Abstract
Background
The low genetic diversity of Upland cotton limits the potential for genetic improvement. Making full use of the genetic resources of Sea-island cotton will facilitate genetic improvement of widely cultivated Upland cotton varieties. The chromosome segments substitution lines (CSSLs) provide an ideal strategy for mapping quantitative trait loci (QTL) in interspecific hybridization.
Results
In this study, a CSSL population was developed by PCR-based markers assisted selection (MAS), derived from the crossing and backcrossing of Gossypium hirsutum (Gh) and G. barbadense (Gb), firstly. Then, by whole genome re-sequencing, 11,653,661 high-quality single nucleotide polymorphisms (SNPs) were identified which ultimately constructed 1211 recombination chromosome introgression segments from Gb. The sequencing-based physical map provided more accurate introgressions than the PCR-based markers. By exploiting CSSLs with mutant morphological traits, the genes responding for leaf shape and fuzz-less mutation in the Gb were identified. Based on a high-resolution recombination bin map to uncover genetic loci determining the phenotypic variance between Gh and Gb, 64 QTLs were identified for 14 agronomic traits with an interval length of 158 kb to 27 Mb. Surprisingly, multiple alleles of Gb showed extremely high value in enhancing cottonseed oil content (SOC).
Conclusions
This study provides guidance for studying interspecific inheritance, especially breeding researchers, for future studies using the traditional PCR-based molecular markers and high-throughput re-sequencing technology in the study of CSSLs. Available resources include candidate position for controlling cotton quality and quantitative traits, and excellent breeding materials. Collectively, our results provide insights into the genetic effects of Gb alleles on the Gh, and provide guidance for the utilization of Gb alleles in interspecific breeding.
Keywords: Cotton, Chromosome substituted segments lines (CSSLs), Quantitative trait loci (QTL), Whole genome re-sequencing, Cottonseed oil content (SOC)
Background
Cotton is one of the most important cash crops, both as the leading natural fiber resource for the textile industry and an important oilseed crop. Approximately 50 species are present in the Gossypium spp., and only 4 species are cultivated worldwide: 2 are diploids (G. herbaceum and G. arboreum), 2 are tetraploids (G. hirsutum and G. barbadense). These two tetraploid (2n = 4x = 52) cotton species both share the common progenitors, which formed by a natural hybridization between A genome and D genome 1–2 million years ago [1–3]. The G. hirsutum (Gh), known as Upland cotton, contributed over 95% of cotton fiber yield by its wide adaptation and high yield [4, 5]. Because of the long process of domestication and selection bottlenecks, the elite Upland cotton has a narrow genetic base and limited genetic diversity [3]. This limitation could be a serious obstacle to improve the fiber quality and maintain continuity in genetic effectiveness [4]. While G. barbadense (Gb), also known as Sea-island cotton or long extra staple cotton, has excellent fiber quality, disease resistance but lower yield [6]. Introgression of interspecific favorable alleles to the Upland cotton can make full use of its high productivity, and it will be an ideal solution for cotton breeding [7, 8]. Although both of their genome sequence shared parts of the homology [9, 10], limited successes have been made in cotton interspecific breeding [6, 11]. Therefore, identifying, cloning, and utilizing beneficial allelic genes from the Gb will be important.
The primary segregating populations such as F2, BC1, have been widely used in genetic analysis for genetic map construction and quantitative trait loci (QTL) mapping. However, several disadvantages such as temporary nature and large deviation for evaluating the small-effect QTL limited their applications in the complex QTL analysis and cloning [12, 13]. In recent years, chromosome segment substitution lines (CSSLs), or referred as introgression lines (ILs), produced by crossing and backcrossing the donor and recipient parents by marker-assisted selection (MAS), provide a useful approach to resolve complex genome and QTL mapping [8]. Each of the CSSLs has one or few homozygous chromosome segments of donor genotype in the genetic background of the recurrent parent [14], which combines the advantages of the near-isogenic lines and backcross inbred lines. Through repeatedly planted in various locations or in different years, CSSLs helped to improve the accurate resolution of the genetic effects in the interspecific genomes [15–18]. Since the pioneering work in tomato [19], several interspecific introgression line libraries have been produced in many crops [20, 21]. Based on traditional molecular markers, such as restriction fragment length polymorphism (RFLP), amplified fragment length polymorphism (AFLP) and simple sequence repeat (SSR), a lot of QTL have been identified. However, limited by low genetic diversity and genetic map density, these molecular markers can identify only a few QTL and cover a wide region in the genome, which reduce the direct application of the QTL in breeding [22, 23].
In recent years, whole-genome re-sequencing technology has been widely used in population genetic analysis [24–26]. The high-throughput genotyping platform of SNP markers has significantly driven the process of genetic mapping and QTL identification [27–29]. Compared with the low density of traditional molecular markers, SNP markers significantly improve the genome coverage and QTL mapping accuracy. Multiple novel QTL for the important agronomic traits have been identified in multiple crops [30–32]. Moreover, high-resolution SNPs are a versatile tool to characterize the relationships between genes and importantly agronomic traits [33].
The prospect of widening the genetic diversity and improving the fiber quality of Upland cotton by accessing the exogenous genes has encouraged interspecific hybridization and introgression efforts for many years [6]. Stunning fiber quality of the Gb promotes it’s widely use in interspecific hybridization. Benefiting from widely range of variations shown in the progeny from Gh × Gb population, a large number of QTL related to multiple traits have been identified (https://www.cottonQTLdb.org). Moreover, some genes controlling specific characteristics of the Gb have been fine-mapped or cloned, such as open-bud floral buds [34], okra leaf [35–37], and naked seed mutant [38, 39]. Other wild Gossypium gene pools also provide a broad genetic diversity for Upland cotton [40–42]. However, none of them used high-throughput sequencing technology for analysis, which partly because there was no ultra-high density genetic map covering the entire genome or high-quality tetraploid cotton reference genome in the public domain. In the last a few years, spells above have been lifted in our lab [10].
Here, a set of interspecific CSSLs derived from a cross between G. hirsutum cv. ‘Emian22’ and G. barbadense acc. 3–79, were developed by using molecular marker selection. Next-generation sequencing technology was used to re-genotype all the lines and their parents by re-sequencing. The CSSLs were evaluated by using PCR-based markers and high-quality SNPs, resulting in a total of 480 introgression segments and 1211 recombination bins, respectively. Fourteen important agronomic traits including yield, fiber quality and oil content traits were measured in five environments to detect QTL. The influence of the Gb chromosome segments in the Gh background was investigated in this study.
Result
Evaluation of introgression chromosome recombination fragments in CSSLs
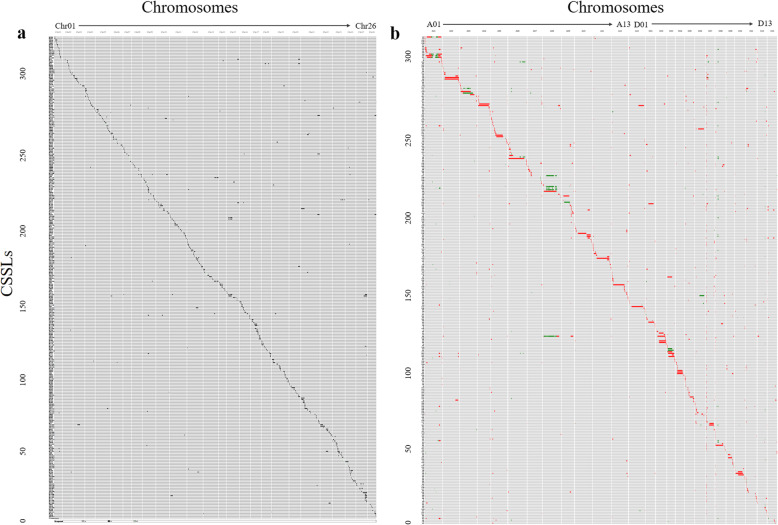
After several generations of self-pollinated, 515 markers were selected to evaluate the locations of introgression segments from donor parent in the lines with multi-segments again. Based on the genotypes of the molecular markers and the basis of the physical locations, the lengths and the locations of the introgression segments in each line were determined (Table 1), and a physical map was constructed (MM-map) (Fig. 1a). A total of 480 introgression segments were identified in the 325 CSSLs using SSR markers, with introgressions ranging from the least 10 ones on chromosome A03, D02 and D04 to the most 30 ones on chromosome D11. Among these, 222 lines carried one introgression segment despite the differences in lengths, and 103 lines were classified into the multi-segments group (Additional file 1: Table S1).
Table 1.
Comparison of genetic map and physical map in the CSSLs
| Chr. | Chromosome length | Number of markers | Average size | Number of segments | Coverage length | Coverage rate | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MM-map (cM) | GR-map (Mb) | MM-map | GR-map | MM-map (marker/cM) | GR-map (SNPs/kb) | MM-map | GR-map | MM-map (cM) | GR-map (Mb) | MM-map | GR-map | |
| A01 | 115.34 | 117.71 | 14 | 361,606 | 8.24 | 3.1 | 19 | 91 | 82.08 | 108.57 | 71.16% | 92.24% |
| A02 | 147.16 | 108.05 | 18 | 604,209 | 8.18 | 5.6 | 19 | 40 | 76.08 | 105.69 | 51.70% | 97.81% |
| A03 | 161.99 | 113.01 | 19 | 596,783 | 8.53 | 5.3 | 10 | 53 | 137.58 | 104.41 | 84.93% | 92.39% |
| A04 | 140.78 | 85.11 | 18 | 466,455 | 7.82 | 5.5 | 20 | 41 | 123.75 | 82.41 | 87.90% | 96.82% |
| A05 | 207.21 | 109.37 | 21 | 524,501 | 9.87 | 4.8 | 20 | 53 | 169.42 | 85.68 | 81.76% | 78.34% |
| A06 | 172.09 | 124.01 | 19 | 662,304 | 9.06 | 5.3 | 16 | 46 | 120.19 | 114.98 | 69.84% | 92.72% |
| A07 | 115.52 | 97.74 | 16 | 528,301 | 7.22 | 5.4 | 14 | 34 | 29.42 | 39.40 | 25.46% | 40.31% |
| A08 | 141.97 | 122.33 | 20 | 673,976 | 7.10 | 5.5 | 13 | 60 | 63.98 | 120.00 | 45.06% | 98.09% |
| A09 | 187.02 | 82.06 | 21 | 432,266 | 8.91 | 5.3 | 23 | 39 | 124.75 | 78.94 | 66.70% | 96.19% |
| A10 | 185.70 | 114.80 | 21 | 616,965 | 8.84 | 5.4 | 16 | 22 | 163.96 | 100.14 | 88.29% | 87.23% |
| A11 | 239.20 | 123.16 | 29 | 665,538 | 8.25 | 5.4 | 23 | 33 | 193.20 | 121.58 | 80.77% | 98.72% |
| A12 | 222.31 | 107.62 | 26 | 600,282 | 8.55 | 5.6 | 29 | 42 | 202.90 | 100.35 | 91.27% | 93.24% |
| A13 | 213.83 | 108.33 | 23 | 611,835 | 9.30 | 5.6 | 14 | 27 | 171.66 | 102.50 | 80.28% | 94.62% |
| At subgenome | 2250.12 | 1413.31 | 265 | 7,345,021 | 8.49 | 5.2 | 236 | 581 | 1658.96 | 1264.64 | 73.73% | 89.48% |
| D01 | 178.95 | 63.18 | 22 | 331,752 | 8.13 | 5.3 | 17 | 27 | 124.09 | 53.84 | 69.34% | 85.21% |
| D02 | 102.25 | 69.81 | 12 | 405,754 | 8.52 | 5.8 | 10 | 14 | 85.99 | 64.76 | 84.10% | 92.76% |
| D03 | 145.67 | 52.68 | 17 | 302,944 | 8.57 | 5.8 | 20 | 27 | 132.24 | 52.67 | 90.78% | 99.98% |
| D04 | 167.36 | 56.41 | 20 | 299,802 | 8.37 | 5.3 | 10 | 17 | 145.07 | 48.92 | 86.68% | 86.73% |
| D05 | 243.38 | 62.90 | 28 | 295,927 | 8.69 | 4.7 | 22 | 35 | 196.42 | 51.82 | 80.70% | 82.38% |
| D06 | 153.96 | 66.84 | 18 | 359,172 | 8.55 | 5.4 | 29 | 36 | 131.23 | 61.67 | 85.24% | 92.26% |
| D07 | 152.94 | 59.23 | 18 | 308,614 | 8.50 | 5.2 | 14 | 126 | 129.79 | 56.28 | 84.86% | 95.02% |
| D08 | 145.99 | 69.01 | 16 | 360,933 | 9.12 | 5.2 | 21 | 116 | 145.99 | 67.78 | 100.00% | 98.22% |
| D09 | 174.52 | 52.80 | 20 | 290,463 | 8.73 | 5.5 | 25 | 29 | 158.55 | 44.98 | 90.85% | 85.19% |
| D10 | 160.88 | 67.98 | 17 | 378,681 | 9.46 | 5.6 | 14 | 36 | 126.42 | 62.95 | 78.58% | 92.61% |
| D11 | 265.89 | 72.91 | 32 | 321,124 | 8.31 | 4.4 | 30 | 62 | 209.45 | 32.85 | 78.77% | 45.06% |
| D12 | 123.72 | 62.67 | 14 | 305,337 | 8.84 | 4.9 | 15 | 73 | 88.12 | 30.38 | 71.22% | 48.48% |
| D13 | 136.87 | 63.32 | 16 | 348,137 | 8.55 | 5.5 | 17 | 32 | 120.31 | 29.40 | 87.90% | 46.43% |
| Dt subgenome | 2152.38 | 819.74 | 250 | 4,308,640 | 8.61 | 5.3 | 244 | 633 | 1793.67 | 658.29 | 83.33% | 80.31% |
| Total | 4402.50 | 2233.05 | 515 | 11,653,661 | 8.5 | 5.2 | 480 | 1211 | 3452.62 | 1922.94 | 78.42% | 86.11% |
MM-map: based on the genetic map constructed with molecular markers
GR-map: based on the physical map constructed by whole-genome resequencing
Fig. 1.
Distribution of introgression segments in the CSSLs on the 26 chromosomes. a Physical map was constructed by SSR markers; b Physical map was constructed by whole-genome re-sequencing SNPs. Each row indicates a CSSL, and each column represents a chromosome. The black and red squares denote the homozygous donor segments from Gb; the light-gray and green represent the heterozygous from Gb; the grey background represents the genetic background of the Gh
Based on SNPs from the sequencing data, 17,992 recombinant bins distributed on the 26 chromosomes were identified, which ultimately constructed 1211 recombination chromosome introgression segments from Gb in the 313 CSSLs (Fig. 1b and Additional file 2: Table S2). None chromosome introgression segments were detected in 10 lines in the CSSLs populations based on SNPs. The physical length of the introgression segments ranged from 97 kb to 104.23 Mb, with an average length of 4.43 Mb. Based on the physical map (GR-map), re-sequencing data significantly reduced the number of SSSLs, only 54 lines carried only one donor segment, and the lines with less than four segments just closed to half of the population (Additional file 1: Table S1). Significant difference of introgressions appeared in Dt-subgenome with 14 one on D02 and 126 ones on D07 (Table 1).
Comparison of the genome coverage between SSR markers and SNPs
Based on the marker position of the genetic map, 6175.33 cM of the total length of the donor segments was counted by SSR markers, with 3462.62 cM of effective coverage length. The whole cotton genome coverage based on the genetic map was 78.42%, and At-subgenome had a lower coverage ratio of 73.73% compared with the 83.33% in Dt-subgenome. The lowest coverage was on chromosome A07 with only 25.46%, and the highest appeared in the Dt-subgenome with no missing on chromosome D08 (Table 1).
The physical map constructed by SNPs covered 2.24 times of the total length of the cotton genome (Additional file 3: Table S3), with 1922.93 Mb of effective coverage length and 86.11% whole genome coverage. Compared to the MM-map, GR-map had a higher percentage of coverage in At-subgenome (89.48% in At-subgenome vs 80.31% in Dt-subgenome). Although the coverage of 16 chromosomes exceeded 90%, there were still 4 chromosomes with coverage of less than 50%. Notably, chromosome A07 had the lowest coverage consistent with the MM-map result, and more than 98 CSSLs detected the same segment on the chromosome D07 located at 5.0–6.5 Mb.
Phenotypic variation in CSSLs
Significant differences were observed between the parents across multiple traits and multiple environments, such as seed cotton weight per boll (BWT), lint percentage (LP), seed oil content (SOC) and all fiber quality traits. Fourteen traits were evaluated in five environments except that SI was just investigated in two environments (Additional file 4: Table S4 and Additional file 5: Table S5), and all traits showed a continuous distribution in the CSSLs. The broad-sense heritability (H2) was lower than 50% for the yield-related traits, indicating that they were easily affected by the environment (Additional file 6: Table S6). Higher H2 value of the lint percentage (LP) (76%), fiber length (FL) (77%) and SOC (87%) indicated that they were more affected by the associated genes coming from the Gb-genome. Fiber quality of Gb was outstanding in all environments, while the mediocre level of the fiber traits was observed in the lots of the CSSLs. Interestingly, recombination of the interspecific genomes also produced various fuzz fiber mutations with different densities and colors (Additional file 7: Figure S1). The N29 line produced fuzz-less phenotype similar to the Gb reported previously [10].
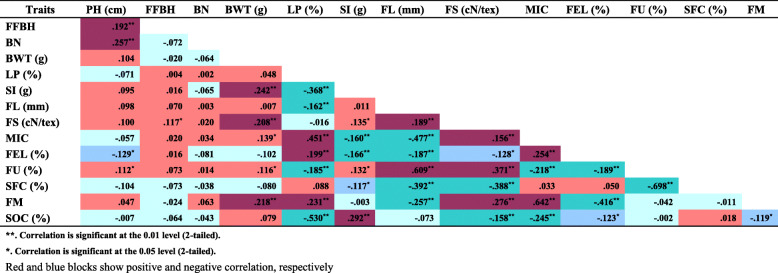
Positive and negative correlations between evaluated traits were calculated (Table 2). Plant height (PH) and first fruit branch height (FFBH) showed weak correlations with each other and with the yield-related traits (BWT and LP). But significant correlations were observed between fiber quality traits. Fiber length (FL) was significant positively correlated with fiber strength (FS) and fiber uniformity (FU), while negatively with micronaire value (MIC), fiber elongation (FEL), short fiber content (SFC) and fiber mature content (FM). The higher value of the SI followed the principle of negative correlation between yield and fiber quality, which may in turn increase of SOC.
Table 2.
Correlation coefficients of 14 traits in the CSSLs over 5 environments.
**. Correlation is significant at the 0.01 level (2-tailed)
*. Correlation is significant at the 0.05 level (2-tailed)
Red and blue blocks show positive and negative correlation, respectively
Genetic basis of the morphological mutation in the CSSLs
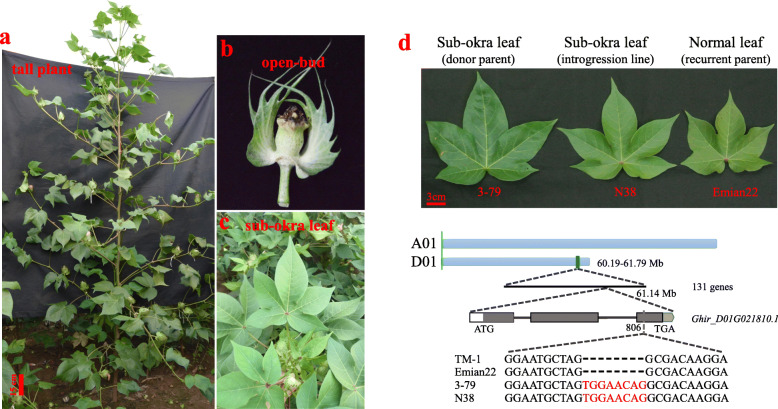
Although the donor parent 3–79, the genetic standard of Sea-island cotton, had undergone artificial selection, cognitive of the plant height type for Sea-island cotton still appeared in the CSSLs (Fig. 2a). The “open-bud” floral buds phenotype was found during the flower development with the exposed stigma and dead anther (Fig. 2b). The associated marker BNL3479 located on chromosome D13 was similar to the former research (Additional file 8: Table S7) [34].
Fig. 2.
Some CSSLs showing morphological variations. a Significant tall mutant plant, b Open-bud mutant with stamen necrosis, c Sub-okra leaf, d Comparison of the LMI1 gene structure in the CSSLs
By using the high resolution of recombination segments, the iconic characteristic of the Gb, sub-okra leaf trait was identified in the CSSLs. Two nearby KNOTTED1-LIKE HOMEOBOX I transcription factors homologous to the LATE MERISTEM IDENTITY1 (LMI1), Ghir_D01G021810.1 and Ghir_D01G021830.1, were located near the 61.14 Mb on chromosome D01. An 8-bp deletion in the third exon of the gene Ghir_D01G021810.1 showed the same mutation as reported previously (Fig. 2c and d) [37]. These examples showed that the high throughput detection methods could confirm an identified locus at a single gene-level resolution in this population.
QTL mapping yield-related and fiber quality traits in the CSSLs
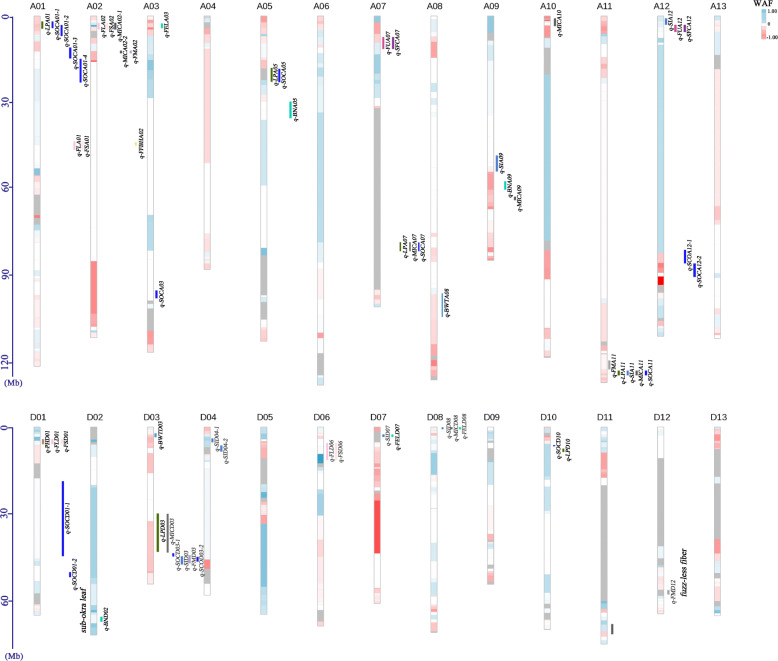
To evaluate the valuable genetic loci of interspecific hybridization that are important in cotton breeding, QTL was mapped based on these CSSLs. The coverage fragments in the genome were divided into 620 blocks, with an average of 3.12 Mb ranging from 29 kb to 69.47 Mb (Additional file 9: Table S8). A total of 64 QTL for 14 traits were mapped on 20 chromosomes with 38 in At-subgenome and 26 in Dt-subgenome (Fig. 3 and Table 3). The phenotypic variation explained by each QTL ranged from 0.73 to 14.67%. There were 19 QTL for four yield-related traits (BN, BWT, LP and SI) and the favorite alleles were from the Gh background. All the QTL for BWT and LP had negative alleles from Gh background, suggesting that the Gh has been domesticated for high yield. While, two QTL had positive alleles for BN indicating that Gb also had the potential to increase yield production. A total of 28 QTL were detected for fiber quality traits, most of which (18/28) had positive alleles from Gb. Of these, completely co-localization was observed for FL and FS, indicating that there was a significant correlation between them. Eight QTL for MIC were detected on seven chromosomes which explained phenotypic variation ranging from 2.54 to 7.09%. Contrary to FEL and FU, the positive alleles of SFC and FM were contributed by Gh. Poor fiber quality phenotype in the CSSLs declined that the genetic recession has occurred in the interspecific hybrids between Gh and Gb.
Fig. 3.
Chromosomal distribution of QTL and WAF value. Colored bars show the location of QTL. Red and blue indicate the additive effects from Gh and Gb, respectively; white and grey represent no effect and gap, respectively
Table 3.
Summary of the QTL in the CSSLs
| Traits | QTL | Blocks | Chr. | LOD | PVE (%) | Additive Effect | Block Interval | Publication | |
|---|---|---|---|---|---|---|---|---|---|
| Start | End | ||||||||
| PH | q-PHD01 | block329 | D01 | 3.33 | 4.78 | 1.02 | 3,186,046 | 4,556,068 | |
| FFBH | q-FFBHA02 | block67 | A02 | 2.81 | 4.04 | 0.34 | 44,641,617 | 45,470,669 | |
| BN | q-BNA05 | block125 | A05 | 3.67 | 4.86 | 0.97 | 31,292,912 | 34,892,387 | |
| q-BNA09 | block224 | A09 | 3.63 | 4.81 | 0.59 | 60,202,506 | 62,601,458 | ||
| q-LPD02 | block352 | D02 | 2.59 | 3.41 | −0.63 | 62,288,058 | 63,106,259 | ||
| BWT | q-BWTA08 | block205 | A08 | 3.04 | 3.97 | −0.35 | 93,642,362 | 102,084,301 | |
| q-BWTD03 | block358 | D03 | 2.81 | 3.65 | − 0.15 | 731,194 | 1,330,783 | ||
| LP | q-LPA01 | block3 | A01 | 6.40 | 6.95 | −1.83 | 2,639,939 | 3,677,553 | |
| q-LPA05 | block122 | A05 | 4.57 | 4.89 | −1.27 | 23,097,331 | 26,299,649 | ||
| q-LPA07 | block171 | A07 | 3.29 | 2.99 | −1.19 | 86,057,988 | 88,357,231 | ||
| q-LPA11 | block278 | A11 | 5.10 | 5.48 | −1.63 | 120,826,316 | 121,562,473 | ||
| q-LPD03 | block371 | D03 | 8.39 | 9.24 | −2.06 | 33,453,595 | 43,516,324 | ||
| q-LPD10 | block536 | D11 | 2.62 | 2.37 | −1.23 | 4,523,812 | 5,385,834 | ||
| SI | q-SIA09 | block222 | A09 | 4.79 | 2.48 | 0.31 | 54,020,891 | 58,352,764 | |
| q-SIA11 | block278 | A11 | 4.02 | 2.06 | 0.28 | 120,826,316 | 121,562,473 | ||
| q-SIA12 | block281 | A12 | 4.31 | 2.21 | 0.33 | 12,595 | 809,856 | ||
| q-SID03 | block373 | D03 | 3.38 | 1.73 | 0.36 | 44,469,168 | 47,084,844 | ||
| q-SID04–1 | block381 | D04 | 17.64 | 10.03 | 1.23 | 2,305,353 | 3,131,469 | ||
| q-SID04–2 | block384 | D04 | 24.46 | 14.67 | −1.05 | 4,507,347 | 6,673,788 | ||
| q-SID07 | block446 | D07 | 4.76 | 2.45 | −0.08 | 5,062,647 | 5,765,634 | ||
| q-SID08 | block471 | D08 | 3.85 | 1.98 | −0.09 | 27,684 | 689,962 | ||
| FL | q-FLA01 | block21 | A01 | 2.53 | 2.89 | 0.48 | 51,165,194 | 53,482,140 | |
| q-FLA02 | block59 | A02 | 8.10 | 9.60 | 0.70 | 2,981,374 | 3,502,835 | ||
| q-FLD01 | block329 | D01 | 3.93 | 4.53 | 0.51 | 3,186,046 | 4,556,068 | ||
| q-FLD06 | block428 | D06 | 2.78 | 3.17 | 0.80 | 8,954,143 | 12,114,917 | ||
| FS | q-FSA01 | block21 | A01 | 2.53 | 2.89 | 0.48 | 51,165,194 | 53,482,140 | |
| q-FSA02 | block59 | A02 | 8.10 | 9.60 | 0.70 | 2,981,374 | 3,502,835 | ||
| q-FSD01 | block329 | D01 | 3.93 | 4.53 | 0.51 | 3,186,046 | 4,556,068 | ||
| q-FSD06 | block428 | D06 | 2.78 | 3.17 | 0.80 | 8,954,143 | 12,114,917 | ||
| MIC | q-MICA02–1 | block59 | A02 | 6.99 | 7.09 | −0.20 | 2,981,374 | 3,502,835 | |
| q-MICA02–2 | block64 | A02 | 3.70 | 3.65 | 0.28 | 14,861,013 | 15,326,651 | ||
| q-MICA07 | block171 | A07 | 2.70 | 2.44 | −0.16 | 86,057,988 | 88,357,231 | ||
| q-MICA09 | block226 | A09 | 6.40 | 6.46 | 0.24 | 63,998,867 | 64,921,374 | ||
| q-MICA10 | block237 | A10 | 2.60 | 2.54 | 0.33 | 1,292,885 | 1,451,802 | ||
| q-MICA11 | block278 | A11 | 3.46 | 3.42 | −0.19 | 120,826,316 | 121,562,473 | ||
| q-MICD03 | block371 | D03 | 3.04 | 2.99 | −0.18 | 33,453,595 | 43,516,324 | ||
| q-MICD08 | block471 | D08 | 4.29 | 4.27 | 0.07 | 27,684 | 689,962 | ||
| q-MICD11 | block580 | D11 | 2.64 | 2.59 | −0.14 | 71,643,126 | 72,910,318 | ||
| FEL | q-FELA03 | block77 | A03 | 4.84 | 6.91 | 0.62 | 1,173,846 | 2,339,035 | |
| q-FELD07 | block446 | D07 | 5.47 | 7.84 | 0.18 | 5,062,647 | 5,765,634 | ||
| q-FELD08 | block471 | D08 | 3.16 | 4.45 | 0.17 | 27,684 | 689,962 | Said et al.2015 | |
| FU | q-FUA07 | block164 | A07 | 3.01 | 5.57 | 0.36 | 8,903,355 | 12,793,020 | |
| q-FUA12 | block282 | A12 | 2.68 | 2.23 | 0.21 | 809,856 | 1,774,576 | ||
| SFC | q-SFCA07 | block164 | A07 | 3.15 | 4.27 | −0.64 | 8,903,355 | 12,793,020 | |
| q-SFCA12 | block282 | A12 | 3.51 | 4.78 | −0.43 | 809,856 | 1,774,576 | ||
| FM | q-FMA02 | block64 | A02 | 3.06 | 3.83 | 0.01 | 14,861,013 | 15,326,651 | |
| q-FMA11 | block276 | A11 | 2.94 | 3.83 | −0.01 | 118,798,683 | 120,093,234 | ||
| q-FMD03 | block373 | D03 | 2.53 | 3.21 | −0.01 | 44,469,168 | 47,084,844 | ||
| q-FMD12 | block593 | D12 | 2.94 | 3.83 | −0.01 | 57,692,428 | 58,105,224 | ||
| SOC | q-SOCA01–1 | block3 | A01 | 21.66 | 5.84 | 2.32 | 2,639,939 | 3,677,553 | |
| q-SOCA01–2 | block4 | A01 | 3.14 | 0.73 | −1.17 | 3,677,553 | 4,874,638 | ||
| q-SOCA01–3 | block12 | A01 | 21.59 | 5.82 | 1.90 | 14,607,137 | 17,693,930 | ||
| q-SOCA01–4 | block13 | A01 | 8.52 | 2.08 | −1.60 | 17,693,930 | 28,547,783 | ||
| q-SOCA03 | block94 | A03 | 4.62 | 1.09 | −1.16 | 96,774,034 | 98,312,105 | ||
| q-SOCA05 | block122 | A05 | 6.73 | 1.62 | 1.00 | 23,097,331 | 26,299,649 | ||
| q-SOCA07 | block171 | A07 | 6.71 | 1.62 | 1.22 | 86,057,988 | 88,357,231 | ||
| q-SOCA11 | block277 | A11 | 6.08 | 1.45 | 1.32 | 120,093,234 | 120,826,316 | ||
| q-SOCA12–1 | block293 | A12 | 28.34 | 8.05 | 3.12 | 82,990,142 | 84,639,641 | ||
| q-SOCA12–2 | block294 | A12 | 17.02 | 4.43 | −2.85 | 84,639,641 | 86,335,298 | Yu et al.2012 | |
| q-SOCD01–1 | block332 | D01 | 34.76 | 10.40 | −4.36 | 17,173,981 | 44,222,294 | ||
| q-SOCD01–2 | block333 | D01 | 44.76 | 14.51 | 4.22 | 44,222,294 | 45,104,476 | Yu et al.2012 | |
| q-SOCD03–1 | block372 | D03 | 8.57 | 2.08 | 1.60 | 43,516,324 | 44,469,168 | ||
| q-SOCD03–2 | block374 | D03 | 4.38 | 1.01 | 1.95 | 47,084,844 | 48,284,786 | ||
| q-SOCD10 | block535 | D10 | 10.08 | 2.49 | 2.14 | 4,155,341 | 4,523,812 | ||
Genetic recession in the CSSLs
Genetic recession was a widespread phenomenon in the distant hybridization population. Fiber quality is one of the primary goals of cotton interspecific breeding. In this study, 7 lines with longer FL and 4 QTL for FL were identified in the CSSLs. Interestingly, two lines (N180 and R88) did not contain the QTL intervals, and two QTL intervals (on A01 and D06) also did not appear in the longer FL lines. The 13 fiber quality QTL identified in the single segment substitute lines (SSSLs) was inconsistent with the results of the same traits in this study except q-FLA02 [10]. So, we designed a weight mean of additive effects of fiber quality (WAF) value to analyze the source of additive effect for minor-effect genetic loci. Based on the correlations among the fiber traits, the additive effect of the genome was calculated (Additional file 10: Table S9). As a result, At-subgenome from Gh showed a higher additive contribution to fiber quality, while D-subgenome from Gb showed opposite results (Additional file 11: Table S10). In the Gb genome, more than 80% regions of chromosome A012, D02 and D12 had an additive effect on fiber quality improvement (Fig. 3). In addition, there was no additive effect from Gb on chromosome D07. More than 90% regions of chromosome A11 showed the effect of Gh. Notably, the non-contribution effect for fiber quality in At-subgenome was signification higher than that in Dt-subgenome. Of these, both chromosome A08 and A12 from Gb or Gh had more than half of the regions contributing no effect for fiber improvement.
QTL mapping for SOC and substitution mapping of QTL locus q-SOCA01–1
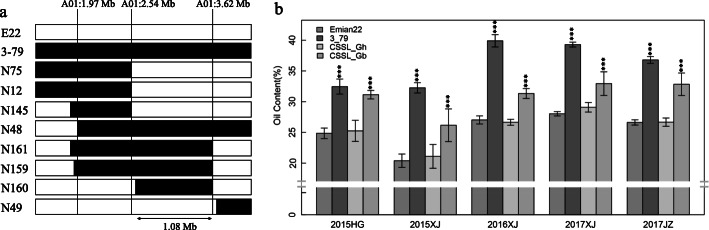
Less concern of the SOC in Gb showed significant difference compared with the recurrent parent ‘Emian22’. A total of 12 lines showed extremely significant (p ≤ 0.001) and stable higher SOC than recurrent parent ‘Emian22’ (Additional file 12: Table S11), and 15 QTL were detected to be related to SOC using BLUPed data; of these QTL, 12 were firstly characterized and only two QTL for SOC have been reported previously in an interspecific population (Table 3) [43]. Fortunately, three SSSLs (N159, N160 and N161) contained the same block (block3) on chromosome A01, providing an excellent materials for further research. Compared with another 7 lines including the parents, these three lines showed extremely significant high SOC properties like the donor parent (Fig. 4). In the associated interval (block 3 ≈ 1.08 Mb), there were 69 and 70 annotated genes in the Gh reference genome TM-1 and Gb reference genome 3–79, respectively. A previously study showed that cottonseed oil accumulates rapidly at the middle-late stages (20 to 30 days post anthesis) [44]. Hence, we focused on the genes that are expressed in gradients in ovules with significantly higher expression levels than other tissues (root, stem, leaf and fiber) [10]. Among these genes, the Gene Ontology (GO) analysis indicated that only six were involved in fatty acid metabolism process in both genome (Additional file 13: Table S12). Unfortunately, it is not significant difference expression of these oil relate genes in ovule between Gh and Gb (Additional file 14: Figure S2). Intringuing, another gene, Gbar_A01G002860.1, encoding a predicted mitochondrial pyruvate dehydrogenase kinase (mtPDK), showed higher expression than its homologous gene Ghir_A01G003150.1. However, previous data from Marillia et al. reported that the seed-specific partial silencing of the mtPDK resulted in increased storage lipid accumulation in developing seeds [45]. Hence, this gene may play an important role in storage lipid accumulation in late developing stage of cotton seeds.
Fig. 4.
Substitution mapping of q-SOC-1 using the 9 introgression lines (ILs) on chromosome. A01 a White and black represent the genotype of ‘Emian22’ and 3–79, respectively. b Seed oil content value are shown for five environments, the CSSL_Gh represent the background of Emian22(include the line of N75, N12, N49 and N145) and the CSSL_Gb represent the background of 3–79(include the line of N159, N160 and N161). One ANOVA analysis for two lines and Dunnett’s multiple comparison for multiple lines. ***. Indicated significantly different at the 0.001 level
Discussion
Cotton is the most important cash crop and contributes to more than 95% of natural textile fiber. Currently, improving the fiber quality by broadening the genetic basis of Upland cotton cultivars has become imperative. Construction of interspecific introgression lines can make full use of the superior fiber quality advantages of Gb on the basis of high yield of Gh, and also provide an ideal strategy for resolving the complex genome and QTL mapping. Several CSSLs with excellent agronomic traits than the Gh were found in this study, which can be directly applied to improve the fiber quality or SOC in cotton breeding.
Development strategy of the cotton introgression lines
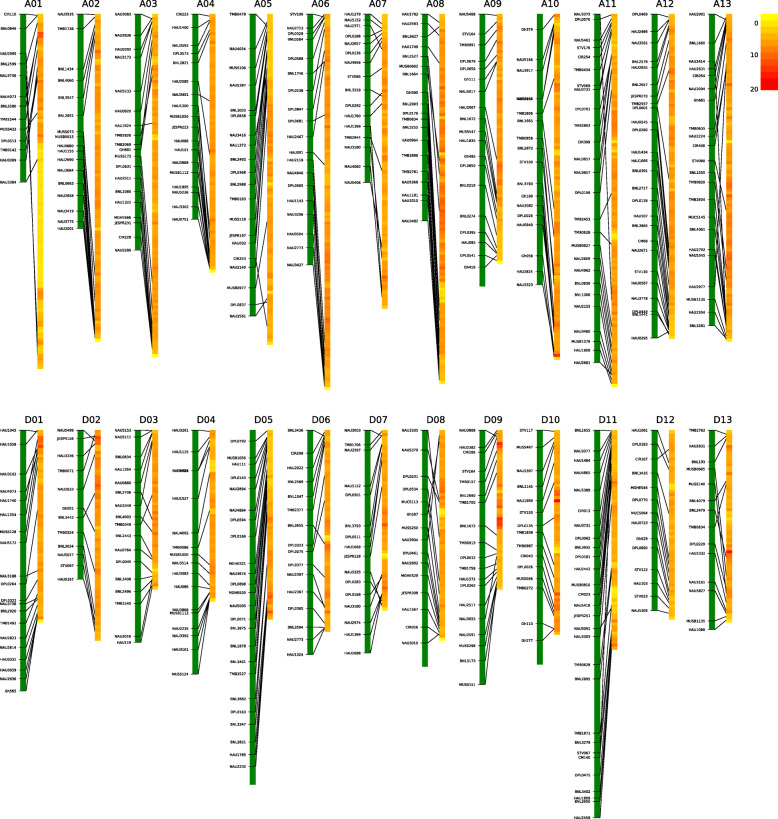
The ideal introgression lines aim to product a series of SSSLs in which all the introgression segments cover the entire donor genome. High cost-effective ratio of PCR-based molecular markers makes it the first choice for tracking the introgression segments due to absence of high quality reference genomic sequence. In this study, a high-density interspecific genetic map between Gh and Gb cotton was constructed and updated. In the early stage, few markers were selected from the primary genetic map to survey introgressions in the early generations, and then new markers were engaged in the advance generations with only targeted region selection after updating the high-density linkage map, which could be significantly reduce the workload during the development of the ILs population. However, identification of false or missing segments cannot be avoided. As a result, a wide range of gaps were found in At-subgenome by aligning the reference genome, especially on chromosome A01, A02, A03 and A06 (Fig. 5). Non-collinear arrangement and clustering of the SSR markers on the physical map significantly reduced the coverage of the genome. Significant clustering of SSR markers appeared at the both ends of multiple chromosomes, such as A02, A03, A06 and A08, which was consisted with that a lot of lines carried a long fragment detected by several sequential markers.
Fig. 5.
Comparison of genetic map and physical map in evaluating the CSSLs. Left and right show SSR markers and SNP markers position on the chromosome, respectively. Colors show the density of the SNPs. Marker’s position is linked by grey lines between two maps
Despite that, the high-density linkage map constructed by our lab still showed a certain advantage in this study. Several SSSLs were confirmed by genome re-sequencing which were identified by PCR-based molecular markers.
High-throughput genotyping technology provides highly reliable introgression
The whole genome re-sequencing technology provides a strategy to understand the entire genomic variations after having a high quality reference genomic sequence, which could help to improve the detection of the donor segments in the whole genome. In this study, the CSSLs were genotyped using next-generation sequencing following the project of the reference genome [10], and an ultrahigh-quality physical map by SNPs was constructed, which was a pioneer study to use this strategy for genotyping CSSLs in cotton. As a result, lots of small segments were newly detected by sequencing, which significantly reduced the number of corresponding chromosomes and candidate confidence intervals for the associated traits. Some segments containing the candidate genes cannot be effectively assessed by SSR markers, although these markers were closely linked with the target trait. For example, the sub-okra leaf shape gene was detected by whole genome re-sequencing, while the MM-map only showed that there was a marker associated with this trait. In this study, none introgression segments were detected in 10 lines by SNPs. The reason is that the introgression fragments in these lines identified by SSR markers are less than 100 kb in length, which were marked as ‘not available’ and filtered. Besides homozygous introgressions, a number of heterozygous fragments were detected on chromosome A01 and A08 after a few rounds of self-fertilization. For example, line R28 carried the heterozygous fragment covering almost the entire chromosome A08, and line R126 carried a wide range of heterozygous fragments on different chromosomes which may result in colorful phenotype of the fuzz fiber (Additional file 7: Figure S1). Consistent with the previous reports [46, 47], we speculate that this may be related to the interspecific segregation distortion.
Based on the above results, we conclude that construction of an ideal introgression population can follow this strategy: (1) PCR markers from high-density genetic map are used to construct the primary introgression lines in the primary generations to decrease the cost; (2) all the lines are genotyped by high-throughput re-sequencing technology to accurately identify the introgression segments; (3) further backcrossing of the lines carrying more than one segment will be performed to achieve the purpose of constructing SSSLs.
CSSLs constructed a platform for resolving the polygene hypothesis
Quantitative traits are usually regulated by multiple minor-efficient genetic loci, which modified by the genetic and external environments [48]. Different QTL for fiber traits were detected between the SSSLs [10] and the whole lines (this study), indicating that the genetic loci for superior fiber quality of the Gb was controlled by multiple genes and dispersed on different chromosomes. A notable evidence appeared in this study was that the CSSLs (N180 and R88) carried multiple donor fragments but did not contain the QTL loci, which means that the genetic effects of these introgression fragments were low enough to be detected as a major QTL. Consistent with the previous study of the introgression population, we aimed at dissecting the donor genome by MAS in this study. However, this strategy may undermine the genetic pattern of quantitative traits such as fiber quality traits, which commonly regulated by multiple genes at different development stages [49]. Hundreds of high expression levels of genes during fiber development also illustrated this view [10]. These co-effector genes derived from Gb donor were segmented and dispersed in different lines, which blocked the regulatory relationship between them. As a result, we summarized that fewer introgression fragments in the SSSLs may effectively block the interaction between different genetic backgrounds and between loci on different chromosomes, which facilitated the detection of the minor-efficient genetic loci [10]. While more introgression segments and higher genomic coverage, especially the long fragments, the noise and epistasis effects were effectively reduced, which improved the reliability of identifying major and stronger effective loci that can be directly applied into breeding in the future. Similar conclusion in previous reports just had a brief description [31, 50, 51]. However, correlations between phenotypes may indicate that complex quantitative traits are controlled by same gene or closely linked genes. Many fiber quality QTL were detected in the interval of block 59 in this study, which indicated that there still existed the single major genetic locus for fiber quality in the Gb genome. Therefore, we can conclude that the genetic locus controlling fiber quality in the Gb genome is the interaction of the major gene with the minor-effect polygenic loci scattered on different chromosomes, and the future breeding for improving fiber quality should try to pyramid more beneficial factors.
Sea-island cotton as an excellent resource for improving cottonseed oil content
Cottonseed oil has a large amount of unsaturated fatty acids [52]. Several lines with higher SOC were identified which could be directly used in oil improvement breeding, connecting with the higher value (87%) of the broad-sense heritability. Multiple QTL for SOC were detected on different chromosomes in this population, which suggested that there should be a network between genes controlling the SOC in the Gb. These results indicate that Sea-island cotton has a high potential in improving the SOC of Gh. In this study, we predicted that a PDK gene may regulated the SOC in Gb, which indicated that the growth advantages of Sea-island cotton may have a more positive influence on regulating other traits than Upland cotton. Complex fatty acid metabolism pathway and the diversity of lipid compositions increase the difficulty to propose the candidate genes in the confidence intervals. However, based on the genomic annotation variation combining with transcriptome and metabolome analysis, the relevant information of the lipid biosynthesis is sufficient to identify candidate genes in the future, which have been proved to be feasible [12, 53].
Conclusions
Plant breeding aims to integrate multiple desirable traits to obtain elite varieties. Introgression between different species is a key process to broaden the genetic basis of the breeding materials. In this study, we developed a CSSLs population carrying introgression segments from Gb in the Gh background. The whole-genome re-sequencing technology was applied to study the CSSLs to construct the high-quality physical map for each line, which provided more accurate introgression than in the map constructed by SSR markers. A total of 64 QTL were mapped for 14 agronomic traits and favorite Gb alleles for fiber quality were identified. Importantly, novel Gb alleles for increasing SOC were found. Our study not only offered guides for future molecular breeding to increase fiber quality and SOC, but also provided a reference basis for fine-mapping and map-based cloning genes to genetic improvement of Upland cotton.
Methods
CSSLs development
In this study, ‘Emian22’ (G. hirsutum) and ‘3–79’ (G. barbadense), were used to develop CSSLs. ‘Emian22’ is an upland cotton cultivar with high yield and moderate fiber quality in Hubei Province. And the ‘3–79’ is a genetic and cytogenetic standard line for G. barbadense with super fiber quality and high resistance to Verticillium wilt. ‘Emian22’ and ‘3–79’ are public available materials and have been kept in our laboratory nearly twenty years. The construction process of this CSSLs population has been brief described in the previous article [10]. In 2006, after four rounds of successive backcrossing, 254 whole-genomic SSR markers were selected to the whole-genome surveying 221 BC4 lines [54] (Additional file 15: Figure S3). The 82 BC4 plants covering the whole donor cotton genome were selected to be further backcrossed with ‘Emian22’, while some of these individuals were selected to be self-pollinated to produce BC4F2. In 2007, target regions were genotyped using the corresponding polymorphic markers in 1686 individual plants derived from 1028 BC4F2 and 658 BC5F1 individual plants. A total of 302 individuals out of them containing less than five, short chromosome segments and possibly covering the donor genome were selected, including 128 individuals with only one donor segment (Additional file 16: Figure S4). In 2008, 515 markers selected from the updated high-density linkage map [55], were used for re-evaluating the plants. About 312 individuals were selected, of which 162 individuals had less than three donor segments (Additional file 17: Figure S5). The plants having only one donor segment were self-pollinated to produce the homozygous CSSLs, and the others were continually backcrossed with ‘Emian22’ to produce the advanced backcrossing generation. In the same way in 2009, corresponding polymorphic markers were executed to identify the target segment in all the lines, including the self-pollinated lines. About 336 individuals containing the target region were selected, including 60 plants with only one donor segment (Additional file 18: Figure S6). In the subsequent process, same steps were executed to select the plants with the target segments. Until 2011, 337 individuals were obtained with 279 plants having less than three target segments, of which 151 plants having only one donor segment (Additional file 19: Figure S7). After two rounds of self-fertilization to ensure the homozygous genotype, a set of 325 CSSLs including 177 SSSLs were ultimately obtained.
Phenotype evaluation
All the CSSLs with their parents were planted in two replicated plots at three different locations which are authorized by local governments: Huanggang (HG), Hubei province and Shihezi (SHZ), Xinjiang province in 2015; Shihezi in 2016; Jingzhou (JZ), Hubei Province and Shihezi in 2017. Field management essentially followed the local agricultural practices. PH, FFBH, and BN were evaluated at blooming stage, including the morphology of the plants (leaf and flower). Twenty bolls from each line were hand-harvested from the internal middle parts of the plants at the mature stage in every year. Yield-related traits, such as BWT, LP, SI, were tested in this CSSLs. And seven fiber quality traits were investigated including FL, FS, MIC, FU, FEL, SFC and FM. The seed phenotypes were scored based on visual inspection; meanwhile, at least 10 g delinted seeds were used to measure for SOC by low field pulsed nuclear magnetic resonance apparatus (NMR) analyzer on a NM-12 (Niumai Analytical Instrument Corporation, China). Best linear unbiased predictions (BLUPs) with broad sense heritability (H2) were used to estimate phenotypic traits across all five environments in R package. Pearson correlation coefficients were calculated to analyse the relationship between traits using BLUPed data by SPSS 17.0 software (SPSS Inc., Chicago, IL, USA).
Estimating the introgression segments in CSSLs using SSR markers
Total genomic DNA of the CSSLs and their parents was extracted from the fresh young leaves at seeding stage using modified CTAB method [56]. A total of 515 SSR markers selected from the high-density interspecific genetic map were used to genotype the CSSLs. The length of Gb introgression segment was estimated by the graphical genotype of the markers. If one marker has the same genotype as the donor parent, this line is considered to carry the introduced fragment from donor parent at this genetic position; otherwise, the genetic background will be considered to be the same as the recipient parent. A segment flanked by two markers with genotype DD, DR, RR, were considered to be 100, 50, 0% of donor type, respectively (Additional file 20: Figure S8). The “D” and “R” represent the donor and recipient genotype, respectively. Thus, the length of the introgression segment was estimated to be the total length of the DD length and two half of DR length [31].
Identification of SNPs and introgression segments in the CSSLs
The CSSLs population was cultivated in the field in Wuhan, China, in 2017. Leaf tissues were collected for plant genome DNA extraction with the Plant Genome Extraction Kit (TIANGEN Biotech). The 177 SSSLs with the parents have been sequenced by Wang et al. [10]. The other 145 CSSLs were sequenced on the same Illumina HiSeq platform with at least 6× coverage (pair-end 150 bp; Additional file 21: Table S13). Meanwhile, the Gh parent line ‘Emian22’ was deep sequenced with 60× coverage. To redo SNP calling, all the clean sequencing reads were mapped on the G. hirsutum reference TM-1 genome using BWA software version 0.7.10 and SNPs were called using GATK software with previously reported method [10].
The CSSLs may had large introduced fragment at the Chromosome recombination interval, so the bin map could be a better strategy to instead consecutive SNPs. A slightly modified sliding windows approach [57] was applied to identify the donor segments from Gb (Additional file 22: Figure S9). Firstly, a total of 11,653,661 SNPs and an average of 5.3 per kb were detected between Gh and Gb, and used to construct the bin. Then, all the alleles represented by SNPs in each CSSL were filtered using SNPs from both parents. And only those having the same allele as one of the parents were retained. The genotype of each window was called with a window size of 50 kb and step size of 5 kb. The ratio of SNPs in the window was calculated (> 80% of SNPs had one parental genotype, the window was called as homozygous of one parent; otherwise, the window was called as heterozygous). Determination of the recombination breakpoints and construction of the bins were performed as described by Han et al. [57]. The regions between two adjacent bins with same genotypes less than 100 kb were defined as the same bin, and bins of less than 100 kb in length were filtered. The recombinant donor chromosome segments for each CSSL were constructed based on the recombinant bins.
QTL mapping and weight mean of additive effects of fiber quality evaluation
To identify the QTL, the Gb introgression segments were divided into several non-overlapping blocks (Additional file 23: Figure S10), ensuring each line carries as smaller overlapping chromosome region as possible. The BLUPed data of the five environments was used as the response variations of the 14 traits. QTL mapping and additive effect calculation were performed using RSETP-LRT-ADD mapping method with QTL IciMapping V4.0 software [58]. The block interval was used as the QTL location, and QTL was named based on the rules of the reporting in the Rosaceae (recommendations for standard QTL nomenclature and reporting in the Rosaceae 2014). To obtain potential candidate genes, the annotated genes were identified for a Gene Ontology (GO) analysis and the transcription profiles for different tissues of TM-1 and 3–79 were employed as a reference [10].
Based on the QTL mapping results, the additive effect of all the fiber traits were calculated. Contributions of the Gb to the fiber quality in the Gh background were estimated using a weight mean model. Based on the correlations between the fiber traits and the broad sense heritability, the WAF model was described by the following formula: t represents the fiber quality traits, Addt is the value of additive effects for each block, rt is the value of positive correlation coefficient and H2t represents the broad sense heritability of the related trait. The distribution of the WAF on chromosome was calculated based on the blocks interval.
Supplementary information
Additional file 1: Table S1. Segments carried by CSSLs.xlsx
Additional file 2: Table S2. Summary of introgression segments.xlsx
Additional file 3: Table S3. Summary of the segments in the CSSLs.xlsx
Additional file 4: Table S4. Description of investigated traits in CSSLs.xlsx
Additional file 5: Table S5. The phenotypic data of CSSLs.xlsx
Additional file 6: Table S6. Broad-sense heritability (H2) of 14 traits in the CSSLs.xlsx
Additional file 7: Figure S1. The fuzz fiber phenotypes in the CSSLs with their parent lines. TIFF
Additional file 8: Table S7. Morphological characteristics of specific introgression lines.xlsx
Additional file 9: Table S8. Summary of the blocks in the genome.xlsx
Additional file 10: Table S9. Additive effects of the fiber length with the positive traits.xlsx
Additional file 11: Table S10. Summary of the Weight mean of fiber Additive effects on chromosome.xlsx
Additional file 12: Table S11. Yield-related and fiber quality traits of specific CSSLs.xlsx
Additional file 13: Table S12. GO enrichment analysis of genes in the candidate chromosome region.xlsx
Additional file 14: Figure S2. Transcript profiles of promising genes for root, stem, leaf, fiber and ovule between Emian22 and 3-79.
Additional file 15: Figure S3. Summary of the introgression segments of the BC4 generation with 221 individuals in 2006.
Additional file 16: Figure S4. Summary of the introgression segments of 302 individuals in 2007. TIFF
Additional file 17: Figure S5. Summary of the introgression segments of 312 individuals in 2008. TIFF
Additional file 18: Figure S6. Summary of the introgression segments of 336 individuals in 2009. TIFF
Additional file 19: Figure S7. Summary of the introgression segments of 337 individuals in 2010. TIFF
Additional file 20: Figure S8. Example of chromosome introduction fragments evaluated by SSR markers. A. Genotype calling based on the graphic of SSR markers on the PAGE. The “DD” and “RR” represent the donor and recipient parent, respectively. B. Introgression fragments evaluating based on the genotype of two near markers: “DD” represents 100% (Marker2 and Marker3); “DR” represents 50% (Marker4 and Marker5); “RR” represents 0% (Marker 5 and Marker6). TIFF
Additional file 21: Table S13. Summary of DNA sequencing data for CSSLs.xlsx
Additional file 22: Figure S9. An overview of the introgression segment identification protocol. A. Schematic diagram on identification of chromosome introduced fragment in CSSLs. First, all of the CSSLs and their parents were sequenced on an Illumina HiSeq platform to produce the genome sequence. All clean data were mapped to the G.hirsutum (TM-1) genome using BWA software and the unique mapping data were retained for further analysis. Then GATK software were applied to identify the SNPs based on the criteria:(1) the quality of SNPs should be over 100; (2) each SNP was supported by at least five reads; and (3) the adjacent SNPs should have a distance of at least 10 bp. To identify the introgression segments in the CSSLs, the SNPs between parents were selected. And a modified sliding-window approach was applied to identify the donor segments from Gb. This approach has been described very clearly by Han et al [57]. All the alleles represented by SNPs in each CSSL were filtered using SNPs from both parents. A bin map was constructed based on the genotype results of the window and consecutive bins with the same genotype were combined into same segments. B. Example of the Genotype calling based on the ratio of the SNPs in the window(>80% of SNPs had one parental genotype, the window was called as homozygous of one parent; otherwise, the window was called as heterozygous). TIFF
Additional file 23: Figure S10. Example diagram of block partition. (A) The diagram show the principle of block partition; (B) The CSSLs carried the introgression segments on the endpoint of the chromosome A01; (C) First five blocks on the chromosome A01. TIFF
Acknowledgements
We thank Dr. Koeun Han from Seoul National University, Korea, for kindly sharing the data processing script. We thank Minghui Meng and Chao Shen for help in bioinformatics analysis. We thank Tianwang Wen and Bin Gao for the help in the experiment. We thank Xinxin Liu, Ruiting Zhang and Xiaojing Li for investigating the phenotypic traits.
Abbreviations
- AFLP
Amplified fragment length polymorphism
- BLUPs
Best linear unbiased predictions
- BN
Boll number per plant
- BWT
Weight per boll
- CSSLs
Chromosome segments substitution lines
- FEL
Fiber elongation
- FFBH
First fruit branch height
- FL
Fiber length
- FM
Fiber mature content
- FS
Fiber strength
- FU
Fiber uniformity
- Gh
Gossypium hirsutum
- Gb
G. barbadense
- GO
Gene Ontology
- LP
Lint percentage
- MAS
Markers assisted selection
- MIC
Micronaire value
- NMR
Nuclear magnetic resonance apparatus
- PH
Plant height
- QTL
Quantitative trait loci
- RFLP
Fragment length polymorphism
- SFC
Short fiber content
- SNPs
Single nucleotide polymorphisms
- SI
Seed index
- SOC
Seed oil content
- SSR
Simple sequence repeat
- SSSLs
Single segment substitute lines (SSSLs)
- WAF
Weight mean of additive effects of fiber quality
Authors’ contributions
ZXL conceived and designed the project. XML and ZWW constructed the introgression lines. DZ (DZ1) conducted the experiments, analyzed the data and wrote the manuscript draft. CYY, XHN and DWZ (DZ2) provided the experimental fields in Xinjiang and collected the phenotypic data. JS performed sequencing and provided the resequencing data of the 145 CSSLs. ZXL and XLZ revised the manuscript. All authors discussed the results and approved the final manuscript.
Funding
The design of the study, field experiment and collection, data analysis, and manuscript writing were financially supported by the Genetically Modified Organisms Breeding Major Project of China (No.2016ZX08009001).
Availability of data and materials
The clean raw sequencing data in this manuscript have been deposited in NCBI Sequence Read Archive under accession number PRJNA433615 and PRJNA543759.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
De Zhu, Email: zhujunde@msn.cn.
Ximei Li, Email: okliximei@163.com.
Zhiwei Wang, Email: wzw_1983@163.com.
Chunyuan You, Email: xjycy99@126.com.
Xinhui Nie, Email: xjnxh2004130@126.com.
Jie Sun, Email: sunjie@shzu.edu.cn.
Xianlong Zhang, Email: xlzhang@mail.hzau.edu.cn.
Dawei Zhang, Email: zbzdw012@126.com.
Zhongxu Lin, Email: linzhongxu@mail.hzau.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12864-020-06800-x.
References
- 1.Senchina DS, Alvarez I, Cronn R, Liu B, Rong J, Noyes RD, Paterson AH, Wing RA, Wilkins TA, Wendel JF. Rate variation among nuclear genes and the age of polyploidy in Gossypium. Mol Biol Evol. 2003;20(4):633–643. doi: 10.1093/molbev/msg065. [DOI] [PubMed] [Google Scholar]
- 2.Wendel JF, Cronn R. Polyploidy and the evolutionary history of cotton. Adv Agron. 2003;78:139–186. [Google Scholar]
- 3.Grover CE, Gallagher JP, Jareczek JJ, Page JT, Udall JA, Gore MA, Wendel JF. Re-evaluating the phylogeny of allopolyploid Gossypium L. Mol Phylogenet Evol. 2015;92:45–52. doi: 10.1016/j.ympev.2015.05.023. [DOI] [PubMed] [Google Scholar]
- 4.Tyagi P, Gore MA, Bowman DT, Campbell BT, Udall JA, Kuraparthy V. Genetic diversity and population structure in the US upland cotton (Gossypium hirsutum L.) Theor Appl Genet. 2014;127(2):283–295. doi: 10.1007/s00122-013-2217-3. [DOI] [PubMed] [Google Scholar]
- 5.Kaur B, Tyagi P, Kuraparthy V. Genetic diversity and population structure in the landrace accessions of Gossypium hirsutum. Crop Sci. 2017;57(5):2457. [Google Scholar]
- 6.Zhang J, Percy RG, McCarty JC. Introgression genetics and breeding between upland and Pima cotton: a review. Euphytica. 2014;198(1):1–12. [Google Scholar]
- 7.Marani A, Avieli E. Heterosis during the early phases of growth in intraspecific and interspecific crosses of cotton. Crop Sci. 1973;13(1):15–18. [Google Scholar]
- 8.Balakrishnan D, Surapaneni M, Mesapogu S, Neelamraju S. Development and use of chromosome segment substitution lines as a genetic resource for crop improvement. Theor Appl Genet. 2019;132(1):1–25. doi: 10.1007/s00122-018-3219-y. [DOI] [PubMed] [Google Scholar]
- 9.Hu Y, Chen J, Fang L, Zhang Z, Ma W, Niu Y, Ju L, Deng J, Zhao T, Lian J, et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat Genet. 2019;51(4):739–748. doi: 10.1038/s41588-019-0371-5. [DOI] [PubMed] [Google Scholar]
- 10.Wang M, Tu L, Yuan D, Zhu D, Shen C, Li J, Liu F, Pei L, Wang P, Zhao G, et al. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat Genet. 2019;51(2):224–229. doi: 10.1038/s41588-018-0282-x. [DOI] [PubMed] [Google Scholar]
- 11.Zhang JF, Percy RG. Improving upland cotton by introducing desirable genes from Pima cotton. World Cotton Res Con. 2007. http://wcrc.confex.com/wcrc/2007/techprogram/P1901.HTM.
- 12.Fernandez-Moreno JP, Levy-Samoha D, Malitsky S, Monforte AJ, Orzaez D, Aharoni A, Granell A. Uncovering tomato quantitative trait loci and candidate genes for fruit cuticular lipid composition using the Solanum pennellii introgression line population. J Exp Bot. 2017;68(11):2703–2716. doi: 10.1093/jxb/erx134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong Q, Zhang Z, Wang L, Zhu Y, Fan Y, Mou T, Ma L, Zhuang J. Dissection and fine-mapping of two QTL for grain size linked in a 460-kb region on chromosome 1 of rice. Rice. 2018;11(1):44. doi: 10.1186/s12284-018-0236-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koumproglou R, Wilkes TM, Townson P, Wang XY, Beynon J, Pooni HS, Newbury HJ, Kearsey MJ. STAIRS: a new genetic resource for functional genomic studies of Arabidopsis. Plant J. 2002;31(3):355–364. doi: 10.1046/j.1365-313x.2002.01353.x. [DOI] [PubMed] [Google Scholar]
- 15.Wan XY, Wan JM, Weng JF, Jiang L, Bi JC, Wang CM, Zhai HQ. Stability of QTLs for rice grain dimension and endosperm chalkiness characteristics across eight environments. Theor Appl Genet. 2005;110(7):1334–1346. doi: 10.1007/s00122-005-1976-x. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J, Zhang J, Liu W, Han H, Lu Y, Yang X, Li X, Li L. Introgression of Agropyron cristatum 6P chromosome segment into common wheat for enhanced thousand-grain weight and spike length. Theor Appl Genet. 2015;128(9):1827–1837. doi: 10.1007/s00122-015-2550-9. [DOI] [PubMed] [Google Scholar]
- 17.Qi L, Sun Y, Li J, Su L, Zheng X, Wang X, Li K, Yang Q, Qiao W. Identify QTLs for grain size and weight in common wild rice using chromosome segment substitution lines across six environments. Breed Sci. 2017;67(5):472–482. doi: 10.1270/jsbbs.16082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Divilov K, Barba P, Cadle-Davidson L, Reisch BI. Single and multiple phenotype QTL analyses of downy mildew resistance in interspecific grapevines. Theor Appl Genet 2018;131(5):1133–43. [DOI] [PMC free article] [PubMed]
- 19.Paterson AH, Deverna JW, Lanini B, Tanksley SD. Fine mapping of quantitative trait loci using selected overlapping recombinant chromosomes, in an interspecies cross of tomato. Genetics. 1990;124(3):735–742. doi: 10.1093/genetics/124.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao J, Liu J, Xu J, Zhao L, Wu Q, Xiao S. Quantitative trait locus mapping and candidate gene analysis for Verticillium wilt resistance using Gossypium barbadense chromosomal segment introgressed line. Front Plant Sci. 2018;9:682. doi: 10.3389/fpls.2018.00682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li X, Wang W, Wang Z, Li K, Lim YP, Piao Z. Construction of chromosome segment substitution lines enables QTL mapping for flowering and morphological traits in Brassica rapa. Front Plant Sci. 2015;6:432. doi: 10.3389/fpls.2015.00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ademe MS, He S, Pan Z, Sun J, Wang Q, Qin H, Liu J, Liu H, Yang J, Xu D, et al. Association mapping analysis of fiber yield and quality traits in upland cotton (Gossypium hirsutum L.) Mol Genet Genomics. 2017;292(6):1267–1280. doi: 10.1007/s00438-017-1346-9. [DOI] [PubMed] [Google Scholar]
- 23.Said JI, Song M, Wang H, Lin Z, Zhang X, Fang DD, Zhang J. A comparative meta-analysis of QTL between intraspecific Gossypium hirsutum and interspecific G. hirsutum × G. barbadense populations. Mol Genet Genomics. 2015;290(3):1003–1025. doi: 10.1007/s00438-014-0963-9. [DOI] [PubMed] [Google Scholar]
- 24.Fang L, Wang Q, Hu Y, Jia Y, Chen J, Liu B, Zhang Z, Guan X, Chen S, Zhou B. Genomic analyses in cotton identify signatures of selection and loci associated with fiber quality and yield traits. Nat Genet. 2017;49(7):1089–1098. doi: 10.1038/ng.3887. [DOI] [PubMed] [Google Scholar]
- 25.Wang M, Tu L, Lin M, Lin Z, Wang P, Yang Q, Ye Z, Shen C, Li J, Zhang L, et al. Asymmetric subgenome selection and cis-regulatory divergence during cotton domestication. Nat Genet. 2017;49(4):579–587. doi: 10.1038/ng.3807. [DOI] [PubMed] [Google Scholar]
- 26.Du X, Huang G, He S, Yang Z, Sun G, Ma X, Li N, Zhang X, Sun J, Liu M, et al. Resequencing of 243 diploid cotton accessions based on an updated a genome identifies the genetic basis of key agronomic traits. Nat Genet. 2018;50(6):796–802. doi: 10.1038/s41588-018-0116-x. [DOI] [PubMed] [Google Scholar]
- 27.Zhang S, Yu H, Wang K, Zheng Z, Liu L, Xu M, Jiao Z, Li R, Liu X, Li J, et al. Detection of major loci associated with the variation of 18 important agronomic traits between Solanum pimpinellifolium and cultivated tomatoes. Plant J. 2018;95(2):312–323. doi: 10.1111/tpj.13952. [DOI] [PubMed] [Google Scholar]
- 28.Ni X, Xia Q, Zhang H, Cheng S, Li H, Fan G, Guo T, Huang P, Xiang H, Chen Q, et al. Updated foxtail millet genome assembly and gene mapping of nine key agronomic traits by resequencing a RIL population. GigaScience. 2017;6(2):1–8. doi: 10.1093/gigascience/giw005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomson MJ, Singh N, Dwiyanti MS, Wang DR, Wright MH, Perez FA, Declerck G, Chin JH, Maliticlayaoen GA, Juanillas VM. Large-scale deployment of a rice 6 K SNP array for genetics and breeding applications. Rice. 2017;10(1):40. doi: 10.1186/s12284-017-0181-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu J, Zhao Q, Du P, Xu C, Wang B, Feng Q, Liu Q, Tang S, Gu M, Han B. Developing high throughput genotyped chromosome segment substitution lines based on population whole-genome re-sequencing in rice (Oryza sativa L.) BMC Genomics. 2010;11(1):656. doi: 10.1186/1471-2164-11-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu J, Niu Y, Tao Y, Wang J, Jian J, Tai S, Li J, Yang J, Zhong W, Zhou Y, et al. Construction of high-throughput genotyped chromosome segment substitution lines in rice (Oryza sativa L.) and QTL mapping for heading date. Plant Breed. 2015;134(2):156–163. [Google Scholar]
- 32.Li Y, Colleoni C, Zhang J, Liang Q, Hu Y, Ruess H, Simon R, Liu Y, Liu H, Yu G, et al. Genomic analyses yield markers for identifying agronomically important genes in potato. Mol Plant. 2018;11(3):473–484. doi: 10.1016/j.molp.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 33.Li X, Wu L, Wang J, Sun J, Xia X, Geng X, Wang X, Xu Z, Xu Q. Genome sequencing of rice subspecies and genetic analysis of recombinant lines reveals regional yield- and quality-associated loci. BMC Biol. 2018;16(1):102. doi: 10.1186/s12915-018-0572-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qian N, Zhang X-W, Guo W-Z, Zhang T-Z. Fine mapping of open-bud duplicate genes in homoelogous chromosomes of tetraploid cotton. Euphytica. 2008;165(2):325–331. [Google Scholar]
- 35.Chang L, Fang L, Zhu Y, Wu H, Zhang Z, Liu C, Li X, Zhang T. Insights into interspecific hybridization events in allotetraploid cotton formation from characterization of a gene-regulating leaf shape. Genetics. 2016;204(2):799–806. doi: 10.1534/genetics.116.193086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andres RJ, Coneva V, Frank MH, Tuttle JR, Samayoa LF, Han SW, Kaur B, Zhu L, Fang H, Bowman DT, et al. Modifications to a LATE MERISTEM IDENTITY1 gene are responsible for the major leaf shapes of upland cotton (Gossypium hirsutum L.) Proc Natl Acad Sci U S A. 2017;114(1):E57–E66. doi: 10.1073/pnas.1613593114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu QH, Zhang J, Liu D, Stiller W, Liu D, Zhang Z, Llewellyn D, Wilson I. Integrated mapping and characterization of the gene underlying the okra leaf trait in Gossypium hirsutum L. J Exp Bot. 2016;67(3):763–774. doi: 10.1093/jxb/erv494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu H, Tian Y, Wan Q, Fang L, Guan X, Chen J, Hu Y, Ye W, Zhang H, Guo W, et al. Genetics and evolution of MIXTA genes regulating cotton lint fiber development. New Phytol. 2018;217(2):883–895. doi: 10.1111/nph.14844. [DOI] [PubMed] [Google Scholar]
- 39.Wan Q, Guan X, Yang N, Wu H, Pan M, Liu B, Fang L, Yang S, Hu Y, Ye W, et al. Small interfering RNAs from bidirectional transcripts of GhMML3_A12 regulate cotton fiber development. New Phytol. 2016;210(4):1298–1310. doi: 10.1111/nph.13860. [DOI] [PubMed] [Google Scholar]
- 40.Wang B, Draye X, Zhuang Z, Zhang Z, Liu M, Lubbers EL, Jones D, May OL, Paterson AH, Chee PW. QTL analysis of cotton fiber length in advanced backcross populations derived from a cross between Gossypium hirsutum and G. mustelinum. Theor Appl Genet. 2017;130(6):1297–1308. doi: 10.1007/s00122-017-2889-1. [DOI] [PubMed] [Google Scholar]
- 41.Saha S, Stelly DM, Makamov AK, Ayubov MS, Raska D, Gutiérrez OA, Manchali S, Jenkins JN, Deng D, Abdurakhmonov IY. Molecular confirmation of Gossypium hirsutum chromosome substitution lines. Euphytica. 2015;205(2):459–473. [Google Scholar]
- 42.Wang B, Nie Y, Lin Z, Zhang X, Liu J, Bai J. Molecular diversity, genomic constitution, and QTL mapping of fiber quality by mapped SSRs in introgression lines derived from Gossypium hirsutum × G. darwinii watt. Theor Appl Genet. 2012;125(6):1263–1274. doi: 10.1007/s00122-012-1911-x. [DOI] [PubMed] [Google Scholar]
- 43.Yu J, Yu S, Fan S, Song M, Zhai H, Li X, Zhang J. Mapping quantitative trait loci for cottonseed oil, protein and gossypol content in a Gossypium hirsutum × Gossypium barbadense backcross inbred line population. Euphytica. 2012;187(2):191–201. [Google Scholar]
- 44.Zhao Y, Wang Y, Huang Y, Cui Y, Hua J. Gene network of oil accumulation reveals expression profiles in developing embryos and fatty acid composition in upland cotton. J Plant Physiol. 2018;228:101–112. doi: 10.1016/j.jplph.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 45.Marillia EF, Micallef BJ, Micallef M, Weninger A, Pedersen KK, Zou J, Taylor DC. Biochemical and physiological studies of Arabidopsis thaliana transgenic lines with repressed expression of the mitochondrial pyruvate dehydrogenase kinase1. J Exp Bot. 2003;54(381):259–270. doi: 10.1093/jxb/erg020. [DOI] [PubMed] [Google Scholar]
- 46.Hulsekemp AM, Lemm J, Plieske J, Ashrafi H, Buyyarapu R, Fang DD, Frelichowski J, Giband M, Hague S, Hinze LL. Development of a 63K SNP array for cotton and high-density mapping of intraspecific and interspecific populations of Gossypium spp. G3. 2015;5(6):1187–1209. doi: 10.1534/g3.115.018416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Z, Qanmber G, Wang Z, Yang Z, Li F. Gossypium genomics: trends, scope, and utilization for cotton improvement. Trends Plant Sci. 2020;25(5):488–500. [DOI] [PubMed]
- 48.Paran I, Zamir D. Quantitative traits in plants: beyond the QTL. Trends Genet. 2003;19(6):303–306. doi: 10.1016/S0168-9525(03)00117-3. [DOI] [PubMed] [Google Scholar]
- 49.Zhao B, Cao JF, Hu GJ, Chen ZW, Wang LY, Shangguan XX, Wang LJ, Mao YB, Zhang TZ, Wendel JF, et al. Core cis-element variation confers subgenome-biased expression of a transcription factor that functions in cotton fiber elongation. New Phytol. 2018;218(3):1061–75. [DOI] [PMC free article] [PubMed]
- 50.Qin G, Nguyen HM, Luu SN, Wang Y, Zhang Z. Construction of introgression lines of Oryza rufipogon and evaluation of important agronomic traits. Theor Appl Genet. 2019;132(2):543–553. doi: 10.1007/s00122-018-3241-0. [DOI] [PubMed] [Google Scholar]
- 51.Watanabe S, Shimizu T, Machita K, Tsubokura Y, Xia Z, Yamada T, Hajika M, Ishimoto M, Katayose Y, Harada K, et al. Development of a high-density linkage map and chromosome segment substitution lines for Japanese soybean cultivar Enrei. DNA Res. 2018;25(2):123–136. doi: 10.1093/dnares/dsx043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Q, Wu M, Zhang B, Shrestha P, Petrie J, Green AG, Singh SP. Genetic enhancement of palmitic acid accumulation in cotton seed oil through RNAi down-regulation of ghKAS2 encoding β-ketoacyl-ACP synthase II (KASII) Plant Biotechnol J. 2017;15(1):132–143. doi: 10.1111/pbi.12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garbowicz K, Liu Z, Alseekh S, Tieman D, Taylor M, Kuhalskaya A, Ofner I, Zamir D, Klee HJ, Fernie AR, et al. Quantitative trait loci analysis identifies a prominent gene involved in the production of fatty acid-derived flavor volatiles in tomato. Mol Plant. 2018;11(9):1147–1165. doi: 10.1016/j.molp.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y, Lin Z, Xia Q, Zhang M, Zhang X. Characteristics and analysis of simple sequence repeats in the cotton genome based on a linkage map constructed from a BC1 population between Gossypium hirsutum and G. barbadense. Genome. 2008;51(7):534–546. doi: 10.1139/G08-033. [DOI] [PubMed] [Google Scholar]
- 55.Yu Y, Yuan D, Liang S, Li X, Wang X, Lin Z, Zhang X. Genome structure of cotton revealed by a genome-wide SSR genetic map constructed from a BC1 population between Gossypium hirsutum and G. barbadense. BMC Genomics. 2011;12(1):15. doi: 10.1186/1471-2164-12-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paterson AH, Brubaker CL, Wendel JF. A rapid method for extraction of cotton (Gossypium spp. ) genomic DNA suitable for RFLP or PCR analysis. Plant Mol Biol Rep. 1993;11(2):122–127. [Google Scholar]
- 57.Han K, Jeong HJ, Yang HB, Kang SM, Kwon JK, Kim S, Choi D, Kang BC. An ultra-high-density bin map facilitates high-throughput QTL mapping of horticultural traits in pepper (Capsicum annuum) DNA Res. 2016;23(2):81–91. doi: 10.1093/dnares/dsv038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang J, Wan X, Crossa J, Crouch J, Weng J, Zhai H, Wan J. QTL mapping of grain length in rice (Oryza sativa L.) using chromosome segment substitution lines. Genet Res. 2006;88(2):93–104. doi: 10.1017/S0016672306008408. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Segments carried by CSSLs.xlsx
Additional file 2: Table S2. Summary of introgression segments.xlsx
Additional file 3: Table S3. Summary of the segments in the CSSLs.xlsx
Additional file 4: Table S4. Description of investigated traits in CSSLs.xlsx
Additional file 5: Table S5. The phenotypic data of CSSLs.xlsx
Additional file 6: Table S6. Broad-sense heritability (H2) of 14 traits in the CSSLs.xlsx
Additional file 7: Figure S1. The fuzz fiber phenotypes in the CSSLs with their parent lines. TIFF
Additional file 8: Table S7. Morphological characteristics of specific introgression lines.xlsx
Additional file 9: Table S8. Summary of the blocks in the genome.xlsx
Additional file 10: Table S9. Additive effects of the fiber length with the positive traits.xlsx
Additional file 11: Table S10. Summary of the Weight mean of fiber Additive effects on chromosome.xlsx
Additional file 12: Table S11. Yield-related and fiber quality traits of specific CSSLs.xlsx
Additional file 13: Table S12. GO enrichment analysis of genes in the candidate chromosome region.xlsx
Additional file 14: Figure S2. Transcript profiles of promising genes for root, stem, leaf, fiber and ovule between Emian22 and 3-79.
Additional file 15: Figure S3. Summary of the introgression segments of the BC4 generation with 221 individuals in 2006.
Additional file 16: Figure S4. Summary of the introgression segments of 302 individuals in 2007. TIFF
Additional file 17: Figure S5. Summary of the introgression segments of 312 individuals in 2008. TIFF
Additional file 18: Figure S6. Summary of the introgression segments of 336 individuals in 2009. TIFF
Additional file 19: Figure S7. Summary of the introgression segments of 337 individuals in 2010. TIFF
Additional file 20: Figure S8. Example of chromosome introduction fragments evaluated by SSR markers. A. Genotype calling based on the graphic of SSR markers on the PAGE. The “DD” and “RR” represent the donor and recipient parent, respectively. B. Introgression fragments evaluating based on the genotype of two near markers: “DD” represents 100% (Marker2 and Marker3); “DR” represents 50% (Marker4 and Marker5); “RR” represents 0% (Marker 5 and Marker6). TIFF
Additional file 21: Table S13. Summary of DNA sequencing data for CSSLs.xlsx
Additional file 22: Figure S9. An overview of the introgression segment identification protocol. A. Schematic diagram on identification of chromosome introduced fragment in CSSLs. First, all of the CSSLs and their parents were sequenced on an Illumina HiSeq platform to produce the genome sequence. All clean data were mapped to the G.hirsutum (TM-1) genome using BWA software and the unique mapping data were retained for further analysis. Then GATK software were applied to identify the SNPs based on the criteria:(1) the quality of SNPs should be over 100; (2) each SNP was supported by at least five reads; and (3) the adjacent SNPs should have a distance of at least 10 bp. To identify the introgression segments in the CSSLs, the SNPs between parents were selected. And a modified sliding-window approach was applied to identify the donor segments from Gb. This approach has been described very clearly by Han et al [57]. All the alleles represented by SNPs in each CSSL were filtered using SNPs from both parents. A bin map was constructed based on the genotype results of the window and consecutive bins with the same genotype were combined into same segments. B. Example of the Genotype calling based on the ratio of the SNPs in the window(>80% of SNPs had one parental genotype, the window was called as homozygous of one parent; otherwise, the window was called as heterozygous). TIFF
Additional file 23: Figure S10. Example diagram of block partition. (A) The diagram show the principle of block partition; (B) The CSSLs carried the introgression segments on the endpoint of the chromosome A01; (C) First five blocks on the chromosome A01. TIFF
Data Availability Statement
The clean raw sequencing data in this manuscript have been deposited in NCBI Sequence Read Archive under accession number PRJNA433615 and PRJNA543759.