Abstract
Background and purpose
CD133 is controversially discussed as putative (surrogate) marker for cancer stem/tumor-initiating cell populations (CSC/TIC) in epithelial tumors including colorectal carcinomas (CRCs). We studied CD133 expression in established CRC cell lines and examined in vitro behavior, radioresponse and in vivo tumor formation of CD133+/− subpopulations of one cell line of interest.
Materials and methods
Ten CRC cell lines were analyzed for CD133 expression using flow cytometry and Western blotting. CD133+ and CD133− HCT-116 subpopulations were separated by FACS and studied in 2-D and 3-D culture and colony formation assays after irradiation. Subcutaneous xenograft formation was monitored in NMRI (nu/nu) mice.
Results and conclusions
CRC cell lines could be classified into three groups: (i) CD133−, (ii) CD133+ and (iii) those with two distinct CD133+ and CD133− subpopulations. Isolated CD133+/− HCT-116 subpopulations were studied relative to the original fraction. No difference was found in 2-D growth, spheroid formation or radioresponse in vitro. Also, tumor formation and growth rate did not differ for the sorted subpopulations. However, a subset of xenografts originated from CD133− HCT-116 showed a striking enrichment in the CD133+ fraction. Our data show that CD133 expression is not selective for sphere forming, tumor-initiating or radioresistant subpopulations in the HCT-116 CRC cell line. This implies that CD133 cannot be regarded as a CSC/TIC marker in all CRC cell lines and that functional measurements of tumor formation have to generally accompany CSC/TIC-directed mechanistic or therapeutic studies.
Keywords: Cancer stem/tumor-initiating cells (CSC/TIC), Colorectal carcinoma (CRC) cell lines, HCT-116, 2-D culture, 3-D culture, Radioresponse
Hallmarks of most cancer cells as emphasized earlier by Hanahan and Weinberg [1] are (a) self-sufficiency for growth signals, (b) insensitivity to growth-inhibitory signals, (c) evasion of apoptosis, (d) unlimited replicative potential, (e) sustained angiogenesis and (f) tissue invasion and metastasis. These features seem inherent in the majority of tumor cells. However, tumors may also contain small subpopulations of cells which are capable of initiating tumor growth due to a unique stem cell-like capability for self-renewal and asymmetric division. These cells are often referred to as cancer stem cells (CSCs) as emphasized in the consensus definition from the AACR Cancer Stem Cells Workshop in 2006 [2], but are also termed tumorigenic or tumor-initiating cells (TICs) as their precise identity and relation to adult somatic stem cells are still uncertain [3–6]. The presence of such small subsets of CSC/TIC in various human epithelial cancers is supported by numerous in vitro and in vivo data. Many related mechanistic studies have used surface markers to identify and enrich tumor cell populations with a CSC/TIC phenotype. However, the hypothesis of those marker-defined cell populations to impair long-term survival after treatment and to be causally related with the relapse of disease due to resistance to therapy is still disputed.
Lack of mechanistic insight is primarily due to the imperfect tool of surface markers without causal evidence used for the identification and isolation of human epithelial CSC/TIC [3,5,7–9]. One of these markers of interest is CD133 (human prominin-1), a cellsurface glycoprotein of 92–110 kDa, five transmembrane domains and two large glycosylated extracellular loops [10–12]. Its function has not been established yet, but it seems to participate in the regulation of membrane topology [10,13–15]. According to the evolving potential of CD133 in cancer research, CD133 has been designated as molecule of the moment in 2008 [16]. It is noted, however, that CD133 antigen detection is quite complex and cell surface expression of the glycosylated form of CD133 may be the more reliable attribute in CSC/TIC than CD133 expression per se [17]. Taking this into account, there is a growing evidence that CD133 is expressed on CSC/TIC in various tumor entities including brain and neural tumors [18–25], melanomas [26] and carcinomas of the lung [27], kidney [28], ovary [29,30], prostate [31,32], liver [33–35], pancreas [36] and CRC as detailed in several recent reports [8,37,38]. In most studies, increased clonogenic survival and/or enhanced tumorigenic potential in immunodeficient mice of CD133+ versus CD133− tumor populations were recorded.
In CRC, CD133-positive primary cancer cell subpopulations with tumor-initiating capacity were described to be maintained and expanded only when grown in a serum-free milieu as 3-D cultures, while addition of serum resulted in adherent growth, downregulation of CD133 expression, tumor cell differentiation and loss of the TIC phenotype [38]. This observation implies that established CRC cell lines which are routinely kept as 2-D cultures in serum-containing media might not express CD133 or have lost their tumor-initiating potential. Human CRC cell lines, however, are known to produce tumors in various xenograft models. Also, HT29 cells with high CD133 immunofluorescence signal were reported to grow faster and have a higher tumorigenic and invasive potential than CD133− HT29 cells [39] indicating that routinely grown cell lines may contain particular CSC/TIC subpopulations within a CD133-expressing cell fraction. We initiated our project to verify the therapeutic relevance of CD133+ versus CD133− cells in CRC cell lines. The aim of the study was to evaluate CD133 expression in various CRC cell lines and to examine one cell line of interest in more detail. Based on an optimized staining and sorting protocol, we identified several cell lines containing two distinct populations with and without expression of CD133 and compared clonogenic survival, 2-D and 3-D culture characteristics, radioresponse and in vivo xenograft formation of CD133+/− HCT-116 subpopulations.
Materials and methods
Cell lines and routine culturing
Human CRC cell lines HT29, WiDr, HCT15, HCT-116, DLD1, Lovo, LS174T, KM12L4A HCC2998 and KM20L2 obtained from the ATCC (Manassas, VA, USA) and the NCI (Bethesda, MA, USA), respectively, were cultured under routine conditions in a standard medium (DMEM supplemented with 10% FCS) as described previously [40,41]. All cell lines were tested free of mycoplasm using a PCR Mycoplasma Kit (Applichem, Germany). Routine verification of cell line purity and clonality was performed by microsatellite analyses at the Institute of Legal Medicine. HCT-116 cells, for example, were studied with the commercial multiplex PCR kits Mentype® NonaplexQS Twin (Biotype AG, Germany) and PowerPlex® 16 (Promega Corporation, USA). Amplicons were detected by capillary electrophoresis in the denaturing polymer POP4 in the ABI 310 sequencer (Perkin-Elmer, USA) following manufacturer’s instructions. Our HCT-116 stock was further investigated by fluorescence in situ hybridization (FISH) using painting probes for chromosomes 8 and 16 and spectral karyotyping (SKY) as previously described [42].
Flow cytometry and fluorescence activated cell sorting (FACS)
Single cell suspensions from monolayers, spheroid cultures and xenografts were prepared for flow cytometric analyses. CD133 cell surface expression was analyzed following staining with anti-CD133/1-PE relative to the respective isotype control (Miltenyi Biotec, Germany). The antibody was diluted 1:100 in PBS containing 0.5% FCS and 2 mM EDTA; 1 × 106 cells in a total volume of 100 μl antibody solution were incubated for 30 min at 4 °C in the dark. Cells were then washed in an adequate volume of washing buffer (PBS containing 2 mM EDTA) and the fluorescence signal was intensified by FASER technology (Fluorescence Amplification by Sequential Employment of Reagents) according to the manufacturer’s instructions in a double amplification process (Miltenyi Biotec). CD133 cell surface expression was detected with a FACScan flow cytometer (BD Biosciences, USA); 2 × 104 events per sample were collected and membrane-defect cells were excluded by the addition of 2 μg/ml propidium iodide per sample. Anti-human CD326-FITC antibody and the respective isotype control (dilution 1:25; Miltenyi Biotec) were applied in addition to anti-CD133 staining to identify human epithelial cells after dissociation of xenograft tumors.
HCT-116 monolayer cell suspensions (density of cell suspensions 107/ml) were sorted according to their CD133 expression with a FACSAria (BD Biosciences) following multicolor staining as described for flow cytometric analyses. Separated subpopulations were reanalyzed for purity.
Western blotting
Whole cell protein was extracted using RIPA buffer (1 × PBS, 1% Nonidet P40, 0.5% sodiumdeoxycholat, 0.1% SDS) after the addition of 0.1 M PMSF plus 1:100 protease inhibitor cocktail (P2714, Sigma–Aldrich, Germany). Protein extract aliquots were stored at −80 °C. Protein content was determined using the BCA Protein Assay Kit (Pierce, Germany) according to the manufacturer’s instructions. After protein separation on 10% SDS–PAGE and transfer of proteins onto a PVDF membrane (Millipore, Germany), the membrane was blocked overnight in PBS containing 0.1% Tween 20 and 5% milk powder. After washing the membrane three times with PBS containing 0.1% Tween 20, CD133 protein was exposed to the rabbit polyclonal anti-human CD133 antibody Ab19898 (Abcam, UK; 1:100 diluted in the same buffer) for 1 h at room temperature (RT). After washing and incubation with secondary anti-rabbit immunoglobulin/HRP (dilution 1:5000, 1 h, RT; Dako, Germany), detection was performed via Western Blotting Luminol Reagent (Santa Cruz, Germany). After an incubation of membrane in luminol reagent for 1 min, film was exposed for 10 min. β-Actin was probed as control using the ab 6276–100 antibody (Abcam, UK) in a TBS buffered system (dilution 1:2000 in TBS containing 3% milk powder).
2-D and 3-D culture setup
Monolayer growth was recorded after seeding of 1 × 104 CD133+, CD133− and mixed HCT-116 cells per well in a 6-well plate. At defined time points, cell numbers of three wells per condition were determined following enzymatic dissociation (0.05% trypsin/0.02% EDTA) using the Casy1 cell analyzer system (Schaerfe System GmbH, Germany). Single cell suspensions were analyzed for their CD133 cell surface expression via flow cytometry (see above).
Spheroid formation capacity of HCT-116 subpopulations after FACS was determined in liquid overlay technique [41,43,44]. 750 CD133+, CD133− and mixed CD133 HCT-116 cells in 200 βl medium were seeded per well in agarose-coated 96-well plates. Spheroid integrity and growth kinetics were monitored by semiautomated measurement of diameters and volumes from phase contrast images as described earlier [41]. A minimum of 16 spheroids per seeded population were analyzed. To determine the distribution of CD133+ cell fractions at defined time points during 3-D culturing, 12–50 spheroids (depending on the respective mean spheroid volume) were collected per condition, dissociated (0.1% trypsin/0.04% EDTA solution) and cell numbers were determined via the Casy1 system. Single cell suspensions were further examined and analyzed by flow cytometry.
Colony formation assay and irradiation
Three hundred cells of CD133+/− sorted or non-separated HCT-116 populations were inoculated per well of a 6-well plate in 2 ml standard medium containing 25 mM HEPES. After an incubation time of 4 h under culture conditions to allow cell adherence, cultures were irradiated at RT (0.5–12 Gy, 1.3 Gy/min, 200 kV X-rays; 0.5 mm Cu filter; YxlonY.TU 320; Yxlon. International, Germany). Ten days later, cells were stained with 0.5% crystal violet and colonies ≥50 cells were manually counted in four wells per condition using an AxioVert200 microscope (Carl Zeiss MicroImaging GmbH, Germany); surviving fractions at 2 Gy were calculated from dose response curves of four individual experiments.
Xenograft formation and analysis
Single cell suspensions of HCT-116 were stained and separated via FACS according to their CD133 cell surface expression as described above. Cell numbers of sorted subpopulations were determined with the Casy1 cell analyzer and 2.5 × 103 or 1 × 104 cells suspended in 100 μl of a 50% Matrigel (BD Biosciences) in PBS solution were injected subcutaneously into the hind limb of 8–10 week old female NMRI (nu/nu) mice (Experimental Center, Medical Faculty, University of Technology Dresden). Tumor growth curves were recorded by measuring tumor axes a (longest tumor axis) and b (perpendicular axis) twice a week using a manual caliper. Tumor volumes were estimated by the formula of a rotational ellipsoid: ν = π/6 × a × b2. Animals were sacrificed when tumors reached a mean diameter of 0.6–0.8 cm or when animals appeared to suffer. Animal facilities and experiments were approved in accordance to institutional guidelines and German animal welfare regulations. Animals were fed with commercial laboratory animal diet and water ad libitum.
Xenografts were dissected at diameters of 0.6–0.8 cm, non-tumor tissue was removed and tumor material was weighted, minced with scalpels, and cell suspensions were generated by incubation in a 0.05% trypsin/0.02% EDTA solution in PBS (PAN Biotech GmbH, Germany). Cell suspensions were filtered (pore size 70 μm) to exclude clusters and cell numbers were determined with the Casy1 system. Single cell suspensions were stained for CD133, CD326 and PI as described and analyzed by flow cytometry; 1 × 105 cells were seeded in a T25 cell culture flask for extended culturing.
Results
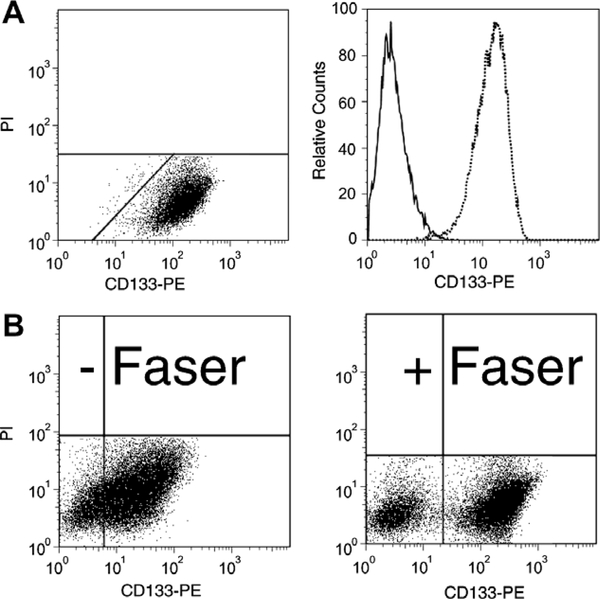
According to literature data on primary colorectal carcinoma cells, the addition of serum to early cultures leads to adherent growth, downregulation of CD133 expression, differentiation and loss of TIC phenotype [38]. Nonetheless, HT29 cells with high CD133 expression grown under serum containing condition were described to possess a higher TIC potential than CD133 low HT29 [39]. To gain insight into this discrepancy we evaluated CD133 cell surface expression in 10 CRC cell lines by flow cytometry using the anti-CD133/1 (AC133) antibody. NTERA-2 teratocarcinoma cells were used as CD133 positive control [11,12] (Fig. 1A). We first optimized the experimental procedure to precisely discriminate CD133-positive (CD133+) and CD133-negative (CD133−) CRC cells as well as to allow fluorescence activated cell sorting with high purities by application of FASER technology (Fig. 1B) to enhance fluorescence signal. Applying this protocol, all cell lines studied herein could be classified into three groups according to their
Fig. 1.
Optimization of flow cytometric detection of CD133 expression in cell lines. (A) The teratocarcinoma cell line NTERA-2 was stained as a control for CD133 cell surface expression analysis by flow cytometry using a PE-conjugated anti-CD133 antibody. Membrane defect propidium iodide (PI) positive cells were excluded. (B) The advantage of the application of Fluorescence Amplification by Sequential Employment of Reagents (FASER) is representatively shown for HCT-116 cells. Fluorescence signal is enhanced and thus allows the clear discrimination of CD133+ and CD133− HCT-116 subpopulations. This protocol was applied for all cell lines studied. The analysis was adapted individually for each cell line.
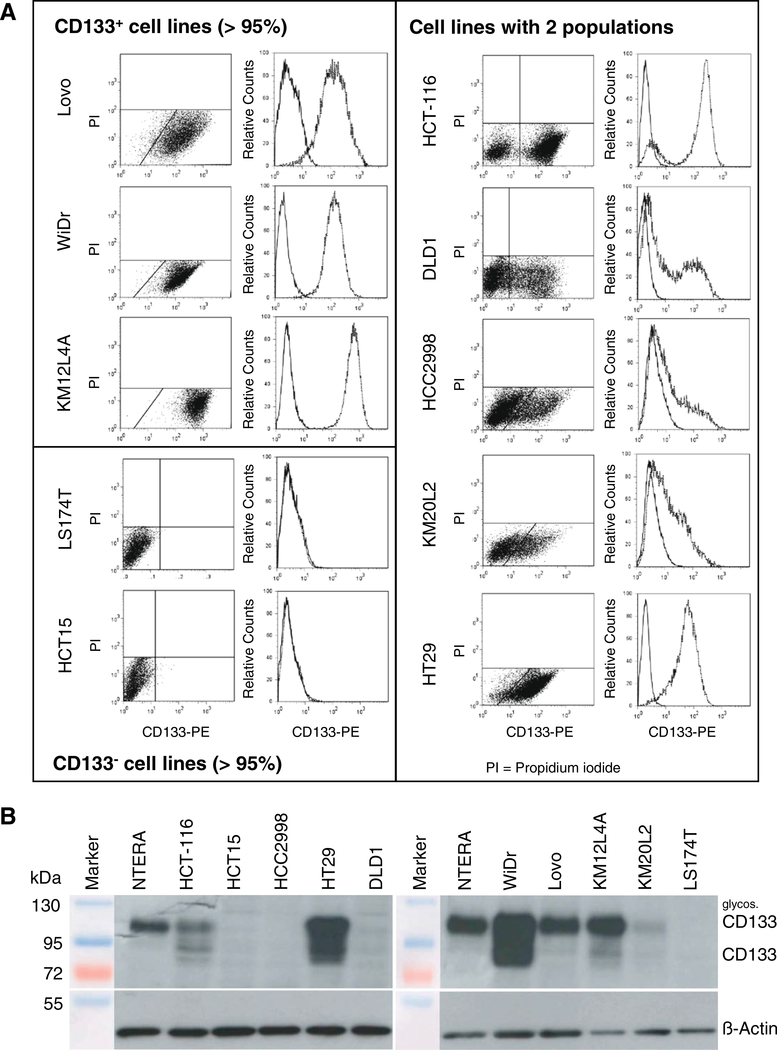
CD133 surface presentation (Fig. 2A): (I) cell lines with roughly all cells (>95%) expressing CD133 on the surface (Lovo, WiDr, KM12L4A); here no distinct CD133− population could be identified; (II) cell lines without CD133 surface expression in monolayer culture (LS174T, HCT-15) and (III) cell lines that contain two distinct populations of CD133+ and CD133− appearing cells (HCT-116, DLD1, HCC2998, KM20L2, HT29). Under 10% serum-supplemented standard conditions, exponential DLD1, HCC2998 and KM20L2 contained 37.0 ± 5.8%, 28.6 ± 2.1% and 20.8 ± 6.6% CD133+ cells, respectively. In HT29 cultures a small CD133− subpopulation could be defined only by using FASER technology to intensify the signal. 90.9 ± 6.4% of the HT29 cells expressed CD133 on the cell surface. Two subpopulations were also clearly distinguishable in HCT-116 cells with a 74.3 ± 6.2% CD133+ population. Western blot analyses using a polyclonal anti-CD133 antibody and whole cell protein lysates confirmed CD133 expression in eight of the 10 CRC cell lines studied by flow cytometry (Fig. 2B). Unexpectedly, no CD133 protein was detected in protein extracts of DLD1 and HCC2998, although these lines clearly showed a CD133+ subpopulation in flow cytometric analyses.
Fig. 2.
Pattern of CD133 expression in 10 different CRC cell lines indicates three different phenotypes. (A) According to the cell surface expression of CD133 in flow cytometry analyses the cell lines can be categorized into three groups, a first group contains cell lines in which >95% of the cells are CD133+, a second group without CD133 cell surface presentation and a third group of lines with two distinct subpopulations, i.e. with and without CD133 cell surface expression. Membrane defect propidium iodide (PI) positive cells were excluded. For every cell line a representative flow cytometric dot plot diagram of CD133 fluorescence versus propidium iodide signal (left panel) and a histogram overlay of the CD133 expression relative to a corresponding isotype control are shown (right panel). (B) The pattern of cell surface presentation was confirmed by Western blot analysis using whole cell protein extracts (50 μg protein per lane) for most of the cell lines with the exception of DLD1 and HCC2998. The predicted size of CD133 is 97 kDa, a higher band of ∼120 kDa indicates a glycosylated CD133 protein. β-Actin is detected as control protein.
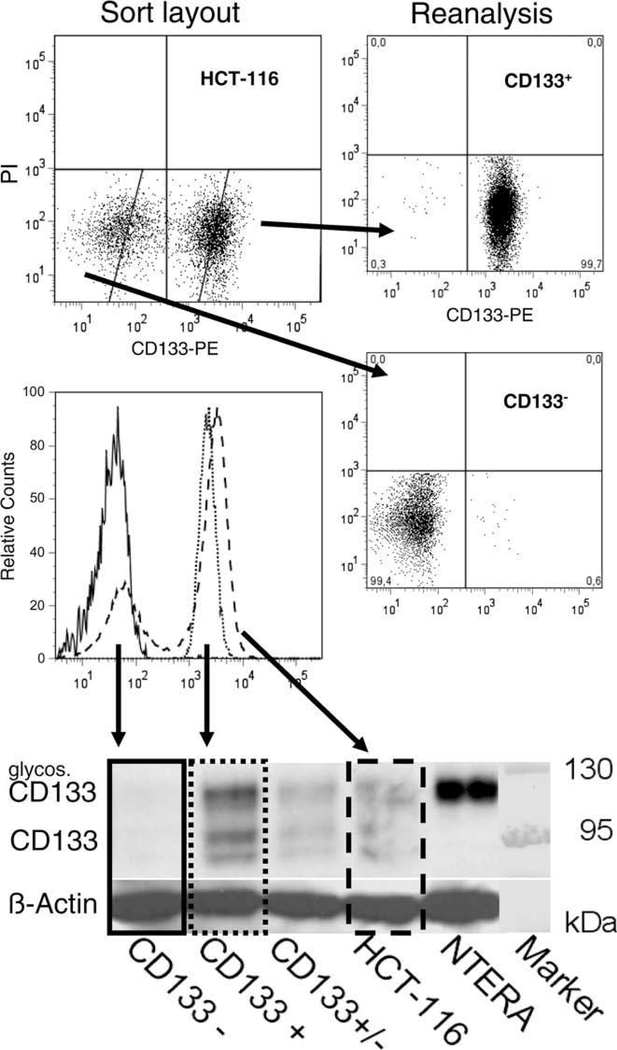
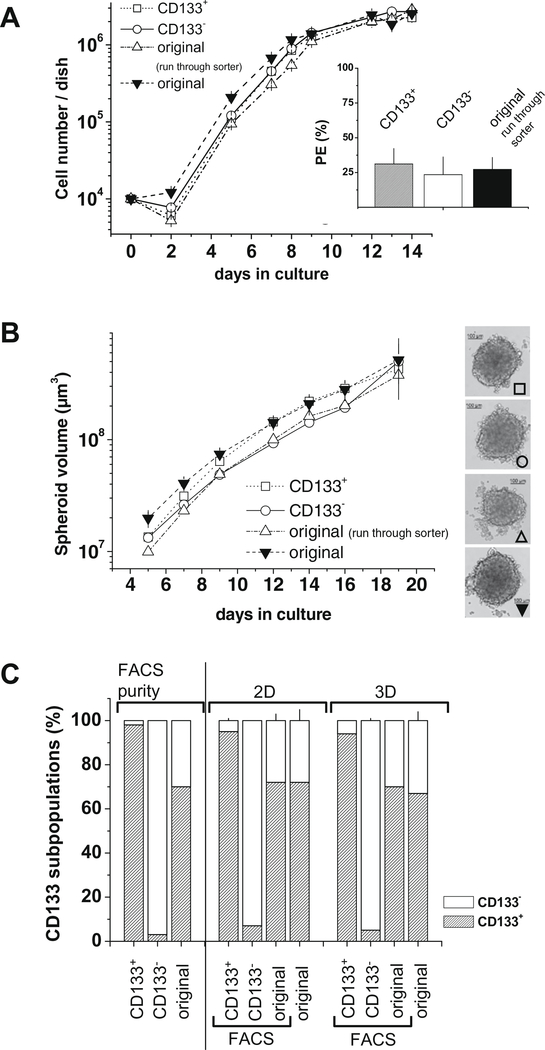
CD133+ CRC cell populations were described to be enriched for TIC/CSC. CD133+ HT29 cells, for example, have been shown to possess a higher tumorigenic potential, higher colony forming abilities and were more proliferative [39]. To investigate these observations for a second cell line, HCT-116 was chosen because this genetically validated cell line is characterized by two clearly distinguishable CD133+/−subpopulations. The sort protocol established to isolate CD133 subpopulations with purities of >98% is documented in Fig. 3. Protein of separated subpopulations of HCT-116 was extracted and Western blotting revealed no CD133 expression in the CD133− sorted subpopulation (Fig. 3). HCT-116 subpopulations were further analyzed according to their CD133 expression in monolayer and in liquid overlay spheroid culture (Fig. 4). Monolayer growth characteristics of HCT-116 CD133+ and CD133− subpopulations did not differ during 14 days of culturing (Fig. 4A). No significant difference in colony forming capacity of CD133+ versus CD133− HCT-116 sorted fractions was recorded (inlay – Fig. 4A). Also, both subpopulations were equally able to form spheroids in liquid overlay. Spheroid growth over a period of 20 days was marginally faster for CD133+ versus CD133− HCT-116 cells as documented in Fig. 4B. During 2-D and 3-D culture experiments, the distribution of CD133+/CD133− HCT-116 subpopulations was determined by flow cytometry. Throughout culturing CD133+ and CD133− subpopulations did not restore the original distribution of non-sorted HCT-116 cells. More than 90% of the cells kept the surface expression profile of the isolated phenotype (Fig. 4C).
Fig. 3.
Protocol to separate CD133 subpopulations by fluorescence activated cell sorting (FACS) with purities >98%. According to flow cytometric setup that includes staining with fluorescence conjugated anti-CD133 antibody, signal enhancement by FASER and the exclusion of membrane defect, PI positive cells, a sort layout for the cell line HCT-116 was defined for FACS isolation of CD133+ and CD133− HCT-116 cell populations. Separated subpopulations were routinely analyzed for purity as shown for a representative experiment (right panel). In addition fluorescence signal intensities of reanalyzed subpopulations and original HCT-116 are visualized in a histogram overlay. Whole cell protein was extracted from the separated HCT-116 subpopulations and the original cell line. The corresponding Western blot analysis reveals that no CD133 protein is expressed in the CD133− HCT-116 subpopulation.
Fig. 4.
CD133+ and CD133− HCT-116 cells do not differ in 2-D and 3-D culture. In vitro culture characteristics were monitored for FACS separated CD133+ and CD133− HCT-116 subpopulations in comparison to the original mixed HCT-116 cells that underwent sort procedure (run through sorter) or not (original). Representative experiments are shown. Experiments were performed at least in triplicate. (A) Monolayer growth kinetics recorded after seeding of separated HCT-116 subpopulations and HCT-116 reveals no differences in 2-D growth. Average plating efficiencies (+SD) from n = 4 independent experiments demonstrate comparable colony forming capacity for HCT-116 cells differing in their CD133 expression in (inlay). (B) Also, spheroid formation capacity of CD133+, CD133− and original HCT-116 cells in liquid overlay technique is comparable. (C) Distribution of CD133 subpopulations was monitored during 14 days of 2-D and 3-D in vitro culturing according to (A) and (B). Fractions of CD133+ and CD133− HCT-116 subpopulations did not change dramatically compared to the original sorted populations shown as FACS purity in the left columns.
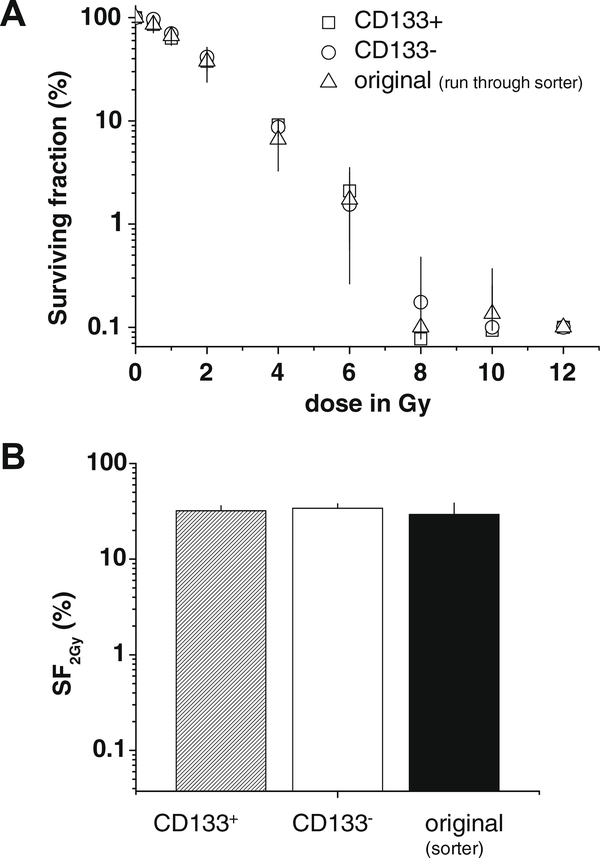
Radioresponse was investigated in colony forming assays by application of single doses ranging from 0.5 to 12 Gy. Separated CD133 HCT-116 subpopulations were seeded in 6-well plates and irradiation was performed after 4 h, when cells were adherent. A representative dose–response curve is shown in Fig. 5A that reveals no difference in radioresponse of CD133+, CD133− and original HCT-116 cells. Calculation of the SF2 Gy value (survival fraction after 2 Gy) from the dose–response curves of four independent experiments confirmed this observation (Fig. 5B). The SF2 Gy was 32.0 ± 4.2% for the CD133+ and 34.1 ± 3.8% for the CD133− HCT-116 fraction, and is therefore independent of the CD133 expression profile.
Fig. 5.
No difference in radioresponse of CD133+ and CD133− HCT-116 subpopulations. Colony formation assays (CFAs) were performed for CD133+, CD133− and originally distributed HCT-116 that underwent sort procedure (run through sorter). CFA set-up: 300 cells/well; culturing in respective media for 10 days. (A) Representative dose response curves of CD133+, CD133− and original HCT-116 cells after irradiation with a single dose regime (0.5–12 Gy) 4 h after plating. Clonogenic survival at 0 Gy (control) for each condition was set to 100% for normalization. (B) Mean clonogenic survival at 2 Gy (SF2 Gy + SD) as determined from n = 4 independent experiments according to (A). Radioresponse of CD133+, CD133− and non-separated HCT-116 cells do not differ.
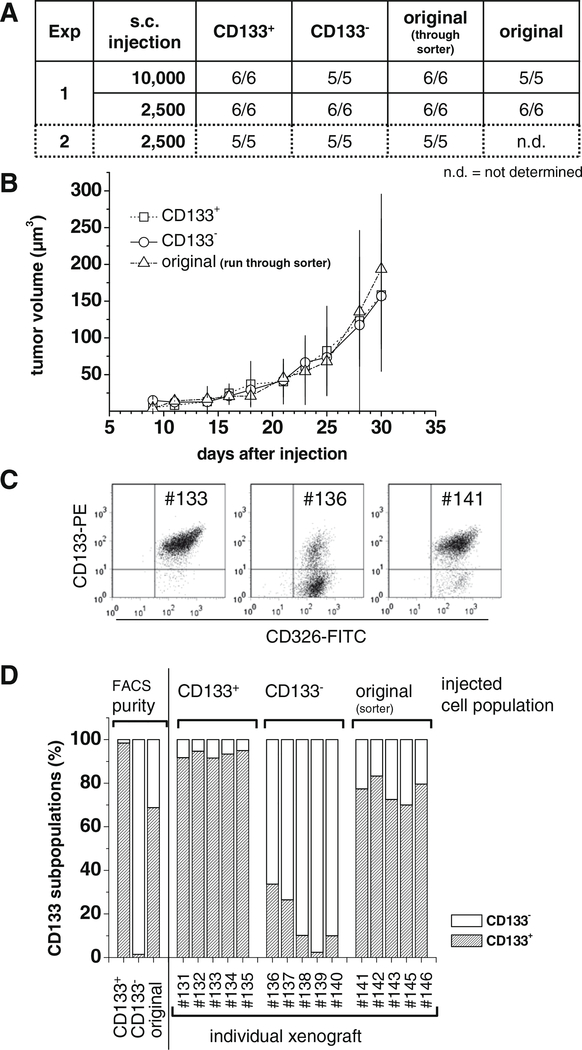
Tumor formation capacity is a major aspect to identify CSC/TIC phenotype. We therefore initiated a first series of xenograft tumor formation experiments in NMRI (nu/nu) mice using FACS separated HCT-116 subpopulations. In two independent experiments 2.5 × 103 or 1 × 104 CD133+ and CD133− HCT-116 cells were injected subcutaneously and tumor formation and growth were analyzed relative to the mixed HCT-116 population. The data documented in Fig. 6 do not indicate a selective CSC/TIC character of the CD133+ HCT-116 cell population as tumor formation and growth rate were comparable for CD133+, CD133− and original HCT-116 cells (Fig. 6A/B). It has been shown in the literature that CD133+ CRC cells differentiate into CD133− cells [38]. Thus, the distribution and a potentially re-expression of CD133 or an enrichment of the CD133 populations in xenograft tumors developed from 2500 CD133+, CD133− and original HCT-116 cells from experiment 2 were analyzed in more detail. Tumors were dissected and dissociated after 30–36 days of growth in NMRI (nu/nu) mice. Single cell suspensions were co-stained for CD133 and CD326 (EpCAM – epithelial cell adhesion molecule) to identify human epithelial cells and analyze their CD133 surface expression. CD326 is an established epithelial cell marker that is often highly expressed in epithelial tumors thus representing a potential target in cancer therapy [45,46]. The CD326-negative fraction in most preparations of HCT-116 xenografts was <3% (data not shown). No or only a marginal reduction of the proportion of CD133+ cells was observed in tumors inoculated with CD133+ cells (Fig. 6C). Surprisingly, some tumors generated from the CD133− HCT-116 subpopulation showed an enrichment of CD133+ cells (Fig. 6D). For the individual xenograft tumor #136 for example, the CD133+ subpopulation in the single cell suspension after dissociation was 34% (Fig. 6C). The enrichment, however, was highly variable but was also seen in xenografts derived from original HCT-116 cells (Fig. 6C/D).
Fig. 6.
No difference in subcutaneous tumor formation of CD133+ and CD133− HCT-116 subpopulations. FACS separated HCT-116 cell subpopulations were injected s.c. into NMRI (nu/nu) mice using different cell numbers according to table (A) in experiments 1 and 2. (A) Xenograft tumors were induced in all mice independent of the HCT-116 subpopulation and the number of cells injected. (B) No difference in xenograft tumor growth was detected as shown for the representative set of tumors from experiment 2 after injection of 2.5 × 103 cells. Tumors were dissected at an average diameter of 0.6–0.8 cm. (C) Tumor material was dissociated and analyzed via flow cytometry after multicolor staining for CD133 (PE), CD326 (FITC; to exclude cells that are not of human epithelial origin) and PI (membrane defect cells). Representative CD133/CD326 dot blot diagrams are shown for tumor samples #133 (CD133+ HCT-116 subpopulation injected), #136 (CD133− HCT-116 subpopulation injected) and #141 (originally distributed HCT-116 injected); membrane defect cells were excluded. The average percentage of CD326 negative cells was 3%. (D) The CD133+:CD133− distributions following sort (FACS purity) and in xenografts derived from injection of the various subpopulations (experiment 2) are documented. Xenograft tumors originated from CD133− HCT-116 cell populations imply a potential increase in CD133+ fraction in vivo, whereas tumor cells derived from CD133+ HCT-116 do not change accordingly with respect to their CD133 expression profile.
Discussion
Colorectal cancer (CRC) is one of the three most common causes of cancer-related deaths with more than 40% of these carcinomas being located in the rectum. While surgery constitutes the primary treatment, preoperative 5-FU (5-fluoruracil)-based chemoradiotherapy (CT/RT) is recommended for locally advanced rectal adenocarcinomas according to the CAO/ARO/AIO-94 trial of the German Rectal Cancer Study Group [47]. However, the response of individual tumors to this established therapeutic approach is not uniform, and complete regressions but also resistant tumors are described which may be related to a discrepancy in the presence and survival of CSC/TIC cell populations in the tumors and patients, respectively, leading to an individual risk of recurrence after treatment even post-operative.
To achieve permanent local control after radio(chemo)therapy it is required that all cells with cancer stem cell or tumor-initiating potential are inactivated [48,49]. These CSC/TIC may be characterized by a distinct phenotype associated with particular cell surface markers which may allow to investigate the response of CSC/TIC accumulated cell population to therapeutic interventions. Even if currently controversial, several investigations suggest that CSC/TIC might be more refractory to conventional agents which kill the rapidly dividing cancer cells constituting the majority of the non-CSC/TIC component in solid tumors [2,5,48,50–52]. The identification, isolation and characterization of tumor cell subpopulations with a CSC/TIC phenotype will thus be beneficial if not essential for the development of new and promising therapeutic strategies for many tumor entities.
Various surface markers have been included in recent studies to identify and enrich CSC/TIC from CRC for extended in vitro and in vivo characterization. O’Brien et al. and Ricci-Vitiani et al. independently documented CD133+ enriched cell populations originated from primary tumor material to be more tumorigenic in vivo than the CD133− counterparts [37,38]. CD133 surface expression level was also described in the established HT29 CRC cell line to define cells with a CRC/TIC character [39] although the respective culture conditions were postulated as inappropriate, differentiation-supportive for primary CD133+ CSC/TIC [38]. And a recent retrospective immunohistochemical in situ study claimed the proportion of CD133-expressing tumor cells in CRC as an independent prognostic marker for poor survival [53] supporting our motivation to study tumorigenic potential and therapeutic response of CD133-positive versus CD133-negative CRC cell line populations. With an optimized experimental protocol, we found five out of 10 cell lines to contain two populations with distinct CD133 cell surface expression. These included the cell line HT29 which was already described by others but showed highly variable CD133+ fractions of 2–90% in the different laboratories [39,54]. Because of this discrepancy and to extend the HT29 studies we chose HCT-116 which showed two clearly distinct, reproducible CD133+/− populations in vitro. Our data on HCT-116 do not indicate a difference in tumorigenic potential of CD133+ versus CD133− subpopulations nor did CD133+ HCT-116 cells differ significantly from their CD133− counterparts with respect to any of the parameters analyzed in vitro in 2-D or 3-D culture including sensitivity to single dose irradiation. While chemotherapy testing is not yet completed for the respective HCT-116 subpopulations, our data clearly show that CD133 cannot be regarded as a CSC/TIC marker in all CRC cell lines. We therefore propose that functional measurements of tumor formation have to generally accompany CSC/TIC-directed mechanistic or therapeutic studies based on surface marker identification. Accordingly, CD133 cannot be used as a marker for radioresistance unless tumor initiation potential for the cell line and particular tumor, respectively, has been proven. This also has clear implications for (individualized) clinical research as CD133 may be a marker for CSC/TIC in some but not all CRCs.
It is recognized that tumor-initiation studies should be designed as in vivo limiting dilution assays potentially up to the single cell level to be most informative. In ongoing xenograft formation experiments in NMRI (nu/nu) mice, we have reduced the injected HCT-116 cell numbers already to 5 × 102 CD133+ and CD133− cells per mouse and still find tumor formation in 100% of the animals (data not shown). These data are indicative since previous CRC studies documented a significant difference in tumor formation capacity of CSC/TIC enriched versus CSC/TIC depleted tumor cell populations at cell numbers as high as (0.5–1) × 104 tumor cells injected per animal independent of the different animal models applied ranging from subcutaneous or renal capsule injection to the use of NOD/SCID, SCID or Balb/c nude mice [37–39,55,56]. However, the discussion of our observations in light of these literature data is much more complicated taking into account that some mouse models such as the classical NOD/SCID xenotransplantation approach may potentially underestimate the frequency of tumorigenic cells as recently indicated in a sophisticated melanoma study using various NOD/SCID mouse strains with different levels of immuno-suppression and injection protocols with vehicle versus matrigel-supplemented cell suspensions [57,58]. Thus, not only the performance and interpretation of limiting dilution experiments are challenging but also there is a clear need for verification of the divergent tumor formation capacity of CD133+ and CD133− HT29 cells at least in the NMRI (nu/nu) mouse model for direct correlation with the HCT-116 data presented herein.
The different behavior of CD133+/− cell populations from HT29 as opposed to HCT-116 cultures may, however, also relate to the two different carcinogenesis pathways reflected by the cell lines. HT29 cells are aneuploid with a chromosome instable (CIN) phenotype [59] whereas HCT-116 are near diploid, microsatellite instable (MSI). The relevance of CD133 as a marker for CSC/TIC might differ in these two types. Interestingly, the two CD133− cell lines LS174T and HCT15 also exhibit an MSI phenotype [59] and are known to produce xenograft tumors supporting our conclusion of CD133 not to define the CSC/TIC phenotype in all CRC cell lines. Extended studies with more cell lines are needed to prove the MSI/CIN hypothesis. Also, implementation of primary material and cells directly isolated from CRC is recommended as cell line model systems have been cultured in vitro for a prolonged time and may have accumulated genetic alterations affecting the expression and down-stream signaling of surface antigens associated with a CSC/TIC phenotype.
CD44 (hyaluronic acid receptor) has been analyzed in recent CD133-related CRC cell line studies [54,56,60] as one of the additional surface markers in the focus of CSC/TIC research. In one of these studies, CD44/CD133 multicolor labeling was also utilized to more precisely confine the CSC/TIC population in primary CRC [56]. CD44+/EpCAMhigh cells within the CD133+ subpopulation originated from primary CRC material have been suggested to be enriched for CSC/TIC as compared to the total CD133+ subpopulation [55]. Dalerba et al. described CD44 expression profile together with other surface proteins, i.e. CD166 (Activated leukocyte cell adhesion molecule, ALCAM), to be more informative and thus a more robust feature in colorectal CSC/TIC [55], a finding that was recently confirmed in extended studies and by others using xenografted cells from primary CRC and/or CD133− negative cell lines [61,62]. Another surface protein that might be referred to is CD24 (heat stable antigen) as CD133+/CD24+ sorted CRC cells revealed a higher potential to self-renew and reconstitute a complete and differentiated carcinoma [63]. The HCT-116 cell line used in the present investigation does not have subpopulations that differ in their CD44 or CD24 presentation; all HCT-116 cells expressed CD44 whereas no CD24 cell surface presentation was found (Kunz-Schughart, unpublished). Sorting according to the expression of these cell surface antigens was therefore not possible.
Various technical difficulties and considerations both when using flow cytometric analyses and sorting strategies as well as immunohistology contribute to the controversial discussion on CD133 in CSC/TIC in CRC [17,64]. The antibody used for flow cytometry recognizes a glycosylated extracellular epitope but may also stain cells carrying a truncated CD133 protein; it does, however, not bind sufficiently to membrane-located non-glycosylated forms. Also, the staining protocol for viable cell separation is not useful to determine intracellular CD133 protein. The antibody applied herein for Western blotting was developed against a peptide of the intracellular C-terminus of CD133 and thus binds to an intracellular domain. The combination of both approaches allowed us to clearly verify lack of CD133 protein in the CD133− sorted HCT-116 and also in cell lines defined as CD133-negative leading to the conclusion that these cells do not show different processing of the protein or defects in membrane presentation. However, flow cytometric and Western blot results differed for two of the 10 cell lines studied (DLD1, HCC2998). For these lines no signal was detected in Western blotting in contrast to flow cytometry. This discrepancy has to be further explored as it may result from the expression and presentation of truncated, phosphorylated or otherwise modified CD133 protein leading to supposedly false positive or negative results which certainly alleviates its marker potential [17,65].
The experimental setup to evaluate CD133 surface presentation in HCT-116 cells included monitoring of the distribution of CD133+/− cell fractions throughout culturing and in xenografts. Interestingly, in contrast to monolayer and spheroid cultures analyzed over a period of 14 days, xenografts derived from sorted cell populations showed CD133+/CD133− distributions that differed more clearly from the originally injected population. In particular, some tumors originated from CD133− sorted HCT-116 contained an enhanced CD133+ cell fraction. This might be due to some enrichment of contaminating CD133+ cells (<2% in the original fraction) which would require a growth advantage of this small population or it may result from restoration of CD133 expression under (patho-) physiological in vivo conditions that are not or insufficiently reflected in the short-time in vitro models. Experiments in brain tumors for example imply that oxygen level impacts the distribution of CD133+/− fractions [66,67]. Reduction of oxygen availability to physiological tissue levels was found to enhance the CD133+ cell pool in glioblastoma cell cultures [67]. In medulloblastoma, a CD133+ CSC niche adjacent to blood vessels was discussed and the oxygen concentration was also shown to affect the distribution of CD133+/− subpopulations in vitro [67,68]. Exploring the impact of physiological and pathophysiological parameters on the expression of putative surface markers for CSC/TIC is thus another important issue for future research. From our data we conclude that the cell surface marker CD133 does not define a spheroid-forming, tumor-initiating or radioresistant cell populations in the CRC cell line HCT-116.
Acknowledgements
The authors gratefully acknowledge the excellent technical assistance of Ms. Marit Wondrak and Ms. Melanie Huether. This work was supported in part by the German Federal Ministry of Education and Research (BMBF) through Grant 01ZZ0502 and the German Research Foundation (DFG) through Grants KU 971/7-1 and GR 3376/2-1, and the KFO 179. OncoRay is funded by the BMBF in the program “Center for Innovation Competence”.
References
- [1].Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000;100:57–70. [DOI] [PubMed] [Google Scholar]
- [2].Clarke MF et al. Cancer stem cells – perspectives on current status and future directions: AACR workshop on cancer stem cells. Cancer Res 2006;66:9339–44. [DOI] [PubMed] [Google Scholar]
- [3].Neuzil J et al. Tumour-initiating cells vs. cancer ‘stem’ cells and CD133: what’s in the name? Biochem Biophys Res Commun 2007;355:855–9. [DOI] [PubMed] [Google Scholar]
- [4].Shipitsin M, Polyak K. The cancer stem cell hypothesis: in search of definitions, markers, and relevance. Lab Invest 2008;88:459–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer 2008;8:755–68. [DOI] [PubMed] [Google Scholar]
- [6].Wicha MS, Liu S, Dontu G. Cancer stem cells: an old idea – a paradigm shift. Cancer Res 2006;66:1883–90 [discussion 1895–6]. [DOI] [PubMed] [Google Scholar]
- [7].Woodward WA, Sulman EP. Cancer stem cells: markers or biomarkers? Cancer Metastasis Rev 2008;27:459–70. [DOI] [PubMed] [Google Scholar]
- [8].Ricci-Vitiani L et al. Colon cancer stem cells. Gut 2008;57:538–48. [DOI] [PubMed] [Google Scholar]
- [9].Fabian A et al. Die hard: are cancer stem cells the Bruce Willises of tumor biology? Cytometry A 2009;75:67–74. [DOI] [PubMed] [Google Scholar]
- [10].Corbeil D et al. Prominin: a story of cholesterol, plasma membrane protrusions and human pathology. Traffic 2001;2:82–91. [DOI] [PubMed] [Google Scholar]
- [11].Miraglia S et al. A novel five-transmembrane hematopoietic stem cell antigen: isolation, characterization, and molecular cloning. Blood 1997;90: 5013–21. [PubMed] [Google Scholar]
- [12].Shmelkov SV et al. Alternative promoters regulate transcription of the gene that encodes stem cell surface protein AC133. Blood 2004;103:2055–61. [DOI] [PubMed] [Google Scholar]
- [13].Jaszai J et al. Focus on molecules: prominin-1 (CD133). Exp Eye Res 2007;85:585–6. [DOI] [PubMed] [Google Scholar]
- [14].Maw MA et al. A frameshift mutation in prominin (mouse)-like 1 causes human retinal degeneration. Hum Mol Genet 2000;9:27–34. [DOI] [PubMed] [Google Scholar]
- [15].Roper K, Corbeil D, Huttner WB. Retention of prominin in microvilli reveals distinct cholesterol-based lipid micro-domains in the apical plasma membrane. Nat Cell Biol 2000;2:582–92. [DOI] [PubMed] [Google Scholar]
- [16].Mizrak D, Brittan M, Alison MR. CD133: molecule of the moment. J Pathol 2008;214:3–9. [DOI] [PubMed] [Google Scholar]
- [17].Bidlingmaier S, Zhu X, Liu B. The utility and limitations of glycosylated human CD133 epitopes in defining cancer stem cells. J Mol Med 2008;86:1025–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bao S et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006;444:756–60. [DOI] [PubMed] [Google Scholar]
- [19].Liu G et al. Analysis of gene expression, chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer 2006;5:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Pfenninger CV et al. CD133 is not present on neurogenic astrocytes in the adult subventricular zone, but on embryonic neural stem cells, ependymal cells, and glioblastoma cells. Cancer Res 2007;67:5727–36. [DOI] [PubMed] [Google Scholar]
- [21].Singh SK et al. Identification of human brain tumour initiating cells. Nature 2004;432:396–401. [DOI] [PubMed] [Google Scholar]
- [22].Singh SK et al. Identification of a cancer stem cell in human brain tumors. Cancer Res 2003;63:5821–8. [PubMed] [Google Scholar]
- [23].Yi L et al. Isolation and characterization of stem cell-like precursor cells from primary human anaplastic oligoastrocytoma. Mod Pathol 2007;20:1061–8. [DOI] [PubMed] [Google Scholar]
- [24].Pallini R et al. Cancer stem cell analysis and clinical outcome in patients with glioblastoma multiforme. Clin Cancer Res 2008;14:8205–12. [DOI] [PubMed] [Google Scholar]
- [25].Shu Q et al. Direct orthotopic transplantation of fresh surgical specimen preserves CD133+ tumor cells in clinically relevant mouse models of medulloblastoma and glioma. Stem Cells 2008;26:1414–24. [DOI] [PubMed] [Google Scholar]
- [26].Monzani E et al. Melanoma contains CD133 and ABCG2 positive cells with enhanced tumourigenic potential. Eur J Cancer 2007;43:935–46. [DOI] [PubMed] [Google Scholar]
- [27].Eramo A et al. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ 2008;15:504–14. [DOI] [PubMed] [Google Scholar]
- [28].Bruno S et al. CD133+ renal progenitor cells contribute to tumor angiogenesis. Am J Pathol 2006;169:2223–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ferrandina G et al. Expression of CD133–1 and CD133–2 in ovarian cancer. Int J Gynecol Cancer 2008;18:506–14. [DOI] [PubMed] [Google Scholar]
- [30].Baba T et al. Epigenetic regulation of CD133 and tumorigenicity of CD133+ ovarian cancer cells. Oncogene 2009;28:209–18. [DOI] [PubMed] [Google Scholar]
- [31].Richardson GD et al. CD133, a novel marker for human prostatic epithelial stem cells. J Cell Sci 2004;117:3539–45. [DOI] [PubMed] [Google Scholar]
- [32].Collins AT et al. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res 2005;65:10946–51. [DOI] [PubMed] [Google Scholar]
- [33].Ma S et al. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology 2007;132:2542–56. [DOI] [PubMed] [Google Scholar]
- [34].Suetsugu A et al. Characterization of CD133+ hepatocellular carcinoma cells as cancer stem/progenitor cells. Biochem Biophys Res Commun 2006;351:820–4. [DOI] [PubMed] [Google Scholar]
- [35].Yin S et al. CD133 positive hepatocellular carcinoma cells possess high capacity for tumorigenicity. Int J Cancer 2007;120:1444–50. [DOI] [PubMed] [Google Scholar]
- [36].Hermann PC et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell 2007;1:313–23. [DOI] [PubMed] [Google Scholar]
- [37].O’Brien CA et al. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 2007;445:106–10. [DOI] [PubMed] [Google Scholar]
- [38].Ricci-Vitiani L et al. Identification and expansion of human colon-cancer-initiating cells. Nature 2007;445:111–5. [DOI] [PubMed] [Google Scholar]
- [39].Ieta K et al. Biological and genetic characteristics of tumor-initiating cells in colon cancer. Ann Surg Oncol 2008;15:638–48. [DOI] [PubMed] [Google Scholar]
- [40].Friedrich J et al. A reliable tool to determine cell viability in complex 3-d culture: the acid phosphatase assay. J Biomol Screen 2007;12:925–37. [DOI] [PubMed] [Google Scholar]
- [41].Friedrich J et al. Spheroid-based drug screen: considerations and practical approach. Nat Protoc 2009;4:309–24. [DOI] [PubMed] [Google Scholar]
- [42].Ghadimi BM et al. Centrosome amplification and instability occurs exclusively in aneuploid, but not in diploid colorectal cancer cell lines, and correlates with numerical chromosomal aberrations. Genes Chromosomes Cancer 2000;27:183–90. [PMC free article] [PubMed] [Google Scholar]
- [43].Carlsson J, Yuhas JM. Liquid-overlay culture of cellular spheroids. Recent Results Cancer Res 1984;95:1–23. [DOI] [PubMed] [Google Scholar]
- [44].Kunz-Schughart LA, Mueller-Klieser W, Masters JRW. Three-dimensional culture In: Animal cell culture. Oxford: Oxford University Press; 2000. p. 123–48. [Google Scholar]
- [45].Trzpis M et al. Epithelial cell adhesion molecule: more than a carcinoma marker and adhesion molecule. Am J Pathol 2007;171:386–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Baeuerle PA, Gires O. EpCAM (CD326) finding its role in cancer. Br J Cancer 2007;96:417–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Sauer R et al. Preoperative versus postoperative chemoradiotherapy for rectal cancer. N Engl J Med 2004;351:1731–40. [DOI] [PubMed] [Google Scholar]
- [48].Baumann M, Krause M, Hill R. Exploring the role of cancer stem cells in radioresistance. Nat Rev Cancer 2008;8:545–54. [DOI] [PubMed] [Google Scholar]
- [49].Baumann M et al. Cancer stem cells and radiotherapy. Int J Radiat Biol 2009:1–12. [DOI] [PubMed] [Google Scholar]
- [50].Donnenberg VS, Landreneau RJ, Donnenberg AD. Tumorigenic stem and progenitor cells: implications for the therapeutic index of anti-cancer agents. J Control Release 2007;122:385–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Giordano A et al. Carcinogenesis and environment: the cancer stem cell hypothesis and implications for the development of novel therapeutics and diagnostics. Front Biosci 2007;12:3475–82. [DOI] [PubMed] [Google Scholar]
- [52].Ishii H et al. Cancer stem cells and chemoradiation resistance. Cancer Sci 2008;99:1871–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Horst D et al. CD133 expression is an independent prognostic marker for low survival in colorectal cancer. Br J Cancer 2008;99:1285–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Dallas NA et al. Chemoresistant colorectal cancer cells, the cancer stem cell phenotype, and increased sensitivity to insulin-like growth factor-I receptor inhibition. Cancer Res 2009;69:1951–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Dalerba P et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci USA 2007;104:10158–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Haraguchi N et al. CD133(+)CD44 (+) population efficiently enriches colon cancer initiating cells. Ann Surg Oncol 2008. [DOI] [PubMed] [Google Scholar]
- [57].Quintana E et al. Efficient tumour formation by single human melanoma cells. Nature 2008;456:593–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Eaves CJ. Cancer stem cells: here, there, everywhere? Nature 2008;456:581–2. [DOI] [PubMed] [Google Scholar]
- [59].Kleivi K et al. Genome signatures of colon carcinoma cell lines. Cancer Genet Cytogenet 2004;155:119–31. [DOI] [PubMed] [Google Scholar]
- [60].Ferrand A et al. Expression of gastrin precursors by CD133-positive colorectal cancer cells is crucial for tumour growth. Biochim Biophys Acta 2009;1793:477–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Dylla SJ et al. Colorectal cancer stem cells are enriched in xenogeneic tumors following chemotherapy. PLoS ONE 2008;3:e2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Chu P et al. Characterization of a subpopulation of colon cancer cells with stem cell-like properties. Int J Cancer 2009;124:1312–21. [DOI] [PubMed] [Google Scholar]
- [63].Vermeulen L et al. Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc Natl Acad Sci USA 2008;105:13427–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].LaBarge MA, Bissell MJ. Is CD133 a marker of metastatic colon cancer stem cells? J Clin Invest 2008;118:2021–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Boivin D et al. The stem cell marker CD133 (prominin-1) is phosphorylated on cytoplasmic tyrosine-828 and tyrosine-852 by Src and Fyn tyrosine kinases. Biochemistry 2009. [DOI] [PubMed] [Google Scholar]
- [66].Platet N et al. Influence of oxygen tension on CD133 phenotype in human glioma cell cultures. Cancer Lett 2007;258:286–90. [DOI] [PubMed] [Google Scholar]
- [67].Blazek ER, Foutch JL, Maki G. Daoy medulloblastoma cells that express CD133 are radioresistant relative to CD133− cells, and the CD133+ sector is enlarged by hypoxia. Int J Radiat Oncol Biol Phys 2007;67:1–5. [DOI] [PubMed] [Google Scholar]
- [68].Hambardzumyan D, Squatrito M, Holland EC. Radiation resistance and stem-like cells in brain tumors. Cancer Cell 2006;10:454–6. [DOI] [PubMed] [Google Scholar]