Abstract
Computational models of the heart are increasingly being used in the development of devices, patient diagnosis and therapy guidance. While software techniques have been developed for simulating single hearts, there remain significant challenges in simulating cohorts of virtual hearts from multiple patients. To facilitate the development of new simulation and model analysis techniques by groups without direct access to medical data, image analysis techniques and meshing tools, we have created the first publicly available virtual cohort of twenty-four four-chamber hearts. Our cohort was built from heart failure patients, age 67±14 years. We segmented four-chamber heart geometries from end-diastolic (ED) CT images and generated linear tetrahedral meshes with an average edge length of 1.1±0.2mm. Ventricular fibres were added in the ventricles with a rule-based method with an orientation of -60° and 80° at the epicardium and endocardium, respectively. We additionally refined the meshes to an average edge length of 0.39±0.10mm to show that all given meshes can be resampled to achieve an arbitrary desired resolution. We ran simulations for ventricular electrical activation and free mechanical contraction on all 1.1mm-resolution meshes to ensure that our meshes are suitable for electro-mechanical simulations. Simulations for electrical activation resulted in a total activation time of 149±16ms. Free mechanical contractions gave an average left ventricular (LV) and right ventricular (RV) ejection fraction (EF) of 35±1% and 30±2%, respectively, and a LV and RV stroke volume (SV) of 95±28mL and 65±11mL, respectively. By making the cohort publicly available, we hope to facilitate large cohort computational studies and to promote the development of cardiac computational electro-mechanics for clinical applications.
Introduction
In the last decades, computational models for cardiac electro-mechanics underwent a rapid development. The increased level of detail included in such models have made them more appealing to the clinical community. Significant progress has been made in the methods and software to simulate single hearts. However, simulating cardiac electro-mechanics on large cohorts still remains a challenge.
Previous cohorts of personalised multi-scale models were focused on one or two cardiac chambers. One of the largest computational studies used forty-six biventricular meshes to simulate ventricular electrical activation and mechanical contraction [1]. The results of the personalised models were then analysed to find novel bio-marker candidates to improve disease severity stratification. Single-patient models [2, 3] and smaller cohorts of biventricular meshes [3–10] also proved the potential of computational modelling for predicting and planning patient treatment, and improving our understanding of cardiac physiology and pathophysiology. Small patient cohorts, however, have a limited capability to capture patient anatomical variability. Furthermore, biventricular models have limitations in representing realistic systolic motion, as anatomical cardiac structures surrounding the ventricles are not included [11].
Recently, computational electro-mechanics started moving towards four-chamber models [11–14]. Simulations including the atria as well as the major vessels allow for a more realistic predicted motion, therefore offering an increased predictive power of the model. Furthermore, the interest in the role of atrial dynamics on ventricular function has been growing in the clinical community [14–16]. Four-chamber heart models are, however, still in their infancy, as existing studies focus on one single heart rather than a cohort of individuals [12–14, 17, 18].
As previously discussed, capturing patient anatomical variability is fundamental for computational electro-mechanics models intended for clinical applications. Representing variation in anatomy across a cohort of individuals is, however, a well-known challenge in medical imaging analysis. In the last decade, cardiac atlases were increasingly used to study variability in shape [19–23] and motion [24–28] in healthy and diseased states. As with simulation studies, most of these atlases are focused on one cardiac chamber. In 2013, Hoogendoorn et al. published an atlas of four-chamber heart models based on 138 subjects [29]. The atlas was made publicly available, and constitutes a powerful tool for the study of population variability and for electro-mechanics computational studies, as it can be registered and warped to available medical images to generate a patient-specific geometry. However, the atlas is made of a surface and not a volumetric mesh and represents summary statistics for a population and not specific meshes of individuals. Generating a patient specific mesh from these data requires medical images, meshing tools to move from a surface to a volumetric mesh and labelling of surfaces and regions to impose spatial varying properties and boundary conditions.
In this paper, we build the first publicly available cohort of four-chamber heart models generated from twenty-four heart failure (HF) patients. We develop a semi-automatic pipeline to build three-dimensional tetrahedral meshes of the whole heart from CT images. We test the usability of the meshes for cardiac electro-mechanics simulations by running electro-mechanical simulation tests on all twenty-four meshes. To facilitate the development of new simulations and model analysis techniques by groups without direct access to medical data, image analysis techniques and meshing tools we make the cohort of meshes available for download (DOI 10.5281/zenodo.3890034 [30]).
Materials and methods
We first describe the image acquisition protocol and the data available for each patient. The pipeline we built to generate four-chamber segmentation’s from high-resolution CT images is then presented, followed by the meshing process and ventricular fibres assignment. Finally, we describe the electro-mechanics simulation tests we used to demonstrate that our meshes are suitable for electro-mechanics simulations.
Ethics statement
Data were gathered as part of two clinical trials: clinical trial REC numbers 14/WM/1069 and 18/LO/0752 approved by the West Midlands Coventry & Warwick ethics committee and by the London-Harrow ethics committee, respectively. Data were analysed anonymously.
Clinical data
Our models were based on twenty-four cardiac resynchronization therapy (CRT) patients recruited for CRT upgrade. Demographics of the cohort are shown in Table 1. Patients underwent ECG-gated CT acquired in 10 or 20 frames over an entire cardiac cycle prior to the CRT upgrade procedure. CT images resolution for each patient is shown in the last column of Table 1. For one patient (case 03), only the ED frame was acquired. Baseline conditions were assessed with 12-lead ECG (QRS duration shown in column 4, Table 1) and ejection fraction (EF) measured with 2D echo (Table 1, column 5). For patients 11 and 22, reported EF was measured with CT because EF echo measurements were not available.
Table 1. Patients cohort demographics.
| Age [y] |
Sex [-] |
QRSd [ms] |
EF [%] |
PP [mmHg] |
EDP [mmHg] |
dP/dtmax
[mmHg ⋅ s-1] |
dP/dtmin
[mmHg ⋅ s-1] |
CT resolution [mm×mm×mm] |
|
|---|---|---|---|---|---|---|---|---|---|
| patient01 | 83 | M | 176 | 23 | 119 | 15 | 934 | -1342 | 0.44×0.44×1.0 |
| patient02 | 66 | M | 168 | 26 | 126 | 17 | 905 | -896 | 0.49×0.49×0.4 |
| patient03 | 72 | M | 178 | 36 | 96 | 18 | 899 | -1016 | 0.42×0.42×0.4 |
| patient04 | 49 | M | 152 | 23 | 92 | 21 | 608 | -938 | 0.48×0.48×0.4 |
| patient05 | 62 | M | 134 | 17 | 130 | 25 | 688 | -795 | 0.49×0.49×0.4 |
| patient06 | 85 | M | 152 | 32 | 90 | 9 | 686 | -779 | 0.38×0.38×0.4 |
| patient07 | 37 | M | 186 | 41 | 113 | 22 | 920 | -992 | 0.38×0.38×0.5 |
| patient08 | 79 | M | 200 | 41 | 125 | 21 | 1092 | -1078 | 0.49×0.49×0.4 |
| patient09 | 40 | M | 180 | 42 | 73 | 7 | 592 | -988 | 0.41×0.41×0.4 |
| patient10 | 76 | M | 180 | 41 | 100 | 11 | 1131 | -1339 | 0.44×0.44×0.5 |
| patient11 ** | 76 | M | 144 | 54* | - | - | - | - | 0.42×0.42×0.4 |
| patient12 | 69 | M | 140 | 15 | 100 | 20 | 620 | -702 | 0.38×0.38×0.5 |
| patient13 | 72 | M | 134 | 35 | 110 | 25 | 829 | -865 | 0.45×0.45×0.5 |
| patient14 ** | 61 | M | 180 | 20 | - | - | - | - | 0.43×0.43×0.5 |
| patient15 | 49 | M | 182 | 28 | 94 | 4 | 1201 | -1756 | 0.39×0.39×0.5 |
| patient16 | 55 | M | 134 | 47 | 114 | 8 | 1600 | -1333 | 0.40×0.40×0.5 |
| patient17 | 76 | M | 120 | 40 | 93 | 14 | 942 | -937 | 0.45×0.45×0.5 |
| patient18 | 82 | M | 132 | 31 | 178 | 9 | 1343 | -1478 | 0.32×0.32×0.5 |
| patient19 ** | 68 | M | 131 | 25 | - | - | - | - | 0.37×0.37×0.5 |
| patient20 ** | 71 | M | 145 | 37 | - | - | - | - | 0.37×0.37×0.5 |
| patient21 | 72 | M | 141 | 42 | 112 | 13 | 1144 | -1252 | 0.39×0.39×0.5 |
| patient22 | 53 | M | 161 | 36* | 133 | 11 | 1126 | -1134 | 0.43×0.43×0.5 |
| patient23 | 79 | F | 186 | 40 | 171 | 17 | 1282 | -1638 | 0.33×0.33×0.4 |
| patient24 | 84 | M | 174 | 43 | 70 | -3 | 873 | -999 | 0.43×0.43×0.4 |
| 67±14 | - | 159 ± 23 | 34 ± 10 | 112 ± 28 | 14 ± 7 | 971 ± 270 | -1113 ± 290 |
*EF measured with CT and not 2D echo as echo data were not available
**Invasive pressure recordings not available
LV pressure was recorded invasively during the upgrade procedure. For each patient, we processed baseline pressure measurements (acquired at the beginning of the upgrade procedure, before pacing) by averaging the acquired trace over five to ten heart beats. Ectopic beats were manually identified and discarded. We were therefore able to measure the baseline peak in systolic pressure, ED pressure, dP/dtmax and dP/dtmin. There data are shown in columns 6 to 9 of Table 1. For four patients (cases 11, 14, 19 and 20), pressure data were not available. We note that the end diastolic pressure for case 24 is -3mmHg. This is clearly not the absolute pressure but potentially reflects artefacts due to either respiration or that calibration occurs outside the patient.
Segmentation
We segmented ED CT images with a semi-automatic pipeline shown in Fig 1. An automatic segmentation method was used to generate labels representing the LV blood pool, LV trabeculations and papillary muscles, LV myocardium, and right ventricular (RV), left atrial (LA), right atrial (RA) and aorta blood pools [31]. The resulting segmentation was post-processed with the free software Seg3D [32]. For cases 13 and 21, the automatic segmentation failed to precisely segment the LV myocardium. Therefore, the LV papillary muscles and the LV blood pool were combined and the resulting region dilated until the segmentation visually matched the epicardium of the LV free wall. In all cases the RV, LA, RA and aortic walls could not be automatically segmented. To estimate the RV wall, the RV blood pool was dilated by 3.5mm, based on values for RV wall thickness reported in the literature [33, 34]. The same procedure was applied to the blood pools of the LA, the RA and the aorta to generate labels representing the LA myocardium, the RA myocardium and the wall thickness of the aorta, with a wall thickness of 2mm [35, 36]. This procedure sometimes caused the labels to overlap. For each of the overlapping regions, the region that was best captured by the image was prioritised. The other label was modified by locally dilating it towards the corresponding blood pool. The LV blood pool, the LV myocardium, the aortic blood pool and wall thickness had highest priority as the automatic segmentation tool performed a better segmentation of these labels. Therefore, those regions were never modified. If any of the other labels overlapped with those, it was adjusted to prevent overlap. In case of overlap between the LA and the RA, the LA blood pool and myocardium were prioritised over the RA, as the image contrast of the LA blood pool was better compared to the contrast at the RA blood pool. The LA segmentation was not modified, while the RA myocardium was adjusted to maintain a constant RA wall thickness.
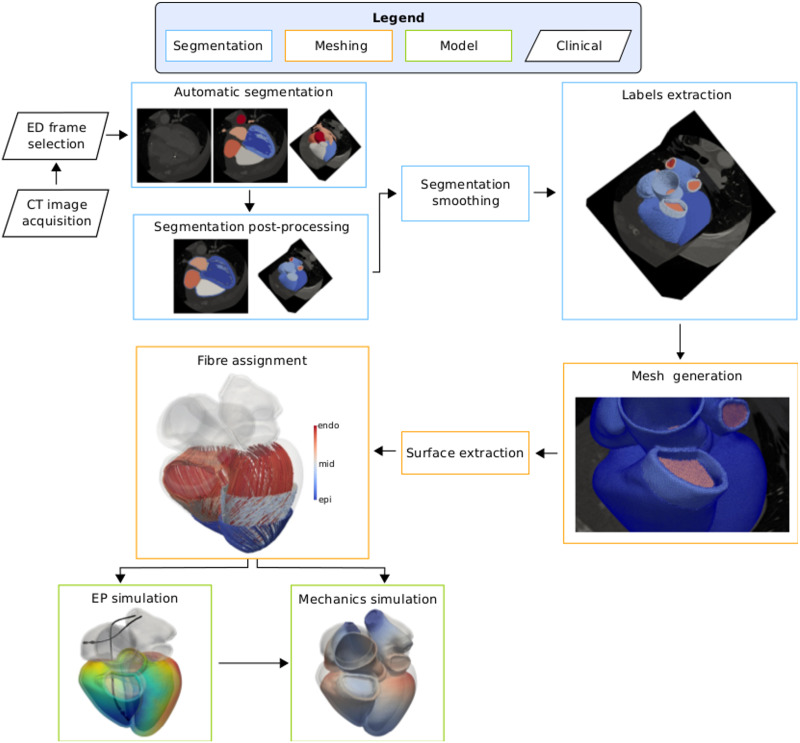
Fig 1. Pipeline.
CT images were acquired in 10 or 20 frames over a whole cardiac cycle. The end-diastolic (ED) geometry was then selected and automatically segmented [31]. The resulting segmentation was post-processed to generate labels representing the RV, LA and RA myocardium, the wall thickness for the aorta and the pulmonary artery, valve planes and rings at the cropped veins. The surface of the segmentation was then smoothed [37] and all the labels but the blood pools and the LV papillary muscles were extracted to generate a tetrahedral mesh. We extracted the endocardial and epicardial surfaces of the atria and ventricles and we assigned fibre orientation to the ventricles with a rule-based method [38]. Finally, we simulated electrical activation and mechanical contraction of the ventricles to test the usability of the meshes.
The pulmonary veins, the superior vena cava and the inferior vena cava were cut from the LA and the RA blood pool perpendicular to the vessel direction. Rings at each of the cut veins were generated to apply boundary conditions in the mechanics simulation. The left atrial appendage (LAA) was only partially covered by the CT image volume for some of the cases. For consistency throughout the cohort, we cropped the LAA from the LA blood pool as we did for the pulmonary veins. Labels representing a small portion of the pulmonary artery blood pool and wall thickness were generated by cutting the RV blood pool and myocardium at the ouflow tract, perpendicular to the vessel direction. The endocardial surface of atria and ventricles were closed with 2mm-thick valve planes at each inflow and outflow [14, 39]. Although this constitutes a simplification of the complex structure of the four cardiac valves, anatomical details of the valve leaflets and of the chordae tendinae of the mitral and tricuspid valves were not visible in the CT images. Therefore, we could not render such structures in the final segmentation.
In order to reduce the staircase effect due to image artifacts or small manual corrections, the resulting four-chamber segmentation was up-sampled and smoothed with an isotropic resolution of 150μm with a variational approach [37]. Prior to the mesh generation step, labels for the LV, RV, LA, RA myocardium, wall thickness of the aorta and the pulmonary artery, all the valve planes and all the rings generated at the cut veins were extracted. We show the segmentation for the twenty-four cases in S1–S6 Figs in the Supporting Information.
Although LV and RV myocardium show intricate trabeculation, we did not include these structures in the segmentation we used to generate the final meshes. Some LV trabeculations and the papillary muscles were grossly segmented by the automatic segmentation tool, but the resolution of the CT images was not high enough to capture them completely. RV trabeculations were not visible in the CT images. Therefore, we excluded these structures from the segmentation and we meshed only the smooth myocardial wall.
Mesh generation
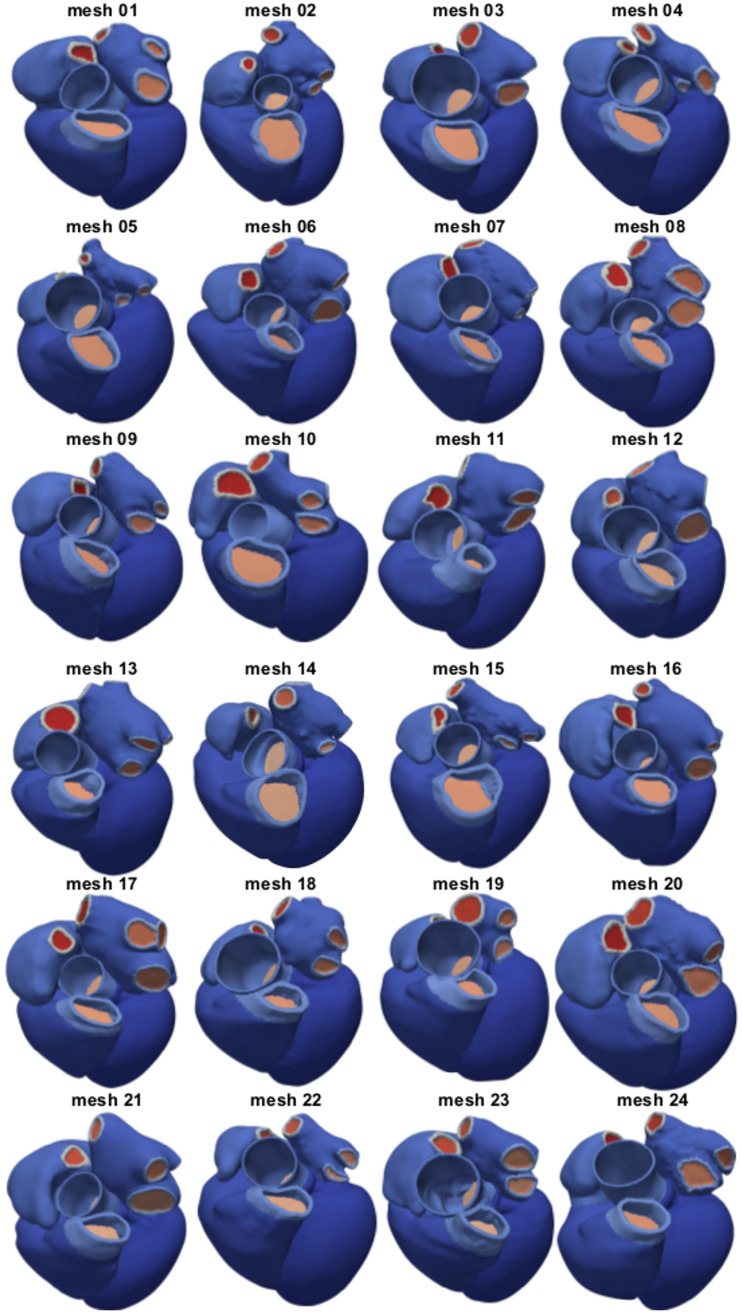
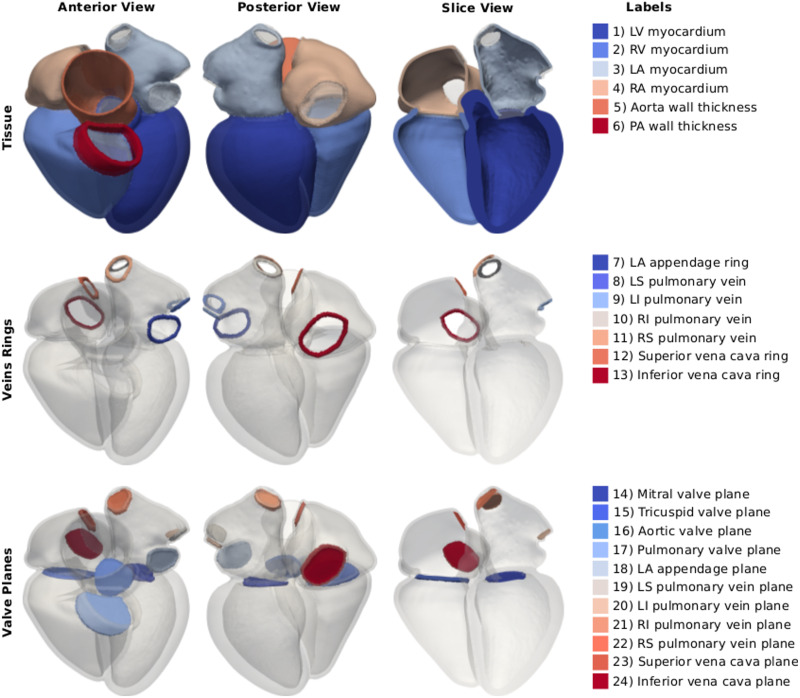
We used the multi-label segmentation to generate tetrahedral meshes with a target resolution of 1mm using CGAL (Computational Geometry Algorithm Library). Fig 2 shows the resulting twenty-four four-chamber heart meshes. Label numbering is shown in Fig 3. The first four labels represent the myocardium of the LV, RV, LA and RA, respectively. Labels five and six correspond to the wall of the aorta and the pulmonary artery. Labels from seven to eleven represent the rings of the cropped LAA and of the four pulmonary veins. Labels twelve and thirteen are the rings at the cropped superior and inferior vena cava. The mitral, tricuspid, aortic and pulmonary valve planes correspond to labels from fourteen to seventeen, while labels from eighteen to twenty-four represent planes for the cut veins, in the same order of the rings. Mesh 12 and 14 had only three pulmonary veins, as the branching between left superior and inferior pulmonary veins occurred further away from the LA and was therefore not covered by the CT image volume. The resulting meshes therefore have only twenty-two labels instead of twenty-four.
Fig 2. Twenty-four four-chamber heart meshes.
The images show an anterior view of the twenty-four meshes generated with the pipeline described in the text.
Fig 3. Labels description.
The images on the left and in the center show an anterior and a posterior view of one of the four-chamber meshes. The image on the right shows an anterior view of a clip of the geometry. On the right, the twenty-four labels of the mesh are listed. The first row shows the myocardium of the LV, RV, LA, RA and the wall of the cropped aorta and pulmonary artery (PA). The second row shows the rings at the cropped veins and at the LAA. The third row shows the valve planes added at all inlets and outlets of the LV, RV, LA and RA.
Fibre and sheet directions in the ventricles were assigned using a rule-based method [38]. The base of the ventricles was defined as the common surface nodes between the ventricles and the mitral and the tricuspid valve planes. The endocardium and the epicardium of the ventricles were extracted as the surface nodes separated by the base. Ventricular endocardium and epicardium were assigned with a fibre orientation of 80° and -60°, respectively [14, 39]. For the sheet direction, we used -65° at the endocardium and 25° at the epicardium [38]. Although myocardial microstructure was reported to change in heart failure [40], this was not accounted for by the used rule-based method. The fibre and sheet directions represent those of a healthy human heart.
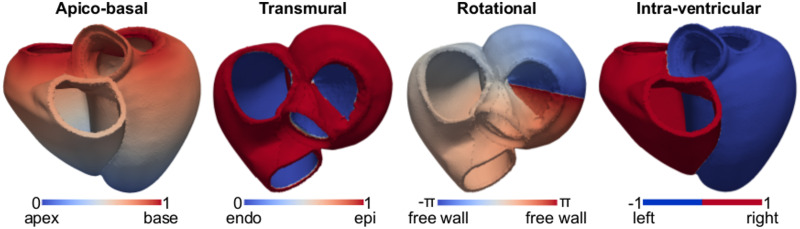
To have a common frame of reference for the twenty-four meshes, we applied a universal ventricular coordinate (UVC) algorithm to all the geometries [41]. This resulted in a system of four coordinates for the ventricles of each mesh, shown in Fig 4: 1) apico-basal coordinate ranging from 0 at the LV apex to 1 at the base; 2) rotational coordinate rotating around the LV, ranging from −π at the LV free wall, 0 at the septum and π back to the LV free wall; 3) a transmural coordinate, ranging from 0 at the endocardium to 1 at the epicardium; 4) an intraventricular coordinate, defined at -1 at the LV +1 at the RV. We assigned ventricular nodes of the meshes with a value for each of these four coordinates. Non-ventricular nodes were assigned with a value of -100.
Fig 4. Universal ventricular coordinates.
From left to right: apico-basal coordinate, ranging between 0 at the apex and 1 at the base (anterior view); transmural coordinate, varying from 0 at the endocardium of the LV and of the RV free wall to 1 at the epicardium (top view); rotational coordinate, ranging between −π at the LV free wall, to 0 at the septum and π back to the LV free wall; intra-ventricular coordinate, defined at -1 at the LV and +1 at the RV.
Mesh refinement
Although an average 1mm edge length is enough to solve for electrical activation times with simplified models (e.g. eikonal), a much finer resolution is needed for more complex models (e.g. monodomain or bidomain). To show that it is possible to refine the given meshes, we resampled all twenty-four original 1mm-resolution meshes to achieve a 0.35mm resolution using meshtool (https://bitbucket.org/aneic/meshtool/src/master/). These high resolution meshes will also be publicly available as part of the database, inclusive of element tags as described above (Fig 3) and ventricular fibres. If the user desires an even higher resolution, the finer meshes provided in the database can be further refined using meshtool.
Database format
We provide a zipped folder for each mesh on zenodo (DOI 10.5281/zenodo.3890034 [30]). Each folder contains the coarse and the finer versions of the same mesh. All twenty-four 1mm-meshes are supplied in case format, readable with paraview (https://www.paraview.org/). All binary files containing the meshes data (ens and geo formats) are provided within the zipped folder. Points coordinates are given in mm. Element tags are assigned to the elements of the mesh (see Fig 3), as well as fibres and sheet directions. Fibres and sheet directions are assigned to the ventricles according to the rule-based method explained above. Non-ventricular elements are assigned with default vectors [1, 0, 0] and [0, 1, 0]. UVCs are assigned to the nodes of the meshes. Non-ventricular nodes are assigned to value of -100 for all four UVCs. We also provide the location of the CRT right-ventricular electrode used to initiate ventricular excitation. This is given as a label on the nodes called electrode_endo_rv, which is 1 at the stimulated nodes. Finer meshes are provided in vtk format, also readable in paraview. For these meshes, we provide element tags, fibres and sheet directions on the ventricles, all in the same file.
Electro-mechanics simulation framework
To ensure that that our mehes are suitable for electro-mechanics simulations, we ran electro-mechanics simulation tests on all twenty-four 1mm-resolution meshes using linear tetrahedral elements. We simulated ventricular electrical activation and free mechanical contraction.
Ventricular electrical activation was simulated with a reaction-eikonal model [42]. Ventricular myocardium was assigned with a longitudinal and transverse conduction velocity of 0.6m s−1 and 0.24 s−1, respectively, consistently with experimental measurements on human ventricular tissue [43]. To simulate early activation of ventricular endocardium, we added a one-element thick endocardial layer to both ventricles and we increased its conduction velocity by two fold [6, 44]. The RV endocardium was stimulated based on the location of the RV lead, segmented by thresholding the ED CT image. The radius of the stimulus site was set to 5mm to approximate the area covered by the tip of the CRT lead. We simulated only one beat for each heart. The RV endocardium was therefore stimulated only once at 0.0ms with a stimulus duration of 2.0ms. To simulate left bundle branch block activation typical of patients with dyssynchrony, the LV was not stimulated [45]. We simulated transmembrane potential changes due to electrical activation using a reaction-eikonal model with diffusion [42]. The activation time computed by the eikonal model triggers the upstroke in transmembrane potential Vm, shown in orange in Fig 5A. As the transmembrane potential overcomes a threshold set at -60mV, the rise in active tension is triggered. Ventricular action potential was determined using the Ten-Tusscher model [46]. As our main interest was in activation time and not reporalisation we used a homogeneous epicardial cell type across the myocardium.
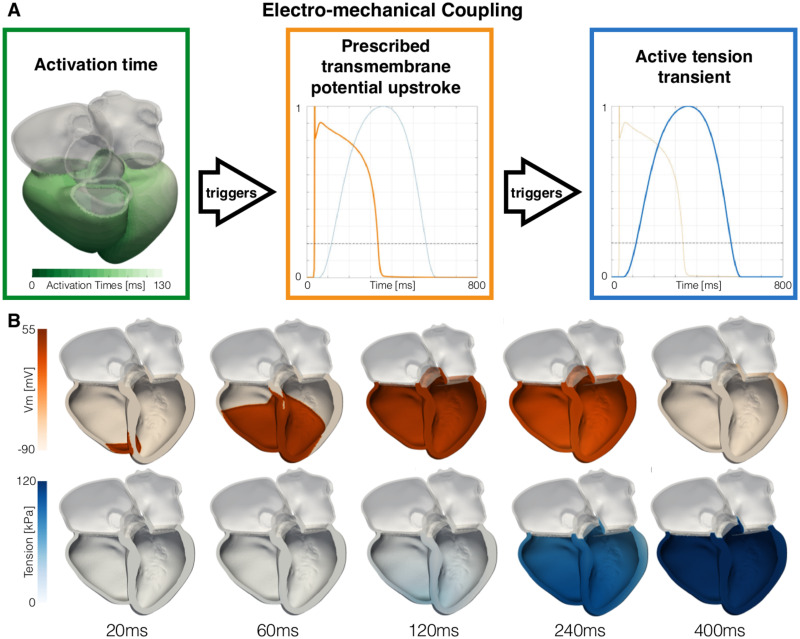
Fig 5. Electro-mechanical coupling.
A The activation times computed by the eikonal model (shown on mesh 01 in the green box) trigger an upstroke in transmembrane potential Vm at each node of the mesh (orange curve in the orange box). When the transmembrane potential overcomes -60mV (black dashed line), the rise in active tension is triggered (blue curve in the blue box). B We show the changes in transmembrane potential (top row) and the active tension (bottom row) over time on mesh 01.
Large deformations of the heart throughout the cardiac cycle were modelled with the finite elasticity equations solved in a Lagrangian reference frame [47]. Cardiac tissue was modelled as a hyperelastic and nearly incompressible material. For ventricular passive mechanics we used the transversely isotropic Guccione law [48]:
where Ψ is the strain energy function and C, bf, bfs and bss are the material parameters. The f, s and n in the Cauchy-Green strain tensor E represent the strain in the fibres, sheet and normal to sheet directions, while J indicates the Jacobian determinant of the deformation gradient tensor. Parameters for ventricular stiffness were set based on parameters estimated in HF patients [49] and are shown in Table 2. Passive mechanics of non-ventricular tissue was modelled with a neo-Hookean model to represent non-linear response of the material to external load:
where c is the only material parameter and I1 denotes the first invariant of the Cauchy-Green strain tensor. For atrial myocardium, wall of the aorta, pulmonary artery and veins, we ignored the effect of the fibres, and we set passive stiffness parameters using the following values from the literature. For the atria, we used the average of the parameter values of the anterior and posterior regions reported in the literature [50], resulting in c = 7.45kPa. For the aorta and the pulmonary artery, we imposed c = 26.66kPa and c = 3.7kPa, the average parameter value of the two cases reported in previous works, respectively [51, 52]. Rings at the cropped pulmonary veins, superior vena cava, inferior vena cava and LAA were assigned with the same parameter value used for the atria. To restrict their deformation, the valve planes were assigned with a parameter c = 1000.0kPa. Tissue incompressibility was enforced with a penalty method [53, 54], with a bulk modulus κ = 1MPa.
Table 2. Passive and active mechanics material parameters.
| Passive Material Parameters | |||||
| C | bf | bfs | bt | κ | |
| kPa | - | - | - | MPa | |
| Ventricles | 3.0 | 19.25 | 8.75 | 7.0 | 1.00 |
| c | k | ||||
| kPa | MPa | ||||
| Atria | 7.45 | 1.00 | |||
| Aorta | 26.66 | 1.00 | |||
| Pulmonary artery | 3.7 | 1.00 | |||
| Valve planes | 1000.0 | 1.00 | |||
| Cropped veins & LAA | 7.45 | 1.00 | |||
| Active Tension Parameters | |||||
| Tpeak | temd | tdur | τr | τd | |
| kPa | ms | ms | ms | ms | |
| Ventricles | 125 | 20 | 550 | 130 | 100 |
Active tension transient of ventricular myocardium was simulated with a phenomenological model [2, 55], discarding the effect of length-dependence. Active tension Ta was computed as follows:
where Tpeak is the peak in active tension, t is time and tact is the activation time computed with the eikonal model. Electro-mechanical delay temd, duration of the twitch tdur, rising time τr and decay time τd were set to 20ms, 550ms, 130ms and 100ms, respectively. Tpeak was set to 125.0kPa to achieve an LV EF of about 35% for all cases. This falls within the range of EF measured in our cohort (34±10%). The active tension transient is shown in Fig 5A (blue curve). We assumed that active contraction occurs only in the fibre direction. Therefore, the active stress computed by the cellular model was added to the passive stress in the fiber direction only [47]. Including transverse active stress was reported to improve simulated strains [56], but was shown to have minimal effects on simulated motion [11]. However, the aim of active contraction test was not to match physiological strains but to show the suitability of the meshes for electro-mechanics simulations. All parameters used for passive and active mechanics are shown Table 2.
We wanted to verify that our meshes element quality was high enough to endure a more significant contraction, with a left ventricular ejection fraction falling within healthy ranges (48%-69%) [57]. Since the meshes of our cohort were generated from HF patients, some ventricles are very dilated with LV EDV sometimes exceeding 400mL. For dilated meshes with left ventricular end-diastolic volume above normal values (>232mL [57]), a 60% ejection fraction would require abnormally large stroke volume between 161mL and 262mL, while normal values range between 59mL and 132mL [57]. For only twelve of our meshes, LV EDV falls within normal and border zone values between 109mL and 232mL, from the UK Biobank database of 804 cases [57]. For these meshes, we ran additional simulations with decreased passive ventricular stiffness to healthy values available in the literature [49]: C = 1.7kPa, bf = 8.0, bfs = 4.0 and bt = 3.0. Parameters for electrical activation and active contraction were left unchanged.
Boundary conditions
The mechanics simulation was constrained by applying omni-directional springs at the right superior and inferior pulmonary veins, and at the superior vena cava [14, 18], with a spring stiffness of 10kPa mm−1. The effect of the pericardium on the ventricles was simulated by applying unidirectional springs to the epicardium to penalise displacement normal to the surface, using the ED state as the reference configuration for the springs [18]. We used a map to scale the penalty for the displacement of ventricular epicardium normal to the surface. The map was based on the average image-derived displacement for all patients. Details about how the penalty map was computed are provided in S2 Appendix.
We did not account for ventricular preload or ventricular afterload. The initial configuration was assumed to be ventricular end-diastole, and we did not perform any unloading of the mesh. The ventricles were assumed to contract against zero pressure and zero resistance. Therefore, we did not include any representation of the aortic and pulmonary valves. The atria were passive, with no electrical excitation and active mechanical contraction. Our model did not include any representation of flow across the atrio-ventricular valves.
Numerical methods
All the electro-mechanical simulations were run with the Cardiac Arrhythmia Research Package (CARP) [12, 58, 59]. The eikonal equation was solved with the fast iterative algorithm [41, 60]. The reaction-eikonal model used to compute transmembrane potential distribution has been described previously [42].
For details of the finite element discretisation of the mechanics equations, we refer to Augustin et al. [61]. The mechanics solver we used was validated with benchmark tests [62].
Results
First, we present results for the general characteristics of the twenty-four meshes. Then, we present results for the electro-mechanics simulations. Finally, we investigate the relationship between LV and RV anatomy and simulation outputs.
Mesh characteristics
To test mesh quality we computed the scaled Jacobian for each element of each mesh to have a measure of the distortion of the tetrahedra [63, 64]. A scaled Jacobian close to 1 indicates a perfectly regular tetrahedron, while a scaled Jacobian close to 0 indicates a highly distorted tetrahedron. Table 3 shows the results for mesh quality for the twenty-four meshes, together with the number of nodes, number of elements and average edge length. We show results for both the coarse and the high resolution meshes.
Table 3. Meshes characteristics.
The table shows the number of nodes, number of elements, average element quality (measured as the scaled Jacobian) and edge length for the twenty-four meshes. For the element quality and the edge length, we also reported the standard deviation within each mesh in brackets.
| Coarse meshes | High-resolution meshes | |||||||
|---|---|---|---|---|---|---|---|---|
| # nodes | # elements | tet quality | edge length [mm] | # nodes | # elements | tet quality | edge length [mm] | |
| mesh01 | 481066 | 2349414 | 0.74 (0.11) | 1.06 (0.15) | 9745931 | 48831236 | 0.51 (0.13) | 0.39 (0.10) |
| mesh02 | 691916 | 3490090 | 0.74 (0.11) | 1.06 (0.15) | 14351113 | 72647039 | 0.51 (0.13) | 0.39 (0.10) |
| mesh03 | 527572 | 2636456 | 0.74 (0.12) | 1.07 (0.15) | 10887723 | 54949789 | 0.51 (0.13) | 0.39 (0.10) |
| mesh04 | 563509 | 2873599 | 0.74 (0.11) | 1.06 (0.15) | 11772640 | 59745743 | 0.52 (0.13) | 0.39 (0.10) |
| mesh05 | 637354 | 3266530 | 0.74 (0.11) | 1.06 (0.15) | 13356834 | 67888242 | 0.52 (0.13) | 0.39 (0.10) |
| mesh06 | 539616 | 2656723 | 0.73 (0.11) | 1.06 (0.15) | 10994447 | 55305234 | 0.51 (0.13) | 0.39 (0.10) |
| mesh07 | 529708 | 2598475 | 0.73 (0.11) | 1.06 (0.15) | 10777223 | 54166600 | 0.51 (0.13) | 0.39 (0.10) |
| mesh08 | 502241 | 2489060 | 0.74 (0.11) | 1.06 (0.15) | 10286788 | 51801290 | 0.51 (0.13) | 0.39 (0.10) |
| mesh09 | 451528 | 2227614 | 0.74 (0.11) | 1.06 (0.15) | 9203067 | 46286710 | 0.51 (0.13) | 0.39 (0.10) |
| mesh10 | 445112 | 2176295 | 0.74 (0.11) | 1.06 (0.15) | 9014841 | 45230037 | 0.51 (0.13) | 0.39 (0.10) |
| mesh11 | 440291 | 2169085 | 0.74 (0.11) | 1.06 (0.15) | 8963833 | 45070335 | 0.51 (0.13) | 0.39 (0.10) |
| mesh12 | 536011 | 2635144 | 0.73 (0.11) | 1.06 (0.15) | 10909862 | 54838853 | 0.51 (0.13) | 0.39 (0.10) |
| mesh13 | 562910 | 2788537 | 0.74 (0.11) | 1.07 (0.15) | 11546318 | 58139068 | 0.51 (0.13) | 0.39 (0.10) |
| mesh14 | 674011 | 3423028 | 0.74 (0.11) | 1.06 (0.15) | 14028329 | 71143503 | 0.52 (0.13) | 0.39 (0.10) |
| mesh15 | 573966 | 2866740 | 0.74 (0.11) | 1.06 (0.15) | 11794981 | 59534694 | 0.51 (0.13) | 0.39 (0.10) |
| mesh16 | 490365 | 2360470 | 0.73 (0.11) | 1.07 (0.15) | 9862653 | 49283398 | 0.51 (0.13) | 0.39 (0.10) |
| mesh17 | 572656 | 2807390 | 0.73 (0.11) | 1.07 (0.15) | 11665496 | 58599804 | 0.51 (0.13) | 0.39 (0.10) |
| mesh18 | 438898 | 2192445 | 0.74 (0.11) | 1.07 (0.15) | 9051204 | 45692821 | 0.51 (0.13) | 0.39 (0.10) |
| mesh19 | 536860 | 2663864 | 0.74 (0.11) | 1.06 (0.15) | 10991686 | 55387888 | 0.51 (0.13) | 0.39 (0.10) |
| mesh20 | 556012 | 2712110 | 0.73 (0.11) | 1.06 (0.15) | 11252728 | 56409760 | 0.51 (0.13) | 0.39 (0.10) |
| mesh21 | 511855 | 2544717 | 0.74 (0.11) | 1.07 (0.15) | 10522190 | 53041672 | 0.51 (0.13) | 0.39 (0.10) |
| mesh22 | 537441 | 2679368 | 0.74 (0.11) | 1.06 (0.15) | 11035397 | 55662824 | 0.51 (0.13) | 0.39 (0.10) |
| mesh23 | 407578 | 1991085 | 0.73 (0.11) | 1.06 (0.15) | 8266657 | 41491489 | 0.51 (0.13) | 0.39 (0.10) |
| mesh24 | 462282 | 2257529 | 0.73 (0.11) | 1.06 (0.15) | 9383315 | 47092793 | 0.51 (0.13) | 0.39 (0.10) |
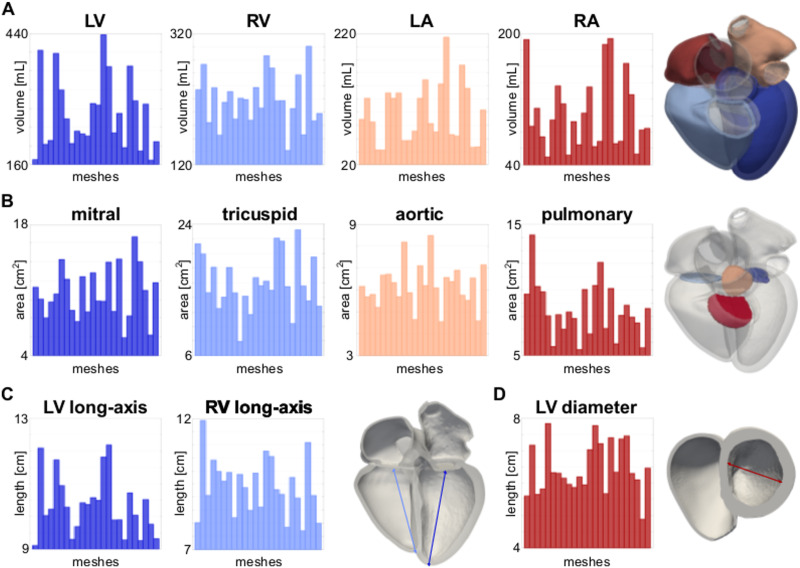
We quantified anatomical differences within the cohort. Fig 6A shows the volume of the LV, RV, LA and RA for the twenty-four meshes. Fig 6B shows the area of the mitral, tricuspid, aortic and pulmonary valve planes. Fig 6C shows the LV and RV long-axis length. The LV long-axis length was computed as the distance between the LV most apical point on the epicardium and the center of mass of the mitral valve plane. The RV long-axis was computed by projecting the LV most apical point onto the RV-LV junction intersected with the RV epicardium and by computing its distance from the center of mass of the tricuspid valve plane.
Fig 6. Anatomical differences.
A LV, RV, LA and RA volumes. B Area of the mitral, tricuspid, aortic and pulmonary valve planes. C LV and RV long-axis lengths. D LV diameter. All quantities are shown for the twenty-four meshes.
Fig 6D shows the LV diameter, computed using the UVC. We defined a plane below the aortic outflow tract by thresholding the apico-basal (or longitudinal) coordinate between 0.78 and 0.82. We chose these values as this approximates the cutting plane below the LV and RV outflow tracts for all the meshes. Then, we took two points on the LV endocardium representing the opposite ends of the chamber. The point on the LV free wall was taken as the point with the rotational coordinate closest to π in UVCs. The point on the side of the septum was taken as the point with the rotational coordinate closest to 0 in UVCs. The LV diameter was computed as the distance between these two points. In addition, we show the variability of the LV wall thickness, and local curvature of the endocardial surface of the LV, RV, LA and RV, which are important quantities for the Laplace law and wall stress estimation in S1 Appendix.
Electro-mechanics simulations
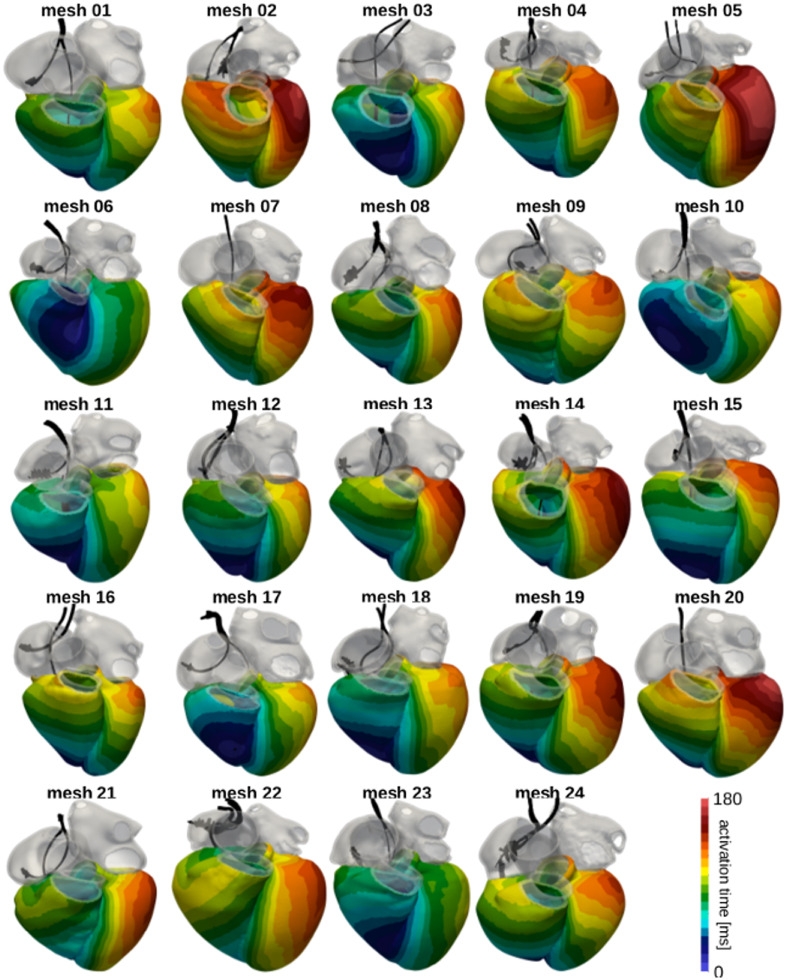
Results for the electro-mechanics simulation tests are summarised in Table 4. Fig 7 shows ventricular activation times resulting from the eikonal model solved on the twenty-four meshes. The black structures represent the CRT leads segmented from the ED CT images. The RV lead was used to define the initial activation site. We also show changes in transmembrane potential Vm (top row) and active tension (bottom row) over time on mesh 01 in Fig 5B.
Table 4. Electro-mechanics test simulations.
The table summarises results for the electro-mechanics simulation tests. For each mesh, we report the LV and RV latest activation times (LAT), the ejection fraction (EF) and the stroke volume (SV). The last row reports the mean ± the standard deviation of these values.
| LV LAT [ms] |
RV LAT [ms] |
LV EF [%] |
RV EF [%] |
LV SV [mL] |
RV SV [mL] |
|
|---|---|---|---|---|---|---|
| mesh01 | 133 | 101 | 37 | 30 | 62 | 71 |
| mesh02 | 174 | 137 | 37 | 25 | 149 | 68 |
| mesh03 | 154 | 105 | 37 | 31 | 75 | 58 |
| mesh04 | 142 | 121 | 37 | 31 | 49 | 73 |
| mesh05 | 183 | 120 | 36 | 30 | 143 | 50 |
| mesh06 | 138 | 115 | 35 | 32 | 110 | 69 |
| mesh07 | 156 | 118 | 36 | 28 | 93 | 64 |
| mesh08 | 140 | 100 | 36 | 31 | 73 | 60 |
| mesh09 | 141 | 120 | 35 | 28 | 81 | 62 |
| mesh10 | 160 | 103 | 34 | 31 | 78 | 68 |
| mesh11 | 132 | 92 | 36 | 32 | 79 | 60 |
| mesh12 | 151 | 118 | 34 | 29 | 99 | 69 |
| mesh13 | 159 | 106 | 35 | 30 | 99 | 62 |
| mesh14 | 174 | 131 | 36 | 28 | 157 | 82 |
| mesh15 | 167 | 113 | 35 | 29 | 130 | 79 |
| mesh16 | 131 | 112 | 35 | 28 | 81 | 61 |
| mesh17 | 144 | 124 | 35 | 29 | 95 | 64 |
| mesh18 | 136 | 83 | 36 | 32 | 71 | 45 |
| mesh19 | 156 | 111 | 34 | 30 | 125 | 62 |
| mesh20 | 156 | 110 | 34 | 30 | 102 | 77 |
| mesh21 | 141 | 98 | 33 | 30 | 63 | 50 |
| mesh22 | 144 | 109 | 35 | 32 | 100 | 96 |
| mesh23 | 116 | 108 | 36 | 30 | 61 | 55 |
| mesh24 | 141 | 108 | 36 | 32 | 74 | 64 |
| 149±16 | 111±12 | 35±1 | 30±2 | 95±28 | 65±11 |
Fig 7. Simulated activation times.
The images show an anterior view of all meshes with the ventricles coloured according to the local activation time computed by the eikonal model. The grey regions were excluded by the eikonal solve and were therefore passive. The black structures represent the CRT leads segmented from the CT images. The RV lead was used as the initial activation point.
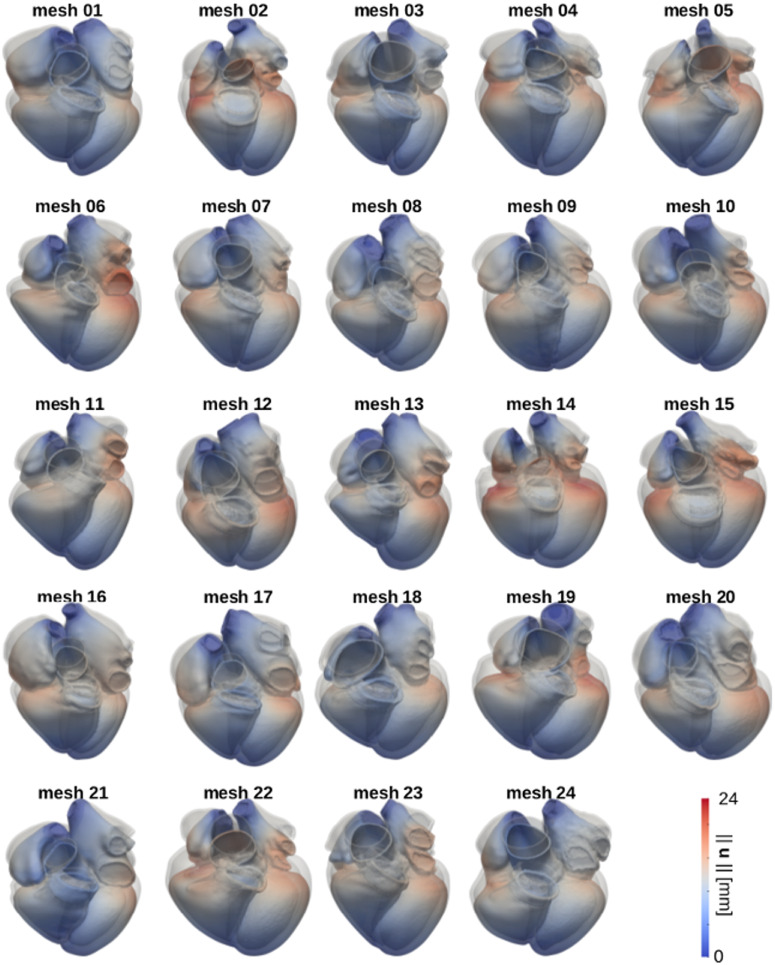
Fig 8 shows the results for the free mechanical contraction simulations on the twenty-four meshes. The three-dimensional geometries represent the end-systolic state (coloured according to the displacement magnitude) and the ED state (grey). S7 Fig in the Supporting Information shows that elements undergo significant distortion, leading to an overall decrease of element quality during ventricular contraction. Nevertheless, free contraction simulation tests ran for a whole cardiac cycle on all twenty-four meshes. We therefore concluded that the starting element quality of all meshes was good enough for the elements to endure deformation due to active contraction.
Fig 8. Simulated free active contraction.
The images show the results for the free contraction simulations run on all meshes. For each mesh, we show the configuration at the end of LV systole (coloured according to the magnitude of the displacement vector ||u|| in mm) and the initial configuration (grey geometry).
We ran additional simulations with healthy passive stiffness in cases with LV EDV within the plausible physiological healthy range to demonstrate that we could achieve a healthy LV EF. These simulations resulted in an average EF of 50% (range: 48%-53%) and 44% (range 42%-46%) for the LV and the RV, respectively. This shows that the element quality of our meshes is high enough to endure an active contraction with LV EF in healthy ranges when LV EDV falls within normal values.
Discussion
We generated the first publicly available cohort of twenty-four four-chamber meshes from HF patients with our semi-automatic pipeline. The elements of the resulting geometries have high quality. We showed that our meshes are suitable for electro-mechanical simulations by successfully running test simulations for electrical activation and free mechanical contraction. We proved that our meshes can be refined to a desired edge length, making our cohort potentially suitable for more complex electrophysiology problems. Both coarse (1.1mm average edge length) and fine (0.39mm average edge length) versions of all twenty-four meshes are publicly available to download, in order to promote the development of computational cardiac electro-mechanics.
Mesh characteristics and usability
Solver convergence in electrophysiology simulations is highly dependent on mesh resolution depending on the model used to simulate electrical activation (for example eikonal, monodomain and bidomain). The average edge length of our meshes was close to the target edge length of 1mm, see Table 3. This element size makes the meshes suitable for electrical activation simulations with the eikonal model [42], and at the same time significantly reduces the computational costs of the simulations. For a more complex electrical activation model (e.g. monodomain or bidomain formulations), a fine mesh resolution is required [42, 62, 65]. We showed that our meshes can be resampled to a desired edge length by generating a finer version of all twenty-four meshes (average edge length 0.39mm) with a publicly available tool. The user can potentially refine our meshes to an even smaller edge length if desired.
As opposed to electrophysiology simulation, in which the most important mesh characteristic for solver convergence is mesh resolution while element distortion does not represent an issue, solver convergence in mechanics simulations is affected by (but not only) element quality. Mesh quality was identified in the past as a metric to determine the stability of the mechanics simulation for different types of elements [53, 64, 66]. The threshold on element distortion is, however, sensitive to the type of elements of the mesh. For cubic-Hermite elements, Lamata et al defined a scaled Jacobian of 0.33 to determine a-priori stability of cardiac mechanics simulations for a given mesh [66, 67]. The use of cubic-Hermite elements can reduce the degrees of freedom of the mesh. However, reduced computational costs gained by a lower number of degrees of freedom comes at the cost of losing accuracy in representing cardiac anatomical structures (e.g. valve planes, LV and RV outflow tract, atria) and an increase in mesh generation complexity [66, 68]. Tetrahedral elements offer a detailed representation of the whole heart. The elements of our meshes had an average scaled Jacobian higher than 0.7. Although we did not test the effect of lower element quality on simulation stability, the quality of our meshes was high enough to run free active contraction successfully on all twenty-four meshes. S7 Fig in the supporting information shows mesh quality of mesh 03 changing over time for a free contraction simulation. Our results show that mesh quality decreases during the simulation due to large deformations. For this reason, it is important for the initial mesh to have high quality elements so that element deformation does not lead to degenerated elements.
Cohort anatomical variability
The size of the heart is used as a determinant of cardiac function, and is identified as an independent predictor of cardiac events in patients with cardiovascular diseases [69–71]. LV ED volumes in our cohort (269±78mL) are consistent with values reported for HF patients in CRT clinical trials (REVERSE: 269±93mL [72]; STARTER: 189±70mL [73]; MIRACLE-ICD: 317±98mL [74]). Despite high variations in both our cohort LV volumes and literature values, these were significantly higher than values reported in the literature for healthy controls (136±21mL [75]). The same observations apply to values for LV diameter. Our cohort had a LV diameter of 64±8mm, similar to values reported in CRT clinical trials (MIRACLE: 70±10mm [76]; REVERSE: 69±9mm [72]; MUSTIC: 71±9mm [77]; MIRACLE-ICD: 76±10mm [74]). As for the LV ED volume, LV diameter values were larger than values reported in the literature for healthy controls (50±8mm [78]). Gibson et al [79] reported a linear correlation of 74% between LV ED volume and LV diameter. We measured a similar correlation of 81% within our cohort. RV, LA and RA size measurements in HF patients are more scarse in the literature compared to LV size measurements. RV ED volume for our cohort (219±39mL) was dilated compared to values reported in the literature for healthy controls (136±24mL [80]). LA volume in our cohort (98±45mL) was also increased compared to values reported for healthy individuals (males: 32±11mL, females: 27±10mL [81]). On the other hand, RA volume in our cohort (102±45mL) falls in ranges for healthy controls (100±20mL [82]). Cardiac chamber volumes indicate that patients in our cohort underwent LV, RV and LA remodelling. Ventricular remodelling is known to happen in HF patients [83]. Ventricular failure in turn causes LA overload, with LA increased pressure and volume [84].
Limitations
Our meshes do not account for intricate and complex microstructures of ventricular endocardium. Highly detailed models of the rabbit [85–88], dog [88] and human [88–90] ventricles were generated in the past. These anatomies were based on ex-vivo imaging data, as in-vivo imaging techniques still do not allow for an enough high resolution to visualise such fine structures. Our four-chamber geometries were generated from in-vivo human CT clinical datasets. Therefore we were not able to include endocardial structures in our ventricular geometries. Ventricular trabeculations were reported to affect ventricular haemodynamics [91, 92]. Ventricular endocardial structures were however found to have a marginal effects on strains [93] and arrhythmias re-entry [94]. Therefore our meshes might not be suitable for detailed haemodynamics simulations, but can be used for mechanics and electrophysiology simulations (when refined if required by the activation model).
Our ventricular models also do not include a Purkinje network. The Purkinje tree plays an important role in ventricular fibrillation [95, 96] and arrhythmias [97–101]. From a modelling point of view, including the ventricular fast conduction system is necessary to simulate physiological ECGs [89]. Purkinje tree morphology was also reported to affect ECG shape [102]. Therefore, if the user intends to use our meshes to investigate ventricular arrhythimas or fibrillation, or to simulate ECG resulting from activation, the Purkinje network can be included using publicly available tools (https://github.com/fsahli/fractal-tree) or by mapping a pre-existing tree using the UVCs [41], also provided for each mesh.
Our geometries do not offer a detailed model of atrial structures. The LAA was not included in the geometries because the CT volume often did not cover it. Other structures such as crista terminalis, pectinate muscles and fossa ovalis were not identified as separate tags in the atrial meshes. However, these can be included based on previous studies offering more detailed representation of LA and RA anatomical regions [5, 103].
Another limitation of our atrial model is that we did not include atrial fibres. Atrial fibers are more challenging to embed in three-dimensional geometries compared to ventricular fibres, due to complex atrial anatomy and scarcity of data about fibre orientation in atrial myocardium. It is however possible for the user to define atrial fibres using a rule-based method [103, 104] or by mapping a pre-existing fibre field using the universal atrial coordinated (UACs) [105–107].
The aim of our study was not to provide a fully detailed model of the heart. We instead provided a reference set of twenty-four four-chamber anatomies which external users can add features (such as Purkinje network, atrial regions, atrial fibres etc…) to. We believe that our database constitutes a significant step forward for the computational modelling community, as it provides the first publicly available database of four-chamber heart models.
Conclusion
We built a cohort of twenty-four four-chamber heart meshes from HF patients CT data. The elements of our meshes have high quality, making them suitable for electro-mechanics simulations. We verified this by running electro-mechanics simulation tests for ventricular electrical activation and free contraction. The twenty-four geometries can be refined and integrated with more purpose-specific features, and used for large cohort computational studies and as a data set for verification of numerical methods for cardiac electro-mechanics.
Supporting information
The image shows the segmentation for the components included in the final tetrahedral mesh for cases 01 to 04 overlapped with the end-diastolic CT image. For each case, we show a four-chamber axial view, two sagittal and two coronal views. Abbreviations: left ventricle (LV), right ventricle (RV), left atrium (LA), right atrium (RA), aorta (Ao) and pulmonary artery (PA).
(PDF)
The image shows the segmentation for the components included in the final tetrahedral mesh for cases 05 to 08 overlapped with the end-diastolic CT image. For each case, we show a four-chamber axial view, two sagittal and two coronal views. Abbreviations: left ventricle (LV), right ventricle (RV), left atrium (LA), right atrium (RA), aorta (Ao) and pulmonary artery (PA).
(PDF)
The image shows the segmentation for the components included in the final tetrahedral mesh for cases 09 to 12 overlapped with the end-diastolic CT image. For each case, we show a four-chamber axial view, two sagittal and two coronal views. Abbreviations: left ventricle (LV), right ventricle (RV), left atrium (LA), right atrium (RA), aorta (Ao) and pulmonary artery (PA).
(PDF)
The image shows the segmentation for the components included in the final tetrahedral mesh for cases 13 to 16 overlapped with the end-diastolic CT image. For each case, we show a four-chamber axial view, two sagittal and two coronal views. Abbreviations: left ventricle (LV), right ventricle (RV), left atrium (LA), right atrium (RA), aorta (Ao) and pulmonary artery (PA).
(PDF)
The image shows the segmentation for the components included in the final tetrahedral mesh for cases 17 to 20 overlapped with the end-diastolic CT image. For each case, we show a four-chamber axial view, two sagittal and two coronal views. Abbreviations: left ventricle (LV), right ventricle (RV), left atrium (LA), right atrium (RA), aorta (Ao) and pulmonary artery (PA).
(PDF)
The image shows the segmentation for the components included in the final tetrahedral mesh for cases 21 to 24 overlapped with the end-diastolic CT image. For each case, we show a four-chamber axial view, two sagittal and two coronal views. Abbreviations: left ventricle (LV), right ventricle (RV), left atrium (LA), right atrium (RA), aorta (Ao) and pulmonary artery (PA).
(PDF)
A The plot shows error bars (mean ± standard deviation) of the mesh quality over time for a free contraction simulation for mesh 03. The other plots show mesh quality variations over simulation time for different regions of the mesh: B LV, C RV, D LA and E RA. Average mesh quality decreases from 0.74 to a minimum of 0.63, reached at maximum ventricular contraction. Change in mesh quality is driven by the LV and the RV, for which the average quality decreases of 16% and 20% from ED to end-systole, respectively. LA and the RA mesh quality, which are not actively contracting in the simulation and are therefore passive, underwent small variations (4% and 3%, respectively). The same applied to all the other labels of the mesh (results not shown).
(PDF)
(PDF)
(PDF)
Data Availability
The data are uploaded to Zenodo and publicly accessible via the following URL: https://doi.org/10.5281/zenodo.3890034.
Funding Statement
S. A. N. received support from the UK Engineering and Physical Sciences Research Council (https://epsrc.ukri.org/), grant numbers: EP/M012492/1, NS/A000049/1, EP/L015226/1 and EP/P01268X/1; from the British Heart Foundation (https://www.bhf.org.uk/), grant numbers: PG/15/91/31812 and PG/13/37/30280; from the Kings Health Partners London National Institute for Health Research (NIHR) Biomedical Research Centre (http://www.guysandstthomasbrc.nihr.ac.uk/). C. M. A. received support from the Marie Sklodowska-Curie fellowship (https://ec.europa.eu/research/mariecurieactions/), grant number 750835. G. P. received support from the Austrian Science Fund (FWF) (https://fwf.ac.at/en/); grant number PI2760-B30. This work made use of ARCHER (http://www.archer.ac.uk/), the UK national high-performance computing service located at the University of Edinburgh and funded by the Office of Science and Technology through Engineering and Physical Sciences Research Councils High End Computing Programme. Miss Strocchi M. was supported by an unrestricted Abbott educational grant through the Centre for Doctoral Training in Medical Imaging at King’s College London. Abbott had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Dr Neic A. is an employee for NumeriCor GmbH and assisted with meshing tools and simulation software development. This does not alter our adherence to PLOS ONE policies on sharing data and materials.
References
- 1. Kayvanpour E, Mansi T, Sedaghat-Hamedani F, Amr A, Neumann D, Georgescu B, et al. Towards personalized cardiology: multi-scale modeling of the failing heart. PLoS One. 2015;10(7):e0134869 10.1371/journal.pone.0134869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Niederer SA, Plank G, Chinchapatnam P, Ginks M, Lamata P, Rhode KS, et al. Length-dependent tension in the failing heart and the efficacy of cardiac resynchronization therapy. Cardiovasc Res. 2010;89(2):336–343. 10.1093/cvr/cvq318 [DOI] [PubMed] [Google Scholar]
- 3. Kerckhoffs RC, Omens JH, McCulloch AD. Mechanical discoordination increases continuously after the onset of left bundle branch block despite constant electrical dyssynchrony in a computational model of cardiac electromechanics and growth. Europace. 2012;14(suppl 5):v65–v72. 10.1093/europace/eus274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Crozier A, Blazevic B, Lamata P, Plank G, Ginks M, Duckett S, et al. The relative role of patient physiology and device optimisation in cardiac resynchronisation therapy: A computational modelling study. J Mol Cell Cardiol. 2016;96:93–100. 10.1016/j.yjmcc.2015.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tobon-Gomez C, Duchateau N, Sebastian R, Marchesseau S, Camara O, Donal E, et al. Understanding the mechanisms amenable to CRT response: from pre-operative multimodal image data to patient-specific computational models. Medical & biological engineering & computing. 2013;51(11):1235–1250. 10.1007/s11517-013-1044-7 [DOI] [PubMed] [Google Scholar]
- 6. Hyde ER, Behar JM, Crozier A, Claridge S, Jackson T, Sohal M, et al. Improvement of right ventricular hemodynamics with left ventricular endocardial pacing during cardiac resynchronization therapy. Pacing Clin Electrophysiol. 2016;39(6):531–541. 10.1111/pace.12854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gsell MA, Augustin CM, Prassl AJ, Karabelas E, Fernandes JF, Kelm M, et al. Assessment of wall stresses and mechanical heart power in the left ventricle: Finite element modeling versus Laplace analysis. International journal for numerical methods in biomedical engineering. 2018;34(12):e3147 10.1002/cnm.3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Prakosa A, Arevalo HJ, Deng D, Boyle PM, Nikolov PP, Ashikaga H, et al. Personalized virtual-heart technology for guiding the ablation of infarct-related ventricular tachycardia. Nature biomedical engineering. 2018;2(10):732 10.1038/s41551-018-0282-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang W, Wang Y, Wu Y, Lin T, Li S, Chen B. Quantification of full left ventricular metrics via deep regression learning with contour-guidance. IEEE Access. 2019;7:47918–47928. 10.1109/ACCESS.2019.2907564 [DOI] [Google Scholar]
- 10. Okada Ji, Washio T, Nakagawa M, Watanabe M, Kadooka Y, Kariya T, et al. Multi-scale, tailor-made heart simulation can predict the effect of cardiac resynchronization therapy. J Mol Cell Cardiol. 2017;108:17–23. 10.1016/j.yjmcc.2017.05.006 [DOI] [PubMed] [Google Scholar]
- 11. Strocchi M, Gsell MA, Augustin CM, Razeghi O, Roney CH, Prassl AJ, et al. Simulating ventricular systolic motion in a four-chamber heart model with spatially varying robin boundary conditions to model the effect of the pericardium. Journal of Biomechanics. 2020;101:109645 10.1016/j.jbiomech.2020.109645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Augustin CM, Neic A, Liebmann M, Prassl AJ, Niederer SA, Haase G, et al. Anatomically accurate high resolution modeling of human whole heart electromechanics: a strongly scalable algebraic multigrid solver method for nonlinear deformation. J Comput Phys. 2016;305:622–646. 10.1016/j.jcp.2015.10.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fritz T, Wieners C, Seemann G, Steen H, Dössel O. Simulation of the contraction of the ventricles in a human heart model including atria and pericardium. Biomech Model Mechanobiol. 2014;13(3):627–641. 10.1007/s10237-013-0523-y [DOI] [PubMed] [Google Scholar]
- 14. Land S, Niederer SA. Influence of atrial contraction dynamics on cardiac function. Int J Numer Method Biomed Eng. 2018;34(3):e2931 10.1002/cnm.2931 [DOI] [PubMed] [Google Scholar]
- 15. Morris DA, Gailani M, Pérez AV, Blaschke F, Dietz R, Haverkamp W, et al. Left atrial systolic and diastolic dysfunction in heart failure with normal left ventricular ejection fraction. Journal of the American Society of Echocardiography. 2011;24(6):651–662. 10.1016/j.echo.2011.02.004 [DOI] [PubMed] [Google Scholar]
- 16. Stefanadis C, Dernellis J, Toutouzas P. A clinical appraisal of left atrial function. European heart journal. 2001;22(1):22–36. 10.1053/euhj.1999.2581 [DOI] [PubMed] [Google Scholar]
- 17. Crozier A, Augustin CM, Neic A, Prassl AJ, Holler M, Fastl TE, et al. Image-Based Personalization of Cardiac Anatomy for Coupled Electromechanical Modeling. Ann Biomed Eng. 2015;44(1):58–70. 10.1007/s10439-015-1474-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pfaller MR, Hörmann JM, Weigl M, Nagler A, Chabiniok R, Bertoglio C, et al. The importance of the pericardium for cardiac biomechanics: From physiology to computational modeling. Biomechanics and modeling in mechanobiology. 2018; p. 1–27. [DOI] [PubMed] [Google Scholar]
- 19. Koulouzis S, Belloum A, Bubak M, Lamata P, Nolte D, Vasyunin D, et al. Distributed data management service for vph applications. IEEE Internet Computing. 2015;20(2):34–41. 10.1109/MIC.2015.71 [DOI] [Google Scholar]
- 20. Medrano-Gracia P, Cowan BR, Ambale-Venkatesh B, Bluemke DA, Eng J, Finn JP, et al. Left ventricular shape variation in asymptomatic populations: the multi-ethnic study of atherosclerosis. Journal of Cardiovascular Magnetic Resonance. 2014;16(1):56 10.1186/s12968-014-0056-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lewandowski AJ, Augustine D, Lamata P, Davis EF, Lazdam M, Francis J, et al. Preterm heart in adult life: cardiovascular magnetic resonance reveals distinct differences in left ventricular mass, geometry, and function. Circulation. 2013;127(2):197–206. 10.1161/CIRCULATIONAHA.112.126920 [DOI] [PubMed] [Google Scholar]
- 22.Zacur E, Wong J, Razavi R, Geva T, Greil G, Lamata P. Revealing Differences in Anatomical Remodelling of the Systemic Right Ventricle. In: International Conference on Functional Imaging and Modeling of the Heart. Springer; 2015. p. 99–107.
- 23. Backhaus M, Chung JD, Cowan BR, Fonseca CG, Tao W, Young AA. In: Kerckhoffs RCP, editor. The Cardiac Atlas Project: Towards a Map of the Heart. New York, NY: Springer New York; 2010. p. 113–129. [Google Scholar]
- 24.Medrano-Gracia P, Cowan BR, Bluemke DA, Finn JP, Kadish AH, Lee DC, et al. Continuous spatio-temporal atlases of the asymptomatic and infarcted hearts. In: International Workshop on Statistical Atlases and Computational Models of the Heart. Springer; 2013. p. 143–151.
- 25. Zhang X, Medrano-Gracia P, Ambale-Venkatesh B, Bluemke DA, Cowan BR, Finn JP, et al. Orthogonal decomposition of left ventricular remodeling in myocardial infarction. GigaScience. 2017;6(3):gix005 10.1093/gigascience/gix005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lötjönen J, Kivistö S, Koikkalainen J, Smutek D, Lauerma K. Statistical shape model of atria, ventricles and epicardium from short-and long-axis MR images. Medical image analysis. 2004;8(3):371–386. 10.1016/j.media.2004.06.013 [DOI] [PubMed] [Google Scholar]
- 27.Chandrashekara R, Rao A, Sanchez-Ortiz GI, Mohiaddin RH, Rueckert D. Construction of a statistical model for cardiac motion analysis using nonrigid image registration. In: Biennial International Conference on Information Processing in Medical Imaging. Springer; 2003. p. 599–610. [DOI] [PubMed]
- 28. Duchateau N, De Craene M, Piella G, Silva E, Doltra A, Sitges M, et al. A spatiotemporal statistical atlas of motion for the quantification of abnormal myocardial tissue velocities. Medical image analysis. 2011;15(3):316–328. 10.1016/j.media.2010.12.006 [DOI] [PubMed] [Google Scholar]
- 29. Hoogendoorn C, Duchateau N, Sánchez-Quintana D, Whitmarsh T, Sukno FM, De Craene M, et al. A high-resolution atlas and statistical model of the human heart from multislice CT. IEEE transactions on medical imaging. 2012;32(1):28–44. 10.1109/TMI.2012.2230015 [DOI] [PubMed] [Google Scholar]
- 30.Strocchi M, Augustin CM, Gsell MAF, Karabelas E, Neic A, Gillette K, et al. A Publicly Available Virtual Cohort of Four- chamber Heart Meshes for Cardiac Electro-mechanics Simulations; 2020. Available from: 10.5281/zenodo.3890034. [DOI] [PMC free article] [PubMed]
- 31. Zheng Y, Barbu A, Georgescu B, Scheuering M, Comaniciu D. Four-chamber heart modeling and automatic segmentation for 3-D cardiac CT volumes using marginal space learning and steerable features. IEEE transactions on medical imaging. 2008;27(11):1668–1681. 10.1109/TMI.2008.2004421 [DOI] [PubMed] [Google Scholar]
- 32.CIBC. Seg3D: Volumetric Image Segmentation and Visualization. Scientific Computing and Imaging Institute (SCI), Download from: http://www.seg3d.org. http://wwwsciutahedu/cibc-software/seg3dhtml. 2016.
- 33. Ho S, Nihoyannopoulos P. Anatomy, echocardiography, and normal right ventricular dimensions. Heart. 2006;92(suppl 1):i2–i13. 10.1136/hrt.2005.077875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Matsukubo H, Matsuura T, Endo N, Asayama J, Watanabe T. Echocardiographic measurement of right ventricular wall thickness. A new application of subxiphoid echocardiography. Circulation. 1977;56(2):278–284. 10.1161/01.CIR.56.2.278 [DOI] [PubMed] [Google Scholar]
- 35. Beinart R, Abbara S, Blum A, Ferencik M, Heist K, Ruskin J, et al. Left atrial wall thickness variability measured by CT scans in patients undergoing pulmonary vein isolation. J Cardiovasc Electrophysiol. 2011;22(11):1232–1236. 10.1111/j.1540-8167.2011.02100.x [DOI] [PubMed] [Google Scholar]
- 36. Mensel B, Kühn JP, Schneider T, Quadrat A, Hegenscheid K. Mean thoracic aortic wall thickness determination by cine MRI with steady-state free precession: validation with dark blood imaging. Acad Radiol. 2013;20(8):1004–1008. 10.1016/j.acra.2013.03.014 [DOI] [PubMed] [Google Scholar]
- 37. Knoll F, Clason C, Bredies K, Uecker M, Stollberger R. Parallel imaging with nonlinear reconstruction using variational penalties. Magn Reson Med. 2012;67(1):34–41. 10.1002/mrm.22964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bayer J, Blake R, Plank G, Trayanova N. A novel rule-based algorithm for assigning myocardial fiber orientation to computational heart models. Ann Biomed Eng. 2012;40(10):2243–2254. 10.1007/s10439-012-0593-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Land S, Park-Holohan SJ, Smith NP, dos Remedios CG, Kentish JC, Niederer SA. A model of cardiac contraction based on novel measurements of tension development in human cardiomyocytes. J Mol Cell Cardiol. 2017;106:68–83. 10.1016/j.yjmcc.2017.03.008 [DOI] [PubMed] [Google Scholar]
- 40. Watson SR, Dormer JD, Fei B. Imaging technologies for cardiac fiber and heart failure: a review. Heart failure reviews. 2018;23(2):273–289. 10.1007/s10741-018-9684-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bayer J, Prassl AJ, Pashaei A, Gomez JF, Frontera A, Neic A, et al. Universal ventricular coordinates: A generic framework for describing position within the heart and transferring data. Medical image analysis. 2018;45:83–93. 10.1016/j.media.2018.01.005 [DOI] [PubMed] [Google Scholar]
- 42. Neic A, Campos FO, Prassl AJ, Niederer SA, Bishop MJ, Vigmond EJ, et al. Efficient computation of electrograms and ECGs in human whole heart simulations using a reaction-eikonal model. J Comput Phys. 2017. 10.1016/j.jcp.2017.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Taggart P, Sutton PM, Opthof T, Coronel R, Trimlett R, Pugsley W, et al. Inhomogeneous transmural conduction during early ischaemia in patients with coronary artery disease. Journal of molecular and cellular cardiology. 2000;32(4):621–630. 10.1006/jmcc.2000.1105 [DOI] [PubMed] [Google Scholar]
- 44. Lee AW. A rule-based method for predicting the electrical activation of the heart with cardiac resynchronization therapy from non-invasive clinical data. Medical Image Analysis. 2019. 10.1016/j.media.2019.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Strik M, Regoli F, Auricchio A, Prinzen F. Electrical and mechanical ventricular activation during left bundle branch block and resynchronization. J Cardiovasc Transl Res. 2012;5(2):117–126. 10.1007/s12265-012-9351-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ten Tusscher KHWJ, Panfilov AV. Alternans and spiral breakup in a human ventricular tissue model. American Journal of Physiology-Heart and Circulatory Physiology. 2006;291(3):H1088–H1100. 10.1152/ajpheart.00109.2006 [DOI] [PubMed] [Google Scholar]
- 47. Nordsletten DA, Niederer SA, Nash MP, Hunter PJ, Smith NP. Coupling multi-physics models to cardiac mechanics. Prog Biophys Mol Biol. 2011;104(1):77–88. 10.1016/j.pbiomolbio.2009.11.001 [DOI] [PubMed] [Google Scholar]
- 48. Guccione JM, McCulloch AD, Waldman LK, et al. Passive material properties of intact ventricular myocardium determined from a cylindrical model. J Biomech Eng. 1991;113(1):42–55. 10.1115/1.2894084 [DOI] [PubMed] [Google Scholar]
- 49. Nasopoulou A, Shetty A, Lee J, Nordsletten D, Rinaldi CA, Lamata P, et al. Improved identifiability of myocardial material parameters by an energy-based cost function. Biomech Model Mechanobiol. 2017;16(3):971–988. 10.1007/s10237-016-0865-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Di Martino ES, Bellini C, Schwartzman DS. In vivo porcine left atrial wall stress: computational model. Journal of biomechanics. 2011;44(15):2589–2594. 10.1016/j.jbiomech.2011.08.023 [DOI] [PubMed] [Google Scholar]
- 51. Hornỳ L, Žitnỳ R, Chlup H, Macková H. Identification of the material parameters of an aortic wall. Bull Appl Mech. 2006;2(8):173–181. [Google Scholar]
- 52. Tian L, Wang Z, Liu Y, Eickhoff JC, Eliceiri KW, Chesler NC. Validation of an arterial constitutive model accounting for collagen content and crosslinking. Acta Biomater. 2016;31:276–287. 10.1016/j.actbio.2015.11.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Flory P. Thermodynamic relations for high elastic materials. Transactions of the Faraday Society. 1961;57:829–838. 10.1039/tf9615700829 [DOI] [Google Scholar]
- 54. Ogden R. Nearly isochoric elastic deformations: application to rubberlike solids. J Mech Phys Solids. 1978;26(1):37–57. 10.1016/0022-5096(78)90012-1 [DOI] [Google Scholar]
- 55. Kerckhoffs RCP, Bovendeerd PHM, Prinzen FW, Smits K, Arts T. Intra and interventricular asynchrony of electromechanics in the ventricularly paced heart. J Eng Math. 2003;47(3-4):201–216. 10.1023/B:ENGI.0000007972.73874.da [DOI] [Google Scholar]
- 56. Walker JC, Ratcliffe MB, Zhang P, Wallace AW, Fata B, Hsu EW, et al. MRI-based finite-element analysis of left ventricular aneurysm. American Journal of Physiology-Heart and Circulatory Physiology. 2005;289(2):H692–H700. 10.1152/ajpheart.01226.2004 [DOI] [PubMed] [Google Scholar]
- 57. Petersen SE, Aung N, Sanghvi MM, Zemrak F, Fung K, Paiva JM, et al. Reference ranges for cardiac structure and function using cardiovascular magnetic resonance (CMR) in Caucasians from the UK Biobank population cohort. Journal of Cardiovascular Magnetic Resonance. 2017;19(1):18 10.1186/s12968-017-0327-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vigmond EJ, Aguel F, Trayanova NA. Computational techniques for solving the bidomain equations in three dimensions. IEEE Transactions on Biomedical Engineering. 2002;49(11):1260–1269. 10.1109/TBME.2002.804597 [DOI] [PubMed] [Google Scholar]
- 59. Vigmond E, Dos Santos RW, Prassl A, Deo M, Plank G. Solvers for the cardiac bidomain equations. Prog Biophys Mol Biol. 2008;96(1-3):3–18. 10.1016/j.pbiomolbio.2007.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fu Z, Kirby RM, Whitaker RT. A fast iterative method for solving the eikonal equation on tetrahedral domains. SIAM Journal on Scientific Computing. 2013;35(5):C473–C494. 10.1137/120881956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Augustin CM, Crozier A, Neic A, Prassl AJ, Karabelas E, Ferreira da Silva T, et al. Patient-specific modeling of left ventricular electromechanics as a driver for haemodynamic analysis. EP Europace. 2016;18(suppl_4):iv121–iv129. 10.1093/europace/euw369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Land S, Gurev V, Arens S, Augustin CM, Baron L, Blake R, et al. Verification of cardiac mechanics software: benchmark problems and solutions for testing active and passive material behaviour. Proceedings of the Royal Society of London A: Mathematical, Physical and Engineering Sciences. 2015;471 (2184). 10.1098/rspa.2015.0641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Knupp PM. Achieving finite element mesh quality via optimization of the Jacobian matrix norm and associated quantities. Part I—a framework for surface mesh optimization. International Journal for Numerical Methods in Engineering. 2000;48(3):401–420. [DOI] [Google Scholar]
- 64. Knupp PM. Achieving finite element mesh quality via optimization of the Jacobian matrix norm and associated quantities. Part II—a framework for volume mesh optimization and the condition number of the Jacobian matrix. International Journal for numerical methods in engineering. 2000;48(8):1165–1185. [DOI] [Google Scholar]
- 65. Niederer S, Mitchell L, Smith N, Plank G. Simulating human cardiac electrophysiology on clinical time-scales. Frontiers in physiology. 2011;2:14 10.3389/fphys.2011.00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lamata P, Roy I, Blazevic B, Crozier A, Land S, Niederer SA, et al. Quality metrics for high order meshes: analysis of the mechanical simulation of the heart beat. IEEE transactions on medical imaging. 2012;32(1):130–138. 10.1109/TMI.2012.2231094 [DOI] [PubMed] [Google Scholar]
- 67. Lamata P, Sinclair M, Kerfoot E, Lee A, Crozier A, Blazevic B, et al. An automatic service for the personalization of ventricular cardiac meshes. Journal of The Royal Society Interface. 2014;11(91):20131023 10.1098/rsif.2013.1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Jafari A, Pszczolkowski E, Krishnamurthy A. A framework for biomechanics simulations using four-chamber cardiac models. Journal of biomechanics. 2019;91:92–101. 10.1016/j.jbiomech.2019.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ivanov A, Mohamed A, Asfour A, Ho J, Khan SA, Chen O, et al. Right atrial volume by cardiovascular magnetic resonance predicts mortality in patients with heart failure with reduced ejection fraction. PloS one. 2017;12(4):e0173245 10.1371/journal.pone.0173245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mousavi N, Tan TC, Ali M, Halpern EF, Wang L, Scherrer-Crosbie M. Echocardiographic parameters of left ventricular size and function as predictors of symptomatic heart failure in patients with a left ventricular ejection fraction of 50–59% treated with anthracyclines. European Heart Journal-Cardiovascular Imaging. 2015;16(9):977–984. [DOI] [PubMed] [Google Scholar]
- 71. Njoku A, Kannabhiran M, Arora R, Reddy P, Gopinathannair R, Lakkireddy D, et al. Left atrial volume predicts atrial fibrillation recurrence after radiofrequency ablation: a meta-analysis. Ep Europace. 2017;20(1):33–42. 10.1093/europace/eux013 [DOI] [PubMed] [Google Scholar]
- 72. Linde C, Abraham WT, Gold MR, St John Sutton M, Ghio S, Daubert C, et al. Randomized trial of cardiac resynchronization in mildly symptomatic heart failure patients and in asymptomatic patients with left ventricular dysfunction and previous heart failure symptoms. Journal of the American College of Cardiology. 2008;52(23):1834–1843. 10.1016/j.jacc.2008.08.027 [DOI] [PubMed] [Google Scholar]
- 73. Saba S, Marek J, Schwartzman D, Jain S, Adelstein E, White P, et al. Echocardiography-Guided Left Ventricular Lead Placement for Cardiac Resynchronization TherapyClinical Perspective. Circulation: Heart Failure. 2013;6(3):427–434. [DOI] [PubMed] [Google Scholar]
- 74. Young JB. Combined Cardiac Resynchronization and Implantable Cardioversion Defibrillation in Advanced Chronic Heart Failure. JAMA. 2003;289(20):2685 10.1001/jama.289.20.2685 [DOI] [PubMed] [Google Scholar]
- 75. Maceira AM, Prasad SK, Khan M, Pennell DJ. Reference right ventricular systolic and diastolic function normalized to age, gender and body surface area from steady-state free precession cardiovascular magnetic resonance. European heart journal. 2006;27(23):2879–2888. 10.1093/eurheartj/ehl336 [DOI] [PubMed] [Google Scholar]
- 76. Abraham WT, Fisher WG, Smith AL. Cardiac resynchronization in chronic heart failure. ACC Current Journal Review. 2002;11(6):75 10.1016/S1062-1458(02)00961-3 [DOI] [PubMed] [Google Scholar]
- 77. Linde C, Leclercq C, Rex S. Long-term benefits of biventricular pacing in congestive heart failure: results from the MUltisite STimulation in cardiomyopathy (MUSTIC) study. ACC Current Journal Review. 2002;11(6):75 10.1016/S1062-1458(02)00962-5 [DOI] [PubMed] [Google Scholar]
- 78. Narayanan K, Reinier K, Teodorescu C, Uy-Evanado A, Aleong R, Chugh H, et al. Left ventricular diameter and risk stratification for sudden cardiac death. Journal of the American Heart Association. 2014;3(5):e001193 10.1161/JAHA.114.001193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Gibson PH, Becher H, Choy JB. Classification of left ventricular size: diameter or volume with contrast echocardiography? Open heart. 2014;1(1):e000147 10.1136/openhrt-2014-000147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Maceira A, Prasad S, Khan M, Pennell D. Normalized left ventricular systolic and diastolic function by steady state free precession cardiovascular magnetic resonance. Journal of Cardiovascular Magnetic Resonance. 2006;8(3):417–426. 10.1080/10976640600572889 [DOI] [PubMed] [Google Scholar]
- 81. Aurigemma GP, Gottdiener JS, Arnold AM, Chinali M, Hill JC, Kitzman D. Left atrial volume and geometry in healthy aging: the Cardiovascular Health Study. Circulation: Cardiovascular Imaging. 2009;2(4):282–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Maceira AM, Cosín-Sales J, Roughton M, Prasad SK, Pennell DJ. Reference right atrial dimensions and volume estimation by steady state free precession cardiovascular magnetic resonance. Journal of Cardiovascular Magnetic Resonance. 2013;15(1):29 10.1186/1532-429X-15-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Konstam MA, Kramer DG, Patel AR, Maron MS, Udelson JE. Left ventricular remodeling in heart failure: current concepts in clinical significance and assessment. JACC: Cardiovascular Imaging. 2011;4(1):98–108. [DOI] [PubMed] [Google Scholar]
- 84. Triposkiadis F, Pieske B, Butler J, Parissis J, Giamouzis G, Skoularigis J, et al. Global left atrial failure in heart failure. European journal of heart failure. 2016;18(11):1307–1320. 10.1002/ejhf.645 [DOI] [PubMed] [Google Scholar]
- 85.Bishop MJ, Hales P, Plank G, Gavaghan DJ, Scheider J, Grau V. Comparison of rule-based and DTMRI-derived fibre architecture in a whole rat ventricular computational model. In: International Conference on Functional Imaging and Modeling of the Heart. Springer; 2009. p. 87–96.
- 86. Bishop MJ, Plank G, Burton RAB, Schneider JE, Gavaghan DJ, Grau V, et al. Development of an anatomically detailed MRI-derived rabbit ventricular model and assessment of its impact on simulations of electrophysiological function. American Journal of Physiology-Heart and Circulatory Physiology. 2010;298(2):H699–H718. 10.1152/ajpheart.00606.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Plank G, Burton RA, Hales P, Bishop M, Mansoori T, Bernabeu MO, et al. Generation of histo-anatomically representative models of the individual heart: tools and application. Philosophical Transactions of the Royal Society A: Mathematical, Physical and Engineering Sciences. 2009;367(1896):2257–2292. 10.1098/rsta.2009.0056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Vadakkumpadan F, Arevalo H, Prassl AJ, Chen J, Kickinger F, Kohl P, et al. Image-based models of cardiac structure in health and disease. Wiley Interdisciplinary Reviews: Systems Biology and Medicine. 2010;2(4):489–506. 10.1002/wsbm.76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Costabal FS, Hurtado DE, Kuhl E. Generating Purkinje networks in the human heart. Journal of biomechanics. 2016;49(12):2455–2465. 10.1016/j.jbiomech.2015.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Baillargeon B, Rebelo N, Fox DD, Taylor RL, Kuhl E. The living heart project: a robust and integrative simulator for human heart function. European Journal of Mechanics-A/Solids. 2014;48:38–47. 10.1016/j.euromechsol.2014.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Sacco F, Paun B, Lehmkuhl O, Iles TL, Iaizzo PA, Houzeaux G, et al. Left ventricular trabeculations decrease the wall shear stress and increase the intra-ventricular pressure drop in CFD simulations. Frontiers in Physiology. 2018;9:458 10.3389/fphys.2018.00458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Vedula V, Seo JH, Lardo AC, Mittal R. Effect of trabeculae and papillary muscles on the hemodynamics of the left ventricle. Theoretical and Computational Fluid Dynamics. 2016;30(1-2):3–21. 10.1007/s00162-015-0349-6 [DOI] [Google Scholar]
- 93.Gurev V, Korte J, Hafez O, Fattebert JL, Richards DF, Rice JJ. Papillary muscles contraction does not change ventricular wall mechanics. In: 2015 Computing in Cardiology Conference (CinC). IEEE; 2015. p. 333–336.
- 94. Bishop MJ, Plank G. The role of fine-scale anatomical structure in the dynamics of reentry in computational models of the rabbit ventricles. The Journal of physiology. 2012;590(18):4515–4535. 10.1113/jphysiol.2012.229062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Huang J, Dosdall DJ, Cheng KA, Li L, Rogers JM, Ideker RE. The importance of Purkinje activation in long duration ventricular fibrillation. Journal of the American Heart Association. 2014;3(1):e000495 10.1161/JAHA.113.000495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Martinez M, Walton R, Haissaguerre M, Hocini M, Bernus O. Role of Purkinje network in maintaining ventricular fibrillation before and after focal catheter ablation of Purkinje fibers in sheep left ventricle. Archives of Cardiovascular Diseases Supplements. 2018;10(2):191 10.1016/j.acvdsp.2018.02.036 [DOI] [Google Scholar]
- 97. Boyden PA, Hirose M, Dun W. Cardiac Purkinje cells. Heart Rhythm. 2010;7(1):127–135. 10.1016/j.hrthm.2009.09.017 [DOI] [PubMed] [Google Scholar]
- 98. Garcia-Bustos V, Sebastian R, Izquierdo M, Rios-Navarro C, Bodí V, Chorro FJ, et al. Changes in the spatial distribution of the Purkinje network after acute myocardial infarction in the pig. PloS one. 2019;14(2):e0212096 10.1371/journal.pone.0212096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Haissaguerre M, Vigmond E, Stuyvers B, Hocini M, Bernus O. Ventricular arrhythmias and the His–Purkinje system. Nature Reviews Cardiology. 2016;13(3):155 10.1038/nrcardio.2015.193 [DOI] [PubMed] [Google Scholar]
- 100. Vigmond EJ, Stuyvers BD. Modeling our understanding of the His-Purkinje system. Progress in biophysics and molecular biology. 2016;120(1-3):179–188. 10.1016/j.pbiomolbio.2015.12.013 [DOI] [PubMed] [Google Scholar]
- 101. Behradfar E, Nygren A, Vigmond EJ. The role of Purkinje-myocardial coupling during ventricular arrhythmia: a modeling study. PloS one. 2014;9(2):e88000 10.1371/journal.pone.0088000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Lyon A, Bueno-Orovio A, Zacur E, Ariga R, Grau V, Neubauer S, et al. Electrocardiogram phenotypes in hypertrophic cardiomyopathy caused by distinct mechanisms: apico-basal repolarization gradients vs. Purkinje-myocardial coupling abnormalities. EP Europace. 2018;20(suppl_3):iii102–iii112. 10.1093/europace/euy226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ferrer A, Sebastián R, Sánchez-Quintana D, Rodríguez JF, Godoy EJ, Martínez L, et al. Detailed anatomical and electrophysiological models of human atria and torso for the simulation of atrial activation. PloS one. 2015;10(11):e0141573 10.1371/journal.pone.0141573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Fastl TE, Tobon-Gomez C, Crozier A, Whitaker J, Rajani R, McCarthy KP, et al. Personalized computational modeling of left atrial geometry and transmural myofiber architecture. Medical image analysis. 2018;47:180–190. 10.1016/j.media.2018.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Labarthe S, Bayer J, Coudière Y, Henry J, Cochet H, Jaïs P, et al. A bilayer model of human atria: mathematical background, construction, and assessment. Europace. 2014;16(suppl_4):iv21–iv29. 10.1093/europace/euu256 [DOI] [PubMed] [Google Scholar]
- 106. Roney CH, Bayer JD, Cochet H, Meo M, Dubois R, Jaïs P, et al. Variability in pulmonary vein electrophysiology and fibrosis determines arrhythmia susceptibility and dynamics. PLoS Comput Biol. 2018;14(5):e1006166 10.1371/journal.pcbi.1006166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Roney CH, Pashaei A, Meo M, Dubois R, Boyle PM, Trayanova NA, et al. Universal atrial coordinates applied to visualisation, registration and construction of patient specific meshes. arXiv preprint arXiv:181006630. 2018. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The image shows the segmentation for the components included in the final tetrahedral mesh for cases 01 to 04 overlapped with the end-diastolic CT image. For each case, we show a four-chamber axial view, two sagittal and two coronal views. Abbreviations: left ventricle (LV), right ventricle (RV), left atrium (LA), right atrium (RA), aorta (Ao) and pulmonary artery (PA).
(PDF)
The image shows the segmentation for the components included in the final tetrahedral mesh for cases 05 to 08 overlapped with the end-diastolic CT image. For each case, we show a four-chamber axial view, two sagittal and two coronal views. Abbreviations: left ventricle (LV), right ventricle (RV), left atrium (LA), right atrium (RA), aorta (Ao) and pulmonary artery (PA).
(PDF)
The image shows the segmentation for the components included in the final tetrahedral mesh for cases 09 to 12 overlapped with the end-diastolic CT image. For each case, we show a four-chamber axial view, two sagittal and two coronal views. Abbreviations: left ventricle (LV), right ventricle (RV), left atrium (LA), right atrium (RA), aorta (Ao) and pulmonary artery (PA).
(PDF)
The image shows the segmentation for the components included in the final tetrahedral mesh for cases 13 to 16 overlapped with the end-diastolic CT image. For each case, we show a four-chamber axial view, two sagittal and two coronal views. Abbreviations: left ventricle (LV), right ventricle (RV), left atrium (LA), right atrium (RA), aorta (Ao) and pulmonary artery (PA).
(PDF)
The image shows the segmentation for the components included in the final tetrahedral mesh for cases 17 to 20 overlapped with the end-diastolic CT image. For each case, we show a four-chamber axial view, two sagittal and two coronal views. Abbreviations: left ventricle (LV), right ventricle (RV), left atrium (LA), right atrium (RA), aorta (Ao) and pulmonary artery (PA).
(PDF)
The image shows the segmentation for the components included in the final tetrahedral mesh for cases 21 to 24 overlapped with the end-diastolic CT image. For each case, we show a four-chamber axial view, two sagittal and two coronal views. Abbreviations: left ventricle (LV), right ventricle (RV), left atrium (LA), right atrium (RA), aorta (Ao) and pulmonary artery (PA).
(PDF)
A The plot shows error bars (mean ± standard deviation) of the mesh quality over time for a free contraction simulation for mesh 03. The other plots show mesh quality variations over simulation time for different regions of the mesh: B LV, C RV, D LA and E RA. Average mesh quality decreases from 0.74 to a minimum of 0.63, reached at maximum ventricular contraction. Change in mesh quality is driven by the LV and the RV, for which the average quality decreases of 16% and 20% from ED to end-systole, respectively. LA and the RA mesh quality, which are not actively contracting in the simulation and are therefore passive, underwent small variations (4% and 3%, respectively). The same applied to all the other labels of the mesh (results not shown).
(PDF)
(PDF)
(PDF)
Data Availability Statement
The data are uploaded to Zenodo and publicly accessible via the following URL: https://doi.org/10.5281/zenodo.3890034.