Abstract
Background & Aims
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infects intestinal cells, and might affect the intestinal microbiota. We investigated changes in the fecal fungal microbiomes (mycobiome) of patients with SARS-CoV-2 infection during hospitalization and on recovery.
Methods
We performed deep shotgun metagenomic sequencing analysis of fecal samples from 30 patients with coronavirus disease 2019 (COVID-19) in Hong Kong, from February 5 through May 12, 2020. Fecal samples were collected 2 to 3 times per week from time of hospitalization until discharge. We compared fecal mycobiome compositions of patients with COVID-19 with those from 9 subjects with community-acquired pneumonia and 30 healthy individuals (controls). We assessed fecal mycobiome profiles throughout time of hospitalization until clearance of SARS-CoV-2 from nasopharyngeal samples.
Results
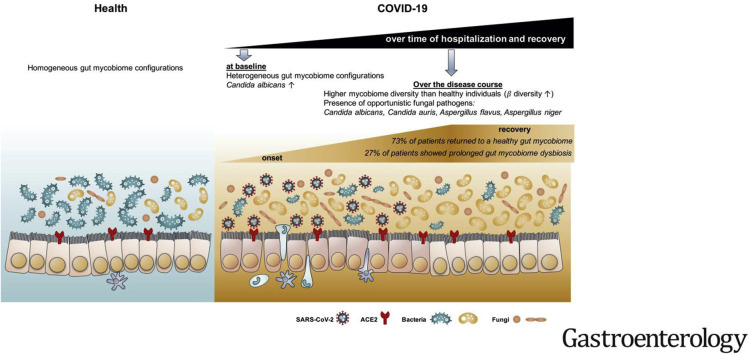
Patients with COVID-19 had significant alterations in their fecal mycobiomes compared with controls, characterized by enrichment of Candia albicans and a highly heterogeneous mycobiome configuration, at time of hospitalization. Although fecal mycobiomes of 22 patients with COVID-19 did not differ significantly from those of controls during times of hospitalization, 8 of 30 patients with COVID-19 had continued significant differences in fecal mycobiome composition, through the last sample collected. The diversity of the fecal mycobiome of the last sample collected from patients with COVID-19 was 2.5-fold higher than that of controls (P < .05). Samples collected at all timepoints from patients with COVID-19 had increased proportions of opportunistic fungal pathogens, Candida albicans, Candida auris, and Aspergillus flavus compared with controls. Two respiratory-associated fungal pathogens, A. flavus and Aspergillus niger, were detected in fecal samples from a subset of patients with COVID-19, even after clearance of SARS-CoV-2 from nasopharyngeal samples and resolution of respiratory symptoms.
Conclusions
In a pilot study, we found heterogeneous configurations of the fecal mycobiome, with enrichment of fungal pathogens from the genera Candida and Aspergillus, during hospitalization of 30 patients with COVID-19 compared with controls. Unstable gut mycobiomes and prolonged dysbiosis persisted in a subset of patients with COVID-19 up to 12 days after nasopharyngeal clearance of SARS-CoV-2. Studies are needed to determine whether alterations in intestinal fungi contribute to or result from SARS-CoV-2 infection, and the effects of these changes in disease progression.
Keywords: Coronovirus, Microbe, Yeast, Intestine
Abbreviations used in this paper: COVID-19, coronavirus disease 2019; GI, gastrointestinal; PCR, polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2
Graphical abstract
See Covering the Cover synopsis on page 1193.
Coronavirus disease 2019 (COVID-19) is caused by a novel coronavirus (severe acute respiratory syndrome coronavirus 2 [SARS-CoV-2]), which primarily affects the respiratory system. Patients with COVID-19 present with variable disease symptoms and severity, some can be severe, resulting in hospitalization, respiratory failure, or even death.1 A recent meta-analysis of 35 COVID-19 studies reported that the pooled prevalence of digestive symptoms was 15% and pooled prevalence of digestive system comorbidities was 4%.2 Patients with gastrointestinal (GI) involvement tended to have a poor disease course.2 In addition, SARS-CoV-2 virus was detected in the feces and anal swabs in patients with COVID-19 and can infect human intestinal epithelium.3, 4, 5 Altogether these data suggest that the GI tract is an important extrapulmonary site for SARS-CoV-2 infection.
Bacterial and fungal infections are common complications of viral pneumonia, which may influence disease course and clinical manifestations, especially in critically ill patients.6 However, data regarding bacterial or fungal coinfections in viral pneumonia led by coronavirus are lacking. Our recent study showed that the gut bacterial microbiome was significantly altered in patients with COVID-19 with a significant expansion of opportunistic pathogens in the gut, which was associated with disease severity and fecal SARS-CoV-2 shedding.7 Beyond bacteria, the GI tract also harbors a large number of fungi, collectively known as the mycobiome. Intestinal fungi have been shown to be causally implicated in microbiome assembly and immune development.8 Accumulating findings highlighted that the gut mycobiota can strongly influence the host immune system and this interaction is linked to bacteria activities.9 , 10 Recent observations of the direct and indirect effects of fungal microbiota on various GI diseases have stimulated further investigation of the mycobiota composition and ways of regulating its diversity.11 Although the magnitude of the number of fungal cells is smaller than that of the bacterial microbiota, their impact on health is significant, especially as a reservoir for blooms of pathogenic microbes when the host is immunocompromised and as a cofactor in driving severe infectious diseases. It is unclear if the gut mycobiome is also altered and whether fungal pathogens cobloom in COVID-19 and underlie disease course.
In this pilot study, we hypothesize that the intestinal fungal microbiome (mycobiome) is altered in SARS-CoV-2 infection and COVID-19 is associated with blooms of certain fungi in the gut. We prospectively included 30 patients with COVID-19 (Table 1 , Figure 1 , Supplementary Figure 1), admitted between February 16, 2020, and April 1, 2020, in Hong Kong, China, followed from hospital admission until discharge. We investigated intestinal fungal compositions in patients with COVID-19 as compared with healthy individuals and patients with community-acquired pneumonia, and their temporal changes over time of hospitalization, via broad-target deep shotgun metagenomics sequencing.
Table 1.
Clinical Characteristics of Study Subjects
| Variables | COVID-19 patients | Pneumonia patients | Healthy controls |
|---|---|---|---|
| Number | 30 | 9 | 30 |
| Male | 16 (53) | 5 (56) | 15 (50) |
| Age, y | 46 (29, 63) | 63 (47,69) | 34 (23, 48) |
| Comorbidities | 11 (37) | 9 (100) | 0 (0) |
| Recent exposure history | |||
| Tzravel to Wuhan City | 0 (0) | 0 (0) | |
| Travel to other cities of Hubei province | 1 (7) | 0 (0) | |
| Contact with person with COVID-19 | 9 (30) | 0 (0) | |
| Have family cluster outbreak | 4 (13) | 0 (0) | |
| Symptoms at admission | |||
| Fever | 17 (57) | 6 (67) | |
| Gastrointestinal symptoms | |||
| Diarrhea | 4 (13) | 2 (22) | |
| Respiratory symptoms | |||
| Cough | 20 (67) | 6 (67) | |
| Sputum | 9 (30) | 5 (56) | |
| Sore throat | 4 (13) | 0 (0) | |
| Rhinorrhea | 5 (17) | 1 (11) | |
| Shortness of breath | 4 (13) | 3 (33) | |
| Blood result | |||
| Lymphocytes (x 109/L, normal range 1.1–2.9) | 1 (0.8, 1.375) | 1 (0.6, 1.2) | |
| Antibiotics therapy | 16 (53) | 100 (100) | |
| 1 type of antibiotics | 6 (20) | 2 (22) | |
| 2 types of antibiotics | 7 (23) | 4 (44) | |
| 3 types of antibiotics | 3 (10) | 3 (33) | |
| Antiviral therapy | 20 (67) | 0 (0) | |
| Kaletra | 19 (63) | 0 (0) | |
| Ribavirin | 12 (40) | 0 (0) | |
| Interferon beta-1b | 3 (10) | 0 (0) | |
| Death | 0 (0) | 0 (0) |
Values are expressed in number (percentage) and median (interquartile range).
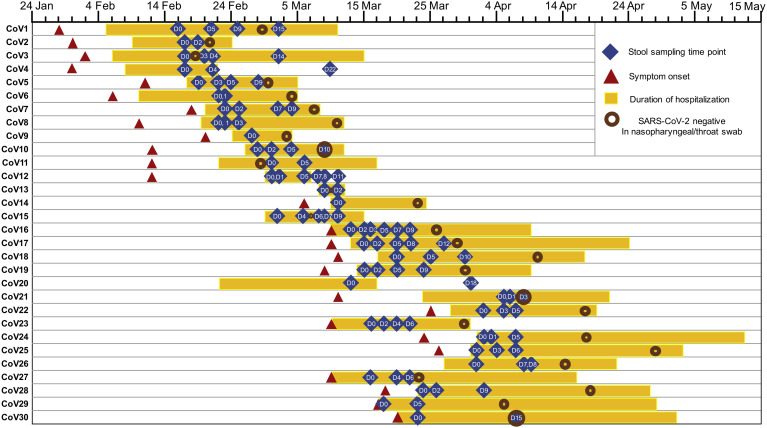
Figure 1.
Schematic diagram of stool specimen collection and duration of hospitalization in patients with COVID-19 (n = 30). “CoV” denotes patient with COVID-19. Stool specimens were serially collected for shotgun metagenomics sequencing. “D0” denotes baseline date when the first stool was collected after hospitalization; the following time points starting with “D” represent days since baseline stool collection. “-ve nasopharyngeal/throat swab”: the first negative result for SARS-CoV-2 virus in 2 consecutive negative nasopharyngeal/throat/pooled swab tests, on which patient was then discharged.
Supplementary Figure 1.
Temporal changes in the diversity of gut mycobiome (fungal) in COVID-19 patients over time of hospitalization.
Methods
Study Subject and Design
This prospective study involved 30 patients with COVID-19 hospitalized with laboratory-confirmed SARS-CoV-2 infection, 9 patients hospitalized with community-acquired pneumonia (pneumonia controls), and 30 healthy individuals (controls) (Table 1). Thirty hospitalized patients with COVID-19 were admitted between February 16, 2020, and April 1, 2020, in Hong Kong, China, and were followed from hospital admission until discharge (Figure 1). SARS-CoV-2 infection was confirmed by 2 consecutive reverse-transcriptase polymerase chain reaction (PCR) tests targeting different regions of the RdRp gene performed by the local hospital and Public Health Laboratory Service. Pneumonia controls were patients admitted with community-acquired pneumonia tested negative for SARS-CoV-2 PCR on 2 respiratory samples. Patients with COVID-19 and pneumonia controls were admitted to the Prince of Wales Hospital or the United Christian Hospital, Hong Kong. Controls were healthy individuals with no past medical history or history of antibiotic intake in the past 3 months recruited via advertisement from the general population and tested negative for SARS-CoV-2. Severity of COVID-19 infection was categorized as (1) mild, if there was no radiographic evidence of pneumonia; (2) moderate, if pneumonia was present along with fever and respiratory tract symptoms; (3) severe, if respiratory rate ≥30/min, oxygen saturation ≤93% when breathing ambient air, or PaO2 / FiO2 ≤300 mm Hg (1 mm Hg = 0.133 kPa); or (4) critical, if there was respiratory failure requiring mechanical ventilation, shock, or organ failure requiring intensive care.12 This study was approved by the Joint Chinese University of Hong Kong–New Territories East Cluster Clinical Research Ethics Committees (2020.076). All patients provided informed consent to participate in this study. Data including demographics, laboratory results, imaging results and drug therapy were obtained from the electronic medical records in the Hong Kong Hospital Authority clinical management system. Fecal samples from patients with COVID-19 were collected serially 2 to 3 times per week until discharge. This study was conducted in accordance with the Declaration of Helsinki.
Fecal DNA Extraction
Approximately 0.1-g fecal sample was prewashed with 1 mL double-distilled H2O and pelleted by centrifugation at 13,000g for 1 minute. The fecal DNA was subsequently extracted from the pellet using Maxwell RSC PureFood GMO and Authentication Kit (Promega, Madison, WI) following the manufacturer’s instructions. Briefly, the fecal pellet was added to 1 mL of CTAB buffer and vortexed for 30 seconds, then the sample was heated at 95°C for 5 minutes. After that, the samples were vortexed thoroughly with beads at maximum speed for 15 minutes. Then 40 μL of proteinase K and 20 μL of RNase A was added into sample and the mixture was incubated at 70°C for 10 minutes. The supernatant was then obtained by centrifuging at 13,000g for 5 minutes and was added to the Maxwell RSC machine for DNA extraction.
Shotgun Metagenomics Sequencing and Fungal Profiling
Extracted DNA was subject DNA libraries construction, completed through the processes of end repairing, adding A to tails, purification and PCR amplification, using Nextera DNA Flex Library Preparation kit (Illumina, San Diego, CA). Libraries were subsequently sequenced on our in-house sequencer Illumina Novaseq 6000 (150 base pairs paired-end) at the Center for Microbiota Research, The Chinese University of Hong Kong.
Raw sequence reads were filtered and quality-trimmed using Trimmomatic v0.3613 as follows: (1) trimming low-quality base (quality score <20); (2) removing reads shorter than 50 base pairs; (3) removing sequencing adapters. Contaminating human reads were filtering using Kneaddata (Reference database: GRCh38 p12) with default parameters. Profiling of the fungal community was performed using MiCoP. The fungal abundance profile of each sample was expressed in relative abundance (%) among the fecal fungal community.14
Statistical Analysis
Data on the relative abundance of fecal fungi was imported into R v3.5.1. Nonmetric multidimensional scaling analyses were performed on all baseline fecal mycobiomes between groups, and serial fecal mycobiomes in each COVID-19 case during the disease course, based on Bray-Curtis dissimilarities using vegan package (v2.5–3). Differential fungal taxa between patients with COVID-19 (at baseline or across all time points during hospitalization) and healthy controls were identified using LefSE.15
Results
Fecal Mycobiome Alterations in COVID-19
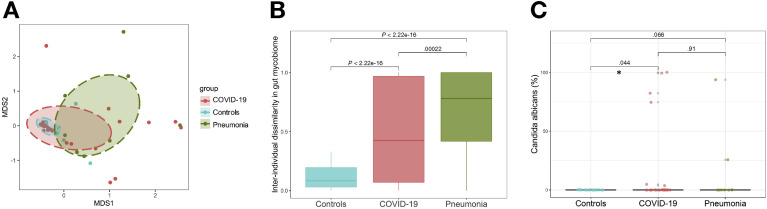
A total of 30 patients hospitalized with COVID-19, 30 healthy controls, and 9 patients hospitalized with community-acquired pneumonia were included ( Table 1; Supplementary Table 1). All patients with COVID-19 presented with respiratory symptoms and 4 also had diarrhea at presentation. To understand alterations of the gut mycobiome that underlies SARS-CoV-2 infection, we compared baseline fecal mycobiome composition (first time point of stool sampling after hospitalization) between patients with COVID-19, healthy controls, and pneumonia controls. At the whole fungal community level, fecal mycobiome of healthy controls densely clustered together, whereas that of patients with COVID-19 formed a substantially broader cluster significantly different from that of controls (PERMANOVA test, P = .001, Figure 2 A). Fecal mycobiome was more heterogeneous across patients with COVID-19 than controls, as demonstrated by an approximately 6-fold increase in interindividual mycobiome dissimilarity in patients with COVID-19 relative to controls (P < 2.22e-16, Figure 2 B). Patients with community-acquired pneumonia also showed heterogeneous fecal mycobiome configurations (Figure 2 B), indicating that both community-acquired pneumonia and COVID-19–associated pneumonia were associated with fungal dysbiosis in the gut.
Figure 2.
Gut mycobiome (fungal community) alterations in patients with COVID-19. (A) Fecal mycobiome alterations in COVID-19, viewed by NMDS (nonmetric multidimensional scaling) plot based on Bray-Curtis dissimilarities. The fecal mycobiome was compared among healthy controls (n = 30), COVID-19 (n = 30), and pneumonia patient controls (n = 9). (B) Interindividual dissimilarities between fecal mycobiomes within each group. The mycobiome dissimilarity was calculated as Bray-Curtis dissimilarity. Between-group comparison was conducted by t test. (C) The relative abundance of Candida albicans in the fecal mycobiome. Between-group comparison was conducted by Wilcoxon rank sum test.
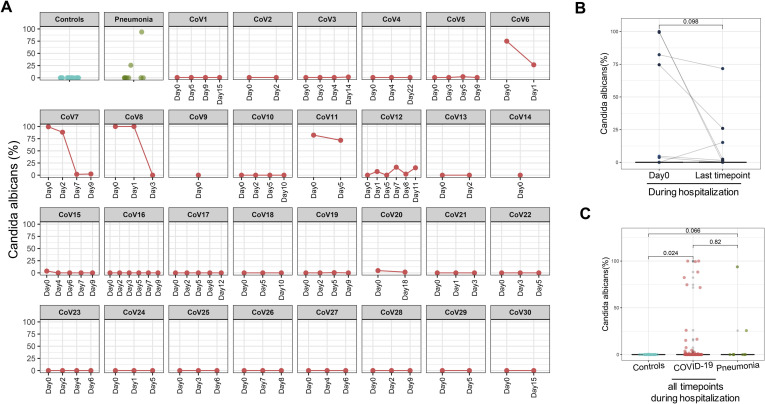
We next identified differential fungal species in fecal samples between patients with COVID-19 and healthy controls adjusting for antibiotics use and comorbidities. We found that Candida albicans was significantly enriched in 6 (20%) of 30 patients with COVID-19 but was absent in healthy controls (false discovery rate P = .04, Figure 2 C). These data suggest that gut mycobiome was significantly altered in patients with COVID-19 with an increase in Candida species.
Temporal Changes in Gut Mycobiome Over Time of Hospitalization
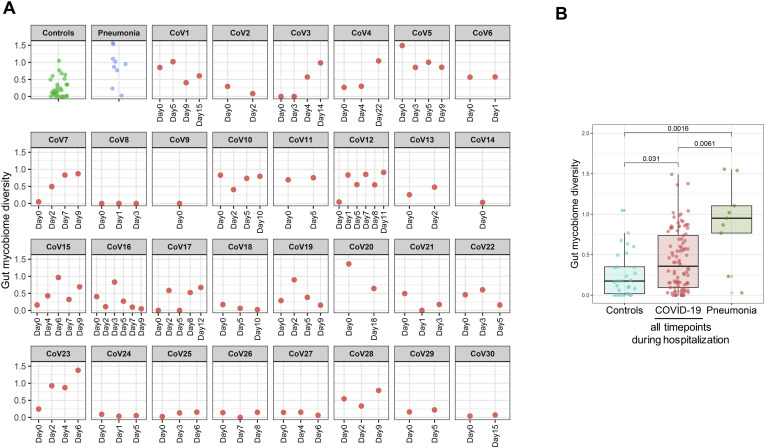
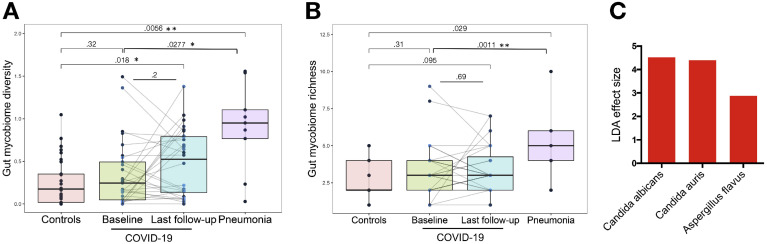
We investigated longitudinal dynamics of the gut mycobiome in COVID-19 over time of hospitalization and explored whether recovery from SAR-CoV-2 infection was associated with restoration of gut mycobiome to a community similar to that of healthy individuals. The gut mycobiome in most (22 of 30) patients with COVID-19 was stable over the course of hospitalization and remained similar to that of healthy controls at the latest follow-up (Figure 3 ). However, fecal mycobiome of 8 of 30 patients (patients CoV3, 7, 8, 10, 11, 12, 17, and 23) showed drastic changes over the course of hospitalization and had a different fecal mycobiome composition compared with that of healthy controls at the last follow-up (Figure 3). The diversity of the fecal mycobiome did not differ between healthy controls and patients with COVID-19 at baseline (Figure 4 A). In contrast, the diversity and richness of the gut mycobiome were both significantly higher in patients with community-acquired pneumonia than patients with COVID-19 at baseline (Figure 4 A and B, P < .05 and < .01, respectively). At the last follow-up after hospitalization, patients with COVID-19 showed a 2.5-fold higher diversity of the fecal mycobiome compared with healthy controls (P < .05, Figure 4 A).
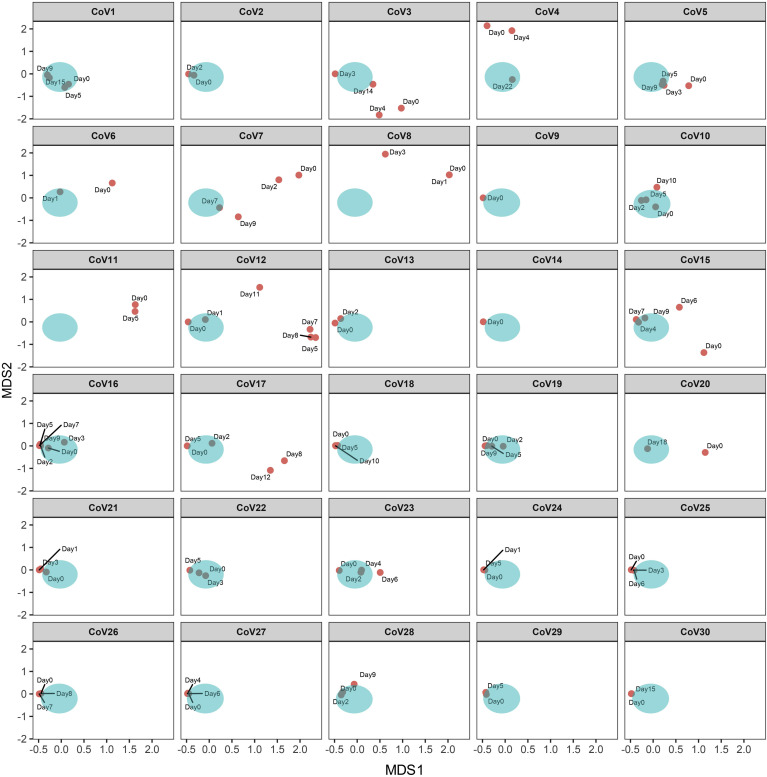
Figure 3.
Gut mycobiome (fungal community) alterations in patients with COVID-19 and longitudinal changes during time of hospitalization. Temporal compositional changes in fecal mycobiome with respect to each COVID-19 case were viewed by NMDS (nonmetric multidimensional scaling) plot based upon Bray-Curtis dissimilarities. The aqua cluster denotes the fecal mycobiome cluster of healthy controls. “CoV” denotes patient with COVID-19. “Day0” denotes baseline date when the first stool was collected after hospitalization; the following time points starting with “Day” represents days since baseline stool collection.
Figure 4.
Bloom of gut fungi in patients with COVID-19 during time of hospitalization. (A) The diversity of fecal mycobiome (fungi) in patients with COVID-19 over time of hospitalization (plotted as the baseline time point and the last follow-up time point after hospitalization), compared with healthy controls (n = 30) and pneumonia patient controls (n = 9). Between-group comparison was conducted by t test, last follow-up versus baseline comparison for the hospitalized patients with COVID-19 were conducted by paired t test. (B) The richness of fecal mycobiome (fungi) in patients with COVID-19 over time of hospitalization (plotted as the baseline time point and the last follow-up time point after hospitalization), compared with healthy controls (n = 30) and pneumonia patient controls (n = 9). Between-group comparison was conducted by t test, last follow-up versus baseline comparison for the hospitalized patients with COVID-19 were conducted by paired t test. (C) Overrepresented fungal species in feces in patients with COVID-19 during time of hospitalization, compared with healthy controls. LefSE analysis was conducted to identify differential species, only species with LDA effect size > 2 and false discovery rate P < .1 were plotted.
Overall, during disease course, the diversity of the fecal mycobiome showed constant changes in 53% (16 of 30) of patients with COVID-19 (Supplementary Figure 1). Altogether these data suggest that the gut mycobiome was unstable in a subset of patients with COVID-19 during the time of hospitalization. Patients CoV3, CoV10, and CoV11 who cleared SARS-CoV-2 virus from nasopharyngeal swab and had resolution of respiratory symptoms continued to display a different gut mycobiome composition with that of healthy controls in later time points up to the last follow-up (12, 0, and 5 days after nasopharyngeal clearance of SARS-CoV-2 respectively for patients CoV3, CoV10, and Cov11, Figure 3). These data indicate persistent gut fungal dysbiosis despite disease resolution in a subset of patients with COVID-19.
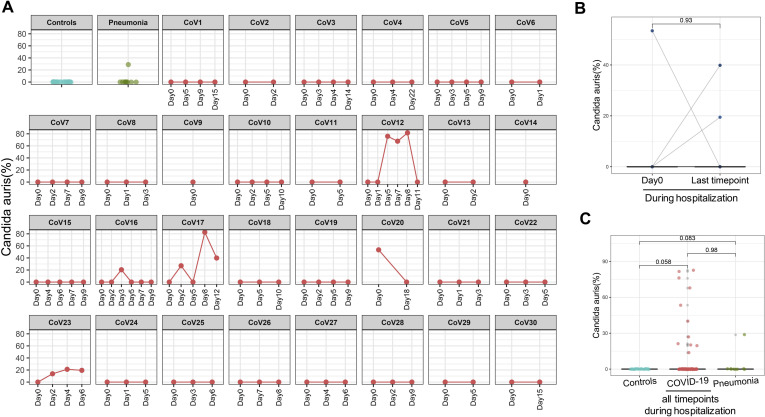
Across all time points during hospitalization, C. albicans, Candida auris and Aspergillus flavus were overrepresented in fecal samples of patients with COVID-19; these species however were absent in healthy controls (Figure 4 C, Supplementary Figures 2 and 3, and Figure 5 A). Patients (CoV6, 7, 8 and 11) who showed high fecal abundance of C. albicans (>50% in relative abundance) had a substantial decrease of C. albicans over time of hospitalization (Supplementary Figure 2 A and B). Blooms of C. albicans, C. auris, and A. flavus, were also seen in patients with community-acquired pneumonia (Figures 2 C and 5, and Supplementary Figures 2 C and 3 C).
Supplementary Figure 2.
Temporal changes in the relative abundance of Candida albicans in the gut mycobiome of COVID-19 patients over time of hospitalization. (A) Longitudinal changes of C. albicans in each COVID-19 case. (B) Alteration in the relative abundance of C. albicans compared between baseline and the last follow-up; statistical comparison was conducted by paired Wilcoxon rank sum test. (C) The relative abundance of C. albicans across all time points in all patients with COVID-19 during hospitalization, as compared with healthy controls. Statistical comparison was conducted by paired Wilcoxon rank sum test.
Supplementary Figure 3.
Temporal changes in the relative abundance of Candida auris in the gut mycobiome of COVID-19 patients over time of hospitalization. (A) Longitudinal changes of C. auris in each COVID-19 case. (B) Alteration in the relative abundance of C. auris compared between baseline and the last follow-up; statistical comparison was conducted by paired Wilcoxon rank sum test. (C) The relative abundance of C. auris across all time points in all patients with COVID-19 during hospitalization, as compared with healthy controls. Statistical comparison was conducted by paired Wilcoxon rank sum test.
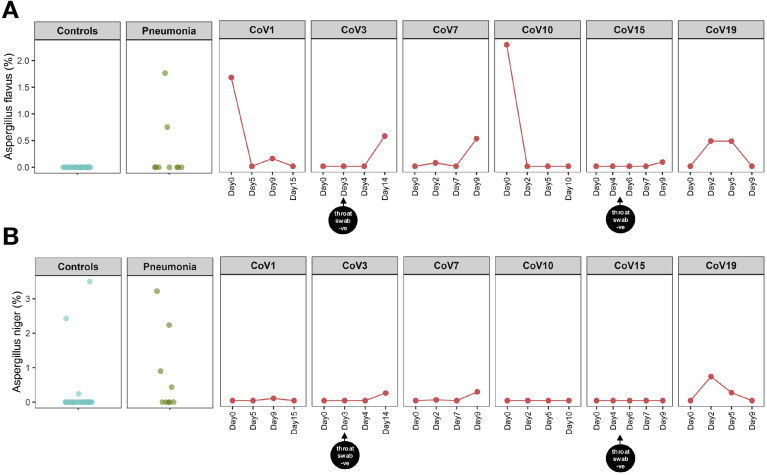
Figure 5.
The presence of Aspergillus flavus (A) and Aspergillus niger (B) in patients with COVID-19 over time of hospitalization. “CoV” denotes patient with COVID-19. “Controls” denotes healthy controls (n = 30). “Pneumonia” denotes patient controls with pneumonia (n = 9). “Day0” denotes baseline date when the first stool was collected after hospitalization; the following time points starting with “Day” represent days since baseline stool collection. “throat swab -ve” indicates the first negative result for SARS-CoV-2 virus in 2 consecutive negative nasopharyngeal/throat/pooled swab tests, on which patient was then discharged.
Two fungal species from the genus Aspergillus, A. flavus and Aspergillus niger, known to cause pulmonary aspergillosis and respiratory illnesses (particularly cough),16 , 17 were detected in serial fecal samples of 6 and 4 patients with COVID-19, respectively, during hospitalization (Figure 5). All 6 patients presented with cough and varying degree of COVID-19 severity. Among the detected Aspergillus species, A. flavus was the most abundant and prevalent member (in 6/30 patients; relative abundance up to 2.28%, Figure 5 A). A. niger was absent at baseline but showed a low relative abundance of <1% in 4 of 30 patients with COVID-19 over time of hospitalization (Figure 5 B). Importantly, patients CoV15 and CoV19 who presented with a mild disease course showed presence of A. flavus and A. niger in fecal samples over time. A. flavus and A. niger were also detected at various time points after nasopharyngeal swab turned negative for SARS-CoV-2 in patients CoV3 and CoV15 (after 11 days and 4 days, respectively, Figure 5). These data indicate cobloom of opportunistic fungal pathogens, Candida species and Aspergillus species, in the gut of patients with COVID-19 over the disease course. Their presence after nasopharyngeal clearance of SARS-CoV-2 may pose a long-term threat to human health beyond SARS-CoV-2 infection.
Discussion
We showed for the first time that the gut mycobiome was disturbed in patients with COVID-19. Patients hospitalized with SARS-CoV-2 infection showed more heterogeneous gut mycobiome configurations (higher interindividual mycobiome dissimilarities) compared with healthy individuals. These data suggest that gut microbiota changes induced by SARS-CoV-2, might at least in part, be stochastic, therefore leading to transition from a stable to unstable microbial community state in COVID-19.18 In line with our findings in patients with COVID-19, a highly heterogeneous assembly of microbial community in response to disease stressors have been reported in the gut of patients with inflammatory bowel disease19 and in the lungs of patients with human immunodeficiency virus/AIDs.18 , 20 , 21 Human immunodeficiency virus/AIDs is associated with decreased CD4+ T-cell counts, which increase an individual’s susceptibility to a wide range of bacterial, viral, and fungal opportunistic pathogens.20 Similarly, SARS-CoV-2 infection was associated with a significant reduction in the number of T cells,22 , 23 which could compromise host immune homeostasis and stability of the microbial communities residing in the human gut. In favor of this hypothesis, the overall gut mycobiome was unstable in a subset of patients with COVID-19 over time of hospitalization, and these individuals ended up with a different gut mycobiome composition compared with that of healthy individuals. In addition, gut mycobiome diversity of patients with COVID-19 at the last follow-up during hospitalization was significantly higher than that of healthy individuals. These data indicate expansion of fungi in the gut of patients with COVID-19 and SARS-CoV-2 infection may be associated with a persistent gut mycobiome dysbiosis in some patients.
Secondary fungal infection or coinfection in patients with COVID-19 during the pandemic is garnering increased attention.24 Specific enrichments of opportunistic fungal pathogens, Candida and Aspergillus lineages, were observed in patients with COVID-19 during the disease course. Among them, C. albicans was overrepresented in COVID-19. C. albicans has been shown to impair holistic gut microbiome assembly in humans and mice.25 , 26 Moreover, gut colonization by C. albicans aggravated inflammation in the gut and non-gut tissues.27 , 28 Aspergillus infections were recently reported in respiratory tract secretions and tracheal aspirates in patients with COVID-19 from 2 studies (detection rate of 10% and 20%, respectively, in cohorts from China and France).29 , 30 Population-based studies have reported 19% to 33% incidence of pulmonary aspergillosis in patients with COVID-19.31, 32, 33 Aspergillus is a genus of ubiquitous fungi that cause a variety of pulmonary and respiratory symptoms.17 Aspergillus may captivate on the immune-compromised host and affect the clinical features, disease course, and prognosis.17 To date, there are a lack of data on whether fungal pathogens exist in the gut of patients with COVID-19. In this case series, we provide the first evidence demonstrating the presence of opportunistic Aspergillus pathogens in the feces of patients with COVID-19; A. flavus were detected in the feces of 20% of patients with COVID-19 via metagenomics sequencing. Cough was previously reported to be more frequent in subjects infected by Aspergillus species than those who were not infected.16 All 6 patients with COVID-19 who had fecal A. flavus presented with cough during hospitalization, suggestive of a systemic effect of Aspergillus infection and the intricate link between the GI and respiratory systems. Two of the 3 patients (CoV1 and 3) who had both fungal species A. flavus and A. niger in their feces presented with critical COVID-19 and were admitted to intensive care unit, and the third case (CoV7) had high fever and moderate COVID-19 severity. Patient CoV19 who had mild COVID-19 showed clearance of A. flavus and A. niger over time of hospitalization. Although the overall gut mycobiome structure in patients with COVID-19 gradually resembled that of healthy individuals over time of hospitalization (Figure 3), Aspergillus species continued to be present in a subset of patients with COVID-19 (CoV3 and CoV15) even after nasopharyngeal clearance of SARS-CoV-2 (Figure 5). The post-recovery existence of Aspergillus species underscores a potential prolonged detrimental effect of secondary fungal infection on host health after SARS-CoV-2 infection, cautioning on long-term monitoring of patients with COVID-19 after recovery.
Pneumonia is one of the leading infectious causes of death.34 Fungal coinfections in the respiratory tract and alterations in the lung mycobiome have been reported in pneumonia and pulmonary diseases.17 , 35 , 36 However, to our knowledge, changes in the gut mycobiome were not reported in patients with pneumonia to date. In the present study, we observed that patients with community-acquired pneumonia also showed a significant alteration in the gut mycobiome compared with healthy controls. Similar to patients with COVID-19, blooms of opportunistic fungal pathogens, Candida species and Aspergillus species, were also seen in patients with community-acquired pneumonia. In contrast, patients with community-acquired pneumonia showed more heterogeneous gut mycobiome configurations than patients with COVID-19, indicating that the gut mycobiome in community-acquired pneumonia may be more “dysbiotic” than that in COVID-19. This could in part relate to the use of antibiotics in these individuals. In addition, the diversity and richness of the gut mycobiome were both significantly lower in patients with COVID-19 than patients with community-acquired pneumonia. These data collectively suggest that patients with COVID-19 have a similar but less severe dysbiosis of gut mycobiome compared with patients with community-acquired pneumonia.
One limitation of this exploratory study is the modest sample size. Although assigning a causative relationship between COVID-19 and gut fungal dysbiosis requires larger validation studies, this pilot study presents the first to examine the influence of SARS-CoV-2 infection on gut mycobiome composition and dynamics. Stool collected after hospitalization for mycobiome analysis does not represent the bona fide baseline mycobiome at COVID-19 onset, nor the baseline mycobiome before disease onset. In addition, it is unclear whether blooms of gut fungi are a result of SARS-CoV-2 infection or of coinfection during hospitalization. The contribution of gut fungi in the pathogenesis and disease progression of COVID-19 is also unknown. Future studies should prospectively include asymptomatic SARS-CoV-2–infected subjects without hospitalization. In addition, patients with symptomatic SARS-CoV-2 infection should be followed from disease onset until after recovery to thoroughly delineate the role of gut fungi during SARS-CoV-2 infection and the effects of these changes in disease progression and long-term health.
In conclusion, our study provides evidence of numerous blooms of fungal species in the gut of patients with COVID-19. These data highlight an important role of gut mycobiome in COVID-19. The effects of gut fungi in disease pathogenesis and long-term health after recovery from SARS-CoV-2 infection warrant further investigation.
Acknowledgments
We thank all health care workers working in isolation wards of Prince of Wales Hospital, Hong Kong, China. We thank Apple C.M. Yeung, Wendy C.S. Ho, Miu L. Chin, Rity Wong, and Vickie Li for their technical contribution in this study. We thank Whitney Tang for her assistance with the graphical abstract.
CRediT Authorship Contributions
Tao Zuo, PhD (Conceptualization: Lead; Formal analysis: Equal; Investigation: Lead; Methodology: Equal; Writing – original draft: Lead). Hui Zhan, PhD (Data curation: Lead; Formal analysis: Equal; Methodology: Lead; Writing – review & editing: Equal). Fen Zhang, PhD (Data curation: Supporting; Formal analysis: Supporting). Qin Liu, PhD (Data curation: Supporting; Formal analysis: Supporting). Eugene Y.K. Tso, MD (Resources: Lead). Grace Lui, Doctor (Resources: Equal). Nan Chen, Master (Methodology: Equal; Validation: Equal). Amy Li, NA (Data curation: Lead; Investigation: Equal). Wenqi Lu, NA (Methodology: Equal). Francis K.L. Chan, MD (Investigation: Supporting; Project administration: Lead). Paul K.S. Chan, PhD (Resources: Supporting). Siew C Ng, PhD (Conceptualization: Supporting; Funding acquisition: Lead; Writing – review & editing: Lead).
Footnotes
Conflict of interest The authors disclose no conflicts.
Funding Hui Hoy & Chow Sin Lan Charity Fund Limited, Pine and Crane Company Limited, and Mr. Hui Ming. Food and Health Bureau, Health and Medical Research Fund from Food and Health Bureau, Hong Kong.
Author names in bold designate shared co-first authorship.
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at https://doi.org/10.1053/j.gastro.2020.06.048.
Supplementary Material
Supplementary Table 1.
Clinical Characteristics of Patients With COVID-19
| Case | Sex | Age | Comorbidities | Recent exposure history | Symptoms at admission |
Admitted to ICU | Medication |
Blood routine lymphocytesa | Chest radiograph findings | COVID-19 severity | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fever and respiratory | GI | Antibiotics | Antiviral | |||||||||
| CoV1 | F | 65 | Hypothyroidism, hypertension, Chronic hepatitis B carrier | No | Fever, cough, sputum | Nil | Yes | Nil | Kaletra, ribavirin | 1.0 | Bilateral LZ haziness | Critical |
| CoV2 | F | 55 | None | Contact with person with COVID-19 | Fever, runny nose | Nil | No | Nil | Kaletra | 1.2 | Bilateral LZ haziness | Moderate |
| CoV3 | M | 42 | None | Travel to Hubei province | Fever, cough | Nil | Yes | Daptomycin meropenem | Nil | 0.6 | Worsening Right LZ haziness, Right LL collapse re-opened | Critical |
| CoV4 | M | 70 | Hyperlipidemia, duodenal ulcer | No | Sputum, shortness of breath | Nil | No | Augmentin, doxycycline | Kaletra | 0.6 | Bilateral lung haziness | Severe |
| CoV5 | M | 58 | None | No | Fever, cough | Diarrhea | No | Ceftriaxone, augmentin, doxycycline | Kaletra, ribavirin | 0.9 | Slight Right LZ haziness | Moderate |
| CoV6 | M | 71 | None | No | Fever, cough, shortness of breath | Nil | No | Nil | Kaletra | 1.0 | Bilateral lung infiltration | Severe |
| CoV7 | M | 48 | Diabetes, hypertension, hyperlipidemia | No | Fever, cough | Nil | No | Augmentin | Kaletra, ribavirin | 1.3 | Left LZ haziness | Moderate |
| CoV8 | F | 38 | None | No | Fever, cough, sputum, runny nose | Nil | No | Ceftriaxone, doxycycline | Kaletra | 0.7 | Bilateral LZ infiltrates | Moderate |
| CoV9 | M | 33 | None | Contact with person with COVID-19 | Fever, cough | Nil | No | Doxycycline | Kaletra, ribavirin | 0.7 | Bilateral LZ haziness | Mild |
| CoV10 | F | 70 | Obesity, hypertension | No | Cough | Nil | No | Ceftriaxone, piperacillin+ tazobactam | Kaletra, ribavirin | 0.8 | Bilateral LZ haziness | Moderate |
| CoV11 | M | 62 | Diabetes, hyperlipidemia, left subclavian artery occlusion | No | Fever, cough, sputum, shortness of breath | Nil | No | Sulperazon, Ceftriaxone Disodium, Doxycycline hyclate | Oseltamivir, Kaletra | 0.9 | Bilateral lung infiltrates | Severe |
| CoV12 | F | 71 | Hypertension, renal impairment, hyperlipidemia | Contact with person with COVID-19 | Cough | Nil | No | Ceftriaxone, azithromycin | Kaletra, ribavirin | 2.2 | Bilateral LZ haziness | Moderate |
| CoV13 | F | 47 | None | Contact with person with COVID-19 | Nil | Nil | No | Nil | Nil | 1.9 | No definite consolidation | Moderate |
| CoV14 | F | 22 | None | Contact with person with COVID-19 | Fever, runny nose | Nil | No | Nil | Nil | 1.8 | Nil | Moderate |
| CoV15 | F | 46 | None | Contact with person with COVID-19 | Cough, shortness of breath | Nil | No | Augmentin | Kaletra, ribavirin, interferon beta-1b | 1.0 | Clear | Mild |
| CoV16 | M | 23 | None | Travel to Egypt | Fever | Nil | No | Nil | Kaletra | 1.4 | Clear | Mild |
| CoV17 | M | 67 | Hypertension | Travel to UK | Fever, cough with white sputum | Diarrhea | No | Augmentin, Azithromycin | Remdesivir, Kaletra, Ribavirin | 1.6 | Right LL infiltrate | Severe |
| CoV18 | F | 34 | None | Travel to UK and Spain | Cough, yellowish sputum and whitish sputum, runny nose, sore throat | Nil | No | Nil | Ribavirin, Kaletra | 1.7 | Left LZ haziness | Mild |
| CoV19 | M | 59 | Ankylosing spondylitis, Bilateral OA hip | Same building with person with COVID-19 | Cough, sore throat, hoarseness, whitish sputum | Nil | No | Augmentin | Ribavirin, Kaletra, Remdesivir | 0.7 | Clear | Mild |
| CoV20 | F | 38 | None | No | Fever, cough with whitish sputum, runny nose | Nil | No | Ceftriaxone Disodium, Doxycycline hyclate | Kaletra, Oseltamivir | 0.7 | Bilateral LZ infiltrates | Moderate |
| CoV21 | F | 18 | None | Travel to USA | Dry cough, loss of olfactory sensation | Nausea | No | Nil | Nil | 1.1 | Bilateral LZ haziness | Mild |
| CoV22 | F | 38 | None | No | Sore throat, cough, fever | Nil | No | Nil | Remdesivir | 1.1 | Bilateral lung infiltrates | Severe |
| CoV23 | F | 63 | None | Contact with person with COVID-19 | Fever, dry cough | Nil | No | Ceftriaxone Disodium | Kaletra, Ribavirin, Interferon Beta-1B | 0.9 | Bilateral LZ haziness | Mild |
| CoV24 | M | 15 | None | Travel to UK | Sore throat, cough with yellowish sputum | Nil | No | Nil | Nil | 1.0 | Clear | Asymptomatic |
| CoV25 | M | 28 | None | Travel to South Africa, Russia, and transit in UK | Nil | Nil | No | Nil | Nil | 1.4 | Clear | Mild |
| CoV26 | M | 66 | Hypertension, hepatoma | Travel to Sydney, contact with Person with COVID-19 | throat discomfort | Diarrhea | No | Piperacillin+ Tazobactam injection | Nil | 0.8 | Mild left LZ infiltrates | Moderate |
| CoV27 | M | 63 | Cervical radiculopathy, bilateral varicose vein with bilateral Trendelenburg operation | Travel to Egypt | Fever | Nil | No | Nil | Interferon Beta-1B, Ribavirin, Kaletra | 1.3 | Clear | Mild |
| CoV28 | F | 16 | None | Travel to UK | Nil | Abdominal pain, nausea, diarrhea | No | Nil | Nil | 1.5 | No pneumonic changes | Mild |
| CoV29 | M | 27 | None | Travel to UK | Fever | Nil | No | Augmentin | Nil | 0.7 | No consolidation | Mild |
| CoV30 | M | 19 | Eczema | Travel to France and Austria, contacted with person with COVID-19 | dry cough, nasal congestion | Nil | No | Nil | Nil | 1.2 | Clear | Mild |
COVID-19, coronavirus disease 2019; GI, gastrointestinal; ICU, intensive care unit; LZ, Lower Zone; LL, Lower Lobe; MZ, Middle Zone
Value in x 109/L, normal range 1.1–2.9.
References
- 1.Onder G., Rezza G., Brusaferro S. Case-fatality rate and characteristics of patients dying in relation to COVID-19 in Italy [printed online ahead of print March 23, 2020]. JAMA. https://doi.org/10.1001/jama.2020.4683 [DOI] [PubMed]
- 2.Mao R., Qiu Y., He J.-S. Manifestations and prognosis of gastrointestinal and liver involvement in patients with COVID-19: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol. 2020;5:667–678. doi: 10.1016/S2468-1253(20)30126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wölfel R., Corman V.M., Guggemos W. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 4.Xiao F., Tang M., Zheng X. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833.e3. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou J., Li C., Liu X. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat Med. 2020;26:1077–1083. doi: 10.1038/s41591-020-0912-6. [DOI] [PubMed] [Google Scholar]
- 6.Zhou P., Liu Z., Chen Y. Bacterial and fungal infections in COVID-19 patients: a matter of concern [published online ahead ofprint April 22, 2020]. Infect Control Hosp Epidemiol. https://doi.org/10.1017/ice.2020.156 [DOI] [PMC free article] [PubMed]
- 7.Zuo T., Zhang F., Lui G.C.Y. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization [published online ahead of print May 19, 2020]. Gastroenterology. https://doi.org/10.1053/j.gastro.2020.05.048 [DOI] [PMC free article] [PubMed]
- 8.van Tilburg Bernardes E., Pettersen V.K., Gutierrez M.W. Intestinal fungi are causally implicated in microbiome assembly and immune development in mice. Nat Commun. 2020;11:2577. doi: 10.1038/s41467-020-16431-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang I., Pletcher S.D., Goldberg A.N. Fungal microbiota in chronic airway inflammatory disease and emerging relationships with the host immune response. Front Microbiol. 2017;8:2477. doi: 10.3389/fmicb.2017.02477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan D., Coughlin L.A., Neubauer M.M. Activation of HIF-1α and LL-37 by commensal bacteria inhibits Candida albicans colonization. Nat Med. 2015;21:808. doi: 10.1038/nm.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richard M.L., Sokol H. The gut mycobiota: insights into analysis, environmental interactions and role in gastrointestinal diseases. Nat Rev Gastroenterol Hepatol. 2019;16:331–345. doi: 10.1038/s41575-019-0121-2. [DOI] [PubMed] [Google Scholar]
- 12.Wu J., Liu J., Zhao X. Clinical characteristics of imported cases of COVID-19 in Jiangsu province: a multicenter descriptive study. Clin Infect Dis. 2020;71:706–712. doi: 10.1093/cid/ciaa199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LaPierre N., Mangul S., Alser M. MiCoP: microbial community profiling method for detecting viral and fungal organisms in metagenomic samples. BMC Genomics. 2019;20:423. doi: 10.1186/s12864-019-5699-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Segata N., Izard J., Waldron L. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baba R, Takaoka H, Kamo T, et al. Clinical interpretations and therapeutic significance of isolating aspergillus species from respiratory specimens. A58. clinical studies in fungal infections: American Thoracic Society, 2020:A2117–A2117.
- 17.Kosmidis C., Denning D.W. The clinical spectrum of pulmonary aspergillosis. Thorax. 2015;70:270–277. doi: 10.1136/thoraxjnl-2014-206291. [DOI] [PubMed] [Google Scholar]
- 18.Zaneveld J.R., McMinds R., Thurber R.V. Stress and stability: applying the Anna Karenina principle to animal microbiomes. Nat Microbiol. 2017;2:1–8. doi: 10.1038/nmicrobiol.2017.121. [DOI] [PubMed] [Google Scholar]
- 19.Zuo T., Lu X.-J., Zhang Y. Gut mucosal virome alterations in ulcerative colitis. Gut. 2019;68:1169–1179. doi: 10.1136/gutjnl-2018-318131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masur H., Brooks J.T., Benson C.A. Prevention and treatment of opportunistic infections in HIV-infected adults and adolescents: Updated Guidelines from the Centers for Disease Control and Prevention, National Institutes of Health, and HIV Medicine Association of the Infectious Diseases Society of America. Clin Infect Dis. 2014;58:1308–1311. doi: 10.1093/cid/ciu094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams B., Landay A., Presti R.M. Microbiome alterations in HIV infection: a review. Cellular microbiology. 2016;18:645–651. doi: 10.1111/cmi.12588. [DOI] [PubMed] [Google Scholar]
- 22.Qin C., Zhou L., Hu Z. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin Infect Dis. 2020;71:762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diao B., Wang C., Tan Y. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox M.J., Loman N., Bogaert D. Co-infections: potentially lethal and unexplored in COVID-19. Lancet Microbe. 2020;1:e11. doi: 10.1016/S2666-5247(20)30009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuo T., Wong S.H., Cheung C.P. Gut fungal dysbiosis correlates with reduced efficacy of fecal microbiota transplantation in Clostridium difficile infection. Nat Commun. 2018;9:3663. doi: 10.1038/s41467-018-06103-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Downward J.R.E., Falkowski N.R., Mason K.L. Modulation of post-antibiotic bacterial community reassembly and host response by Candida albicans. Sci Rep. 2013;3:2191. doi: 10.1038/srep02191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sonoyama K., Miki A., Sugita R. Gut colonization by Candida albicans aggravates inflammation in the gut and extra-gut tissues in mice. Med Mycol. 2011;49:237–247. doi: 10.3109/13693786.2010.511284. [DOI] [PubMed] [Google Scholar]
- 28.Kumamoto C.A. Inflammation and gastrointestinal Candida colonization. Curr Opin Microbiol. 2011;14:386–391. doi: 10.1016/j.mib.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang X., Yu Y., Xu J., Liu H. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lescure F.-X., Bouadma L., Nguyen D. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect Dis. 2020;20:697–706. doi: 10.1016/S1473-3099(20)30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Arkel A.L., Rijpstra T.A., Belderbos H.N. COVID-19 associated pulmonary aspergillosis. Am J Respir Crit Care Med. 2020;202:132–135. doi: 10.1164/rccm.202004-1038LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alanio A., Dellière S., Fodil S. Prevalence of putative invasive pulmonary aspergillosis in critically ill patients with COVID-19. Lancet Respir Med. 2020;8:e48–e49. doi: 10.1016/S2213-2600(20)30237-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koehler P., Cornely O.A., Böttiger B.W. COVID-19 associated pulmonary aspergillosis. Mycoses. 2020;63:528–534. doi: 10.1111/myc.13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mortensen E.M., Coley C.M., Singer D.E. Causes of death for patients with community-acquired pneumonia: results from the Pneumonia Patient Outcomes Research Team cohort study. Arch Intern Med. 2002;162:1059–1064. doi: 10.1001/archinte.162.9.1059. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen L.D.N., Viscogliosi E., Delhaes L. The lung mycobiome: an emerging field of the human respiratory microbiome. Front Microbiol. 2015;6:89. doi: 10.3389/fmicb.2015.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Z., Lu G., Meng G. Pathogenic fungal infection in the lung. Front Immunol. 2019;10:1524. doi: 10.3389/fimmu.2019.01524. [DOI] [PMC free article] [PubMed] [Google Scholar]