Highlights
-
•
The temporal trends of the COVID-19 pandemic and the 1918–19 influenza pandemic in the United Kingdom were compared.
-
•
It was found that the ongoing COVID-19 wave of infection had matched the major wave of the 1918–19 influenza pandemic surprisingly well over the previous 2 months.
-
•
The similar characteristics of these two pandemics were discussed.
-
•
We also showed the years of life lost (YLL) due to 1918–19 pandemic. A comparison based on YLL would be more appropriate.
Keywords: COVID-19, 1918–19 influenza, United Kingdom, Multiple waves, Years of life lost
Abstract
We compared the COVID-19 and 1918–19 influenza pandemics in the United Kingdom. We found that the ongoing COVID-19 wave of infection matched the major wave of the 1918–19 influenza pandemic surprisingly well, with both reaching similar magnitudes (in terms of estimated weekly new infections) and spending the same duration with over five cases per 1000 inhabitants over the previous two months. We also discussed the similarities in epidemiological characteristics between these two pandemics.
Introduction
The fast spread and high fatality rate of coronavirus disease 2019 (COVID-19) remind us of the first pandemic in the last century — the 1918–19 influenza pandemic. Indeed, the SARS-CoV-2 and the 1918 A/H1N1 influenza virus share some common properties:
-
•
Similar basic reproductive number (R 0), ranging from 2 to 4.
-
•
Similar patterns of viral shedding from infectious patients (Zou et al., 2020, Wölfel et al., 2020), and thus presumably comparable generation intervals. Zou et al. (2020) reported ‘Our analysis suggested that the viral nucleic acid shedding pattern of patients infected with SARS-CoV-2 resembles that of patients with influenza and appears different from that seen in patients infected with SARS-CoV’. In particular, COVID-19 may have a similar latent period to that of influenza.
-
•
Comparable dispersion parameter, k defined (Lloyd-Smith et al., 2005), which controls the variance in distribution of the number of secondary cases caused by a typical primary case. A smaller k value implies a bigger contribution to total infections from super-spreaders. For instance, 1918 influenza A/H1N1 had a relatively large k (= 0.94) (Fraser et al., 2011) compared with severe acute respiratory syndrome (SARS, k = 0.16) and Middle-East respiratory syndrome (MERS, k = 0.26). It was found that that k for COVID-19 may be 0.8 with 95%CI from 0.63, 0.98 (He et al., 2020), thus closer to that for 1918 influenza A/H1N1. However, the study designs behind these estimates may be different (household only versus household and non-household) and confidence intervals are large in some cases. A summary is given in Table 1 .
-
•
Comparable case fatality rates (CFR) in some situations. It was conventionally accepted that the CFR for 1918–19 influenza was 2%. For COVID-19, the crude CFR shows a wide range, but covering 2%. The actual infection fatality rate (IFR) could be as low as 0.5% if the medical system does not break down. Here crude CFR means the number of reported deaths divided by the number of reported cases. The IFR means the number of reported deaths divided by the actual number of those infected.
Table 1.
Summary of dispersion parameter k values from empirical offspring distribution.
| Virus | Study design | Number of cases/location | Estimates of k (95% confidence interval) | Reference |
|---|---|---|---|---|
| SARS-CoV-2, 2019 | Household and non-household | 9120/mainland China | 0.8 (0.63, 0.98) | He et al. (2020) |
| SARS-CoV-2, 2019 | Household and non-household | 1038/Hong Kong, China | 0.45 (0.31, 0.76) | Adam et al. (2020) |
| SARS-CoV-2, 2019 | Household and non-household | 391/Shenzhen, China | 0.58 (0.35, 1.18) | Bi et al. (2020) |
| A/H1N1, 1918 | Household | 7140/Baltimore, USA | 0.94 (0.59, 1.72) | Fraser et al. (2011) |
| SARS-CoV, 2003 | Household and non-household | 238/Singapore | 0.16 (0.11, 0.64) | Lloyd-Smith et al. (2014) |
Therefore, it is reasonable to compare the temporal patterns of the 1918–19 influenza and the ongoing COVID-19 pandemics in places where both data are available, e.g., the United Kingdom (UK).
Pneumonia and influenza deaths in London, UK
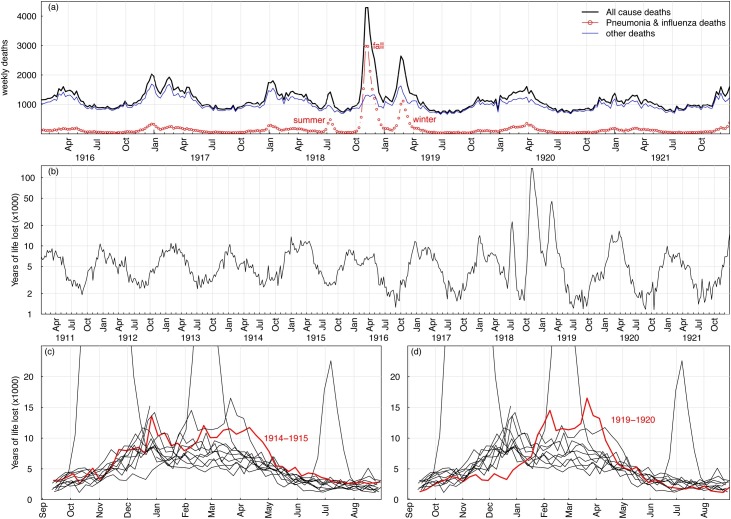
We retrieved weekly mortality data for pneumonia and influenza (P&I) and for all causes in the London boroughs, UK, between January 1911 and December 1921, from our earlier study (He et al., 2011). These data covered three waves (summer, fall, and winter) of the 1918–19 pandemic, and also 2 years of the pre- and post-pandemic periods, as shown in Figure 1 a. Deaths from P&I and all-causes resumed the usual seasonal pattern after the third wave in early 1919.
Figure 1.
The impact of A/H1N1 1918 influenza in the UK from 1918 to 1919, the pre-pandemic era, and the post-pandemic era. Panel (a) showed the weekly deaths from all causes (black) and pneumonia-influenza-associated deaths (red) in London, UK between January 1916 and December 1921. Panel (b) showed the years of life lost (YLL) between 1911 and 1921, related to pneumonia and influenza (P&I) deaths. (c) The YLL distribution was abnormal in 1914–15 when the First World War (WWI) started. (d) The YLL distribution was also abnormal in 1919–20, which could have been a lasting effect of the 1918–19 pandemic. In all panels, the three waves (summer, fall, and winter) stand out.
It is well known that most of the P&I deaths during the 1918–19 pandemic were from the 20–40 years age group, which was different from seasonal influenza. Hence, it would be appropriate to look at the years of life lost (YLL) data (Miller et al., 2008). Figure 1b shows the YLL due to P&I deaths from 1911 to 1921. The three waves stand out (on a logarithmic scale).
Figure 1c shows that the YLL data were abnormal in 1914–15 when the First World War (WWI) started (on July 28, 194). Figure 1d shows abnormal YLL data for 1919–20, which was a lasting effect of the 1918–19 pandemic.
Overall, Figure 1 gives a picture of the course of the 1918–19 pandemic, as well as the pre- and post-pandemic periods. It would be useful for researchers interested in carrying out a comparison study. The WWI occurred between July 28, 1914 and November 11, 1918, facilitating the spreading of the virus.
Age-grouped P&I data were chosen for our study, with an assumed maximum age of 80. A comparison of YLL for COVID-19 and 1918–19 influenza should be conducted, since YLL is an important indicator of the severity of a pandemic.
Comparing the epidemic curves for COVID-19 and A/H1N1 1918
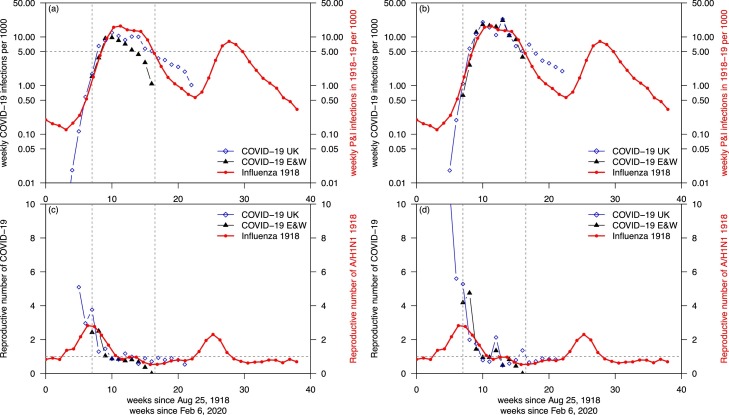
There are many reasons to compare the current pandemic with the 1918–19 pandemic. We attempted such a preliminary comparison, shown in Figure 2 .
Figure 2.
Comparisons of COVID-19 and A/H1N1 1918. (a) Comparing COVID-19 estimated infections based on reported cases (assuming 5% reporting ratio, black) with 1918-19 influenza estimated infections (assuming 2% CFR, red) in E&W. (b) Comparing COVID-19 estimated infections based on reported deaths (assuming 0.5% CFR, black) with 1918–19 influenza estimated infections (red) in E&W. The two matched surprisingly well between weeks 7 and 16, and above five cases per 1000 inhabitants in (b). The time-varying crude reproductive numbers are shown in (c,d), based on data in (a,b), respectively.
Figure 2a compares weekly numbers of reported COVID-19 cases from February 6 to May 25, 2020 with estimated P&I cases from August 1918 to May 1919 in England and Wales (E&W). City-level COVID-19 data were not available to us, but data for E&W were available for both. Reported cases of COVID-19 form only a fraction of actual infections, and thus in order to match the two data sets we assumed a reporting ratio of 5%, i.e., the weekly infections of COVID-19 in Figure 2a were 20 times of the weekly reported cases. The weekly infections of A/H1N1 1918 were estimated using weekly recorded P&I deaths divided by a CFR at 2%. A shortcoming is the uncertainty of the 5% (artificial) reporting ratio. Many serological studies have shown that reported cases of COVID-19 are only a small proportion of the actual infections.
We found a ‘match’ between the COVID-19 wave (in the UK) and the major wave during October 1918 (in E&W), if we aligned the two waves with the same time scale. The week zero started on August 25, 1918 for the 1918 influenza, and February 6, 2020 for the COVID-19. However, this match was artificial without strong evidence because we chose a 5% reporting ratio. We chose to match the ongoing COVID-19 situation to the fall wave in 1918 because the summer wave was very minor and obviously not comparable.
The influenza A/H1N1 epidemic in 1918 had a conventionally accepted CFR of 2% (Mills et al., 2004); here we treated cases as infections. The infection attack rate (IAR, i.e., the proportion of population being infected) could be estimated given the P&I deaths, and was around 25% from 1918 to 1919, and thus roughly 25% of the E&W population was infected between June 1918 and May 1919, based on the recorded P&I deaths and a CFR at 2%.
The E&W population was 44 million in 1918, compared with 59 million in 2020. By assuming a 0.5% IFR for COVID-19 in 2020 and a 2% IFR for A/H1N1 in 1918, we could calculate and compare the infections based on reported deaths, which should be more reliable than reported cases. Figure 2b shows such a comparison, with the ‘match’ being unexpected.
Here the 0.5% IFR for COVID-19 is a reasonable guess based on a serological study in Gangelt, Germany (Streeck et al., 2020) and observed infection fatality rates in Hong Kong (Hong Kong SAR Government, 2020) and Singapore (Singapore Government, 2020), where testing was extensive. Furthermore, according to Faust and del Rio (Faust and del Rio, 2020), the CFR on the Diamond Princess cruise ship outbreak was 0.5% after age standardization. The estimated infections per 1000 population are shown in Figure 2b. So far, the current COVID-19 wave in E&W matches the major wave of 1918–19 influenza between week 7 and week 16 (indicated by the vertical dotted lines).
Figures 2c and 2d show the estimated reproductive number (R e) based on data in Figures 2a and 2b, respectively. Here, we used a simple way to estimate R e, which equals the number of cases in week i divided by number of cases in week i-1 (preceeding week). This rule of thumb should be acceptable given that the serial intervals of the two viruses are roughly 1 week (5 or 6 days) for COVID-19 (Xu et al., 2020) and probably the same for the A/H1N1 1918.
The mechanisms (factors) behind the multiple mortality waves in 1918–19 were shown in two previous studies (Bootsma and Ferguson, 2007; He et al., 2011, 2013). Namely, the on-and-off of public health measures (such as bans on public gatherings) and public behavioral reactions accounted for the following waves. A similar model framework has been applied to COVID-19 in Wuhan, China (Lin et al., 2020). Hence, it is reasonable to argue that public health measures and public behavioral reactions would have acted in a similar fashion in both pandemics. Although Figure 2a may be limited without strong evidence in applying a reporting ratio of 5% without strong evidence, Figure 2b was based on an IFR at 0.5%, in line with some references. Nevertheless, the matching of magnitudes (in terms of weekly infections, based on the highest week so far) was unexpected. The matching of durations of the two pandemics spent above a threshold of five infections per 1000 population would appear to be nontrivial. Using a model (simulation) to match COVID-19 would require several assumptions. Here, we used minimal assumptions that included a 0.5% IFR for COVID-19 and a 2% CFR for the influenza epidemic in 1918, and a time shift representing the introduction time of COVID-19 into the UK.
The aforementioned similarities between COVID-19 and the 1918–19 influenza may be partly responsible for the unexpected matching between week 7 and week 16, as shown in Figure 2b.
Besides their similarities, 1918–19 influenza and COVID-19 also have differences, including age patterns for infection and mortality, the pre-existing immunity, and vastly different conditions. For example, the age structure for P&I deaths in 1918–19 comprised three peaks, the famous W shape, which were shown in (He et al., 2013). The age structures for infections and deaths of COVID-19 were presented in (Sun et al., 2020).
Declarations
Ethical approval and consent to participate are not applicable.
Consent for publication
Not applicable.
Availability of data and materials
All data used are from the public domains.
Conflict of interests
DH was supported by an Alibaba (China) - Hong Kong Polytechnic University Collaborative Research Project. Other authors declared no conflict of interest.
Funding
DH was supported by a General Research Fund (grant number 15205119) from the Research Grants Council (RGC) of Hong Kong, China and an Alibaba (China) Co., Ltd. - Hong Kong Polytechnic University Collaborative Research Project (P0031768).
Author contributions
DH conceived and conducted the research, and wrote the draft. All authors critically revised the manuscript, and all authors approved the submission.
Acknowledgements
DH would like to thank Jonathan Dushoff, Lewi Stone, and David Earn for insightful discussion. The authors would like to thank the International Infectious Disease Data Archive (IIDDA) for influenza data.
References
- Adam D., Wu P., Wong J., Lau E., Tsang T., Cauchemez S. Clustering and superspreading potential of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections in Hong Kong. Research Square. 2020 doi: 10.1038/s41591-020-1092-0. https://www.researchsquare.com/article/rs-29548/v1 [DOI] [PubMed] [Google Scholar]
- Bi Q., Wu Y., Mei S., Ye C. Epidemiology and transmission of COVID-19 in 391 cases and 1286 of their close contacts in Shenzhen, China: a retrospective cohort study. Lancet Infect Dis. 2020 doi: 10.1016/S1473-3099(20)30287-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bootsma M.C., Ferguson N.M. The effect of public health measures on the 1918 influenza pandemic in US cities. Proc Nat Acad Sci. 2007;104(18):7588–7593. doi: 10.1073/pnas.0611071104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust J.S., del Rio C. Assessment of deaths from COVID-19 and from seasonal influenza. JAMA Int Med. 2020 doi: 10.1001/jamainternmed.2020.2306. [DOI] [PubMed] [Google Scholar]
- Fraser C., Cummings D.A., Klinkenberg D., Burke D.S., Ferguson N.M. Influenza transmission in households during the 1918 pandemic. Am J Epidemiol. 2011;174(5):505–514. doi: 10.1093/aje/kwr122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D., Dushoff J., Day T., Ma J., Earn D.J. Mechanistic modelling of the three waves of the 1918 influenza pandemic. Theor Ecol. 2011;4(2):283–288. [Google Scholar]
- He D., Dushoff J., Day T., Ma J., Earn D. Inferring the causes of the three waves of the 1918 influenza pandemic in England and Wales. Proc Roy Soc B. 2013;280(1766):20131345. doi: 10.1098/rspb.2013.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D., Zhao S., Xu X., Zhuang Z., Cao P., Wang M.H. Individual variation in infectiousness of coronavirus 2019 implies difficulty in control. SSRN. 2020 doi: 10.2139/ssrn.3559370. [DOI] [Google Scholar]
- Lin Q., Zhao S., Gao D., Lou Y., Yang S., Musa S.S. A conceptual model for the outbreak of Coronavirus disease 2019 (COVID-19) in Wuhan, China with individual reaction and governmental action. Int J Infect Dis. 2020;93:211–216. doi: 10.1016/j.ijid.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Smith J.O., Schreiber S.J., Kopp P.E., Getz W.M. Superspreading and the effect of individual variation on disease emergence. Nature. 2005;438(7066):355–359. doi: 10.1038/nature04153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M.A., Viboud C., Olson D.R., Grais R.F., Rabaa M.A., Simonsen L. Prioritization of influenza pandemic vaccination to minimize years of life lost. J Infect Dis. 2008;198(3):305–311. doi: 10.1086/589716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills C.E., Robins J.M., Lipsitch M. Transmissibility of 1918 pandemic influenza. Nature. 2004;432:904–906. doi: 10.1038/nature03063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Kong SAR Government, https://www.coronavirus.gov.hk/eng/index.html; 2020. [accessed 1 June 2020].
- Singapore Government, https://www.moh.gov.sg/covid-19; 2020. [accessed 1 June 2020].
- https://www.land.nrw/sites/default/files/asset/document/zwischenergebnis_covid19_case_study_gangelt_0.pdf; 2020. [accessed 1 June 2020].
- Sun K., Chen J., Viboud C. Early epidemiological analysis of the coronavirus disease 2019 outbreak based on crowdsourced data: a population-level observational study. Lancet Digital Health. 2020;2(4):E201–E208. doi: 10.1016/S2589-7500(20)30026-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. 1-0. [DOI] [PubMed] [Google Scholar]
- Xu X., Liu X., Wu Y., Ali S.T., Du Z., Bosetti P. Reconstruction of transmission pairs for novel coronavirus disease 2019 (COVID-19) in mainland China: estimation of super-spreading events, serial interval, and hazard of infection. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Guo Q. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382(12):1177–1179. doi: 10.1056/NEJMc2001737. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data used are from the public domains.