Abstract
To improve drug discovery yield, a method which is implemented at the beginning of drug discovery that accurately predicts drug side effects, indications, efficacy, and mode of action based solely on the input of the drug’s chemical structure is needed. In contrast, extant predictive methods do not comprehensively address these aspects of drug discovery and rely on features derived from extensive, often unavailable experimental information for novel molecules. To address these issues, we developed MEDICASCY, a multi-label based boosted random forest machine learning method that only requires the small molecule’s chemical structure for drug side effect, indication, efficacy, and probable mode of action target predictions, yet, it has comparable or even significantly better performance than existing approaches requiring far more information. In retrospective benchmarking on high confidence predictions, MEDICASCY shows about 78% precision and recall for predicting at least one severe side effect, and 72% precision drug efficacy. Experimental validation of MEDICASCY’s efficacy predictions on novel molecules shows close to 80% precision for the inhibition of growth in ovarian, breast and prostate cancer cell lines. Thus, MEDICASCY should improve the success rate for new drug approval. A web service for academic users is available at http://pwp.gatech.edu/cssb/MEDICASCY.
Keywords: machine learning, drug side effect prediction, drug efficacy prediction, indication, mode of action protein, drug clinical trials
INTRODUCTION
The cost of developing a new drug for complex diseases has been estimated to be about 1.4 billion dollars 1. Moreover, the likelihood of a drug being approved is quite low 2, 3. For example, the overall success rate for a drug candidate to pass a Phase 1 trial is 13.8% 3. The success rates from Phase 1 to 2, Phase 2 to 3 and Phase 3 to final approval are 66.4%, 58.3% and 59.0%, respectively. There can be many reasons for drug failure. While lack of safety is the major cause of Phase 1 failures, lack of efficacy is responsible for over 50% of Phase 2 and 3 drug failures 4. To reduce the low success rates for new drug development, computational methods that accurately predict severe drug side effects and appropriate drug indications are sorely needed. In that regard, there have been a number of methods developed for predicting drug side effects 5–15 or efficacy 16, 17; these treatments address drug side effects and efficacy independently and often do not provide insight into the mode of action of a successful or failed drug. Moreover, except for the empirical drug side effect prediction method developed in our earlier work, DR. PRODIS 7, whose drug coverage and precision for severe side effect prediction was limited, all alternative approaches require very extensively curated experimental or/and bioinformatics data as input for the given drug. For new drug development, these kinds of data are mostly unavailable, and the acquisition of such data for a large ligand library whose translational relevance is unknown is prohibitively expensive. Thus, their practical application is limited to the repurposing of well-studied drugs 16, 18; as such, they are not much help in new drug development.
To begin to address this critical need, Zhou et al developed DR. PRODIS 7, an empirical side effect prediction method that can be applied to new drugs and which merely requires the drug’s chemical structure as its input. As a first step, DR. PRODIS employs the virtual ligand screening algorithm FINDSITEcomb 19 to predict its binding human protein targets. Then, using the drug’s predicted protein targets, it’s possible side effects on humans are predicted by an empirical drug inference method. In practice, DR. PRODIS was applied to all small molecules in DrugBank 20. In testing on the 996 SIDER2 drug set 21, DR. PRODIS gives an average precision and recall per drug of 56.5% and 23.6%, respectively. This performance is better than the 30% precision at a 24% recall rate of the earlier machine learning based method that required substantial experimental and bioinformatics data as input10.
In this paper, to improve the accuracy for predicting side effects, especially severe ones, provided by DR. PRODIS whose precision for predicting at least one severe side effect is 58.7% and which could neither predict drug efficacy, nor drug mode of action, we have developed MEDICASCY (Machine lEarning approach for preDICting mode of Action, Side effects, indications efficaCY of small molecule drugs). MEDICASCY employs sophisticated Boosted Random Forest (BRF) machine learning approaches where small molecule ligand-protein partners are predicted from a significantly improved version of FINDSITEcomb 19, FINDSITEcomb2.0 22. MEDICASCY has two models, one predicts side effects and the other predicts indications. Efficacy and modes of action are then derived from the side effect and indication predictions. The advantage of these simultaneous predictions is that by predicting side effects and indications within the same framework, MEDICASCY can filter out those drugs having severe side effects and distinguish efficacious indications from non-efficacious indications and from side effects that might cause the disease indication. For example, a drug with the predicted side effect of cancer might cause cancer, whereas a drug with the cancer indication but without the cancer side effect might be useful for treating cancer. Moreover, when combined with the predicted human protein targets, MEDICASCY can be utilized to infer probable mode of action targets responsible for the side effect and/or indication. Thus, it can also provide deeper insights into how the drug works that go beyond the single disease-causing protein target-drug interaction paradigm of drug discovery. This combined ability to predict drug side effects, indications, efficacy and mode of action within one framework is, to the best of our knowledge, a unique feature of MEDICASCY. Here, we benchmark this new method against the state-of-the-art methods and show that by just using the drug’s chemical structure, MEDICASCY performs comparably to or is often better than the state-of-the-art methods 5, 6, 12, 16 that require extensive experimental and/or bioinformatics data as input. Thus, MEDICASCY is particularly useful for new drug development when such experimental and/or bioinformatics data are rarely available.
MATERIALS AND METHODS
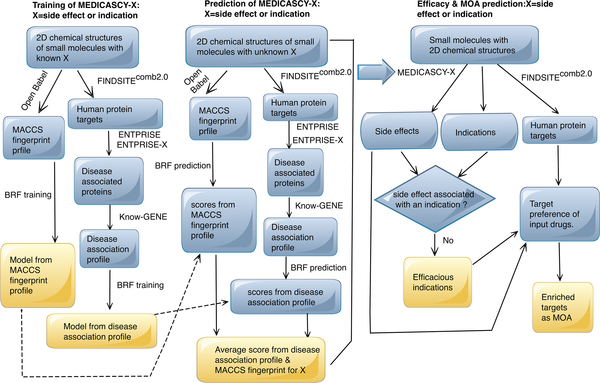
The flowchart of MEDICASCY is shown in Figure 1. The training and prediction procedures are the same for both side effects and indications. They only differ in the dataset used for training. For a drug to be predicted to be efficacious for a given indication, it must be predicted as having that indication but not have the side effect associated with the indication. For example, a given drug can have the indication as cancer, but also have the same side effect as cancer. We would then predict that the drug could cause cancer. Conversely, if a drug has the indication as cancer but not the side of effect of cancer, then it would be predicted to be efficacious in cancer treatment.
Figure 1:
Flowchart of the MEDICASCY algorithm.
For training, the inputs are the known side effects or indications of a library of drugs and their corresponding two-dimensional (2D) chemical structures. The training objective function (label) values are multi-dimensional vectors. For example, for SIDER4 side effects, the dimension is 4251 representing 4251 types of side effects. If a drug has a given type of side effect, the value of the coordinate representing the side effect is set to 1, otherwise, it is 0. The dimension of the indication label vector is 3608. Side effects and indications are trained independently of each other. We compute two classes of machine learning features: the predicted disease association profiles of the protein targets and the fingerprints of the drugs (see below for details) which is based on the idea that similar drugs might have similar side effects and indications. A Boosted Random Forest (BRF) multi-label regression approach learns from known side effect or disease indication data sets to generate models. Each class of features is used independently to generate the requisite models. For the prediction of drug side effects or drug disease indications, the only input required for the drug is its 2D chemical structure. Again, the predicted disease association profiles and fingerprints of the drugs are computed. The BRF uses the corresponding models trained from the two different feature classes to make two predictions. The predicted label vector has the same dimension as the training labels. However, the coordinate can assume any value between 0 and 1. The final prediction label score is the average of the two disparate predictions, i.e. the average label vector of the two label vectors.
Machine learning method
A newly developed, Boosted Random Forest approach for multiple label regression is employed for learning and prediction. A Random Forest (RF) is an ensemble learning method for classification and regression23. Here, we utilize a RF machine learning methodology because we found that it performs better than alternatives such as k-nearest neighbors (kNN)24 or multi-layer perceptron methods5,when boosting is applied on the RF. A Boosted Random Forest is an iterated application of Random Forests: each time the residual function value of all previous fittings is fit:
| (1a), |
where δ is a learning rate parameter. In this work, we set δ = 0.02 and the total number of iterations to be NRF=50. The final regression function is
| (1b). |
We implement the BRF using the standard Python package scikit-learn (https://scikit-learn.org/) with the default setting (n_estimators=10) of the multi-value regression function RandomForestRegressor(). It should be noted that the performance of a single random forest using the default setting is worse than some other methods5. However, during the development of the BRF method, we found that a RF regression with a larger value of n_estimators, say 500, can achieve the same accuracy as a BRF with NRF=50, but it would require the use of computers with very large memory and much longer computational times. Thus, BRF is the better practical choice.
Machine Learning Features
One set of features comes directly from the chemical structure converted to MACCS fingerprints using the Open Babel software (http://openbabel.org/wiki/Main_Page). During the development of MEDICASY, we have tried other types of fingerprint, e.g. Open Babel FP2, FP3 & FP4, and find the MACCS fingerprint is slightly better than others. The MACCS fingerprint is pattern based and when Open Babel with the default setting is used, it produces a 256-bit fingerprint. Each bit has a value of 0 or 1 and is a dimension of feature space. Thus, the MACCS fingerprint feature is a 256-dimensional vector.
The other type of feature is generated from a drug’s predicted human protein target. The latest version of FINDSITEcomb2.0 22 predicts the possible human targets of the drug. FINDSITEcomb2.0 screens the given drug against the 97% of human proteins with pre-computed pockets25 for which appropriately accurate predicted structures from the TASSERVMT structure prediction approach26 are available. For each protein, a precision score between 0 and 1 that characterizes the likelihood of the protein binding to the drug is obtained. We then remove proteins that are unlikely to be relevant for diseases (see below for cutoffs). To characterize the importance of human proteins in diseases, for each protein, we employ the ENTPRISE27 and ENTPRISE-X28 methods for predicting the disease association of all its possible missense (amino acid substitution) and nonsense (stop or frameshift) mutations. If less than 5% of its amino acid sequence positions have a disease-associated mutation based on ENTPRISE’s and ENTPRISE-X’s cutoffs of 0.5, the protein is not considered. We denote the list of proteins that are disease associated based on ENTPRISE27 and ENTPRISE-X28 and with predicted precision xi of binding to the given drug as P(x1,…,xN). For each human protein, we also pre-computed its disease association status for 960 diseases using the Know-GENE method that was previously developed29. While ENTPRISE27 and ENTPRISE-X28 indicate whether a protein is likely to be disease associated or not, the Know-GENE method predicts which of the 960 diseases it is likely to be associated with using a cutoff of 0.5. Thus, for each disease-associated protein i, a vector Di=(di,1,…,di,960) of 960 dimensions with values of either 0 or 1 to characterize its probability of association to a given disease is generated. The feature vector is calculated as a 960-dimensional vector:
| (2), |
where i runs through all disease associated proteins. Notice that the component values of F range continuously from 0 to 1. This stands in contrast to the MACCS fingerprint feature that has either discrete values of 0 or 1. In practice, the majority of the components of F are non-zero, resulting in longer regression times than that when only the binary values of 0 or 1 are considered.
Training and testing datasets
For drug side effect training and testing, we used two sets: The first is the SIDER4 set from the SIDER4 (version 4.1) database21 downloaded on Jan 19, 2017. This set has 1426 small molecule drugs (excluding antibody drugs) and 4251 unique side effects with PTs (preferred terms). The second set is the Zhang set derived from an earlier version of the SIDER4 database that is divided into training and testing subsets6. This set has 771 training drugs, 309 testing drugs, and 2260 unique side effects with greater than 3 drugs having the given side effect. The Zhang set also includes six types of features: 1) chemical substructure (dimension=881); 2) protein targets (dimension=1046); 3) pathways (dimension=268); 4) enzymes (dimension=160); 5) transporters (dimension=96); 6) treatments (dimension=2537).
For efficacy training, we collected drug indication data from three different sources: (1) the approved drug subset from the Therapeutic Target Database (TTD version Sept 12, 2017)30; (2) the above SIDER4 indication set21; (3) all the clinical trial drug sets in ClinicalTrial collected from ClinicalTrials.gov and mapped to DrugBank20 by Himmelstein et al16. All of their disease indications are converted to the IDs as defined by Human Disease Ontology31–33 (releases/2018–12-17) and merged if the Tanimoto Coefficient (Tc)34 of the two drugs is one. The merged dataset has 2,059 drugs with 3,608 unique indication terms and 123,146 drug-indication pairs.
For testing efficacy predictions in comparison to other methods, we used three datasets used by Himmelstein et al16 downloaded from https://github.com/dhimmel/:
The DrugCentral set obtained from the DrugCentral database(http://drugcentral.org). This set has 671 drug-indication pairs between 454 drugs and 68 indications.
The ClinicalTrialSlim set from ClinicalTrials.gov which has 6382 pairs of drug-indications between 794 drugs and 130 indications.
The Symptomatic set collected from PharmacotherapyDB (https://think-lab.github.io/d/182/) which has 390 drug-indication pairs between 221 drugs and 50 indications.
A summary of all the data sets is given in Table 1. In Table S1, to show the clear advantages of this work, we also summarize the sources of data for deriving the features for a given drug by state-of-the-art methods. MEDICASCY uses only the chemical structure of the drug, whereas the other methods need multiple sources that might not be available for new drugs: the drug’s target, pathways, enzymes transporter and indications. To facilitate the use of MEDICASCY, we have implemented a free web service for academic users at http://pwp.gatech.edu/cssb/MEDICASCY. It takes the SMILES string of a molecule and returns the predictions of efficacy and side effects to the user provided email address.
Table 1.
Summary of training and testing data sets
| Side Effect Data Sets | |||
| Training | Testing | ||
| data | description | data | description |
| SIDER4 (4.1) | 1426 drugs 4,251 side effects 145,020 pairsa |
SIDER4 (4.1) for cross-validation | Same as training |
| Zhang set training subset | 771 drugs 2,260 side effects 92,012 pairs |
Zhang set testing subset | 309 drugs 2,260 side effects 20,131 pairs |
| Efficacy Data Sets | |||
| Training | Testing | ||
| Combined TTD, SIDER4 (4.1), ClinicalTrial | 2,059 drugs 3,608 indications 123,146 pairs |
DrugCentral | 454 drugs, 68 indications, 671 pairs |
| ClinicalTrialSlim | 794 drugs, 130 indications, 6,382 pairs | ||
| Symptomatic | 221 drugs, 50 indications, 390 pairs | ||
Drug-side effect or drug-indication positive samples.
Mode-of-Action targets
The procedure for inferring probable mode-of-action targets, MOA, for any given side effect or indication is also depicted in Figure 1. As indicated above, MEDICASCY predicts the human protein targets of a drug as well as its side effects and efficacy. These predictions relate the human protein targets to a given side effect or indication. Thus, MEDICASCY predictions can be utilized for inference of the probable mode-of-action protein targets for a given side effect or indication. To find the potential MOA targets for a given side effect or indication, we examine the target distribution between drugs having the side-effect/efficacy and drugs having not the side-effect/efficacy as predicted by MEDICASCY. Protein targets having a larger preference of binding to the drugs having the given side effect or indication than that to the drugs having not the given side effect or indication are probable MOA targets. To characterize the preference of a target T for a given side effect or indication D, we define an enrichment factor EF(T,D) as:
| (3) |
An EF(T,D) > 1 indicates that the target T has the potential (a larger value means a higher probability) of being a MOA target of the side effect or indication D. Notice that these targets with EF(T,D) > 1 are potential MOA targets of a given side effect/indication. This work does not pin-point the specific targets of a given drug for a given side effect/indication. However, based on the predicted targets of a given drug, we can narrow down the list of possible MOA targets for the drug.
To apply equation (3) and ensure enough statistics to calculate the fractions, a large set of drugs is needed. In this work, we use the 2,095 FDA approved drugs from DrugBank20 version 5.09 and applied MEDICASCY to predict their side effects and efficacy in prediction mode (molecules identical to the input drugs are excluded from the training libraries).
Assessment of methods
For drug side effect prediction, we employ a cutoff dependent assessment of side effects predictions for individual drug. We define a predicted side effect for a given drug when its score > cutoff. Using the predicted side effects, we can define an overall drug coverage of the prediction:
| (4). |
Then, for each of those drugs having predicted (killing) side effects, we define the precision and recall:
| (5a), |
| (5b) |
When assessing a drug that has at least one killing side effect correctly predicted, we use:
| (5c) |
It should be noted that precision and recall are defined on individual drugs for assessing its side effects. For an individual drug, when the cutoff increases, we will expect the precision to increase and the recall to decrease. However, since we report only the averages over all the assessed drugs, an increased cutoff value will result in smaller drug coverage, i.e. a smaller set of drugs having predictions. This smaller set could have a higher average recall even if they have a smaller individual recall because the higher cutoff removes those drugs with even smaller recalls from assessment.
We also evaluate the various methods by two commonly used metrics that do not depend on cutoffs14 for comparison to other methods: the AUPR-area under precision-recall curve and AUC-ROC-area under the curve of Receiver Operating Characteristics curve. While the AUC-ROC curves tells how much a model is capable of distinguishing between classes, AUPR gives the more informative and relevant picture of the model’s performance when true positive classes are rare35. AUPR depends very sensitively on the number of top ranked true positives among overwhelmingly more true negatives cases.
Computational requirements for benchmarking
We will gladly provide information for researchers that are interested in checking our benchmarking results and in the use of our method. The methods used in computing MEDICASCY features from the chemical structure are either Georgia Tech’s licensed properties (FINDSITEcomb2.0, ENTPRISE & ENTPRISE-X, Know-Gene algorithms) with distribution limitations or are openly available (Open Babel). Thus, we only provide the pre-computed MEDICASCY features and the six types of features that come with the Zhang dataset6 and the necessary scripts for reproducing our benchmarking results at https://github.com/hzhou3ga/MEDICASCY/. They are available for local downloading and running. With all pre-prepared features, the computational requirements for each benchmarking set is summarized in Table S2. For cross-validation of the SIDER4 set, cross-validation of the efficacy training set, testing of the ClinicalTrialSlim, DrugCentral, Symptomatic sets a model has to be trained for each drug, and thus parallel computing is required to simultaneously compute multiple drugs. For the Zhang set, both with our predicted features and the original six types of features, a single model is trained on the training subset and used for all drugs on the testing subset, thus sequential computing is feasible for users to reproduce the training and testing processes.
Cell culture and Tox-8 assay
The NCI molecules and cancer cell lines were obtained from the National Cancer Institute (NCI)/Division of Cancer Treatment and Diagnosis (DCTD)/Developmental Therapeutics Program (DTP) (http://dtp.cancer.gov). Ten NCI molecules without prior knowledge of any cancer indication (NSC # 68116, 330796, 213708, 238010, 341902, 101777, 372499, 123797, 107582, 370387), and the positive control (Plicamycin, NSC # 24559, with known inhibition effects on breast, ovarian and prostate cancer types) were administered as drug treatments to assess their effects on growth of cancer cell lines. NSC # 68116, 330796, 213708, 238010 were tested against OVCAR3 ovarian and PC3 prostate cell lines. NSC # 24559, 341902, 101777, 372499, 123797, 107582, 370387 were tested against OVCAR3 ovarian, PC3 prostate and MCF7 breast cancer cell lines. For each cell line, the cell culture stocks frozen in RPMI 1640 media (Mediatech, Manassas, VA) containing 10% DMSO were stored in liquid nitrogen. After being thawed, the cells were cultured in RPMI 1640 media with 10% Fetal Bovine Serum (FBS; Atlanta Biologicals, Lawrenceville, GA) at 37°C in a 5% CO2 atmosphere and passaged upon near confluence.
To set up cell cultures for the Tox-8 assays, the cells were applied onto a Corning™ Falcon™ 96-Well Imaging Microplate (Corning Ref# 353219), with 3,000 cells per well. After 24 hours of incubation at 37oC in a 5% CO2 atmosphere, the cells were treated with 0, 10 nM, 100 nM, 1 μM, 10 μM, and 50 μM of each drug of interest. Each concentration condition had three to four replicates. The drug stock solutions were made by serial dilution in FBS supplemented RPMI media. After 48 hours of drug treatment, the cells were analyzed for their viability using the Tox-8 assay kit (Sigma, USA) following the manufacturer’s protocol36. In particular, the metabolic activity of the living cells was assessed based on their bioreduction of the exogenously introduced Resazurin fluorescent dye. The fluorescent signals with excitation and emission wavelengths at 560 nm and 590 nm were analyzed using a BioTek Synergy4 fluorescence microplate reader following the default Tox8 assay protocol provided by Sigma, USA.
RESULTS
Cross-validation on SIDER4 set for side effect prediction
We performed a modified leave one out cross-validation (MLOOCV) of MEDICASCY on the 1426 drug SIDER4 set21 for side effect prediction and compared the results to our previous empirical approach, DR. PRODIS 25 that was, to the best of our knowledge, the only existing method for predicting side effects of new drugs from their chemical structures. Here, MLOOCV is different from usual leave one out cross-validation (LOOCV) in that we do not use all the drugs after excluding the left one for training, instead we use only those drugs that have Tanimoto Coefficient34 (Tc) less than a cutoff value to the tested drug for training. This is to make the test more challenging than just LOOCV due to the possible presence of similar drugs in the training set. To examine the effect of similarity between a tested drug and the training drugs, we also implemented a baseline approach called Baseline-Tc that simply uses the side effects of the closest training drug based on Tc calculated from the default FP2 fingerprints of Open Babel software (http://openbabel.org/wiki/Main_Page) as the prediction for the tested drug. In addition to evaluating all 4251 side effects described in SIDER4, we pay special attention to 9 serious side effects that are potential causes of drug disapproval: Neoplasm malignant, Sudden death, Cardiac failure, Cardiac failure congestive, Sudden cardiac death, Cardiac death, Haemorrhagic stroke, Death, Cerebral haemorrhage. Notice that Neoplasm malignant stands for any kind of cancer without referring to the specific type. This is often seen on drug labels. Neoplasm malignant is curated as such in the SIDER4 database21. We call these “killing” side effects. In evaluation, we rank side effect predictions (whose value ranges from 0 to 1, with 1 the highest rank) and compute its recall and precision based on cutoff scores.
We then compare the methods based on their average per drug recall and precision (defined in the Materials and Methods) in Table 2 with a Tc cutoff of 0.7. To examine the contributions of the individual component of MEDICASCY, we also present the results for the algorithm termed by MEDICASCY-MACCS using just the MACCS fingerprint and MEDICASCY-Knowgene for the disease profile but without use of the MACCS fingerprint. In Table 2, we also applied the empirical DR. PRODIS method described in our earlier work 25 to the SIDER4 set rather than previously used SIDER2 set for comparison. Using a cutoff score of 0.5 that approximately gives the same recall rate of 23% as the empirical DR. PRODIS method for all side effects (most of the 4,251 side effects are not especially important), MEDICASCY has a precision of 62.2% as compared to 51.3% by the empirical method. A Student’s-t test of their common 1105 drugs (both methods have predictions) shows that the p-value of their difference in precision is 3.5 × 10−38. Thus, the improvement of MEDICASCY over the empirical DR. PRODIS method for all side effects is significant. MEDICASCY has slightly better precision than those by its component methods. It is understandable that including MACCS fingerprint component does not improve so much because the similarity between testing drugs and training drugs is very low (Tc < 0.7). With a precision of 31.4%, Baseline-Tc is much worse than both DR. PRODIS and MEDICASCY. Although both Baseline-Tc and MEDICASCY-MACCS depend on the similarity between testing drug and training drugs, MEDICASCY-MACCS performs much better than Baseline-Tc. This could be due to the fact that the machine learning approach in MEDICASCY-MACCS can learn from many drugs’ structural patterns in connection with their side effects whereas Baseline-Tc uses only the closest drug’s side effect for prediction.
Table 2.
Cross-validation testing with a Tc cutoff 0.7 on the 1426 drug SIDER4 set
| All Drug Side Effects | |||
| All 1426 drug set | |||
| Method | Number of drugs/Drug coverage | Recall a | Precision a |
| Baseline-Tc | 1426/100% | 31.9% | 31.4% |
| DR. PRODIS empirical 25 | 1123/78.8% | 22.7% | 51.3% |
| MEDICASCY (0.5) a | 1408/98.7% | 23.1% | 62.2% |
| MEDICASCY-MACCS (0.5) | 1401/98.2% | 24.6% | 59.4% |
| MEDICASCY-Knowgene (0.5) | 1419/99.5% | 23.2% | 61.9% |
| Killing Drug Side Effects | |||
| All 456 killing drug set | |||
| Method | Number of drugs/Drug coverage | Recall/Precision all 9 side effects | Recall=Precision when at least one side effect is correct |
| Baseline-Tc | 152/33.3% | 44.8%/43.3% | 60.5% |
| DR. PRODIS empirical 25 | 99/21.7% | 37.1%/39.4% | 47.5% |
| MEDICASCY (0.5) | 0/0.0% | - | - |
| MEDICASCY (0.3) | 27/5.9% | 57.4%/67.3% | 77.8% |
| MEDICASCY (0.2) | 169/37.1% | 50.8%/51.7% | 66.3% |
| MEDICASCY-MACCS (0.2) | 210/46.1% | 45.0%/46.7% | 59.5% |
| MEDICASCY-Knowgene (0.2) | 154/33.8% | 58.2%/51.8% | 73.4% |
| Consensus 70 drugs: both MEDICASCY (0.2) and Baseline-Tc have predictions | |||
| Baseline-Tc | - | 45.2%/45.7% | 62.9% |
| MEDICASCY (0.2) | - | 57.1%/61.2% | 74.3% |
Numbers in parenthesis are the cutoff scores.
For killing side effects, with a cutoff score of 0.5, MEDICASCY has no predictions. However, we can lower the cutoff score to lower values, say 0.2. The improvement on the 9 killing side effects is obvious, for at least one correct killing side effect and a cutoff score 0.2, MEDICASCY has a precision and recall of 66.3% compared to 47.5% by DR. PRODIS. With a precision of 60.5%, Baseline-Tc performs better than DR. PRODIS, but worse than MEDICASCY. This could be due to the lack of sufficient killing side effects in the training set for inference by DR. PRODIS. The improvement of MEDICASCY over Baseline-Tc is even larger when evaluating on the consensus 70 drugs (the precision 74.3% vs. 62.9% for at least one correct killing side effect). Compared to its component methods for killing side effects, MEDICASCY has lower drug coverage, and better recall and precision than MEDICASCY-MACCS, but at higher drug coverage, worse recall and precision than MEDICASCY-Knowgene. This is again due to the difficulty of the testing drugs and the similarity-based MACCS component slightly drags down its performance. When a larger Tc cutoff, say 0.95, is applied, MEDICASCY has a better precision than MEDICASCY-Knowgene (60.3% vs. 55.4%, see Figure S1). The detailed dependence of the precision of MEDICASCY (cutoff score = 0.2) and Baseline-Tc for killing side effects on the Tc cutoff is given in Figure S1 in the Supplementary Information.
Comparison to other methods for side effect prediction
We next compare MEDICASCY to other state-of-the-art methods for the Zhang dataset6 which is a derivative of an earlier version of the SIDER4 database21. These methods are only feasible for well-studied drugs such as those in the SIDER4 database. Since the methods by Zhang et. al.12 and Cheng et. al.15 were designed to predict missing or undetected side effects of known drugs with annotated side effects and MEDICASCY aims to predict side effects for drugs without a priori knowledge of their side effects, it is not appropriate to compare MEDICASCY to them. To directly compare the machine learning BRF component with alternative approaches5, 6, 14, we also implemented an adapted version of MEDICASCY called BRF-ORG (Boosted Random Forest using the ORiGinal six types of features; see dataset section for feature types). Each feature is used independently in training and prediction and gives six scores for each drug, with the final prediction score the average of these six scores.
The results are compiled in Table 3 which shows that using the dataset provided six types of features, MEDICASCY (BRF-ORG) achieves the best AUPR=0.394. The next best AUPR=0.391 is from LNSM-SMI13. We also noted that the Random Forest (RF) method5 has AUPR of 0.300 that is much worse than that of our BRF based method. This indicates that the BRF machine learning method is the best choice amongst these methods. Using our two types of predicted features from only the input of the drug’s chemical structure, i.e. using the 256 dimensional MACCS fingerprints and the 960 dimensional disease associations, MEDICASCY has an AUPR=0.385. This is the third best performance among all methods, but importantly, it uses far less data. This also suggests that there is not too much room for improvement of MEDICASCY by just improving its features, as its performance with known or predicted features is very close. However, this does not preclude improvement by better methods using the same or better features. Both component methods MEDICASCY-MACCS and MEDICASCY-Knowgene are worse than full MEDICASCY.
Table 3.
Performance of different methods using the Zhang dataset
| Methods | AUPRa | AUC-ROC |
|---|---|---|
| MEDICASCY: BRF-ORG | 0.394 | 0.902 |
| LNSM-SMI13 | 0.391 | 0.879 |
| MEDICASCY | 0.385 | 0.895 |
| LNSM-CMI13 | 0.380 | 0.885 |
| MEDICASCY-Knowgene | 0.375 | 0.890 |
| MEDICASCY-MACCS | 0.374 | 0.884 |
| FS-MLKNN13 | 0.365 | 0.872 |
| Multi-layer perceptron5 | 0.355 | 0.894 |
| KNN5 | 0.343 | 0.891 |
| KG-SIM-PROP46 | 0.338 | 0.889 |
| DrugClust: K-Means14 | 0.334 | 0.914 |
| Random forests5 | 0.300 | 0.824 |
By definition, the average precision (AP) is equivalent to the AUPR (see e.g. https://sanchom.wordpress.com/tag/average-precision/). In practice, their values are very close. We use the AP values from Refs. 5, 13, 46 since their AUPR values differ significantly from the respective AP values. In fact, using the same multi-layer perceptron prediction data from author of Ref. 5, we obtained an AUPR=0.353 that is very close to its reported value of AP=0.355.
Cross-validation on training set for drug efficacy prediction
The training set for drug efficacy prediction has 2,059 drugs with a total of 123,146 indications. On average, experimentally each drug has ~60 out of a total of 3,608 possible indications, with a median number of indications of 10. The promiscuousness of drugs is consistent with our earlier findings that a drug can have many targets.25 A detailed distribution of the number of both experimental and predicted indications is given in Figure S2. The experimental indications are collected from (1) the approved drug subset from the Therapeutic Target Database (TTD version Sept 12, 2017)30; (2) the SIDER4 indication set21; (3) all of the clinical trial drug sets in ClinicalTrial collected from ClinicalTrials.gov and mapped to DrugBank20 by Himmelstein et al16. As was the case for the SIDER4 set for side effects, it is not only a useful dataset for training efficacy prediction methods but is also useful for the large scale benchmarking for efficacy prediction by the modified leave one out cross-validation that applies a similarity cutoff for the training drugs as measured by Tc. We also implemented an empirical approach in the spirit of DR. PRODIS25 for efficacy prediction, termed DR. PRODIS-E, by replacing the side effects with indications and compared its performance to MEDICASCY. Similar to side effect prediction, a baseline approach called Baseline-Tc-E is also implemented. An alternative method MEDICASCY-RF using only RF instead of BRF is also implemented for comparison. The results are compiled in Table 4 for Tc cutoffs of 0.7 and 0.9. With a Tc cutoff of 0.7 (i.e. drugs having Tc >= 0.7 to the tested drug are excluded from its training set), the whole dataset is hard. For the overall set, using a cutoff score of 0.5, MEDICASCY has around 6 times the recall and precision of DR. PRODIS-E empirical (58.2% and 71.8% vs. 9.6% and 13.5%, respectively). The baseline method Baseline-Tc-E has only slightly better precision of 14.6% than DR. PRODIS-E. However, MEDICASCY has a much smaller drug coverage (5.3% vs. 24.5% and 100% respectively). To increase the drug coverage, a smaller cutoff can be used. For example, with a cutoff of 0.2, MEDICASCY has a drug coverage, recall and precision of 75.4%, 15.7% and 26.3%. These are still significantly better than the 24.5%, 9.6% and 13.5% by DR. PRODIS-E. The precision is also much better than the baseline approach (26.3% vs. 14.6%, 16.3% for consensus drugs that both methods have predictions). Thus, MEDICASCY has significantly better performance than both the empirical DR. PRODIS-E and baseline approaches. Both the component methods MEDICASCY-MACCS (cutoff 0.2) and MEDICASCY-Knowgene (cutoff 0.2) are better than baseline and DR. PRODIS approaches and are worse than full MEDICASCY. Consistent with side effect prediction, using RF instead of BRF results in worse performance (see Tables 3 & 4). MEDICASCY-RF(cutoff=0.2) has larger drug coverage (99.2% vs. 75.4%) and recall (22.9% vs.15.7%), but much worse precision (11.6% vs. 26.3%) than MEDICASCY. With a higher Tc cutoff of 0.9, the increased performance of Baseline-Tc-E and DR. PRODIS-E is expected, from ~14% for a Tc cutoff of 0.7 to ~23% for 0.9. Still they are much worse than MEDICASCY’s precisions of 64% at a 0.5 cutoff score and 32% at a 0.2 cutoff score. Evaluation on the consensus drugs that both MEDICASCY and Baseline-Tc-E have predictions shows the same trend that MEDICASCY has better precision than Baseline-Tc-E. A more detailed comparison of MEDICASCY’s (at a cutoff 0.2) and Baseline-Tc-E’s precision dependence on the similarity as measured by the Tc between tested and training drugs is in Figure S3. The distribution of the number of predicted indications using a cutoff score of 0.2 can be found in Figure S2. The less than 100% drug coverage of MEDICASCY indicates it under-predicts at cutoff score 0.2.
Table 4.
Cross-validation testing with Tc cutoffs of 0.7, 0.9 on the 2059 efficacy training set
| Method | Drug coverage | Recall | Precision |
|---|---|---|---|
| Tc cutoff = 0.7 | |||
| All drugs | |||
| Baseline-Tc-E | 100% | 14.6% | 14.6% |
| DR. PRODIS-E 25 | 24.5% | 9.6% | 13.5% |
| MEDICASCY (0.5)a | 5.3% | 58.2% | 71.8% |
| MEDICASCY (0.4) | 9.8% | 41.1% | 55.8% |
| MEDICASCY (0.3) | 28.4% | 20.7% | 38.9% |
| MEDICASCY (0.2) | 75.4% | 15.7% | 26.3% |
| MEDICASCY-MACCS (0.2) | 87.7% | 16.2% | 20.1% |
| MEDICASCY-Knowgene (0.2) | 67.9% | 17.6% | 24.7% |
| MEDICASCY-RF (0.2) | 99.2% | 22.9% | 11.6% |
| Consensus 1552 drugs: both MEDICASCY (0.2) and Baseline-Tc-E have predictions | |||
| Baseline-Tc-E | - | 16.7% | 16.3% |
| MEDICASCY (0.2) | - | 15.7% | 26.3% |
| Consensus 110 drugs: both MEDICASCY (0.5) and Baseline-Tc-E have predictions | |||
| Baseline-Tc-E | - | 63.9% | 59.4% |
| MEDICASCY (0.5) | - | 58.2% | 71.8% |
| Tc cutoff = 0.9 | |||
| All drugs | |||
| Baseline-Tc-E | 100% | 24.3% | 22.9% |
| DR. PRODIS-E 25 | 14.8% | 8.9% | 22.7% |
| MEDICASCY (0.5) | 13.0% | 39.2% | 64.0% |
| MEDICASCY (0.4) | 23.4% | 32.2% | 54.1% |
| MEDICASCY (0.3) | 44.7% | 27.1% | 46.2% |
| MEDICASCY (0.2) | 81.9% | 25.5% | 32.1% |
| MEDICASCY-MACCS (0.2) | 92.0% | 25.9% | 25.8% |
| MEDICASCY-Knowgene (0.2) | 70.5% | 24.4% | 30.2% |
| MEDICASCY-RF (0.2) | 99.3% | 26.5% | 14.4% |
| Consensus 1687 drugs: both MEDICASCY (0.2) and Baseline-Tc-E have predictions | |||
| Baseline-Tc-E | - | 26.7% | 25.0% |
| MEDICASCY (0.2) | - | 25.5% | 32.1% |
| Consensus 268 drugs: both MEDICASCY (0.5) and Baseline-Tc-E have predictions | |||
| Baseline-Tc-E | - | 51.4% | 47.4% |
| MEDICASCY (0.5) | - | 39.2% | 64.0% |
Numbers in parenthesis are the cutoff scores.
Comparison to other methods for drug efficacy prediction
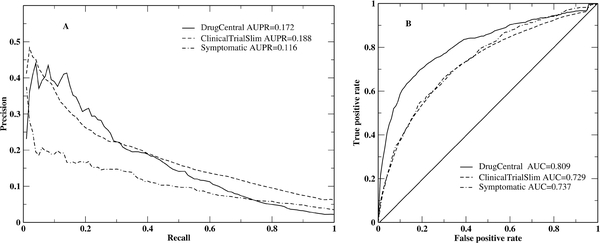
MEDICASCY was tested on three datasets (see Dataset section) for efficacy prediction to compare it to other methods. For each testing dataset, we exclude drugs with Tc=1 to the testing drug. As in Reference 16, we evaluate the combined predictions for all drugs in a given testing set. The precision-recall and ROC curves are given in Figure 2. Compared to the method in Reference 16 where a systematic integration of biomedical knowledge was used for computing drug features and a logistic regression machine learning was employed for learning and prediction, MEDICASCY has a significantly better AUPR for all three sets: 0.172 for DrugCentral, 0.188 for ClinicalTrialSlim and 0.116 for Symptomatic set, as compared to 0.056, 0.093, and 0.005, respectively by Project Rephetio16. MEDICASCY has a slightly worse AUC=0.81 compared to Project Rephetio’s 16 AUC=0.86 for the DrugCentral set. For ClinicalTrialSlim and Symptomatic sets, MEDICASCY has a slightly better AUC=0.73 and 0.74 compared to 0.70 and 0.70 by Project Rephetio16, respectively. Thus, MEDICASCY has much better performance in terms of ranking the true positives at the very top rank that are required in practice for efficacy prediction.
Figure 2:
Precision-recall (A) and ROC (B) curves of MEDICASCY for the DrugCentral, ClinicalTrialSlim and Symptomatic datasets for efficacy prediction.
Experimental validation of efficacy prediction
Since experimental validation of efficacy is not trivial, to validate our efficacy predictions, we first utilized the experimental data from the NCI-60 program (https://dtp.cancer.gov/databases_tools/default.htm) acquired via community efforts for large scale “postdiction type” of validation. We also utilized the bioactivity data on human disease cell lines from ChEMBL37 and PubChem38 for further validations. Having seen promising results, we then performed our own experimental tests.
To assess the level of success on efficacy predictions of the NCI-60 set, we downloaded the DOSE_RESPONSE (June 2016) experimental data. The data shows the dose response of 53,215 molecules characterized by cell line growth rate against 10 common cancers: breast, colon, central nervous system(cns), leukemia, melanoma, non-small cell lung, ovarian, prostate, renal, small cell lung. Thus, these data allow for large scale experimental validation. It should be noted that these validations are based on cell line experiments. They are not necessarily equivalent to or a replacement of actual clinical trial outcomes of efficacy. Furthermore, we have not performed any additional training other than on the above 2059 training set. In addition to the 2,095 FDA approved drugs from DrugBank20, we also applied MEDICASCY to 1,597 NCI (http://dtp.nci.nih.gov/branches/dscb/repo_open.html) diversity molecules from the same NCI-60 site downloaded earlier (2012). All predictions were carried out by excluding molecules having a Tc=1 to the given molecule from the training data. However, we note, that no additional efficacy training was done as part of this evaluation. For each cancer indication, we assessed the top 20 molecules predicted to have an indication against the NCI-60 experimental data. If a molecule at a concentration <~10−3 molar causes negative growth of the cell line, we consider it to be a correct prediction that has the effect of killing the cancer cell line. The detailed results are compiled in Tables 5 and 6 for FDA approved drugs and NCI diversity set molecules, respectively. Since only subsets of FDA approved drugs and the NCI diversity set have experimental data in the DOSE_RESPONSE data set, not all of the top 20 ranked molecules could be assessed. Nevertheless, we list them all for future assessment when additional experimental data becomes available. For the FDA approved drug set, the mean success rate across the 10 cancer types is 85.2%. For the NCI diversity set, the mean success rate is 82.2%. As an example, drug DB06810 (Plicamycin) is predicted to have breast, colon, cns, leukemia, melanoma, ovarian and prostate cancer indications. NCI-60 data shows that it has effective concentration as low as 10−13 molar against colon, melanoma & ovarian cancers whereas the previously known indications of DB06810 from DrugBank20 are: for the treatment of testicular cancer, as well as hypercalcemia and hypercalciuria associated with a variety of advanced forms of cancer. While most of the tested FDA drugs have previously known indications of other types of cancer (see Table 5), a drug having indications for one type of cancer does not imply it should have indications for other cancers. For example, DB00531 (Cyclophosphamide) has no effect for small cell lung cancer even though it has been used in the treatment of lymphoma and leukemia20. On the other hand, there are two tested FDA drugs that show a cancer killing effect with no previously known cancer indications. They are good examples of new applications for old drugs. One is DB00162 (Vitamin A) for the treatment of vitamin A deficiency20. MEDICASCY predicts it has killing effects for non-small cell lung cancer which it does at 10−4 M and renal carcinoma at 10−4 M. The other is DB01103 (Quinacrine) that is approved to treat giardiasis and cutaneous leishmaniasis and the management of malignant effusions20. MEDICASCY predicts that DB01103 inhibits the growth of small cell lung cancer which it does at 10−5 M. Thus, MEDICASCY is shown to have excellent performance in efficacy prediction.
Table 5.
Experimental validation of the top 20 predictions for the FDA approved drugs on the NCI-60 data.
| Breast: 4/4 correct | Colon: 5/6 correct | Central Nervous System: 4/4 correct | ||||||
|---|---|---|---|---|---|---|---|---|
| ID(score) | NSC# | negative growth concentration (molar): cell line¶ | ID(score) | NSC# | negative growth concentration (molar): cell line | ID(score) | NSC# | negative growth concentration (molar): cell line |
| DB06772(0.85) | DB06772(0.84) | DB06772(0.84) | ||||||
| DB01248(0.82) | 628503 | 10−7:HS578T, MDA-MB-231/ATCC | DB01248(0.81) | 628503 | 10−8:COLO205, HCC-2998,HT29, KM12,KM20L2 | DB00309(0.67) | ||
| DB00666(0.74) | DB01229(0.67) | 125973 | 10−9:KM12, Ht29 | DB00603(0.62) | ||||
| DB00116(0.73) | DB00116(0.66) | DB04465(0.62) | ||||||
| DB00644(0.73) | DB11256(0.61) | DB00570(0.59) | ||||||
| DB00282(0.72) | DB04465(0.61) | DB06810(0.58) | 24559 | 10−12:SF-295 | ||||
| DB00432(0.72) | 75520 | 10−5:MCF7 | DB00398(0.57) | DB11256(0.55) | ||||
| DB00603(0.71) | DB06810(0.56) | 24559 | 10−13:COLO205 | DB00361(0.52) | ||||
| DB01229(0.71) | 125973 | 10−9:HS578T | DB00228(0.54) | DB00480(0.51) | ||||
| DB06287(0.70) | DB11737(0.51) | DB00318(0.50) | ||||||
| DB00624(0.70) | DB01189(0.51) | DB11737(0.50) | ||||||
| DB00977(0.70) | DB00351(0.49) | DB01181(0.50) | 109724 | 10−7.3:U251 | ||||
| DB00309(0.69) | DB01181(0.48) | 109724 | 10−4.3:SW-620 | DB09462(0.60) | ||||
| DB00519(0.68) | DB00432(0.47) | 75520 | No effect | DB08896(0.50) | ||||
| DB01189(0.67) | DB01028(0.47) | DB01177(0.48) | 256438 | 10−8:SNB-75 | ||||
| DB06789(0.65) | DB01177(0.46) | 256438 | 10−7:COLO205,HCC-2998,HT29 | DB00541(0.48) | ||||
| DB06810(0.64) | 24559 | 10−9:MCF7 | DB00541(0.46) | DB00515(0.47) | 119875 | 10−7.3:SF-767 | ||
| DB01156(0.64) | DB00515(0.45) | DB00531(0.47) | ||||||
| DB04465(0.62) | DB00531(0.45) | DB09070(0.47) | ||||||
| DB00928(0.61) | DB09079(0.44) | DB06261(0.46) | ||||||
| Leukemia: 1/2 | Melanoma: 8/8 | Non-small cell lung: 4/4 | ||||||
| DB06772(0.85) | DB01229(0.67) | 125973 | 10−10:MDA-MB-435,MDA-N | DB06772(0.83) | ||||
| DB00570(0.70) | DB06772(0.62) | DB08910(0.68) | ||||||
| DB08910(0.70) | DB04465(0.61) | DB00541(0.60) | ||||||
| DB00309(0.69) | DB06810(0.56) | 24559 | 10−13:SK-MEL-5 | DB00608(0.50) | ||||
| DB00541(0.69) | DB01248(0.53) | 628503 | 10−8:MDA-MB-435,M14,MDA-N,SK-MEL-2 | DB08896(0.50) | ||||
| DB00860(0.68) | DB11737(0.50) | DB01181(0.50) | 109724 | 10−7.3:A549/ATCC,HOP-18,HOP-92 | ||||
| DB00116(0.68) | DB08896(0.48) | DB00531(0.49) | 26271 | 10−7.6:HOP-18 | ||||
| DB00631(0.67) | DB00432(0.46) | 75520 | 10−4:M14,UACC-62 | DB11737(0.48) | ||||
| DB00577(0.66) | DB00361(0.45) | DB00339(0.47) | ||||||
| DB11256(0.64) | DB01177(0.45) | 256438 | 10−7:SK-MEL-5,LOXIMVI,MALME-3M,UACC-62 | DB01412(0.46) | ||||
| DB01073(0.63) | DB01181(0.45) | 109724 | 10−7.3:SK-MEL-5 | DB00175(0.42) | ||||
| DB06810(0.63) | 24559 | 10−9:MOLT-4 | DB00541(0.45) | DB01229(0.42) | 125973 | 10−11:NCI-H522 | ||
| DB00928(0.62) | DB06825(0.44) | DB00162(0.41)* | 122759 | 10−4:HOP-62,HOP-92,A549/ATCC,NCI-H522,NCI-H322M,NCI-H23,NCI-H226 | ||||
| DB01041(0.61) | 66847 | No effect | DB00531(0.43) | 26271 | 10−4.6:M19-MEL | DB04573(0.40) | ||
| DB00361(0.61) | DB09070(0.42) | DB01201(0.38) | ||||||
| DB04465(0.61) | DB01204(0.41) | 279836 | 10−6:UACC-62,LOXIMVI,SK-MEL-5,SK-MEL-2 | DB00201(0.38) | ||||
| DB01004(0.60) | DB06261(0.41) | DB06813(0.38) | ||||||
| DB06809(0.60) | DB00515(0.41) | DB01613(0.38) | ||||||
| DB01610(0.59) | DB01708(0.39) | DB00583(0.37) | ||||||
| DB01181(0.58) | DB00385(0.38) | DB00824(0.36) | ||||||
| Ovarian: 6/8 | Prostate: 3/5 | Renal: 4/4 | ||||||
| DB06772(0.84) | DB06772(0.84) | DB06772(0.82) | ||||||
| DB01248(0.82) | 628503 | 10−8:OVCAR-3,SK-OV-3 | DB01248(0.82) | 628503 | 10−4:DU-145 | DB08910(0.66) | ||
| DB08910(0.68) | DB00666(0.81) | DB01590(0.63) | ||||||
| DB01229(0.67) | 125973 | 10−8.6:OVCAR-3,OVCAR-5,OVCAR-8,IGROV1 | DB00644(0.80) | DB06287(0.61) | ||||
| DB00116(0.66) | DB00977(0.73) | DB00877(0.52) | 226080 | 10−10:A498,RXF 393 | ||||
| DB11256(0.62) | DB08910(0.70) | DB00309(0.52) | ||||||
| DB04465(0.61) | DB01229(0.67) | 125973 | 10−9:DU-145 | DB00318(0.50) | ||||
| DB06825(0.61) | DB00603(0.67) | DB08896(0.48) | ||||||
| DB06810(0.57) | 24559 | 10−13:OVCAR-8 | DB01041(0.66) | 66847 | No effect | DB00570(0.46) | ||
| DB00050(0.56) | 373964 | No effect | DB01412(0.65) | DB00608(0.45) | ||||
| DB08896(0.56) | DB06825(0.63) | DB01466(0.41) | ||||||
| DB11737(0.56) | DB06810(0.62) | 24559 | 10−6.9:PC-3 | DB00162(0.41) | 122759 | 10−4:UO-31,786-0,A498,RXF 393,SN12C | ||
| DB01181(0.55) | 109724 | 10−7.3:IGROV1 | DB04465(0.62) | DB00541(0.41) | ||||
| DB00515(0.53) | 119875 | 10−5.1:IGROV1 | DB11737(0.60) | DB00175(0.41) | ||||
| DB09079(0.52) | DB04574(0.60) | DB01229(0.40) | 125973 | 10−8.6:SN12K1,SN12C,RXF 393,TK-10,UO-31,CAKI-1,ACHN,A498,786-0 | ||||
| DB00531(0.51) | 26271 | 10−4.6:OVCAR-3 | DB01181(0.59) | 109724 | No effect | DB01181(0.39) | 109724 | 10−7.3:CAKI-1 |
| DB00339(0.51) | DB05273(0.59) | DB00337(0.39) | ||||||
| DB01685(0.50) | DB08896(0.57) | DB00531(0.38) | ||||||
| DB00175(0.49) | DB00531(0.57) | DB01173(0.37) | ||||||
| DB00432(0.49) | 75520 | No effect | DB09509(0.57) | DB06789(0.36) | ||||
| Small cell lung: 5/6 | ||||||||
| DB06772(0.83) | ||||||||
| DB00541(0.59) | ||||||||
| DB00570(0.53) | ||||||||
| DB00309(0.50) | ||||||||
| DB08896(0.46) | ||||||||
| DB01103(0.40)* | 14229 | 10−5:DMS114,DMS273 | ||||||
| DB01229(0.40) | 125973 | 10−8.6:DMS114, DMS273 | ||||||
| DB00175(0.37) | ||||||||
| DB00162(0.37)* | ||||||||
| DB00444(0.37) | 122819 | 10−5.6:DMS114, DMS273 | ||||||
| DB01181(0.36) | 109724 | 10−3.3:DMS273 | ||||||
| DB00531(0.35) | 26271 | No effect | ||||||
| DB01177(0.32) | 256438 | 10−8:DMS273 | ||||||
| DB00339(0.32) | ||||||||
| DB01590(0.31) | ||||||||
| DB00227(0.28) | ||||||||
| DB00337(0.28) | ||||||||
| DB08910(0.27) | ||||||||
| DB00959(0.27) | ||||||||
| DB01083(0.27) | ||||||||
Cell lines having the lowest concentrations with negative growth effects are reported.
Bold and italic indicate tested drugs having no previously known cancer indications.
Table 6.
Experimental validation of the top 20 predictions for the NCI diverse molecules on the NCI-60 data
| Breast: 8/8 correct | Colon: 6/7 correct | Central Nervous System: 6/6 correct | Leukemia: 7/8 correct | ||||
|---|---|---|---|---|---|---|---|
| NSC#(score) | negative growth concentration (molar): cell line¶ | NSC#(score) | negative growth concentration (molar): cell line | NSC#(score) | negative growth concentration (molar): cell line | NSC#(score) | negative growth concentration (molar): cell line |
| 665497(0.59) | 10−6:MAXF401,HS578T | 317003(0.47) | 10−5:HCC-2998,HCT-116,HCT-15,HT29,KM12,SW-620 | 68116(0.47) | 121182(0.74) | No effect | |
| 68116(0.57) | 68116(0.45) | 317003(0.47) | 10−5:U251,U251/H.Fine,U373/H.Fine,U-78/H.Fine,U118/H.Fine,SNB-75,SF-539,SF-268,LN-229/H.Fine,A-172/H.Fine | 122819(0.57) | 10−6.6:MOLT-4,CCRF-CEM,HL-60(TB),RPMI-8226,SR | ||
| 341902(0.55) | 665497(0.40) | 10−5:HCC-2998,HT29,COLO205,HCT-116,KM12 | 665497(0.43) | 10−6:SF-539 | 68116(0.57) | ||
| 317003(0.52) | 10−5:MCF7,HS578T,MDA-MB-231/ATCC | 372275(0.39) | 372275(0.40) | 343256(0.57) | 10−6:HL-60(TB),RPMI-8226,SR | ||
| 727038(0.48) | 10−7:MDA-MB-231/ATCC,BT-549 | 101777(0.37) | 330796(0.40) | 370383(0.57) | |||
| 372275(0.48) | 330796(0.37) | 341902(0.40) | 318799(0.56) | ||||
| 343256(0.48) | 10−6:MCF7,T-47D,MDA-MB-231/ATCC,MDA-MB-468 | 108972(0.36) | 101777(0.40) | 25740(0.56) | |||
| 101777(0.47) | 156565(0.36) | 10−4:HT29,KM12,SW-620,HCT-15,HCC-2998,COLO205 | 156565(0.40) | 10−4:SF-539,SF-295,SNB-19,SNB-75,U251 | 341902(0.54) | ||
| 372499(0.47) | 341902(0.36) | 372499(0.38) | 665497(0.53) | 10−6:HL-60(TB) | |||
| 123797(0.46) | 343256(0.36) | 10−6:COLO205,HT29 | 213708(0.37) | 105827(0.50) | 10−6:RPMI-8226,MOLT-4,K-562,CCRF-CEM,SR | ||
| 330796(0.46) | 122819(0.34) | 10−5.6:COLO205,HCC-2998 | 122819(0.37) | 10−5.6:SF-539,SF-268,SF-295,SNB-19,SNB-75,SNB-78,U251,XF498 | 317003(0.49) | 10−5:K-562,CCRF-CEM,HL-60(TB),MOLT-4,RPMI-8226,SR | |
| 59620(0.45) | 10−4:BT-549,HS578T | 79582(0.34) | 353451(0.36) | 10−4:SF-268 | 101777(0.48) | ||
| 13248(0.45) | 10−4:MDA-MB-468,HS578T | 372063(0.34) | 343256(0.36) | 10−6:SF-295,SF-539,SNB-75,U251 | 16416(0.47) | ||
| 108972(0.45) | 372499(0.34) | 318799(0.36) | 29193(0.47) | ||||
| 372063(0.44) | 211356(0.33) | 83345(0.35) | 156565(0.46) | 10−4:RPMI-8226,CCRF-CEM | |||
| 353451(0.44) | 10−2.9:BT-549,HS578TMCF7,MDA-MB-231/ATCC,T-47D | 22070(0.32) | 10−5:SW-620,COLO205,DLD-1 | 99796(0.34) | 173103(0.45) | ||
| 83345(0.44) | 83345(0.31) | 215585(0.34) | 373535(0.45) | 10−5:HL-60(TB) | |||
| 380279(0.44) | 87084(0.31) | No effect | 108972(0.34) | 19063(0.45) | |||
| 156565(0.44) | 10−4:MDA-MB-231/ATCC,MDA-MB-468,BT-549,HS578T,MCF7,T-47D | 374703(0.30) | 374703(0.33) | 330796(0.45) | |||
| 318799(0.44) | 17796(0.28) | 129260(0.33) | 50405(0.45) | ||||
| Melanoma: 6/7 | Non-small cell lung: 6/7 | Ovarian: 5/6 | Prostate: 4/8 | ||||
| 317003(0.47) | 10−5:M14,LOXIMVI,MALME-3M,MDA-MB-435,MDA-N,SK-MEL-28,SK-MEL-5,UACC-62 | 68116(0.48) | 68116(0.53) | 328403(0.70) | No effect | ||
| 68116(0.43) | 317003(0.41) | 10−5:NCI-H522,NCI-H460,NCI-H226,HOP-62,A549/ATCC | 317003(0.49) | 10−5:OVCAR-8,OVCAR-4,SK-OV-3 | 665497(0.60) | 10−5:PC-3 | |
| 372275(0.37) | 353451(0.39) | 10−6.9:HOP-62 | 341902(0.49) | 68116(0.57) | |||
| 156565(0.35) | 10−4:LOXIMVI,M14,MALME-3M,SK-MEL-28,SK-MEL-5,UACC-257,UACC-62 | 35534(0.34) | 665497(0.47) | 10−6:OVCAR-3,NCI/ADR-ES | 343256(0.57) | 10−5:PC-3,DU-145 | |
| 101777(0.35) | 123797(0.34) | 370387(0.46) | 341902(0.54) | ||||
| 330796(0.35) | 8813(0.34) | 101777(0.45) | 317003(0.53) | 10−4:PC-3,DU-145 | |||
| 122819(0.33) | 10−6.6:UACC-62 | 108972(0.33) | 372275(0.45) | 372275(0.51) | |||
| 372499(0.32) | 329065(0.33) | 156565(0.43) | 10−4:IGROV1,NCI/ADR-RES,OVCAR-5,SK-OV-3 | 87084(0.49) | No effect | ||
| 665497(0.32) | 10−6:MDA-MB-435,MDA-N | 338205(0.31) | 107582(0.43) | 101777(0.49) | |||
| 22070(0.32) | 10−5:LOXIMVI,M14,MALME-3M | 98363(0.31) | 10−5:NCI-H522 | 372063(0.42) | 328010(0.48) | ||
| 83345(0.31) | 51093(0.31) | 330796(0.42) | 108972(0.47) | ||||
| 341902(0.30) | 261037(0.31) | No effect | 83345(0.41) | 372499(0.47) | |||
| 211356(0.30) | 116565(0.31) | 108972(0.41) | 318799(0.47) | ||||
| 343256(0.30) | 10−6:SK-MEL-5,SK-MEL-2,MALME-3M,UACC-257 | 11128(0.31) | 372499(0.40) | 156565(0.47) | 10−4:PC-3,DU-145 | ||
| 374703(0.29) | 665497(0.31) | 10−5:NCI-H522 | 123797(0.39) | 373535(0.47) | No effect | ||
| 108972(0.28) | 87084(0.31) | 10−4:HOP-92 | 343256(0.38) | 10−6:OVCAR-8,OVCAR-3,OVCAR-4,NCI/ADR-RES | 123797(0.46) | ||
| 57890(0.28) | 9441(0.31) | 211356(0.38) | 107582(0.46) | ||||
| 30622(0.27) | No effect | 99796(0.31) | 288686(0.38) | 213708(0.46) | |||
| 372063(0.27) | 122819(0.30) | 10−7.6:EKVX | 727038(0.37) | 10−8:IGROV1,OVCAR-8 | 88795(0.46) | No effect | |
| 326422(0.27) | 330796(0.30) | 190336(0.37) | No effect | 92849(0.46) | |||
| Renal: 7/11 | Small cell lung: 4/5 | ||||||
| 121182(0.42) | No effect | 122819(0.39) | 10−5.6:DMS114,DMS273 | ||||
| 68116(0.39) | 68116(0.36) | ||||||
| 665497(0.34) | 10−6:A498 | 13248(0.35) | |||||
| 105827(0.29) | 10−6:RXF393,TK-10,CAKI-1,UO-31 | 665497(0.27) | |||||
| 88795(0.27) | No effect | 94600(0.25) | 10−8:DMS273 | ||||
| 94600(0.26) | 10−8:SN12C,CAKI-1,RXF393,UO-31 | 88795(0.23) | |||||
| 373535(0.26) | No effect | 353451(0.23) | 10−2.9:DMS114, DMS273 | ||||
| 122819(0.26) | 10−8.6:SN12K1,A498 | 123797(0.22) | |||||
| 13248(0.25) | 10−4:UO-31 | 30041(0.22) | |||||
| 352890(0.25) | 10−8:ACHN | 101345(0.21) | |||||
| 30041(0.24) | No effect | 261037(0.20) | No effect | ||||
| 31712(0.24) | 639174(0.19) | ||||||
| 59776(0.24) | 294150(0.19) | ||||||
| 372063(0.24) | 107582(0.19) | ||||||
| 48231(0.23) | 373535(0.18) | ||||||
| 51093(0.23) | 279834(0.18) | ||||||
| 97920(0.22) | 603071(0.17) | 10−7:DMS273,DMS114 | |||||
| 177407(0.22) | 10−5:UO-1,A498,ACHN,RXF 393 | 380279(0.17) | |||||
| 175415(0.22) | 40817(0.17) | ||||||
| 215585(0.22) | 97920(0.17) | ||||||
Cell lines having the lowest concentrations with negative growth effects are reported.
To further validate our method in a postdiction scenario for drug molecules derived from traditional Chinese medicine, we obtained a set of traditional Chinese medicine ingredients from the SYMMAP database39. Since the SYMMAP database has no direct connections between ingredients and symptoms/diseases, for validation, we opted to test the predictions for these molecules using the bioactivity data on human disease cell lines from ChEMBL and PubChem for validation. We mapped the names of the ingredients to their ChEMBL and PubChem IDs and obtained the respective ingredients’ SMILES string using the PubChem ID exchange service. After excluding those ingredients having a TC=1 to our training drugs, we obtained a set of 3,994 ingredients and applied MEDICASCY to predict the above 10 cancer indications. For each indication, the 3,994 molecules are ranked by their machine learning scores. Then, the top 20 molecules are investigated against the bioactivity data on human cell lines of the corresponding cancer in the ChEMBL and PubChem databases. The detailed results are compiled in Supporting Information Table S5. If a successful prediction is defined as when a molecule is indicated “active” with an IC50/GI50 value < 50 μM, the average success rate per indication is 61.8%. This result is entirely consistent with the value we previously obtained of 61.2% for NCI molecules when the same 50 μM cutoff is applied. However, with a cutoff 50 μM, the FDA drug set has a slightly higher success rate of 75.3% per indication. These results indicate that MEDICASCY has consistent performance across different sets of drugs.
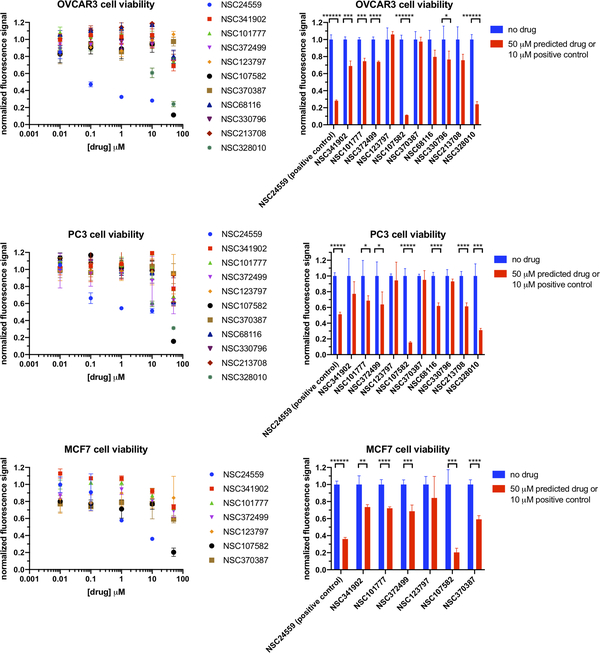
In addition to validation using the above postdiction data from the NCI-60, ChEMBL and PubChem cell line data, we further performed experimental verification of our computational predictions of NCI drug indications on human ovarian, prostate, and breast cancers. Using the Tox-8 assay 36, 40, we assessed cancer cell viability in response to 48 hours of drug treatment. Cancer inhibition effects were examined for MEDICASCY’s top ranked 10 drugs from the NCI diversity set with novel indication predictions along with a positive control with known anticancer activity, NSC24559 (Plicamycin). The prediction of drug indications is ruled correct if there is a statistically significant inhibition effect on the corresponding cancer type with 50μM treatment of the molecule vs no treatment. The results are summarized in Figure 3 and Tables 7 and 8. Table 7 shows a mean success rate of 75% in agreement with the above postdictions on existing NCI-60 data. Not only does our method have a high success rate on novel indication predictions when agnostic against specific cancer types, but it also proved to be useful to predict cancer type specificity with a cross-type precision of up to 0.5 (Table 8). In particular, our method correctly predicted that NSC330796 inhibits ovarian but not prostate cancer and NSC213708 vice versa (Table 7).
Figure 3:
Cell viability in response to treatment of predicted and positive control drugs. The Tox-8 assay was used to assess cell viability for OVCAR3, PC3 and MCF7 human cancer cells. The fluorescent signal of each condition was normalized against the corresponding signal with no drug treatment. The detailed of cell culture and assay conditions are described in methods. Statistical significance when present is indicated as *p<0.05. **p<0.01, ***p<0.001, ****p<0.0001, *****p<0.00001, ******p<0.000001, based on the unpaired t-test. Each data point represents an averaged value of n=3 or 4 independent measurements.
Table 7.
Computational predictions vs. experimental observations for individual drugs.
| Ovarian cancer | Prostate cancer | Breast cancer | |
|---|---|---|---|
| NSC24559 (positive control) | yes (yes) | yes (yes) | yes (yes) |
| NSC341902 | yes (yes) | yes (no) | yes (yes) |
| NSC101777 | yes (yes) | yes (yes) | yes (yes) |
| NSC372499 | yes (yes) | yes (yes) | yes (yes) |
| NSC123797 | yes (no) | yes (no) | yes (no) |
| NSC107582 | yes (yes) | yes (yes) | |
| NSC370387 | yes (no) | ||
| NSC68116* | yes (no) | yes (yes) | |
| NSC330796* | yes (yes) | ||
| NSC213708* | yes (yes) | ||
| NSC328010* | yes (yes) | ||
| Success rate§ | 6/9 | 7/9 | 4/5 |
Computational predictions are listed for each cancer type in this study. Experimental observed indications based on statistically significant cancer inhibition with 48 hours of 50μM drug treatment are listed in parentheses. The positive control NSC24559 (plicamycin) is an FDA approved cancer drug and its known indications are from the NCI-60 data. All the other 10 drugs in this study were predicted to have novel cancer indications without any prior knowledge or known documentation of cancer indications (machine learning prediction scores are in the range of 0.38–0.57, corresponding to expected precisions of 0.35–0.57 based on benchmarking results.)
NSC68116, NSC330796, NSC213708, NSC328010 were experimentally tested against ovarian and prostate cancers but not breast cancer in this study.
Success rate is defined as the ratio of the # of tested molecules having experimental inhibition to the # of tested molecules predicted to have inhibition (including the control molecule because it is predicted to have inhibition).
Table 8.
Precision and recall of predictions assessed by specific type of cancer inhibition by NCI diversity molecules at a 50μM concentration
| Precision | Recall | |
|---|---|---|
| OVCAR3 ovarian cancer | 0.6 (6/10) | 0.625 (5/8) |
| PC3 prostate cancer | 0.8 (8/10) | 0.75 (6/8) |
| MCF7 breast cancer | 0.5 (3/6) | 0.75 (3/4) |
| Average over all three cancer types | 0.654 (17/26) | 0.7 (14/20) |
| Cancer specificity (OVCAR3 vs. PC3) | 0.5 (5/10) | |
| Cancer specificity (OVCAR3 vs. MCF7) | 0.5 (3/6) | |
| Cancer specificity (PC3 vs. MCF7) | 0.333 (2/6) | |
| Cancer specificity (OVCAR3 vs. PC3 vs. MCF7) | 0.333 (2/6) | |
| Indication of at least one cancer type | 0.9 (9/10) |
The exact number of predictions used to calculate each precision or recall value is in parentheses. Pairwise or cross-cancer type comparison of prediction precision was calculated based on the rate of correct predictions of indications for all the cancer types under comparison.
Inference of probable mode-of-action targets
We next show another advantage of MEDICASCY beyond the fact that it merely requires the drug’s chemical structure over other methods for drug side effect and efficacy prediction: MEDICASCY can be utilized for the inference of mode-of-action (MOA) targets for a given side effect or efficacious indication (see MATERIALS AND METHODS).
As an example, using eq. (3), we infer the most likely MOA targets for the side effect neoplasm malignant or cancer. With a cutoff of 0.25 (the value of the average score+2× standard deviation which is equivalent to a Z-score of 2 defined as the [score – average score of all 2095 drugs/standard deviation of all 2095 scores]), MEDICASCY predicts 91 (2004) drugs with (without) this side effect. There are 537 proteins having an EF(T,D) > 2, and each protein binds to at least 5 (~5% of 91) drugs with the “side effect” of cancer. For example, the top ranked MAPK13 gene (mitogen-activated protein kinase 13) is predicted to have an EF(T,D) of 27.5, and is known to be involved in cancer41. The second ranked gene EPHA7 (ephrin type-A receptor 7 precursor) with an EF(T,D) of 22.0 is also associated with cancer42. In Table S3 (see Supplementary Information), we list the top 20 proteins based on their EF(T,D). All proteins have literature evidence that they are associated with one or multiple cancers.
Next, we show the example of inferring the MOA for the indication Crohn’s Disease (CD). Given a minimum Z-score cutoff of 2.0, 74 (2021) drugs are predicted to have (not have) this indication. In total, 430 protein targets have an EF > 2, and each protein binds to at least 4 (or ~5% of 74) drugs with a CD indication. The resulting top 20 protein targets are listed in Table S4. Except for MCOLN3, we found literature evidence that all of the other 19 genes either are directly associated with CD (or the related disease ulcerative colitis) or interact with genes associated with CD. Thus, these top ranked genes are potential MOA targets for CD.
We next show examples of the possible repurposing of FDA approved drugs for new indications, e.g. Crohn’s Disease. Depending on whether a drug is an agonist or antagonist of the protein target (or group of targets), it can either treat the disease or exacerbate it. Thus, in order for a drug to be repurposed for CD, it should have an indication of CD, but not have the CD side effect. Among the 74 FDA approved drugs that MEDICASCY predicts have CD as their indication, 64 are not known to our training library for treating CD and are not predicted to have the side effect of CD. One example is DB00337 (pimecrolimus), a topical cream used in the treatment of atopic dermatitis (eczema). MEDICASCY predicts that it binds to many top ranked targets in Table S4: ILE1 (EF=54.6), FKBP6(54.6), MLK1 (36.4), FKBP4(36.4), IL2RA(36.4), DCAF4L2(36.4), FKBP5(36.4), FKBP1A(27.3), FKBP2(27.3), WDR33(27.3), FKBP1B(27.3), and WDR89(22.8) that all have evidence of being involving in CD. Indeed, there is a literature report that pimecrolimus might be effective for CD 43. Another example is DB01252, (mitiglinide), for which there are no reports in the literature for its treating CD. Its primary indication is for the treatment of type 2 diabetes. It may stimulate insulin secretion in beta-cells by closing off ATP dependent potassium ion channels. MEDICASCY predicts that it binds to WDR13 (EF=21.8, interacting with NRF144 ) that could be its MOA target for CD. A third example is drug DB01615 (aceprometazine) that is used to treat sleep disorders. MEDICASCY predicts it binds to S100A4 with an EF=6.2. A recent study finds that S100A4 is related to CD45.
DISCUSSION
We have shown that MEDICASCY performs comparably to the state-of-the-art methods for side effect prediction just given the small molecule’s chemical structure as opposed to requiring extensive prior knowledge including the drug’s targets, pathways, carriers, enzyme interference and transporters. However, with the same feature inputs as other methods, MEDICASCY performs better than these alternatives based on the AUPR metric. More importantly, when only the chemical structure of the small molecule drug is employed in efficacy prediction, MEDICASCY has a much higher value of the more informative AUPR metric than the competing approaches. Moreover, experimental validation using the NCI-60 data shows that its success rate is more than 80% for efficacy prediction.
It should be noted that drug effects (indications, side effects) are not solely determined by a drug’s structure. They also depend on other factors like dosage, patient’s genetics and epigenetics, disease history, and so on. Because our machine learning method is trained on the outcome of the drug effects for a general patient, the effects of factors other than the chemical structure of a given drug on an average patient with average dosage and average internal conditions are implicitly taken into account and encoded in the model parameters. Thus, our sole structural based prediction model should be useful for average drug dosage and patient conditions.
MEDICASCY employs the state-of-the-art target screening method FINDSITEcomb2.0 to predict possible human protein targets of the drug. Based on these predicted human targets and employing updated and improved Know-GENE, ENTPRISE, ENTPRISE-X 27–29 methods, MEDICASCY computes a 960 dimensional feature of disease associations in addition to the directly computed MACCS fingerprint feature. While experimental or bioinformatics data might be available for well-investigated drugs, they are usually not available for new drug hits or leads. Thus, MEDICASCY has the very appealing advantage that it can be applied to new drug development as well as to repurposing of FDA approved drugs. Furthermore, it provides a way of linking side effects/indications to probable MOA protein targets. With these useful advantages, there is also a caveat that for a given drug, the whole prediction process takes longer than some of its peers5, 13.e.g. the multi-layer perceptron method5 takes ~2 hours for the Zhang dataset. In contrast, with pre-computed features, MEDICASCY takes roughly 1 week for the entire Zhang dataset. However, MEDICASCY is still more efficient than the FS-MLKNN method13 that according to Refernce5 takes > 2 weeks run time on a single core machine. Once the model is trained, MEDICASCY takes 15 minutes for the target prediction of a single molecule and 10 minutes for side effect or efficacy prediction. It also requires around 50 gigabytes to store the trained models. One way of speeding up the prediction is to use parallel computation of the RF and then summing the terms in equation (1b). If one uses a single RF with larger n_estimators, e.g. 500, then the memory required will be prohibitive, and there is no easy way of using parallel computing to reduce the time to obtain a prediction.
By using the experimental or bioinformatics features as provided by the Zhang side effect dataset, MEDICASCY has only a slightly better AUPR compared to using predicted features from the drug’s chemical structure (0.395 vs. 0.385). As mentioned above, this indicates that MEDICASCY has acceptable accuracy for its predicted features. It also implies that there is not much room for improvement of side effect prediction by using better features. However, since current indication data for a given drug is not complete, when more indication data becomes available for training, we would expect more accurate predictions (e.g. improving AUPRs) for efficacy.
We have also shown that MEDICASCY is also better, most especially for predicting killing side effects, than our previous empirical method where side effects for individual human proteins are inferred and the side effects of a drug are the union of side effects from all its binding protein targets. The fact that machine learning based MEDICASCY performs better indicates that side effects are collective effects of many protein targets of the drug. This could also be true for drug indications. Furthermore, the fact that there are many protein targets having an enrichment factor EF(T,D) > 2 for a given side effect or indication also supports the view that the one target-one disease paradigm might not be ground truth in many cases. In other words, drug side effects and efficacy are not, in general, Mendelian features that are dictated uniquely by interactions with a single protein.
For practical applications, MEDICASCY can serve as a pre-filter to de-risk and increase efficacy for lead discovery by pre-screening a small molecule library to select those without killing side effects and higher probability of preferred indications. For FDA approved drugs, efficacy predictions are useful for drug repurposing. More generally, MEDICASCY is the beginning of a general genotype to phenotype function prediction engine. Whether a drug-protein interaction induces a side effect or is merely a repurposing indication is just a matter of the use of the phenotypical response, not the underlying biology. Similarly, MEDICASCY unifies protein-protein interacting disease module information (through Know-GENE29) and the relevance of a given protein to collective responses as assessed by the disease association of the protein’s genetic variations (through ENTPRISE27 and ENTPRISE-X28). If a given protein can be deleted due to a frame shift variation with no deleterious phenotype, this implies that the loss of function of the protein cannot be a driver of disease. Similarly, small molecule inhibition of a protein (or set of proteins) that causes a side effect should have a similar phenotypical outcome as that when a protein’s function is entirely eliminated due to genetic variations. Thus, unification of genomic, side effect and efficacy information, all of which have a strong collective component that is captured by the BRF, also provides insights into the underlying modes of action of the given disease as well as how proteins function in the context of cells. These ideas will be expanded upon in future work.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grant No. 1R35GM-118039 of the Division of General Medical Sciences of the National Institutes of Health. We thank the National Cancer Institute (NCI)/Division of Cancer Treatment and Diagnosis (DCTD)/Developmental Therapeutics Program (DTP) (http://dtp.cancer.gov) for generously providing the NCI compounds used for experimental validation in this study. The authors also thank Dr. Bartosz Ilkowski for managing the cluster on which this work was conducted.
Footnotes
Supporting Information
Additional figures and tables presenting dependence of precision on Tc cutoffs; distributions of the number of indications of known and predicted for drugs; data for deriving features by various methods; computational time of MEDICASCY on a single node CPU;predicted mode of action targets for side effect cancers and indication Crohn’s Disease; detailed results for SYMMAP traditional Chinese medicine herb ingredients.
Training and Benchmarking Information
Scripts and data for reproducing all benchmarking results can be found at https://github.com/hzhou3ga/MEDICASCY/
REFERENCES
- 1.DiMasi J; Grabowsky H; Hansen R, Innovation in the pharmaceutical industry: New estimates of R&D costs. Journal of Health Economics 2016, 47, 20–33. [DOI] [PubMed] [Google Scholar]
- 2.Hay M; Thomas D; Craighead J, Clinical development success rates for investigational drugs. Nature Biotechnology 2014, 32, 40–51. [DOI] [PubMed] [Google Scholar]
- 3.Wong CH; Siah KW; Lo AW, Estimation of clinical trial success rates and related parameters. Biostatistics 2018, 20, 273–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sacks L; Shamsuddin H; Yasinskaya Y; Bouri K; Lanthier M; Sherman R, Scientific and regulatory reasons for delay and denial of FDA approval of initial applications for new drugs, 2000–2012. JAMA 2014, 311, 378–384. [DOI] [PubMed] [Google Scholar]
- 5.Muñoz E; Novácek V; Vandenbussche P, Facilitating prediction of adverse drug reactions by using knowledge graphs and multi-label learning models. Brief Bioinform. 2019, 20, 190–202. [DOI] [PubMed] [Google Scholar]
- 6.Zhang W; Liu F; Luo L; Zhang J, Predicting drug side effects by multi-label learning and ensemble learning. BMC Bioinformatics 2015, 16, 365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou H; Gao M; Skolnick J, Comprehensive prediction of drug-protein interactions and side effects for the human proteome. Scientific Reports 2015, 5, 11090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowes J; Brown A; Hamon J; Jarolimek W; Sridhar A; Waldron G; Whitebread S, Reducing safety-related drug attrition: the use of in vitro pharmacological profiling. Nat Rev Drug Discov. 2012, 11, 909–922. [DOI] [PubMed] [Google Scholar]
- 9.Tan Y; Hu Y; Liu X; Yin Z; Chen X; Liu M, Improving drug safety: From adverse drug reaction knowledge discovery to clinical implementation. Methods 2016, 110, 14–25. [DOI] [PubMed] [Google Scholar]
- 10.Yamanishi Y; Pauwels E; Kotera M, Drug side-effect prediction based on the integration of chemical and biological spaces. J Chem Inf Model. 2012, 52. [DOI] [PubMed] [Google Scholar]
- 11.Liu M; Wu Y; Chen Y; Sun J; Zhao Z; Chen X; Matheny M; Xu H, Large-scale prediction of adverse drug reactions using chemical, biological, and phenotypic properties of drugs. J Am Med Inform Assoc. 2012, 19, e28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang W; Zou H; Luo L; Liu Q; Wu W; Xiao W, Predicting potential side effects of drugs by recommender methods and ensemble learning. Neurocomputing 2016, 173. [Google Scholar]
- 13.Zhang W; Chen Y; Tu S; Liu F; Qu Q Drug side effect prediction through linear neighborhoods and multiple data source integration. In 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) 2016; IEEE: 2016; pp 427–434. [Google Scholar]
- 14.Dimitri GM; Lió P, DrugClust: A machine learning approach for drugs side effects prediction. Computational Biology and Chemistry 2017, 68, 204–210. [DOI] [PubMed] [Google Scholar]
- 15.Cheng F; Li W; Wang X; Zhou Y; Wu Z; Shen J; Tang Y, Adverse drug events: database construction and in silico prediction. J. Chem. Inf. Model. 2013, 53, 744–752. [DOI] [PubMed] [Google Scholar]
- 16.Himmelstein DS; Lizee A; Hessler C; Leo Brueggeman; Chen SL; Hadley D; Green A; Khankhanian P; Baranzini SE, Systematic integration of biomedical knowledge prioritizes drugs for repurposing. eLife 2017, 6, e26726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Menshykau D, Emerging technologies for prediction of drug candidate efficacy in the preclinical pipeline. Drug Discov Today 2017, 22, 1598–1603. [DOI] [PubMed] [Google Scholar]
- 18.Sleigh S; Barton C, Repurposing Strategies for Therapeutics. Pharm Med 2010, 24, 151–159. [Google Scholar]
- 19.Zhou H; Skolnick J, FINDSITEcomb: A Threading/Structure-Based, Proteomic-Scale Virtual Ligand Screening Approach. J. Chem. Inf. Model. 2013, 53, 230–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wishart D; Knox C; Guo A; Shrivastava S; Hassanali M; Stothard P; Chang Z; Woolsey J, DrugBank: a comprehensive resource for in silico drug discovery and exploration.. Nucl. Acid. Res. 2006, 34, D668–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhn M; Campillos M; Letunic I; Jensen L; Bork P, A side effect resource to capture phenotypic effects of drugs. Mol Syst Biol. 2010, 6, 343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou H; Cao H; Skolnick J, FINDSITEcomb2.0: A New Approach for Virtual Ligand Screening of Proteins and Virtual Target Screening of Biomolecules. Journal of Chemical Information and Modeling 2018, 58, 2343–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ho TK Random Decision Forests. In Proceedings of the 3rd International Conference on Document Analysis and Recognition, Montreal, QC, 1995; Montreal, QC, 1995; pp 278–282. [Google Scholar]
- 24.Altman NS, An Introduction to Kernel and Nearest-Neighbor Nonparametric Regression. The American Statistician 1992, 46, 175–185. [Google Scholar]
- 25.Zhou H; Gao M; Skolnick J, Comprehensive prediction of drug-protein interactions and side effects for the human proteome. Nature Scientific Reports 2015, 5, 11090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou H; Skolnick J, Template-Based Protein Structure Modeling Using TASSERVMT. Proteins 2011, 80, 352–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou H; Gao M; Skolnick J, ENTPRISE: An Algorithm for Predicting Human Disease-Associated Amino Acid Substitutions from Sequence Entropy and Predicted Protein Structures. PLOS ONE 2016, 11, e0150965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou H; Gao M; Skolnick J, ENTPRISE-X: Predicting disease-associated frameshift and nonsense mutations. PLOS ONE 2018, 13, e0196849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou H; Skolnick J, A knowledge-based approach for predicting gene-disease associations. Bioinformatics 2016, 32, 2831–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li YH; Yu CY; Li XX; Zhang P; Tang J; Yang Q; Fu T; Zhang X; Cui X; Tu G; Zhang Y; Li S; Yang F; Sun Q; Qin C; Zeng X; Chen Z; Chen YZ; Zhu F, Therapeutic target database update 2018: enriched resource for facilitating bench-to-clinic research of targeted therapeutics. Nucleic Acids Res. 2018, 46, D1121–D1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schriml L; Arze C; Nadendla S; Chang Y; Mazaitis M; Felix V; Feng G; Kibbe W, Disease Ontology: a backbone for disease semantic integration. Nucleic Acids Research 2012, 40, D940–D946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kibbe W; Arze C; Felix V; Mitraka E; Bolton E; Fu G; Mungall C; Binder J; Malone J; Vasant D; Parkinson H; Schriml L, Disease Ontology 2015 update: an expanded and updated database of human diseases for linking biomedical knowledge through disease data. Nucleic Acids Research 2015, 43, D1071–D1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schriml LM; Mitraka E; Munro J; Tauber B; Schor M; Nickle L; Felix V; Jeng L; Bearer C; Lichenstein R; Bisordi K; Campion N; Hyman B; Kurland D; Oates CP; Kibbey S; Sreekumar P; Le C; Giglio M; Greene C, Human Disease Ontology 2018 update: classification, content and workflow expansion Nucleic Acids Res. 2019, 47, D955–D962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanimoto TT, An Elementary Mathematical Theory of Classification and Prediction. IBM Interanl Report 1958. [Google Scholar]
- 35.Davis J; Goadrich M The Relationship Between Precision-Recall and ROC Curves. In ICML ‘06 Proceedings of the 23rd International Conference on Machine Learning, Pittsburgh, 2006; ACM New York, NY, USA Pittsburgh, 2006; pp 233–240. [Google Scholar]
- 36.Mezencev R; Updegrove T; Kutschy P; Repovská M; McDonald J, Camalexin induces apoptosis in T-leukemia Jurkat cells by increased concentration of reactive oxygen species and activation of caspase-8 and caspase-9. J. Nat. Med. 2011, 65, 488–499. [DOI] [PubMed] [Google Scholar]
- 37.Gaulton A; Bellis L; Bento A; Chambers J; Davies M; Hersey A; Light Y; McGlinchey S; Michalovich D; Al-Lazikani B; Overington J, ChEMBL: a large-scale bioactivity database for drug discovery. Nucl. Acid. Res. 2012, 40, D1100–07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim S; Chen J; Cheng T; Gindulyte A; He J; He S; Li Q; Shoemaker BA; Thiessen PA; Yu B; Zaslavsky L; Zhang J; Bolton EE, PubChem 2019 update: improved access to chemical data. Nucleic acids research 2019, 47, D1102–D1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu Y; Zhang F; Yang K; Fang S; Bu D; Li H; Sun L; Hu H; Gao K; Wang W; Zhou X; Zhao Y; Chen J, SymMap: an integrative database of traditional Chinese medicine enhanced by symptom mapping. Nucleic Acids Research 2018, 47, D1110–D1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Brien J; Wilson I; Orton T; Pognan F, Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mammalian cell cytotoxicity. Eur J Biochem 2000, 267, 5421–5426. [DOI] [PubMed] [Google Scholar]
- 41.Tan FLS; Ooi A; Huang D; Wong JC; Qian CN; Chao C; Ooi L; Tan YM; Chung A; Cheow PC; Zhang Z; Petillo D; Yang XJ; Teh BT, p38delta/MAPK13 as a diagnostic marker for cholangiocarcinoma and its involvement in cell motility and invasion. Int. J. Cancer 2010, 126, 2353–2361. [DOI] [PubMed] [Google Scholar]
- 42.Li S; Wu Z; Ma P; Xu Y; Chen Y; Wang H; He P; Kang Z; Yin L; Zhao Y; Zhang X; Xu X; Ma X; Guan M, Ligand-dependent EphA7 signaling inhibits prostate tumor growth and progression. Cell Death & Disease 2017, 8, e3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cassona DH; Eltumic M; Tomlinb S; Walker-Smitha JA; Murcha SH, Topical tacrolimus may be effective in the treatment of oral and perineal Crohn’s disease. Gut. 2000, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baillie JK; Arner E; Daub C; Hoon MD; Itoh M; Kawaji H; Lassmann T; Carninci P; Forrest ARR; Hayashizaki Y; Consortium F; Faulkner GJ; Wells CA; Rehli M; Pavli P; Summers KM; Hume DA, Analysis of the human monocyte-derived macrophage transcriptome and response to lipopolysaccharide provides new insights into genetic aetiology of inflammatory bowel disease. PloS Genet. 2017, 13, e1006641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang J; Hou S; Gu J; Tian T; Yuan Q; Jia J; Qin Z; Chen Z, S100A4 promotes colon inflammation and colitis-associated colon tumorigenesis. Oncoimmunology. 2018, 7, e1461301. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.