ratio never exceeded 1.0. Additionally, all particles analyzed in this work had at least 2.5 s of emission before photobleaching. These empirical observations combined with our simulation data (Figure 3e) suggest that the error on our measurement of the dipole moment angle rarely, if ever, exceeded 1° in magnitude. The distributions of orientation angle for the C12 and DHPE samples are shown in Figure 4f. The average orientation angle of the C12 samples is 53.1° ± 1.7° (mean ± SD), and the DHPE average is 51.2° ± 2.0° (mean ± SD). Note SD represents the distribution of the orientation angle of the measured molecules. Whereas BODIPY FL C12 and BODIPY FL DHPE show similar orientation angle, they both orient distinctly differently from DiO. These results agree with our previous bulk fluorescence-detected linear dichroism measurements of DiI and BODIPY-labeled lipids in Nanodiscs.28 While a detailed analysis of the exact position of the transition dipole relative to the constituent chemical bonds in the dye is beyond the scope of this work, the results support a model in which the BODIPY-labeled lipids in Nanodiscs are oriented parallel to the lipid tails, in contrast to DiO which is parallel to the bilayer surface.
Figure 4.
Resolving the molecular orientation of BODIPY in Nanodiscs. (a) Snapshots showing BODIPY-labeled lipids incorporated into DMPC Nanodiscs. Top row: BODIPY FL-C12 (BODIPY label on the end of the fatty acid within the bilayer). Bottom row: BODIPY FL-DHPE (BODIPY label conjugated to an ethanolamine headgroup on a phospholipid). Samples are shown illuminated with p- (left) and s- (right) polarized light. Imaging parameters are the same as described in Figure 3a. Scale bar: 5 μm. (b) Single-molecule fluorescence of BODIPY-labeled lipids. Emission intensity vs time for a single BODIPY FL-C12 (top) and a single BODIPY FL-DHPE (bottom) incorporated into Nanodiscs. (c) BODIPY fluorescence under continuous polarization modulation. The averaged intensity of 100 BODIPY-labeled Nanodiscs (top, BODIPY FL-C12; bottom, BODIPY FL-DHPE). The maxima appear at different times compared with DiO molecules (see Figure 3c). (d) The same molecules as in panel b plotted in the frequency domain. (e) Noise statistics for BODIPY-Nanodiscs. The same noise statistics as described in Figure 3f computed from all BODIPY FL-C12 particles (top) and all BODIPY FL-DHPE particles (bottom). (f) The probability distribution function of the orientation angle of BODIPY-labeled lipids in Nanodiscs. Normalized histograms of BODIPY FL-C12 (top, N = 113) and BODIPY FL-DHPE (bottom, N = 131) orientation angles recovered by SOLVING.
Besides polarization modulation, there are many other single-molecule detection approaches to resolve molecular orientation, such as those based on defocusing,29,30 point spread function (PSF) engineering,31 annular illumination,32 or direct imaging of emission patterns at the back aperture plane of the objective.33 While PSF or aberration-based imaging can determine a molecule’s 3D orientation,29,34-37 these methods require fitting a spatial intensity pattern to a model, and the camera’s pixel size will influence the resolution of these methods. In contrast, the resolution of our method does not depend on pixel size nor does it require defocusing. Instead, our method uses only the total intensity at the focal plane, resulting in higher contrast and simplified data analysis. Biological membranes also display lateral symmetry, and the most physiologically relevant information is likely contained in the molecules’ 2D orientation. Compared with the bulk assay, SOLVING provides a means to directly measure the probability distribution of molecular orientation within single Nanodiscs. Although we demonstrated SOLVING with an objective-based TIRF setup, a similar measurement can be done on a prism-based TIRF microscope as well.
■ CONCLUSION
In summary, we combine single-molecule detection with Nanodisc technology to resolve the molecular orientation of molecules in lipids. Taking advantages of the Nanodisc technology, we expect SOLVING can be conveniently generalized to other applications. For instance, SOLVING could be used to determine the effects of lipid composition on molecular orientation. By constructing Nanodiscs with different lipid components, one can visualize how charge (e.g., by doping phospholipids) or hydrophobicity affects the molecular orientation of a membrane-bound protein. Such a capacity is critical in assessing membrane’s role in modifying the activation state (or signaling outcome) of signaling molecules, such as Ras, a GTPase at the center of major cellular growth and differentiation pathways, which has gained renewed interest as a potential drug target in cancer treatment.38
Another potential application involves the determination of molecular motions and kinetics within the membrane. Although it has been postulated, and shown by computation, that membrane-bound molecules undergo constant movement, direct visualization of molecular orientation within the membrane at the single-molecule level have been challenging. Single-molecule Forster resonant energy transfer (smFRET) has recently been used to determine molecular motions39 but does not provide the molecular orientation. Compared with FRET, which requires simultaneous labeling of two fluorophores, SOLVING uses one fluorophore with the easier design of instrumentation. Of course, SOLVING can be combined with multicolor labeling and smFRET to achieve multimodal analysis that could provide kinetic measurement of both intramolecular conformational changes and molecular orientation.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Dr. Mark McLean for consultation and materials for the TAMRA-labeled Nanodisc experiments. This work was supported by NIH grant number NIH R35 GM118145 (S.S.) and R01GM132438 (K.Z.), as well as the School of Molecular and Cellular Biology at the University of Illinois at Urbana–Champaign (K.Z.)
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.9b04950.
Scheme for the conversion of incident polarization and the direction of the electric field at the sample plane, supplementary methods including reagents, calculation of the dichroic ratio, absorbance-based linear dichroism, the relationship between the dichroic ratio and evanescent electric field components, and computer simulation of sampling rate (PDF)
The authors declare no competing financial interest.
■ REFERENCES
- (1).Bayburt TH; Sligar SG FEBS Lett. 2010, 584, 1721–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Ritchie TK; Grinkova YV; Bayburt TH; Denisov IG; Zolnerciks JK; Atkins WM; Sligar SG, Chapter Eleven - Reconstitution of Membrane Proteins in Phospholipid Bilayer Nanodiscs In Methods in Enzymology; Duzgunes N, Ed.; Academic Press, 2009; Vol. 464, pp 211–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Denisov IG; Sligar SG Chem. Rev 2017, 117, 4669–4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Denisov IG; Sligar SG Nat. Struct. Mol. Biol 2016, 23, 481–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Zwaal RF; Comfurius P; Bevers EM Biochim. Biophys. Acta, Rev. Biomembr 1998, 1376, 433–453. [DOI] [PubMed] [Google Scholar]
- (6).Morrissey JH; Davis-Harrison RL; Tavoosi N; Ke K; Pureza V; Boettcher JM; Clay MC; Rienstra CM; Ohkubo YZ; Pogorelov TV; Tajkhorshid E Thromb. Res 2010, 125 (Suppl 1), S23–S25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Sunshine H; Iruela-Arispe ML Curr. Opin. Lipidol 2017, 28, 408–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Ye X; McLean MA; Sligar SG Biochim. Biophys. Acta, Biomembr 2016, 1858, 1833–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Gregory MC; McLean MA; Sligar SG Biochem. Biophys. Res. Commun 2017, 487, 351–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Mustafa G; Nandekar PP; Camp TJ; Bruce NJ; Gregory MC; Sligar SG; Wade RC Biophys. J 2019, 116, 419–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Baylon JL; Lenov IL; Sligar SG; Tajkhorshid EJ Am. Chem. Soc 2013, 135, 8542–8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Guttler F; Sepiol J; Plakhotnik T; Mitterdorfer A; Renn A; Wild UP J. Lumin 1993, 56, 29–38. [Google Scholar]
- (13).Xie XS; Dunn RC Science 1994, 265, 361–364. [DOI] [PubMed] [Google Scholar]
- (14).Ha T; Laurence TA; Chemla DS; Weiss SJ Phys. Chem. B 1999, 103, 6839–6850. [Google Scholar]
- (15).Ha T; Enderle T; Chemla S; Selvin R; Weiss S Phys. Rev. Lett 1996, 77, 3979–3982. [DOI] [PubMed] [Google Scholar]
- (16).Forkey JN; Quinlan ME; Shaw MA; Corrie JE; Goldman YE Nature 2003, 422, 399–404. [DOI] [PubMed] [Google Scholar]
- (17).Peterman EJG; Sosa H; Moerner WE Annu. Rev. Phys. Chem 2004, 55, 79–96. [DOI] [PubMed] [Google Scholar]
- (18).Sase I; Miyata H; Ishiwata S; Kinosita K Proc. Natl. Acad. Sci. U. S. A 1997, 94, 5646–5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Knight JD; Lerner MG; Marcano-Velazquez JG; Pastor RW; Falke JJ Biophys. J 2010, 99, 2879–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Thompson NL; Drake AW; Chen L; Vanden Broek W Photochem. Photobiol 1997, 65, 39–46. [DOI] [PubMed] [Google Scholar]
- (21).Nath A; Trexler AJ; Koo P; Miranker AD; Atkins WM; Rhoades E, Single-Molecule Fluorescence Spectroscopy Using Phospholipid Bilayer Nanodiscs. Methods in Enzymology: Single Molecule Tools, Pt A: Fluorescence Based Approaches, 2010; Vol. 472, pp 89–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Liu Y; Liu Y; He L; Zhao Y; Zhang XC Biophysics Reports 4, 153–165. [Google Scholar]
- (23).Ritchie TK; Grinkova YV; Bayburt TH; Denisov IG; Zolnerciks JK; Atkins WM; Sligar SG Methods Enzymol. 2009, 464, 211–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Li H; Yang HJ Phys. Chem. B 2019, 123, 689–701. [DOI] [PubMed] [Google Scholar]
- (25).Watkins LP; Yang HJ Phys. Chem. B 2005, 109, 617–628. [DOI] [PubMed] [Google Scholar]
- (26).Axelrod D Biophys. J 1979, 26, 557–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Cropek DM; Bohn PW J. Phys. Chem 1990, 94, 6452–6457. [Google Scholar]
- (28).Bayburt TH; Sligar SG Protein Sci. 2003, 12, 2476–24781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Toprak E; Enderlein J; Syed S; McKinney SA; Petschek RG; Ha T; Goldman YE; Selvin PR Proc. Natl. Acad. Sci. U. S. A 2006, 103, 6495–6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Patra D; Gregor I; Enderlein J J. Phys. Chem. A 2004, 108, 6836–6841. [Google Scholar]
- (31).Pavani SRP; Thompson MA; Biteen JS; Lord SJ; Liu N; Twieg RJ; Piestun R; Moerner WE Proc. Natl. Acad. Sci. U. S. A 2009, 106, 2995–2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Sick B; Hecht B; Novotny L Phys. Rev. Lett 2000, 85, 4482–4485. [DOI] [PubMed] [Google Scholar]
- (33).Lieb MA; Zavislan JM; Novotny L J. Opt. Soc. Am. B 2004, 21, 1210–1215. [Google Scholar]
- (34).Armendariz KP; Huckabay HA; Livanec PW; Dunn RC Analyst 2012, 137, 1402–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Huckabay HA; Dunn RC Langmuir 2011, 27, 2658–66. [DOI] [PubMed] [Google Scholar]
- (36).Livanec PW; Dunn RC Langmuir 2008, 24, 14066–14073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Livanec PW; Huckabay HA; Dunn RC J. Phys. Chem. B 2009, 113, 10240–10248. [DOI] [PubMed] [Google Scholar]
- (38).Stephen AG; Esposito D; Bagni RK; McCormick F Cancer Cell 2014, 25, 272–281. [DOI] [PubMed] [Google Scholar]
- (39).Prakash P; Litwin D; Liang H; Sarkar-Banerjee S; Dolino D; Zhou Y; Hancock JF; Jayaraman V; Gorfe AA Biophys. J 2019, 116, 179–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.