Abstract
Small cell lung cancer (SCLC) is an aggressive malignancy in which inhibitors of poly (ADP-ribose) polymerase (PARP) have modest single-agent activity. We performed a phase I/II trial of combination olaparib tablets and temozolomide (OT) in previously treated SCLC. We established a recommended phase 2 dose (RP2D) of olaparib 200 mg PO BID with temozolomide 75 mg/m2 daily, both on days 1–7 of a 21-day cycle, and expanded to a total of 50 patients. The confirmed overall response rate (ORR) was 41.7% (20/48 evaluable); median progression free survival (mPFS) was 4.2 months (95% CI 2.8–5.7); and median overall survival (mOS) was 8.5 months (95% CI 5.1–11.3). Patient-derived xenografts (PDXs) from trial patients recapitulated clinical OT responses, enabling a 32-PDX co-clinical trial. This revealed a correlation between low basal expression of inflammatory response genes and cross-resistance to both OT and standard first-line chemotherapy (etoposide/platinum, EP). These results demonstrate a promising new therapeutic strategy in SCLC and uncover a molecular signature of those tumors most likely to respond.
Keywords: SCLC, PARP inhibitor, PDX, cross-resistance, innate immunity
INTRODUCTION
Small cell lung cancer (SCLC), which accounts for approximately 15% of all lung cancers, is a high-grade neuroendocrine carcinoma with high metastatic potential and poor clinical outcomes. The most common genetic alterations in SCLC are inactivation of TP53 and RB1, and no clear targetable alterations have been described [1–3]. Untreated SCLCs tend to be highly sensitive to cytotoxic chemotherapy; response rates to first-line EP are 50–70% [4, 5]. The duration of time from completion of first-line chemotherapy (generally 4–6 cycles are given) to disease progression, often referred to as the degree of “platinum sensitivity”, predicts likelihood of response to second-line cytotoxics [6]. However, response rates in the second-line setting are significantly lower, ranging approximately 5–30% [6–10].
Recently, targeting DNA damage repair by inhibition of poly [ADP-ribose] polymerase (PARP) has emerged as a potential therapeutic strategy in SCLC [11]. Although SCLCs are not characterized by homologous recombination (HR) deficiency or mutations in BRCA1/2, PARP inhibitors have shown some activity in SCLC preclinical models and early phase trials [12–15]. However, the single agent activity of PARP inhibitors in SCLC appears to be minimal. Notably, the UK STOMP trial failed to show an improvement in PFS for patients treated with olaparib in the maintenance setting [16]. PARP inhibitors may synergize with agents that increase the prevalence of single stranded (ss) DNA breaks [17]. Mechanistically, this combination strategy is supported even in HR- and BRCA-intact tumors by the observation that trapping of PARP complexes to sites of ssDNA breaks leads to failure of repair and induction of double stranded (ds) breaks [15, 18]. Addition of the PARP inhibitor veliparib to temozolomide in a randomized phase II trial led to improved response rate (39% in the veliparib/temozolomide arm vs 14% in the placebo/temozolomide arm), though no significant improvement in the 4-month PFS or mPFS [13]. The ECOG-ACRIN 2511 study demonstrated a modest improvement in mPFS with the addition of veliparib to cisplatin and etoposide (mPFS 6.1 vs 5.3 months; HR 0.75, one-sided P = 0.01), though no significant improvement in overall survival [14]. Olaparib, which has stronger PARP trapping activity than veliparib [18], may offer significant anti-tumor efficacy while maintaining a manageable therapeutic window in combination with temozolomide.
As a field, SCLC research suffers from a paucity of available tissue for preclinical and co-clinical investigations. Initial diagnoses are often made from scant aspirate tissue, and repeat biopsies after initial treatment are typically outside of standard of care. We have previously reported generation of a panel of patient-derived xenograft (PDX) models of SCLC that recapitulate patient tumor genomic features and sensitivity to EP [19]. Here, we develop and test the clinical activity of combination olaparib and temozolomide (OT) in patients with SCLC, and then apply a PDX panel to discover molecular signatures predictive of response to this treatment.
RESULTS
Study design, enrollment and patient demographics
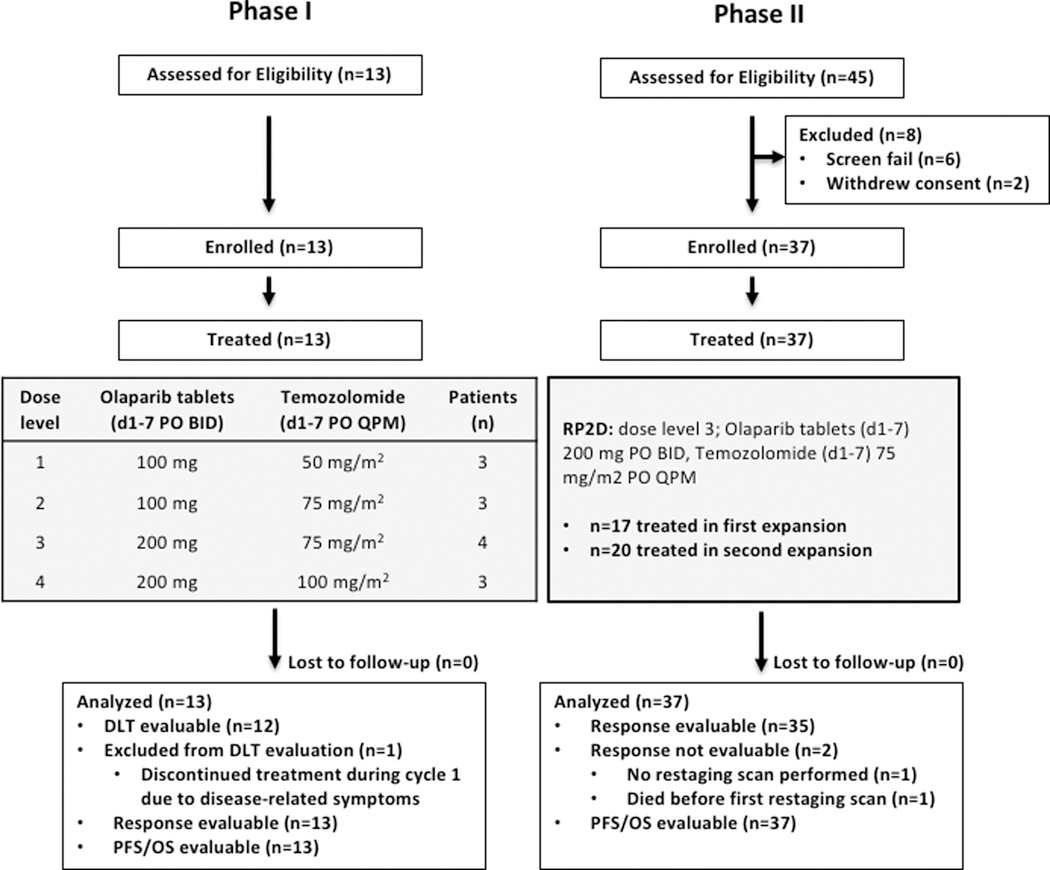
The trial consisted of a phase I dose escalation portion and a phase II multi-stage portion. The phase I portion (Figure 1, left) was a conventional 3+3 dose escalation design with a primary objective of determining the recommended phase II dose (RP2D) of combination olaparib tablets and temozolomide, both dosed days 1–7 of each 21-day cycle. Dose limiting toxicities (DLTs) were monitored during cycle 1. One patient enrolled at dose level 3 was not DLT evaluable and was therefore replaced, per protocol. The phase II portion of the study (Figure 1, right) was a dose expansion at the RP2D, dose level 3, with the primary objective to assess efficacy as measured by ORR. This was a multi-stage optimal design to allow for early termination due to lack of efficacy.
Figure 1. Study Design.
The phase I portion of the study (left) was a 3+3 dose escalation schema with a primary objective of determining the RP2D of combination olaparib tablets and temozolomide, both dosed days 1–7 of each 21-day cycle. The phase 2 portion of the study (right) was a dose expansion at dose level 3, with the primary objective of determine overall response rate.
Between October 2015 and April 2018, we enrolled 50 patients with previously treated SCLC to the clinical trial. The baseline demographics of patients enrolled study-wide are shown in Table 1. The majority of patients (86%) had an Eastern Cooperative Oncology Group Performance Status (ECOG PS) of 1, and the number of prior lines of cancer therapy ranged from 1 to 7 (median 2). 72% were platinum sensitive, defined here as having a chemotherapy-free interval from completion of first-line therapy to initiation of second-line therapy of at least 90 days. The study required baseline brain imaging, and asymptomatic untreated brain metastases measuring < 1 cm were allowed for eligibility. Twenty patients (40%) had metastases at baseline, 12 of which were untreated at study entry. Demographics of the subset of patients treated at dose level 3/RP2D are shown in Supplementary Table 1.
Table 1. Baseline patient demographics.
Shown are data for all patients in the phase I and II portions. *Chemotherapy-free interval, defined as time from last date of first-line platinum-based chemotherapy to first date of second-line systemic therapy.
| Patient demographics | Phases I & II (n=50) |
|---|---|
| Age, years, median (range) | 63 (39 – 85) |
| Sex, male/female (%) | 20 (40) / 30 (60) |
| ECOG performance status, n (%) | |
| 0 | 6 (12) |
| 1 | 43 (86) |
| 2 | 1 (2) |
| Prior lines of SCLC therapy, n (%) | |
| 1 | 23 (46) |
| 2 | 17 (34) |
| 3 | 4 (8) |
| >3 | 6 (12) |
| Median (range) | 2 (1–7) |
| Chemotherapy-free interval* | |
| ≥ 90 days (“platinum sensitive”) (%) | 36 (72) |
| < 90 days (“platinum resistant”) (%) | 14 (28) |
| Baseline brain metastases present (%) | 20 (40) |
| Treated | 8 (16) |
| Untreated | 12 (24) |
Safety and tolerability
At the four dose levels tested in the phase I portion, there were no DLTs, serious adverse events (SAEs) or grade 4 or 5 treatment related toxicities. We observed increasing neutropenia, anemia and thrombocytopenia in dose levels 3 and 4 compared to dose levels 1 and 2. Additionally, one patient treated at dose level 4 experienced grade 3 vomiting. Although none of these adverse events met DLT criteria, dose level 3 was selected as the RP2D based on the phase I experience.
Summaries of treatment related adverse events (TRAEs) study-wide and among patients treated at dose level 3/RP2D are shown in Supplementary Table 2. Study-wide, the most common TRAEs were thrombocytopenia, anemia and neutropenia (occurring in 68%, 68%, and 54% of patients, respectively), though the majority were grades 1–2. Rates of fatigue, nausea, and vomiting were 50%, 42% and 22%, respectively, and again the majority were grade 1 or 2. Notable AEs included two grade 5 events which occurred during the phase II portion and that were deemed possibly related to study drugs. One was due to pneumonia and occurred during cycle 1. The other was due to neutropenic sepsis and occurred during cycle 3. There was one case of pneumonitis, which was grade 3 and thought to be related to study drugs. This event occurred after two cycles of treatment and resolved with steroids.
Dose reductions were performed per protocol and at the investigator’s discretion. Among 41 patients treated at the RP2D, dose reductions occurred in 44% overall and 64% of those who received at least 3 cycles (Supplementary Table 3).
Efficacy
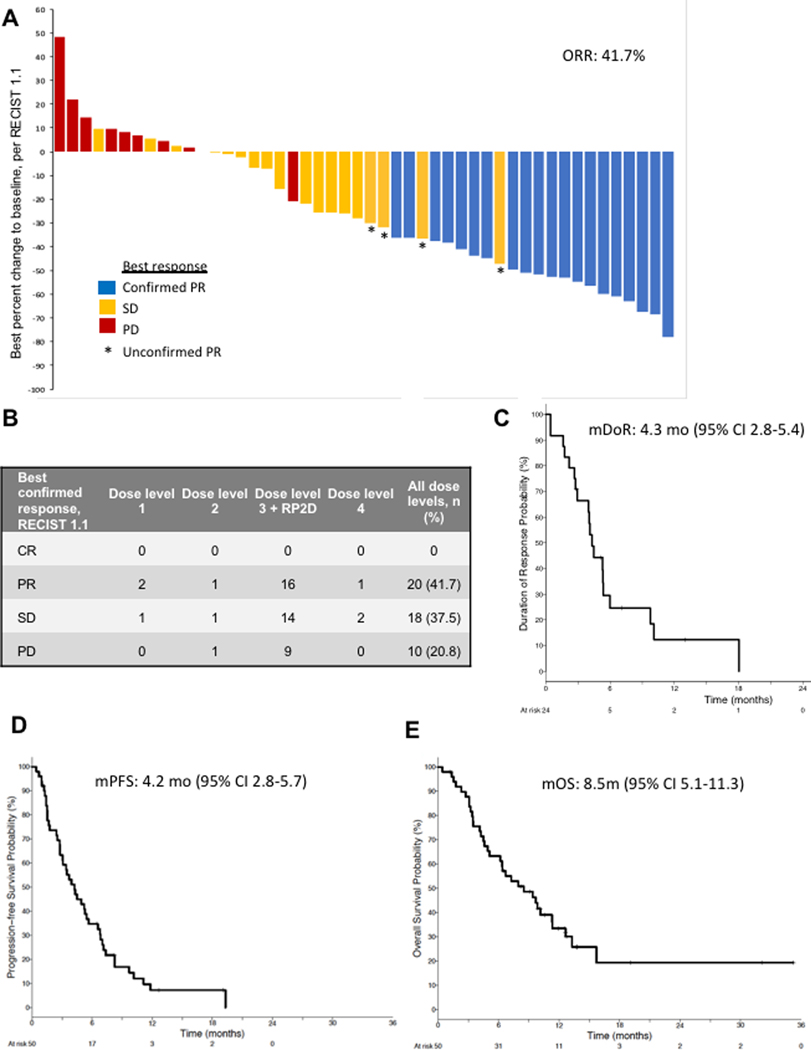
Study-wide, 48 patients were evaluable for response assessment. The confirmed ORR was 41.7% (20/48; Figure 2A, B). Four additional patients had unconfirmed partial response (PR); two had PR on the first scan and then progressive disease (PD) on the subsequent scan, and two had a PR on the first scan and then expired prior to a confirmatory scan. Among patients treated at dose level 3/RP2D, confirmed ORR was 44.4% (16/36). Responses were seen at all dose levels (Figure 2B). Among the responders and non-responders, the median number of prior lines of therapy were 2 and 1 (mean 2.1 and 2.0), respectively. The median duration of response (mDOR) was 4.3 months (Figure 2C). After a median follow up of 7.1 months, the mPFS was 4.2 months (95% CI 2.8–5.7; Figure 2D) and the mOS was 8.5 months (95% CI 5.1–11.3; Figure 2E). Among patients treated at dose level 3/RP2D, the mDOR was 5.3 months (95% CI 2.7–5.8), mPFS was 4.2 months (95% CI 2.5–5.7) and mOS was 6.7 months (95% CI 4.6–12.6). Nine patients continued on treatment post-progression because of ongoing clinical benefit in the opinion of the treating investigator, with the duration of treatment post-progression ranging from 3 weeks to 12 months.
Figure 2. Clinical efficacy of olaparib/temozolomide.
(A) Best objective responses using RECIST 1.1 criteria are shown in waterfall plot. (B) Best confirmed responses per dose level and overall. (C-E) Kaplan-Meier curves showing duration of response (C), progression-free survival (D) and overall survival (E), with medians (m) indicated at the top. CR, complete response; PR, partial response; SD, stable disease; PD, progressive disease.
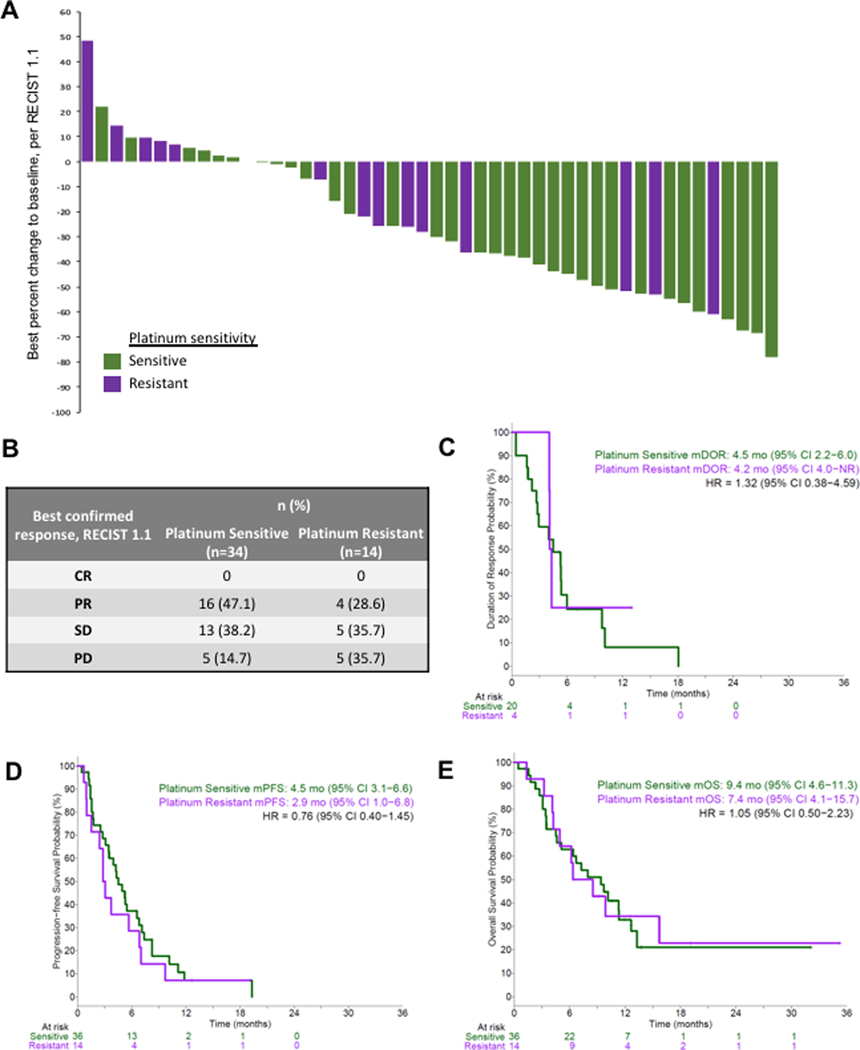
In an exploratory analysis, we further assessed efficacy based on platinum sensitivity (Figure 3). The confirmed ORR was 47.1% among the 34 patients with platinum sensitive disease, and 28.6% among the 14 patients with platinum resistant disease (Figure 3A, B). The median duration of response among patients with at least one time point demonstrating PR was 4.2 months for platinum sensitive (20 patients) and 4.5 months for platinum resistant (4 patients) (Figure 3C). The mPFS was 4.5 months and 2.9 months, and mOS was 9.4 months and 7.4 months, among platinum sensitive and resistant patients, respectively (Figure 3D, E). There was no statistically significant difference observed in PFS (HR=0.76, p=0.400) or OS (HR=1.05, p=0.898) when comparing outcomes among platinum sensitive and platinum resistant patients, though the small size of these cohorts limits the power to detect a significant difference in this study.
Figure 3. Clinical efficacy of olaparib/temozolomide according to platinum sensitivity.
(A) Best objective responses using RECIST 1.1 criteria are shown in waterfall plot. (B) Best confirmed responses. (C-E) Kaplan-Meier curves showing duration of response (C), progression-free survival (D) and overall survival (E), with medians (m) indicated at the top. CR, complete response; PR, partial response; SD, stable disease; PD, progressive disease.
Co-clinical trial in PDX models
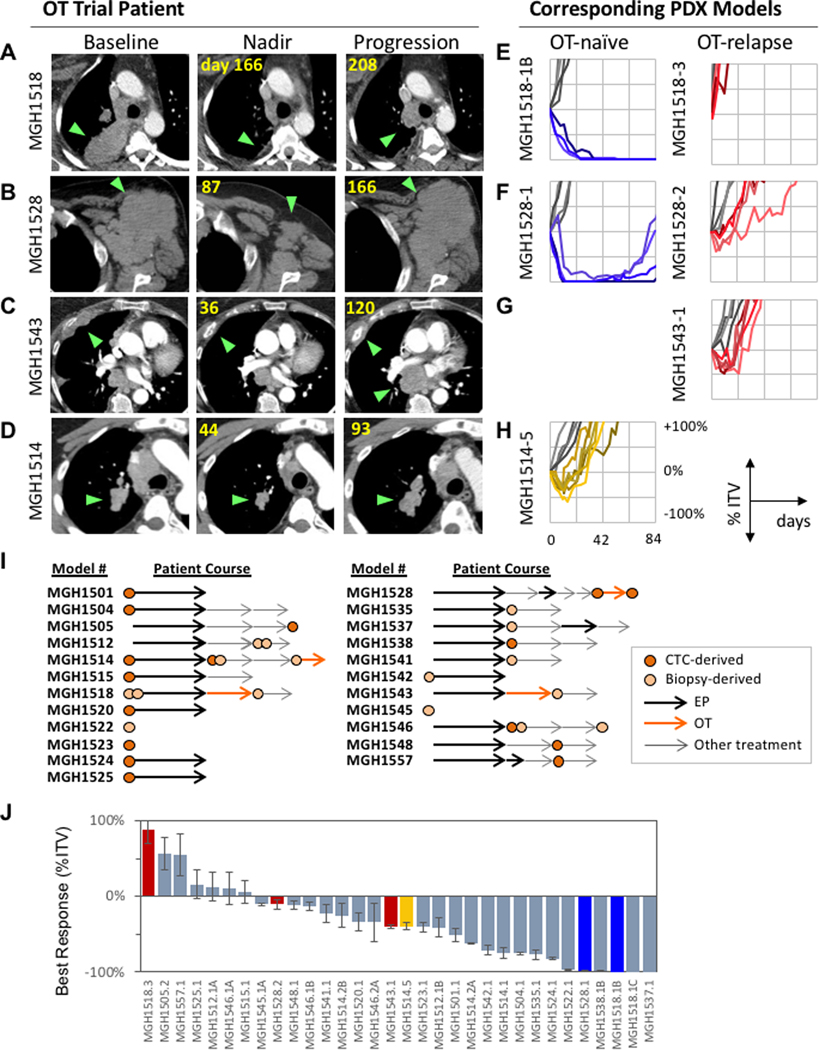
To model clinical OT sensitivity and resistance in the laboratory setting, we generated a series of six PDX models from four patients treated during the phase I portion of the trial. All four patients, MGH1518, MGH1528, MGH1543, and MGH1514 had clinical benefit on OT, with RECIST 1.1 best responses of −61%, −50%, −41% and −28%, and time to progression of 6.8, 5.5, 3.9, and 3.1 months, respectively (Figure 4A–D). These patients had received 1, 7, 1 and 4 prior lines of therapy, respectively. PDX models were developed from these patients prior to receiving OT (MGH1518–1B, MGH1528–1, and MGH1514–5) and at the time of disease progression (MGH1518–3, MGH1528–2, and MGH1543–1). These 6 PDX models were treated with a single 5-day cycle of twice-daily olaparib plus daily temozolomide. Following treatment, PDX tumor responses were compared with those of their donor patients (Figure 4E–H). To quantify tumor responses, we measured maximum tumor regression after treatment (best response) and days to 200% initial tumor volume (time to progression, TTP). By both metrics, PDX sensitivity mirrored patient tumor sensitivity to OT at the time of model generation. Specifically, PDXs from OT-naïve patients who went on to have durable partial responses, MGH1518–1B and MGH1528–1, regressed completely in all mice (100% response) and did not relapse for at least 80 days (Figure 4E, F, blue curves). Patient MGH1514 had a modest tumor response (−28%) and a brief PFS (3.1 months); the corresponding PDX MGH1514–5 derived at the time this patient enrolled on the OT study showed transient partial regression (−40% ± 4.4% days SEM) followed by progression (42.5 ± 3.1 days SEM) (Figure 4H, yellow curves). By contrast, in PDXs derived from three patients at the time of progression on OT, tumors showed minimal responses and progressed at nearly the same paces as vehicle treated controls (Figure 4E–G, red curves). Collectively, these data gave us confidence that the PDX tumors accurately recapitulate the sensitivity and resistance of their donor patient tumors.
Figure 4. Co-clinical trial of OT in SCLC PDX models.
PDX models were derived from four SCLC patients on the OT trial: MGH1518 (A), MGH1528 (B), MGH1543 (C) and MGH1514 (D). (A-D) Axial CT scan images of patients represent baseline, nadir and progression on OT, with green arrows indicating tumors. (E-H) PDXs generated from corresponding patients, with tumor volume curves after a single 5-day cycle of OT, represented as % initial tumor volume (ITV) vs. days after start of treatment. Timing of model derivation relative to OT initiation and progression is indicated. For patients MGH1518 (E) and MGH1528 (F), serial models were derived before (blue) and after (red) durable responses. For MGH1514 (G), a model was derived before brief disease stabilization (yellow), and for MGH1543 (H) a model was derived after progression (red). Untreated tumor volume curves in gray. (I) Panel of PDX models treated with OT, with abstracted patient clinical courses. Models derived from either CTCs (dark orange circles) or biopsies/effusions (light orange circles) were generated either prior to chemotherapy or between lines of therapy (arrows). Arrows are not drawn to scale with respect to time on treatments. (J) Waterfall plot of PDX best response to OT, defined as minimum % ITV between days 7–28. Models derived from OT trial patients (colored) serve to calibrate model response to clinical history: models derived from OT trial patients upon progression are red, pre-treatment models from trial patients with durable responses are blue, and pre-treatment model from patient with stable disease (MGH1514–5) in yellow.
We extended the co-clinical trial to a total of 32 PDX models derived from 22 patients (Figure 4I, Supplementary Figure S1A). The additional 26 models in this panel were not derived from patients who had been treated with OT, but were an unselected cohort of consecutively derived models, with the goal of representing the biologic diversity seen among patients with SCLC. This panel included 13 models from chemotherapy-naïve patients and 19 models generated from patients after at least one prior line of therapy. Two to six replicate xenografts per model were assessed for OT sensitivity using the 5-day single-cycle treatment protocol, with best response and TTP measured to assess efficacy (Figure 4J, Supplementary Figure S2A). We observed a range of responses across the models (Figure 4J), consistent with the range of responses seen in our patient cohort (Figure 2A). Replicate xenografts demonstrated highly concordant tumor volume curves following OT treatment (average SEM for model response = ± 8.4% and TTP = ± 3.1 days), and across the 32-model panel, the best response and TTP metrics were tightly inversely correlated (Pearson r = −0.87, Supplementary Figure S2B). OT-sensitive models were substantially less sensitive to single-agent olaparib or temozolomide (Supplementary Figure S2C), indicating that PDX responses are dependent upon the combined activity.
Correlation between OT and EP sensitivity
The clinical course of SCLC is generally characterized by initial sensitivity to DNA damaging therapies such as EP, followed by relapse and broad cross-resistance to second-line agents [6]. Acquisition of cross-resistance remains an ongoing clinical challenge, and yet it is difficult to study because direct comparison of different regimens is not possible in the same patient. The SCLC PDX panel captures intertumoral heterogeneity, and provides a unique opportunity to study cross-resistance. We have previously demonstrated that PDX models faithfully recapitulate clinical sensitivity to EP [19]. A direct comparison of the patient platinum sensitivity and corresponding PDX model EP sensitivity (Supplementary Figure S3A–D) further supports this conclusion. For OT, we also observe a high degree of clinical fidelity in models derived from trial patients (Figure 4A–H). We therefore applied the models to investigate patterns of cross-resistance between EP and OT.
We compared in vivo drug response to each regimen with both PDX clinical history and gene expression levels across 32 models (Supplementary Figure S1A). PDX TTP on EP was superior for those models derived from treatment-naive patients compared to those derived from previously treated patients, and a similar trend was observed for OT in the same models (Figure 5A). We then directly compared in vivo sensitivity for each regimen in each model and observed a moderate correlation (Pearson r=0.56, Figure 5B). Both results suggest partial cross-resistance to these regimens. To interrogate the molecular underpinnings of these observations, paired-end transcriptome sequencing was performed in untreated replicate xenografts for each model, and replicates were averaged for transcript abundance (Supplementary Table 4). The transcriptional profiles associated with EP sensitivity correlated well with the profiles associated with OT sensitivity (Pearson r=0.68, Figure 5C). Furthermore, there was a notable absence of genes whose expression could stratify the regimens by marking sensitivity to one but not the other. These results argue that clinical resistance to EP may also predict resistance to OT, and no clear marker(s) emerge from this data set to distinguish tumors sensitive to one regimen but not the other.
Figure 5. Transcriptional signatures of EP/OT cross-resistance.
(A) Kaplan-Meier curves showing duration of response (TTP = days to 2x ITV) following in vivo treatment with EP or OT of PDX models derived before (13) or after (19) first-line EP, with log-rank test p-values. (B) Direct comparison of mean TTP for 32 PDX models (2–6 mice/model) following EP and OT, with Pearson correlation (r). (C) Correlation of gene expression (Pearson) with mean EP TTP vs. correlation with OT TTP across 32 PDX models. Transcript abundance for 16,100 genes measured by paired-end RNA-sequencing on untreated xenografts in biologic duplicate for each model. Significant correlation at FDR < 10%. (D) Hallmark gene set enrichment (normalized enrichment scores) of transcript abundance for EP TTP vs. OT TTP across the PDX panel. Significantly enriched gene sets (FDR < 25%) are labeled. (E-F) Signature scores overlaid onto mean EP TTP vs. OT TTP for PDX panel as shown in (B). Scores are mean transcript z-scores of leading edge genes present in significantly enriched gene sets in both EP and OT. (E) Positively enriched gene sets (sensitivity) were related by participation in the inflammatory response, with an 82-gene overlap between EP and OT. (F) MYC transcriptional targets led the negatively enriched gene sets (resistance), with a 65-gene overlap.
To identify candidate molecular pathways associated with EP and OT sensitivity and resistance, gene set enrichment analysis (GSEA) was performed across PDX models using the transcription data. When we compared enrichment scores for the regimens, we found that significantly enriched gene sets for both EP and OT were closely related (Figure 5D). Interestingly, inflammatory response gene sets (interferons α and γ, inflammation, TNFα and TGFβ) enriched for sensitivity to both EP and OT across regimens (Figure 5D). By contrast, MYC-regulated transcripts correlated with resistance to the regimens (Figure 5D). Leading edge analysis revealed a high degree of overlapping genes between the members of each set responsible for the enrichment scores. These were combined into expression signatures for MYC targets (65 genes) and inflammatory response (82 genes), and when mapped to the PDX panel these signatures labeled models that were most resistant to both EP and OT (MYC-target high, inflammatory low, Figure 5E, F). We further assessed whether the recently described sub-classification of SCLC tumors by expression of the transcriptional regulators ASCL1, NEUROD1, YAP1, POU2F3 could identify EP/OT-sensitive or resistant models (Supplementary Figure S4A–C). All four categories were represented by at least one model, but the number of models in each category was limited and did not significantly correlate with cross-resistance [20, 21]. This analysis reveals the potential of clinically representative PDX models to interrogate molecular signatures associated with cross-resistance to therapies.
Discovery of candidate biomarkers for OT sensitivity
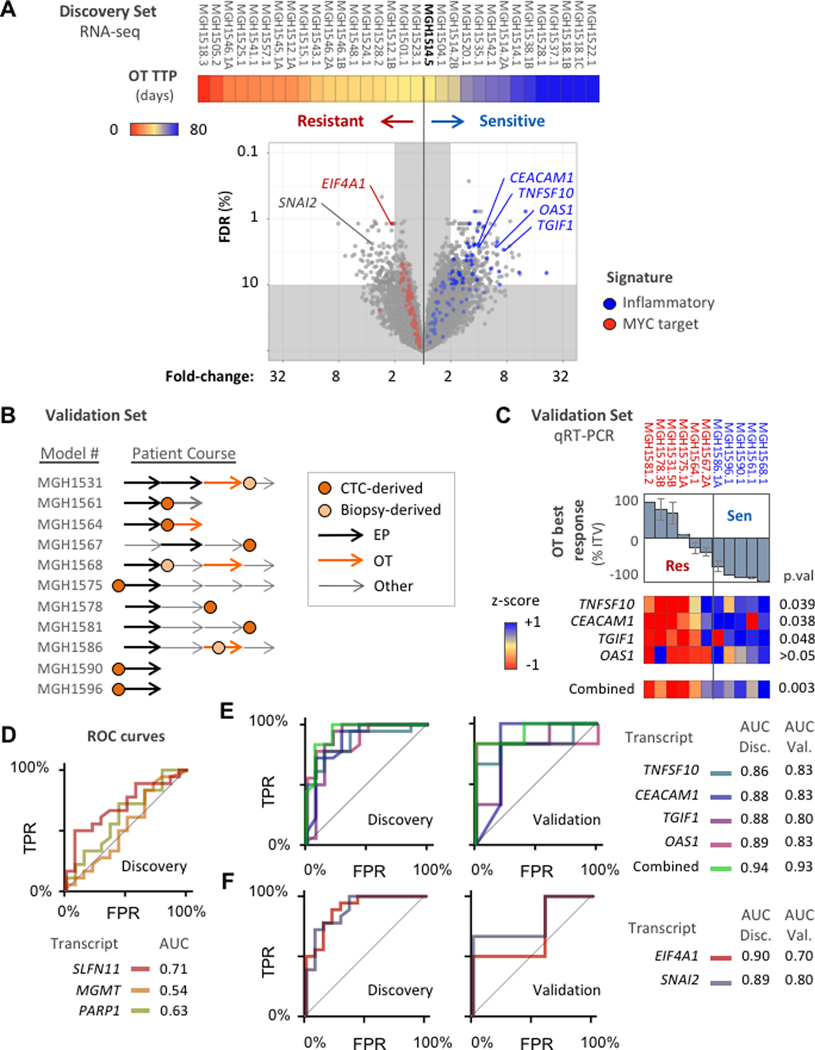
While our data suggest that EP resistance may predict OT resistance, this can be a difficult metric to apply clinically, and there may still be a valuable role for a molecular biomarker in further clinical development of OT. We therefore analyzed the relationships between OT sensitivity and gene expression to identify candidates predictive of response to this regimen. The six models derived from patients treated with OT on the clinical trial (Figure 4A–H) were used as calibration points to divide the 32-model discovery set into OT-sensitive and resistant cohorts. MGH1514–5, which was derived pre-treatment from a patient with RECIST SD (−28% best response), and which had a deeper best response and longer TTP than the post-relapse models MGH1528–2 and MGH1543–1, was selected to mark the boundary delineating sensitive and resistant cohorts in the PDX panel (Figure 4J, Figure 6A, Supplementary Figure S1A). Each transcript was scored for differential expression between the sensitive and resistant cohorts (Figure 6A, Supplementary Table 5). Genes with significant differential expression (>2-fold, FDR <10%) were further evaluated for (1) performance as a classifier (ROC curve AUC > 0.8), and (2) ease of distinguishing expression states (e.g. on/off, Bimodality Index > 1.1) [22]. In total, 216 genes satisfied these criteria for marking OT sensitivity, and 79 genes for OT resistance (Supplementary Table 5).
Figure 6. OT expression biomarker analysis.
(A) Volcano plot compares gene expression levels between OT-sensitive and OT-resistant models, Models ordered left-to-right by increasing TTP with OT (color bar, days) and threshold at MGH1514–5. Significance of difference (y-axis) = FDR-adjusted Welch’s t-test p-value (Benjamini-Hochberg method). Magnitude of difference (x-axis) = fold-difference in mean expression values between cohorts. Genes from the cross-resistance signatures in Figure 5E–F are colored: red = MYC target signature, blue = inflammatory signature. (B) Validation set of 11 PDX models treated with OT, with abstracted patient clinical courses as in Figure 4I. (C) Candidate transcripts labeled in (A) from inflammatory signature were measured by qRT-PCR across the validation set and compared with OT response. Top: waterfall plot of model best response (mean), with threshold from discovery set model MGH1514–5 (−40%). Bottom: heatmap of candidate gene expression z-scores, and aggregate z-scores, with unpaired t-test p-values. (D-F) ROC curves and AUC for candidate biomarkers to distinguish OT-sensitive and resistant models based on measurement of transcript abundance. TPR = true positive rate, FPR = false positive rate. (D) Hypothesis-driven biomarker candidates, discovery set only. (E) Inflammatory response signature candidates for OT sensitivity, discovery and validation sets. (F) OT resistance candidates, discovery and validation sets.
We treated an additional 11 SCLC PDX models for which RNA sequencing had not been performed with OT in vivo to serve as a validation set for candidate biomarkers (Figure 6B, Supplementary Figure S1B). The validation set was separated into sensitive and resistant cohorts by best response (−40% threshold from MGH1514–5), which correlates tightly with TTP (Supplementary Figure S2B). We hypothesized that genes that were members of underlying pathways upregulated in sensitive or resistant tumors would be more likely to validate in unknown tumors. Therefore we used the EP/OT cross-resistance signatures (Figure 5E, F) to guide candidate selection (Figure 6A, blue and red). 24 of the 82 members of the inflammatory response signature satisfied all statistical criteria for enrichment in the discovery set (Supplementary Table 5). We tested four of these candidates by quantitative RT-PCR in the validation set: CEACAM1 (CD66a), TNFSF10 (TRAIL), TGIF1 (TGFβ-induced factor I), and OAS1. Although our analysis represents basal gene expression across the PDX panel, interferon-induced expression has been well characterized for three candidates (CEACAM1, TNFSF10, OAS1) and TGFβ-induced expression for TGIF1 [23–29]. CEACAM1, TNFSF10, and TGIF1 were expressed at significantly higher levels in the 5 OT-sensitive validation models, and OAS1 performs as a complementary biomarker that enhances the combined specificity for OT sensitivity (Figure 6C, Supplementary Figure S5A). Aggregate expression of these four candidates strongly distinguished sensitive from resistant models in both the discovery and validation sets (Figure 6C, Supplementary Figure S5A).
Although the MYC target signature marked OT resistance, the dynamic range for its component transcripts was low (Figure 6A, Supplementary Table 4), with only EIF4A1 demonstrating significant differential expression in resistant models. Even for EIF4A1 basal expression across the panel was high, and distributions in resistant versus sensitive cohorts showed small differences and tight variation consistently in the discovery and validation sets (Supplementary Figure S5B). The MYC-target signature represents an aggregate of moderate differences in gene expression, rather than the standout individual transcripts in the inflammatory response set. Interestingly, SNAI2 (SLUG), a regulator of the epithelial-to-mesenchymal transition (EMT) and neural crest development [30–33], was among the best transcriptional classifiers of resistance outside the MYC signature (Figure 6A), and a role for EMT has been proposed for chemo-resistance in both SCLC and other solid tumors [12, 31, 34]. High SNAI2 expression also marked OT resistance in the validation set, and classifier performance was maintained at the protein level (Supplementary Figure S5C, D).
Expression biomarker candidates that emerged from the RNAseq screen were compared with genes with strong preclinical rationales: PARP1, MGMT and SLFN11 [11–13, 15, 35]. In the discovery set, SLFN11 expression performed as a classifier for OT (AUC = 0.71, 95% CI = 0.53–0.89), with optimal performance observed at thresholds that would distinguish the most resistant PDX models (Figure 6D and Supplementary Figure S5D, E). Neither PARP1 nor MGMT significantly distinguished OT-sensitive and resistant cohorts, as shown in ROC curve AUC values (Figure 6D). In contrast, the inflammatory response genes (CEACAM1, TNFSF10, OAS1, TGIF1) significantly outperformed the hypothesis-driven candidates as markers of OT sensitivity, and SNAI2 as a marker of resistance, in both the discovery and validation sets regardless of threshold (Figure 6E, F; Supplementary Figure S5A). These candidate biomarkers emerged from an analysis of cross-resistance and are unlikely to differentiate EP from OT, but instead may identify SCLC tumors with broad vulnerability to DNA damaging regimens.
DISCUSSION
SCLC remains one of the most aggressive and lethal malignancies, with a minimal role for curative surgery, no targetable driver mutations, and a median overall survival for metastatic disease generally under 1 year. Nonetheless, SCLC is initially highly sensitive to DNA damage, and combinations that pair DNA damaging agents with inhibitors of DNA damage checkpoints and repair, such as inhibitors of PARP, WEE1, CHK1 and ATR, have recently emerged as promising new strategies to treat relapsed SCLC [13–15, 18, 36–39]. Two challenges unify these approaches: (1) development of biomarkers to identify sensitive cases, and (2) determination of the common features of tumors that are cross-resistant to EP and subsequent DDR inhibitors. Recent progress has been made on both fronts, with the identification of SLFN11 as a marker of sensitivity for both PARP inhibition and EP [12, 13, 40], though unbiased screens for clinically relevant biomarkers have not been achieved. Here we describe a single-arm phase I/II study of OT in relapsed SCLC, with an integrated co-clinical trial in PDX models to discover candidate biomarkers and dissect cross-resistance to chemotherapy.
Among 50 patients with relapsed SCLC, combination OT shows a strong signal of clinical efficacy. Although cross-study comparisons are difficult to interpret, the confirmed ORR of 41.7% and mPFS of 4.2 months are numerically superior to several recent second- and third-line SCLC studies [9, 41, 42]. The addition of veliparib to temozolomide also showed improved responses in a placebo-controlled randomized phase 2 study, further supporting this strategy [13]. In contrast, monotherapy with the PARP inhibitor talazoparib led to only 2 partial responses among 23 SCLC patients treated in a phase 1 expansion study [43], and monotherapy olaparib did not confer a PFS benefit compared to placebo when used as a single agent in the maintenance setting after first-line platinum/etoposide [16]. The superiority of the combination over monotherapy was also evident in our PDX models, where OT was significantly more active than either drug alone (Supplementary Figure 2C). Collectively, these data support the further development of strategies that combine induction of DNA damage with PARP inhibition. Such an approach in the SCLC second-line setting is timely, given that standard of care is shifting to incorporate immune checkpoint blockade in combination with EP in the first-line setting [5].
A recent preclinical study also demonstrated striking synergy when DNA damage repair inhibitors (either olaparib or the CHK1 inhibitor prexasertib) were combined with PD-L1 inhibition [44]. Unfortunately, combination durvalumab and olaparib showed minimal clinical activity (ORR 10.5% among 19 evaluable patients) in an early phase clinical trial [45]. Although the low response rate to durvalumab/olaparib is disappointing, it should not stop further investigation of similar combinations. Mechanistically, if combination PARPi + temozolomide more potently induces DNA damage than single agent PARPi, as appears to be the case in clinical studies, OT may also be more robustly potentiated than olaparib alone when combined with an immune checkpoint inhibitor. A follow-on study combining OT with a PD-(L)1 inhibitor may be warranted. We would argue that this strategy could be beneficial even in SCLCs that have progressed after first-line chemo/IO [5], if for example cGAS and STING pathways were more robustly activated by OT than by carboplatin/etoposide.
Cross-resistance to DNA damaging regimens remains a major challenge in the management of relapsed SCLC. We established an experimental system tailored to study cross-resistance: a large collection of PDX models with diverse clinical histories in which the models derived from patients resistant to either EP or OT recapitulated this behavior in vivo. We compared EP and OT efficacy with gene expression across the panel, and found that the transcriptional profiles of drug resistance were highly similar, permitting an initial description of cross-resistant SCLC: low basal expression of mediators of innate immunity, and increased expression of MYC-regulated transcripts. The high MYC target signature echoes our previous findings for EP alone across a smaller panel of PDX models [19], but the low inflammatory signature was both robust and novel. Although the SCLC PDXs accurately recapitulate clinical sensitivity to EP and OT, they are tested in NSG mice, with near-total impairment of adaptive immunity. A similar signature of 38 interferon-stimulated genes (ISGs) was recently observed in a pan-cancer analysis of The Cancer Genome Atlas (TCGA), though not in the context of drug resistance [46]. This signature could not be attributed to immune or stromal infiltrates, but instead to chronic interferon production from the cancer cells themselves. Furthermore, the same ISG signature was observed in a large collection of 379 PDX models of diverse tumors grown in immunocompromised nude mice, and in the complete absence of an immune system cancer cell lines grown in vitro [46]. The SCLC PDX inflammatory signature in drug-sensitive models may represent a marker of increased endogenous DNA damage, as through replication stress, and this may decrease cellular tolerance of exogenous DNA damaging agents and inhibitors of repair. In this case, transcription of inflammatory response genes could result from low levels of cytosolic dsDNA detected via the cGAS-STING pathway [47–50], and it has recently been demonstrated that treatment with olaparib can activate the pathway in SCLC genetically engineered mouse (GEM) models [44]. Alternatively, components of the inflammatory response may play an active role in mediating the cytotoxicity of DNA damage, independent of CD8 T-cell or NK cell function. The cross-resistant PDX models identified here will provide valuable tools for investigating these possibilities, and uncovering the mechanistic underpinnings of a clinical phenotype that has a profound impact on survival in SCLC.
We were able to generate six PDX models from four patients on the OT trial. While additional PDX models from patients on trial may have further enhanced our translational studies, these six models nonetheless facilitated a search for dichotomous biomarkers for OT by forging a direct connection between the clinical and co-clinical trials (Figure 4A–H). These patient:PDX anchor points were used to calibrate in vivo model responses according to clinical history, allowing a rational division of the 32-model discovery set into sensitive/resistant cohorts. From the comparison of EP and OT transcriptional profiles, we recognized that the strongest connection between drug response and gene expression was the low inflammatory signature for cross-resistance. Four components of this signature that have been well-characterized as genes upregulated in response to either interferon signaling or TGFβ were tested in a validation set of 11 additional PDX models. These genes significantly outperformed current hypothesis-driven biomarker candidates for PARP inhibitor combinations in SCLC that have emerged from recent studies [12, 15, 40]: SLFN11, MGMT, PARP1, ATM, and EMT signature. The reasons for the differences across these studies may be due to differences in model systems, specific PARP inhibitor drugs and dosing strategies, study designs, or other factors. Ultimately, the optimal strategy for clinical application may be to combine candidates, as aggregate expression of CEACAM1 (CD66b), OAS1, TNFSF10 (TRAIL), and TGIF1 strongly distinguished OT sensitive from resistant models. The performance of these interferon response genes as classifiers in the validation set further supports the inflammatory response signature as a marker of sensitivity to OT, and likely other DNA damaging regimens, in SCLC. One limitation of our study is that the clinical trial did not require archival or fresh tissue for eligibility, and we had insufficient archival tissue available from patients on the study to be able to perform further validation of proposed biomarkers. This experience highlights the potential added value of requiring tissue samples from patients enrolling onto studies in the future.
While SCLC has historically been treated as a homogenous disease, this paradigm is poised to shift with identification of functionally distinct subgroups [20] that may benefit from different therapies. Our PDX co-clinical trial strategy can be applied to early phase SCLC trials to accelerate this progress. The anchor-point trial design leverages the size of the PDX panel without sacrificing clinical context, presenting advantages over other laboratory models such as established cell lines as well as scarce archival patient samples. Each model within this diverse and well-annotated panel can be assessed for sensitivity to multiple therapies, permitting direct comparisons to segregate SCLC into functional as well as molecular subgroups. Predictive biomarker discovery is not the only application of the co-clinical trial strategy; well-validated PDX models from trial patients can be used to optimize dosing schedules, and serial models derived before and after therapy represent a powerful tool for uncovering mechanisms of acquired resistance. We anticipate that PDX co-clinical trials will provide critical contributions to breaking the monolithic view of SCLC and establishing personalized medicine for this recalcitrant disease. This approach may be further applied more broadly to refine patient selection and drug development strategies for therapies with a signal of clinical activity in early-phase trials.
METHODS
Clinical trial eligibility criteria
This was a single-arm, open-label, single-institution phase I/II study (NCT 02446704). The study was reviewed and approved by the Dana-Farber/Harvard Cancer Center Institutional Review Board. Written informed consent was provided by all participants and the study was performed in accordance with the Declaration of Helsinki. Key inclusion criteria included patients ≥ 18 years old with histologically or cytologically confirmed SCLC who were not candidates for curative therapy, and had received prior first-line platinum/etoposide with subsequent radiographic progression. Any number of interval prior therapies were allowed. Eligibility criteria were the same for the phase I and phase II portions of the study, as detailed in the clinical protocol (Supplementary Data).
Clinical trial treatment
In the Phase I portion of the study, patients were treated using a 3+3 dose escalation study design. The starting dose level was olaparib tablets 100 mg PO BID and temozolomide capsules 50 mg/m2 PO QPM, both administered days 1–7 of a 21-day cycle. Subsequent dose levels escalated one drug at a time, with the same days 1–7 administration schedule (Figure 1). Treatment was continued until disease progression, concurrent illness preventing further administration of treatment, or DLT not resolving within 7 days, or at the discretion of the participant or investigator. Continuation of treatment post-progression was allowed in cases of ongoing clinical benefit. Cycle length could be extended to 28 days at the investigator’s discretion in cases where additional time was needed to recover from treatment related adverse events. Growth factor support was not allowed.
The phase II portion of the study was a dose expansion at dose level 3, which was selected as the recommended phase 2 dose (RP2D) (Figure 1), with the primary objective to determine overall response rate. During the initial 20 patient expansion, patients requiring dose reductions could be reduced by > 1 dose level at a time (i.e., dose level 3 to 1) at the treating physician’s discretion. A second 20 patient expansion was added in a protocol amendment. In the second expansion group, if dose reductions were required below the RP2D, patients were reduced first to a new dose level 2A (olaparib 200 mg PO BID and temozolomide 50 mg/m2 PO QPM) for at least 1 cycle, and then to dose level 1 if further reduction was required.
Clinical trial study evaluations
Clinical and laboratory evaluations were required weekly during treatment cycle 1 and on day 1 of each subsequent cycle. Dose limiting toxicities (DLTs) were monitored during cycle 1. Toxicity assessments were performed using the NCI Common Terminology for Adverse Events (CTCAE v4.0). Response assessment by CT of the chest, abdomen and pelvis was performed every 6 weeks. Brain imaging by MRI or CT was required at baseline and subsequently at the discretion of the investigator. Best objective response was determined using RECIST 1.1 criteria [51].
Clinical trial statistical analysis
The primary endpoint of the phase I portion was presence of DLT, with the objective of determining the MTD and RP2D of combination olaparib and temozolomide. The primary endpoint of the phase II portion was overall response rate. For the phase II portion, a multi-stage optimal design was used to allow early termination due to lack of efficacy. After the first 9 evaluable patients were treated at the RP2D, accrual was continued only if at least one partial or complete best overall response had been observed, in which case the protocol then allowed enrollment of an additional 11 patients for a total accrual of 20 at the RP2D. If at least 4 responses were observed among these 20 patients, the protocol proceeded to enroll an additional 20 patients at the RP2D. The multi-stage design provided 87% power to determine that the RP2D combination of combination olaparib and temozolomide was associated with a 30% overall response rate if at least 6 patients achieved a response in total. The study design is associated with type 1 error of 9% if the underlying rate of overall response were truly 10% suggesting a lack of efficacy. Best objective response was determined using RECIST 1.1 criteria [51]. All patients who had at least one response assessment performed were included in the response evaluation. All enrolled patients were included in the PFS and OS analyses. Duration of response (DOR), PFS and OS rates were estimated by the Kaplan-Meier method, and platinum sensitive and resistant groups were compared using the score test of the proportional hazards model. Data analysis was performed using SAS 9.4 (SAS Inst Inc, Cary, NC), and p-values were based on a 2-sided hypothesis. A data cutoff of May 31 2018 was used for toxicity analysis and November 6 2018 was used for efficacy analysis.
PDX model generation, treatment and evaluation.
All tissue and blood samples from patients were collected per Institutional Review Board (IRB) approved protocols with written informed consent from the patients and in accordance with the Declaration of Helsinki. All mouse studies were conducted through IACUC approved animal protocols in accordance with Massachusetts General Hospital institutional guidelines. The source of tumor material for model generation was either core needle biopsy or circulating tumor cells. PDX model generation and treatment with EP and OT were as previously described [19]. Notably, for models previously reported [19], additional replicate xenografts have been added. Furthermore, new models not treated with either regimen are included in this data set. Briefly, treatment studies were initiated at xenograft volumes = 400–600 mm3 for 2–6 mice per model per treatment arm, and tumors were measured 2–3x weekly. OT: olaparib 50 mg/kg oral gavage (OG) d1–5 + temozolomide 25 mg/kg OG d1–5. EP: cisplatin 7 mg/kg intraperitoneal (IP) d1,8 + etoposide 10 mg/kg IP d1–3,8–10. Trial tumor metrics: TTP = days from start of treatment to 2x initial tumor volume (ITV), response = change in tumor volume between ITV and d7–28 minimum, Endpoints: tumor volume > 2x ITV or 100 days after start of treatment.
PDX model RNA sequencing and downstream analysis
Paired-end transcriptome sequencing was performed in biologic duplicate for each model. Two untreated mice per model were selected with xenograft volumes 800–1200 mm3, and tumor fragments were flash-frozen in liquid nitrogen within 5 minutes of euthanasia. Frozen tissue fragments were lysed and homogenized with the Tissue Lyzer (Qiagen), and RNA extractions were performed with the Qiagen RNAeasy Mini Kit. RNA quality was assessed with a Bioanalyzer 2100 DNA Chip 7500 (Agilent Technologies) and samples with an RNA integrity number (RIN) of over 8 were selected for library construction. cDNA libraries were prepared with the Illumina “truseq-stranded-mrna-sample-prep” kit and sequenced with a paired-end 2×75 bp protocol on an Illumina HiSeq 4000 instrument at the Novartis Institute for Biomedical Research. Sequence alignments were performed against a chimeric transcriptome index consisting of human and mouse reference genomes (hg38 and mm10) to sort PDX- and mouse-specific reads. Transcript abundance values, transcripts per million (TPM), were estimated by salmon (0.8.2) [52] using the following options (-l ISR, --seqBias, --gcBias, --useVBOpt). Gene level expression was generated using the R tximport package [53]. As expression values approximated a log-normal distribution, expression values (TPM) were log-transformed (log2(1+TPM)). Model-level expression values were calculated by averaging gene expression across biological replicates. Low expression genes (max log2(1+TPM) <1) and expressed genes with more than 25% average variation between replicates (mean replicate CV > 0.25) were removed.
Untreated PDX transcript abundance (log2(1+TPM)) was compared with mean PDX TTP following treatment with EP vs. OT for each of the 32 models in the discovery set by Pearson correlation, and the threshold for significant correlation was assigned at an FDR-adjusted p-value of 10% (Benjamini-Hochberg method). Gene set enrichment analysis (GSEA) [54] was performed for transcript abundance vs. EP and OT TTP (treated as continuous variables, Pearson method), using the Hallmark gene set collection (MSigDB v6.2, 5000 permutations). Normalized enrichment scores (NES) were compared and significant enrichment was assigned for FDR <25%. Leading edge analysis was performed for each significantly enriched gene set for EP or OT, and genes present in the leading edge of at least one gene set (union) for both regimens (intersect) were assigned to one of two expression signatures: inflammatory response (positively correlated with EP/OT TTP, 82 genes) or MYC target (negatively correlated with EP/OT TTP, 65 genes). Geneset signature scores were calculated as the average transcript abundance z-scores of the signature genes for each model versus the PDX panel.
Transcript expression levels were compared in OT-sensitive versus resistant cohorts, with model MGH1514–5 demarcating the two cohorts. Genes with at least 2-fold change in expression between the cohorts and an FDR-adjusted Welch’s t-test p-value of less than 10% (Benjamini-Hochberg method) were selected for further analysis. Each differentially expressed transcripts was assessed for biomarker suitability using two criteria, performance as a two-population classifier, and bimodal distribution of expression levels. Bimodal expression was estimated using the bimodality index [22] (BimodaIndex R package), a measure of the degree to which gene expression across the PDX panel fits a two-component mixture model, and classifier performance was estimated by the receiver-operator characteristic area-under the curve (ROC AUC, pROC R package) [55].
PDX model quantitative RT-PCR
Total cell RNA was extracted from PDX snap frozen tissue using the Qiagen RNAeasy Mini Kit (Qiagen, Cat. No. 74104). cDNA was synthesized from 1 μg total RNA, using oligo-dT priming and the TaqMan Reverse Transcription kit (Applied biosystems, Cat. No. N8080234) following manufacturer’s instructions. Relative expression of transcripts was quantified by real-time PCR, using the FastStart Universal SYBR Green Master mix (Roche, Cat. No. 4913914001) and the LightCycler 480 System (Roche). mRNA levels were normalized to ACTB. Primer sets are listed in Supplementary Methods.
Supplementary Material
STATEMENT OF SIGNIFICANCE.
We demonstrate substantial clinical activity of combination olaparib/temozolomide in relapsed SCLC, revealing a promising new therapeutic strategy for this highly recalcitrant malignancy. Through an integrated co-clinical trial in PDXs, we then identify a molecular signature predictive of response to OT, and describe the common molecular features of cross-resistant SCLC.
ACKNOWLEDGEMENTS:
We are grateful to the patients and families who participated in these research studies. We thank the members of the MGH thoracic oncology group and other MGH Cancer Center staff for assistance with recruitment of patients and collection of samples, and Edwin Choy for his guidance around initial protocol design. We thank I. Sanidas, A. Guarner-Peralta, B. Krishnan, P. Rumde, V. Kamesan, J. Grinnell, and current and former members of the Dyson and Farago research groups; and L. Zou, R. Corcoran and C. Benes for critical discussions and scientific input. We thank Jeffrey Engelman for his vision and guidance in launching this project.
Financial Support: This work has been supported by AstraZeneca (ISS22810111 to A.F.F), National Cancer Institute grant no. U01CA220323-A1 (N.J.D., A.F.F) and grant no. U24CA213274 (A.F.F.), the V Foundation translational grant no. T2016–003 (N.J.D.), NIH career development award no. K12CA087723 (A.F.F.), career development awards from Uniting Against Lung Cancer (A.F.F.) and the Lung Cancer Research Foundation (B.J.D.), the ASCO Young Investigator Award (B.J.D.), the research fellowship grant from the Deutsche Forschungsgemeinschaft (M.S.), NIH RO1 CA1299933 (D.A.H.), the Howard Hughes Medical Institute and National Foundation for Cancer Research (D.A.H.), and a research agreement through the Novartis-MGH Alliance.
Footnotes
Conflict of interest disclosure statement: The authors declare the following competing interests: A.F.F. consulting/advisory role for PharmaMar, Abbvie, Loxo, Stemcentrx, Genentech/Roche, Bayer, AstraZeneca, Bristol-Myers Squibb, and Boerhinger Ingelheim; research funding from PharmaMar, AbbVie, AstraZeneca, Bristol-Myers Squibb, Merck, Loxo, Ignyta, Amgen, Genentech/Roche, Bayer and Novartis. R.S.H consulting/advisory role for Boehringer Ingelheim, Novartis, Tarveda Therapeutics, and Apollomics; research funding from AbbVie, Novartis, Roche, Incyte, Celgene, Mirati Therapeutics, Peregrine Pharmaceuticals, Millenium, Debiopharm Group, Corvus Pharmaceuticals, Daiichi Sankyo, Agios, Exelixis, and Pfizer. D.R. research funding from Bristol-Myers Squibb. D.A.B. consulting/advisory role for N of One and Tango Therapeutics; honoraria from Loxo Oncology and Madalon Consulting; research funding from Novartis and Bristol-Myers Squibb. S.R.D. provides independent image analysis for clinical trials sponsored by Merck, Pfizer, Bristol-Myers Squibb, Novartis, Roche, Polaris, Cascadian, Abbvie, Gradalis, Clinical Bay, Zai Laboratories, and received honorarium from Siemens Medical Solutions. L.V.S. consulting/advisory role for Merrimack, Genentech, Blueprint, Pfizer, Janssen and AstraZeneca; research funding from Boerhinger Ingelheim, AstraZeneca, Novartis, Blueprint, LOXO; honoraria from AstraZeneca. A.N.H. research funding from Amgen, Pfizer, Relay Therapeutics, Novartis. D.A.H. is cofounder of TorpedoDx, which is developing the CTC-iChip technology, and serves on the SAB for Janssen Pharma. A.T.S. consultant/advisory role for Pfizer, Novartis, Genentech/Roche, Ariad/Takeda, Ignyta, Loxo, Blueprint Medicines, KSQ Therapeutics, Daiichi Sankyo, EMD Serono, Taiho Pharmaceutical, TP Therapeutics, Foundation Medicine, Natera, and Guardant; research funding from Pfizer, Novartis, and Roche/Genentech. M.M.K. has as consulting/advisory role for Merrimack Pharmaceuticals and H3 biomedicine. N.J.D. and B.J.D. receive research funding from Novartis, AbbVie, Merck and AstraZeneca. The remaining authors declare no competing interests.
REFERENCES
- [1].George J, Lim JS, Jang SJ, Cun Y, Ozretic L, Kong G, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524:47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Rudin CM, Durinck S, Stawiski EW, Poirier JT, Modrusan Z, Shames DS, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet. 2012;44:1111–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Peifer M, Fernandez-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012;44:1104–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Farago AF, Keane FK. Current standards for clinical management of small cell lung cancer. Transl Lung Cancer Res. 2018;7:69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Horn L, Mansfield AS, Szczesna A, Havel L, Krzakowski M, Hochmair MJ, et al. First-Line Atezolizumab plus Chemotherapy in Extensive-Stage Small-Cell Lung Cancer. N Engl J Med. 2018;379:2220–9. [DOI] [PubMed] [Google Scholar]
- [6].Owonikoko TK, Behera M, Chen Z, Bhimani C, Curran WJ, Khuri FR, et al. A systematic analysis of efficacy of second-line chemotherapy in sensitive and refractory small-cell lung cancer. J Thorac Oncol. 2012;7:866–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ardizzoni A, Hansen H, Dombernowsky P, Gamucci T, Kaplan S, Postmus P, et al. Topotecan, a new active drug in the second-line treatment of small-cell lung cancer: a phase II study in patients with refractory and sensitive disease. The European Organization for Research and Treatment of Cancer Early Clinical Studies Group and New Drug Development Office, and the Lung Cancer Cooperative Group. J Clin Oncol. 1997;15:2090–6. [DOI] [PubMed] [Google Scholar]
- [8].Eckardt JR, von Pawel J, Pujol JL, Papai Z, Quoix E, Ardizzoni A, et al. Phase III study of oral compared with intravenous topotecan as second-line therapy in small-cell lung cancer. J Clin Oncol. 2007;25:2086–92. [DOI] [PubMed] [Google Scholar]
- [9].von Pawel J, Jotte R, Spigel DR, O’Brien ME, Socinski MA, Mezger J, et al. Randomized phase III trial of amrubicin versus topotecan as second-line treatment for patients with small-cell lung cancer. J Clin Oncol. 2014;32:4012–9. [DOI] [PubMed] [Google Scholar]
- [10].von Pawel J, Schiller JH, Shepherd FA, Fields SZ, Kleisbauer JP, Chrysson NG, et al. Topotecan versus cyclophosphamide, doxorubicin, and vincristine for the treatment of recurrent small-cell lung cancer. J Clin Oncol. 1999;17:658–67. [DOI] [PubMed] [Google Scholar]
- [11].Byers LA, Wang J, Nilsson MB, Fujimoto J, Saintigny P, Yordy J, et al. Proteomic profiling identifies dysregulated pathways in small cell lung cancer and novel therapeutic targets including PARP1. Cancer Discov. 2012;2:798–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Stewart AC, Tong P, Cardnell RJ, Sen T, Li L, Gay CM, et al. Dynamic variations in epithelial-to-mesenchymal transition (EMT), ATM, and SLFN11 govern response to PARP inhibitors and cisplatin in small cell lung cancer. Oncotarget. 2017;8:28575–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Pietanza MC, Waqar SN, Krug LM, Dowlati A, Hann CL, Chiappori A, et al. Randomized, Double-Blind, Phase II Study of Temozolomide in Combination With Either Veliparib or Placebo in Patients With Relapsed-Sensitive or Refractory Small-Cell Lung Cancer. J Clin Oncol. 2018;36:2386–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Owonikoko TK, Dahlberg SE, Sica GL, Wagner LI, Wade JL 3rd, Srkalovic G, et al. Randomized Phase II Trial of Cisplatin and Etoposide in Combination With Veliparib or Placebo for Extensive-Stage Small-Cell Lung Cancer: ECOG-ACRIN 2511 Study. J Clin Oncol 2019;37:222–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lok BH, Gardner EE, Schneeberger VE, Ni A, Desmeules P, Rekhtman N, et al. PARP Inhibitor Activity Correlates with SLFN11 Expression and Demonstrates Synergy with Temozolomide in Small Cell Lung Cancer. Clin Cancer Res. 2017;23:523–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Woll P, Gaunt P, Steele N, Ahmed S, Mulatero C, Shah R, et al. P1.07–015 STOMP: A UK National Cancer Research Network Randomised, Double Blind, Multicentre Phase II Trial of Olaparib as Maintenance Therapy in SCLC. Journal of Thoracic Oncology. 2017;12:S704–S5. [Google Scholar]
- [17].Hopkins TA, Shi Y, Rodriguez LE, Solomon LR, Donawho CK, DiGiammarino EL, et al. Mechanistic Dissection of PARP1 Trapping and the Impact on In Vivo Tolerability and Efficacy of PARP Inhibitors. Mol Cancer Res. 2015;13:1465–77. [DOI] [PubMed] [Google Scholar]
- [18].Murai J, Zhang Y, Morris J, Ji J, Takeda S, Doroshow JH, et al. Rationale for poly(ADP-ribose) polymerase (PARP) inhibitors in combination therapy with camptothecins or temozolomide based on PARP trapping versus catalytic inhibition. J Pharmacol Exp Ther. 2014;349:408–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Drapkin BJ, George J, Christensen CL, Mino-Kenudson M, Dries R, Sundaresan T, et al. Genomic and Functional Fidelity of Small Cell Lung Cancer Patient-Derived Xenografts. Cancer Discov. 2018;8:600–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Rudin CM, Poirier JT, Byers LA, Dive C, Dowlati A, George J, et al. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat Rev Cancer. 2019;19:289–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zhang W, Girard L, Zhang YA, Haruki T, Papari-Zareei M, Stastny V, et al. Small cell lung cancer tumors and preclinical models display heterogeneity of neuroendocrine phenotypes. Transl Lung Cancer Res. 2018;7:32–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wang J, Wen S, Symmans WF, Pusztai L, Coombes KR. The bimodality index: a criterion for discovering and ranking bimodal signatures from cancer gene expression profiling data. Cancer Inform. 2009;7:199–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Attallah AM, Needy CF, Noguchi PD, Elisberg BL. Enhancement of carcinoembryonic antigen expression by interferon. Int J Cancer. 1979;24:49–52. [DOI] [PubMed] [Google Scholar]
- [24].Takahashi H, Okai Y, Paxton RJ, Hefta LJ, Shively JE. Differential regulation of carcinoembryonic antigen and biliary glycoprotein by gamma-interferon. Cancer Res. 1993;53:1612–9. [PubMed] [Google Scholar]
- [25].Chen CJ, Lin TT, Shively JE. Role of interferon regulatory factor-1 in the induction of biliary glycoprotein (cell CAM-1) by interferon-gamma. J Biol Chem. 1996;271:28181–8. [DOI] [PubMed] [Google Scholar]
- [26].Kayagaki N, Yamaguchi N, Nakayama M, Eto H, Okumura K, Yagita H. Type I interferons (IFNs) regulate tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) expression on human T cells: A novel mechanism for the antitumor effects of type I IFNs. J Exp Med. 1999;189:1451–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wang Q, Ji Y, Wang X, Evers BM. Isolation and molecular characterization of the 5’-upstream region of the human TRAIL gene. Biochem Biophys Res Commun. 2000;276:466–71. [DOI] [PubMed] [Google Scholar]
- [28].Hovnanian A, Rebouillat D, Mattei MG, Levy ER, Marie I, Monaco AP, et al. The human 2’,5’-oligoadenylate synthetase locus is composed of three distinct genes clustered on chromosome 12q24.2 encoding the 100-, 69-, and 40-kDa forms. Genomics. 1998;52:267–77. [DOI] [PubMed] [Google Scholar]
- [29].Bridge AJ, Pebernard S, Ducraux A, Nicoulaz AL, Iggo R. Induction of an interferon response by RNAi vectors in mammalian cells. Nat Genet. 2003;34:263–4. [DOI] [PubMed] [Google Scholar]
- [30].Ye X, Tam WL, Shibue T, Kaygusuz Y, Reinhardt F, Ng Eaton E, et al. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature. 2015;525:256–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol. 2017;14:611–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sanchez-Martin M, Rodriguez-Garcia A, Perez-Losada J, Sagrera A, Read AP, Sanchez-Garcia I. SLUG (SNAI2) deletions in patients with Waardenburg disease. Hum Mol Genet. 2002;11:3231–6. [DOI] [PubMed] [Google Scholar]
- [33].Cheung M, Chaboissier MC, Mynett A, Hirst E, Schedl A, Briscoe J. The transcriptional control of trunk neural crest induction, survival, and delamination. Dev Cell. 2005;8:179–92. [DOI] [PubMed] [Google Scholar]
- [34].Wu WS, Heinrichs S, Xu D, Garrison SP, Zambetti GP, Adams JM, et al. Slug antagonizes p53-mediated apoptosis of hematopoietic progenitors by repressing puma. Cell. 2005;123:641–53. [DOI] [PubMed] [Google Scholar]
- [35].Erice O, Smith MP, White R, Goicoechea I, Barriuso J, Jones C, et al. MGMT Expression Predicts PARP-Mediated Resistance to Temozolomide. Mol Cancer Ther. 2015;14:1236–46. [DOI] [PubMed] [Google Scholar]
- [36].Lallo A, Frese KK, Morrow CJ, Sloane R, Gulati S, Schenk MW, et al. The Combination of the PARP Inhibitor Olaparib and the WEE1 Inhibitor AZD1775 as a New Therapeutic Option for Small Cell Lung Cancer. Clin Cancer Res. 2018;24:5153–64. [DOI] [PubMed] [Google Scholar]
- [37].Laird JH, Lok BH, Ma J, Bell A, de Stanchina E, Poirier JT, et al. Talazoparib Is a Potent Radiosensitizer in Small Cell Lung Cancer Cell Lines and Xenografts. Clin Cancer Res. 2018;24:5143–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Thomas A, Redon CE, Sciuto L, Padiernos E, Ji J, Lee MJ, et al. Phase I Study of ATR Inhibitor M6620 in Combination With Topotecan in Patients With Advanced Solid Tumors. J Clin Oncol. 2018;36:1594–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sen T, Tong P, Stewart CA, Cristea S, Valliani A, Shames DS, et al. CHK1 Inhibition in Small-Cell Lung Cancer Produces Single-Agent Activity in Biomarker-Defined Disease Subsets and Combination Activity with Cisplatin or Olaparib. Cancer Res. 2017;77:3870–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Gardner EE, Lok BH, Schneeberger VE, Desmeules P, Miles LA, Arnold PK, et al. Chemosensitive Relapse in Small Cell Lung Cancer Proceeds through an EZH2-SLFN11 Axis. Cancer Cell. 2017;31:286–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ready N, Farago AF, de Braud F, Atmaca A, Hellmann MD, Schneider JG, et al. Third-Line Nivolumab Monotherapy in Recurrent SCLC: CheckMate 032. J Thorac Oncol. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Evans TL, Cho BC, Udud K, Fischer JR, Shepherd FA, Martinez P, et al. Cabazitaxel Versus Topotecan in Patients with Small-Cell Lung Cancer with Progressive Disease During or After First-Line Platinum-Based Chemotherapy. J Thorac Oncol. 2015;10:1221–8. [DOI] [PubMed] [Google Scholar]
- [43].de Bono J, Ramanathan RK, Mina L, Chugh R, Glaspy J, Rafii S, et al. Phase I, Dose-Escalation, Two-Part Trial of the PARP Inhibitor Talazoparib in Patients with Advanced Germline BRCA1/2 Mutations and Selected Sporadic Cancers. Cancer Discov. 2017;7:620–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Sen T, Rodriguez BL, Chen L, Corte CMD, Morikawa N, Fujimoto J, et al. Targeting DNA Damage Response Promotes Antitumor Immunity through STING-Mediated T-cell Activation in Small Cell Lung Cancer. Cancer Discov. 2019;9:646–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Thomas A, Vilimas R, Trindade C, Erwin-Cohen R, Roper N, Xi L, et al. Durvalumab in Combination with Olaparib in Patients with Relapsed Small Cell Lung Cancer: Results from a Phase II Study. J Thorac Oncol. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Liu H, Golji J, Brodeur LK, Chung FS, Chen JT, deBeaumont RS, et al. Tumor-derived IFN triggers chronic pathway agonism and sensitivity to ADAR loss. Nat Med. 2019;25:95–102. [DOI] [PubMed] [Google Scholar]
- [47].Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Wu J, Sun L, Chen X, Du F, Shi H, Chen C, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45:228–47. [DOI] [PubMed] [Google Scholar]
- [52].Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 2015;4:1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.