Abstract
Background
Sex-related genes play a crucial role in gonadal differentiation into testes or ovaries. However, the genetic control of gonadal differentiation in Muscovy ducks remains unknown. Therefore, the objective of our study was to screen new candidate genes associated with ovarian and testicular development.
Results
In this study, 24 males before gonadal differentiation (MB), 24 females before gonadal differentiation (FB), 24 males after gonadal differentiation (MA) and 24 females after gonadal differentiation (FA) were selected from Putian Muscovy ducks, forming 4 groups. RNA-Seq revealed 101.76 Gb of clean reads and 2800 differentially expressed genes (DEGs), including 46 in MB vs FB, 609 in MA vs FA, 1027 in FA vs FB, and 1118 in MA vs MB. A total of 146 signalling pathways were enriched by KEGG analysis, among which 20, 108, 108 and 116 signalling pathways were obtained in MB vs FB, MA vs MB, MA vs FA and FA vs FB, respectively. In further GO and KEGG analyses, a total of 21 candidate genes related to gonad differentiation and development in Muscovy ducks were screened. Among these, 9 genes were involved in the differentiation and development of the testes, and 12 genes were involved in the differentiation and development of the ovaries. In addition, RNA-Seq data revealed 2744 novel genes.
Conclusions
RNA-Seq data revealed 21 genes related to gonadal differentiation and development in Muscovy ducks. We further identified 12 genes, namely, WNT5B, HTRA3, RSPO3, BMP3, HNRNPK, NIPBL, CREB3L4, DKK3, UBE2R2, UBPL3KCMF1, ANXA2, and OSR1, involved in the differentiation and development of ovaries. Moreover, 9 genes, namely, TTN, ATP5A1, DMRT1, DMRT3, AMH, MAP3K1, PIK3R1, AGT and ADAMTSL1, were related to the differentiation and development of testes. Moreover, after gonadal differentiation, DMRT3, AMH, PIK3R1, ADAMTSL1, AGT and TTN were specifically highly expressed in males. WNT5B, ANXA2 and OSR1 were specifically highly expressed in females. These results provide valuable information for studies on the sex control of Muscovy ducks and reveal novel candidate genes for the differentiation and development of testes and ovaries.
Keywords: Muscovy ducks, Sex differentiation, Transcriptome, Sex-related genes, Gonad
Background
Sex in birds and mammals is governed by sex chromosomes [1], and sex determination genes on sex chromosomes initiate sex differentiation and regulate the differentiation of gonads into ovaries or testes [2]. A host of critical gonadal differentiation genes have been detected [3, 4]. However, screening for novel regulators of male and female embryos is highly necessary to enhance our understanding of gonadal differentiation. The duck embryo is an excellent model for studying gonadal development because development takes place in ovo, and we can intervene directly in gonadal development [5]. In ducks and other avian species, there is a female heterogametic system of ZZ/ZW sex chromosomes [6], and their sex may be governed by genes on the W chromosome [7]. Studies have found that the key Z-linked regulator of testicular morphogenesis is doublesex and mab-3 related transcription (DMRT1) in birds. In addition, studies have found that the mechanism of sex differentiation in birds is different from that in mammals [6, 8]. In mammals, there is a male heterogametic system of XX/XY sex chromosomes. The male is determined by a dominant gene on Y (SRY), which triggers a series of events that lead to testicular development [9]. Nevertheless, the testis-determining gene, Sex-determining Region on the Y Chromosome (SRY), is absent in ducks [9]. The W chromosome contains few genes and seems to have undergone independent degradation in different bird populations [10, 11]. The exact role of avian sex-determining genes on Z and W has yet to be completely resolved.
In chickens, aromatase is a key developmental switch in sex determination [12], and oestrogen synthesis by the gonad is female-specific [13]. Various enzymes are related to the biosynthesis of oestrogen [14]. At the rate-limiting step, the synthesis of oestrogens occurs via the aromatization of androgens. This reaction is catalysed by a complex of enzymes including the cytochrome P450 aromatase, the product of the cytochrome P450 family 19 subfamily A member 1 (CYP19A1) gene, and a NADPH-dependent cytochrome P450 reductase in a wide variety of tissues, including ovary and testis tissue [15–17]. In addition, aromatase inhibitor can induce female conversion into males in Muscovy ducks [18]. Thus, aromatase may play a role in testicular and ovarian differentiation in Muscovy ducks.
In Muscovy duck embryos, the nephridium appears in a transverse section at E4. In that area, 3–6 layers of cells are observed from the splanchnic layers to the coelom corner. The genital ridge is observed at E6. At E7 and E8, the initial shape of the gonad emerges. At E9, gonads initiate differentiation into spermatogonia or spermatocytes. In addition, several identified genes show sexually dimorphic expression, such as DMRT1, anti-Mullerian hormone (AMH), and oestrogen receptor-α (ESR-α) [19]. However, in Muscovy ducks, the sex differentiation mechanisms underlying reproductive regulation remain unknown.
RNA-Seq provides general knowledge about the complete set of transcripts in a particular cell and identifies a number of phenotype-associated DEGs. Research in chickens has shown that the robust sexually dimorphic gene expression of both sex-linked and autosomal genes was detected by RNA-Seq in tissues pre-dating gonadogenesis [20]. In domestic ducks, several candidate genes involved in the development of plumulaceous feather and flight feather structures were obtained by RNA-Seq [21]. Research in Peking ducks has revealed that a key regulator of duck follicle development in the laying period [3]. In addition, RNA-Seq is also widely used to explore the mechanism of sex differentiation. For example, research in chickens revealed that the left-right asymmetry mechanism could be detected in female gonads by RNA-Seq [22]. Researchers have focused on sex differentiation in chickens, but transcriptomic analysis of the sex differentiation mechanism in Muscovy ducks has rarely been reported. Therefore, we used RNA-Seq to study the sex differentiation mechanism of Muscovy ducks and analysed the mRNA expression levels of gonadal tissues at two developmental time points in female and male Muscovy ducks. This study allowed us to further unravel the transcriptome of Muscovy ducks and provided valuable information for studying sex control in Muscovy duck.
Results
Overview of RNA-Seq results
In order to identify gonadal differentiation genes in Muscovy ducks, transcriptome analysis of Muscovy duck gonads was performed on MB, FB, MA, and FA. The main RNA-Seq results are listed in Tables 1, Tables 2, and Fig. 1. RNA-Seq yielded from 47,282,572 to 70,602,322 clean reads with more than 89% Q30 bases from each library; 47.36–52.117% of the clean reads were uniquely mapped to the Anas platyrhynchos genome (URL:https://www.ncbi.nlm.nih.gov/genome/?term=Anas+platyrhynchos). The mapping rate was much lower than that of chickens (79%) [23]. It was similar to Peking duck (42.27%) [24] and Jinding duck (55.9%) [25]. Given that RNA-Seq produced 47,282,572 to 70,602,322 clean reads, as shown by the percentage of Q30 bases (Table 1), we suspected that the main reason for the low mapping rates might be the poor quality of the currently published duck reference genome assembly [26]. However, on the basis of the criteria reported by Conesa [27] and Martin [28], the number of mapped reads was still sufficient to reconstruct intact transcripts and reliably quantify expression levels for most medium-abundance and high-abundance genes [25]. The gene structure of each library was analysed, and approximately 87% of the mapped reads were aligned to the exon region of the Anas platyrhynchos genome, followed by the intergenic and intron regions (Table 2).
Table 1.
Summary of the RNA-Seq data collected from MB, FB, MA and FA
| Sample name | Raw reads | Clean reads | Clean bases | Error rate (%) | Q20 (%) | Q30 (%) | GC content (%) |
|---|---|---|---|---|---|---|---|
| FB1 | 73,297,772 | 70,602,322 | 10.59G | 0.02 | 95.93 | 90.23 | 49.71 |
| FB2 | 60,975,078 | 58,772,554 | 8.82G | 0.02 | 95.84 | 89.93 | 50.49 |
| FB3 | 59,397,884 | 57,157,288 | 8.57G | 0.02 | 95.50 | 89.34 | 49.85 |
| MB1 | 52,554,762 | 51,693,516 | 7.75G | 0.02 | 96.64 | 91.85 | 51.42 |
| MB2 | 62,138,482 | 59,945,236 | 8.99G | 0.02 | 95.04 | 88.28 | 50.22 |
| MB3 | 55,490,528 | 54,592,060 | 8.19G | 0.02 | 96.71 | 91.92 | 50.50 |
| FA1 | 58,175,840 | 56,009,556 | 8.40G | 0.02 | 95.68 | 89.64 | 51.50 |
| FA2 | 50,554,446 | 48,793,882 | 7.32G | 0.02 | 95.61 | 89.44 | 51.29 |
| FA3 | 61,510,464 | 59,329,652 | 8.90G | 0.02 | 95.52 | 89.26 | 50.95 |
| MA1 | 48,763,588 | 47,282,572 | 7.09G | 0.02 | 95.57 | 89.32 | 51.52 |
| MA2 | 58,272,482 | 57,132,880 | 8.57G | 0.02 | 97.03 | 92.59 | 51.42 |
| MA3 | 58,878,702 | 57,142,740 | 8.57G | 0.02 | 95.52 | 89.25 | 50.96 |
Table 2.
Summary of clean reads mapped from MB, FB, MA and FA to the reference genome
| Sample name | Total reads | Total mapped | Multiple mapped | Uniquely mapped | Reads mapped to ‘+’ | Reads mapped to ‘-’ |
|---|---|---|---|---|---|---|
| FB1 | 70,602,322 | 34,506,109 (48.87%) | 593,713 (0.84%) | 33,912,396 (48.03%) | 17,038,856 (24.13%) | 16,873,540 (23.9%) |
| FB2 | 58,772,554 | 29,828,795 (50.75%) | 519,611 (0.88%) | 29,309,184 (49.87%) | 14,715,210 (25.04%) | 14,593,974 (24.83%) |
| FB3 | 57,157,288 | 27,739,072 (48.53%) | 471,088 (0.82%) | 27,267,984 (47.71%) | 13,689,588 (23.95%) | 13,578,396 (23.76%) |
| MB1 | 51,693,516 | 25,653,470 (49.63%) | 426,958 (0.83%) | 25,226,512 (48.8%) | 12,670,333 (24.51%) | 12,556,179 (24.29%) |
| MB2 | 59,945,236 | 30,339,732 (50.61%) | 510,473 (0.85%) | 29,829,259 (49.76%) | 14,955,979 (24.95%) | 14,873,280 (24.81%) |
| MB3 | 54,592,060 | 28,392,840 (52.01%) | 473,745 (0.87%) | 27,919,095 (51.14%) | 14,024,427 (25.69%) | 13,894,668 (25.45%) |
| FA1 | 56,009,556 | 27,021,161 (48.24%) | 493,654 (0.88%) | 26,527,507 (47.36%) | 13,317,028 (23.78%) | 13,210,479 (23.59%) |
| FA2 | 48,793,882 | 24,294,041 (49.79%) | 453,668 (0.93%) | 23,840,373 (48.86%) | 11,963,536 (24.52%) | 11,876,837 (24.34%) |
| FA3 | 59,329,652 | 28,914,663 (48.74%) | 525,602 (0.89%) | 28,389,061 (47.85%) | 14,257,697 (24.03%) | 14,131,364 (23.82%) |
| MA1 | 47,282,572 | 24,015,491 (50.79%) | 433,496 (0.92%) | 23,581,995 (49.87%) | 11,827,474 (25.01%) | 11,754,521 (24.86%) |
| MA2 | 57,132,880 | 30,326,910 (53.08%) | 553,848 (0.97%) | 29,773,062 (52.11%) | 14,950,119 (26.17%) | 14,822,943 (25.94%) |
| MA3 | 57,142,740 | 28,317,706 (49.56%) | 498,305 (0.87%) | 27,819,401 (48.68%) | 13,965,413 (24.44%) | 13,853,988 (24.24%) |
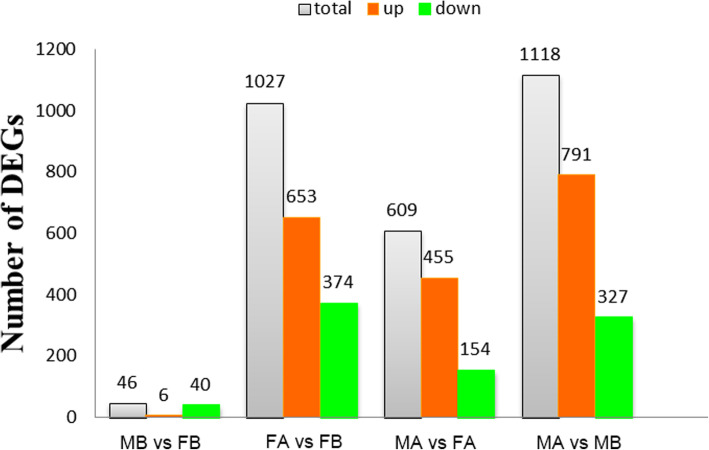
Fig. 1.
The number of differentially expressed genes between the comparison libraries. Total DEGs (grey), up-regulated genes (red), and down-regulated genes (green) are presented by a histogram
Significantly differentially expressed transcripts and clustering
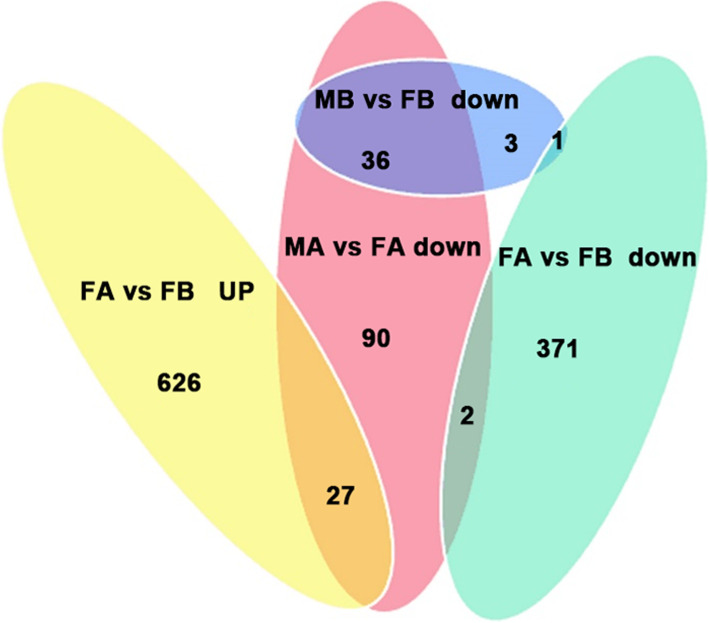
In the comparison of MB vs FB, 40 DEGs were down-regulated and 6 DEGs were up-regulated. In the comparison of FA vs FB, 374 DEGs were down-regulated, and 653 DEGs were up-regulated. In the comparison of MA vs FA, 154 DEGs were down-regulated, and 455 DEGs were up-regulated. In the comparison of MA vs MB, 327 DEGs were down-regulated, and 791 DEGs were up-regulated. This result may be due to the dimorphic expression of sex differentiation genes after gonadal differentiation (Fig. 1). To more precisely examine DEGs, a Venn diagram analysis revealed a total of 350 upregulated genes in MA vs FA, such as ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1 (ATP5A1), doublesex and mab-3 related transcription factor 3 (DMRT3), AMH, a disintegrin and metalloproteinase with thrombospondin motif like 1 (ADAMTSL1), titin (TTN), phosphoinositide-3-kinase regulatory subunit 1 (PIK3R1), angiotensinogen (AGT), DMRT1 and mitogen-activated protein kinase kinase kinase 1 (MAP3K1). Among these, ATP5A1 was upregulated in both the MB vs FB and MA vs FA comparisons. A total of 96 genes were upregulated in both the MA vs FA and MA vs MB comparisons, including DMRT3, AMH, ADAMTSL1, PIK3R1 and TTN (Fig. 2). By comparing the downregulated genes in MB vs FB with those in MA vs FA, 36 identical genes were identified, including heterogeneous nuclear ribonucleoprotein K (HNRNPK), nipped-B-like protein (NIPBL), cyclic AMP-responsive element-binding protein 3-like protein 4 (CREB3L4), ubiquitin-conjugating enzyme E2 R2(UBE2R2) and E3 ubiquitin-protein ligase KCMF1 (UBL3KCMF1). In addition, the comparison between the downregulated genes in MA vs FA and the upregulated genes in FA vs FB revealed a total of 27 identical genes, among which HtrA serine peptidase 3 (HTRA3), odd-skipped related transcription factor 1 (OSR1), Wnt family member 5B (WNT5B), and annexin A2 (ANXA2) were identified (Fig. 3). Hierarchical clustering was performed in MB, FB, MA and FA. The expression level of DEGs did not greatly change between FB and MB. This finding indicates that many sex-related genes have not yet been expressed prior to sex differentiation. Interestingly, the DEGs produced different clustering results in MA vs FA, which showed sexually dimorphic expression (Fig. 4).
Fig. 2.
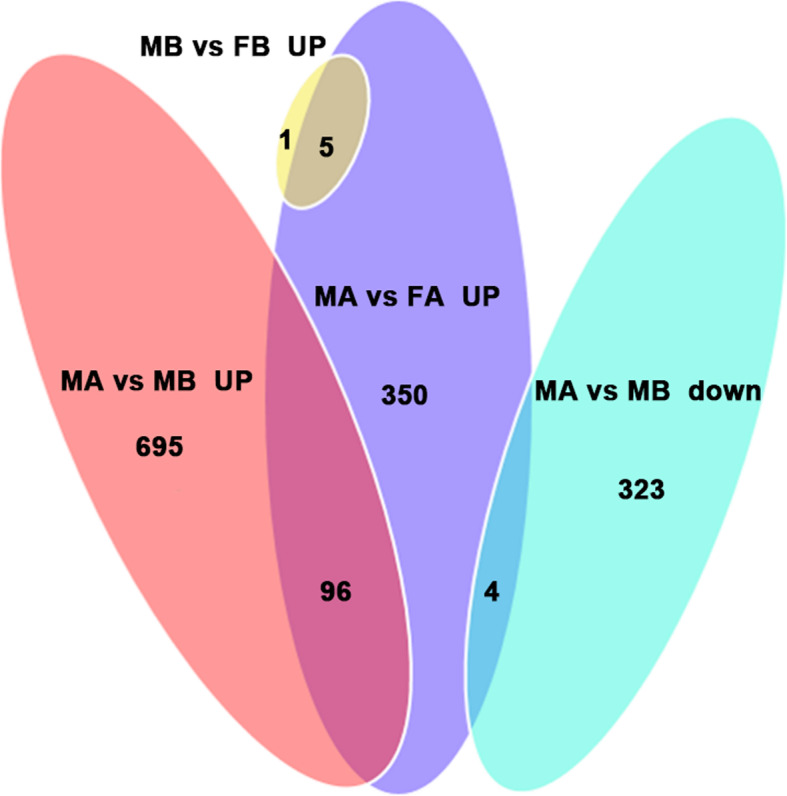
Venn diagram of male-related differentially expressed genes
Fig. 3.
Venn diagram of female-related differentially expressed genes
Fig. 4.
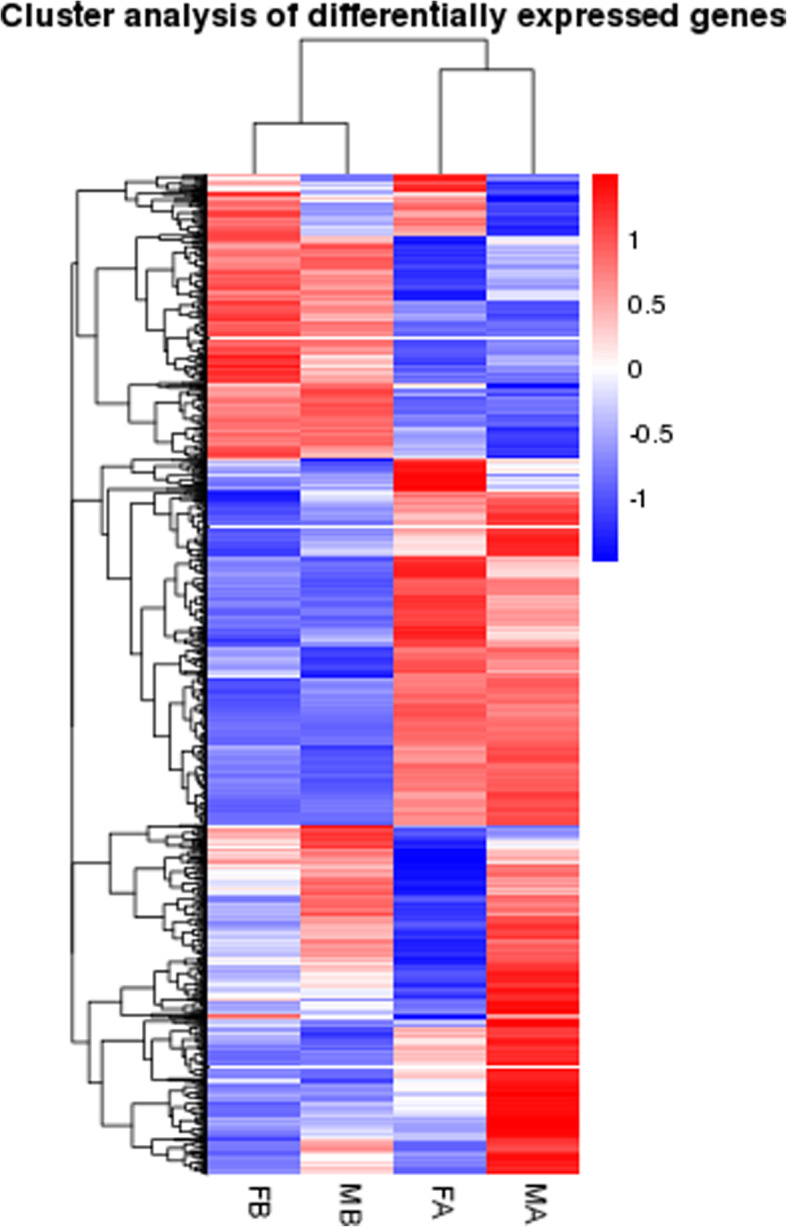
Heat map analysis is used to classify gene expression patterns at MB, FB, MA, and FA. Genes with similar expression patterns were clustered, as shown in the heat map. Red represents genes with high expression levels and blue represents genes with low expression levels
GO and KEGG enrichment analysis of DEGs
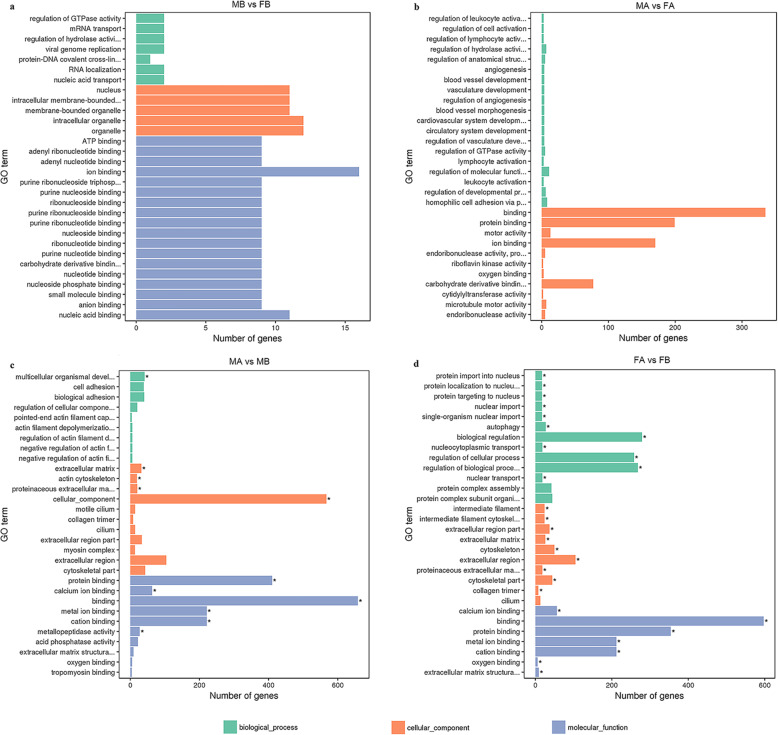
To better understand the possible functions involved in gonad development in Muscovy ducks, the DEGs from MB, FB, MA, and FA were annotated with the GO database, and the genes fell into three categories: biological process (BP), cellular component (CC) and molecular function (MF). According to the GO enriched results, dickkopf WNT signalling pathway inhibitor 3 (DKK3) was enriched in the regulation of the female receptivity process, bone morphogenetic protein 3 (BMP3) was enriched in germ cell development, oogenesis, and the female gamete generation process. TTN was enriched in the male gamete generation, spermatogenesis, sperm individualization process, and germ cell development process. AMH was enriched in the sex differentiation process. AGT was enriched in the fertilization process. OSR1 and ATP5A1 were enriched in the cellular aromatic compound metabolic process. NIPBL, HNRNPK, ANXA2, UBL3KCMF1 and CREB3L4 were enriched in the aromatic compound biosynthetic process. To identify genes useful for further study, 30 significant GO terms are listed in Fig. 5.
Fig. 5.
GO enrichment terms of differentially expressed genes. a MB-vs-FB; b. MA-vs-FA; c. MA-vs-MB; d. FA-vs-FB. The results are summarized in three main categories: biological process (BP), cellular component (CC) and molecular function (MF). The x-axis indicates the number of genes, and the y-axis indicates the second-level GO term
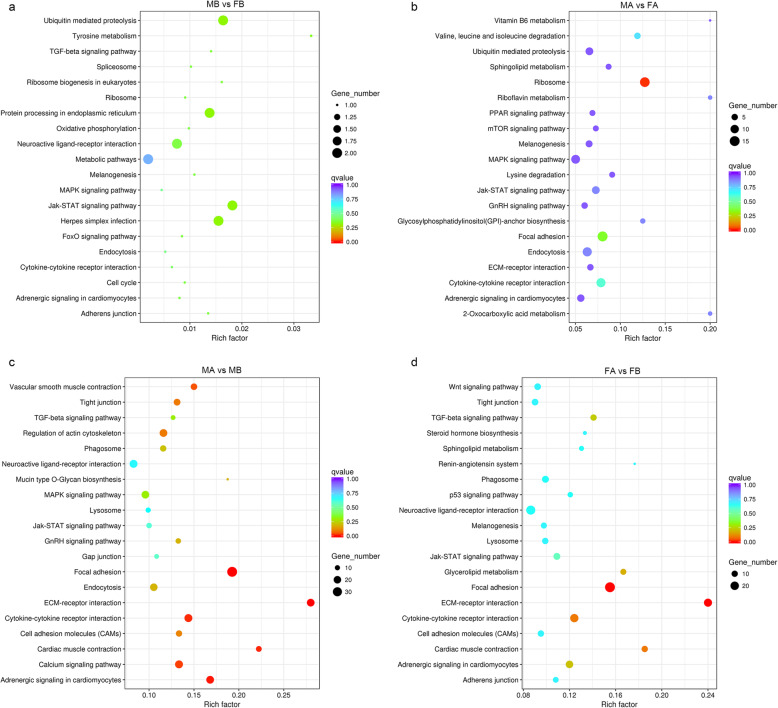
Subsequently, KEGG pathway analysis was performed, revealing 20, 108, 108 and 116 signalling pathways enriched in MB vs FB, MA vs MB, MA vs FA, and FA vs FB, respectively. The most enriched top 20 KEGG pathways are listed in Fig. 6. In MB vs FB, the most enriched KEGG pathways were ubiquitin mediated proteolysis, herpes simplex infection, the Jak-STAT signalling pathway, protein processing in the endoplasmic reticulum, neuroactive ligand-receptor interaction, and metabolic pathways (Fig. 6a). In MA vs FA, the six most enriched pathways were metabolic pathways, cytokine-cytokine receptor interaction, focal adhesion, ribosome, endocytosis, and MAPK signalling pathway (Fig. 6b). Most of the top enriched KEGG pathways were focal adhesion, adrenergic signalling in cardiomyocytes, cytokine-cytokine receptor interaction ECM-receptor interaction, and cardiac muscle contraction in MA vs MB (Fig. 6c). In FA vs FB, the six most enriched pathways were ECM-receptor interaction, metabolic pathways, cytokine-cytokine receptor interaction, adrenergic signalling in cardiomyocytes, focal adhesion, and neuroactive ligand-receptor interaction (Fig. 6d). BMP3 was enriched in the cytokine-cytokine receptor interaction signalling pathway. AMH was enriched in the TGF-β signalling pathway and the cytokine-cytokine receptor interaction. AGT was enriched in adrenergic signalling in cardiomyocytes, the neuroactive ligand-receptor interaction and the vascular smooth muscle contraction signalling pathway. ATP5A1 was enriched in the oxidative phosphorylation and metabolic pathways, and MAP3K1 was enriched in the MAPK signalling pathway.
Fig. 6.
Top 20 pathways in KEGG enrichment by QValue. a. MB-vs-FB; b. MA-vs-FA; c. MA-vs-MB; d. FA-vs-FB. The Rich factor is the ratio of the differentially expressed number of genes in the pathway and the total number of genes in the pathway. The higher the Rich factor, the higher is the degree of enrichment. The QValue is the P-value after multiple hypothesis test correction, in the range from 0 to 1; the closer the QValue is to zero, the more significant is the enrichment
Screening of gonadal differentiation genes
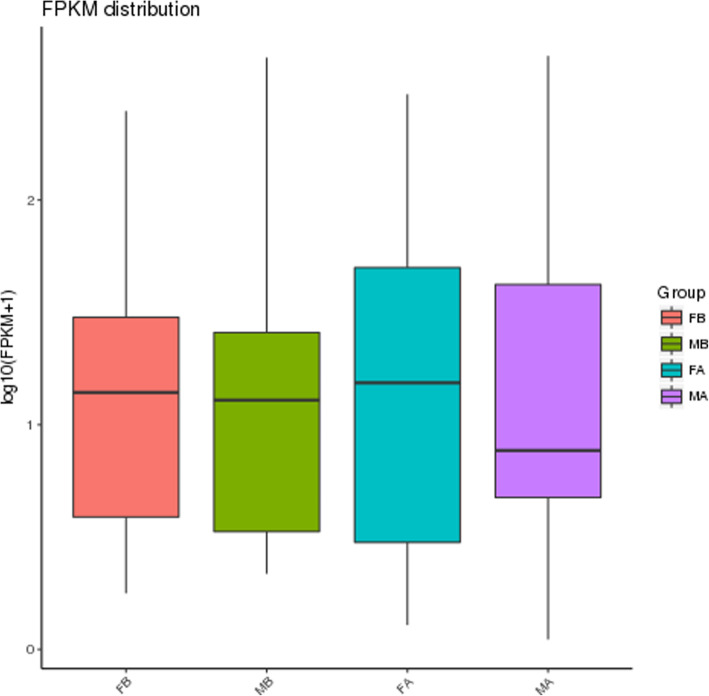
Of the DEGs, 21 DEGs were identified as sex-related genes according to clustering, GO analyses, and the published literature. The majority were unknown in Muscovy ducks, but most had already been identified in other organisms. According to the clustering diagram (Fig. 4), considerable changes in gene expression occurred in MA vs FA. We further excavated DEGs from the MA and FA groups, and we revealed that DMRT1, DMRT3, AMH, MAP3K1, ADAMTSL1, ATP5A1, AGT, PIK3R1 and TTN were significantly upregulated in MA vs FA. In addition, NIPBL, HNRNPK, UBE2R2, UBL3KCMF1, CREB3L4, R-spondin 3(RSPO3), WNT5B, HTRA3, ANXA2 and OSR1 were significantly downregulated in MA vs FA. These genes were categorized into 10 groups putatively involved in oogenesis, gametogenesis, sex differentiation, aromatic compound biosynthetic process, female receptivity, spermatogenesis, sperm individualization process, cellular aromatic compound metabolic process, fertilization, and germ cell development process. In addition, DKK3 and BMP3 were significantly upregulated in FA vs FB, and DKK3 was enriched in the regulation of female receptivity process. BMP3 was enriched in the oogenesis and female gamete generation process. Among these DEGs, DMRT1, DMRT3, AMH, PIK3R1, MAP3K1, ADAMTSL1, ATP5A1, AGT and TTN were involved in the differentiation and development of the testes, and NIPBL, HNRNPK, UBE2R2, UBL3KCMF1, CREB3L4, RSPO3, WNT5B, HTRA3, ANXA2, DKK3, BMP3 and OSR1 were involved in the differentiation and development of the ovaries. Hierarchical clustering was performed to compare these genes in FA, FB, MA, and MB (Fig. 7), and genes with similar expression patterns were classified into the same group. DMRT3, AMH, PIK3R1, ADAMTSL1, AGT and TTN were classified into MA, and WNT5B, ANXA2 and OSR1 were classified into FA. Therefore, after gonadal differentiation, DMRT3, AMH, PIK3R1, ADAMTSL1, AGT and TTN were specifically highly expressed in males. WNT5B, ANXA2 and OSR1 were specifically highly expressed in females. The distribution of gene expression levels between different groups is shown by boxplots (Fig. 8).
Fig. 7.
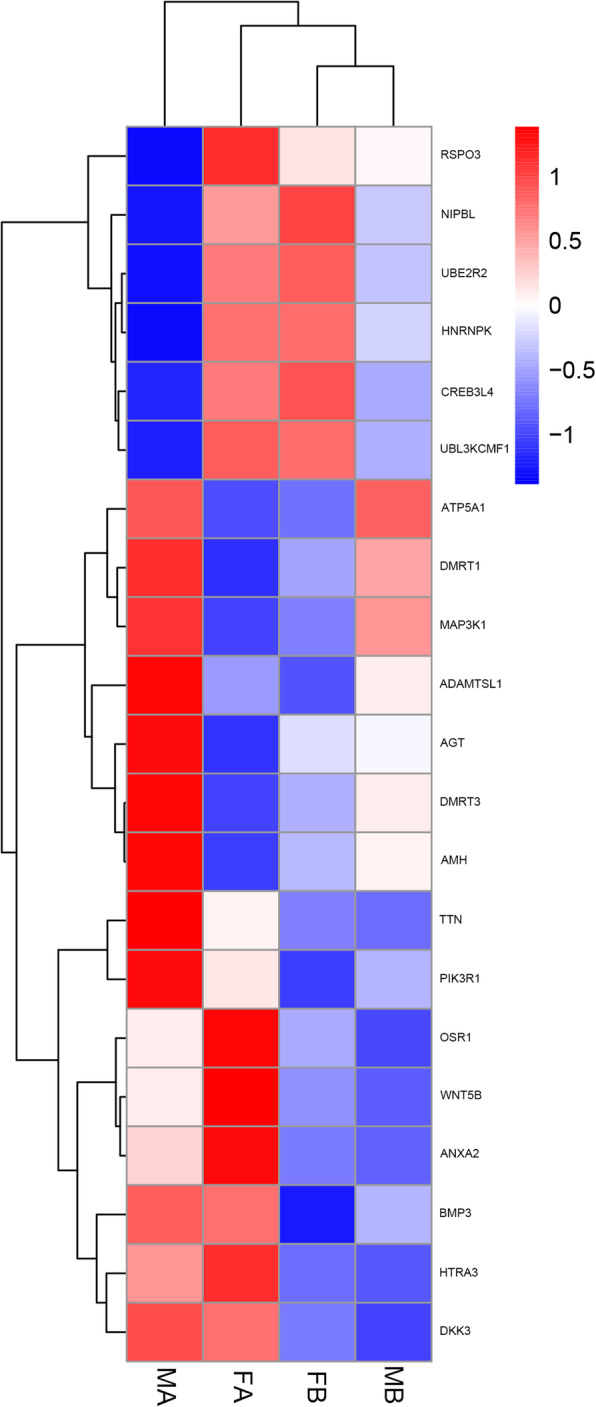
Heat map diagram of expression levels for selected sex-related genes. Red represents a high expression level, and blue indicates a low expression level. The columns and rows in the heat map represent samples and genes, respectively. Sample names are displayed below the heat map. The colour scale indicates fold changes in gene expression
Fig. 8.
The x-axis represents the group name, whereas the y-axis represents log10 (FPKM+ 1) values. Box plots indicate the maximum, top quartile, median, bottom quartile, and minimum values from top to bottom
Verification of DEGs by qPCR
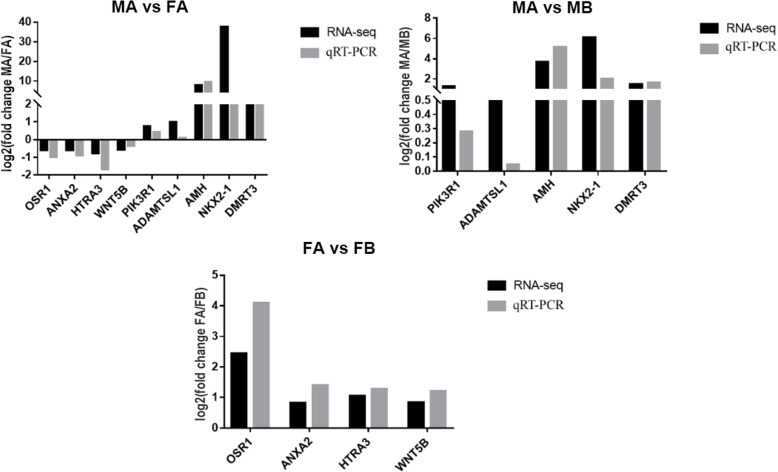
Nine genes were selected for RT-qPCR validation to verify the reproducibility of RNA-Seq results. Among these, 5 genes were upregulated in both the MA vs FA and MA vs MB comparisons, and 4 genes were downregulated in the MA vs FA comparison but upregulated in the FA vs FB comparison. The expression profiles of the nine candidate genes generated from RT-qPCR corresponded to the RNA-Seq results (Fig. 9). The primers used for the 9 genes are listed in Table 3.
Fig. 9.
qRT-PCR validation of differentially expressed genes identified by RNA-Seq
Table 3.
PCR primers used in this study
| Genes | Sequence (5′-3′) | Product size (bp) | GenBank No. |
|---|---|---|---|
| OSR1 |
ACCGATGAACGACCCTACAC CCTTTCCCACATTCTTGACAC |
125 | XM_027455274.1 |
| ANXA2 |
AATCATCTGCTCACGGACAAAT GTCGATCACAGAAGTATCTTCACAT |
172 | XM_027466033.1 |
| HTRA3 |
CCTCTAGTTAATCTGGATGGTGAAG TGATGCGGTCTGATGGAATAG |
91 | XM_021270005.2 |
| PIK3R1 |
GGAACGTGGGAAATATCAACAG ACTTCTCCATCCACCACAACA |
130 | XM_027446161.1 |
| ADAMTSL1 |
GCAGGAAAGCCGCTAATAAAG GCAGGTGGTGAGCAGAGGAT |
199 | XM_027446720.1 |
| AMH |
AGTGCGGCTGTCGGTAGAG GGGAAAAGACGTGCGGAG |
99 | AY904047.1 |
| NKX2–1 |
ACCCCGACTCAGGTCAAAAT CATTGGTAGCCACTGTGATTGT |
253 | XM_027458955.1 |
| DMRT3 |
CTGTCCTTCCTTCTCGCTCAT GCTGCACTGCAAATATCACTTAG |
197 | XM_027446949.1 |
| WNT5B |
CTGCTTTCACCTATGCTGTCA ACCCGTACTCCACGTTATCC |
165 | XM_027467202.1 |
| β-actin |
GCTATGTCGCCCTGGATTT CCACAGGACTCCATACCCAA |
169 | EF667345.1 |
Prediction of novel transcripts
From the sequencing data, 2744 novel genes were detected by genome assembly and compared with the reference genome. A list of results is shown in Additional file 1.
Discussion
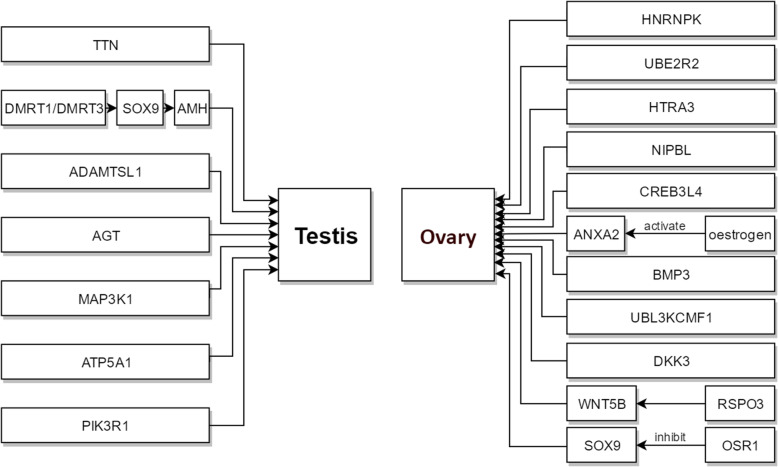
Little information has been reported regarding the reproductive activity and molecular aspects of gonadal development in Muscovy ducks. The transcriptome data generated herein provide valuable genetic resources to understand sex differentiation mechanisms and pathways underlying Muscovy duck reproductive regulation. A total of 315 DEGs were obtained, and a total of 146 signalling pathways were enriched from MB, FB, MA, and FA. DEGs were screened by clustering, GO analyses, and the published literature. We obtained 21 sex-related genes (Fig. 10).
Fig. 10.
A speculation explaining the gene expression pattern in testes and ovaries
Sex differentiation is suggested to be controlled by a major gene. AMH was found to be involved in gonad development and sex differentiation in this study. The anti-Mullerian hormone type 2 receptor (AMHR2) (the first AMH receptor identified in birds) is expressed in the early stage of sex differentiation and regulated by DMRT1, and AMH plays a decisive role downstream of DMRT1 during sexual differentiation in birds [29]. In addition, AMH is significantly upregulated in MA vs FA and MA vs MB, that is, AMH is expressed in males after gonadal differentiation, so it is a candidate gene for the testis in Muscovy ducks. DMRT1 was significantly upregulated in MA vs FA. Previous studies in chickens have identified DMRT1 as a potential male-determining gene, which is expressed prior to SRY-box transcription factor 9 (Sox9) [30]. DMRT1 is a requirement for male-specific expression and testicular development in vertebrates [31, 32]. Research in Takifugu rubripes [33] revealed that the expression levels of DMRT1 and DMRT3 is significantly higher in testis than in ovaries. Hence, DMRT1 is associated with normal testicular development in Muscovy ducks. DMRT3 was significantly upregulated in MA vs FA and MA vs MB. Previous studies in chickens have shown that DMRT1 and DMRT3 may bind to the enhancer of SOX9 [30]. In addition, studies have found that hemogen (HEMGN) may activate SOX9. HEMGN, as a developmental window, is expressed after DMRT1 and before SOX9 [8]. We speculated that DMRT1 and DMRT3 induce gonads to develop as males in Muscovy ducks. Additionally, the expression of DMRT1 and DMRT3 induces SOX9, which then induces AMH.
In animals, spermatogenesis is a conserved process in which male diploid spermatogonia develop to maturity and haploid spermatozoa capable of fertilizing an oocyte [34]. Sperm individualization is one of the last stages of spermatogenesis, which separates spermatids from syncytial male germline cysts [35]. TTN was involved in male gamete generation, spermatogenesis, the sperm individualization process, and the germ cell development process in this study. Titin (TTN) plays key structural, developmental, mechanical, and regulatory roles in cardiac and skeletal muscles [36]. In Bombyx mori, Bmkettin (TTN) is expressed at higher levels in males than females [37, 38]. TTN was significantly upregulated in MA vs FA and MA vs MB. Thus, TTN was expressed in males after gonadal differentiation in Muscovy ducks.
Fertilization is a multi-step process that culminates in the fusion of the sperm and oocyte plasma membranes [39]. Herein, AGT was involved in fertilization and the acrosome reaction process. In the kidney cortex, AGT gene expression was higher in males than females [40, 41]. Moreover, both SRY and SRY-box transcription factor 3 (SOX3) upregulated the promoter of AGT [42]. In addition, AGT was significantly upregulated in MA vs FA. We speculated that AGT was associated with normal testicular development in Muscovy ducks.
During oogenesis, gene expression of oocytes is dynamically regulated by a series of well-coordinated transcription factors that are active in germ cell lines and somatic cells. Many somatic-expressed and germ cell-specific transcriptional regulators play key roles in folliculogenesis and ovarian formation [43]. Here, BMP3 was involved in oogenesis, the female gamete generation and germ cell development process. Gametogenesis begins with an early germline distribution in the embryo [43]. During development, both male and female germ cells undergo a round of meiotic cell division to reduce the ploidy of the gametes, and thus maintain the ploidy after fertilization [44]. In the chicken embryo, BMP3 is preferentially expressed in the developing ovary [45]. BMP3 was significantly upregulated in FA vs FB. Therefore, in Muscovy ducks, BMP3 induced differentiated gonads to develop as females.
The aromatase enzyme is a key enzyme in the regulation of oestrogen levels [46]. Oestrogens not only appear in the form of female hormones [47], but also as steroids that are necessary for the endocrine balance in males [48]. Our present results showed that ATP5A1 and OSR1 were involved in cellular aromatic compound metabolic processes, and NIPBL, HNRNPK, ANXA2 and CREB3L4 were involved in aromatic compound biosynthetic processes. ATP5A1 exists in a copy on the non-recombinant region of the Z and W chromosomes [10, 49, 50]. ATP5A1 is a Z-linked gene in chickens [51]. Thus, ATP5A1 was expressed in male Muscovy ducks. OSR1 and ANXA2 were upregulated both in FA vs MA and FA vs FB. Thus, OSR1 and ANXA2 were expressed in males after gonadal differentiation. As a member of the OSR family, OSR1 is a zinc finger transcription factor that is required for embryonic development [52]. OSR1 is enriched in the pregranular cell population of females, indicating that the supporting cell progenitors are female-primed before gonadal sex differentiation in the chicken embryo [14]. OSR1 can bind to the promoter region of SOX9, and overexpression of OSR1 can inhibit the expression of SOX9 [53]. Therefore, we speculated that although OSR1 was not directly involved in sex differentiation, it affected sex differentiation by inhibiting the expression of SOX9 in ovaries. ANXA2 plays a role in the development of chicken ovaries and follicles [54]. Relevant studies have reported that oestrogen may stimulate ANXA2 expression [55]. In brief, OSR1 and ANXA2 were associated with the differentiation of ovaries in Muscovy ducks. HNRNPK interacts with DNA, RNA, and various proteins to participate in transcription, translation, RNA splicing, DNA repair, and chromatin remodelling, and it plays key roles in the nervous system. HNRNPK, as an oestrogen-induced transcription factor, is involved in ovary development [56–60]. NIPBL, as a sex-related gene in chickens, is assembled on the W chromosome that has homologous sequences on chromosome Z [61]. CREB3L4 is characterized by a unique putative transmembrane domain with its C-terminus arranged in the bZIP region, which is assigned to the CREB/ATF subfamily transcription factors [62, 63]. In addition, in mice, CREB3L4 is expressed in a stage-specific manner during sperm cell differentiation, and the lack of CREB3L4 does not dramatically affect mouse spermatogenesis [64, 65]. NIPBL, HNRNPK and CREB3L4 were significantly upregulated in FB vs MB and FA vs MA, and we analysed sex differentiation under this condition. We found that NIPBL, HNRNPK and CREB3L4 were expressed in female Muscovy ducks.
In our study, ADAMTSL1 and PIK3R1 were significantly upregulated in MA vs FA and MA vs MB. Previous studies have indicated that ADAMTSL1 lies on the Z chromosome and is highly expressed in the testis during sex differentiation in chicken embryos [45]. PIK3R1, an essential regulatory gene in the phosphatidylinositol 3-kinase/AKT (PI3K/AKT) signalling pathway, engages in various biological activities and metabolic pathways in organisms [66, 67]. In adult half-smooth tongue sole, the expression of PIK3R1 in male gonads has been reported to be significantly higher than that in other tissues [68]. Therefore, ADAMTSL1 and PIK3R1 were expressed in males after gonadal differentiation and related to testicular development in Muscovy ducks. MAP 3 K1 was significantly upregulated in MA vs FA. previous studies have shown that knocking out MAP 3 K1 results in testicular abnormalities in developing mice [69]. Additionally, mutations in MAP 3 K1 downregulate testicular decisive factors such as SOX9 and SRY [70]. Therefore, MAP 3 K1 is associated with normal testicular development in Muscovy ducks.
In this assay, HTRA3 was upregulated both in FA vs MA and FA vs FB. DKK3 was significantly upregulated in FA vs FB, and RSPO3 was significantly upregulated in FA vs MA. HTRA3 is an important reproduction-related gene and regulates ovarian development [71–74]. HTRA3 inhibits TGF-β signalling during mouse embryonic development [75]. RSPO3 is a member of the RSPO protein family. Previous studies have shown that four members of the RSPO protein family can activate the WNT/β-catenin signalling pathway [76]. Furthermore, studies on mammals have also suggested that the WNT signalling pathway plays an significant role in ovarian development and hormone secretion [77, 78]. Moreover, RSPO3 is more effective in activating the WNT/β-catenin signalling pathway [79]. Dkk3 is abundant in the liver and brain [80]. Moreover, as a soluble Wnt inhibitor, most knowledge about Dkk3 involves molecular cancer therapy.Dkk3 may be an activator or inhibitor of the canonical pathway [81–89], which is strictly context-dependent [90]. In this study, DKK3 can positively or negatively regulate the Wnt signalling pathway. Additionally, DKK3 was enriched in biological process with the regulation of female receptivity. The functions and expression of DKK3 were also associated with ovary development [91]. Therefore, HTRA3, DKK3 and RSPO3 were associated with the differentiation of ovaries in Muscovy ducks.
In addition to known sexual regulators based on previous studies in other species, WNT5B, UBE2R2 and UBL3KCMF1 were identified. WNT5B was significantly upregulated in FA vs MA and FA vs FB. Thus, WNT5B was expressed in females after gonadal differentiation. Moreover, WNT5B was enriched in the WNT/β-catenin signalling pathway, and research has shown that RSPO3 is more effective in activating the WNT/β-catenin signalling pathway [79]. As a result, we hypothesized that RSPO3 is involved in the differentiation of ovaries in Muscovy ducks by activating WNT5B in the WNT signalling pathway. UBE2R2 and UBL3KCMF1 were significantly upregulated in both FB vs MB and FA vs MA. Furthermore, these genes were associated with the ubiquitin-binding enzyme E2 and ubiquitin protein ligase E3, respectively. It has been reported that the expression level of ubiquitin-binding enzyme E2 is significantly different at different developmental stages of the testes and ovaries [92]. Previous studies have demonstrated that ubiquitin conjugating enzyme 9 (Ubc9), a member of the ubiquitin-binding protein E2 family, is implicated in the regulation of oogenesis [93]. These two genes showed higher expression in female than male gonads before and after sex differentiation. UBE2R2 was assigned to the W and Z chromosome in chicken [94, 95]. As a result, UBE2R2, UBL3KCMF1 and WNT5B were associated with the differentiation of ovaries in Muscovy ducks.
Conclusions
In this study, RNA-Seq revealed 101.76 Gb of clean reads and 2800 DEGs, including 46 in MB vs FB, 609 in MA vs FA, 1027 in FA vs FB, and 1118 in MA vs MB. A total of 146 signalling pathways were enriched by KEGG analysis. Furthermore, a total of 21 candidate genes related to sex differentiation and development in Muscovy ducks were screened by clustering and GO analyses. Among these, 12 genes, namely, UBE2R2, UBL3KCMF1 RSPO3, WNT5B, BMP3, HNRNPK, NIPBL, CREB3L4, HTRA3, DKK3, ANXA2 and OSR1, were involved in the differentiation and development of ovaries. Moreover, 9 genes, namely, DMRT1, DMRT3, AMH, MAP3K1, PIK3R1, TTN, ATP5A1, AGT and ADAMTSL1, were related to the differentiation and development of testes. Furthermore, after gonadal differentiation, DMRT3, AMH, PIK3R1, ADAMTSL1, AGT and TTN were especially highly expressed in males, and WNT5B, ANXA2 and OSR1 were especially highly expressed in female. These results provide a reference for subsequent research on the mechanism of sex differentiation in Muscovy ducks and significant information for the study of sex control in Muscovy ducks.
Methods
Ethics statement
This study adhered to the recommendations of the “Regulations for the Management of Affairs Concerning Experimental Animals” (Ministry of Science and Technology, China, revised in June 2004). The Muscovy duck shed and feeding conditions were reviewed and approved by the Institutional Animal Care and Use Committee of the College of Animal Science, Fujian Agriculture and Forestry University. In this assay, all ducks were permitted continuous access to standard commercial feed rations and water. All efforts were made to minimize animal suffering.
Animals and tissue collection
A total of 96 Muscovy duck eggs were provided by China Putian Guangdong Wenshi Poultry Co., Ltd. (Putian, China). Four groups were assessed (n = 3 per group): males and females at 8 days and 12 days of incubation, respectively. Eight paired gonads were pooled for each replicate. Muscovy eggs in good condition were examined at 8 days and 12 days of incubation. The selected Muscovy duck eggs were all from the same population and hatched at approximately 37 °C with a humidity of 60–70%. First, the selected duck egg was gently tapped with tweezers at the air chamber side, and the eggshell and shell membranes were removed to expose the embryo. The embryo was removed with a spoon, and then the embryonic gonad of the duck (including the kidney and gonads) was excised, labelled, and placed in liquid nitrogen. After all the samples were collected, they were stored at − 80 °C. The embryo head and tail were collected, and DNA was extracted for sex determination by PCR [12].
RNA extraction and sequencing
RNA extraction was performed using TRIzol reagent in accordance with the manufacturer’s protocol. Then, 1% agarose gels were used to evaluate RNA degradation and contamination. RNA concentration and integrity were checked with an RNA Assay Kit and a Qubit® 2.0 Fluorometer and the RNA Nano 6000 Assay Kit for the Bioanalyzer 2100 system, respectively. Gonad tissue was pooled based on sex and stage of sex differentiation, eight gonads of the same genetic sex were mixed in a library, and a total of 12 libraries from the MB, FB, MA, and FA groups were sequenced. First, twelve libraries were constructed. Briefly, purification of mRNA and cleave the mRNA into fragments were performed. Then reverse transcription, cDNA synthesis, and cluster generation were carried out. The libraries were then sequenced on an Illumina HiSeq platform. These data are available at the NCBI Sequence Read Archive (accession number PRJNA625194, SRX8108890, SRX8108889, SRX8108888, SRX8108887, SRX8108886, SRX8108885, SRX8108884, SRX8108883, SRX8108882, SRX8108881, SRX8108880 and SRX8108879).
Bioinformatic analysis
First, after removing reads containing adaptors, poly-N sequences and low-quality reads from the raw data through in-house Perl scripts., clean reads were obtained. Next, all the high-quality clean reads were aligned to the Anas platyrhynchos genome using Hisat2 v2.0.4. Subsequently, HTSeq v0.9.1 and the fragments per kilobase of transcript per million mapped reads (FPKM) were used to quantify gene expression level [96, 97]. In addition, differential expression analysis of MB vs FB, FA vs FB, MA vs FA, and MA vs MB was performed using the DEGSeq R package (1.18.0). We used the Benjamini & Hochberg method corrected P-value < 0.05 as the threshold to consider to be the differentially expressed genes. The GO and KEGG enrichment analyses were then conducted according to a P-value cut-off of 0.05 using GOSeq Release2.12 and KOBAS v2.0 [98, 99]. Finally, Hisat2 v2.0.4 was used as the mapping tool, and reads without annotation information were assembled with the Cufflinks v2.1.1 reference annotation-based transcript (RABT). Cuffcompare was compared with known genetic models to obtain new genes [97, 100].
Real-time PCR verification
Nine significantly DEGs were selected for real-time quantitative PCR (qRT-PCR) to validate the RNA-Seq results. The PrimeScript™ RT reagent Kit with gDNA Eraser was used to cDNA synthesis following the manufacturer’s instructions. The primer information for these genes and the endogenous reference gene are available in Table 3. β-actin was employed as the endogenous reference gene. The qPCR mixture consisted of 10 μL of 2 × Master Mix, 2 μL of cDNA, 0.5 μL of each primer and 7 μL of ddH2O under the following conditions to collect fluorescence: denaturation at 95 °C for 30 min, followed by 40 cycles of amplification at 95 °C for 5 s and 60 °C for 40 s. To establish the melting curve of the PCR product, the reaction was performed at 95 °C for 10 s, 60 °C for 60 s, and 95 °C for 15 s, after which it was slowly heated from 60 °C to 99 °C. The relative expression levels of target genes in each sample were individually determined using the 2-ΔΔCT method [101].
Supplementary information
Acknowledgments
We thanks China Putian Guangdong Wenshi Poultry Co., Ltd. afford Muscovy duck eggs and technical support, and Thanks AJE Editing Services.
Abbreviations
- MB
Male before gonadal differentiation
- FB
Female before gonadal differentiation
- MA
Male after gonadal differentiation
- FA
Female after gonadal differentiation
- DEGs
Differentially expressed genes
- DMRT1
Doublesex and mab-3 related transcription
- SRY
Sex-determining Region on the Y Chromosome
- CYP19A1
The cytochrome P450 family 19 subfamily A member
- AMH
Anti-Mullerian hormone
- E
Embryonic day
- ESR-α
Oestrogen receptor-α
- ATP5A1
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1
- TTN
Titin
- NIPBL
Nipped-B-like protein
- CREB3L4
Cyclic AMP-responsive element-binding protein 3-like protein 4
- HNRNPK
Heterogeneous nuclear ribonucleoprotein K
- HEMGN
Hemogen
- SOX9
SRY-box transcription factor 9
- SOX3
SRY-box transcription factor 3
- Go
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- BP
Biological process
- CC
Cellular component
- MF
Molecular function
- MAPK
The mitogen-activated protein kinases
- TGF-β
Transforming growth factor-β
- DMRT3
Doublesex and mab-3 related transcription factor 3
- HTRA
high-temperature requirement factor A
- HTRA3
HtrA serine peptidase 3
- OSR1
Odd-skipped related transcription factor 1
- WNT5B
Wnt family member 5B
- NKX2-1
NK2 homeobox 1
- AMHR2
Anti-Mullerian hormone type 2 receptor
- MAP3K1
Mitogen-activated protein kinase kinase kinase 1
- PI3K
Phosphatidylinositol 3-kinase
- PIK3R1
phosphoinositide-3-kinase regulatory subunit 1
- ADAMTSL1
A disintegrin and metalloproteinase with thrombospondin motif like 1
- UBE2R2
Ubiquitin-conjugating enzyme E2 R2
- UBL3KCMF1
E3 ubiquitin-protein ligase KCMF1
- Ubc9
Ubiquitin conjugating enzyme 9
- RSPO3
R-spondin 3
- ANXA2
Annexin A2
- RIN
RNA integrity number
- FPKM
Fragments per kilobase of transcript per million mapped reads
- RABT
Reference Annotation Based Transcript
- qRT-PCR
real-time quantitative PCR
Authors’ contributions
DPB, AL, and YQH designed the research framework and carried out the experiments. YC drafted the manuscript. YC and WFH performed the bioinformatics analyses and qPCR verification. YZS, RTL, and QMF performed manuscript formatting and editing. YQH collected and prepared the experimental materials. DPB contributed to the research design and reviewed the manuscript. All authors read and approved the final manuscript.
Funding
This project was supported by the Earmarked Fund for Modern Agro-industry Technology Research System of China (CaRS-43), the Discipline Development Grant from the College of Animal Sciences FAFU (2018DK006) and the Natural Science Foundation of Fujian Province of China (2019 J01377). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
The datasets generated and analysed during the current study are available in the NCBI Sequence Read Archive database (https://www.ncbi.nlm.nih.gov/sra), under the accession number PRJNA625194 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA625194), SRX8108890, SRX8108889, SRX8108888, SRX8108887, SRX8108886, SRX8108885, SRX8108884, SRX8108883, SRX8108882, SRX8108881, SRX8108880 and SRX8108879. The name of the bioproject is Muscovy ducks sequencing. The link to the Anas platyrhynchos genome is https://www.ncbi.nlm.nih.gov/genome/?term=Anas+platyrhynchos. The primer information for β-actin (GeneBank accession number EF667345.1) is available in Table 3.
Ethics approval and consent to participate
All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee of the College of Animal Science, Fujian Agriculture and Forestry University.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ding-Ping Bai and Yue Chen contributed equally to this work.
Contributor Information
Ding-Ping Bai, Email: bdpnx@163.com.
Yue Chen, Email: 1176844830@qq.com.
Yu-Qiong Hu, Email: 932847881@qq.com.
Wen-Feng He, Email: 1169781280@qq.com.
Yu-Zhu Shi, Email: 1402024360@qq.com.
Qin-Ming Fan, Email: 2582093700@qq.com.
Ru-Tang Luo, Email: 853835112@qq.com.
Ang Li, Email: poul@fafu.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12864-020-06852-z.
References
- 1.Major AT, Smith CA. Sex reversal in birds. Sex Dev. 2016;10:288–300. doi: 10.1159/000448365. [DOI] [PubMed] [Google Scholar]
- 2.Smith CA, Sinclair AH. Sex determination: insights from the chicken. Bioessays. 2004;26:120–132. doi: 10.1002/bies.10400. [DOI] [PubMed] [Google Scholar]
- 3.Ren J, Sun C, Chen L, Hu J, Huang X, Liu X, et al. Exploring differentially expressed key genes related to development of follicle by RNA-seq in Peking ducks (Anas Platyrhynchos) PLoS One. 2019;14:e0209061. doi: 10.1371/journal.pone.0209061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ayers KL, Lambeth LS, Davidson NM, Sinclair AH, Oshlack A, Smith CA. Identification of candidate gonadal sex differentiation genes in the chicken embryo using RNA-seq. BMC Genomics. 2015;16:704. doi: 10.1186/s12864-015-1886-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R. Male development of chromosomally female mice transgenic for Sry. Nature. 1991;351:117–121. doi: 10.1038/351117a0. [DOI] [PubMed] [Google Scholar]
- 6.Smith CA, Roeszler KN, Ohnesorg T, Cummins DM, Farlie PG, Doran TJ, et al. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature. 2009;461:267. doi: 10.1038/nature08298. [DOI] [PubMed] [Google Scholar]
- 7.Eggers S, Sinclair A. Mammalian sex determination—insights from humans and mice. Chromosom Res. 2012;20:215–238. doi: 10.1007/s10577-012-9274-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lambeth LS, Raymond CS, Roeszler KN, Kuroiwa A, Nakata T, Zarkower D, et al. Over-expression of DMRT1 induces the male pathway in embryonic chicken gonads. Dev Biol. 2014;389:160–172. doi: 10.1016/j.ydbio.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wallis M, Waters P, Graves J. Sex determination in mammals—before and after the evolution of SRY. Cell Mol Life Sci. 2008;65:3182. doi: 10.1007/s00018-008-8109-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fridolfsson A-K, Cheng H, Copeland NG, Jenkins NA, Liu H-C, Raudsepp T, et al. Evolution of the avian sex chromosomes from an ancestral pair of autosomes. Proc Natl Acad Sci. 1998;95:8147–8152. doi: 10.1073/pnas.95.14.8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellegren H. Evolution of the avian sex chromosomes and their role in sex determination. Trends Ecol Evol. 2000;15:188–192. doi: 10.1016/s0169-5347(00)01821-8. [DOI] [PubMed] [Google Scholar]
- 12.Elbrecht A, Smith RG. Aromatase enzyme activity and sex determination in chickens. Science. 1992;255:467–470. doi: 10.1126/science.1734525. [DOI] [PubMed] [Google Scholar]
- 13.Andrews JE, Smith CA, Sinclair AH. Sites of estrogen receptor and aromatase expression in the chicken embryo. Gen Comp Endocrinol. 1997;108:182–190. doi: 10.1006/gcen.1997.6978. [DOI] [PubMed] [Google Scholar]
- 14.Estermann MA, Williams S, Hirst CE, Roly ZY, Serralbo O, Adhikari D, et al. Insights into gonadal sex differentiation provided by single-cell transcriptomics in the chicken embryo. Cell Rep. 2020;31:107491. doi: 10.1016/j.celrep.2020.03.055. [DOI] [PubMed] [Google Scholar]
- 15.Simpson ER, Mahendroo MS, Means GD, Kilgore MW, Hinshelwood MM, Graham-Lorence S, et al. Aromatase cytochrome P450, the enzyme responsible for estrogen biosynthesis. Endocr Rev. 1994;15:342–355. doi: 10.1210/edrv-15-3-342. [DOI] [PubMed] [Google Scholar]
- 16.Thompson EA, Siiteri PK. Utilization of oxygen and reduced nicotinamide adenine dinucleotide phosphate by human placental microsomes during aromatization of androstenedione. J Biol Chem. 1974;249:5364–5372. [PubMed] [Google Scholar]
- 17.Morishima A, Grumbach MM, Simpson ER, Fisher C, Qin K. Aromatase deficiency in male and female siblings caused by a novel mutation and the physiological role of estrogens. J Clin Endocrinol Metab. 1995;80:3689–3698. doi: 10.1210/jcem.80.12.8530621. [DOI] [PubMed] [Google Scholar]
- 18.Hu Y-Q, Bai D-P, Chen Y, Lu Z-X, Zheng H-B, Xu F-Q, et al. The degree of sex reversal in Muscovy ducks (Cairina moschata domestica) induced by an aromatase inhibitor. Sex Dev. 2019;13:137–142. doi: 10.1159/000502195. [DOI] [PubMed] [Google Scholar]
- 19.Wang Q, Weng H, Chen Y, Wang C, Lian S, Wu X, et al. Early development of gonads in Muscovy duck embryos. Br Poult Sci. 2015;56:390–397. doi: 10.1080/00071668.2015.1027172. [DOI] [PubMed] [Google Scholar]
- 20.Ayers KL, Davidson NM, Demiyah D, Roeszler KN, Grützner F, Sinclair AH, et al. RNA sequencing reveals sexually dimorphic gene expression before gonadal differentiation in chicken and allows comprehensive annotation of the W-chromosome. Genome Biol. 2013;14:R26. doi: 10.1186/gb-2013-14-3-r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J, Qu Y, Huang Y, Lei F. Dynamic transcriptome profiling towards understanding the morphogenesis and development of diverse feather in domestic duck. BMC Genomics. 2018;19:391. doi: 10.1186/s12864-018-4778-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wan Z, Lu Y, Rui L, Yu X, Yang F, Tu C, et al. Gene expression profiling reveals potential players of left-right asymmetry in female chicken gonads. Int J Mol Sci. 2017;18:1299. doi: 10.3390/ijms18061299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Truong AD, Hong YH, Lillehoj HS. RNA-seq profiles of immune related genes in the spleen of necrotic enteritis-afflicted chicken lines. Asian-Australas J Anim Sci. 2015;28(10):1496. doi: 10.5713/ajas.15.0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu T, Gu L, Michael SK, Liu X, Wei H, Ming X, et al. Identification of differentially expressed genes in breast muscle and skin fat of postnatal Pekin duck. PLoS One. 2014;9:e107574. doi: 10.1371/journal.pone.0107574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Z, Meng G, Bai Y, Liu R, Du Y, Su L. Comparative transcriptome analysis provides clues to molecular mechanisms underlying blue-green eggshell color in the Jinding duck (Anas platyrhynchos) BMC Genomics. 2017;18:725. doi: 10.1186/s12864-017-4135-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu F, Yuan J-M, Zhang Z-H, Hao J-P, Yang Y-z, Hu S-Q, et al. De novo transcriptome assembly and identification of genes associated with feed conversion ratio and breast muscle yield in domestic ducks. Anim Genet. 2015;46:636–645. doi: 10.1111/age.12361. [DOI] [PubMed] [Google Scholar]
- 27.Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, Mcpherson A, et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016;17:1–19. doi: 10.1186/s13059-016-0881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin JA, Wang Z. Next-generation transcriptome assembly. Nat Rev Genet. 2011;12:671–682. doi: 10.1038/nrg3068. [DOI] [PubMed] [Google Scholar]
- 29.Cutting AD, Ayers K, Davidson N, Oshlack A, Doran T, Sinclair AH, et al. Identification, expression, and regulation of anti-Müllerian hormone type-II receptor in the embryonic chicken gonad. Biol Reprod. 2014;90(106):101–112. doi: 10.1095/biolreprod.113.116491. [DOI] [PubMed] [Google Scholar]
- 30.Bagheri-Fam S, Sinclair AH, Koopman P, Harley VR. Conserved regulatory modules in the Sox9 testis-specific enhancer predict roles for SOX, TCF/LEF, Forkhead, DMRT, and GATA proteins in vertebrate sex determination. Int J Biochem Cell Biol. 2010;42:472–477. doi: 10.1016/j.biocel.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 31.Raymond CS, Shamu CE, Shen MM, Seifert KJ, Hirsch B, Hodgkin J, et al. Evidence for evolutionary conservation of sex-determining genes. Nature. 1998;391:691–695. doi: 10.1038/35618. [DOI] [PubMed] [Google Scholar]
- 32.Zarkower D. DMRT genes in vertebrate gametogenesis. Curr Top Dev Biol. 2013;102:327–356. doi: 10.1016/B978-0-12-416024-8.00012-X. [DOI] [PubMed] [Google Scholar]
- 33.Yan H, Shen X, Cui X, Wu Y, Wang L, Zhang L, et al. Identification of genes involved in gonadal sex differentiation and the dimorphic expression pattern in Takifugu rubripes gonad at the early stage of sex differentiation. Fish Physiol Biochem. 2018;44:1275–1290. doi: 10.1007/s10695-018-0519-8. [DOI] [PubMed] [Google Scholar]
- 34.Roosen-Runge EC. Kinetics of spermatogenesis in mammals. Ann N Y Acad Sci. 1952;55:574–584. doi: 10.1111/j.1749-6632.1952.tb26577.x. [DOI] [PubMed] [Google Scholar]
- 35.Fabrizio JJ, Hime G, Lemmon SK, Bazinet C. Genetic dissection of sperm individualization in Drosophila melanogaster. Development. 1998;125:1833–1843. doi: 10.1242/dev.125.10.1833. [DOI] [PubMed] [Google Scholar]
- 36.Chauveau C, Rowell J, Ferreiro A. A rising titan: review and mutation update. Hum Mutat. 2014;35:1046–1059. doi: 10.1002/humu.22611. [DOI] [PubMed] [Google Scholar]
- 37.Koike Y, Mita K, Suzuki MG, Maeda S, Abe H, Osoegawa K, et al. Genomic sequence of a 320-kb segment of the Z chromosome Ofbombyx moricontaining akettinortholog. Mol Gen Genomics. 2003;269:137–149. doi: 10.1007/s00438-003-0822-6. [DOI] [PubMed] [Google Scholar]
- 38.Suzuki MG, Shimada T, Kobayashi M. Bm kettin, homologue of the drosophila kettin gene, is located on the Z chromosome in Bombyx mori and is not dosage compensated. Heredity. 1999;82:170–179. doi: 10.1038/sj.hdy.6884570. [DOI] [PubMed] [Google Scholar]
- 39.Primakoff P, Myles DG. Penetration, adhesion, and fusion in mammalian sperm-egg interaction. Science. 2002;296:2183–2185. doi: 10.1126/science.1072029. [DOI] [PubMed] [Google Scholar]
- 40.Rands VF, Seth DM, Kobori H, Prieto MC. Sexual dimorphism in urinary angiotensinogen excretion during chronic angiotensin II− salt hypertension. Gender Med. 2012;9:207–218. doi: 10.1016/j.genm.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bourgeois CT, Satou R, Prieto MC. HDAC9 is an epigenetic repressor of kidney angiotensinogen establishing a sex difference. Biol Sex Differ. 2017;8:18. doi: 10.1186/s13293-017-0140-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Araujo FC, Milsted A, Watanabe IKM, Del Puerto HL, Santos RAS, Lazar J, et al. Similarities and differences of X and Y chromosome homologous genes, SRY and SOX3, in regulating the renin-angiotensin system promoters. Physiol Genomics. 2015;47:177–186. doi: 10.1152/physiolgenomics.00138.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jagarlamudi K, Rajkovic A. Oogenesis: transcriptional regulators and mouse models. Mol Cell Endocrinol. 2012;356:31–39. doi: 10.1016/j.mce.2011.07.049. [DOI] [PubMed] [Google Scholar]
- 44.MacLennan M, Crichton JH, Playfoot CJ, Adams IR. Oocyte development, meiosis and aneuploidy. Semin Cell Dev Biol. 2015;45:68–76. doi: 10.1016/j.semcdb.2015.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.GAL C, Couty I, Hennequet-Antier C, Govoroun MS. Gene expression profiling reveals new potential players of gonad differentiation in the chicken embryo. PLoS One. 2011;6:e23959. doi: 10.1371/journal.pone.0023959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gyllenhammar I, Eriksson H, Söderqvist A, Lindberg RH, Fick J, Berg C. Clotrimazole exposure modulates aromatase activity in gonads and brain during gonadal differentiation in Xenopus tropicalis frogs. Aquat Toxicol. 2009;91:0–109. doi: 10.1016/j.aquatox.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 47.Hess RA, Bunick D, Lee KH, Bahr J, Lubahn DB. A role for Oestrogens in the male reproductive system. Nature. 1997;390:509–512. doi: 10.1038/37352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Séralini GE, Moslemi S. Aromatase inhibitors: past, present and future. Mol Cell Endocrinol. 2001;178:117–131. doi: 10.1016/s0303-7207(01)00433-6. [DOI] [PubMed] [Google Scholar]
- 49.Dvorak J, Halverson JL, Gulick P, Rauen KA, Abbott UK, Kelly BJ, et al. cDNA cloning of a Z- and W-linked gene in gallinaceous birds. J Hered. 1992;1:1. [Google Scholar]
- 50.Bantock TM, PRYS-JONES RP, Lee PL. New and improved molecular sexing methods for museum bird specimens. Mol Ecol Resour. 2008;8:519–528. doi: 10.1111/j.1471-8286.2007.01999.x. [DOI] [PubMed] [Google Scholar]
- 51.Nanda I, Zend-Ajusch E, Shan Z, Grützner F, Schartl M, Burt D, et al. Conserved synteny between the chicken Z sex chromosome and human chromosome 9 includes the male regulatory gene DMRT1: a comparative (re) view on avian sex determination. Cytogenet Genom Res. 2000;89:67–78. doi: 10.1159/000015567. [DOI] [PubMed] [Google Scholar]
- 52.Yamauchi M, Kawai S, Kato T, Ooshima T, Amano A. Odd-skipped related 1 gene expression is regulated by Runx2 and Ikzf1 transcription factors. Gene. 2008;426:81–90. doi: 10.1016/j.gene.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 53.Han L, Yu L, Jingyue X, Ching-Fang C, Brugmann SA, Rulang J. Odd-skipped related-1 controls neural crest chondrogenesis during tongue development. Pnas. 2013;110:18555–18560. doi: 10.1073/pnas.1306495110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gui-yu Zhu. The Expression and Regulation of ANXA2 Gene in the Chicken Ovary. J Shandong Agric Univ (Natural Science Edition). 2014;3:5.
- 55.Zhu G, Chen X, Mao Y, Kang L, Ma X, Jiang Y. Characterization of annexin A2 in chicken follicle development: evidence for its involvement in angiogenesis. Anim Reprod Sci. 2015;161:104–111. doi: 10.1016/j.anireprosci.2015.08.011. [DOI] [PubMed] [Google Scholar]
- 56.Barboro P, Ferrari N, Balbi C. Emerging roles of heterogeneous nuclear ribonucleoprotein K (hnRNP K) in cancer progression. Cancer Lett. 2014;352:152–159. doi: 10.1016/j.canlet.2014.06.019. [DOI] [PubMed] [Google Scholar]
- 57.Gallardo M, Hornbaker MJ, Zhang X, Hu P, Bueso-Ramos C, Post SM. Aberrant hnRNP K expression: all roads lead to cancer. Cell Cycle. 2016;15(12):1552. doi: 10.1080/15384101.2016.1164372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He L, Xue X, Wang Z, Hou E, Liu Y, Liang M, et al. Transcriptional regulation of heterogeneous nuclear ribonucleoprotein K gene expression. Biochimie. 2015;109:27–35. doi: 10.1016/j.biochi.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ping Z, Ningling W, Xianhua L, Li J, Hong X. Expression and localization of heterogeneous nuclear ribonucleoprotein K in mouse ovaries and preimplantation embryos. Biochem Biophys Res Commun. 2016;471(1):260. doi: 10.1016/j.bbrc.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 60.Euteneuer AM, Seeger-Nukpezah T, Nolte H, Henjakovic M. Estrogen receptor α (ERα) indirectly induces transcription of human renal organic anion transporter 1 (OAT1) Phys Rep. 2019;7:e14229. doi: 10.14814/phy2.14229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nam K, Ellegren H. The chicken (Gallus gallus) Z chromosome contains at least three nonlinear evolutionary strata. Genetics. 2008;180:1131–1136. doi: 10.1534/genetics.108.090324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qi YM, Lei T, Zhou L, Chen XD, Long QQ, Long H, et al. Genomic organization, alternative splicing and tissues expression of porcine CREB3L4 gene. Mol Biol Rep. 2009;36:1881–1888. doi: 10.1007/s11033-008-9394-1. [DOI] [PubMed] [Google Scholar]
- 63.Labrie C, Lessard J, Aicha SB, Savard M-P, Pelletier M, Fournier A, et al. Androgen-regulated transcription factor AIbZIP in prostate cancer. J Steroid Biochem Mol Biol. 2008;108:0–244. doi: 10.1016/j.jsbmb.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 64.El-Alfy M. Stage-specific expression of the Atce1/Tisp40? Isoform of CREB3L4 in mouse spermatids. J Androl. 2006;27:686–694. doi: 10.2164/jandrol.106.000596. [DOI] [PubMed] [Google Scholar]
- 65.Adham IM, Eck TJ, Mierau K, Müller N, Engel W. Reduction of spermatogenesis but not fertility in Creb3l4-deficient mice. Mol Cell Biol. 2005;25:7657–7664. doi: 10.1128/MCB.25.17.7657-7664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cizkova M, Vacher S, Meseure D, Trassard M, Susini A, Mlcuchova D, et al. PIK3R1 underexpression is an independent prognostic marker in breast cancer. BMC Cancer. 2013;13:545. doi: 10.1186/1471-2407-13-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thauvin-Robinet C, Auclair M, Duplomb L, Caron-Debarle M, Avila M, St-Onge J, et al. PIK3R1 mutations cause syndromic insulin resistance with lipoatrophy. Am J Hum Genet. 2013;93:141–149. doi: 10.1016/j.ajhg.2013.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang L, Liu W, Shao C, Zhang N, Li H, Liu K, et al. Cloning, expression and methylation analysis of piwil2 in half-smooth tongue sole ( Cynoglossus semilaevis ) Mar Genomics. 2014;18:45–54. doi: 10.1016/j.margen.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 69.Warr N, Bogani D, Siggers P, Brixey R, Tateossian H, Dopplapudi A, et al. Minor abnormalities of testis development in mice lacking the gene encoding the MAPK signalling component, MAP 3K1. PLoS One. 2011;6:e19572. doi: 10.1371/journal.pone.0019572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Loke J, Pearlman A, Radi O, Zuffardi O, Giussani U, Pallotta R, et al. Mutations in MAP 3K1 tilt the balance from SOX9/FGF9 to WNT/β-catenin signaling. Hum Mol Genet. 2013;23:1073–1083. doi: 10.1093/hmg/ddt502. [DOI] [PubMed] [Google Scholar]
- 71.Nie G. Serine peptidase HTRA3 is closely associated with human placental development and is elevated in pregnancy serum. Biol Reprod. 2006;74:366–374. doi: 10.1095/biolreprod.105.047324. [DOI] [PubMed] [Google Scholar]
- 72.Bowden MA, Li Y, Liu Y-X, Findlay JK, Salamonsen LA, Nie G. HTRA3 expression in non-pregnant rhesus monkey ovary and endometrium, and at the maternal-fetal interface during early pregnancy. Reprod Biol Endocrinol. 2008;6:22. doi: 10.1186/1477-7827-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bowden M, Drummond AE, Salamonsen LA, Findlay JK, Nie G. Evolutionary conservation of mammalian HTRA3 and its developmental regulation in the rat ovary. J Exp Zool Part B Mol Dev Evol. 2009;312:701–713. doi: 10.1002/jez.b.21288. [DOI] [PubMed] [Google Scholar]
- 74.Xudong W, Guiying Z, Guowen F, Yonggang L. Molecular cloning of the porcine HTRA3 gene and association of a SNP with litter size traits. Folia Biol. 2017;63:217–221. doi: 10.14712/fb2017063050217. [DOI] [PubMed] [Google Scholar]
- 75.Tocharus J, Tsuchiya A, Kajikawa M, Ueta Y, Kawaichi M. Developmentally regulated expression of mouse HtrA3 and its role as an inhibitor of TGF-β signaling. Develop Growth Differ. 2004;46:257–274. doi: 10.1111/j.1440-169X.2004.00743.x. [DOI] [PubMed] [Google Scholar]
- 76.Knight MN, Hankenson KD. R-spondins: novel matricellular regulators of the skeleton. Matrix Biol J Int Soc Matrix Biol. 2014;37:157–161. doi: 10.1016/j.matbio.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 77.Fan H-Y, O'Connor A, Shitanaka M, Shimada M, Liu Z, Richards JS. β-Catenin (CTNNB1) promotes preovulatory follicular development but represses LH-mediated ovulation and luteinization. Mol Endocrinol. 2010;24:1529–1542. doi: 10.1210/me.2010-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sanchez AM, Viganò P, Quattrone F, Pagliardini L, Papaleo E, Candiani M, et al. The WNT/β-catenin signaling pathway and expression of survival promoting genes in luteinized granulosa cells: endometriosis as a paradigm for a dysregulated apoptosis pathway. Fertil Steril. 2014;101:1688–1696. doi: 10.1016/j.fertnstert.2014.02.040. [DOI] [PubMed] [Google Scholar]
- 79.Kazanskaya O, Ohkawara B, Heroult M, Wu W, Maltry N, Augustin HG, et al. The Wnt signaling regulator R-spondin 3 promotes angioblast and vascular development. Development. 2008;135:3655–3664. doi: 10.1242/dev.027284. [DOI] [PubMed] [Google Scholar]
- 80.Zhang K, Watanabe M, Kashiwakura Y, Li S-A, Edamura K, Huang P, et al. Expression pattern of REIC/Dkk-3 in various cell types and the implications of the soluble form in prostatic acinar development. Int J Oncol. 2010;37:1495–1501. doi: 10.3892/ijo_00000802. [DOI] [PubMed] [Google Scholar]
- 81.Mizobuchi Y, Matsuzaki K, Kuwayama K, Kitazato K, Mure H, Kageji T, et al. REIC/Dkk-3 induces cell death in human malignant glioma. Neuro-oncology. 2008;10:244–253. doi: 10.1215/15228517-2008-016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dellinger TH, Planutis K, Jandial DD, Eskander RN, Martinez ME, Zi X, et al. Expression of the Wnt antagonist Dickkopf-3 is associated with prognostic clinicopathologic characteristics and impairs proliferation and invasion in endometrial cancer. Gynecol Oncol. 2012;126:259–267. doi: 10.1016/j.ygyno.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nakamura RE, Hunter DD, Yi H, Brunken WJ, Hackam AS. Identification of two novel activities of the Wnt signaling regulator Dickkopf 3 and characterization of its expression in the mouse retina. BMC Cell Biol. 2007;8:1–10. doi: 10.1186/1471-2121-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Caricasole A, Ferraro T, Iacovelli L, Barletta E, Caruso A, Melchiorri D, et al. Functional characterization of WNT7A signaling in PC12 cells interaction with a fzd5 lrp6 receptor complex and modulation by dickkopf proteins. J Biol Chem. 2003;278:37024–37031. doi: 10.1074/jbc.M300191200. [DOI] [PubMed] [Google Scholar]
- 85.Hoang BH, Kubo T, Healey JH, Yang R, Nathan SS, Kolb EA, et al. Dickkopf 3 inhibits invasion and motility of Saos-2 osteosarcoma cells by modulating the Wnt-β-catenin pathway. Cancer Res. 2004;64:2734–2739. doi: 10.1158/0008-5472.can-03-1952. [DOI] [PubMed] [Google Scholar]
- 86.Kawano Y, Kitaoka M, Hamada Y, Walker MM, Waxman J, Kypta RM. Regulation of prostate cell growth and morphogenesis by Dickkopf-3. Oncogene. 2006;25:6528–6537. doi: 10.1038/sj.onc.1209661. [DOI] [PubMed] [Google Scholar]
- 87.Lee EJ, Jo M, Rho SB, Park K, Yoo YN, Park J, et al. Dkk3, downregulated in cervical cancer, functions as a negative regulator of beta-catenin. Int J Cancer. 2010;124:287–297. doi: 10.1002/ijc.23913. [DOI] [PubMed] [Google Scholar]
- 88.Wang XY, Yin Y, Yuan H, Sakamaki T, Okano H, Glazer RI. Musashi1 modulates mammary progenitor cell expansion through Proliferin-mediated activation of the Wnt and notch pathways. Mol Cell Biol. 2008;28:3589–3599. doi: 10.1128/MCB.00040-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wen Y, Sun Q, Sanja D, Landreneau RJ, Siegfried JM, Jian Y, et al. Downregulation of Dkk3 activates β-catenin/TCF-4 signaling in lung cancer. Carcinogenesis. 2007;1:84. doi: 10.1093/carcin/bgm267. [DOI] [PubMed] [Google Scholar]
- 90.Lu D, Bao D, Dong W, Liu N, Zhang X, Gao S, et al. Dkk3 prevents familial dilated cardiomyopathy development through Wnt pathway. Lab Investig. 2016;96(2):239. doi: 10.1038/labinvest.2015.145. [DOI] [PubMed] [Google Scholar]
- 91.Xu Y, Li X, Wang H, Xie P, Yan X, Bai Y, et al. Hypermethylation of CDH13, DKK3 and FOXL2 promoters and the expression of EZH2 in ovary granulosa cell tumors. Mol Med Rep. 2008;14:2739–2745. doi: 10.3892/mmr.2016.5521. [DOI] [PubMed] [Google Scholar]
- 92.Shen B, Zhang Z, Wang Y, Wang G, Chen Y, Lin P, et al. Differential expression of ubiquitin-conjugating enzyme E2r in the developing ovary and testis of penaeid shrimp Marsupenaeus japonicus. Mol Biol Rep. 2009;36:1149–1157. doi: 10.1007/s11033-008-9291-7. [DOI] [PubMed] [Google Scholar]
- 93.Zhang F, Chen L, Wu P, Zhao W, Li E, Qin J. cDNA cloning and expression of Ubc9 in the developing embryo and ovary of oriental river prawn, Macrobrachium nipponense. Comp Biochem Physiol B Biochem Mol Biol. 2010;155:288–293. doi: 10.1016/j.cbpb.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 94.Itoh Y, Kampf K, Arnold AP. Comparison of the chicken and zebra finch Z chromosomes shows evolutionary rearrangements. Chromosom Res. 2006;14:805–815. doi: 10.1007/s10577-006-1082-1. [DOI] [PubMed] [Google Scholar]
- 95.Scholz B, Kultima K, Mattsson A, Axelsson J, Brunström B, Halldin K, et al. Sex-dependent gene expression in early brain development of chicken embryos. BMC Neurosci. 2006;7:12. doi: 10.1186/1471-2202-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Simon A, Pyl PT, Wolfgang H. HTSeq--a python framework to work with high-throughput sequencing data. Bioinformatics (Oxford, England) 2015;31(2):166. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, Van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2007;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analysed during the current study are available in the NCBI Sequence Read Archive database (https://www.ncbi.nlm.nih.gov/sra), under the accession number PRJNA625194 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA625194), SRX8108890, SRX8108889, SRX8108888, SRX8108887, SRX8108886, SRX8108885, SRX8108884, SRX8108883, SRX8108882, SRX8108881, SRX8108880 and SRX8108879. The name of the bioproject is Muscovy ducks sequencing. The link to the Anas platyrhynchos genome is https://www.ncbi.nlm.nih.gov/genome/?term=Anas+platyrhynchos. The primer information for β-actin (GeneBank accession number EF667345.1) is available in Table 3.