Abstract
Background
Identification of correlates of protection against human influenza A virus infection is important in development of broadly protective (“universal”) influenza vaccines. Certain assumptions underlie current vaccine developmental strategies, including that infection with a particular influenza A virus should offer long-term or lifelong protection against that strain, preventing reinfection. In this study we report observations made when 7 volunteers participated in sequential influenza challenge studies where they were challenged intranasally using the identical influenza A(H1N1)pdm09 virus approximately 1 year apart. We evaluate and describe the outcomes of these 7 rechallenge participants and discuss what these results may suggest about correlates of protection and development of more broadly protective influenza vaccines.
Methods
Seven participants were enrolled in 2 viral challenge studies at 7.5- to 18.5-month intervals. Both challenge studies used the identical lot of influenza A (H1N1)pdm09 virus administered intranasally. We evaluated pre- and postchallenge hemagglutination inhibition, neuraminidase inhibition, and stalk antibody titers; peripheral blood leukocyte host gene expression response profiles; daily viral detection via nasal wash; and clinical signs and symptoms.
Results
At least 3 of 7 participants demonstrated confirmed laboratory evidence of sequential infection, with 5 of 7 demonstrating clinical evidence.
Conclusions
The data presented in this report demonstrate that sequential infection with the identical influenza A virus can occur and suggest it may not be rare. These data raise questions about immune memory responses in an acute superficial respiratory mucosal infection and their implications in development of broadly protective influenza vaccines. Further investigation of these observations is warranted.
Clinical Trials Registration
Keywords: influenza A, healthy volunteer, challenge, CHIM, vaccine
Sequential influenza A challenge studies in humans demonstrate reinfection with an identical A(H1N1)pdm09 virus. This raises important questions regarding correlates of protection and vaccine development.
Identifying correlates of protection against human influenza A virus infection is a necessary step in developing more broadly protective (“universal”) influenza vaccines [1]. Although true correlates of protection have never been fully established, certain assumptions underlie vaccine developmental strategies. All currently licensed influenza vaccines were designed to induce serum antibody responses against the influenza virus hemagglutinin (HA) head, and many newer vaccine candidates are being developed to induce serum antibody responses to additional epitopes as well, including those on the HA stalk [2–6]. Serum antibodies induced by vaccines can be measured using neutralization, hemagglutination inhibition (HAI), and enzyme-linked immunosorbent assays, among others. Such serologic assays have been used as markers of protection because they are easy to perform [7]. Antibodies at or above certain titers (eg, HAI ≥1:40) are considered rough correlates of protection, based largely upon population or aggregate data, even though natural infections take place in the presence of substantial serum anti-HA antibody titers and ≥10% of naturally exposed individuals may not generate detectable serum antibodies against influenza HA, whether or not they become infected [8–10].
Human influenza challenge studies performed at the National Institutes of Health Clinical Center, using a wild-type influenza A(H1N1)pdm09 virus, feature a controlled setting in which to evaluate influenza natural history, pathogenesis, and immune responses [11], including correlates of protection between prechallenge serum antibody titers and subsequent infection and disease.
As part of a dose-finding and validation study, 46 volunteers with variably high and low preexisting HAI titers against the A(H1N1)pdm09 virus were challenged intranasally [10]. In a subsequent challenge study using the identical challenge virus, 7 of those original 46 volunteers were rechallenged approximately 1 year later [11]. In this substudy, we evaluate and describe the outcomes of these 7 rechallenge participants and discuss what these results may suggest about correlates of protection and development of more broadly protective influenza vaccines.
MATERIALS AND METHODS
Clinical Studies
Two healthy volunteer influenza challenge studies took place independently of one another [10, 11]. Continuing recruitment of subjects for influenza studies, which spanned the years of these studies, resulted in a pool of both earlier- and later-recruited volunteers. Seven of the 46 participants from the first study were among those who also volunteered for the second study; these subjects were enrolled and challenged in the second set of studies 7.5–18.5 months after their participation in the first study (Table 1). These challenge studies used the identical lot of influenza A (H1N1)pdm09 virus produced in Vero cells and administered intranasally [10, 11]. All 7 subjects described herein volunteered on their own, independently of each other, and at separate times, to participate in both challenge studies, and were not selected by the investigators for rechallenge. None of the 7 participants reported having had influenza in the interval between challenges 1 and 2 and none had received influenza vaccine within 1 year of challenge during either study. Inclusion and exclusion criteria were identical except for HAI titer requirements for each study [10, 11]. All volunteers provided written informed consent before any protocol procedures were carried out.
Table 1.
Multiple Influenza Challenge Exposure Timing and Outcomes
| Participant (Sex, Age, y) | Challenge Date | Viral Dose (TCID50) | Nasal Viral Detection (Duration, d) | Days of Symptoms | No. of Symptoms | Severity Scorea | D0 and W8 HAI Titerb | D0 and W8 NAI Titerb | D0 and W8 HA Stalk Titerb | PBL Gene Expressionc |
|---|---|---|---|---|---|---|---|---|---|---|
| Challenge 1 | ||||||||||
| 1 (F, 21) | 9 Oct 2012 | 106 | + (4) | 5 | 11 | 55 | 0–40 | 0–1280 | 44 084–80 480 | + |
| 2 (M, 19) | 25 Sept 2012 | 106 | – (0) | 6 | 1 | 6 | 10–10 | 1280–2560 | NA | – |
| 3 (M, 24) | 21 Jan 2013 | 107 | + (1) | 8 | 6 | 48 | 20–160 | 160–2560 | 43 048–58 892 | + |
| 4 (M, 21) | 19 Mar 2013 | 107 | – (0) | 2 | 1 | 2 | 0–160 | 2560–1280 | 65 165–81 987 | + |
| 5 (F, 24) | 2 Apr 2013 | 107 | + (7) | 11 | 12 | 132 | 0–40 | 0–80 | 27 422–86 008 | + |
| 6 (F, 24) | 10 Dec 2012 | 106 | – (0) | 6 | 2 | 12 | 0–80 | 80–160 | NA | – |
| 7 (F, 28) | 5 Jun 2012 | 103 | – (0) | 1 | 2 | 2 | 0–10 | 0–40 | NA | NA |
| Challenge 2 | ||||||||||
| 1 (F, 22) | 15 Oct 2013 | 107 | + (2) | 0 | 0 | 0 | 10–80 | 80–640 | 60 221–83 250 | + |
| 2 (M, 21) | 15 Oct 2013 | 107 | + (1) | 0 | 0 | 0 | 0–80 | 640–2560 | 42 016–96 331 | – |
| 3 (M, 24) | 12 Nov 2013 | 107 | + (1) | 11 | 5 | 55 | 160–160 | 1280–1280 | 86 034–67 954 | + |
| 4 (M, 21) | 12 Nov 2013 | 107 | – (0) | 4 | 3 | 12 | 10–20 | 1280–1280 | NA | – |
| 5 (F, 24) | 12 Nov 2013 | 107 | – (0) | 4 | 5 | 20 | 20–80 | 320–320 | 54 703–55 000 | – |
| 6 (F, 25) | 12 Nov 2013 | 107 | – (0) | 5 | 3 | 15 | 40–160 | 80–80 | 24 681–22 228 | – |
| 7 (F, 30) | 9 Dec 2013 | 107 | + (6) | 7 | 8 | 56 | 0–10 | 0–320 | 18 486–60 623 | NA |
Abbreviations: –, negative; +, positive; D0, prechallenge baseline day 0; F, female; HA, hemagglutinin; HAI, hemagglutination inhibition; M, male; NA, not available for testing; NAI, neuraminidase inhibition; PBL, peripheral blood leukocyte; TCID50, 50% tissue culture infectious dose; W8, 8 weeks after challenge.
aMean and median symptom scores across both challenge trials were 35.5 and 10, respectively. Severity scores ranged from 0 to 750. Symptom scores represent the totality of the disease experienced after challenge.
bSerum antibody titers against the hemagglutinin head, NAI, and HA stalk were measured at D0 and at W8.
cPeripheral blood leukocyte expression of antiviral and immune-related genes is shown in Figure 1.
We evaluated pre- and postchallenge serum HAI, neuraminidase inhibition (NAI), and anti–HA stalk antibody titers; peripheral blood leukocyte (PBL) host gene expression response profiles; daily viral detection via nasal wash, and clinical signs and symptoms using a validated scoring tool [10–12]. Pre- and postchallenge HAI titers from both studies were titered in parallel in a single assay (Table 1). In both studies, participants remained isolated for a minimum of 9 days. Clinical illness was evaluated multiple ways [10, 11]: (1) identifying the presence or absence of mild to moderate influenza disease (MMID; defined as viral shedding detected by US Food and Drug Administration–approved molecular testing, plus the onset of at least 1 acute influenza-like illness sign/symptom after intranasal challenge); (2) counting the number of signs/symptoms; (3) measuring the duration of signs/symptoms; and (4) calculating a clinical score of disease severity. All subjects placed in hospital isolation were tested daily for 20 different respiratory pathogens by the BioFire FilmArray RP (BioFire Diagnostics, Utah). Both clinical studies (ClinicalTrials.gov identifiers NCT01646138 and NCT01971255) were approved by the National Institute of Allergy and Infectious Diseases Institutional Review Board and were conducted in accordance with the provisions of the Declaration of Helsinki and Good Clinical Practice guidelines.
Host RNA Isolation and Expression Microarray Analysis
Total RNA was isolated from whole blood using the PAXgene Blood RNA Kit IVD (Qiagen, Gaithersburg, Maryland). RNA quality was assessed using a BioAnalyzer (Agilent Technologies, California). Gene expression profiling was performed using Agilent Human Whole Genome 44K microarrays. Fluorescent probes were prepared using Agilent QuickAmp Labeling Kit according to the manufacturer’s instructions. Each RNA sample was labeled and hybridized to individual arrays. Spot quantitation was performed using Agilent’s Feature Extractor software; all data were then entered into a custom-designed database, SLIMarray (http://slimarray.systemsbiology.net), and uploaded into Genedata Analyst 9.0 (Genedata, Basel, Switzerland). Data normalization was performed in Genedata Analyst using central tendency, followed by relative normalization, using each individual participant’s baseline (day 0) as a reference for each of the challenge studies. The complete minimum information about a microarray experiment–compliant [13] microarray dataset was deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) [14] and is accessible through GEO Series accession number GSE118223.
RESULTS
Two sequential challenge studies, conducted approximately 1 year apart, referred to here as challenge 1 and challenge 2, were carried out using the identical influenza A(H1N1)pdm09 challenge virus [10, 11]. The median time between the 2 challenge exposures was just under 1 year (337 days). Seven of the 46 participants from challenge 1, all of whom had remained in the growing recruitment pool, independently volunteered to be enrolled in challenge 2. During challenge 1 (a dose-finding study), these 7 participants received varying doses of virus, ranging from 103 to 107 the 50% tissue culture infectious dose (TCID50) (Table 1). In challenge 2, all 7 participants had received 107 TCID50 intranasally (Table 1). All participants tested negative for 19 other respiratory pathogens daily during both challenges 1 and 2 (BioFire FilmArray, BioFire Diagnostics, Salt Lake City, Utah).
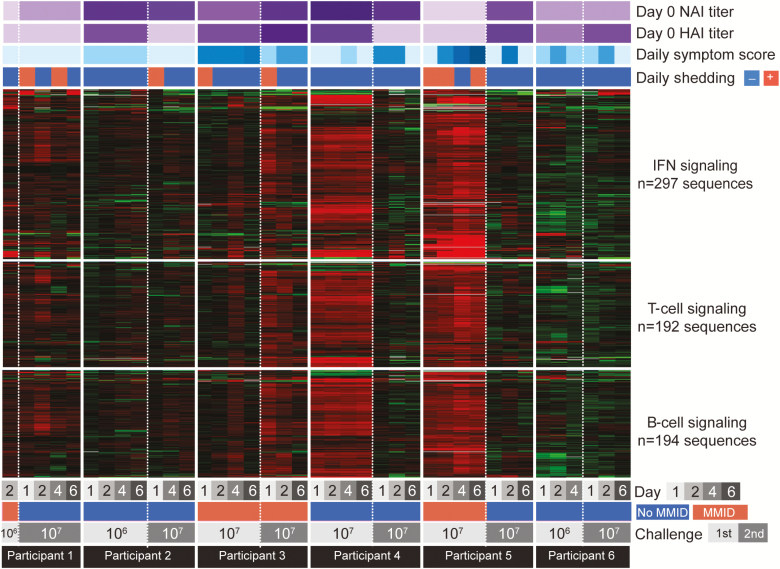
Clinical and laboratory data collected on the 7 participants are shown in Table 1 and in Figure 1. In challenge 1, all 7 participants developed signs/symptoms consistent with experimental influenza infection. Five of the 7 demonstrated objective laboratory evidence of experimental influenza infection: 3 of the 5 had detectable viral shedding, all 5 demonstrated HAI seroconversion, and 4 of the 5 demonstrated NAI seroconversion. Four participants also demonstrated rises in anti-HA stalk antibody titer, including one of the participants who did not demonstrate other laboratory evidence of infection, but the significance of antistalk titer increases is not yet fully understood [11]. Two challenge subjects with very mild or moderate symptom scores (participants 2 and 7, Table 1), had neither viral detection nor seroconversion. Of note, the 3 challenge 1 participants with both viral shedding and seroconversion also showed the strongest increases in PBL antiviral, T-cell, and B-cell gene expression acutely (Figure 1), and developed the most significant influenza-related symptoms over a 5- to 11-day period (Table 1), with tabulated symptom scores above the mean for challenge 1 [10, 11].
Figure 1.
Expression of type I interferon (IFN), T-cell, and B-cell signaling genes during A(H1N1)pdm09 influenza challenge 1 and challenge 2. Heatmap diagram showing expression profiles of type I IFN, T-cell, and B-cell signaling immune response pathway genes. For each participant, gene expression analysis was performed by expression microarray, and gene expression on days post–viral challenge was normalized to each participant’s baseline (day 0) expression values. Expression of genes shown in red was increased, and expression of genes shown in green was decreased relative to each participant’s day 0 expression, with black indicating no relative change in expression following A(H1N1)pdm09 virus inoculation. Each participant’s preexisting neuraminidase inhibition (NAI) and hemagglutination inhibition (HAI) titers, daily symptom score, viral shedding status, and clinical outcome (mild to moderate influenza disease [MMID] or no MMID) and challenge 50% tissue culture infectious dose (TCID50) are also shown. Global gene expression analysis was not performed in challenge 1 for participant 7 who received a 103 TCID50 dose due to the absence of samples.
At the time of challenge 2, prechallenge HAI titers had declined from the 8-week post–challenge 1 titers in 4 of the 7 participants (Table 1). Six of the 7 participants had lower pre–challenge 2 NAI titers than had been observed at 8 weeks post–challenge 1; 2 of these had lower NAI titers compared to titers before challenge 1 (Table 1).
During challenge 2, 5 of 7 patients developed signs/symptoms consistent with influenza infection (Table 1), and 6 developed objective laboratory evidence of infection. Four of the 7 participants (1, 2, 3, and 7) demonstrated detectable viral shedding of the identical challenge virus to which they had been previously inoculated in challenge 1. Two of these 4 participants also demonstrated ≥4-fold increases in challenge 2 postexposure HAI titer (participants 1 and 2), and 3 (participants 1, 2, and 7) had ≥4-fold increases in challenge 2 postexposure NAI titer, as well as increases over preexposure challenge 2 baseline of anti-HA stalk antibody titer (Table 1). Two other participants (5 and 6) developed 4-fold rises in HAI titer 8 weeks after inoculation despite not having virus detected. Two of the 4 participants (1 and 3) who had detectable nasal wash influenza viral shedding in challenge 2 also showed increases in PBL antiviral, T-cell, and B-cell gene expression acutely (Figure 1). While 5 of the 7 challenge 2 participants exhibited influenza-related symptoms (participants 3–7), the remaining 2 participants, who both had detectable viral shedding, were asymptomatic (participants 1 and 2). Of the 5 participants who had objective laboratory evidence of influenza infection in challenge 1, 4 demonstrated similar objective evidence of infection in challenge 2, as documented by viral shedding and/or seroconversion. One additional subject (participant 4), who had clear laboratory evidence of infection in challenge 1, experienced 4 days of mild influenza-like illness without detectable viral shedding or seroconversion.
Virologic evidence for reinfection with the identical influenza A virus after both challenge 1 and challenge 2, was found in 2 of the 7 individuals studied here: participants 1 and 3 (Table 1). Although postchallenge clinical illness in the absence of viral shedding or seroconversion is not typically considered proof of influenza infection, it is interesting to note that all participants in challenge 1 had clinical signs/symptoms indicative of influenza in this controlled environment after challenge, including those in challenge 1 who did not have detectable viral shedding or seroconversion. One of the latter participants (participant 4) also had PBL gene expression similar to those who shed influenza virus (Figure 1).
DISCUSSION
The 7 influenza subjects evaluated here represent a group of healthy individuals who were exposed to the identical influenza A(H1N1)pdm09 virus in 2 consecutive challenges. This is a unique circumstance: In nature, most individuals are probably relatively protected from reinfection with the same or different influenza strains and subtypes for several months after infection [15] and may not typically be reexposed to identical or closely related influenza viruses until a subsequent influenza season, at which time circulating viruses may be antigenically drifted.
While it has long been known that vaccine-induced protection wanes after 6–12 months [9, 16], it has seemed reasonable to many observers that homotypic protection against a natural influenza virus infection should be long-lasting. However, the observations reported here, that persons can be infected with a specific influenza A virus, mount an immune response against that virus with or without influenza signs and symptoms, and then be reinfected with the genetically identical virus after about 1 year’s time, raises questions about the existence, nature, and kinetics of long-term protective immunity and correlates of protection.
The clearest example of reinfection may be participant 3, who developed nearly equivalent clinical disease and detectable viral shedding after both challenges (Table 1). One other subject (participant 1) also demonstrated viral evidence of sequential infection, but with much less clinical illness during the second infection. The other 5 individuals (participants 2, 4, 5, 6, and 7) also demonstrated clinical signs and symptoms supportive of sequential infection (Table 1).
In our experience with influenza challenge, viral shedding is intermittent and may be easily missed when assays are only obtained daily, providing one possible explanation for lack of viral detection in all participants. In some cases, viral detection is only observed at 24 hours postchallenge. It is unlikely that this represents detection of residual inoculum, as experimental data suggest that detection of residual virus is unlikely after about 3 hours postchallenge [17], and in our experience virus is virtually never detected at 24 hours in participants who do not become ill. It is also known that high levels of preexisting anti-HA and anti-NA serum antibody titers have been negatively correlated with viral replication [10], potentially reducing the chance of viral detection. The 5 participants noted above who lacked viral detection in both of the challenges nevertheless demonstrated clear signs of infection such as 4-fold rises in anti-HA or NA antibody titer, or clear clinical disease. Participant 7, for example, did not have detectable viral shedding but did have a ≥4-fold increase in NAI titer during challenge 1, and was able to be reinfected in challenge 2, experiencing symptoms and viral shedding.
Given that natural or experimental reinfection with the same or a closely related (nondrifted) influenza virus in a short time interval is not a well-documented phenomenon, these results were somewhat unexpected, although there have been recent case studies and mathematical models of short-interval sequential infections that have suggested this is a possible explanation for multiple-wave influenza outbreaks of H3N2 [18, 19]. In animal models, at least 2 studies have demonstrated reinfection with the same influenza A(H3N2) or A(H1N1) strain, and one human challenge study of a partially attenuated influenza B virus also demonstrated evidence of reinfection [17, 20, 21]. On the other hand, epidemiologic evidence for immunoprotection following long-interval reexposures to antigenically related human H1N1 viruses include data derived from the reappearance of a 1950s lineage H1N1virus in 1977 [22], and from the emergence of the 2009 A(H1N1)pdm09 pandemic virus, which expressed an H1 protein antigenically similar to 1918 H1N1 lineage human viruses that circulated prior to 1957 [23]. However, these are population-level data that probably reflect not only actual individual protection from infection, but also infection of reduced or asymptomatic severity, as well as herd-incidence protection from exposure, and thus are not directly applicable to the interpretation of experimental challenge data.
Other unrelated human mucosal viruses, such as respiratory syncytial virus and norovirus, have also been associated with natural or experimental reinfection in association with factors such as genetic susceptibility, rapidity of mucosal immune responses, waning immunity, and other factors [24–26]. Therefore, it is important to question the virologic/immunologic bases for this observed phenomenon of reinfection. In addition to studying a much larger number of subjects, it will be important in future studies to challenge and rechallenge with influenza A(H3N2) viruses to examine the possibility that the current results are subtype-specific. In addition, we must consider aspects of the challenge study itself including route of inoculation of the virus, but we are unaware of any mechanism by which this type of challenge might overcome otherwise protective immunity. The virus itself is phenotypically identical to the original wild-type virus it is derived from, and in validating dose-finding experiments we found that a 107 dose produced only marginally higher rates of infection than the 106 and 105 doses, without altering clinical illness or incubation period in those who were infected. The 107 challenge dose we used was in line with other human and experimental influenza challenge studies. Such studies have in general found that higher doses within this range are associated with increased percentages of infections without substantially altering the course of illness or overcoming immune protection [27].
After 6 passages in Vero cells, the challenge virus was already a nonclonal quasispecies, which expanded quickly in the first few days of infection to become a more complex quasispecies similar to those associated with natural infection (unpublished data). An age-cohort or prior exposure effect seems unlikely but is possible, as does a “volunteer effect,” in which individuals with recurrent natural influenza might be more likely to volunteer. Nevertheless, in future studies it will be important to examine all variables associated with the challenge milieu in order to rule out experimental effects. This will include enrolling participants with any level of prechallenge titers to examine challenge and rechallenge in persons with a spectrum of preexisting immunologic measurements, as is found in open populations. The possibility that reinfection reflects unidentified genetic susceptibilities is of interest, but cannot be adequately studied at this time.
In this challenge model, it is clear that after a 7- to 18-month interval, at least 2 of 7 individuals were able to be reinfected by the identical A(H1N1)pdm09 influenza virus, and that previous infection experience did not in these cases afford sterilizing protective immunity. Although this is a small substudy with only 7 individuals, we saw clinical evidence of sequential infection in almost all participants and clear laboratory-confirmed evidence in at least 5 participants (participants 1, 3, 5, 6, and 7), suggesting this is not a rare phenomenon. Of note is the observation that some challenge participants who neither shed virus nor seroconverted postchallenge nevertheless developed typical, and sometimes multiday, influenza-like illnesses after challenge, often in association with a PBL gene expression response. While these illnesses might have been caused by unrelated viruses, the participants were in hospital isolation before challenge and remained negative to a large panel of other respiratory viruses during the course of the studies. The most likely explanations for these illnesses are either experimental infection or an inflammatory/immune response to the challenge virus producing influenza-like symptoms.
CONCLUSIONS
Sequential challenge performed with an identical influenza A virus in a small set of participants demonstrated that reinfection with the same virus can occur, in association with a diversity of susceptibility, illnesses, and immune responses. There were no obvious correlates of protection against reinfection, but many more individuals would need to be sequentially challenged to determine if such correlates exist.
The data presented here raise questions that must be investigated further as they could impact the development of broadly protective or universal vaccines. If infection following a high-dose viral challenge does not raise fully protective homotypic immunity, can we expect vaccines to be made that will do so, let alone provide heterologous protection? Moreover, the individuality of outcomes observed in this small set of healthy volunteers is likely to be amplified greatly when vaccinating populations at large. Identifying additional correlates of protection will be a critical first step, including evaluation of mucosal immunity mediated by secretory immunoglobulin G and immunoglobulin A, cellular immunity, and other effectors. It is also possible that unknown genetic variations among the participants may also account for the varied responses described here. It is likely that strategies may need to be employed that target multiple viral antigens to produce new generations of influenza vaccines that induce strong and durable protection. Clearly, much more needs to be learned about the natural history of human influenza infection, including variables associated with protection from infection and illness.
Notes
Acknowledgments. The authors acknowledge Dr H. Clifford Lane, Dr Richard T. Davey, the National Institutes of Health (NIH) Clinical Center Special Clinical Studies Unit staff, the Department of Laboratory Medicine, and the Clinical Center Pharmacy for their support of the clinical protocols. The authors also acknowledge Kim Green, PhD, for observations on reinfections with enteric viruses.
Financial support. This work was supported by intramural funds from the NIH and the National Institute of Allergy and Infectious Diseases, and by the Biomedical Advanced Research and Development Authority.
Potential conflicts of interest. The authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Erbelding EJ, Post DJ, Stemmy EJ, et al. A universal influenza vaccine: the strategic plan for the National Institute of Allergy and Infectious Diseases. J Infect Dis 2018; 218:347–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yassine HM, Boyington JC, McTamney PM, et al. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat Med 2015; 21:1065–70. [DOI] [PubMed] [Google Scholar]
- 3. Neu KE, Henry Dunand CJ, Wilson PC. Heads, stalks and everything else: how can antibodies eradicate influenza as a human disease? Curr Opin Immunol 2016; 42:48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nachbagauer R, Krammer F. Universal influenza virus vaccines and therapeutic antibodies. Clin Microbiol Infect 2017; 23:222–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nabel GJ, Fauci AS. Induction of unnatural immunity: prospects for a broadly protective universal influenza vaccine. Nat Med 2010; 16:1389–91. [DOI] [PubMed] [Google Scholar]
- 6. Impagliazzo A, Milder F, Kuipers H, et al. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science 2015; 349:1301–6. [DOI] [PubMed] [Google Scholar]
- 7. Hirst GK. The quantitative determination of influenza virus and antibodies by means of red cell agglutination. J Exp Med 1942; 75:49–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ohmit SE, Petrie JG, Cross RT, Johnson E, Monto AS. Influenza hemagglutination-inhibition antibody titer as a correlate of vaccine-induced protection. J Infect Dis 2011; 204:1879–85. [DOI] [PubMed] [Google Scholar]
- 9. Jacobson RM, Grill DE, Oberg AL, Tosh PK, Ovsyannikova IG, Poland GA. Profiles of influenza A/H1N1 vaccine response using hemagglutination-inhibition titers. Hum Vaccin Immunother 2015; 11:961–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Memoli MJ, Shaw PA, Han A, et al. Evaluation of antihemagglutinin and antineuraminidase antibodies as correlates of protection in an influenza A/H1N1 virus healthy human challenge model. MBio 2016; 7:e00417–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Memoli MJ, Czajkowski L, Reed S, et al. Validation of the wild-type influenza A human challenge model H1N1pdMIST: an A(H1N1)pdm09 dose-finding investigational new drug study. Clin Infect Dis 2015; 60:693–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Park JK, Han A, Czajkowski L, et al. Evaluation of preexisting anti-hemagglutinin stalk antibody as a correlate of protection in a healthy volunteer challenge with influenza A/H1N1pdm virus. MBio 2018; 9. doi: 10.1128/mBio.02284-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brazma A, Hingamp P, Quackenbush J, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet 2001; 29:365–71. [DOI] [PubMed] [Google Scholar]
- 14. Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002; 30:207–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev 1992; 56:152–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Petrie JG, Ohmit SE, Johnson E, Truscon R, Monto AS. Persistence of antibodies to influenza hemagglutinin and neuraminidase following one or two years of influenza vaccination. J Infect Dis 2015; 212:1914–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Keitel WA, Couch RB, Cate TR, Six HR, Baxter BD. Cold recombinant influenza B/Texas/1/84 vaccine virus (CRB 87): attenuation, immunogenicity, and efficacy against homotypic challenge. J Infect Dis 1990; 161:22–6. [DOI] [PubMed] [Google Scholar]
- 18. Camacho A, Cazelles B. Does homologous reinfection drive multiple-wave influenza outbreaks? Accounting for immunodynamics in epidemiological models. Epidemics 2013; 5:187–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Camacho A, Ballesteros S, Graham AL, Carrat F, Ratmann O, Cazelles B. Explaining rapid reinfections in multiple-wave influenza outbreaks: Tristan da Cunha 1971 epidemic as a case study. Proc Biol Sci 2011; 278:3635–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Berendt RF. Simian model for the evaluation of immunity to influenza. Infect Immun 1974; 9:101–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Carroll TD, Matzinger SR, Fritts L, McChesney MB, Miller CJ. Memory B cells and CD8+ lymphocytes do not control seasonal influenza A virus replication after homologous re-challenge of rhesus macaques. PLoS One 2011; 6:e21756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nakajima K, Desselberger U, Palese P. Recent human influenza A (H1N1) viruses are closely related genetically to strains isolated in 1950. Nature 1978; 274:334–9. [DOI] [PubMed] [Google Scholar]
- 23. Glezen WP, Keitel WA, Taber LH, Piedra PA, Clover RD, Couch RB. Age distribution of patients with medically-attended illnesses caused by sequential variants of influenza A/H1N1: comparison to age-specific infection rates, 1978–1989. Am J Epidemiol 1991; 133:296–304. [DOI] [PubMed] [Google Scholar]
- 24. Lindesmith L, Moe C, Marionneau S, et al. Human susceptibility and resistance to Norwalk virus infection. Nat Med 2003; 9:548–53. [DOI] [PubMed] [Google Scholar]
- 25. Ascough S, Paterson S, Chiu C. Induction and subversion of human protective immunity: contrasting influenza and respiratory syncytial virus. Front Immunol 2018; 9:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kutsaya A, Teros-Jaakkola T, Kakkola L, et al. Prospective clinical and serological follow-up in early childhood reveals a high rate of subclinical RSV infection and a relatively high reinfection rate within the first 3 years of life. Epidemiol Infect 2016; 144:1622–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Carrat F, Vergu E, Ferguson NM, et al. Time lines of infection and disease in human influenza: a review of volunteer challenge studies. Am J Epidemiol 2008; 167:775–85. [DOI] [PubMed] [Google Scholar]