Abstract
Background
Medulloblastoma (MB) is one of the most common malignant cancers in children. MB is primarily classified into four subgroups based on molecular and clinical characteristics as (1) WNT (2) Sonic-hedgehog (SHH) (3) Group 3 (4) Group 4. Molecular characteristics used for MB classification are based on genomic and mRNAs profiles. MB subgroups share genomic and mRNA profiles and require multiple molecular markers for differentiation from each other. Long non-coding RNAs (lncRNAs) are more than 200 nucleotide long RNAs and primarily involve in gene regulation at epigenetic and post-transcriptional levels. LncRNAs have been recognized as diagnostic and prognostic markers in several cancers. However, the lncRNA expression profile of MB is unknown.
Methods
We used the publicly available gene expression datasets for the profiling of lncRNA expression across MB subgroups. Functional analysis of differentially expressed lncRNAs was accomplished by Ingenuity pathway analysis (IPA).
Results
In the current study, we have identified and validated the lncRNA expression profile across pediatric MB subgroups and associated molecular pathways. We have also identified the prognostic significance of lncRNAs and unique lncRNAs associated with each MB subgroup.
Conclusions
Identified lncRNAs can be used as single biomarkers for molecular identification of MB subgroups that warrant further investigation and functional validation.
Keywords: Long non-coding RNA, Pediatric Medulloblastoma, Cancer biomarkers, Gene expression and pathways, Therapeutic targets
Background
Medulloblastoma (MB), the most common pediatric brain tumor, constitutes nearly 20% of newly diagnosed brain tumors in children [1, 2]. Treatment of MB involves radiation therapy, chemotherapy and surgical resection. These strategies have improved the survival by 70–80% but also lead to serious morbidities [3, 4]. MB are classified into four major molecular subgroups as WNT, Sonic hedgehog (SHH), Group 3 and Group 4. The WNT subgroup is least common among all 4 subgroups and present in only 10% of cases. Genetic changes in genes: CTNNB1, DDX3X, SMARCA4 and DKK1 are frequently observed in the WNT subgroup. WNT has the best prognosis among all types of MB. SHH is second most common subgroup with abnormalities in SHH signaling pathway and accounts for ~ 30% of total MB cases. Genetic anomalies in genes: MYCN, GLI1, PTCH1, SUFU, MLL2, SMO, TP53, BCOR1, GAB1, GABRG1 and LDB1 are frequently seen in the SHH subgroup. The SHH subgroup has an intermediate prognosis among MB subgroups. Group 3 is the third most common subgroup with 25% of the total MB cases. Group 3 is mainly MYC-driven and genetic aberrations are seen in genes: MYC, PVT1, OTX2, MLL2, SMARCA4, and CHD7 in this subgroup. The prognosis of the Group 3 is very poor and 5 year overall survival is less than 50%. Group 4 is the most common subgroup of MB and accounts for 35% of total cases. The prognosis of the Group 4 is intermediate and genetic aberrations are commonly present in genes: OTX2, DDX31, CHD7, NCAIP, MYCN, CDK6, GFI1/GFI1B, MLL2, KDM6A, MLL3, and ZMYM3 [5–9]. Molecular markers used for WNT identification are CTNNB1 (nuclear), FLIA, YAP1 and DKK1; for SHH are SFRP1, GLI1, FLIA, YAP1 and GAB1; for Group 3, NPR3; and for Group 4, KCNA1. Identification of new molecular markers for drug targeting, diagnosis and prognosis are important due to need for improved molecular profiling of MB [10].
Long non-coding RNAs (LncRNAs) are RNAs of more than 200 bp in length and can be transcribed from an intergenic region, genic regions or super enhancer regions in the genome. LncRNAs can modulate chromatin structure, gene regulation via interactions with epigenetic modifiers and transcriptional co-factors, and also have post-translation effects via affecting the stability of mRNA or proteins [11, 12]. Deregulated lncRNA expression is associated with many cancers [13]. LncRNA signatures have been used to classify different types of cancer as biomarkers for diagnosis, prognosis and therapy [14–18]. LncRNAs are secreted in serum, plasma, and CSF in a stable form protected from endogenous RNAase and can be used for non-invasive analysis from patient samples [19, 20]. The role of lncRNA in brain development is well studied [21–26]. However, there is not much known about role of lncRNAs in MB. LncRNA LOXL1-AS1 promotes the proliferation and metastasis of MB by activating the PI3K-AKT pathway [27]. LncRNA CCAT1 promotes cell proliferation and metastasis in human MB by regulating the MAPK pathway [28]. Silencing of ANRIL in MB cell lines significantly lowered cell viability and migration. ANRIL promoted the apoptosis of MB cell lines through miR-323-mediated regulation of BRI3, which activates p38 MAPK, ERK, and AKT as well as the WNT signaling pathway [29]. LINC-NeD125 expression is upregulated in Group 4 MB and after interacting to miRNA-induced silencing complex(MISC), it directly binds to miR-19a-3p, miR-19b-3p and miR-106a-5p. Functionally, LINC-NeD125 acts by sequestering the three miRNAs, which leads to the de-repression of major driver genes (CDK6, MYCN, SNCAIP, and KDM6A) of Group 4 MB [30]. LncRNA CRNDE expression is elevated in MB and knockdown of CRNDE significantly reduced cell proliferation and inhibited colony formation in MB cell lines, Daoy and D341 [31].
In the current study, we have identified the lncRNAs expression profile of pediatric MB subgroups and associated molecular pathways. We have also identified the unique lncRNAs associated with each subgroup.
Methods
We searched the Gene Expression Omnibus (GEO) database for MB related microarray datasets and found two relevant studies, GSE37418 [for pediatric MB subgroups expression data] and GSM1094863, GSM1094864, GSM1094865, GSM1094866, GSM1094867 [for pediatric primary cerebellum expression data from GSE44971] for our analyses. We further used large GSE124814 datasets for the validation of lncRNAs expression profiles of MB subgroups obtained from our original analyses. We selected the age < 18 years as an inclusion criteria for selecting pediatric MB samples. We selected the datasets which used the Affymetrix U133 Plus2 array for probe level RNA expression studies. For data analyses, we first did background correction, normalization (RMA), quality control checks, intensity and batch effect corrections of each dataset. Following that, we did probe level differential analyses of datasets using the limma package (ANOVA with eBayes) with criteria of p < 0.001 and fold change greater than two folds. We then annotated the probe sets with the Affymetrix U133 Plus2 library and filtered out lncRNA genes. The lncRNA gene database used is verified and approved by HGNC. Functional analysis of differentially expressed lncRNAs was done by Ingenuity pathway analysis (IPA) software from BioRad, Inc. We used default parameters and checked all the node types, all species (except uncharted), and all tissue types for core analysis in IPA.
Results
Differentially expressed lncRNAs in the WNT subgroup and their functional roles
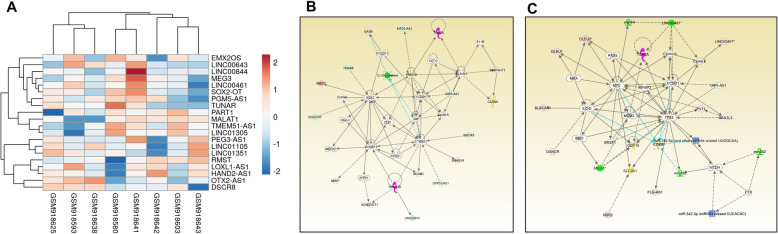
Comparative analyses of WNT MB (N = 8) and normal cerebellum tissue (N = 5) datasets with p < 0.05 and fold changes > 2 provided 199 differentially expressed lncRNAs with approved status. Tables 1 and 2 show the fold change in the top 10 upregulated and downregulated lncRNAs. Heatmap of top 10 upregulated and downregulated lncRNAs is shown in Fig. 1a. The complete list of lncRNAs can be seen in Additional file 1. We found 73% overlap with lncRNAs in validation datasets [WNT N = 31, Control = 5] (Additional file 2). We found all the top 10 upregulated and downregulated lncRNAs present in validation datasets. We mostly see non-overlap in lncRNAs at lower expression values.
Table 1.
Top 10 up-regulated lncRNAs in WNT subgroup of MB
| Gene Symbol | Fold Change | P-val | FDR P-val |
|---|---|---|---|
| EMX2OS | 38.18 | 9.01E-14 | 4.92E-10 |
| OTX2-AS1 | 37.84 | 1.14E-09 | 3.93E-07 |
| PGM5-AS1 | 30.26 | 9.54E-09 | 1.63E-06 |
| DSCR8 | 24.56 | 0.0001 | 0.0013 |
| LOXL1-AS1 | 21.06 | 1.03E-08 | 1.73E-06 |
| HAND2-AS1 | 18.51 | 9.37E-07 | 3.59E-05 |
| TMEM51-AS1 | 16.9 | 3.88E-09 | 8.82E-07 |
| RMST | 13.92 | 1.14E-07 | 8.49E-06 |
| LINC01305 | 11.11 | 0.0001 | 0.001 |
| PART1 | 10.94 | 1.89E-05 | 0.0003 |
Table 2.
Top 10 down-regulated lncRNAs in WNT subgroup of MB
| Gene Symbol | Fold Change | P-val | FDR P-val |
|---|---|---|---|
| LINC00461 | −62.39 | 1.16E-06 | 4.17E-05 |
| MEG3 | −58.9 | 9.41E-07 | 3.60E-05 |
| LINC00844 | −24.94 | 0.0003 | 0.0024 |
| LINC00643 | −13.3 | 6.95E-06 | 0.0001 |
| SOX2-OT | −12.13 | 0.0003 | 0.0024 |
| PEG3-AS1 | −10.02 | 3.49E-07 | 1.84E-05 |
| TUNAR | −7.72 | 2.04E-12 | 5.87E-09 |
| MALAT1 | −7.39 | 1.65E-07 | 1.10E-05 |
| LINC01105 | −7.39 | 3.77E-05 | 0.0005 |
| LINC01351 | −6.48 | 2.24E-05 | 0.0003 |
Fig. 1.
a: Heatmap of top 10 upregulated and downregulated lncRNAs in WNT subgroup. Expression value of different lncRNAs was clustered using correlation distance method. b: Differentially expressed lncRNAs in a non-canonical biological network in WNT subgroup. The important nodes in this biological network are CCND1, AKT, SOX2, POU5F1, DNMT3B, and CTNNB1. c: Differentially expressed lncRNAs in another non-canonical biological network in WNT subgroup. The important nodes in this biological network are TP53, MYC, EZH2, and MDM2. Green indicates downregulated and red indicates upregulated lncRNAs
We did functional analysis of differentially expressed (DE) lncRNAs of the WNT subgroup using IPA. We identified different functional parameters involved in this subgroup. MAX (a MYC interacting partner), miR-150, miR-133a, FOLR1, E2F NCAM1, GAS2L3 and ATF5 are the most significantly associated upstream regulators, while cancer, neurogenesis, metastasis and cellular development are the most important biological functions affected in this subgroup (Tables 3 and 4). Heatmap of 5 upstream regulators is shown in supplementary Fig. 1 (Additional file 3). The two most important non-canonical networks enriched with DE lncRNAs are shown in Fig. 1b and c. In networks 1; CCND1, AKT, SOX2, POU5F1, DNMT3B, and CTNNB1, in network 2; TP53, MYC, EZH2, and MDM2 are the central regulators linked with DE lncRNAs.
Table 3.
Top 10 upstream regulators involved in DE lncRNAs in WNT subgroup
| Upstream Regulator | Molecule Type | P-val of overlap | Target molecules in dataset |
|---|---|---|---|
| MAX | transcription regulator | 5.53E-03 | DLEU1,DLEU2 |
| miR-150-5p (and other miRNAs w/seed CUCCCAA) | mature microRNA | 5.54E-03 | MIAT |
| miR-133a-3p (and other miRNAs w/seed UUGGUCC) | mature microRNA | 7.38E-03 | MALAT1 |
| mir-133 | microRNA | 9.22E-03 | MALAT1 |
| FOLR1 | transporter | 1.31E-02 | GAS5,PVT1 |
| E2f | group | 1.37E-02 | DLEU1,DLEU2 |
| ATF5 | transcription regulator | 1.47E-02 | GAS5 |
| NCAM1 | other | 1.84E-02 | MALAT1 |
| mir-150 | microRNA | 2.56E-02 | MIAT |
| GAS2L3 | other | 2.92E-02 | PVT1 |
Table 4.
Top 10 disease and function identified by IPA from DE lncRNAs in WNT subgroup
| Categories | Diseases or Functions Annotation | P-val | Activation z-score |
|---|---|---|---|
| Cellular Development, Cellular Growth and Proliferation, Nervous System Development and Function | Neurogenesis of nervous tissue cell lines | 3.38E-06 | |
| Cellular Movement | Cell movement of tumor cell lines | 1.12E-05 | 1.324 |
| Cellular Movement | Migration of tumor cell lines | 1.14E-05 | 1.498 |
| Cellular Movement | Invasion of tumor cell lines | 1.55E-04 | 1.083 |
| Cell Cycle | Arrest in G0 phase of tumor cell lines | 3.83E-04 | |
| Cancer, Organismal Injury and Abnormalities | Metastasis of tumor cell lines | 4.26E-04 | −0.277 |
| Cell Death and Survival | Cell death of eye cell lines | 5.07E-04 | |
| Cellular Movement | Migration of cells | 6.27E-04 | 0.573 |
| Cellular Movement | Cell movement | 6.75E-04 | 0.453 |
| Cellular Movement | Migration of hepatoma cell lines | 1.34E-03 |
Differentially expressed lncRNAs in the SHH subgroup and their functional roles
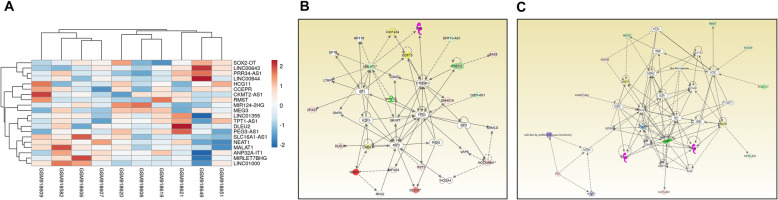
Comparative analyses of the SHH subgroup (N = 10) and normal cerebellum tissue (N = 5) datasets with p < 0.05 and fold change > 2 provided 145 differentially expressed lncRNAs with approved status. Tables 5 and 6 show the fold change in the top 10 upregulated and downregulated lncRNAs. Heatmap of top 10 upregulated and downregulated lncRNAs is shown in Fig. 2a. The complete list of lncRNAs can be seen in Additional file 1. We found 50% overlap with lncRNAs in validation datasets [SHH N = 65, Control = 5] (Additional file 2). We found all the top 10, upregulated and downregulated lncRNAs, present in validation datasets except DLEU2 and PRR34-AS1.
Table 5.
Top 10 up-regulated lncRNAs in SHH subgroup of MB
| Gene Symbol | Fold Change | P-val | FDR P-val |
|---|---|---|---|
| NEAT1 | 23.48 | 0.0003 | 0.0022 |
| DLEU2 | 13.24 | 5.79E-11 | 2.41E-08 |
| PRR34-AS1 | 8.07 | 1.58E-07 | 8.49E-06 |
| LINC01355 | 8.05 | 2.93E-09 | 5.15E-07 |
| MIRLET7BHG | 7.49 | 1.79E-07 | 9.34E-06 |
| CKMT2-AS1 | 6.23 | 1.86E-09 | 3.59E-07 |
| SLC16A1-AS1 | 5.65 | 8.13E-08 | 5.36E-06 |
| TPT1-AS1 | 5.28 | 4.44E-08 | 3.50E-06 |
| LINC01000 | 4.96 | 1.10E-08 | 1.32E-06 |
| ANP32A-IT1 | 4.94 | 9.36E-07 | 3.07E-05 |
Table 6.
Top 10 down-regulated lncRNAs in SHH subgroup of MB
| Gene Symbol | Fold Change | P-val | FDR P-val |
|---|---|---|---|
| LINC00844 | −33.36 | 1.08E-08 | 1.32E-06 |
| MIR124-2HG | −28.13 | 0.0005 | 0.0032 |
| SOX2-OT | −13.83 | 2.39E-07 | 1.15E-05 |
| PEG3-AS1 | −12.94 | 5.12E-08 | 3.88E-06 |
| LINC00643 | −11.76 | 3.97E-06 | 8.83E-05 |
| HCG11 | −11.26 | 0.0012 | 0.0065 |
| RMST | −9.43 | 0.0036 | 0.0155 |
| CCEPR | −8.93 | 1.49E-06 | 4.27E-05 |
| MEG3 | −8.53 | 0.0002 | 0.0018 |
| MALAT1 | −8.25 | 2.82E-06 | 6.86E-05 |
Fig. 2.
a: Heatmap of top 10 upregulated and downregulated lncRNAs in SHH subgroup. Expression value of different lncRNAs was clustered using correlation distance method. b: Differentially expressed lncRNAs in a non-canonical biological network in SHH subgroup. The important nodes in this biological network are CCND1, TP53, MYC, MALAT1, CTNNB1, and SP1. c: Differentially expressed lncRNAs in another non-canonical biological network in SHH subgroup. The important nodes in this biological network are Histone H3, MDM2, CCNA2, SOX2, POU2F1, SP1, and ESR1. Green indicates downregulated and red indicates upregulated lncRNAs
Functional analysis of DE lncRNAs of SHH MB subgroup using IPA predicts, MAX (a MYC interacting partner), miR-133a, FOLR1, E2F, ATF5, AM1, E2F3, GAS2L3 and ACSL5 as most significantly associated upstream regulators, while cancer, neurogenesis, cell proliferation, metastasis and cellular development are the most important biological functions affected in this subgroup (Tables 7 and 8). Heatmap of 5 upstream regulators is shown in supplementary Fig. 1 (Additional file 3). The two most important non-canonical networks enriched with DE lncRNAs are shown in Fig. 2b and c. In network 1; CCND1, TP53, MYC, MALAT1, CTNNB1, and SP1, in network 2; Histone H3, MDM2, CCNA2, SOX2, POU2F1, SP1, and ESR1 are the central regulators linked with DE lncRNAs.
Table 7.
Top upstream regulators involved in DE lncRNAs in SHH subgroup
| Upstream Regulator | Molecule Type | P-val of overlap | Target molecules in dataset |
|---|---|---|---|
| MAX | transcription regulator | 2.74E-03 | DLEU1,DLEU2 |
| miR-133a-3p (and other miRNAs w/seed UUGGUCC) | mature microRNA | 5.17E-03 | MALAT1 |
| mir-133 | microRNA | 6.46E-03 | MALAT1 |
| FOLR1 | transporter | 6.58E-03 | GAS5,PVT1 |
| E2f | group | 6.85E-03 | DLEU1,DLEU2 |
| ATF5 | transcription regulator | 1.03E-02 | GAS5 |
| NCAM1 | other | 1.29E-02 | MALAT1 |
| E2F3 | transcription regulator | 1.49E-02 | MALAT1,NEAT1 |
| GAS2L3 | other | 2.05E-02 | PVT1 |
| ACSL5 | enzyme | 2.18E-02 | ST7-AS1 |
Table 8.
Top 10 disease and function identified by IPA from DE lncRNAs in SHH subgroup
| Categories | Diseases or Functions Annotation | P-val | Activation z-score |
|---|---|---|---|
| Cellular Development, Cellular Growth and Proliferation, Nervous System Development and Function | Neurogenesis of nervous tissue cell lines | 1.79E-06 | |
| Cellular Development, Cellular Growth and Proliferation | Proliferation of kidney cancer cell lines | 3.01E-06 | −0.095 |
| Cellular Development, Cellular Growth and Proliferation | Cell proliferation of tumor cell lines | 3.90E-04 | 0.933 |
| Cellular Movement | Migration of carcinoma cell lines | 5.25E-04 | 0.762 |
| Cellular Movement | Migration of kidney cancer cell lines | 6.63E-04 | |
| Cellular Movement | Cell movement of tumor cell lines | 6.65E-04 | 0.751 |
| Cellular Movement | Migration of tumor cell lines | 1.14E-03 | 1.033 |
| Cellular Development, Cellular Growth and Proliferation | Cell proliferation of carcinoma cell lines | 1.30E-03 | 0.277 |
| Cellular Development, Connective Tissue Development and Function, Tissue Development | Osteogenic differentiation of nucleus pulposus cells | 1.36E-03 | |
| Cancer, Gastrointestinal Disease, Organismal Injury and Abnormalities | Stage I colorectal adenocarcinoma | 1.36E-03 |
Differentially expressed lncRNAs in the Group 3 subgroup and their functional roles
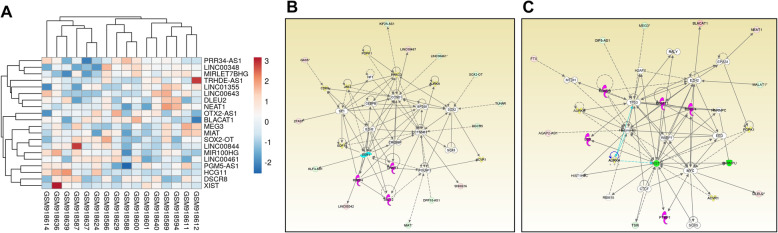
Comparative analyses of the Group 3 MB (N = 16) and normal cerebellum tissue (N = 5) datasets with p < 0.05 and fold change > 2 provided 149 differentially expressed lncRNAs with approved status. Tables 9 and 10 show the fold change in the top 10 upregulated and downregulated lncRNAs. Heatmap of top 10 upregulated and downregulated lncRNAs is shown in Fig. 3a. The complete list of lncRNAs can be seen in Additional file 1. We found 86% overlap with lncRNAs in validation datasets [Group 3 N = 46, Control N = 5] (Additional file 2). We found all the top 10 upregulated and downregulated lncRNAs in the validation dataset, except NEAT1.
Table 9.
Top 10 up-regulated lncRNAs in Group 3 of MB
| Gene Symbol | Fold Change | P-val | FDR P-val |
|---|---|---|---|
| OTX2-AS1 | 80.96 | 1.03E-14 | 1.38E-11 |
| BLACAT1 | 24.59 | 6.32E-08 | 3.32E-06 |
| LINC00348 | 19.06 | 0.0013 | 0.0077 |
| LINC01355 | 9.91 | 1.54E-08 | 1.09E-06 |
| DLEU2 | 8.97 | 2.31E-09 | 2.49E-07 |
| PGM5-AS1 | 7.73 | 4.32E-05 | 0.0005 |
| NEAT1 | 7.27 | 0.0082 | 0.0315 |
| DSCR8 | 6.73 | 0.0104 | 0.0378 |
| PRR34-AS1 | 6.63 | 1.37E-06 | 3.58E-05 |
| MIRLET7BHG | 6.32 | 0.0004 | 0.003 |
Table 10.
Top 10 down-regulated lncRNAs in Group 3 of MB
| Gene Symbol | Fold Change | P-val | FDR P-val |
|---|---|---|---|
| XIST | −315.1 | 0.0066 | 0.0267 |
| MEG3 | −58.01 | 0.0001 | 0.0011 |
| SOX2-OT | −50.3 | 1.15E-14 | 1.44E-11 |
| LINC00844 | −37.6 | 6.71E-08 | 3.47E-06 |
| MIR100HG | −16.61 | 1.16E-06 | 3.15E-05 |
| HCG11 | −12.99 | 1.93E-05 | 0.0003 |
| MIAT | −10.36 | 0.0002 | 0.0016 |
| LINC00461 | −9.38 | 0.002 | 0.0105 |
| LINC00643 | −9.34 | 0.0005 | 0.0036 |
| TRHDE-AS1 | −7.59 | 7.27E-09 | 6.14E-07 |
Fig. 3.
a: Heatmap of top 10 upregulated and downregulated lncRNAs in Group 3 MB. Expression value of different lncRNAs was clustered using correlation distance method. b: Differentially expressed lncRNAs in a non-canonical biological network in Group 3 MB. The important nodes in this biological network are CCND1, EP300, CREBBP, ESR1, CTNNB1, and PRKCD. c: Differentially expressed lncRNAs in another non-canonical biological network in Group 3 MB. The important nodes in this biological network are Histone H3, TP53, MYC, XIST, and EZH2. Green indicates downregulated and red indicates upregulated lncRNAs
Functional analysis of DE lncRNAs of Group 3 MB using IPA predicted C17orf98, ZNF426, RNF165, FBXO8, CTCF, LAYN, PYGO1, Firre, TSIX and miR-150-5pa as most significantly associated upstream regulators, while activation/inactivation of X-chromosome, cell movement, and metastasis are the most important biological functions affected in this subgroup (Tables 11 and 12). Heatmap of 5 upstream regulators is shown in supplementary Fig. 2 (Additional file 3). The two most important non-canonical networks enriched with DE lncRNAs are shown in Fig. 3b and c. In network 1; CCND1, EP300, CREBBP, ESR1, CTNNB1, and PRKCD, in network 2; Histone H3, TP53, MYC, XIST, and EZH2 are the central regulators linked with DE lncRNAs.
Table 11.
Top 10 upstream regulators involved in DE lncRNAs in Group 3 MB
| Upstream Regulator | Molecule Type | P-val of overlap | Target molecules in dataset |
|---|---|---|---|
| C17orf98 | other | 1.11E-03 | XIST |
| ZNF426 | transcription regulator | 1.11E-03 | XIST |
| RNF165 | enzyme | 1.11E-03 | XIST |
| FBXO8 | other | 1.11E-03 | XIST |
| CTCF | transcription regulator | 2.08E-03 | TSIX,XIST |
| LAYN | other | 2.22E-03 | XIST |
| PYGO1 | other | 2.22E-03 | XIST |
| Firre | other | 3.33E-03 | XIST |
| TSIX | other | 3.33E-03 | XIST |
| miR-150-5p (and other miRNAs w/seed CUCCCAA) | mature microRNA | 3.33E-03 | MIAT |
Table 12.
Top 10 disease and function identified by IPA from DE lncRNAs in Group 3 MB
| Categories | Diseases or Functions Annotation | P-val | Activation z-score |
|---|---|---|---|
| Gene Expression | Inactivation of mouse X chromosome | 5.74E-06 | |
| Gene Expression | Activation of mouse X chromosome | 5.74E-06 | |
| Cellular Movement | Cell movement of tumor cell lines | 2.36E-05 | 0.783 |
| Cellular Movement | Migration of tumor cell lines | 3.60E-05 | 0.955 |
| Cell Cycle | Arrest in G0 phase of tumor cell lines | 1.65E-04 | |
| Cellular Movement | Invasion of tumor cell lines | 6.99E-04 | 0.495 |
| Cellular Movement | Cell movement | 1.30E-03 | 0.804 |
| Gene Expression | Imprinting | 1.31E-03 | |
| Cancer, Organismal Injury and Abnormalities | Metastasis of tumor cell lines | 1.32E-03 | 0.152 |
| Hereditary Disorder, Organismal Injury and Abnormalities | Familial skewed X inactivation | 1.41E-03 |
Differentially expressed lncRNAs in the Group 4 MB and their functional roles
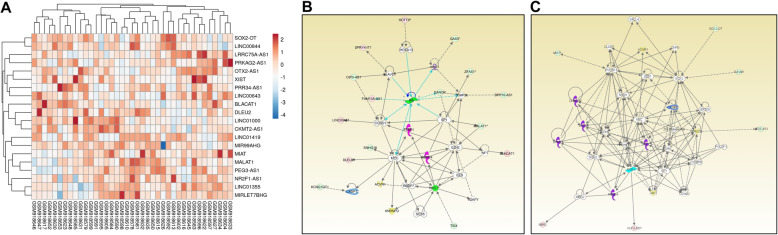
Comparative analyses of Group 4 MB (N = 39) and normal cerebellar tissue (N = 5) datasets with p < 0.05 and fold change > 2 provided 150 differentially expressed lncRNAs with approved status. Tables 13 and 14 show the fold change in the top 10 upregulated and downregulated lncRNAs. Heatmap of top 10 upregulated and downregulated lncRNAs is shown in Fig. 4a. The complete list of lncRNAs can be seen in Supplementary file 1. We found 82% overlap with lncRNAs in validation datasets [Group 4 N = 95, Control = 5] (Additional file 2). We found all the top 10 upregulated and downregulated lncRNAs in validation datasets.
Table 13.
Top 10 up-regulated lncRNAs in Group 4 of MB
| Gene Symbol | Fold Change | P-val | FDR P-val |
|---|---|---|---|
| LINC01419 | 139.78 | 0.0047 | 0.0175 |
| OTX2-AS1 | 60.12 | 9.95E-16 | 2.03E-13 |
| BLACAT1 | 27.67 | 1.13E-18 | 4.59E-16 |
| DLEU2 | 11.58 | 2.25E-15 | 4.16E-13 |
| LINC01355 | 7.09 | 2.23E-07 | 3.85E-06 |
| MIRLET7BHG | 7.01 | 2.03E-06 | 2.55E-05 |
| PRR34-AS1 | 6.82 | 8.84E-12 | 6.11E-10 |
| LINC01000 | 6.29 | 4.10E-12 | 3.13E-10 |
| CKMT2-AS1 | 6.19 | 5.04E-11 | 2.82E-09 |
| MIR99AHG | 5.27 | 9.82E-07 | 1.38E-05 |
Table 14.
Top 10 down-regulated lncRNAs in Group 4 of MB
| Gene Symbol | Fold Change | P-val | FDR P-val |
|---|---|---|---|
| XIST | − 343.06 | 0.0287 | 0.0745 |
| SOX2-OT | −31.6 | 1.90E-13 | 2.06E-11 |
| MALAT1 | −13.08 | 5.80E-10 | 2.34E-08 |
| LINC00643 | −11.64 | 4.39E-13 | 4.32E-11 |
| LINC00844 | −9.89 | 1.26E-05 | 0.0001 |
| LRRC75A-AS1 | −9.53 | 1.52E-08 | 3.87E-07 |
| MIAT | −7.87 | 2.15E-10 | 9.90E-09 |
| PRKAG2-AS1 | −7.8 | 4.29E-07 | 6.75E-06 |
| NR2F1-AS1 | −5.98 | 8.89E-11 | 4.63E-09 |
| PEG3-AS1 | −5.74 | 1.92E-08 | 4.73E-07 |
Fig. 4.
a: Heatmap of top 10 upregulated and downregulated lncRNAs in Group 4 MB. Expression value of different lncRNAs was clustered using correlation distance method. b: Differentially expressed lncRNAs in a non-canonical biological network in Group 4 MB. The important nodes in this biological network are AR, MYC, XIST, SP1, CCND1, and EZH2. c: Differentially expressed lncRNAs in another non-canonical biological network in Group 4 MB. The important nodes in this biological network are Histone H3, SP1, ESR1, MYC, SOX2, POU5F1, CDH1, and CEBPB. Green indicates downregulated and red indicates upregulated lncRNAs
Functional analysis of DE lncRNAs of Group 4 MB using IPA predicted C17orf98, ZNF426, RNF165, FBXO8, CTCF, LAYN, PYGO1, Firre, TSIX and miR-150-5pa as most significantly associated upstream regulators, while activation/inactivation of X-chromosomes, cell movement, methylation of DNA and metastasis are the most important biological functions affected in this subgroup (Tables 15 and 16). Heatmap of 5 upstream regulators is shown in supplementary Fig. 2 (Additional file 3). The two important non-canonical networks enriched with DE lncRNAs are shown in Fig. 4b and c. In network 1; AR, MYC, XIST, SP1, CCND1, and EZH2, in network 2; Histone H3, SP1, ESR1, MYC, SOX2, POU5F1, CDH1, and CEBPB are the central regulators linked with DE lncRNAs.
Table 15.
Top 10 upstream regulators involved in DE lncRNAs in Group 4 MB
| Upstream Regulator | Molecule Type | P-val of overlap | Target molecules in dataset |
|---|---|---|---|
| C17orf98 | other | 1.34E-03 | XIST |
| ZNF426 | transcription regulator | 1.34E-03 | XIST |
| RNF165 | enzyme | 1.34E-03 | XIST |
| FBXO8 | other | 1.34E-03 | XIST |
| LAYN | other | 2.68E-03 | XIST |
| PYGO1 | other | 2.68E-03 | XIST |
| CTCF | transcription regulator | 3.03E-03 | TSIX,XIST |
| Firre | other | 4.02E-03 | XIST |
| TSIX | other | 4.02E-03 | XIST |
| miR-150-5p (and other miRNAs w/seed CUCCCAA) | mature microRNA | 4.02E-03 | MIAT |
Table 16.
Top 10 disease and function identified by IPA from DE lncRNAs in Group 4 MB
| Categories | Diseases or Functions Annotation | P-val | Activation z-score |
|---|---|---|---|
| Cellular Movement | Cell movement of tumor cell lines | 4.56E-06 | −0.938 |
| Gene Expression | Inactivation of mouse X chromosome | 6.13E-06 | |
| Gene Expression | Activation of mouse X chromosome | 6.13E-06 | |
| Cellular Movement | Migration of tumor cell lines | 6.36E-06 | −0.877 |
| Gene Expression | Imprinting | 2.27E-05 | |
| Cell Death and Survival | Apoptosis of kidney cancer cell lines | 3.30E-05 | |
| Cancer, Organismal Injury and Abnormalities | Metastasis of tumor cell lines | 1.28E-04 | 0.555 |
| Cellular Movement | Invasion of tumor cell lines | 1.36E-04 | 0.031 |
| Cellular Development, Cellular Growth and Proliferation | Proliferation of kidney cancer cell lines | 1.68E-04 | |
| DNA Replication, Recombination, and Repair, Gene Expression | Methylation of DNA | 1.82E-04 | |
| Cell Death and Survival | Cell death of eye cell lines | 3.08E-04 |
Prognostic significance of lncRNAs in different subgroups of MB
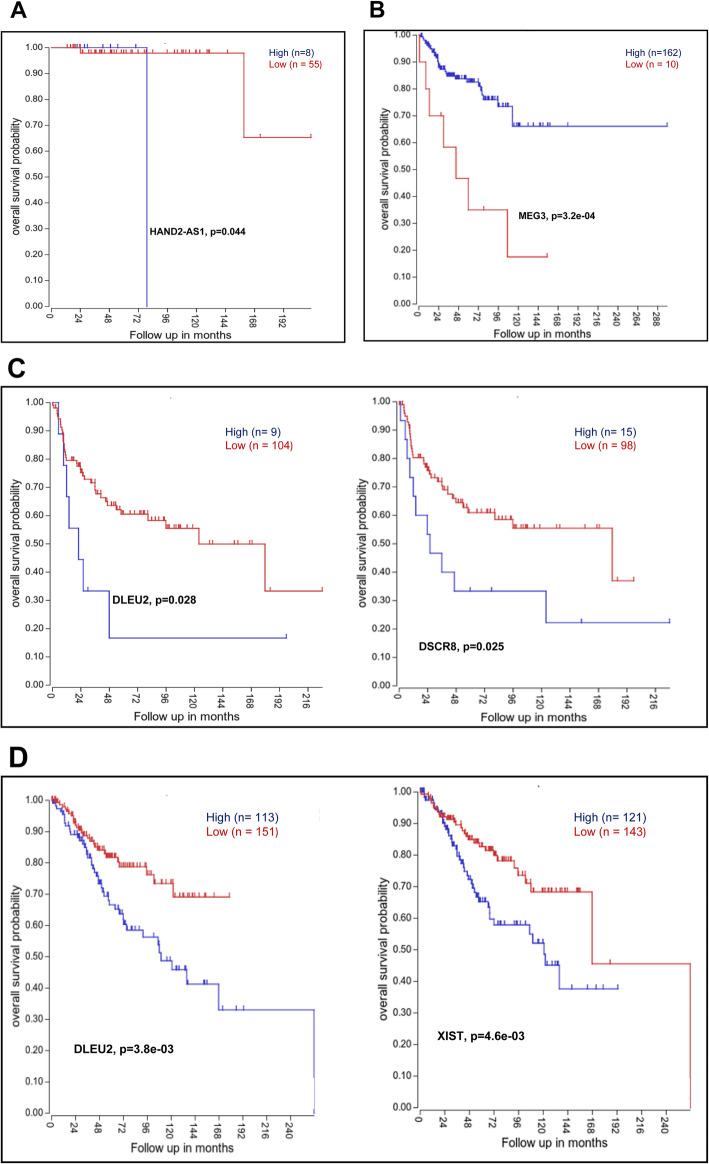
We used a publicly available dataset GSE85217 (Cavalli dataset) to understand the prognostic significance of DE lncRNAs of different MB subgroups. As shown in Fig. 5, high expression of HAND2-AS1 is associated with poor prognosis in WNT MB. Similarly, low expression of MEG3 in SHH, high expression of DLEU2 and DSCR8 in Group 3 and high expression of DLEU2 and low expression of XIST in Group 4 are associated with poor prognosis in MB (p < 0.05).
Fig. 5.
Kaplan Meier survival curves of different lncRNAs expressed in different subgroups of MB (Cavalli dataset) obtained using scan cut-off method on hgserver (https://hgserver1.amc.nl). a: High expression of HAND2-AS1 is associated with poor prognosis in WNT MB. b: Low expression of MEG3 is associated with poor prognosis in SHH MB. c: High expression of DLEU2 and DSCR8 are associated with poor prognosis in Group 3 MB. d: High expression of DLEU2 and low expression of XIST in Group 4 MB are associated with poor prognosis (p < 0.05)
Discussion
LncRNAs are known regulators of gene expression. Disruptions in gene regulatory pathways in cancers dictate the aberrant LncRNAs expression [11–13]. Notably, almost 40% of lncRNAs are aberrantly expressed in the brain-related disorders including brain tumors. However, lncRNA expression profile in MB is largely unexplored. In this study, we have identified the lncRNA expression profile of pediatric MB subgroups and associated molecular pathways. The identified key lncRNAs require further functional validation in vitro and in vivo to explore their potential role in MB subgroup-specific manner. Here, we discuss the known cancer-relevant function of the key lncRNAs identified in MB subgroups.
EMX2OS is the most differentially expressed lncRNA in the WNT subgroup. This lncRNA is known to regulate EMX gene expression in the brain development [32, 33]. OTX2-AS1 (antisense strand of the OTX2 gene) is predominantly involved in eye development [34]. High PGM5-AS1 (antisense strand of the PGM5 gene) expression is associated with development and poor prognosis of colorectal cancer (CRC) [35]. Increased expression of DSCR8 is associated to malignant pathology and poor survival in hepatocellular carcinoma (HCC) patients [36]. LOXL1-AS1 (antisense strand of the LOXL1 gene) is involved in the progression and metastasis of MB by regulating the PI3K-AKT signaling [27]. In addition, it is also known to play roles in the proliferation and survival of prostate cancer (PC) cells via miR-541-3p and cell cycle gene CCND1 [37] as well as aggressive nature of glioblastoma by activating NF-kB pathway [38]. HAND2-AS1 (antisense strand of the HAND2 gene) is overexpressed in esophageal squamous cell carcinoma (ESCC) [39] while it is downregulated in non-small cell lung cancer (NSCLC) cells [40]. TMEM51-AS1 (antisense strand of the TMEM51 gene) is associated with renal cell carcinoma (RCC) [41]. RMST acts as a tumor suppressor in triple-negative breast cancer (TNBC) by inducing apoptosis and inhibiting proliferation/invasion and migration [42]. PART1 promotes gefitinib-resistance in ESCC by regulating the miR-129/Bcl-2 pathway [43] and also associated with PC tumorigenesis [44]. LINC00461 is involved in glioma tumorigenesis via MAPK/ERK and PI3K/AKT signaling pathways [45]. Downregulation of MEG3 is involved in the proliferation and apoptosis of PC cells by regulating miR-9-5p and its target gene QKI-5 [46]. Downregulation of LINC00844 is associated with poor clinical outcomes and suppressed tumor progression/metastasis in PC [47]. SOX2-OT is overexpressed and promotes tumorigenesis by upregulating SOX2 gene and activating PI3K/AKT signaling pathway in cholangiocarcinoma (CCA) [48]. SOX2-OT is also a prognostic biomarker for osteosarcoma (OS) and involved in cell survival and cancer stem cells [49]. TUNAR plays a tumor suppressive role in glioma cells by upregulating miR-200a and inhibiting Rac1 [50]. MALAT1 promotes the chemo-resistance of cervical cancer via BRWD1-PI3K/AKT pathway [51]. MALAT1 is a well-studied lncRNA in several solid and hematological cancers [52].
NEAT1 is overexpressed in most cancer types, except leukemia and myeloma, where it is down-regulated [53–55]. DLEU2 exhibits role in the proliferation and survival of laryngeal cancer cells via miR-16-1 [56]. DLEU2 is also significantly overexpressed in gastric cancer and contributes to cell proliferation [57]. TPT1-AS1 (antisense strand of the TPT1 gene) expression is upregulated in cervical cancer and has influence on proliferation and migration [58]. HCG11 is significantly overexpressed in hepatocellular carcinoma (HCC) and genetic-silencing of HCG11 in HCC cells leads to decreased proliferation [59]. HCG11 expression is downregulated in PC and associated with poor prognosis of patients [60]. CCEPR contributes significantly in promoting cell proliferation and inhibiting apoptosis in bladder cancer [61].
BLACAT1 is overexpressed in chemo-resistant NSCLC and induces autophagy by regulating miR-17 and ATG7 pathway [62]. It also triggers proliferation/survival by regulating WNT signaling in cervical cancer [63].
XIST is elevated in bladder cancer and inhibits p53 function via binding to TET1 [64]. XIST also binds to miR-34a and elicits proliferation and tumor development in thyroid cancer [65]. XIST is an important regulator of progression and oxaliplatin-resistance in malignant melanoma [66]. MIR100HG is known to be involved in cetuximab-resistance in CRC via the β-catenin cellular pathway [67]. In addition, elevated expression of MIR100HG is correlated with poor prognosis of osteosarcoma [68]. MIAT is overexpressed in clear cell renal cell carcinoma (CCRCC) and associated with poor prognosis [69]. MIAT associates with miR-133 and contributes a role in the progression pancreatic cancer development [70]. MIAT also plays a key role in CRC tumorigenesis via miR-132/Derlin-1 axis [71]. NR2F1-AS1 (antisense strand of the NR2F1 gene) promotes chemotherapy-resistance in HCC by regulating miR-363-ABCC1 drug-transporter pathway [72].
Conclusions
We propose that the majority of DE lncRNAs in MB might have oncogenic properties as seen in other cancers (Supplementary Table S1 in Additional file 3) [73–82]. We found approximately 25% of these DE lncRNAs in MB are tumor suppressive. Also, each MB subgroup has unique and common lncRNAs in their expression profile (Fig. 6). We performed a unique lncRNAs analysis in both original datasets and validation datasets (Additional files 1 and 2). Unique lncRNAs can be validated for differential diagnosis and prognosis of MB subgroups. Common lncRNAs and associated molecules in pathways can be important therapeutic targets. We identified important lncRNAs DELU2, CASC15, LINC01355 and GAS5 are present in each subgroup and can be further explored for functional analyses in different MB subgroups. We also found SOX2, Protein kinase C delta (PRKCD), and EZH2 associated with functional networks of each subgroup and could be important drug targets. We also identified the prognostic significance of lncRNAs in different subgroups of MB.
Fig. 6.
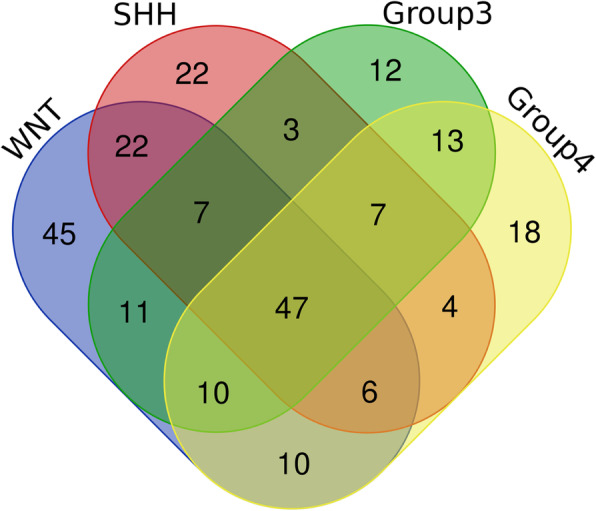
Venn diagram shows unique and overlapping lncRNAs profile in different MB subgroups. A complete list of unique lncRNAs in each subgroup can be viewed in supplementary file (highlighted in purple in Additional file 1)
Supplementary information
Acknowledgements
The authors wish to acknowledge the support of Bioinformatics Core Facility for providing access to IPA for functional analysis of datasets. The authors thank the State of Nebraska for their financial support of the UNMC/CHRI/Children’s Hospital Pediatric Cancer Research Program.
Abbreviations
- MB
Medulloblastoma
- LncRNAs
Long-non-coding RNAs
- GEO
Gene Expression Omnibus
- DE
Differentially expressed
- mRNA
Messenger ribonucleic acid
- DNA
Deoxyribonucleic acid
- IPA
Ingenuity pathway analysis
- ANOVA
Analysis of variance
Authors’ contributions
VK and NKC conceived and designed the study. VK, MS and NKC analyzed and interpreted the data. DC, JGS and SSJ critically interpreted the data. VK and NKC wrote the manuscript. All authors read and approved the final version of the manuscript.
Funding
This work was supported by the State of Nebraska through the Pediatric Cancer Research Grant Funds (LB905) awarded to D. W. Coulter, MD. This funding had no role in the study design, data collection and analysis, interpretation of the data, decision to publish, or writing the manuscript.
Availability of data and materials
We used publicly available GEO datasets (https://www.ncbi.nlm.nih.gov/geo/) GSE37418, GSM1094863, GSM1094864, GSM1094865, GSM1094866, GSM1094867, GSE124814, and GSE85217 for our analyses. The gene expression data GSE85217 (Cavalli dataset) was used for survival analyses in the R2-Genomics Analysis and Visualization Platform (https://hgserver1.amc.nl/cgi-bin/r2/main.cgi).
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12920-020-00744-7.
References
- 1.Cho YJ, Tsherniak A, Tamayo P, Santagata S, Ligon A, Greulich H, et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J Clin Oncol. 2011;29:1424–1430. doi: 10.1200/JCO.2010.28.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kijima N, Kanemura Y. Molecular Classification of Medulloblastoma. Neurol Med Chir (Tokyo) 2016;56:687–697. doi: 10.2176/nmc.ra.2016-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spiegler BJ, Bouffet E, Greenberg ML, Rutka JT, Mabbott DJ. Change in neurocognitive functioning after treatment with cranial radiation in childhood. J Clin Oncol. 2004;22:706–713. doi: 10.1200/JCO.2004.05.186. [DOI] [PubMed] [Google Scholar]
- 4.Zeltzer LK, Recklitis C, Buchbinder D, Zebrack B, Casillas J, Tsao JC, et al. Psychological status in childhood cancer survivors: a report from the childhood Cancer survivor study. J Clin Oncol. 2009;27:2396–2404. doi: 10.1200/JCO.2008.21.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Northcott PA, Korshunov A, Witt H, Hielscher T, Eberhart CG, Mack S, et al. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol. 2011;29:1408–1414. doi: 10.1200/JCO.2009.27.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Northcott PA, Shih DJ, Peacock J, Garzia L, Morrissy AS, Zichner T, et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature. 2012;488:49–56. doi: 10.1038/nature11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones DT, Jager N, Kool M, Zichner T, Hutter B, Sultan M, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L, et al. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488:43–48. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kool M, Korshunov A, Remke M, Jones DT, Schlanstein M, Northcott PA, et al. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, group 3, and group 4 medulloblastomas. Acta Neuropathol. 2012;123:473–484. doi: 10.1007/s00401-012-0958-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor MD, Northcott PA, Korshunov A, Remke M, Cho YJ, Clifford SC, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123:465–472. doi: 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, et al. GENCODE: the reference human genome annotation for the ENCODE project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 13.Shen XH, Qi P, Du X. Long non-coding RNAs in cancer invasion and metastasis. Mod Pathol. 2015;28:4–13. doi: 10.1038/modpathol.2014.75. [DOI] [PubMed] [Google Scholar]
- 14.Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum Mol Genet. 2010;19:R152–R161. doi: 10.1093/hmg/ddq353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grossi E, Sanchez Y, Huarte M. Expanding the p53 regulatory network: LncRNAs take up the challenge. Biochim Biophys Acta. 1859;2016:200–208. doi: 10.1016/j.bbagrm.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 16.Qi P, Zhou XY, Du X. Circulating long non-coding RNAs in cancer: current status and future perspectives. Mol Cancer. 2016;15:39. doi: 10.1186/s12943-016-0524-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Do H, Kim W. Roles of oncogenic Long non-coding RNAs in Cancer development. Genomics Inform. 2018;16:e18. doi: 10.5808/GI.2018.16.4.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Oliveira JC, Oliveira LC, Mathias C, Pedroso GA, Lemos DS, Salviano-Silva A, et al. Long non-coding RNAs in cancer: Another layer of complexity. J Gene Med. 2018:e3065. [DOI] [PubMed]
- 19.Tinzl M, Marberger M, Horvath S, Chypre C. DD3PCA3 RNA analysis in urine--a new perspective for detecting prostate cancer. Eur Urol. 2004;46:182–186. doi: 10.1016/j.eururo.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 20.Isin M, Ozgur E, Cetin G, Erten N, Aktan M, Gezer U, et al. Investigation of circulating lncRNAs in B-cell neoplasms. Clin Chim Acta. 2014;431:255–259. doi: 10.1016/j.cca.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 21.Roberts TC, Morris KV, Wood MJ. The role of long non-coding RNAs in neurodevelopment, brain function and neurological disease. Philos Trans R Soc Lond Ser B Biol Sci. 2014;369. [DOI] [PMC free article] [PubMed]
- 22.Pavlaki I, Alammari F, Sun B, Clark N, Sirey T, Lee S, et al. The long non-coding RNA Paupar promotes KAP1-dependent chromatin changes and regulates olfactory bulb neurogenesis. EMBO J. 2018;37. [DOI] [PMC free article] [PubMed]
- 23.Wu P, Zuo X, Deng H, Liu X, Liu L, Ji A. Roles of long noncoding RNAs in brain development, functional diversification and neurodegenerative diseases. Brain Res Bull. 2013;97:69–80. doi: 10.1016/j.brainresbull.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 24.Cho KO, Hsieh J. The lncRNA Pnky in the brain. Cell Stem Cell. 2015;16:344–345. doi: 10.1016/j.stem.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 25.Ramos AD, Andersen RE, Liu SJ, Nowakowski TJ, Hong SJ, Gertz C, et al. The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell. 2015;16:439–447. doi: 10.1016/j.stem.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Briggs JA, Wolvetang EJ, Mattick JS, Rinn JL, Barry G. Mechanisms of Long non-coding RNAs in mammalian nervous system development, plasticity, disease, and evolution. Neuron. 2015;88:861–877. doi: 10.1016/j.neuron.2015.09.045. [DOI] [PubMed] [Google Scholar]
- 27.Gao R, Zhang R, Zhang C, Liang Y, Tang W. LncRNA LOXL1-AS1 Promotes the Proliferation and Metastasis of Medulloblastoma by Activating the PI3K/AKT Pathway. Anal Cell Pathol (Amst). 2018;2018:9275685. [DOI] [PMC free article] [PubMed]
- 28.Gao R, Zhang R, Zhang C, Zhao L, Zhang Y. Long noncoding RNA CCAT1 promotes cell proliferation and metastasis in human medulloblastoma via MAPK pathway. Tumori. 2018;104:43–50. doi: 10.5301/tj.5000662. [DOI] [PubMed] [Google Scholar]
- 29.Zhang H, Wang X, Chen X. Potential role of Long non-coding RNA ANRIL in pediatric Medulloblastoma through promotion on proliferation and migration by targeting miR-323. J Cell Biochem. 2017;118:4735–4744. doi: 10.1002/jcb.26141. [DOI] [PubMed] [Google Scholar]
- 30.Laneve P, Po A, Favia A, Legnini I, Alfano V, Rea J, et al. The long noncoding RNA linc-NeD125 controls the expression of medulloblastoma driver genes by microRNA sponge activity. Oncotarget. 2017;8:31003–31015. doi: 10.18632/oncotarget.16049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song H, Han LM, Gao Q, Sun Y. Long non-coding RNA CRNDE promotes tumor growth in medulloblastoma. Eur Rev Med Pharmacol Sci. 2016;20:2588–2597. [PubMed] [Google Scholar]
- 32.Noonan FC, Goodfellow PJ, Staloch LJ, Mutch DG, Simon TC. Antisense transcripts at the EMX2 locus in human and mouse. Genomics. 2003;81:58–66. doi: 10.1016/s0888-7543(02)00023-x. [DOI] [PubMed] [Google Scholar]
- 33.Cecchi C, Boncinelli E. Emx homeogenes and mouse brain development. Trends Neurosci. 2000;23:347–352. doi: 10.1016/s0166-2236(00)01608-8. [DOI] [PubMed] [Google Scholar]
- 34.Alfano G, Vitiello C, Caccioppoli C, Caramico T, Carola A, Szego MJ, et al. Natural antisense transcripts associated with genes involved in eye development. Hum Mol Genet. 2005;14:913–923. doi: 10.1093/hmg/ddi084. [DOI] [PubMed] [Google Scholar]
- 35.Zhu H, Yu J, Zhu H, Guo Y, Feng S. Identification of key lncRNAs in colorectal cancer progression based on associated protein-protein interaction analysis. World J Surg Oncol. 2017;15:153. doi: 10.1186/s12957-017-1211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y, Sun L, Wang L, Liu Z, Li Q, Yao B, et al. Long non-coding RNA DSCR8 acts as a molecular sponge for miR-485-5p to activate Wnt/beta-851. [DOI] [PMC free article] [PubMed]
- 37.Long B, Li N, Xu XX, Li XX, Xu XJ, Liu JY, et al. Long noncoding RNA LOXL1-AS1 regulates prostate cancer cell proliferation and cell cycle progression through miR-541-3p and CCND1. Biochem Biophys Res Commun. 2018;505:561–568. doi: 10.1016/j.bbrc.2018.09.160. [DOI] [PubMed] [Google Scholar]
- 38.Wang H, Li L, Yin L. Silencing LncRNA LOXL1-AS1 attenuates mesenchymal characteristics of glioblastoma via NF-kappaB pathway. Biochem Biophys Res Commun. 2018;500:518–524. doi: 10.1016/j.bbrc.2018.04.133. [DOI] [PubMed] [Google Scholar]
- 39.Yan Y, Li S, Wang S, Rubegni P, Tognetti L, Zhang J, et al. Long noncoding RNA HAND2-AS1 inhibits cancer cell proliferation, migration, and invasion in esophagus squamous cell carcinoma by regulating microRNA-21. J Cell Biochem. 2019;120:9564–9571. doi: 10.1002/jcb.28233. [DOI] [PubMed] [Google Scholar]
- 40.Miao F, Chen J, Shi M, Song Y, Chen Z, Pang L. LncRNA HAND2-AS1 inhibits non-small cell lung cancer migration, invasion and maintains cell stemness through the interactions with TGF-beta1. Biosci Rep. 2019;39:BSR20181525. doi: 10.1042/BSR20181525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He HT, Xu M, Kuang Y, Han XY, Wang MQ, Yang Q. Biomarker and competing endogenous RNA potential of tumor-specific long noncoding RNA in chromophobe renal cell carcinoma. Onco Targets Ther. 2016;9:6399–6406. doi: 10.2147/OTT.S116392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang L, Liu D, Wu X, Zeng Y, Li L, Hou Y, et al. Long non-coding RNA (LncRNA) RMST in triple-negative breast cancer (TNBC): expression analysis and biological roles research. J Cell Physiol. 2018;233:6603–6612. doi: 10.1002/jcp.26311. [DOI] [PubMed] [Google Scholar]
- 43.Kang M, Ren M, Li Y, Fu Y, Deng M, Li C. Exosome-mediated transfer of lncRNA PART1 induces gefitinib resistance in esophageal squamous cell carcinoma via functioning as a competing endogenous RNA. J Exp Clin Cancer Res. 2018;37:171. doi: 10.1186/s13046-018-0845-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Sun M, Geng D, Li S, Chen Z, Zhao W. LncRNA PART1 modulates toll-like receptor pathways to influence cell proliferation and apoptosis in prostate cancer cells. Biol Chem. 2018;399:387–395. doi: 10.1515/hsz-2017-0255. [DOI] [PubMed] [Google Scholar]
- 45.Yang Y, Ren M, Song C, Li D, Soomro SH, Xiong Y, et al. LINC00461, a long non-coding RNA, is important for the proliferation and migration of glioma cells. Oncotarget. 2017;8:84123–84139. doi: 10.18632/oncotarget.20340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu M, Huang Y, Chen T, Wang W, Yang S, Ye Z, et al. LncRNA MEG3 inhibits the progression of prostate cancer by modulating miR-9-5p/QKI-5 axis. J Cell Mol Med. 2019;23:29–38. doi: 10.1111/jcmm.13658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lingadahalli S, Jadhao S, Sung YY, Chen M, Hu L, Chen X, et al. Novel lncRNA LINC00844 regulates prostate Cancer cell migration and invasion through AR signaling. Mol Cancer Res. 2018;16:1865–1878. doi: 10.1158/1541-7786.MCR-18-0087. [DOI] [PubMed] [Google Scholar]
- 48.Wei CX, Wong H, Xu F, Liu Z, Ran L, Jiang RD. IRF4-induced upregulation of lncRNA SOX2-OT promotes cell proliferation and metastasis in cholangiocarcinoma by regulating SOX2 and PI3K/AKT signaling. Eur Rev Med Pharmacol Sci. 2018;22:8169–8178. doi: 10.26355/eurrev_201812_16509. [DOI] [PubMed] [Google Scholar]
- 49.Wang Z, Tan M, Chen G, Li Z, Lu X. LncRNA SOX2-OT is a novel prognostic biomarker for osteosarcoma patients and regulates osteosarcoma cells proliferation and motility through modulating SOX2. IUBMB Life. 2017;69:867–876. doi: 10.1002/iub.1681. [DOI] [PubMed] [Google Scholar]
- 50.Dai J, Ma J, Yu B, Zhu Z, Hu Y. Long noncoding RNA TUNAR represses growth, migration, and invasion of human Glioma cells through regulating miR-200a and Rac1. Oncol Res. 2018;27:107–115. doi: 10.3727/096504018X15205622257163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang N, Hou MS, Zhan Y, Shen XB, Xue HY. MALAT1 promotes cisplatin resistance in cervical cancer by activating the PI3K/AKT pathway. Eur Rev Med Pharmacol Sci. 2018;22:7653–7659. doi: 10.26355/eurrev_201811_16382. [DOI] [PubMed] [Google Scholar]
- 52.Li ZX, Zhu QN, Zhang HB, Hu Y, Wang G, Zhu YS. MALAT1: a potential biomarker in cancer. Cancer Manag Res. 2018;10:6757–6768. doi: 10.2147/CMAR.S169406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Z, Wu K, Wu J, Tian D, Chen Y, Yang Z, et al. NEAT1 is a potential prognostic biomarker for patients with nasopharyngeal carcinoma. J Cell Biochem. 2019;120:9831–9838. doi: 10.1002/jcb.28263. [DOI] [PubMed] [Google Scholar]
- 54.Ghaforui-Fard S, Taheri M. Nuclear enriched abundant transcript 1 (NEAT1): a long non-coding RNA with diverse functions in tumorigenesis. Biomed Pharmacother. 2018;111:51–59. doi: 10.1016/j.biopha.2018.12.070. [DOI] [PubMed] [Google Scholar]
- 55.Klec C, Prinz F, Pichler M. Involvement of the long noncoding RNA NEAT1 in carcinogenesis. Mol Oncol. 2019;13:46–60. doi: 10.1002/1878-0261.12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xie ZZ, Xiao ZC, Song YX, Li W, Tan GL. Long non-coding RNA Dleu2 affects proliferation, migration and invasion ability of laryngeal carcinoma cells through triggering miR-16-1 pathway. Eur Rev Med Pharmacol Sci. 2018;22:1963–1970. doi: 10.26355/eurrev_201804_14723. [DOI] [PubMed] [Google Scholar]
- 57.Liu H, Zhang Z, Wu N, Guo H, Zhang H, Fan D, et al. Integrative Analysis of Dysregulated lncRNA-Associated ceRNA Network Reveals Functional lncRNAs in Gastric Cancer. Genes (Basel). 2018:9. [DOI] [PMC free article] [PubMed]
- 58.Jiang H, Huang G, Zhao N, Zhang T, Jiang M, He Y, et al. Long non-coding RNA TPT1-AS1 promotes cell growth and metastasis in cervical cancer via acting AS a sponge for miR-324-5p. J Exp Clin Cancer Res. 2018;37:169. doi: 10.1186/s13046-018-0846-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 59.Xu Y, Zheng Y, Liu H, Li T. Modulation of IGF2BP1 by long non-coding RNA HCG11 suppresses apoptosis of hepatocellular carcinoma cells via MAPK signaling transduction. Int J Oncol. 2017;51:791–800. doi: 10.3892/ijo.2017.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y, Zhang P, Wan X, Su X, Kong Z, Zhai Q, et al. Downregulation of long non-coding RNA HCG11 predicts a poor prognosis in prostate cancer. Biomed Pharmacother. 2016;83:936–941. doi: 10.1016/j.biopha.2016.08.013. [DOI] [PubMed] [Google Scholar]
- 61.Zhan Y, Li Y, Guan B, Chen X, Chen Z, He A, et al. Increased expression of long non-coding RNA CCEPR is associated with poor prognosis and promotes tumorigenesis in urothelial bladder carcinoma. Oncotarget. 2017;8:44326–44334. doi: 10.18632/oncotarget.17872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang FX, Chen HJ, Zheng FX, Gao ZY, Sun PF, Peng Q, et al. LncRNA BLACAT1 is involved in chemoresistance of nonsmall cell lung cancer cells by regulating autophagy. Int J Oncol. 2019;54:339–347. doi: 10.3892/ijo.2018.4614. [DOI] [PubMed] [Google Scholar]
- 63.Wang CH, Li YH, Tian HL, Bao XX, Wang ZM. Long non-coding RNA BLACAT1 promotes cell proliferation, migration and invasion in cervical cancer through activation of Wnt/beta-catenin signaling pathway. Eur Rev Med Pharmacol Sci. 2018;22:3002–3009. doi: 10.26355/eurrev_201805_15057. [DOI] [PubMed] [Google Scholar]
- 64.Hu B, Shi G, Li Q, Li W, Zhou H. Long noncoding RNA XIST participates in bladder cancer by downregulating p53 via binding to TET1. J Cell Biochem. 2019;120:6330–6338. doi: 10.1002/jcb.27920. [DOI] [PubMed] [Google Scholar]
- 65.Liu H, Deng H, Zhao Y, Li C, Liang Y. LncRNA XIST/miR-34a axis modulates the cell proliferation and tumor growth of thyroid cancer through MET-PI3K-AKT signaling. J Exp Clin Cancer Res. 2018;37:279. doi: 10.1186/s13046-018-0950-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pan B, Lin X, Zhang L, Hong W, Zhang Y. Long noncoding RNA X-inactive specific transcript promotes malignant melanoma progression and oxaliplatin resistance. Melanoma Res. 2019;29:254–262. doi: 10.1097/CMR.0000000000000560. [DOI] [PubMed] [Google Scholar]
- 67.Lu Y, Zhao X, Liu Q, Li C, Graves-Deal R, Cao Z, et al. lncRNA MIR100HG-derived miR-100 and miR-125b mediate cetuximab resistance via Wnt/beta-catenin signaling. Nat Med. 2017;23:1331–1341. doi: 10.1038/nm.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Su X, Teng J, Jin G, Li J, Zhao Z, Cao X, et al. ELK1-induced upregulation of long non-coding RNA MIR100HG predicts poor prognosis and promotes the progression of osteosarcoma by epigenetically silencing LATS1 and LATS2. Biomed Pharmacother. 2019;109:788–797. doi: 10.1016/j.biopha.2018.10.029. [DOI] [PubMed] [Google Scholar]
- 69.Qu Y, Xiao H, Xiao W, Xiong Z, Hu W, Gao Y, et al. Upregulation of MIAT regulates LOXL2 expression by competitively binding MiR-29c in clear cell renal cell carcinoma. Cell Physiol Biochem. 2018;48:1075–1087. doi: 10.1159/000491974. [DOI] [PubMed] [Google Scholar]
- 70.Li TF, Liu J, Fu SJ. The interaction of long non-coding RNA MIAT and miR-133 play a role in the proliferation and metastasis of pancreatic carcinoma. Biomed Pharmacother. 2018;104:145–150. doi: 10.1016/j.biopha.2018.05.043. [DOI] [PubMed] [Google Scholar]
- 71.Liu Z, Wang H, Cai H, Hong Y, Li Y, Su D, et al. Long non-coding RNA MIAT promotes growth and metastasis of colorectal cancer cells through regulation of miR-132/Derlin-1 pathway. Cancer Cell Int. 2018;18:59. doi: 10.1186/s12935-017-0477-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang H, Chen J, Ding CM, Jin X, Jia ZM, Peng J. LncRNA NR2F1-AS1 regulates hepatocellular carcinoma oxaliplatin resistance by targeting ABCC1 via miR-363. J Cell Mol Med. 2018;22:3238–3245. doi: 10.1111/jcmm.13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Duan M, Fang M, Wang C, Wang H, Li M. LncRNA EMX2OS induces proliferation, invasion and sphere formation of ovarian Cancer cells via regulating the miR-654-3p/AKT3/PD-L1 Axis. Cancer Manag Res. 2020;12:2141–2154. doi: 10.2147/CMAR.S229013. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 74.Yan SP, Chu DX, Qiu HF, Xie Y, Wang CF, Zhang JY, et al. LncRNA LINC01305 silencing inhibits cell epithelial-mesenchymal transition in cervical cancer by inhibiting TNXB-mediated PI3K/Akt signalling pathway. J Cell Mol Med. 2019;23:2656–2666. doi: 10.1111/jcmm.14161. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 75.Hsu CM, Lin PM, Lin HC, Lai CC, Yang CH, Lin SF, et al. Altered expression of imprinted genes in squamous cell carcinoma of the head and neck. Anticancer Res. 2016;36:2251–2258. [PubMed] [Google Scholar]
- 76.Ye M, Ma J, Liu B, Liu X, Ma D, Dong K. Linc01105 acts as an oncogene in the development of neuroblastoma. Oncol Rep. 2019. 10.3892/or.2019.7257. [DOI] [PubMed]
- 77.Ai B, Kong X, Wang X, Zhang K, Yang X, Zhai J, et al. LINC01355 suppresses breast cancer growth through FOXO3-mediated transcriptional repression of CCND1. Cell Death Dis. 2019;10:502. doi: 10.1038/s41419-019-1741-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu HY, Lu SR, Guo ZH, Zhang ZS, Ye X, Du Q, et al. lncRNA SLC16A1-AS1 as a novel prognostic biomarker in non-small cell lung cancer. J Investig Med. 2020;68:52–59. doi: 10.1136/jim-2019-001080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhuan B, Lu Y, Chen Q, Zhao X, Li P, Yuan Q, et al. Overexpression of the long noncoding RNA TRHDE-AS1 inhibits the progression of lung cancer via the miRNA-103/KLF4 axis. J Cell Biochem. 2019;120:17616–17624. doi: 10.1002/jcb.29029. [DOI] [PubMed] [Google Scholar]
- 80.Wang LL, Zhang L, Cui XF. Downregulation of long noncoding RNA LINC01419 inhibits cell migration, invasion, and tumor growth and promotes autophagy via inactivation of the PI3K/Akt1/mTOR pathway in gastric cancer. Ther Adv Med Oncol. 2019;11:1758835919874651. doi: 10.1177/1758835919874651. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 81.Emmrich S, Streltsov A, Schmidt F, Thangapandi VR, Reinhardt D, Klusmann JH. LincRNAs MONC and MIR100HG act as oncogenes in acute megakaryoblastic leukemia. Mol Cancer. 2014;13:171. doi: 10.1186/1476-4598-13-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen J, Lan J, Ye Z, Duan S, Hu Y, Zou Y, et al. Long noncoding RNA LRRC75A-AS1 inhibits cell proliferation and migration in colorectal carcinoma. Exp Biol Med (Maywood) 2019;244:1137–1143. doi: 10.1177/1535370219874339. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We used publicly available GEO datasets (https://www.ncbi.nlm.nih.gov/geo/) GSE37418, GSM1094863, GSM1094864, GSM1094865, GSM1094866, GSM1094867, GSE124814, and GSE85217 for our analyses. The gene expression data GSE85217 (Cavalli dataset) was used for survival analyses in the R2-Genomics Analysis and Visualization Platform (https://hgserver1.amc.nl/cgi-bin/r2/main.cgi).