Highlights
-
•
The 2019 Novel Coronavirus Detection Kit (nCoV-DK) halves the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) detection time by eliminating RNA extraction and purification.
-
•
nCoV-DK detects SARS-CoV-2 as effectively as the direct PCR.
-
•
Saliva is a reliable sample for the detection of SARS-CoV-2 by nCoV-DK.
Keywords: COVID-19, SARS-CoV-2, PCR, Saliva
Abstract
Rapid detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is critical for the diagnosis of coronavirus disease 2019 (COVID-19) and preventing the spread of the virus. A novel detection kit – the 2019 Novel Coronavirus Detection Kit (nCoV-DK) – halves the detection time by eliminating the steps of RNA extraction and purification. We evaluated the concordance between the nCoV-DK and direct PCR. The virus was detected in 53/71 specimens (74.6%) by direct PCR and in 55/71 specimens (77.5%) by nCoV-DK; the overall concordance rate was 94.4%: 95.2% for nasopharyngeal swab, 95.5% for saliva, and 85.7% for sputum. The nCoV-DK test effectively detects SARS-CoV-2 in all types of sample including saliva, while reducing the time required for detection, labor, and the risk of human error.
Introduction
Rapid and accurate detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is critical for the prevention of outbreaks of coronavirus disease 2019 (COVID-19) in communities and hospitals. The diagnosis of COVID-19 is made by real-time quantitative PCR (RT-qPCR) testing of specimens collected by nasopharyngeal swab (Wang et al., 2020, Zou et al., 2020). However, swab sample collection poses a risk of viral transmission to healthcare workers. Self-collection of saliva reduces the risk to healthcare workers. We and others have shown the efficacy of saliva as a diagnostic tool (Azzi et al., 2020, Iwasaki et al., 2020, To et al., 2020, Williams et al., 2020, Wyllie et al., 2020).
The 2019 Novel Coronavirus Detection Kit (nCoV-DK; Shimadzu Corporation, Kyoto, Japan) eliminates the steps of RNA extraction and purification by using the Ampdirect technology (Nishimura et al., 2010), thus significantly reducing the time required for sample preparation and PCR detection from more than 2 h to about 1 h. In addition, the risk of human error during RNA extraction can be reduced. However, there is a need to elucidate whether saliva samples can be applied to the nCoV-DK, since saliva has high RNase (Pandit et al., 2013). This study was performed to compare the efficacy of the nCoV-DK with direct PCR requiring RNA extraction and purification.
Methods
Samples and PCR
Samples were collected from nine patients with COVID-19, as described previously (Iwasaki et al., 2020). A total of 71 frozen stock samples were available from these patients, with a median of 8 samples (range 2–15 samples) per patient. This study was approved by the institutional ethics board and informed consent was obtained from all patients.
Total RNA was extracted and direct RT-qPCR was performed as described previously (Iwasaki et al., 2020). The nCoV-DK PCR was performed using the corresponding frozen specimens.
Statistical analysis
Agreement between the two methods was assessed using Cohen’s kappa. Pearson’s correlation coefficient test was performed to identify the relationship of the cycle threshold (Ct) values between the methods. Statistical analyses were performed with EZR (Jichi Medical University, Saitama, Japan). A p-value of 0.05 was the cut-off for statistical significance.
Results
It was first examined whether the freeze–thaw step could affect the availability of viral RNA for detection. The nCoV-DK PCR was performed on three fresh samples and the corresponding freeze–thaw specimens. Ct values did not change significantly after the freeze–thaw steps.
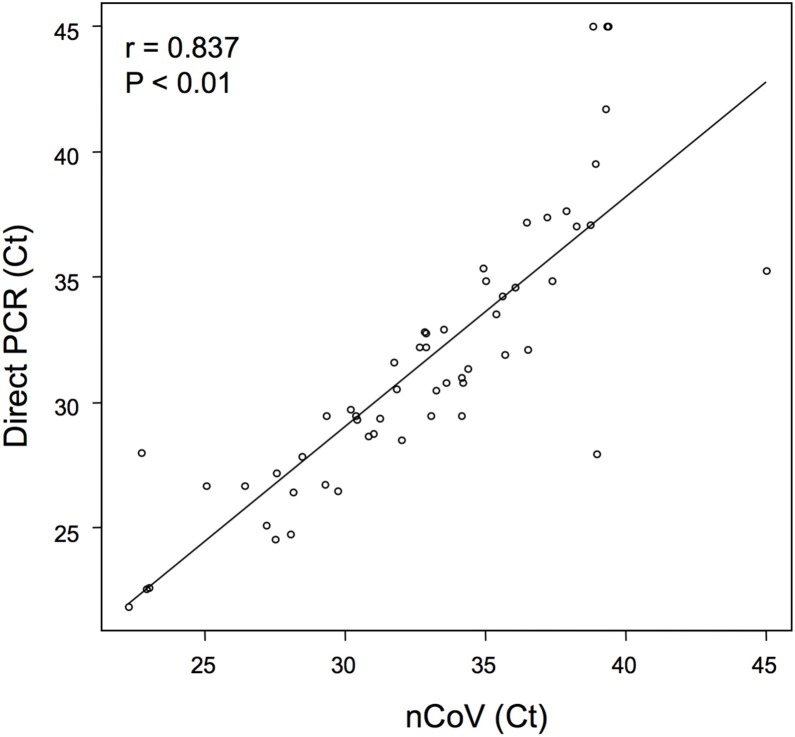
The viral detection rates were then evaluated in 71 specimens. The virus was detected in 53 (74.6%) fresh samples by the direct PCR and in 55 (77.5%) of the corresponding frozen samples by the nCoV-DK (Table 1 ). The overall concordance rate of virus detection between the two methods was 94.4% (95% confidence interval (CI) 86.2–98.4%). Inter-rater reliability of the two methods was strong (κ = 0.85), as judged by Cohen’s kappa analysis. The concordance rate was 95.2% (95% CI 83.8–99.4%) for nasopharyngeal swab samples, 95.5% (95% CI 77.2–99.9%) for saliva samples, and 85.7% (95% CI 42.1–99.6%) for sputum samples. Figure 1 shows a scatter plot presenting a comparison of Ct values for each sample between the two methods. There was a strong correlation between the two methods (r = 0.837, 95% CI 0.736–0.902, p < 0.01). Significant correlations were also demonstrated for each sample type (swab, r = 0.82, 95% CI 0.673–0.905, p < 0.01; saliva, r = 0.818, 95% CI 0.507–0.94, p < 0.01; sputum, r = 0.945, 95% CI 0.574–0.994, p < 0.01).
Table 1.
Comparison of SARS-CoV-2 detection by direct PCR and nCoV-DK method.
| Direct PCR |
||||
|---|---|---|---|---|
| nCoV-DK | Positive | Negative | Kappa (95% CI) | |
| Total | Positive | 52 | 3 | 0.85 (0.70–0.99) |
| Negative | 1 | 15 | ||
| Swab | Positive | 34 | 2 | 0.83 (0.60–1) |
| Negative | 0 | 6 | ||
| Saliva | Positive | 13 | 0 | 0.90 (0.72–1) |
| Negative | 1 | 8 | ||
| Sputum | Positive | 5 | 1 | 0.59 (0–1) |
| Negative | 0 | 1 | ||
SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; nCoV-DK, 2019 Novel Coronavirus Detection Kit; CI, confidence interval.
Figure 1.
Correlation of Ct values between the direct PCR and nCoV-DK methods. The scatter plot shows the comparison of Ct values between the two methods. Negative samples are those with a Ct of 45, which is the limit of detection.
Discussion
This study demonstrated that a novel SARS-CoV-2 detection kit – nCoV-DK – is as effective as direct PCR in detecting SARS-CoV-2 in all types of sample. Particularly, it should be noted that saliva is a reliable sample for virus detection by the nCoV-DK even without the process of RNA extraction and purification. There were some discordant results between the two methods. The virus was detected only by direct PCR in one sample, while the virus was detected only by the nCoV-DK in three samples. It is unclear whether these were false-positive or true-positive, since the PCR primers in the two methods are not the same.
In conclusion, the nCoV-DK has advantages over direct PCR, including a shorter detection time by eliminating the steps of RNA extraction and purification, without impairing diagnostic accuracy.
Conflict of interest
The authors declare that they have no competing interests.
Author contributions
Study design: SI, SF, TT. Data analysis: TF, SI, SF, KH, KS, SO, JS, MN, TT, KT. Sample collection: SN, KK, YY, SK. Writing: TF, SI, SF, JS, TT.
Acknowledgement
This work was supported in part by grants from the Japan Medical Association Policy Research Organization (JMARI).
References
- Azzi L., Carcano G., Gianfagna F., Grossi P., Gasperina D.D., Genoni A. Saliva is a reliable tool to detect SARS-CoV-2. J Infect. 2020;81(1):e45–e50. doi: 10.1016/j.jinf.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki S., Fujisawa S., Nakakubo S., Kamada K., Yamashita Y., Fukumoto T. Comparison of SARS-CoV-2 detection in nasopharyngeal swab and saliva. J Infect. 2020 doi: 10.1016/j.jinf.2020.05.071. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N., Nakayama H., Yoshizumi S., Miyoshi M., Tonoike H., Shirasaki Y. Detection of noroviruses in fecal specimens by direct RT-PCR without RNA purification. J Virol Methods. 2010;163(2):282–286. doi: 10.1016/j.jviromet.2009.10.011. [DOI] [PubMed] [Google Scholar]
- Pandit P., Cooper-White J., Punyadeera C. High-yield RNA-extraction method for saliva. Clin Chem. 2013;59(7):1118–1122. doi: 10.1373/clinchem.2012.197863. [DOI] [PubMed] [Google Scholar]
- To K.K., Tsang O.T., Chik-Yan Yip C., Chan K.H., Wu T.C., Chan J.M.C. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa149. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams E., Bond K., Zhang B., Putland M., Williamson D.A. Saliva as a non-invasive specimen for detection of SARS-CoV-2. J Clin Microbiol. 2020 doi: 10.1128/JCM.00776-20. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyllie A.L., Fourmier J., Casanovas-Massana A., Campbell M., Tokuyama M., Vijayakumar P. Saliva is more sensitive for SARS-CoV-2 detectionin COVID-19 patients than nasopharyngeal swabs. medRxiv. 2020 doi: 10.1101/2020.04.16.20067835. [DOI] [Google Scholar]
- Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382(12):1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]