Abstract
Background
Whether human immunodeficiency virus (HIV) infection impacts gut microbial α-diversity is controversial. We reanalyzed raw 16S ribosomal RNA (rRNA) gene sequences and metadata from published studies to examine α-diversity measures between HIV-uninfected (HIV–) and HIV-infected (HIV+) individuals.
Methods
We conducted a systematic review and individual level meta-analysis by searching Embase, Medline, and Scopus for original research studies (inception to 31 December 2017). Included studies reported 16S rRNA gene sequences of fecal samples from HIV+ patients. Raw sequence reads and metadata were obtained from public databases or from study authors. Raw reads were processed through standardized pipelines with use of a high-resolution taxonomic classifier. The χ2 test, paired t tests, and generalized linear mixed models were used to relate α-diversity measures and clinical metadata.
Results
Twenty-two studies were identified with 17 datasets available for analysis, yielding 1032 samples (311 HIV–, 721 HIV+). HIV status was associated with a decrease in measures of α-diversity (P < .001). However, in stratified analysis, HIV status was associated with decreased α-diversity only in women and in men who have sex with women (MSW) but not in men who have sex with men (MSM). In analyses limited to women and MSW, controlling for HIV status, women displayed increased α-diversity compared with MSW.
Conclusions
Our study suggests that HIV status, sexual risk category, and gender impact gut microbial community α-diversity. Future studies should consider MSM status in gut microbiome analyses.
Keywords: HIV, AIDS, microbiome
Gut microbial α-diversity decreases in HIV+ compared with HIV– women and men who have sex with women (MSW), but not in men who have sex with men. Women have increased α-diversity as compared to MSW.
Potent antiretroviral therapy (ART) has dramatically increased the lifespan of people infected with human immunodeficiency virus (HIV+). Despite receiving effective ART, the average life expectancy of HIV+ individuals remains lower than that of uninfected individuals (HIV–) [1]. This appears to be driven by inflammation-related clinical diseases (eg, cardiovascular disease [CVD], stroke, cancer, long-bone fractures, and renal dysfunction), for which HIV+ patients are disproportionately at risk [2–4]. While mechanisms are incompletely understood, recent studies raise the possibility that gut microbial dysbiosis contributes.
HIV infection promotes a chronic systemic proinflammatory state that is only partially reversed by ART-induced HIV viral load (VL) suppression [2]. Research suggests that, even with HIV virologic control, gut microbiome alterations, combined with decreased intestinal barrier function and increased bacterial translocation from the intestine, drive systemic inflammation, promoting CVD and other chronic complications of HIV disease [5–10]. However, causality remains speculative.
These findings are intriguing. Yet, the field is hampered by lack of consensus on what characterizes the gut microbiota in HIV+ individuals and distinguishes the HIV+ and HIV– gut microbiota. Gut microbial α-diversity is of interest because increased diversity is generally considered a marker of health. In contrast, decreased diversity associates with several disease states (eg, obesity, inflammatory bowel disease, recurrent Clostridioides difficile infection) and predicts mortality in select populations (eg, hematopoietic stem cell recipients) [11–14].
Most studies in the HIV literature have compared α-diversity microbiome measures in HIV+ and HIV– subjects. However, studies have been heterogeneous (in sampled populations, sequencing techniques, and statistical analyses [including the measures of α-diversity examined]), often small in size, and, most importantly, have yielded inconsistent results. While a decrease in α-diversity is often associated with HIV [15–25], no difference [26–29] or even an increase [30] in fecal α-diversity measures in HIV+ individuals is reported. Most studies did not control for sexual preference; only one examined sexual activity [21]. However, recent evidence suggests that status as a man who has sex with men (MSM) impacts gut microbiome measures, perhaps relating to receptive anal intercourse or other behaviors [17, 31–34]. Finally, limited data suggest that CD4 cell count [17, 24], HIV VL [17], and elite controller status [25] impact α-diversity in HIV+ patients. The impact of ART on α-diversity is inconsistent [15, 18–20, 30, 35].
A more definitive understanding of α-diversity measures in HIV+ compared to HIV– individuals may inform future studies examining the relationship between the gut microbiota and long-term complications (such as CVD, stroke, and cancer) in persons with HIV, as well as microbiota-based interventions to improve the health of these patients. Thus, herein, we conducted an individual level meta-analysis to identify differences in α-diversity in HIV+ as compared with HIV– individuals using available 16S ribosomal RNA (rRNA) gene sequence data from published studies through December 2017. We reanalyzed these data using rigorous bioinformatics methods including a novel classification tool to define α-diversity measures and conducted stratified analyses incorporating key metadata such as gender, sexual orientation, and HIV treatment measures.
MATERIALS AND METHODS
Search Strategy and Selection Criteria
This was a systematic review and individual-level meta-analysis. Embase, Medline, and Scopus were searched with keyword and controlled vocabulary terms for HIV and the gastrointestinal microbiome (inception to 31 December 2017). Two independent reviewers (S. A. T., W. L. A. K.) assessed each article; differences were resolved by consensus. Unpublished data, reviews, studies lacking HIV+ participants (exception, below), stool or rectal swab samples, 16S rRNA gene sequencing, and studies with <10 HIV+ patients were excluded. Raw 16S rRNA gene sequence and metadata were downloaded from publicly available databases or obtained from study authors. In a sensitivity analysis, we incorporated stool samples from healthy HIV– individuals (no antibiotics for ≥3 months, immunosuppressants for the last month, and without a C. difficile diagnosis [ever]). Male and female pairs were matched on age and race [36] (see Supplementary Table 1 for search protocol).
Data Analysis
Raw 16S rRNA gene sequence data were preprocessed using one of 6 standardized protocols (Supplementary Table 2). In brief, paired-end 16S rRNA reads from the Illumina platform were merged into consensus sequences using FLASH [37] and filtered for quality and length using Trimmomatic [38] and QIIME [39, 40]. Sequences from the PhiX control genome were identified using BLASTN and removed. Roche/454 raw sequences were error corrected using ACACIA [41]. Passing sequences were trimmed of primers (when present), screened for chimeras using UCLUST (de novo mode) [42], and filtered for human-genome contaminant using Bowtie2 [43]. Chloroplast and mitochondrial contaminants were detected and filtered using the RDP classifier [44]. High-quality 16S rRNA gene sequences were assigned to a high-resolution taxonomic lineage using Resphera Insight [36, 45, 46], which generates a set of operational taxonomic units (OTUs) approximating the species-level composition of each sample. OTU profiles were rarified to an even level of coverage per sample for each study (mean, 9111 [range, 1000–20 000] reads; Supplementary Table 2); we intended to maximize the depth of coverage per sample while minimizing sample loss due to insufficient coverage. Meta-data variables including HIV status, age, race, gender, sexual orientation, ART use, CD4 count, and HIV VL were merged and reconciled between studies.
Four measures of α-diversity were calculated using QIIME. Observed species reports the total species observed and reflects sample richness [47]. Chao 1 also reflects richness, particularly in settings with many low-abundance classes [48]. It may not perform well in settings with low or different sequencing depths.[49]. In contrast, the Shannon index [50] and the inverse Simpson index [51] estimate both richness and evenness (Supplementary Table 3).
Generalized linear mixed models were used to relate raw and log-transformed α-diversity measures to clinical metadata. For all models, a random intercept for each study was included to account for study-specific variations in α-diversity. All P values were corrected for multiple comparisons through false discovery rate (FDR) across the 4 α-diversity measurements. Results in stratified analysis were validated with a nonparametric test (Wilcoxon rank sum). For regression coefficients to be comparable across α-diversity measurements, we divided the regression coefficients by their corresponding pooled standard deviation across all studies to generate stβ, which estimated effect size. To explore study-related heterogeneity, boxplots were generated using the ggplot2 package in R (version 3.3.0), after transformation of the α-diversity values through mean centering to zero (within each study) and scaling to unit variance. We additionally constructed Forest plots (requiring at least 5 patients/categories), using Hedge’s G statistic, and calculated I2 as a measure of heterogeneity for each subanalysis.
Sensitivity analyses removing the largest studies, studies with outlying results, studies conducted in non-European/non-US sites [19, 24, 29]; including HIV– samples from healthy women and men assumed to be heterosexual [36]; and including age in all models were conducted. Stata version 14 and R (version 3.3.0) software were utilized for all analyses. This research was deemed nonhuman subjects research by the Johns Hopkins Institutional Review Board (IRB00133905).
RESULTS
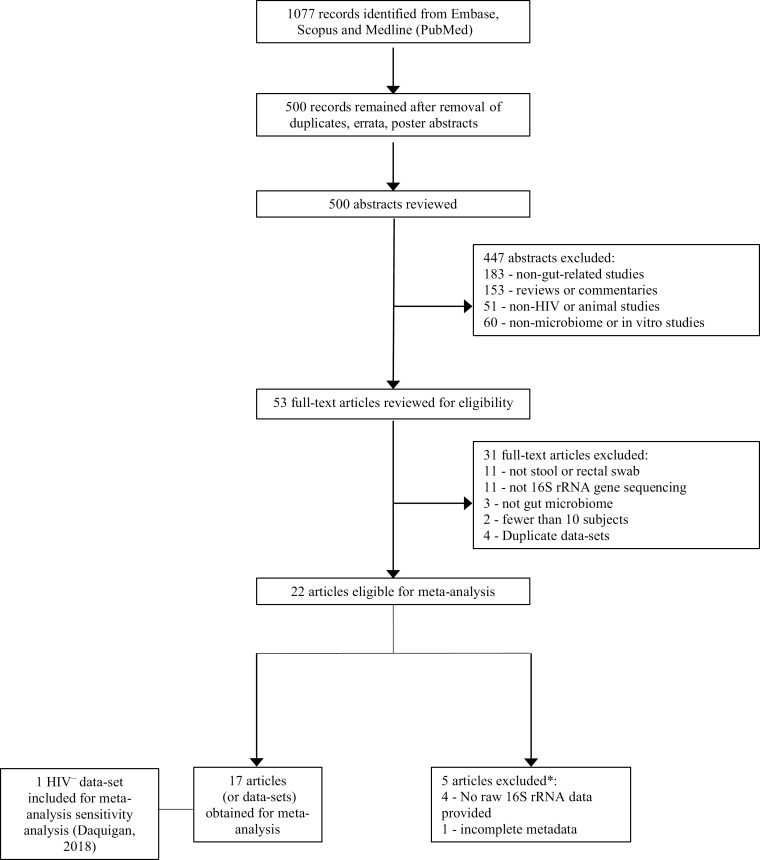
A total of 500 articles were identified after duplicate, erratum, and poster abstract removal (Figure 1). After review, 22 relevant articles were identified [15–22, 24–30, 35, 52–57]. We obtained data from 17 articles [15–21, 24, 25, 28–30, 52–55, 57] representing 17 separate datasets (5 datasets removed due to unavailable or incomplete data [22, 26, 27, 35, 56]) (Table 1 and Supplementary Table 4 present details of studies included and excluded, respectively). Due to low numbers of HIV– individuals (particularly HIV– men who have sex with women [MSW]), we obtained an additional 120 HIV– samples (60 women and 60 men) [36]. These samples were not included in the main analysis; however, they were included in a sensitivity analysis as described above.
Figure 1.
Summary of evidence search and selection. *See Supplementary Table 4. Abbreviations: HIV, human immunodeficiency virus; rRNA, ribosomal RNA.
Table 1.
Characteristics of Studies Included
| Author, Year [Ref] | Country | Study Period | Study Design | Study Setting | Study Population | Sample Type | 16S rRNA Variable Region, Sequencing Platform | Diversity Measuresa | Results: α-Diversity in HIV+ c/w HIV– | Limitations/ Potential Bias |
|---|---|---|---|---|---|---|---|---|---|---|
| Dillon 2014 [52] | US | ND | Cross-sectional study | University of Colorado Anschutz Medical Campus | 18 HIV+, 14 HIV– | Rectal swabs | V4, Illumina MiSeq | ND | ND | Small sample sizeb, lacked MSMc |
| Dinh 2015 [28] | US | ND | Prospective cohort | ID Clinic at Tufts Medical Center, Massachusetts | 21 HIV+, 16 HIV– | Stool samples | V3–V5, Roche 454 | Equitability, OTU, Shannon, Chao 1, PD | No significant differences. | Small sample sizeb, lacked MSMc |
| Dubourg 2016 [15] | France | 2012–2014 | Case-control study | ID Departments in Conception and North Hospitals, Marseille | 31 HIV+, 27 HIV– | Stool samples | V3–V4, Illumina MiSeq | Shannon | Decreased in HIV+ c/w HIV–. | Small sample sizeb, lacked MSMc |
| Lozupone 2013 [30] | US | ND | Prospective cohort | University of Colorado Hospital ID Group Practice | 25 HIV+, 13 HIV– | Rectal swabs | V4, Illumina MiSeq | OTU, Shannon, PD | Increased in HIV+ untreated c/w HIV– (Shannon, PD). | Small sample sizeb, lacked MSMc |
| Monaco 2016 [24] | Uganda | ND | Prospective cohort | Uganda AIDS Rural Treatment Outcomes study at Mbarara Regional Referral Hospital | 82 HIV+, 40 HIV– | Stool samples | V4, Illumina MiSeq | Chao 1, PD | Near significant decrease (Chao 1) and significant decrease (PD) in HIV+ CD4 <200 c/w HIV–. | Lacked MSMc |
| Mutlu 2014 [16] | US | ND | Retrospective cohort | Tissue bank at Rush University Medical Center, Illinois | 21 HIV+, 22 HIV– | Fecal sample from colond | V1–V3, Roche 454 | OTU, Chao 1, PD | Reduced in HIV+ c/w HIV– (OTU, Chao 1, PD) | Small sample sizeb, lacked MSMc |
| Noguera-Julian 2016 [17] | Spain, Sweden | 2014 | Cross-sectional study | HIV clinics at University Hospitals, community-based center. | Barcelona: 129 HIV+, 27 HIV– Stockholm: 77 HIV+, 7 HIV– |
Stool samples | V3–V4, Illumina MiSeq | OTU, Chao 1, ACE, Shannon, Simpson | Reduced in HIV+ c/w HIV–. MSM with increased diversity compared to non-MSM. | |
| Nowak 2015 [18] | Sweden | ND | Prospective cohort | HIV clinic at Karolinska University Hospital | 31 HIV+, 9 HIV– | Stool samples | V3–V4, Illumina MiSeq | OTU, Shannon, Reciprocal Simpson | Reduced in HIV+ c/w to HIV– by all measures. | Small sample sizeb, lacked MSMc |
| Nowak 2017 [29] | Nigeria | ND | Cross-sectional study | Nested in the parent cohort (TRUST/ RV368) | 75 HIV+, 55 HIV– | Rectal swabs | V3–V4, Illumina MiSeq | Shannon | No significant differences. | |
| Perez-Santiago 2013 [53] | US | ND | Prospective study | San Diego Primary Infection Cohort | 13 HIV+ | Rectal swabs | V6, Illumina MiSeq | ND | No HIV– group. | Small sample sizeb, no HIV– |
| Pinto-Cardoso 2017 [19] | Mexico | ND | Cross-sectional study | National Institute of Respiratory Diseases, Mexico City | 33 HIV+, 10 HIV– | Stool samples | V3–V4, Illumina MiSeq | OTU, Chao 1, PD, Shannon | Reduced in HIV+ treated c/w HIV– by all measures. | Small sample sizeb, lacked MSMc |
| Serrano-Villar 2017 [54] | Spain | ND | Prospective study | University Hospitals Clinico San Carlos and Ramón y Cajal | 35 HIV+, 9 HIV– | Stool samples | V1–V3, Roche 454 | ACE, Chao 1, OTU, Shannon | HIV+ immune nonresponders less diverse c/w HIV– by all measures | Small sample sizeb, lacked MSMc |
| Serrano-Villar 2017 [55] | Spain | ND | Cross-sectional study | High-resolution anoscopy clinic | 42 HIV+ | Stool samples | V3–V4, Illumina MiSeq | Chao 1, Shannon | No HIV– group. | No HIV– controlse |
| Vesterbacka 2017 [25] | Sweden | ND | Cross-sectional study | HIV clinic at Karolinska University Hospital | 48 HIV+, 16 HIV– | Stool samples | V3–V4, Illumina MiSeq | OTU, Chao 1, ACE, Shannon, Simpson | Reduced in HIV+ untreated c/w HIV– (Chao 1, ACE, Shannon). | Few MSMc |
| Villanueva-Millan 2017 [20] | Spain | ND | Cross-sectional study | San Pedro’s Hospital, Logrono | 50 HIV+, 21 HIV– | Stool samples | V4, Illumina MiSeq | OTU, Chao1, α-index, Margalef diversity index | Reduced in HIV+ c/w HIV– by all measures. | Lacked MSMc |
| Villar-Garcia 2017 [57] | Spain | Aug 2012– July 2013 | Prospective study | Hospital del Mar Medical Research Institute | 44 HIV+ | Stool samples | V3–V4, Illumina MiSeq | ND | ND | No HIV– controls |
| Yu 2014 [21] | US | 1982–1999 | Prospective study | Outpatient primary care clinics in Washington, DC and New York City | 25 HIV+, 51 HIV– | Rectal swabs | V3–V4, Illumina MiSeq | OTU, Shannon, Chao 1, PD | Reduced in HIV+ c/w HIV– by all measures; attenuated when adjusted for age, race, and smoking. | Small sample sizeb |
| Used only in sensitivity analysis | ||||||||||
| Daquigan 2017 [36] | US | ND | Retrospective study | Multiple studies in US | 120 HIV– | Stool samples | V4 | ND | ND | Lacked MSMc |
Abbreviations: ACE, abundance-based coverage estimator; c/w, compared with; HIV–, human immunodeficiency virus uninfected; HIV+, human immunodeficiency virus infected; ID, infectious diseases; MSM, men who have sex with men; ND, not described; OTU, operational taxonomic unit; PD, phylogenetic diversity; rRNA, ribosomal RNA.
aSee Supplementary Table 3.
bThirty-five or fewer HIV+ patients.
cLacked information on MSM status, did not include MSM controls, or did not control for MSM status in analysis.
dObtained during colonoscopy.
eTwo additional HIV– patients were included in the dataset provided by authors.
A total of 1032 individual samples were available, including samples from 311 HIV– and 721 HIV+ participants, including 114 HIV– and 323 HIV+ MSM (Supplementary Table 5). For all models, analyses were conducted with and without controlling for age; however, age did not significantly affect results (data not shown). All models were also run using log-transformed data (data not shown). Results did not change substantially over models using raw data; therefore, results were reported using raw data for ease of interpretation.
HIV and α-Diversity: All Samples
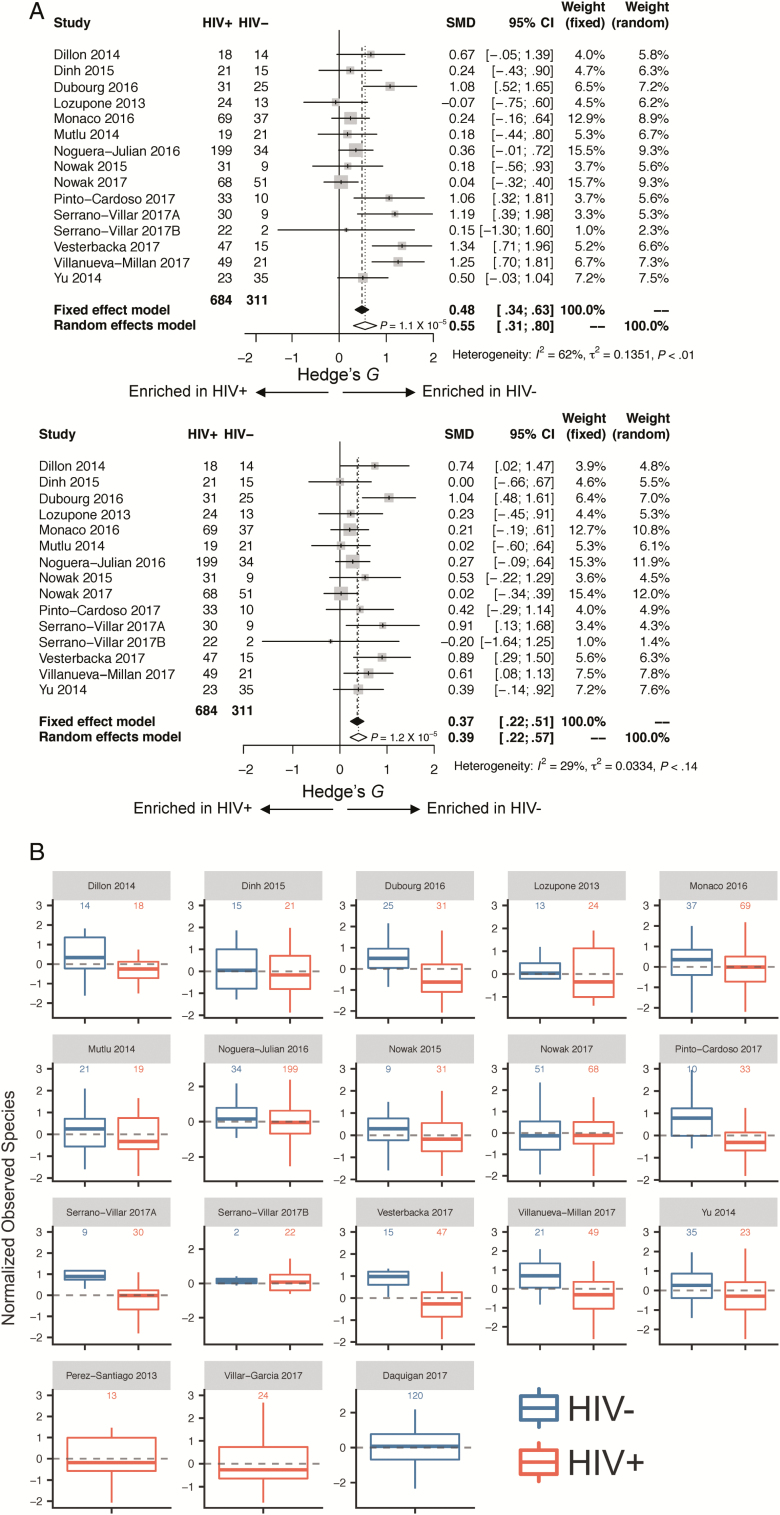
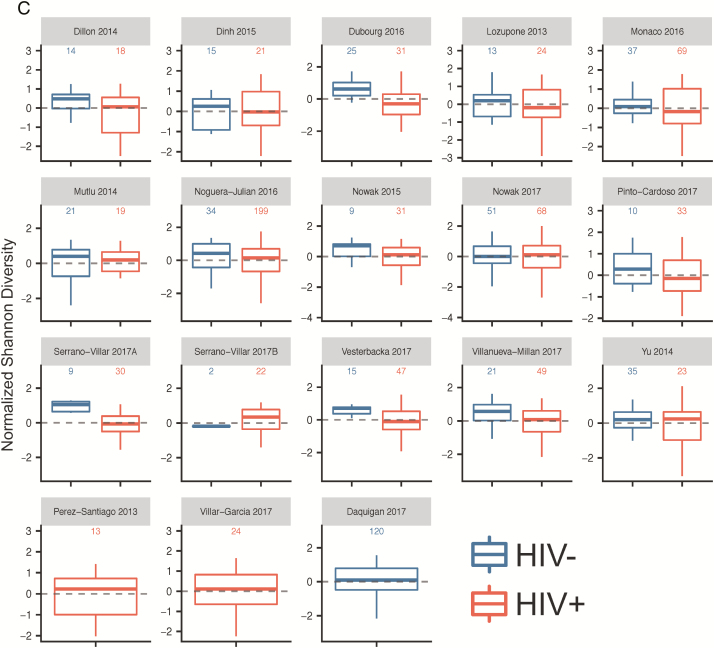
HIV+ patients were older (mean age, 40.9 years) than HIV– patients (mean age, 38.4 years) (P = .002). Before controlling for other factors, HIV status was strongly associated with a decrease in all measures of α-diversity, including observed species (FDR P < .0001, stβ = –0.48) and Chao 1 (FDR P < .0001, stβ = –0.38), Shannon (FDR P < .0001, stβ = –0.38) and inverse Simpson (FDR P = .001, stβ = –0.23) indices (Supplementary Table 6A). Forest plots of the 15 studies including both HIV+ and HIV– samples (Figure 2A; Supplementary Figure 1A) and box plots (Figure 2B and 2C and Supplementary Figure 1B and 1C) show these same trends within most, though not all individual studies. Results were consistent in sensitivity analysis when an additional 120 HIV– samples [36] were added and after removal of the largest study [17], after removal of a study that was observed to be an outlier in mean observed species values [15], and after removal of the 3 non-European/non-US studies [19, 24, 29] (Supplementary Table 6A).
Figure 2.
A, Forest plots utilizing all samples, comparing human immunodeficiency virus infected (HIV+) to human immunodeficiency virus uninfected (HIV–): observed species (above) and Shannon index (below). Associations between gut microbial α-diversity and HIV status. Hedge’s G difference statistic is shown on the x-axis. Fixed effects models (black diamonds) and random effects models (white diamonds) with 95% CI above or below 0 were considered statistically significant. The fixed effects model assumes there exists a single effect size shared by all included studies, while the random effects model allows for variation in the effect size from study to study. Heterogeneity analysis includes estimates of I2 (percentage of variation reflecting true heterogeneity), τ2 (random effects between study variance), and P value from Cochran Q test for heterogeneity. Top panel of A: Based on observed species, gut microbial α-diversity is increased in HIV– as compared with HIV+ patients. There is significant heterogeneity between studies (I2 = 62%, P < .01). Bottom panel of A: Based on Shannon index, gut microbial α-diversity is increased in HIV– as compared with HIV+ patients. Heterogeneity between studies is not statistically significant (I2 = 29%, P = .14). B and C, Boxplots showing α-diversity in terms of observed species (B) and Shannon index (C) by study and HIV status (dark blue = HIV–, red = HIV+). α-Diversity is centered within study and scaled to unit variance. Most studies show decreased α-diversity in HIV+ patients as compared with HIV– patients. Abbreviations: CI, confidence interval; HIV, human immunodeficiency virus; SMD, standardized mean difference.
HIV and α-Diversity: MSM
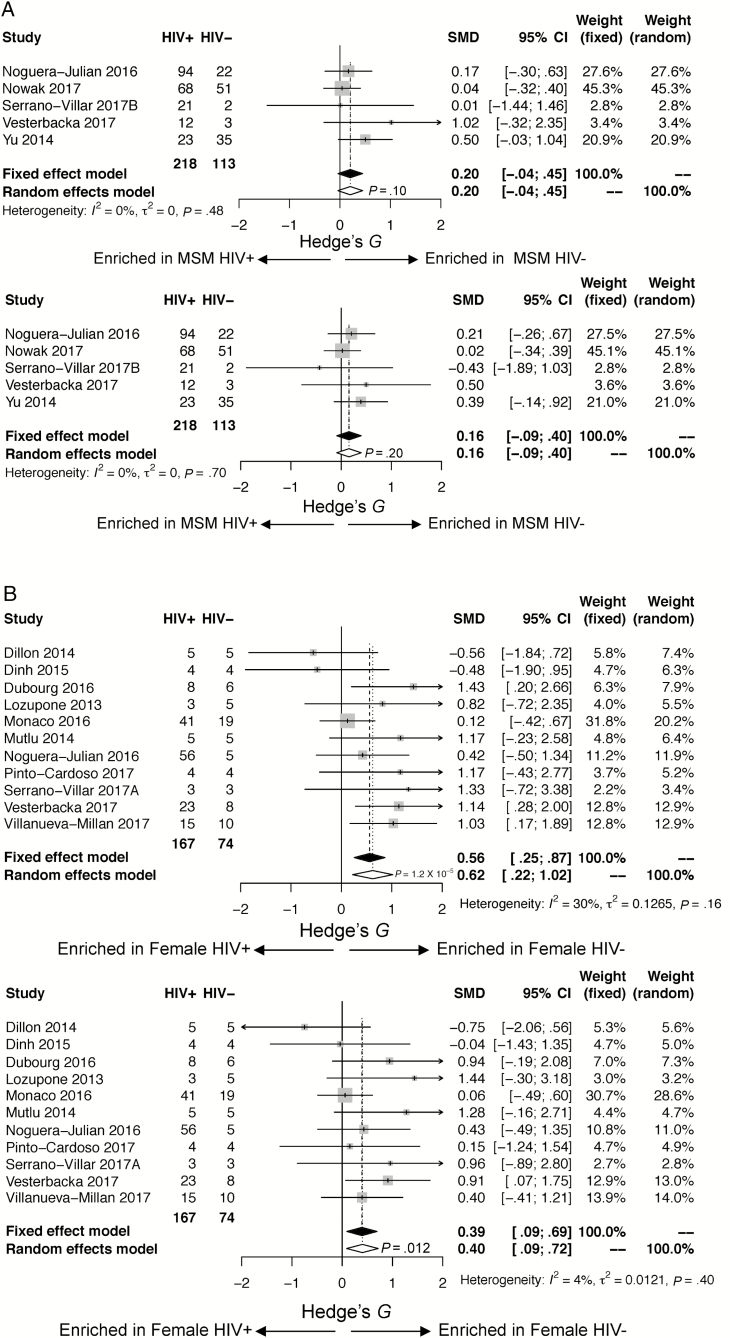
When restricting the analysis to MSM only (n = 323 HIV+ and n = 114 HIV–), there was no significant association between HIV+ status and α-diversity (observed species [FDR P = .377, stβ = –0.18], Chao 1 [FDR P = .548, stβ = –0.10], Shannon [FDR P = .377, stβ = –0.16], inverse Simpson [FDR P = .565, stβ = –0.07]). The results were fairly consistent across studies (see Forest plots in Figure 3A and Supplementary Figure 2 and boxplots in Supplementary Figure 3A–D). Results remained unchanged in sensitivity analysis after removal of the largest study [17] and the non-European/non-US studies [19, 29] (Supplementary Table 6B).
Figure 3.
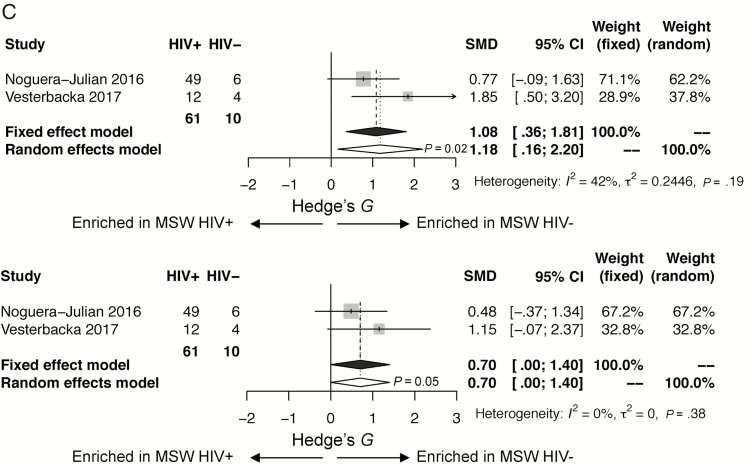
A, Forest plots restricted to men who have sex with men (MSM), comparing human immunodeficiency virus infected (HIV+) to human immunodeficiency virus uninfected (HIV–): observed species (above), Shannon (below). Associations between gut microbial α-diversity and HIV status in stratified analysis restricted to MSM. Hedge’s G difference statistic is shown on the x-axis. Fixed effects models (black diamonds) and random effects models (white diamonds) with 95% confidence interval (CI) above or below 0 were considered statistically significant. The fixed effects model assumes there exists a single effect size shared by all included studies, while the random effects model allows for variation in the effect size from study to study. Heterogeneity analysis includes estimates of I2 (percentage of variation reflecting true heterogeneity), τ2 (random effects between study variance), and P value from Cochran Q test for heterogeneity. Top panel: Based on observed species, gut microbial α-diversity is not statistically significantly different in HIV– compared with HIV+ MSM. There is little heterogeneity between studies (I2 = 0%, P = .48). Bottom panel: Based on Shannon index, gut microbial α-diversity is not statistically significantly different in HIV– compared with HIV+ MSM. There is little heterogeneity between studies (I2 = 0%, P = .70). B, Forest plots restricted to women, comparing HIV+ to HIV–: observed species (above), Shannon (below). Associations between gut microbial α-diversity and HIV status in stratified analysis restricted to women. Hedge’s G difference statistic is shown on the x axis. Fixed effects models (black diamonds) and random effects models (white diamonds) with 95% CI above or below 0 were considered statistically significant. The fixed effects model assumes there exists a single effect size shared by all included studies, while the random effects model allows for variation in the effect size from study to study. Heterogeneity analysis includes estimates of I2 (percentage of variation reflecting true heterogeneity), τ2 (random effects between study variance), and P value from Cochran Q test for heterogeneity. Top panel: Based on observed species, gut microbial α-diversity is increased in HIV– compared with HIV+ women (P <.0001). There is little heterogeneity between studies (I2 = 30%, P = .16). Bottom panel: Based on Shannon index, gut microbial α-diversity is increased in HIV– as compared with HIV+ women (P = .012). There is little heterogeneity between studies (I2 = 29%, P = .40). C, Forest plots restricted to men who have sex with women (MSW), comparing HIV+ to HIV–: observed species (above), Shannon (below). Associations between gut microbial α-diversity and HIV status in stratified analysis restricted to MSW. Hedge’s G difference statistic is shown on the x-axis. Fixed effects models (black diamonds) and random effects models (white diamonds) with 95% CI above or below 0 were considered statistically significant. The fixed effects model assumes there exists a single effect size shared by all included studies, while the random effects model allows for variation in the effect size from study to study. Heterogeneity analysis includes estimates of I2 (percentage of variation reflecting true heterogeneity), τ2 (random effects between study variance), and P value from Cochran Q test for heterogeneity. Of note, there were only 10 HIV– MSW. Top panel: Based on observed species, gut microbial α-diversity is increased in HIV– compared with HIV+ MSW (P = .02). There is little heterogeneity between studies (I2 = 42%, P = .19). Bottom panel: Based on Shannon index, there is a trend toward gut microbial α-diversity being increased in HIV– compared with HIV+ MSW (P = .05). There is little heterogeneity between the 2 studies (I2 = 0%, P = .38).
HIV and α-Diversity: Women
When restricting the analysis to women only (171 HIV+ and 74 HIV–), HIV+ status was significantly associated with a decrease in α-diversity as measured by observed species (FDR P = .0001, stβ = –0.61), Chao 1 (FDR P = .0003, stβ = –0.55), and Shannon (FDR P = .0093, stβ = –0.40), with a trend toward a decreased diversity by inverse Simpson (FDR P = .130, stβ = –0.22). The results were fairly consistent across studies (Figure 3B, Supplementary Figures 3A–D and 4). Results were similar when additional HIV– samples [36] were added. HIV+ status was associated with a decrease in all measures of α-diversity when the largest study [24] and the non-European/non-US studies [19, 24] were removed in sensitivity analysis (Supplementary Table 6C).
HIV and α-Diversity: MSW
When restricting the analysis to MSW (107 HIV+ and 10 HIV–) only, HIV+ status was associated with a statistically significant decrease in observed species (FDR P = .036, stβ = –0.90), and a trend toward a decrease in Chao 1 (FDR P = .073, stβ = –0.66), Shannon (FDR P = .073, stβ = –0.68) and inverse Simpson (FDR P = .073, stβ = –0.61). Very few studies included HIV– MSW (Figure 3C, Supplementary Figures 3A–D and 5). In sensitivity analysis, these results remained consistent when a non-European/non-US study [19] was removed. When the 60 additional (presumed MSW) HIV– men were added to the analysis, HIV+ status was associated with a statistically significant decrease in all measures of diversity (Supplementary Table 6D).
HIV, Gender, and α-Diversity: Women and MSW
When restricting the analysis to MSW and women, HIV+ status was associated with a statistically significant decrease in all measures of diversity (Supplementary Table 6E). When restricting to MSW and women but adjusting for gender and HIV, HIV+ status remained statistically significantly associated with a decrease in all measures of diversity (observed species [FDR P < .0001, stβ = –0.70], Chao 1 [FDR P < .0001, stβ = –0.59], Shannon [FDR P = .0004, stβ = –0.48], inverse Simpson [FDR P = .020, stβ = –0.30]). Controlling for HIV status, heterosexual men had decreased diversity compared with women in terms of the Shannon (FDR P = .0055, stβ = –0.38) and inverse Simpson (FDR P = .005, stβ = –0.40) indices, with a trend toward decreased diversity in Chao 1 (FDR P = .371, stβ = –0.11) and observed species (FDR P = .172, stβ = –0.19). These results remained consistent when additional HIV– samples were added in sensitivity analysis (Supplementary Table 6E).
HIV+ Individuals
Demographic, Clinical Factors, and HIV
Among HIV+ participants, gender and, in men, sexual preference, were available for 601 individuals: 323 MSM, 107 MSW, and 171 women. MSW were older than women, who were older than MSM. There were no significant differences in CD4 count between any of the groups in pairwise comparisons. However, MSM were more likely to have a detectable viral load (>400 copies/mL) than MSW and women. Women were more likely to have a detectable viral load than MSW. There was a trend toward MSM and MSW being more likely to be on ART than women (Table 2).
Table 2.
Summary Patient Characteristics of Included Studies
| Characteristic | MSM | MSW | Women | Men With Unknown MSM Status | Unknown Gender, MSM Status | Statistical Significance (P Value)a | ||
|---|---|---|---|---|---|---|---|---|
| MSM vs MSW | MSM vs Women | MSW vs Women | ||||||
| Total HIV negative | 114 (36.7) | 10 (3.2) | 74 (23.8) | 104 (33.4) | 9 (2.9) | |||
| Age, y, mean (SD) | 33.7 (11.0) | 46.1 (5.7) | 40.1 (9.3)a | 42.1 (13.1)b | Unknownc | <.01 | <.01 | .05 |
| Race | ||||||||
| White | 53 (46.5) | 8 (80.0) | 30 (40.5) | 29 (27.9) | 0 (0.0) | .17 | .19 | .31 |
| Black | 51 (44.7) | 2 (20.0) | 24 (32.4) | 27 (26.0) | 0 (0.0) | |||
| Latino | 4 (3.5) | 0 (0.0) | 6 (8.1) | 10 (9.6) | 0 (0.0) | |||
| Asian | 0 (0.0) | 0 (0.0) | 1 (1.4) | 2 (1.9) | 0 (0.0) | |||
| Other | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |||
| Unknown | 6 (5.3) | 0 (0.0) | 13 (17.6) | 36 (34.6) | 9 (100) | |||
| Total HIV positive | 323 (44.8) | 107 (14.8) | 171 (23.7) | 89 (12.3) | 31 (4.3) | |||
| Age, y, mean (SD) | 37.7 (11.3)d | 48.3 (7.9)e | 41.8 (10.6)a | 40.9 (9.9)f | Unknownc | <.001 | <.001 | <.001 |
| Race | ||||||||
| White | 145 (44.9) | 83 (77.6) | 59 (34.5) | 24 (27.0) | 0 (0.0) | <.001 | <.001 | <.001 |
| Black | 74 (22.9) | 12 (11.2) | 80 (46.8) | 32 (36.0) | 0 (0.0) | |||
| Latino | 51 (15.8) | 8 (7.5) | 11 (6.4) | 3 (3.4) | 0 (0.0) | |||
| Asian | 0 (0.0) | 0 (0.0) | 4 (2.3) | 1 (1.1) | 0 (0.0) | |||
| Other | 1 (0.3) | 1 (0.9) | 2 (1.2) | 1 (1.1) | 0 (0.0) | |||
| Unknown | 52 (16.1) | 3 (2.8) | 15 (8.8) | 28 (31.5) | 31 (100) | |||
| CD4 count, cells/µL | ||||||||
| <200 | 24 (7.4) | 11 (10.3) | 21 (12.3) | 15 (16.9) | 0 (0.0) | .36 | .08 | .61 |
| ≥200 | 297 (92.0) | 96 (89.7) | 150 (87.7) | 74 (83.1) | 0 (0.0) | |||
| Unknown | 2 (0.6) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 31 (100) | |||
| Viral load | ||||||||
| <400 | 157 (48.6) | 71 (66.4) | 91 (53.2) | 46 (51.7) | 0 (0.0) | .001 | .28 | .02 |
| ≥400 | 163 (50.5) | 33 (30.8) | 77 (45.0) | 31 (35.8) | 0 (0.0) | |||
| Unknown | 3 (0.9) | 3 (2.8) | 3 (1.8) | 12 (13.5) | 31 (100) | |||
| ART use | ||||||||
| No | 136 (42.1) | 46 (43.0) | 94 (55.0) | 30 (33.7) | 0 (0.0) | .68 | .04 | .05 |
| Yes | 164 (50.8) | 61 (57.0) | 77 (45.0) | 59 (66.3) | 0 (0.0) | |||
| Unknown | 23 (7.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 31 (100) |
Data are presented as no. (%) unless otherwise indicated. P values represent χ2 analysis. Based on Bonferroni correction, statistical significance set at P <.02.
Abbreviations: ART, antiretroviral therapy; HIV, human immunodeficiency virus; MSM, men who have sex with men; MSW, men who have sex with women; SD, standard deviation.
aMissing age on 5 individuals.
bMissing age on 16 individuals.
cMissing age on all individuals.
dMissing age on 25 individuals.
eMissing age on 1 individual.
fMissing age on 3 individuals.
gExcludes unknown categories.
HIV, Demographic, Clinical Factors, and α-Diversity
Among HIV+ individuals, controlling for MSM status and gender, CD4 count (dichotomized as <200 or >200 cells/µL), VL (dichotomized as <400 or >400 copies/mL), and ART use were not statistically significantly associated with α-diversity of observed species or Chao1. However, controlling for MSM status, HIV+ men had decreased diversity (Shannon and inverse Simpson indices) compared with HIV+ women (Table 3). In sensitivity analysis, removing non-European/non-US studies [19, 24, 29], results were largely similar, with a trend toward decreased Shannon diversity in HIV+ men compared with HIV+ women. Finally, when 2 studies [53, 54] were removed in which modest inconsistencies were noted between the published and provided data in terms of CD4 count and VL, results were unchanged (Supplementary Table 7A). In stratified analyses examining HIV+ MSM only, ART use, CD4 count, and VL were not significantly associated with α-diversity. In a stratified analysis restricted to MSW, ART use was associated with decreased α-diversity as measured by observed species, Shannon, and inverse Simpson (Supplementary Table 7B).
Table 3.
Human Immunodeficiency Virus–infected Patients: Multivariate Model Including Men Who Have Sex With Men Status, Gender, CD4 Cell Count, Viral Load, and Antiretroviral Therapy Use
| Variable | Measures of α-Diversity | |||
|---|---|---|---|---|
| Observed Speciesa | Chao 1a | Shannona | Inverse Simpsona | |
| All samples | ||||
| Menb (Ref: Women) | –0.18 (0.146) | –0.14 (0.233) | –0.34 (0.023) | –0.40 (0.011) |
| MSM (Ref: non-MSMc) | 0.15 (0.362) | 0.15 (0.362) | 0.10 (0.560) | –0.03 (0.800) |
| CD4 count (Ref: <200 cells/µL) | 0.10 (0.594) | 0.09 (0.594) | 0.13 (0.594) | 0.08 (0.594) |
| HIV viral load (Ref: <400 copies/mL) | –0.25 (0.143) | –0.16 (0.182) | –0.23 (0.143) | –0.24 (0.143) |
| ART use (Ref: no ART) | –0.19 (0.282) | –0.16 (0.282) | –0.14 (0.334) | –0.21 (0.282) |
Abbreviations: ART, antiretroviral therapy; HIV, human immunodeficiency virus; MSM, men who have sex with men.
astβ (false discovery rate adjusted P value).
bMen includes MSM and men who have sex with women (MSW).
cNon-MSM includes women and MSW.
DISCUSSION
Herein, we assembled the largest dataset available to date to evaluate α-diversity in the gut microbiome of HIV+ compared within HIV– individuals. Overall, HIV+ status was significantly associated with decreased α-diversity, but only among MSW and women. When controlling for HIV, women had increased diversity as compared to MSW, consistent with recently presented data from another cohort [58]. Among HIV+ individuals, we did not find overall associations with CD4 count, viral load, or ART use, albeit these parameters (along with gender and MSM status) were available in a more limited subset of individuals (79%). Taken together, our results imply that HIV status, gender, and sexual risk category impact α-diversity.
Reports of HIV-associated dysbiosis, including decreases in α-diversity, have raised significant interest in the development of microbiota-based interventions to alter the structure of the gut microbiota and improve the health of patients with HIV. In a recent small study, 6 HIV+ individuals received fecal microbial transplantation (FMT) [59], and at least 2 clinical trials of FMT are planned, with the aim of improving dysbiosis and inflammation. Yet, as our study demonstrates, interactions between gender, MSM status, and HIV affect the microbiome and may significantly impact the outcome of such interventions.
There are significant limitations to our study. First, we were not able to obtain data from 5 studies. However, the majority of these studies were small (Supplementary Table 4). Second, patient metadata were collected using different approaches (some retrospective, some prospective) for each study, and multiple studies did not collect the full set of characteristics of interest across all subjects. We attempted to reconcile these variables, but the heterogeneous ways in which metadata were collected could introduce bias and potential misclassification. We were unable to account for type or duration of ART, both of which could affect α-diversity. Importantly, increased diversity has been seen in gut microbiota of individuals living in agrarian African societies as compared to urban, European controls [60], with diet and antibiotic use further impacting the gut microbial composition [61, 62]; associations between race and the gut microbiota are less robust [63]. Populations in our study came from differing geographic regions, and unmeasured factors, such as diet, smoking, recent or distant antibiotic use, and race could have influenced results. To address this possibility, we conducted sensitivity analyses in which the largest studies and non-European/non-US studies were removed from our analyses, which led to broadly unchanged results.
Finally, cohort design, sample collection techniques, DNA extraction protocols, primer sets, and sequencing platforms differed between studies. We found (see Forest plots) that studies utilizing rectal swabs [21, 29, 30, 52, 53] fell within range of those using stool samples. Additionally, we designed 6 tailored preprocessing protocols that adapted to subtle differences in the underlying raw data with the goal of minimizing bias and maximizing high-quality sequences for downstream analysis. Sequence data from each study that passed preestablished quality metrics were subjected to a taxonomic assignment algorithm that prioritized classification of each individual sequence to avoid potential biases associated with 16S rRNA gene region and clustering based on sequence similarity. Importantly, to normalize within and across studies, we subsampled to an even level of coverage within each study prior to downstream α-diversity calculations and included a random effects term in our statistical models to account for study-to-study variation. However, despite our efforts, it remains possible that differing techniques at every stage (including choice of 16S rRNA gene [V] region; Table 1) could have introduced bias.
In these 17 datasets, only 10 samples from HIV– MSW were available for analysis, which limited power, particularly for stratified analyses restricted to MSW. To address this, we obtained samples from a study including only HIV– men and women [36]. Although information on MSM status was not available, we found that with inclusion of these samples in a sensitivity analysis, trends in Chao1, Shannon, and inverse Simpson indices, suggesting decreased α-diversity in the fecal samples of HIV+ MSW as compared to HIV– MSW, became statistically significant.
Despite these limitations, our findings clarify and extend the reported findings regarding α-diversity in HIV+ vs HIV– individuals. Within the prolific research on the microbiome, studies often report on results with small numbers of patient samples and use heterogeneous analytic techniques that together likely contribute to the conflicting results reported. Thus, one approach applicable, not only within HIV research, but also within the broader microbiome research field, is to try to resolve study differences by compiling and reanalyzing data in a standardized fashion, with the goal of identifying more definitive patterns. Results from our overall HIV+ vs HIV– analysis were broadly consistent with the original study results (Figure 2A and Supplementary Figure 1A); however, compiling data enabled us to conduct stratified analyses, which revealed additional nuances. Our individual level meta-analysis, along with similar prior efforts by Drewes et al [64], illustrates the potential to refine knowledge using this approach and to inform future study design and research questions. Herein, we observed that gender and sexual risk category impact the relationship between HIV status and α-diversity. Future studies should collect and consider these variables in study designs to identify associations between clinical outcomes and gut microbiota features.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Collaborating Group—HIV Microbiome Re-analysis Consortium authors. Stephanie Dillon, Cara Wilson, and Catherine Lozupone (University of Colorado Denver, Aurora); Honorine Ward and Christine Wanke (Tufts University, Boston, Massachusetts); Gregory Dubourg (Institut Hospitalo-Universitaire Méditerranée Infection, Marseille, France); Didier Raoult (Aix-Marseille Universite, Marseille, France); Brent Palmer (University of Colorado Anschutz, Aurora); Cynthia Monaco (University of Rochester Medical Center,); Douglas Kwon (Ragon Institute of MGH, MIT, and Harvard, Cambridge, Massachusetts); Ece Mutlu and Alan Landay (Rush University Medical Center, Chicago, Illinois); Roger Paredes and Marc Noguera-Julian (IrsiCaixa AIDS Research Institute, Barcelona, Spain); Anders Sönnerborg and Ujjwal Neogi (Karolinska Institutet, Stockholm, Sweden); Rebecca Nowak and Jacques Ravel (University of Maryland, Baltimore); Josué Pérez-Santiago (University of Puerto Rico, San Juan), Davey M. Smith (University of California, San Diego, La Jolla); Sandra Pinto-Cardoso and Gustavo Reyes-Terán (National Institute of Respiratory Diseases, Mexico City, Mexico); Sergio Serrano-Villar (Hospital Universitario Ramón y Cajal, Madrid, Spain); Maria Jose Gosalbes Soler (Universidad de Valencia, Spain); Jan Vesterbacka and Piotr Nowak (Karolinska University Hospital, Stockholm, Sweden); P. Pérez-Matute and José A. Oteo (Center for Biomedical Research of La Rioja [CIBIR], Logroño, La Rioja, Spain); Giuseppe D’Auria (Fundación para el Fomento de la Investigacion sanitaria y Biomédica [FISABIO], Valencia, Spain); Judit Villar-García (Hospital del Mar Medical Research Institute, Barcelona, Spain); Guoqin Yu (University of Kansas Medical Center, Kansas City); and James J. Goedert (National Cancer Institute, Bethesda, Maryland).
Author contributions. C. S. conceived of the study design, guided the statistical analysis, and revised the manuscript; S. T. and W. K. conducted the literature review, obtained data, reconciled the metadata, guided the analysis, and wrote the manuscript; J. R. W. compiled the data, processed samples through the bioinformatics pipeline, conducted statistical analysis, and contributed to the manuscript; N. Z. conducted the statistical analysis and contributed to the manuscript; K. G. reviewed and revised the manuscript. Members of the collaborating group contributed sequence and metadata. S. T. and W. K. contributed equally to the manuscript.
Acknowledgments. The authors thank Daquigan et al [36] study for use of their data, and authors for the Sun et al [22] study for their response, although raw 16S rRNA sequences were not available. The 16S rRNA gene reads and Resphera Insight taxonomic assignments utilized for our analyses can be accessed at https://www.synapse.org/#!Synapse:syn18406803. Data from Mutlu et al [16] are not included because Institutional Review Board rules currently limit release.
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health (NIH).
Financial support. This publication resulted from research supported by a Willowcroft Foundation grant (to S. T.) and by the Johns Hopkins University Center for AIDS Research, a NIH-funded program (P30AI094189), which is supported by the following NIH cofunding and participating institutes and centers: National Institute of Allergy and Infectious Diseases, National Cancer Institute, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Heart, Lung, and Blood Institute, National Institute on Drug Abuse, National Institute of Mental Health, National Institute on Aging, Fogarty International Center, National Institute of General Medical Sciences, National Institute of Diabetes and Digestive and Kidney Diseases, and Office of AIDS Research. S. T. and W. K. are supported by the NIH (award numbers K23AI125715 and T32AI052071, respectively).
Potential conflicts of interest. J. R. W. discloses ownership of Resphera Biosciences. C. L. S. has received research funding from Bristol-Myers Squibb for studies on the microbiome and cancer therapy. S. S. V. has received grants and personal fees from Gilead and MSD, and personal fees from Janssen. All other authors report no potential conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Contributor Information
HIV Microbiome Re-analysis Consortium:
Stephanie Dillon, Cara Wilson, Catherine Lozupone, Honorine Ward, Christine Wanke, Gregory Dubourg, Didier Raoult, Brent Palmer, Cynthia Monaco, Douglas Kwon, Ece Mutlu, Alan Landay, Roger Paredes, Marc Noguera-Julian, Anders Sönnerborg, Ujjwal Neogi, Rebecca Nowak, Jacques Ravel, Josué Pérez-Santiago, Davey M Smith, Sandra Pinto-Cardoso, Gustavo Reyes-Terán, Sergio Serrano-Villar, Maria Jose Gosalbes Soler, Jan Vesterbacka, Piotr Nowak, P Pérez-Matute, José A Oteo, Giuseppe D’Auria, Judit Villar-García, Guoqin Yu, and James J Goedert
References
- 1. Egger M, May M, Chêne G, et al. . ART Cohort Collaboration Prognosis of HIV-1-infected patients starting highly active antiretroviral therapy: a collaborative analysis of prospective studies. Lancet 2002; 360:119–29. [DOI] [PubMed] [Google Scholar]
- 2. Gootenberg DB, Paer JM, Luevano JM, Kwon DS. HIV-associated changes in the enteric microbial community: potential role in loss of homeostasis and development of systemic inflammation. Curr Opin Infect Dis 2017; 30:31–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Glesby MJ. Cardiovascular complications of HIV infection. Top Antivir Med 2017; 24:127–31. [PMC free article] [PubMed] [Google Scholar]
- 4. Coghill AE, Pfeiffer RM, Shiels MS, Engels EA. Excess mortality among HIV-infected individuals with cancer in the United States. Cancer Epidemiol Biomarkers Prev 2017; 26:1027–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kelesidis T, Kendall MA, Yang OO, Hodis HN, Currier JS. Biomarkers of microbial translocation and macrophage activation: association with progression of subclinical atherosclerosis in HIV-1 infection. J Infect Dis 2012; 206:1558–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Klatt NR, Funderburg NT, Brenchley JM. Microbial translocation, immune activation, and HIV disease. Trends Microbiol 2013; 21:6–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Manner IW, Baekken M, Kvale D, et al. . Markers of microbial translocation predict hypertension in HIV-infected individuals. HIV Med 2013; 14:354–61. [DOI] [PubMed] [Google Scholar]
- 8. Pedersen KK, Pedersen M, Trøseid M, et al. . Microbial translocation in HIV infection is associated with dyslipidemia, insulin resistance, and risk of myocardial infarction. J Acquir Immune Defic Syndr 2013; 64:425–33. [DOI] [PubMed] [Google Scholar]
- 9. Peters L, Neuhaus J, Duprez D, et al. . INSIGHT SMART Study Group Biomarkers of inflammation, coagulation and microbial translocation in HIV/HCV co-infected patients in the SMART study. J Clin Virol 2014; 60:295–300. [DOI] [PubMed] [Google Scholar]
- 10. Vujkovic-Cvijin I, Dunham RM, Iwai S, et al. . Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci Transl Med 2013; 5:193ra91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chang JY, Antonopoulos DA, Kalra A, et al. . Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis 2008; 197:435–8. [DOI] [PubMed] [Google Scholar]
- 12. Ni J, Wu GD, Albenberg L, Tomov VT. Gut microbiota and IBD: causation or correlation? Nat Rev Gastroenterol Hepatol 2017; 14:573–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Taur Y, Jenq RR, Perales MA, et al. . The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood 2014; 124:1174–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Turnbaugh PJ, Hamady M, Yatsunenko T, et al. . A core gut microbiome in obese and lean twins. Nature 2009; 457:480–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dubourg G, Lagier JC, Hüe S, et al. . Gut microbiota associated with HIV infection is significantly enriched in bacteria tolerant to oxygen. BMJ Open Gastroenterol 2016; 3:e000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mutlu EA, Keshavarzian A, Losurdo J, et al. . A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV infected subjects. PLoS Pathog 2014; 10:e1003829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Noguera-Julian M, Rocafort M, Guillén Y, et al. . Gut microbiota linked to sexual preference and HIV infection. EBioMedicine 2016; 5:135–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nowak P, Troseid M, Avershina E, et al. . Gut microbiota diversity predicts immune status in HIV-1 infection. AIDS 2015; 29:2409–18. [DOI] [PubMed] [Google Scholar]
- 19. Pinto-Cardoso S, Lozupone C, Briceño O, et al. . Fecal bacterial communities in treated HIV infected individuals on two antiretroviral regimens. Sci Rep 2017; 7:43741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Villanueva-Millán MJ, Pérez-Matute P, Recio-Fernández E, Lezana Rosales JM, Oteo JA. Differential effects of antiretrovirals on microbial translocation and gut microbiota composition of HIV-infected patients. J Int AIDS Soc 2017; 20:21526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yu G, Fadrosh D, Ma B, Ravel J, Goedert JJ. Anal microbiota profiles in HIV-positive and HIV-negative MSM. AIDS 2014; 28:753–60. [DOI] [PubMed] [Google Scholar]
- 22. Sun Y, Ma Y, Lin P, et al. . Fecal bacterial microbiome diversity in chronic HIV-infected patients in China. Emerg Microbes Infect 2016; 5:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vázquez-Castellanos JF, Serrano-Villar S, Latorre A, et al. . Altered metabolism of gut microbiota contributes to chronic immune activation in HIV-infected individuals. Mucosal Immunol 2015; 8:760–72. [DOI] [PubMed] [Google Scholar]
- 24. Monaco CL, Gootenberg DB, Zhao G, et al. . Altered virome and bacterial microbiome in human immunodeficiency virus-associated acquired immunodeficiency syndrome. Cell Host Microbe 2016; 19:311–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vesterbacka J, Rivera J, Noyan K, et al. . Richer gut microbiota with distinct metabolic profile in HIV infected elite controllers. Sci Rep 2017; 7:6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Volpe GE, Ward H, Mwamburi M, et al. . Associations of cocaine use and HIV infection with the intestinal microbiota, microbial translocation, and inflammation. J Stud Alcohol Drugs 2014; 75:347–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ling Z, Jin C, Xie T, Cheng Y, Li L, Wu N. Alterations in the fecal microbiota of patients with HIV-1 infection: an observational study in a Chinese population. Sci Rep 2016; 6:30673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dinh DM, Volpe GE, Duffalo C, et al. . Intestinal microbiota, microbial translocation, and systemic inflammation in chronic HIV infection. J Infect Dis 2015; 211:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nowak RG, Bentzen SM, Ravel J, et al. . TRUSTRV368 Study Group Rectal microbiota among HIV-uninfected, untreated HIV, and treated HIV-infected in Nigeria. AIDS 2017; 31:857–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lozupone CA, Li M, Campbell TB, et al. . Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe 2013; 14:329–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kelley CF, Kraft CS, de Man TJ, et al. . The rectal mucosa and condomless receptive anal intercourse in HIV-negative MSM: implications for HIV transmission and prevention. Mucosal Immunol 2017; 10:996–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fulcher JA, Hussain SK, Cook R, et al. . Effects of substance use and sex practices on the intestinal microbiome during HIV-1 infection. J Infect Dis 2018; 218:1560–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Armstrong AJS, Shaffer M, Nusbacher NM, et al. . An exploration of Prevotella-rich microbiomes in HIV and men who have sex with men. Microbiome 2018; 6:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kehrmann J, Menzel J, Saeedghalati M, et al. . HIV-HEART Study Group Gut microbiota in human immunodeficiency virus-infected individuals linked to coronary heart disease. J Infect Dis 2019; 219:497–508. [DOI] [PubMed] [Google Scholar]
- 35. Lozupone CA, Rhodes ME, Neff CP, Fontenot AP, Campbell TB, Palmer BE. HIV-induced alteration in gut microbiota: driving factors, consequences, and effects of antiretroviral therapy. Gut Microbes 2014; 5:562–70. [DOI] [PubMed] [Google Scholar]
- 36. Daquigan N, Seekatz AM, Greathouse KL, Young VB, White JR. High-resolution profiling of the gut microbiome reveals the extent of Clostridium difficile burden. NPJ Biofilms Microbiomes 2017; 3:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011; 27:2957–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kuczynski J, Stombaugh J, Walters WA, Gonzalez A, Caporaso JG, Knight R. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr Protoc Bioinformatics 2011; Chapter 10: Unit 10 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 2010; 26:266–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bragg L, Stone G, Imelfort M, Hugenholtz P, Tyson GW. Fast, accurate error-correction of amplicon pyrosequences using Acacia. Nat Methods 2012; 9:425–6. [DOI] [PubMed] [Google Scholar]
- 42. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011; 27:2194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012; 9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 2007; 73:5261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Drewes JL, White JR, Dejea CM, et al. . High-resolution bacterial 16S rRNA gene profile meta-analysis and biofilm status reveal common colorectal cancer consortia. NPJ Biofilms Microbiomes 2017; 3:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Grim CJ, Daquigan N, Lusk Pfefer TS, Ottesen AR, White JR, Jarvis KG. High-resolution microbiome profiling for detection and tracking of Salmonella enterica. Front Microbiol 2017; 8:1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Whittaker RH. Evolution and measurement of species diversity. Taxon 1972; 21: 213–51. [Google Scholar]
- 48. Chao A. Nonparametric estimation of the number of classes in a population. Scand J Stat 1984; 11: 265–70. [Google Scholar]
- 49. Lemos LN, Fulthorpe RR, Triplett EW, Roesch LF. Rethinking microbial diversity analysis in the high throughput sequencing era. J Microbiol Methods 2011; 86:42–51. [DOI] [PubMed] [Google Scholar]
- 50. Shannon CE. A mathematical theory of communication. Bell System Technical Journal 1948; 27: 379–423. [Google Scholar]
- 51. Simpson EH. Measurement of diversity. Nature 1949; 163:688. [Google Scholar]
- 52. Dillon SM, Lee EJ, Kotter CV, et al. . An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol 2014; 7:983–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pérez-Santiago J, Gianella S, Massanella M, et al. . Gut Lactobacillales are associated with higher CD4 and less microbial translocation during HIV infection. AIDS 2013; 27:1921–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Serrano-Villar S, Vázquez-Castellanos JF, Vallejo A, et al. . The effects of prebiotics on microbial dysbiosis, butyrate production and immunity in HIV-infected subjects. Mucosal Immunol 2017; 10:1279–93. [DOI] [PubMed] [Google Scholar]
- 55. Serrano-Villar S, Vásquez-Domínguez E, Pérez-Molina JA, et al. . HIV, HPV, and microbiota: partners in crime? AIDS 2017; 31:591–4. [DOI] [PubMed] [Google Scholar]
- 56. Stiksrud B, Nowak P, Nwosu FC, et al. . Reduced levels of D-dimer and changes in gut microbiota composition after probiotic intervention in HIV-infected individuals on stable ART. J Acquir Immune Defic Syndr 2015; 70:329–37. [DOI] [PubMed] [Google Scholar]
- 57. Villar-García J, Güerri-Fernández R, Moya A, et al. . Impact of probiotic Saccharomyces boulardii on the gut microbiome composition in HIV-treated patients: a double-blind, randomised, placebo-controlled trial. PLoS One 2017; 12:e0173802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cuesta-Zuluaga JMN, Corrales-Agudelo V, Abad JM, Alvarez R, Durango KM, Escobar JM. Gender differences in the gut microbiota composition and formation of fecal short-chain fatty acids. In: Obesity Week, Washington, DC, 2017. [Google Scholar]
- 59. Vujkovic-Cvijin I, Rutishauser RL, Pao M, et al. . Limited engraftment of donor microbiome via one-time fecal microbial transplantation in treated HIV-infected individuals. Gut Microbes 2017; 8:440–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schnorr SL, Candela M, Rampelli S, et al. . Gut microbiome of the Hadza hunter-gatherers. Nat Commun 2014; 5:3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. David LA, Maurice CF, Carmody RN, et al. . Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014; 505:559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zaura E, Brandt BW, Teixeira de Mattos MJ, et al. . Same exposure but two radically different responses to antibiotics: resilience of the salivary microbiome versus long-term microbial shifts in feces. MBio 2015; 6:e01693–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Walejko JM, Kim S, Goel R, et al. . Gut microbiota and serum metabolite differences in African Americans and white Americans with high blood pressure. Int J Cardiol 2018; 271:336–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Drewes JL, White JR, Dejea CM, et al. . High-resolution bacterial 16S rRNA gene profile meta-analysis and biofilm status reveal common colorectal cancer consortia. NPJ Biofilms Microbiomes 2017; 3:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.