SUMMARY
HIV-1 Rev protein mediates nuclear export of un-spliced and partially spliced viral RNAs for production of viral genomes and structural proteins. Rev assembles on a 351-nt Rev response element (RRE) within viral transcripts and recruits host export machinery. Small (<40-nt) RNA aptamers that compete with the RRE for Rev binding inhibit HIV-1 viral replication. We determined the X-ray crystal structure of a potential anti-HIV-1 aptamer that binds Rev with high affinity (Kd = 5.9 nM). The aptamer is structurally similar to the RRE high-affinity site but forms additional contacts with Rev unique to its sequence. Exposed bases of the aptamer interleave with the guanidinium groups of two arginines of Rev, forming stacking interactions and hydrogen bonds. The aptamer also obstructs an oligomerization interface of Rev, blocking Rev self-assembly. We propose that this aptamer can inhibit HIV-1 replication by interfering with Rev-RRE, Rev-Rev, and possibly Rev-host protein interactions.
Graphical Abstract
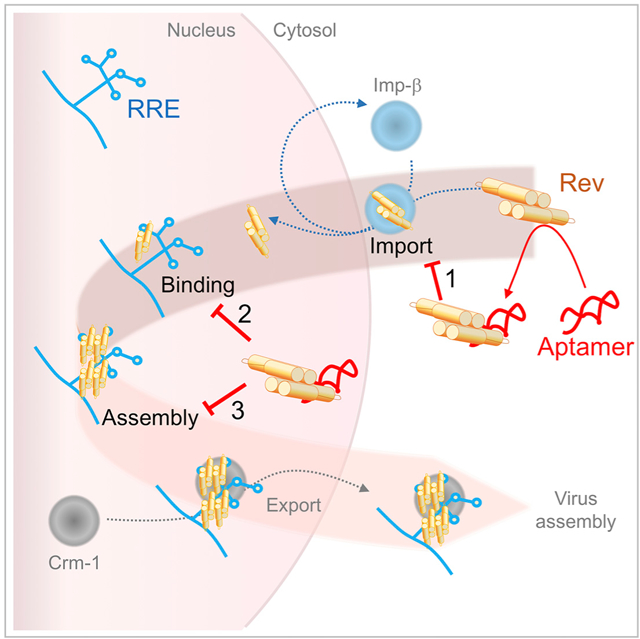
In Brief
With age, people living with HIV-1 accrue co-morbidities that may affect anti-retroviral drug tolerance. Also, as anti-retroviral drugs are used, resistant viral strains enter circulation, making the development of additional anti-retroviral drugs desirable. We have determined the structure of a potential RNA-based HIV-1 inhibitor, bound to its target: HIV-1 Rev.
INTRODUCTION
On a global scale, more than 36 million people are living with an HIV infection (UNAIDS, 2016). Of the main group subtypes, A–K, subtype C is responsible for half of these cases but subtype B is overwhelmingly responsible for cases in North America (Buonaguro et al., 2007). With early diagnosis and combinatorial drug therapy, near-normal life expectancy can be achieved (Anti-retroviral Therapy Cohort Collaboration, 2017). Treatment using three anti-retroviral drugs, inhibiting at least two steps in the viral life cycle, drastically reduces the likelihood that drug resistance will arise (AIDSinfo, 2017). The development of additional therapeutic options would help curtail off-target effects, particularly as HIV-infected people accrue co-morbidities with advancing age (Arhel and Kirchhoff, 2010). In vitro selected RNA aptamers avoid immunogenicity concerns and are well tolerated at therapeutically relevant concentrations (Bunka et al., 2010). Furthermore, they can have specificities and affinities equivalent to therapeutic antibodies (Bunka et al., 2010), suggesting that the development of anti-viral aptamers could be an attractive approach to meet this medical challenge.
HIV-1 Rev (116 residues, 13 kDa) is an essential regulatory protein expressed early in viral replication (Sodroski et al., 1986). Rev accumulates in the nucleus due to its nuclear localization sequence (NLS). In the nucleus, 5–13 copies of the protein assemble on the Rev response element (RRE) (Rausch and Le Grice, 2015), an RNA structure located in an intron that coincides with the bulk of the gene encoding the envelope protein (Sodroski et al., 1986). Rev self-assembly on the RRE is critical for virus replication (Pollard and Malim, 1998). This complex then engages the Crm1-exportin system to ferry RRE-containing viral transcripts to the cytoplasm (Malim et al., 1988). From these transcripts, structural proteins are expressed and progeny virus particles are assembled.
In solution, Rev is a dimeric protein that rapidly forms tetrameric assemblies, which slowly associate into filaments (DiMattia et al., 2016). The crystal structure of the Rev dimer was previously solved by blocking self-assembly with a high-affinity Fab fragment (DiMattia et al., 2010). The N-terminal half of the protein forms an α-helical hairpin (Figures 1 and S1), whereas the C-terminal half, which includes the nuclear-export signal (NES), was not observed in electron density maps, suggesting that it is disordered in protein crystals (DiMattia et al., 2010). However, for polymeric Rev to assemble, the C-terminal 23 residues are required (Zhuang et al., 2014). Furthermore, cryoelectron microscopy (cryo-EM) reconstructions of tubular Rev filaments showed a partially resolved C-terminal domain, indicating that it is transiently ordered (DiMattia et al., 2016). Rev self-assembles through reciprocal interaction of A, B, and C surfaces in the N-terminal half of the protein (Figure 1). While the A and B surfaces occupy opposite sides of the oligomerization domain (Figure 1A), the C surface is formed by the poly-proline motif that accommodates the length disparity between the two helices (Figures 1B and S1). Rotational freedom at the A-A, B-B, and C-C interfaces permits Rev to form dimers with various crossing angles and filaments of variable diameter (Daugherty et al., 2010; DiMattia et al., 2016; Jayaraman et al., 2014; Watts et al., 2018). Rev binds to at least two sites on the RRE, using an arginine-rich motif (ARM) that coincides with the NLS (Figures 1 and S1) (Fang et al., 2013). Based on the A shape of the RRE, it was proposed that each subunit of dimeric Rev separately engages the two RNA-binding sites (IIB and IA) (Fang et al., 2013). This model was later modified to one wherein each of the two sites engages a dimer of Rev with the C-C interface bridging and stabilizing a tetrameric Rev:RRE complex (DiMattia et al., 2016). Once this checkpoint on Rev assembly has been met, additional Rev dimers are thought to add, unmasking the NES for recruitment of host nuclear-export factors (Neville et al., 1997; Watts et al., 2018).
Figure 1. Schematic Representations of Rev.
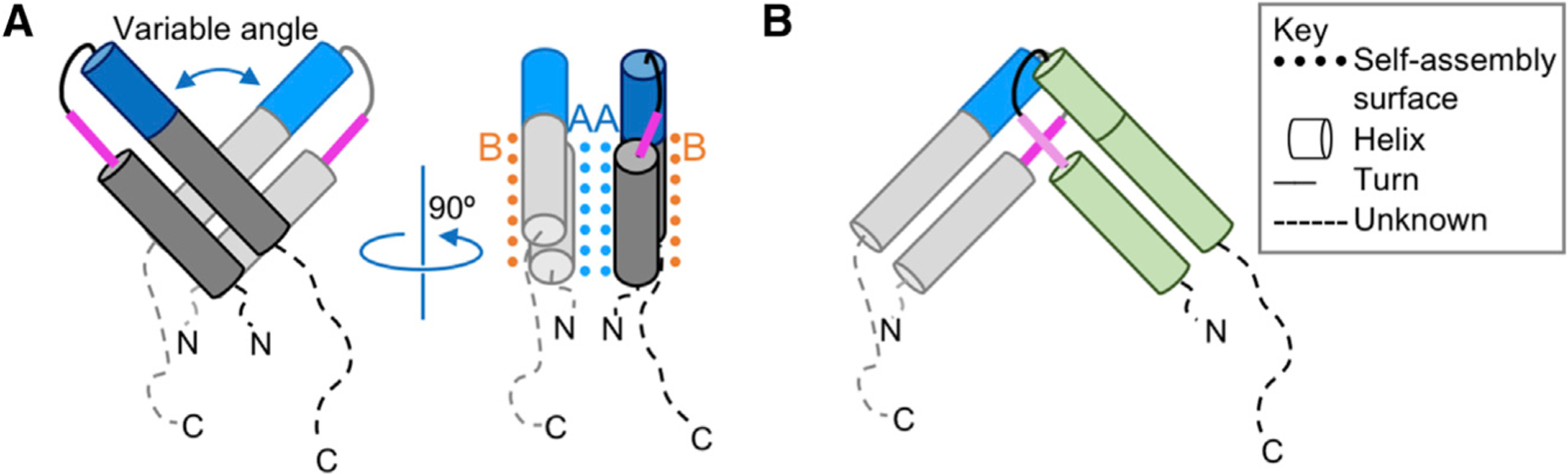
(A) An A-A dimer of Rev with A and B-surfaces (blue and orange, respectively). The ARM (blue) and proline-rich motif (pink) are indicated.
(B) A C-C dimer of Rev involves the proline-rich motif (pink, dark pink).
The binding and efficacy of RNA-based Rev inhibitors has been studied previously, although the mechanism of binding in the context of folded Rev remains unclear. A series of Selex-derived RNA aptamers (35–41 nt) were developed and using gel-retardation assays shown to have up to 10-fold higher affinity for Rev than a 234-nt fragment of RRE (Giver et al., 1993). The aptamers have a widened major groove, a CUC-GAG stretch of duplex, and an adjacent bulge of two to three bases with a penultimate uracil (Figure S2A, dashed lines, blue box and bold, respectively) (Giver et al., 1993). Similarly, Stem IIB has a widened major groove and CGC-GCG duplex in equivalent locations (Figure S2B, dashed lines and orange box, respectively) but no equivalent bulge (Jayaraman et al., 2014). Nuclear expression of several of these competitors was shown to reduce late viral protein expression by 60%– 90% (Good et al., 1997). These RNA aptamers represent the first examples of prospective Rev inhibitors and a promising approach to targeted therapeutics. To facilitate structural analysis, a GC clamp was added to Selex aptamer 79.9–14, which had a 5.4-fold higher affinity for Rev than the RRE (Giver et al., 1993; Ye et al., 1996). NMR structural analysis was performed on the modified aptamer, herein referred to as Rev-Binding Aptamer 14 (RBA-14) (Figure S2A), bound to a Rev-derived peptide (residues 34–50) that comprise the ARM (Figure 1A, blue) (Ye et al., 1996). The Rev peptide exhibited conformational shifts when bound to either RBA-14 or another aptamer, indicating that the structural constraints present in the full-Rev molecule will be required to fully decipher interface interactions (Ye et al., 1996, 1999).
In the present study, we determined the X-ray crystal structure of an RNA aptamer (RBA-14), bound to a C-terminally truncated variant of Rev (Rev93), and a single-chain variable fragment antibody (scFv) crystallization chaperone (DiMattia et al., 2016). We observe that the aptamer binds with its major groove to the arginine-rich helix of Rev, making several contacts additional to those formed by the wild-type RNA. When bound, the aptamer would block Rev oligomerization at that surface.
RESULTS
RBA-14 Has High Affinity for Rev
Previous determinations of the Rev-binding affinity to RBA-14 (unmodified) (Giver et al., 1993) and to Stem-loop IIB (Pond et al., 2009) were made using very different experimental approaches (gel retardation or fluorescence resonance energy transfer competition with different lengths of RRE sequence). To make a direct comparison, we employed surface plasmon resonance (SPR). To display the ARM of Rev uniformly, mutant Rev116 (the ligand) was immobilized on the detection chip by thiol linkage to a unique cysteine appended to the C terminus (Table 1 and Figure S1); the intervening CTD serves as a long, flexible linker. Thirty-five-nucleotide RNAs (analytes) were bound to the immobilized ligand (Figures 2 and S2). The SPR data were best fitted to a two-state model (residuals < 20 response units [RU] or ~10% of signal), consistent with a lure-and-lock mechanism common to RNA aptamers (Table S1) (Law et al., 2006). RBA-14 (Figure S2A) binds to Rev with high affinity (Kd = 5.9 nM) (Table S1 and Figure 2A). The 35-nt hairpin with the Stem IIB sequence (Figure S2B) binds to Rev with a similar affinity (Kd = 12.9 nM) (Table S1 and Figure 2B). Because RBA-14 binds Rev with about the same affinity as the high-affinity site on RRE, it is a potential competitive inhibitor, consistent with its anti-HIV activity (Good et al., 1997).
Table 1.
Description of Rev Proteins
| Name | Residue Range | Type | Reference |
|---|---|---|---|
| Rev69 | 1–69 | assembly domain only | Watts et al., 2018 |
| Rev93 | 1–93 | non-filamentous | Zhuang et al., 2014 |
| Rev116 | 1–116 | full-length wild-type |
DiMattia et al., 2010 |
| Rev116(Leu64Ala) | 1–116 | A-surface mutant | DiMattia et al., 2016 |
| Rev70(Leu12Ser,Glu47Ala,Leu60Arg) | 1–70 | A-surface mutant | Jayaraman et al., 2014 |
| Rev116(Pro28Ala) | 1–116 | C-surface mutant | DiMattia et al., 2016 |
| Rev117(Cys85Ala,Cys89Ala,117Cys) | 1–117 | unique, C-terminal cysteine | This work |
Rev without modifier is used for generic statements regarding any Rev variant retaining the property mentioned in that particular statement.
Figure 2. SPR Analysis of Aptamers Binding to Rev.
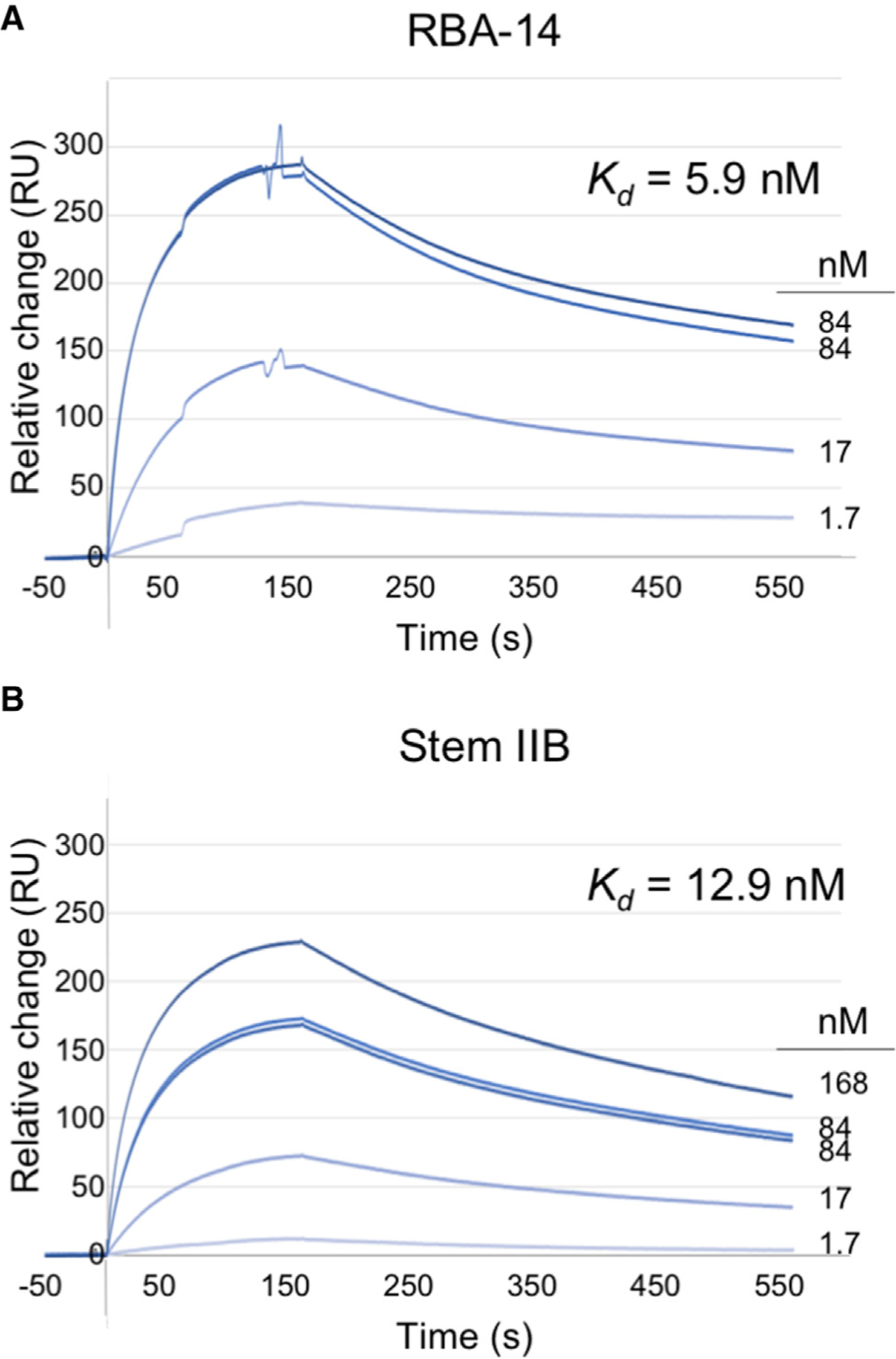
Baseline normalized SPR of (A) RBA-14 and (B) Stem IIB binding to C-terminally thiol-linked Rev117(Cys85Ala, Cys89Ala, 117Cys), residuals <10% of signal. Concentrations range from 0.02 to 3 μg/mL RNA and are labeled by molar concentration for direct comparison with affinities (light dark to blue lines).
Structure of the Rev93:scFv:RBA-14 Ternary Complex
The structure of the Rev116:scFv immune complex showed that the Rev epitope included the B-surface of the N-terminal oligomerization domain and that the nucleic acid-binding ARM/NLS region was unobstructed (DiMattia et al., 2016). Hence, we used Rev:scFv as a crystallization platform for studying nucleic acid binding. We made a ternary complex of Rev93, scFv, and RBA-14, where the single-chain antibody functions as an assembly inhibitor, crystallization chaperone, and initial model for molecular replacement (Table 1). The binding affinity of the aptamer to the Rev93:scFv complex was not determined directly, but it was assumed to be of the same high affinity as established for the full-length Rev (Figure 2). This seems reasonable as the Rev93:scFv:RBA-14 complex was extremely stable during gel filtration and handling, and its structure (especially the interface analysis; Figures 4 and 5 and Table S2) exhibited many of the contacts and interactions of the analogous Rev70:Stem IIBC complex. Crystals grew as three-dimensional (3D) prisms and belong to the I2 space group. The structure was solved to 3.04Å resolution (Rwork 0.19/Rfree 0.26) (Table 2). Within the crystal lattice, the asymmetric unit comprises one Rev93 (gray/blue), one scFv antibody (green), and one hairpin-shaped RBA-14 (pink) (Figure 3A). The Rev A-A interface forms at one of the crystal contacts (Figure S3A) with a dimerization angle of 128.5° (Figure S3B), which is within the reported range of 96°–140° (DiMattia et al., 2016). Also, two RBA-14 molecules stack end to end to form a crystal contact (Figure S3C). RBA-14 has two bulges (bases U23, G24, and A29) (Figure S2A, arrows and Figure 3A, pink), consistent with the NMR structure of RBA-14 bound to an ARM peptide (PDB :1ULL) (Ye et al., 1996). The scFv is composed of a pair of eight-stranded, IgG β-sandwich domains (green) joined by a 20-residue flexible linker (Figure 3A) (DiMattia et al., 2016). Overall, the scFv superimposes with the previous Rev116:scFv crystal structure that lacks RBA-14 (PDB: 5DHV) (DiMattia et al., 2016) with a root-mean-square deviation (RMSD) of 0.67Å (Figure 3A, yellow).
Figure 4. Comparison of Rev Position in the Major Groove of RBA-14 and Stem IIB.
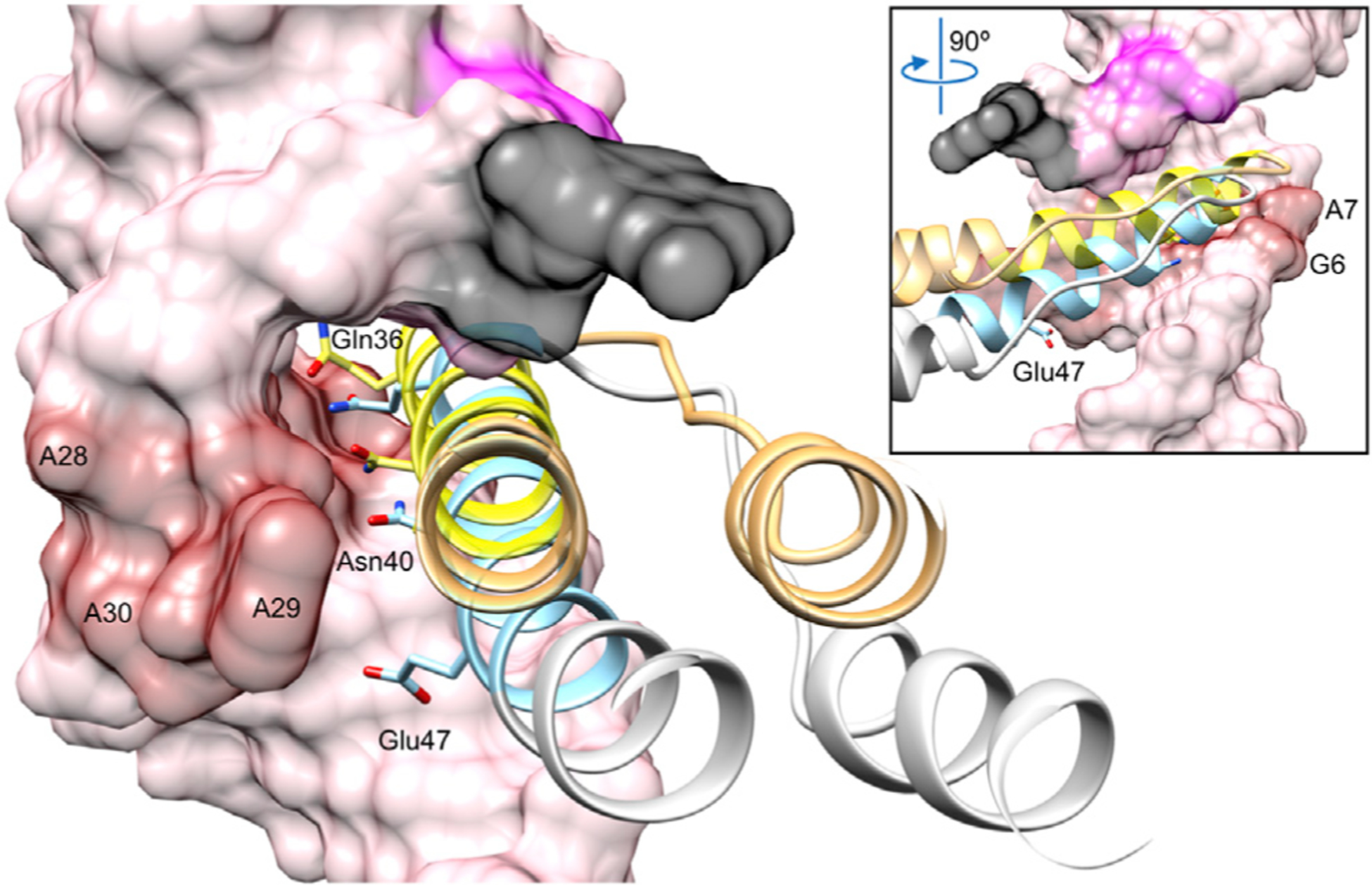
Rev (ribbon) positioned relative to the widened major groove of RBA-14 (surface) and Stem IIB (from PDB: 4PMI, not shown) (Jayaraman et al., 2014) colored blue/gray and yellow/orange, respectively. Purine-purine base pairs of RBA-14 (red surface) were used for alignment. Previously discussed bases are colored as in Figure 3C for relative orientation. Residues that convey Rev specificity, Gln36, Asn40, and Glu/Ala47 occupy one face of the ARM helix (blue and yellow, respectively). Inset view rotated 90°.
Figure 5. Comparison of Rev Bound to RBA-14 and Stem IIB (PDB: 4PMI).
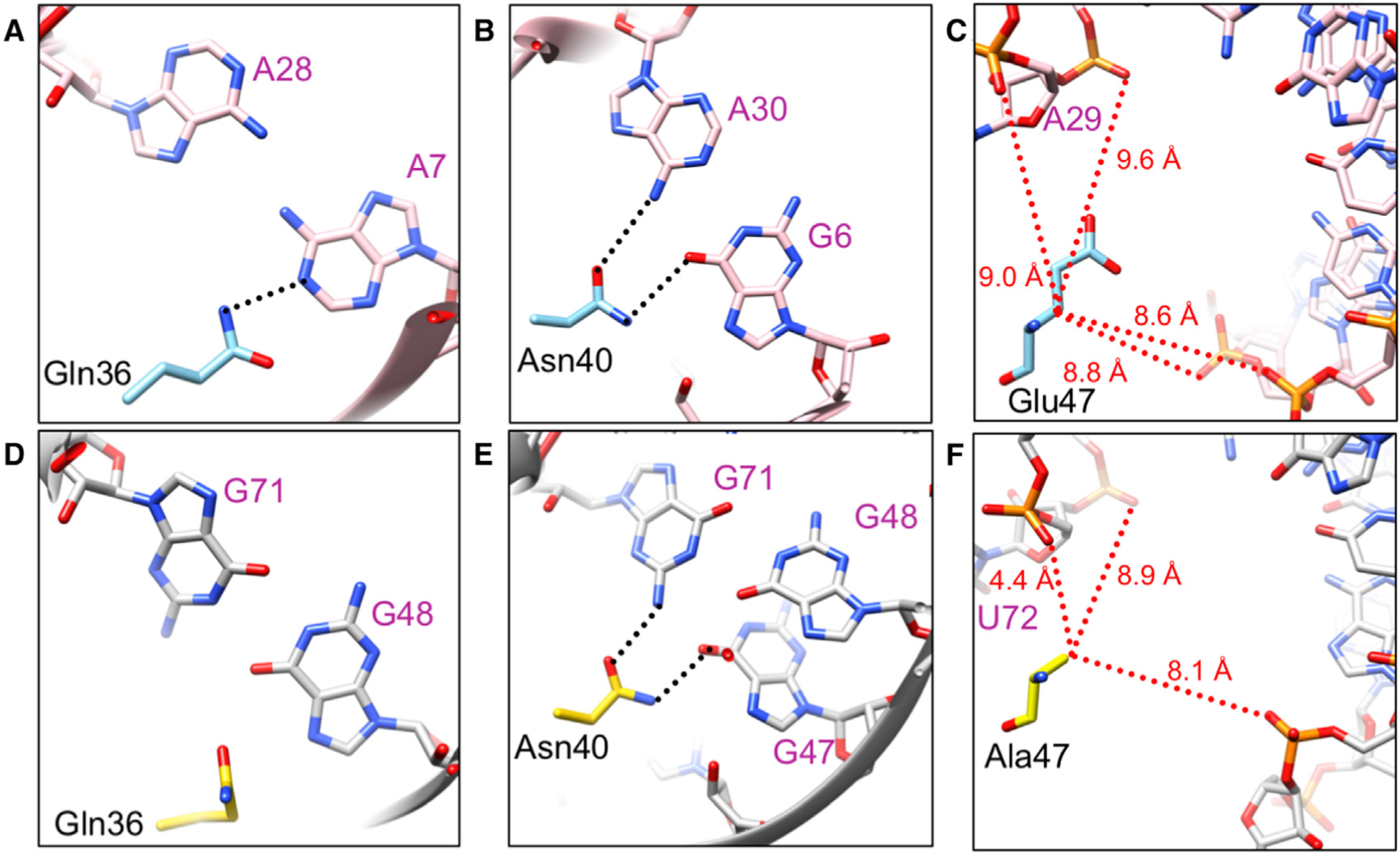
(A–C) RBA-14 (pink) and (D–F) Stem IIB (gray) near residues 36 (A and D), 40 (B and E), and 47 (C and F) of Rev (A–C, blue and D–F, yellow), respectively. Sequence-specific hydrogen bonds (black dotted lines) and distances to Cβ of the residue (red dotted lines) are indicated (Jayaraman et al., 2014).
Table 2.
X-Ray Diffraction Data Collection and Refinement Statistics
| Data Collection | |
|---|---|
| Beamline | Advanced Photon Source 22-ID-D |
| Wavelength | 1.0000 |
| Space group | I2 |
| Cell dimensions | |
| a, b, c (Å) | 64.2, 97.7, 87.8 |
| A, β, ϒ (°) | 90, 109.7, 90 |
| Resolution (Å) | 38.0–3.04 (3.15–3.04)a |
| Multiplicity | 2.0 (2.0) |
| Completeness (%) | 99.1 (98.6) |
| l/σ | 7.4 (1.9) |
| R-meas | 13.5 (79) |
| R-merge | 9.6 (55.8) |
| R-pim | 9.6 (55.8) |
| CC1/2 | 96.5 (50.9) |
| Refinement | |
| Resolution (Å) | 38.0–3.04 |
| No. reflections | 10,171 |
| Reflections used for Rfree (%) | 10 |
| Rwork/Rfree | 0.208/0.241 |
| No. atoms | |
| Protein | 2,185 |
| RNA | 749 |
| B factors | |
| Protein | 56.8 |
| RNA | 65.5 |
| RMSDs | |
| Bond lengths (Å) | 0.004 |
| Bond angles (°) | 0.8 |
| Ramachandran % (favored/allowed/outliers) | 97.1/2.9/0.0 |
Statistics for the highest-resolution shell are shown in parentheses.
Figure 3. The Crystal Structure of the Rev93:scFv:RBA-14 Ternary Complex.
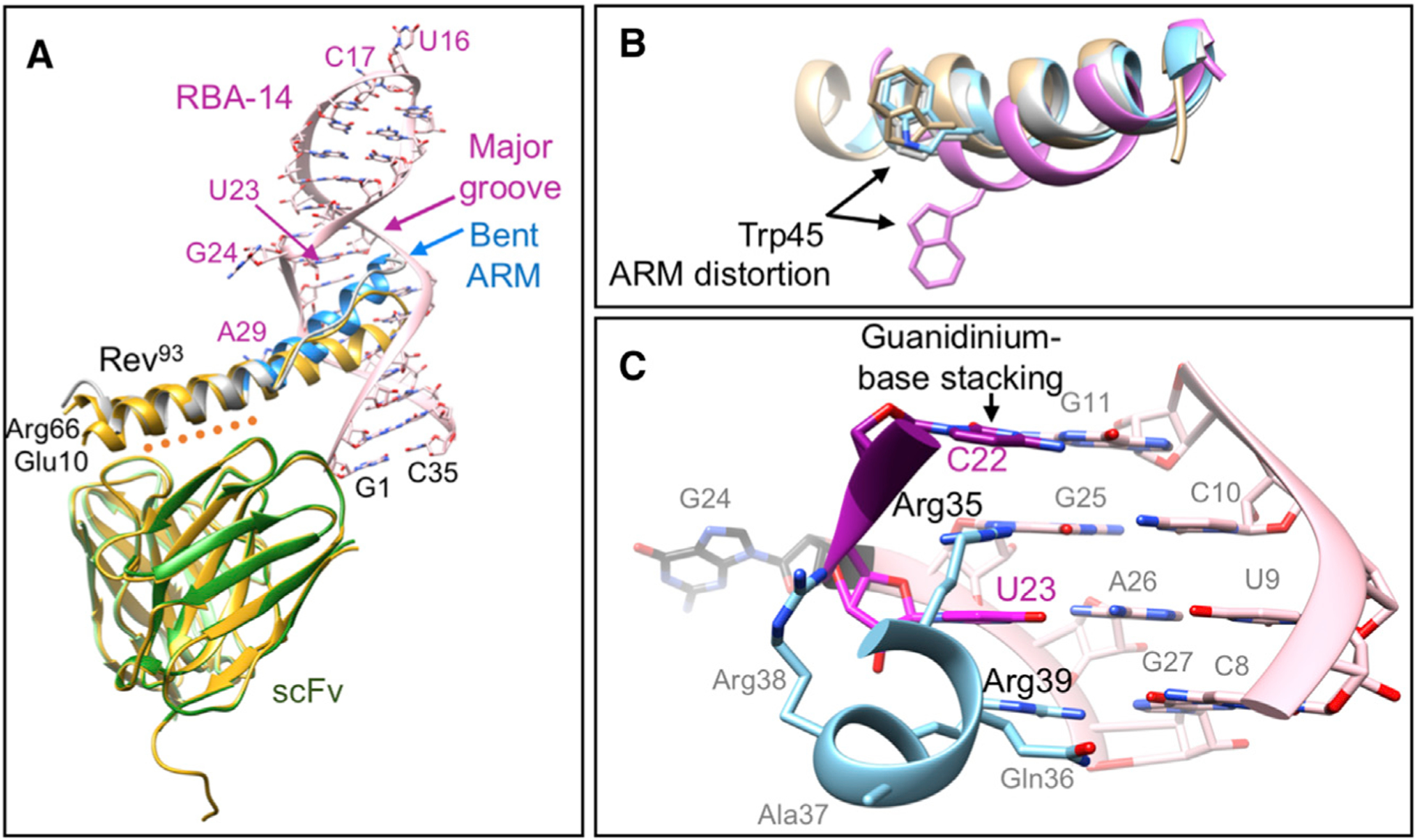
(A) The asymmetric unit with scFv light (light green) and heavy (green) chains interacting with the B-surface of Rev (orange dotted line). A crystal structure of the Rev116:scFv binary complex (PDB: 5DHV) (DiMattia et al., 2016) is superimposed (yellow). Rev93 (gray) with ARM (blue) residue numbers mark the ends of the ordered structure. The ARM is located in the major groove of RBA-14 and is kinked relative to the oligomerization domain. Base numbers mark the ends (black) of RBA-14 (pink) and those bases that are not stacked (dark pink).
(B) Superposition of the ARM (blue) backbone with PDB: 4PMI (gray), 1ETF (tan), and 1ULL (lavender) (Battiste et al., 1996; Jayaraman et al., 2014; Ye et al., 1996) (RMSD = 0.51, 1.02, and 1.86Å, respectively). The side chain of Trp45 is shown for each.
(C) Guanidinium group stacking by Arg35 and Arg39 in the first helical turn (blue) of the ARM caps the exposed bases C22 (purple) and U23 (dark pink) of RBA-14 (pink). An unpaired G24 (black) completes the bulge in RBA-14 (pink).
Only residues 10–66 of Rev93 were ordered in the crystal structure (Figure 3A). Large disordered regions typically preclude crystallization, but that did not occur here or with Rev116 (DiMattia et al., 2010, 2016). Each of these crystals included an antibody, either Fab or scFv, as a crystallization chaperone. Crystallization of Rev without an antibody has succeeded for a truncated Rev69 (Watts et al., 2018) and for a truncated and mutated Rev70(L12S, L60R, E47A) (Daugherty et al., 2010; Jayaraman et al., 2014). The presumption has been that, in addition to eliminating the need for mutation by blocking one of the dimerization surfaces, the antibody promotes sufficient channel volume in the crystal lattice to accommodate the disordered portion of Rev.
The two a helices of Rev, residues 10–26 and 35–66, form a helical hairpin with an extended poly-proline motif compensating for the disparity in helix length (Figure 3A). There is a kink in the second helix of Rev, between the ARM (residues 35–50, blue) and the second half of the oligomerization motif (residues 51–66, gray) that is not present in the aptamer-free structure (Figure 3A, yellow) and is likely due to crystal packing. The oligomerization motifs of the aptamer-bound Rev closely match those of the aptamer-free structure (Figure 3A gray and yellow) with RMSD of 0.70 and 1.80Å, respectively. Due to the kinked helix, the ARM differed, with an RMSD of 4.89Å (Figure 3A blue and yellow). The ARM of Rev binds in the major groove of RBA-14 (Figure 3A) but forms a more regular α helix than in the NMR structure (Ye et al., 1996) where the ARM peptide is distorted after the first helical turn (Figure 3B). This is best illustrated by the relative position of Trp45: RBA-14 distorts the peptide (lavender) in a way not permitted by Rev93 (blue) (Figure 3B). Rather, the protein structure closely matches the ARM in the crystal structure of Stem IIB with a B-B dimer of Rev (PDB: 4PMI) (Jayaraman et al., 2014) and the NMR structure of Stem IIB with an ARM peptide (PDB: 1ETF) (Battiste et al., 1996) (Figure 3B, gray and tan, respectively). This distortion is most likely due to the absence of structural constraints imposed by the protein.
Rev-RBA-14: High-Affinity Interactions and Specificity
The major groove of RBA-14 forms extensive high-affinity interactions with the helically arranged arginine residues of the ARM domain. RBA-14 and other aptamers of its class (Giver et al., 1993) are characterized by a CUC-GAG duplex, adjacent to a 2–3 nt bulge that contains a penultimate uracil. In RBA-14, this bulge is composed of U23 and G24 (Figure 3C, dark pink and black, respectively) and creates a break between stretches of duplex defined on one side by the G11-C22 base pair and on the other by the CUC-GAG duplex (Figure 3C). This break leaves the ring face of C22 (purple) exposed. The first helical turn of the ARM (blue) is bound to this sequence through a series of hydrogen bonds and stacking interactions of Arg35 and Arg39 (Figure 3C). The guanidinium group of Arg35 is stabilized by hydrogen bonds to G25 and to the phosphate backbone, and is positioned to stack with C22. The spacer nucleotide, in this sequence G24, has sufficient flexibility to allow U23 to flip into the major groove and form a Hoogsteen base pair with A26, which is base paired with U9. The guanidinium group of Arg39 (located on the next helical turn) stacks on the remaining face of U23, and Arg38 forms a hydrogen bond with the phosphate backbone (Figure 3C and Table S2). A similar Hoogsteen base pair and stacking of arginines was observed in the binding of HIV-1 Tat to its RNA target (Puglisi et al., 1993), but this feature is absent in the Stem IIB structure (Jayaraman et al., 2014). Additional stacking interactions between Arg44 and U4 and between Arg50 and A29, as well as various arginine residues hydrogen bonding with the phosphate backbone, complete the network of contacts (Table S2).
In order to accommodate the ARM of Rev, with its bulky side chains, the major grooves of both RBA-14 and Stem IIB are widened by two purine-purine base pairs (Figure S2). To compare the relative orientation of Rev in the major grooves of RBA-14 and Stem IIB, the RNA molecules were superimposed, aligning their purine-purine base pairs (Figure S4). The bases in RBA-14 (G6, A7, Figure 4 inset, red surface; and A28, A29, and A30, Figure 4, red surface) were aligned with the bases in Stem 11B (G47, G48, G71, U72, and A73, respectively) (Figure S4). Rev (Figure 4 blue/gray helix), when bound to RBA-14, is shifted and rotated within the major groove relative to Rev (Figure 4 yellow/orange helix), which is bound to Stem IIB.
For anti-HIV aptamers to be useful therapeutically, their interaction with Rev must have elements of specificity to avoid interaction with a broad spectrum of arginine-rich host RNA-binding proteins and consequent cytotoxicity. The residues of the ARM suggested to confer binding specificity on the RRE are Gln36, Asn40, Glu47, and perhaps Gln51 (sequence specificity of Gln51 is only affected by mutation to arginine, indicating a gain of affinity for any RNA attributable to the arginine rather than to the importance of Gln51) (Jain and Belasco, 1996). All these residues occupy one side of the ARM α helix (Figure 4). In our structure, Gln36 and Asn40 form hydrogen bonds with the bases of two sequential purine-purine base pairs, A7:A28 and G6:A30, respectively (Figures 5A and 5B). The acidic residue Glu47 occupies a gap between phosphate backbones (Figure 5C). Subtype-B viral isolates vary at this position; for example, Ala, Gln, Arg, and Lys (Figure S5A). The most common mutation, Ala, reduces affinity (10-fold) for wild-type Stem IIB but increases affinity for mutants Stem IIB (G47A) and Stem IIB (G71U) (Jain and Belasco, 1996). As neither one of these base mutations is proximal and other Glu47 variants are bulky with varying charge, steric accommodation rather than charge appears to be key in maintaining the register of the ARM within the major groove (Figure 5C). Outside of subtype B, a 2-nt mutation is common, encoding Rev (Glu47Ala) as well as Env (Arg740Gln). This mutation correlates with an upstream mutation that is silent in Rev and Env but missense in the stop codon of Tat. Together these mutations give rise to Tat100 with mutations Arg92Ser and Glu93Lys (Figure S5B) and Tat100 increases transactivation and lowers apoptotic activity (Campbell et al., 2005). These mutations confer a considerable change in Tat activity, indicating that the effect on Rev is a consequence of utilizing overlapping coding regions.
In a mutant Rev70 bound to Stem IIB (PDB: 4PMI) (Jayaraman et al., 2014), Gln36, Asn40, and Ala47 interactions are shifted ~1.3Å, approximately half a base pair, relative to the equivalent purine-purine base pairs in the RBA-14-binding site. In Stem IIB, these base pairs are G47:A73 and G48:G71 and the adjacent duplex, CGC-GCG, whereas the equivalent bases in RBA-14 are G6:A30, A7:A28, and CUC-GAG, respectively (Figures S2 and S4). Because of this shift, Gln36 does not make sequence-specific contacts to the Stem IIB RNA (Figure 5D) and Asn40 forms hydrogen bonds to G71 and G47, bridging the two purine-purine base pairs (Figure 5E). This difference in register may be due to the mutation of Glu47 to alanine, ostensibly made to reduce surface entropy but it is common outside of subtype B (Foley et al., 2017; Jayaraman et al., 2014). The Cβ of Ala47 comes as close as 4.4Å to the phosphate backbone of Stem IIB (Figure 5F), whereas the Cβ of Glu47 is more than 8Å from the backbone of RBA-14 (Figure 5C). This difference in register leads to Asn40 forming a hydrogen bond to the G:A base pair (G6:A30 in RBA-14 and G47:A73 in Stem IIB) (Figure 5B), rather than bridging the two purine-purine base pairs (Figure 5E), and is more consistent with its direct contact with A73 of Stem IIB and the 10-fold reduction in affinity for Stem IIB when Glu47 is mutated to Ala (Jain and Belasco, 1996). Although Glu to Ala is the most common mutation at position 47 (Figure S5A) and it increases the affinity of Rev for Stem IIB mutants (Jain and Belasco, 1996), there are no correlated mutations to the Stem IIB among viral isolates. Therefore, this mutation should not be considered a compensatory mutation for changes to the Stem IIB sequence but rather subtype variation in Tat or Env that causes mild, collateral impairment of Rev.
RNA Binding Blocks C-C Dimerization of Rev
The C-C interface of Rev is mediated by the poly-proline motif running parallel to the ARM that binds to RNA, RBA-14 included (Figure 1B, dark pink lines) (DiMattia et al., 2016). We compared a C-C dimer model with our crystal structure to see if RNA binding and C-C interface formation could occur simultaneously. Our ARM structure (blue) was superimposed on one subunit of Rev derived from the cryo-EM reconstruction of Rev filament V (DiMattia et al., 2016) (Figure 6A). Inspection of the C-associated Rev subunit (green) clearly indicates steric hindrance between this subunit and the phosphate backbone of RBA-14 near bases G24 (black) and G25 (purple) (Figure 6A). Hence, a Rev subunit can bind to RNA or form a C-C interface but not both. To test this directly, we studied the reversible, temperature-induced polymerization of Rev, which has the hallmarks of classic protein assembly (Andreu and Timasheff, 1986). Thermal cycling of Rev116 (5°C −35°C) indicates the reversible formation of polymeric protein detected by light scattering with a critical concentration (Cr) of ~11 μM at 30°C (Figures 6B and 6C) (Wingfield et al., 1991). This indicates a specific association process as opposed to random aggregation. It is noteworthy that the Cr (essentially the concentration of tetramer in equilibrium with polymer) is close in value to association constants (Kd ~ 1 μM) describing unlimited isodesmic association of Rev (Cole et al., 1993).
Figure 6. RNA Binding and Formation of the C-C dimer Interface Are Mutually Exclusive.
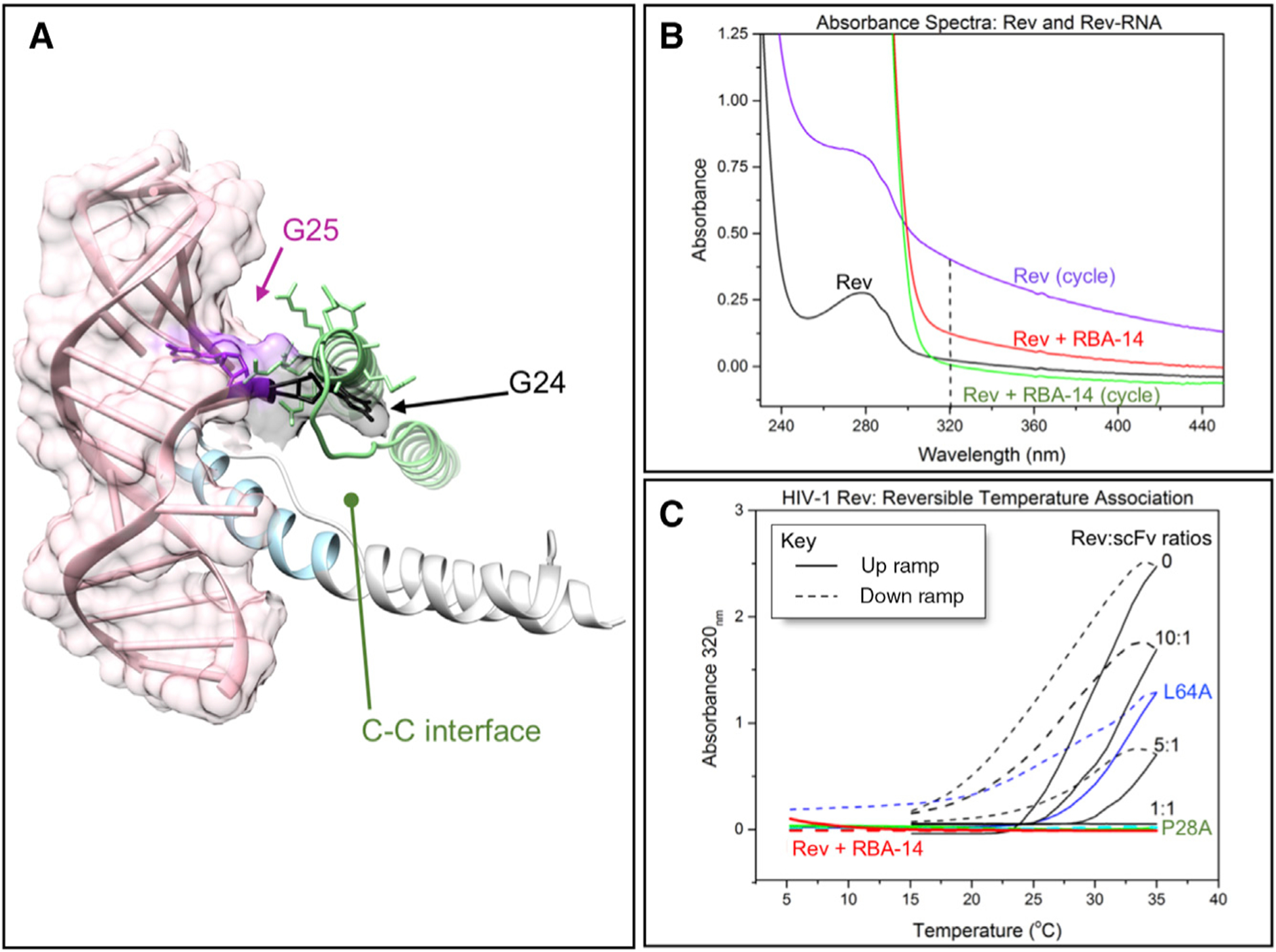
(A) Superposition of Rev (gray/blue) with RBA-14 (pink and surface) crystal structure with the C-associated subunit model (green) from cryo-EM reconstruction (DiMattia et al., 2016). G24 (black) and G25 (purple) clash with the C-associated subunit.
(B and C) Light scattering of thermally induced Rev assembly. (B) UV and visible light absorption spectra of Rev116 before thermally induced assembly and after three cycles between 5°C and 35°C, immediately following addition of RBA-14 to the assembled Rev, and after three additional cycles between 5°C and 35°C. Measured at 5°C, dashed line indicates optimal scattering signal at 320 nm. (C) A320 light scatter of assemblies upon thermal cycling of Rev116 (black), Rev116 with equimolar RBA-14 added (red), Rev (Leu64Ala) an A-surface mutant (blue), Rev116 titrated with various molar ratios of scFv to block the B-surface (black), and Rev (Pro28Ala) a C-surface mutant (green).
The titration of Rev116 with an scFv that targets the B-B interface requires an equimolar ratio to completely block polymerization events (Figure 6C). The residue Leu64 was identified as integral to the resilience of the A-A interface and the mutant Rev116(Leu64Ala) completely blocks formation of Rev filaments (DiMattia et al., 2016). As shown here, however, there is little or no effect on the formation of temperature-induced polymers (Figure 6C). The C-C interface mutant Rev116(Pro28Ala) (DiMattia et al., 2016) had minimal effects on the overall Rev structure but did reduce the propensity for filament formation. However, its effects on the reversible polymer formation are more dramatic, almost completely abrogating the light-scattering response (Figure 6C). Because RBA-14 appears to block the C-C interface (Figure 6A), we hypothesized that the addition of RBA-14 would cause Rev116 to lose its temperature effect similar to the C-C interface mutant Rev116(Pro28Ala). Following three cycles of temperature cycling there is a build-up of high-molecular-weight Rev (Figure 6B), which will slowly dissociate at 20°C (Wingfield et al., 1991). We observed that addition of RBA-14 disassembles previously formed Rev116 polymer (Figure 6B), and the Rev116:RBA-14 complex that is formed exhibits no temperature effects (Figure 6C).
The size distribution of Rev116 and the Rev116:RBA-14 complex were examined by sedimentation velocity. Rev sediments as a main boundary with a sedimentation coefficient (s) of ~5 (~42 kDa), indicating tetrameric association (Figure S6A). The Rev116:RBA-14 complex was resolved into several protein:RNA species with apparent masses of ~60, 130, and >250 kDa (Figure S6B). Although we have not attempted rigorous determination of the protein:RNA stoichiometries of these complexes, the RBA-14 clearly limits the assembly competence of Rev.
Titration of RBA-14 or an equivalent molar quantity of 30-nt single-stranded RNA (ssRNA) over subsequent thermal cycles showed that one molecule of RBA-14 for every 10 molecules of Rev is sufficient to eliminate assemblies that are large enough to detect by turbidity. Even with three heating cycles to generate the most aggregate, the scattering signal drops to the baseline as the amount of RBA-14 is increased (Figure S7A). Titration of Rev with equivalent quantities of ssRNA has no effect and the iterative thermal cycles gradually flatten the scattering signal at a maximal signal, indicating that the Rev assemblies lose reversibility on the timescale of these experiments (Figure S7B). Taken together, these results show that the major groove of RNA obstructs the C-C interface and prevents the unchecked assembly that leads to Rev filament formation.
DISCUSSION
We have solved the structure of a potential anti-HIV-1 RNA aptamer in complex with Rev93:scFv. The Rev binds to the aptamer with a similar high affinity as to the Stem IIB sequence from the RRE (Figure 2). Contributing to the high-affinity binding is the presence of a bulge of 2 or 3 nt, which exposes the face of the adjacent base (Figure 3C). Arginine side chains stack against this face and with the Hoogsteen base from the bulge and form a dovetail interface that is lacking in the Stem IIB site in the RRE (Figure 3C). Arg35 occupies a particularly snug pocket in RBA-14, interleaved between two bases and forming two hydrogen bonds, and may be useful as an affinity hot spot.
The purine-purine base pairing (similar to the Stem IIB site) conveys sequence specificity for Rev and supports the contacts identified between Rev and the Stem IIB sequence by biochemical and mutational studies (Jain and Belasco, 1996). The relative position of the ARM bound to RBA-14 and to Stem IIB (Jayaraman et al., 2014) is shifted (Figures 4 and 5), which may be due to the mutant Rev(Glu47Ala) that was crystallized with Stem IIB (Jayaraman et al., 2014). While subtype B has a glutamate at this position, others, including globally dominant subtype C, use alanine, so comparison of these two structures elucidates subtype-specific differences of RNA binding. This difference could affect the binding affinity and efficacy of Rev inhibitors across subtypes and will need to be explored further. This mutation also affects tat and env, and preference among viral isolates for a particular dinucleotide change indicates selective pressure on those reading frames (Figure S5B).
Aptamer binding to Rev clashes with the C-C oligomerization interface and, thus, blocks protein self-assembly. This was shown directly by titration studies with a Rev mutant and RBA-14 (Figure 6). In order for Rev to recruit host export factors to the RRE it must assemble on the RNA. In one scheme, a dimer of Rev binds to (high-affinity) Stem IIB, a second Rev dimer binds to (lower-affinity) Stem IA, and then the tetrameric assembly is stabilized by dimer-dimer interactions via the C-C interface (DiMattia et al., 2016). Further addition of Rev dimers, essential to form an active export complex (Pollard and Malim, 1998), occurs through protein-protein contacts. If an ARM were proximal to the major groove of the RNA, then the rotational latitude inherent to the protein-protein interfaces can accommodate non-specific RNA binding (Jayaraman et al., 2014). Without RNA, Rev can assemble through these contacts into filaments unlimited in length (Wingfield et al., 1991).
The inhibitory effects of aptamers are likely to be more nuanced than simple competitive binding to RRE, as the nuclear milieu is complex where local concentrations, steric availability, multiple interactions sites, etc., must all come into play. Furthermore, Rev-binding aptamers would also be expected to exert an anti-HIV effect in the cytoplasm. The Rev ARM-aptamer interaction would shield the coincident NLS, thus interfering with the engagement of nuclear import factors such as, for example, importin β (Pollard and Malim, 1998).
We propose that high-affinity aptamers analogous to RBA-14 can inhibit HIV-1 replication by interfering with Rev-RRE, Rev-Rev, and possibly Rev-host protein interactions. Future studies will test these mechanisms by developing cell delivery systems and by following the cellular effects.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Paul T. Wingfield (wingfiep@mail.nih.gov).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All proteins were expressed in E. coli BL21 (DE3) cells (NEB) in Luria broth in a 2 L fermenter (Sartorius Stedim) operated at 37°C.
METHOD DETAILS
Cloning Expression and Purification of Rev Proteins and Antibodies
Recombinant Rev116, Rev93 (amino acids 1–93), Rev116(Leu64Ala), and Rev116(Pro28Ala) were expressed and purified as previously reported (DiMattia et al., 2016; Wingfield et al., 1991; Zhuang et al., 2014). Briefly, the proteins were denatured by 2 M urea and purified by ammonium sulfate precipitation and ion-exchange chromatography. The resulting fractions of purified protein were pooled and stored frozen at −80°C. The plasmid encoding Rev117(Cys85Ala, Cys89Ala, 117Cys) was purchased (Genscript) and was expressed and purified in essentially the same way.
Recombinant, His6-tagged scFv was expressed and purified as previously reported (Stahl et al., 2010). Briefly, the protein was collected from the periplasm by freeze-thaw lysis of the outer membrane and purified by nickel-affinity chromatography in 1x phosphate buffered saline.
Synthetic RNA
Synthetic RNA was purchased (IDTDNA) with standard desalting. The RNA was dissolved in commercial buffer, THE Ambion RNA Storage Solution (ThermoFisher) (1 mM sodium citrate, pH 6.4) to a concentration of 10 mg/mL (839 μM) and was stored frozen in 10-mL aliquots until use. Concentrations were determined by UV/Vis spectrophotometry using a Nanodrop-1000 spectrophotometer (ThermoFisher Scientific) with default RNA settings. For surface plasmon resonance, crystallography, and thermal assembly, the 35-nt RBA-14 aptamer has the sequence: 5’- GGC UGG ACU CGU ACU UCG GUA CUG GAG AAA CAG CC −3’ (Ye et al., 1996). For surface plasmon resonance, the 35-nt Stem IIB aptamer has the sequence: 5’- CCU GGG CGC AGU GUC AUU GAC GCU GAC GGU ACA GG −3’ and represents bases 45–75 of the complete RRE (Foley et al., 2017). This sequence differs from the Stem IIBC RNA reported by Jayaraman and colleagues in that it does not include Stem IIC (Jayaraman et al., 2014). The predicted structures of the RBA-14 and Stem loop 11B complexes, with the lowest δG (−17.17 and −17.67 kcal mol−1, respectively), match the observed crystal structures and have melting temperatures (Tm) of 75.6 and 77.1°C, respectively (IDTDNA OligoAnalyzer 3.1). Because the duplex portion of RBA-14 and Stem IIB are 14- and 15-bp, respectively, the ssRNA used as a control for thermally-induced Rev assembly is comprised of two 15-nt synthetic RNAs with complementary but not reversed sequences 5’- AUG CCU UAG CCA UCG −3’ and 5’-UAC GGA AUC GGU AGC −3’. These give rise to base content equivalent to 15-bp of duplex, but with no propensity for forming dsRNA. The two are predicted to form no more than two basepairs with a dG of −3.61 kcal mol−1, and so are expected to be single stranded (IDTDNA OligoAnalyzer 3.1).
Crystallization of Rev93:scFv:RBA-14
C-terminally truncated HIV-1 Rev93 protein was purified as described above (Wingfield et al., 1991). Protein expression, purification and folding were carried out essentially as described previously for the wild-type protein, using an S-Sepharose column. To prevent oligomerization, Rev93 was kept in 20 mM sodium phosphate, 400 mM sodium chloride and 2 M urea, pH: 7.4. Prior to protein:RNA complex formation, Rev93 was mixed with chimeric rabbit/human scFv SJS-R1 (Stahl et al., 2010) in 1:1 ratio and dialyzed against 20 mM Tris, 50 mM sodium chloride, pH 8.0, overnight at 4°C. Rev93:scFv was concentrated to ~5 mg/mL and RNA aptamer, RBA-14, was added to the concentrated protein complex in 2:1 ratio. Ȉ
Rev93:scFv:RBA-14 initial crystallization trials were set up at 16–18°C in hanging drops containing 200 nL of protein:RNA complex and 200 nL precipitant solution equilibrated against 70 μL of reservoir solution in 96-well plates. 3-D crystals appeared overnight in 2% v/v 1,4-dioxane, 0.1 M Tris, pH 8.0, 15% (w/v) polyethylene glycol (PEG) 3350. Crystals (space group I2) were extremely fragile and lost diffraction upon protection with glycerol, ethylene glycol, various alcohols, sucrose and mineral oil. Subsequently, the crystals were cryo-protected by swiping through corn oil (refractive index 1.7) and were frozen in liquid nitrogen. Data was collected at the Advanced Photon Source (APS, Lemont, IL) beam line 22-ID-D from a single crystal which diffracted to 3.04Å.
Sequence Alignment
Viral isolate sequences were extracted from the AIDS database (Foley et al., 2017), and aligned using Clustal Omega (EMBL-EBI) (Goujon et al., 2010; Sievers et al., 2011).
Surface Plasmon Resonance
Surface plasmon resonance was performed on a Biacore ×100 (GE Healthcare), in 30 μL/minute HBS-EP+ buffer (10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.4, 150 mM sodium chloride, 3 mM ethylenediaminetetraacetic acid, 0.05 % polysorbate 20) (GE Healthcare) at 25°C.
To immobilize Rev116 homogeneously at the C-terminal end of the protein and thereby use the disordered C-terminal half of the protein as a flexible linker, we mutated the native cysteine residues to alanine (at positions 85 and 89) and appended a now unique cysteine to the end of the sequence (at position 117). Rev117(Cys85Ala, Cys89Ala, 117Cys) (0.2 mg/mL in 20 mM sodium phosphate, pH 6, 1 M sodium chloride, 2 mM dithiothreitol, 2 M urea) was concentrated in a 3 kDa MWCO centrifuge concentrator, centrifuged at 3,000 × g, to 1 mL of 1 mg/mL and then desalted in a PD10 column equilibrated with 20 mM sodium phosphate, pH 6.6, 20 mM sodium chloride, 2 M urea). The 0.5 mg/mL eluent was diluted 10-fold in 10 mM sodium acetate, pH 4.8 and immobilized on the experimental cell of a CM5 sensor chip (GE Healthcare) using a thiol coupling kit (GE Healthcare) according to manufacturer instructions.
The experiments shown were performed on the same CM5 chip with an immobilization level of ~8700 response units (RU), using concentrations of RNA analyte 0.02–2 μg/mL (serially diluted in HBS-EP+ buffer) to elicit signals up to ~250 RU. After each injection, the CM5 sensor chip was regenerated with a 30-second injection of 50 mM sodium hydroxide. Ȉ
Thermally-Induced Rev Assembly
Rev116 (1.6 mg/mL in 20 mM sodium phosphate, pH 6.5, 1 M sodium chloride, 1 mM dithiothreitol, 2 M urea) was thawed and dialyzed in 6 kDa MWCO dialysis membrane against buffer (50 mM sodium phosphate, pH 7.4, 150 mM sodium chloride). In the experiment shown, 100 μL of 32 μM Rev116 was thermally cycled. When complete, 3.8 μL of 839 μM or RBA-14 was added directly to the cuvette to generate an equimolar ratio of 31 μM, and was cycled immediately. Rev (Pro28Ala) (0.5 mg/mL in 20 mM sodium phosphate, pH 6.0, 750 mM sodium chloride, 2 mM dithiothreitol, 5 mM ammonium sulfate, 2 M urea) and Rev (Leu64Ala) (2.2 mg/mL in 20 mM sodium phosphate, pH 6.0, 900 mM sodium chloride, 2 M urea) were thawed, dialyzed, diluted, and thermally cycled as above. Titrations (with 50 μM Rev) were performed as above with RNA additions following every third cycle.
Based on previous experiments (Wingfield et al., 1991), thermally-induced assembly of Rev was observed in a 100 μL-capacity quartz cuvette by light scatter at 320 nm in an HP G1103A (Hewlett Packard) spectrophotometer equipped with an HP 89090A (Hewlett Packard) Peltier and a custom nitrogen jacket to control temperature and condensation, respectively. Automated acquisition was programed to execute three consecutive cycles. In each cycle, the cuvette was heated from 5 to 35°C and then cooled to 5°C at a rate of approximately 1°C/minute. A 10-minute hold step was approximated by heating then cooling the cuvette from 5 to 6°C and back at a rate of 0.2°C/minute. The resulting curves were plotted in Origin (OriginLab, Northampton, MA).
Analytical Ultracentrifugation
A Beckman Optima XL-1 analytical ultracentrifuge, absorption optics, an An-60 Ti rotor, and standard, double-sector centerpiece cells were used. Sedimentation velocity runs were made at 15°C at 40,000 rpm. Absorbance scans at 260 and 280 were performed every 8 minutes for 3 hours. Protein and RNA samples were in 50 mM sodium phosphate, pH 7.5, 150 mM sodium chloride.
QUANTIFICATION AND STATISTICAL ANALYSIS
Structure Determination of Rev93:scFv:RBA-14
Data processing and integration was done with iMosflm (Battye et al., 2011) and scaling was done with Aimless (Evans and Murshudov, 2013). Molecular replacement was carried out by Phaser (McCoy et al., 2007) using the HIV-Rev69 structure (PDB ID: 5DHV), scFv SJS-R1 structure (PDB ID: 5DHV) and 8 base-long (ACUC—GAGA) of the RNA aptamer structure (PDB ID: 1ULL) as search models (DiMattia et al., 2016; Ye et al., 1996). The rest of the RNA was built manually inside the apparent nucleic acid density after one round of refinement by phenix refine (Afonine et al., 2012). The final structure was obtained after multiple rounds of model building with Coot (Emsley and Cowtan, 2004) and refinement with phenix refine. Interface analysis was performed using jsPISA (Krissinel, 2015) and Entangle (Allers and Shamoo, 2001).
Surface Plasmon Resonance
Curves were base-line corrected and fitted best to a two-state binding model with residuals < 10% of signal. The association and dissociation rate constants (ka and kd, respectively) were used to calculate the equilibrium dissociation constant (Kd). The Kd for the basic 1:1 binding model was calculated from the ratio (kd/ka). The overall Kd for the two-state model was calculated from (kd1/ka1)(kd2/ka2). The rate constants used are given in Table S1.
Analytical Ultracentrifugation
The partial specific volume (n-bar) of Rev (0.724 g/cc) was calculated from the amino acid composition, and a value of 0.53 g/cc was used for the RNA. Solvent densities were estimated using the program: SEDNTERP (http://www.rasmb.bbri.org/). Data analysis was done using DCDT+ 2.4.3 (Philo, 2006).
DATA AND SOFTWARE AVAILABILITY
The accession number for the HIV-1 Rev (residues 1–93)-RNA aptamer complex X-ray structure reported in this paper is [PDB]: [6CF2].
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| scFv(Rev) | Paul T. Wingfield (DiMattia et al., 2016) | scFv(Rev) |
| Bacterial and Virus Strains | ||
| BL21 (DE3) | NEB | Catalog # C2527l |
| NEB 5alpha | NEB | Catalog # C2987l |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Rev93 | Paul T. Wingfield (Zhuang et al., 2014) | Rev93 |
| Rev116 | Paul T. Wingfield (DiMattia et al., 2010) | Rev116 |
| Rev116(Leu64Ala) | Paul T. Wingfield (DiMattia et al.,2016) | Rev116(L64A) |
| Rev116(Pro28Ala) | Paul T. Wingfield (DiMattia et al., 2016) | Rev116(P28A) |
| Rev117(Cys85Ala,Cys89Ala,117Cys) | This manuscript | Rev2CA117C |
| Deposited Data | ||
| RNA aptamer complexed with HIV-1 Rev peptide, NMR, 7 structures | RCSB PDB (Ye et al., 1996) | PDB ID: 1ULL |
| Rev Response Element (RRE) RNA complexed with Rev peptide, NMR, minimized average structure | RCSB PDB (Battiste et al., 1996) | PDB ID: 1ETF |
| Crystal structure of Rev and Rev-response- element RNA complex | RCSB PDB (Jayaraman et al., 2014) | PDB ID: 4PMI |
| Molecular model from helical cryo-EM reconstruction of HIV Rev filament V | Alasdair C. Steven (DiMattia et al., 2016) | HIV-1 Rev filament V |
| Crystal structure of HIV-1 Rev (residues 1–93)-RNA aptamer complex | This manuscript | PDB ID: 6CF2 |
| Oligonucleotides | ||
| RBA-14 RNA 5’- GGC UGG ACU CGU ACU UCG GUA CUG GAG AAA CAG CC −3’ | IDTDNA | N/A |
| Stem IIB RNA 5’- CCU GGG CGC AGU GUC AUU GAC GCU GAC GGU ACA GG −3’ | IDTDNA | N/A |
| Recombinant DNA | ||
| pET11a Rev 1–93 | Paul T. Wingfield (Genscript) | plasmid # 1979 |
| pET11a Rev 1–116 | Paul T. Wingfield (Genscript) | plasmid # 1978 |
| pLmu Rev 1–116 L64A | Paul T. Wingfield (DiMattia et al., 2016) | plasmid # 1626 |
| pET11a Rev 1–116 P28A | Paul T. Wingfield (Genscript) | plasmid # 1925 |
| pET11a Rev 1–116 2CA117C | Paul T. Wingfield (Genscript) | plasmid # 1939 |
| pET11a Rev scFv long polyHis(2) | Paul T. Wingfield (DiMattia et al., 2016) | plasmid # 1398 |
| Software and Algorithms | ||
| OligoAnalyzer 3.1 | IDT DNA | https://www.idtdna.com/calc/analyzer |
| iMosflm | Battye et al., 2011 | https://www.mrc-lmb.cam.ac.uk/harry/imosflm/ver722/introduction.html |
| Aimless | Evans and Murshudov, 2013 | http://www.ccp4.ac.uk/html/aimless.html |
| Phaser | McCoy et al., 2007 | http://www.ccp4.ac.uk/html/phaser.html |
| Phenix Refine | Afonine et al., 2012 | https://www.phenix-online.org/documentation/reference/refinement.html |
| Coot | Emsley and Cowtan, 2004 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| jsPISA | Krissinel, 2015 | http://www.ccp4.ac.uk/pisa/ |
| Entangle | Allers and Shamoo, 2001 | http://www.bioc.rice.edu/~shamoo/entangle.html |
| Clustal Omega | EMBL-EBI (Goujon et al., 2010; Sievers etal., 2011) | https://www.ebi.ac.uk/Tools/msa/clustalo/ |
| Biacore X100 Control Software 2.0.1 | GE Healthcare | https://www.biacore.com/lifesciences/index.html |
| Biacore X100 Evaluation Software 2.0.1 | GE Healthcare | https://www.biacore.com/lifesciences/index.html |
| SEDENTERP | RASMB-BBRI | http://www.rasmb.bbri.org/ |
| DCDT+ 2.4.3 | Philo, 2006 | http://www.jphilo.mailway.com/dcdt+.htm |
| Other | ||
| AIDS database | LANL (Foley et al., 2017) | https://www.hiv.lanl.gov/content/index |
Highlights.
X-ray crystal structure of potential anti-HIV-1 RNA aptamer
The aptamer binds to the arginine-rich motif of Rev
The aptamer blocks Rev self-assembly
ACKNOWLEDGMENTS
We would like to thank Ms. Keyanna Franklin for her help in protein preparation, and Dr. Jinwei Zhang for helpful discussion. This work was supported by the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases, and by the NIH Intramural AIDS Targeted Antiviral Program (IATAP).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and two tables and can be found with this article online at https://doi.org/10.1016/j.str.2018.06.001.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, and Adams PD (2012). Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr 68, 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AIDSinfo. (2017). Panel on Antiretroviral Guidelines for Adults and Adolescents. Guidelines for the Use of Antiretroviral Agents in Adults and Adolescents Living with HIV (Department of Health and Human Services), F-1, F-4. [Google Scholar]
- Allers J, and Shamoo Y (2001). Structure-based analysis of protein-RNA interactions using the program ENTANGLE. J. Mol. Biol 311, 75–86. [DOI] [PubMed] [Google Scholar]
- Andreu JM, and Timasheff SN (1986). The measurement of cooperative protein self-assembly by turbidity and other techniques. Methods Enzymol. 130, 47–59. [DOI] [PubMed] [Google Scholar]
- Antiretroviral Therapy Cohort Collaboration (2017). Survival of HIV-positive patients starting antiretroviral therapy between 1996 and 2013: a collaborative analysis of cohort studies. Lancet HIV 4, e349–e356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arhel N, and Kirchhoff F (2010). Host proteins involved in HIV infection: new therapeutic targets. Biochim. Biophys. Acta 1802, 313–321. [DOI] [PubMed] [Google Scholar]
- Battiste JL, Mao H, Rao NS, Tan R, Muhandiram DR, Kay LE, Frankel AD, and Williamson JR (1996). Alpha helix-RNA major groove recognition in an HIV-1 rev peptide-RRE RNA complex. Science 273, 1547–1551. [DOI] [PubMed] [Google Scholar]
- Battye TG, Kontogiannis L, Johnson O, Powell HR, and Leslie AG (2011). iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr 67, 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunka DH, Platonova O, and Stockley PG (2010). Development of aptamer therapeutics. Curr. Opin. Pharmacol 10, 557–562. [DOI] [PubMed] [Google Scholar]
- Buonaguro L, Tornesello ML, and Buonaguro FM (2007). Human immunodeficiency virus type 1 subtype distribution in the worldwide epidemic: path-ogenetic and therapeutic implications. J. Virol 81, 10209–10219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell GR, Watkins JD, Esquieu D, Pasquier E, Loret EP, and Spector SA (2005). The C terminus of HIV-1 Tat modulates the extent of CD178-mediated apoptosis of T cells. J. Biol. Chem 280, 38376–38382. [DOI] [PubMed] [Google Scholar]
- Cole JL, Gehman JD, Shafer JA, and Kuo LC (1993). Solution oligomerization of the rev protein of HIV-1: implications for function. Biochemistry 32, 11769–11775. [DOI] [PubMed] [Google Scholar]
- Daugherty MD, Liu B, and Frankel AD (2010). Structural basis for cooperative RNA binding and export complex assembly by HIV Rev. Nat. Struct. Mol. Biol 17, 1337–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMattia MA, Watts NR, Cheng N, Huang R, Heymann JB, Grimes JM, Wingfield PT, Stuart DI, and Steven AC (2016). The structure of HIV-1 rev filaments suggests a bilateral model for Rev-RRE assembly. Structure 24, 1068–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMattia MA, Watts NR, Stahl SJ, Rader C, Wingfield PT, Stuart DI, Steven AC, and Grimes JM (2010). Implications of the HIV-1 Rev dimer structure at 3.2 A resolution for multimeric binding to the Rev response element. Proc. Natl. Acad. Sci. USA 107, 5810–5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, and Cowtan K (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Evans PR, and Murshudov GN (2013). How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr 69, 1204–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Wang J, O’Carroll IP, Mitchell M, Zuo X, Wang Y, Yu P, Liu Y, Rausch JW, Dyba MA, et al. (2013). An unusual topological structure of the HIV-1 Rev response element. Cell 155, 594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley B, Leitner T, Apetrei C, Hahn B, Mizrachi I, Mullins J, Rambaut A, Wolinsky S, and Korber B (2017). HIV Sequence Compendium 2017 (Theoretical Biology and Biophysics Group, Los Alamos National Laboratory). [Google Scholar]
- Giver L, Bartel D, Zapp M, Pawul A, Green M, and Ellington AD (1993). Selective optimization of the Rev-binding element of HIV-1. Nucleic Acids Res. 21, 5509–5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good PD, Krikos AJ, Li SX, Bertrand E, Lee NS, Giver L, Ellington A, Zaia JA, Rossi JJ, and Engelke DR (1997). Expression of small, therapeutic RNAs in human cell nuclei. Gene Ther. 4, 45–54. [DOI] [PubMed] [Google Scholar]
- Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, and Lopez R (2010). A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38, W695–W699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain C, and Belasco JG (1996). A structural model for the HIV-1 Rev-RRE complex deduced from altered-specificity rev variants isolated by a rapid genetic strategy. Cell 87, 115–125. [DOI] [PubMed] [Google Scholar]
- Jayaraman B, Crosby DC, Homer C, Ribeiro I, Mavor D, and Frankel AD (2014). RNA-directed remodeling of the HIV-1 protein Rev orchestrates assembly of the Rev-Rev response element complex. Elife 3, e04120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E (2015). Stock-based detection of protein oligomeric states in jsPISA. Nucleic Acids Res. 43, W314–W319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law MJ, Linde ME, Chambers EJ, Oubridge C, Katsamba PS, Nilsson L, Haworth IS, and Laird-Offringa IA (2006). The role of positively charged amino acids and electrostatic interactions in the complex of U1A protein and U1 hairpin II RNA. Nucleic Acids Res. 34, 275–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim MH, Hauber J, Fenrick R, and Cullen BR (1988). Immunodeficiency virus rev trans-activator modulates the expression of the viral regulatory genes. Nature 335, 181–183. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ (2007). Phaser crystallographic software. J. Appl. Crystallogr 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neville M, Stutz F, Lee L, Davis LI, and Rosbash M (1997). The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr. Biol 7, 767–775. [DOI] [PubMed] [Google Scholar]
- Philo JS (2006). Improved methods for fitting sedimentation coefficient distributions derived by time-derivative techniques. Anal. Biochem 354, 238–246. [DOI] [PubMed] [Google Scholar]
- Pollard VW, and Malim MH (1998). The HIV-1 Rev protein. Annu. Rev. Microbiol 52, 491–532. [DOI] [PubMed] [Google Scholar]
- Pond SJ, Ridgeway WK, Robertson R, Wang J, and Millar DP (2009). HIV-1 Rev protein assembles on viral RNA one molecule at a time. Proc. Natl. Acad. Sci. USA 106, 1404–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglisi JD, Chen L, Frankel AD, and Williamson JR (1993). Role of RNA structure in arginine recognition of TAR RNA. Proc. Natl. Acad. Sci. USA 90, 3680–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch JW, and Le Grice SF (2015). HIV rev assembly on the rev response element (RRE): a structural perspective. Viruses 7, 3053–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodroski J, Goh WC, Rosen C, Dayton A, Terwilliger E, and Haseltine W (1986). A second post-transcriptional trans-activator gene required for HTLV-III replication. Nature 321, 412–417. [DOI] [PubMed] [Google Scholar]
- Stahl SJ, Watts NR, Rader C, DiMattia MA, Mage RG, Palmer I, Kaufman JD, Grimes JM, Stuart DI, Steven AC, et al. (2010). Generation and characterization of a chimeric rabbit/human Fab for co-crystallization of HIV-1 Rev. J. Mol. Biol 397, 697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNAIDS (2016). Global AIDS Update 2016. http://www.unaids.org/sites/default/files/media_asset/global-AIDS-update-2016_en.pdf.
- Watts NR, Eren E, Zhuang X, Wang YX, Steven AC, and Wingfield PT (2018). A new HIV-1 Rev structure optimizes interaction with target RNA (RRE) for nuclear export. J. Struct. Biol 10.1016/j.jsb.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingfield PT, Stahl SJ, Payton MA, Venkatesan S, Misra M, and Steven AC (1991). HIV-1 Rev expressed in recombinant Escherichia coli: purification, polymerization, and conformational properties. Biochemistry 30, 7527–7534. [DOI] [PubMed] [Google Scholar]
- Ye X, Gorin A, Ellington AD, and Patel DJ (1996). Deep penetration of an alpha-helix into a widened RNA major groove in the HIV-1 rev peptide-RNA aptamer complex. Nat. Struct. Biol 3, 1026–1033. [DOI] [PubMed] [Google Scholar]
- Ye X, Gorin A, Frederick R, Hu W, Majumdar A, Xu W, McLendon G, Ellington A, and Patel DJ (1999). RNA architecture dictates the conformations of a bound peptide. Chem. Biol 6, 657–669. [DOI] [PubMed] [Google Scholar]
- Zhuang X, Stahl SJ, Watts NR, DiMattia MA, Steven AC, and Wingfield PT (2014). A cell-penetrating antibody fragment against HIV-1 Rev has high antiviral activity: characterization of the paratope. J. Biol. Chem 289, 20222–20233. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the HIV-1 Rev (residues 1–93)-RNA aptamer complex X-ray structure reported in this paper is [PDB]: [6CF2].


