Abstract
Zebra2 is a highly automated web-tool to search for subfamily-specific and conserved positions (i.e. the determinants of functional diversity as well as the key catalytic and structural residues) in protein superfamilies. The bioinformatic analysis is facilitated by Mustguseal—a companion web-server to automatically collect and superimpose a large representative set of functionally diverse homologs with high structure similarity but low sequence identity to the selected query protein. The results are automatically prioritized and provided at four information levels to facilitate the knowledge-driven expert selection of the most promising positions on-line: as a sequence similarity network; interfaces to sequence-based and 3D-structure-based analysis of conservation and variability; and accompanied by the detailed annotation of proteins accumulated from the integrated databases with links to the external resources. The integration of Zebra2 and Mustguseal web-tools provides the first of its kind out-of-the-box open-access solution to conduct a systematic analysis of evolutionarily related proteins implementing different functions within a shared 3D-structure of the superfamily, determine common and specific patterns of function-associated local structural elements, assist to select hot-spots for rational design and to prepare focused libraries for directed evolution. The web-servers are free and open to all users at https://biokinet.belozersky.msu.ru/zebra2, no login required.
INTRODUCTION
During evolution of proteins from a common ancestor, one functional property may be preserved, while others may vary as a result of mutations accumulated within a shared 3D-structure, leading to functional diversity. Comparative bioinformatic analysis of homologs implementing different properties within the superfamily can help to understand the relationship between protein sequence/structure and its biological function. Highly conserved positions are not subjected to evolution and play the functional and/or structural role common for the entire superfamily. The specific positions – also known as ‘subfamily-specific’ (1) or ‘family-specific’ (2) positions (SSPs, FSPs), ‘specificity-determining residues/sites/positions’ or SDRs/SDSs/SDPs (3) etc.—that are conserved only within families/subfamilies, but are different between them, seem to play an important role in functional diversity of homologs. Information on both conserved and specific positions can help to understand how the enzyme performs its inherent function, while the latter can also be selected as hotspots for mutation in an attempt to improve the wild-type protein for a particular purpose (4–7). Identification of the conserved positions is relatively straightforward and represents an efficient approach to predict the key functionally important residues from sequence information alone (8). Selection of the specific positions is more complicated. Several algorithms to identify SSPs/SDPs in a multiple sequence alignment of homologs have been developed (3,7,9–13). These were advertised as helpful tools to study functional diversity and eventually, as one way of practical application, to select hot-spots for protein engineering (4,5,8). The rationale for using the SSPs/SDPs for protein design was very promising and far reaching: first, these positions highlight hot-spots in protein structures that can accommodate different amino acid types (i.e. as in homologs sharing a common structural fold); second, they present a particular experimentally testable hypotheses—i.e. how to switch residues in these positions between functionally diverse protein families/subfamilies to implement the respective properties (5,6,14). In this context, the original Zebra algorithm/web-server to identify SSPs/SDPs (9,10) was recognized as a tool (15,16) to help studying the structure-function relationship in various protein superfamilies and used to assist the design of improved enzymes (e.g. 2,17–20). Despite the potential of bioinformatic analysis in protein studies and engineering (6,21), the stochastic approaches (e.g. the ‘directed evolution’) that require a very little prior knowledge of the protein system of interest remain the most widely used methods to engineer novel enzymes with improved properties as of today (22). Probably, the three main reasons for this can be summarized as follows. First, the available SSP/SDP-prediction tools are provided either as web-servers with a rather limited functionality or standalone command-line programs that require specialized training and skills in computational biology. All these tools put the burden of constructing the input multiple alignment of diverse protein families on the end-user, what presents a significant methodological (23,24) and computational (25) challenges. Second, although the bioinformatic tools implement advanced algorithms and perform well in benchmarking, their predictions (i.e. the point mutations in SSPs/SDPs suggested for experimental evaluation) do not always work in practice, highlighting the fact that complex structure–function relationship in proteins is still poorly understood. Such computationally designed enzymes can display poor catalytic activities, and have to be further improved by means of stochastic techniques such as directed evolution (26). A constructive solution would be a knowledge-driven expert selection of the most promising positions based on the automatically prioritized list of ‘raw’ predictions prepared by the web-server. To facilitate such analysis the results should be provided in a way that would be most convenient for the researcher. However, in practice, the output from the available tools is not presented in an intuitive way, e.g. as plain text files or simple HTML tables with a list of identified SSPs/SDPs and their statistical scores. Finally, all currently available web-methods were designed to handle relatively small alignments, and thus are of limited productivity at large-scale analysis of families/superfamilies. The available SSP/SDP-prediction tools have made a significant contribution in the field, but as the protein sequence and structure databases demonstrate a continuous growth, the new bioinformatic solutions have to be developed to address new challenges and opportunities.
We have developed Zebra2—a major upgrade from the original Zebra web-server (10)—focused on improving usability by integrating with the companion web-server to automatically construct large alignments of functionally diverse protein families and improving the quality of bioinformatic predictions by implementing a versatile analysis toolkit to study the results on-line.
MATERIALS AND METHODS
Algorithm and parameters
The Zebra2 web-server implements the original Zebra algorithm (9) featuring the relative entropy-based specificity scoring function and random shuffling (by default, 1000 random permutations are performed by shuffling amino acid content within each column and retaining subfamilies in their original proportions) to calculate the statistical significance P-values using the Bernoulli ‘B-cutoff’ estimator (3). The sequence conservation is assessed using the Valdar and Thornton measure (27). The initially calculated specificity scores for each position are corrected to improve the ranking of those SSPs that assemble into clusters with other subfamily-specific and conserved positions in the representative protein 3D-structure (i.e. the ‘3D-mode’; by default, all neighboring positions within 4 Å are considered). The Zebra can automatically assign proteins to functional subfamilies by graph-based clustering at different sequence similarity levels that are selected to produce the most different classifications (9). Alternatively, the CD-HIT clustering algorithm can be implemented for a particular purpose to classify proteins into subfamilies by 'cutting’ the sequence identity graph at the user-selected level (28). The SSP/SDP prediction accuracy of Zebra was previously evaluated on a set of 435 enzyme families with diverse functional properties (9). The precision–recall analysis manifested that Zebra is competitive with other tools. The Zebra2 parameters are further discussed on-line at https://biokinet.belozersky.msu.ru/zebra-parameters.
Input
The default input to Zebra2 (i.e. the Mode 1) is (i) a PDB file of the query protein and (ii) a FASTA alignment of its homologs. The choice of the query should be based on the research objective, e.g. you can select a protein that is available for the experimental site-directed mutagenesis to improve its properties or to study the key functional residues. The input for Zebra2 can be prepared automatically and on-line using the Mustguseal tool (24), as further discussed below and on-line at https://biokinet.belozersky.msu.ru/zebra-input. Availability of a 3D-structure is not mandatory when using Zebra2. In the Mode 2, the web-server accepts a multiple sequence alignment in the FASTA format as the sole input. In this mode, the on-line analysis toolkit will be partially disabled (see ‘Output’). Generally, the Zebra2 analysis is provided on a one-chain-at-a-time basis, i.e. unequal subunits of heteromeric proteins and corresponding multiple alignments should be submitted as independent tasks. There are two reasons for this: (i) specificity/conservation is a characteristic of an individual alignment column, thus simultaneous analysis of all protein chains at once is not required, but significantly complicates the input data preparation; (ii) unequal chains of heteromeric proteins can represent domains with different evolutionary history among homologs; therefore, it is most reasonable to study their functional specificity independently.
The Mustguseal web-server can be used to automatically collect and superimpose a large representative set of functionally diverse homologs with high structural, but low sequence similarity to the selected query protein. Click on the ‘Mustguseal it NOW’ button at the Zebra2 input page to load the Mustguseal web-interface with a predefined set of parameters (i.e. the Scenario 2). Provide the PDB code (29) and chain identifier of the selected query protein to start the alignment construction process. In the first step, the evolutionarily distant relatives of the query are selected by searching for similar 3D-structures in the PDB database (i.e., the representative proteins, see details below). In the second step of the Mustguseal protocol, the sequence similarity search is used to reveal close homologs of the collected representative proteins in the UniProtKB/Swiss-Prot+TrEMBL databases. At most 500 proteins with at most 90% pairwise sequence identity are collected per each sequence similarity search, followed by the ‘Dissimilarity filter’ to eliminate outliers sharing less than 0.5 bit-score-per-column with the corresponding representative protein. The collected non-redundant set of sequences of functionally diverse evolutionarily distant homologs is finally superimposed according to the 3D-alignment of representative proteins, as recently discussed (24).
The representative proteins are selected by the Mustguseal in the first step of the protocol based on the percentage of secondary structure shared by the query protein and each entry in the PDB database, and define the scope and diversity of the final alignment. Selection of threshold values for this step should be based on the research objective and structural organization of the protein family of interest. By default, a general-purpose ‘70%–70%’ pair of thresholds is applied (30) to collect evolutionarily distant proteins representing functionally diverse families, yet similar enough for alignment, as recently discussed (24). By setting the thresholds below or above the default values the user can consider evolutionarily more distant proteins or limit the scope of the alignment only to close homologs, respectively. Non-symmetric thresholds should be used to study protein superfamilies featuring non-uniform domain organization (e.g. modular neuraminidases, as recently discussed (31)).
Click on the ‘Submit to Zebra2’ button at the Mustguseal results page to submit the output alignment and the PDB structure of the selected query to the Zebra2 web-server for further bioinformatic analysis. The accession numbers of the collected proteins will be embedded into the alignment file for further use by the Zebra2 to retrieve the corresponding annotation data from the internally stored PDB (29) and UniProt (32), as well as to generate hyper-links to the related pages in PDB (29), UniProt (32), BRENDA (33) and BacDive (34).
Output
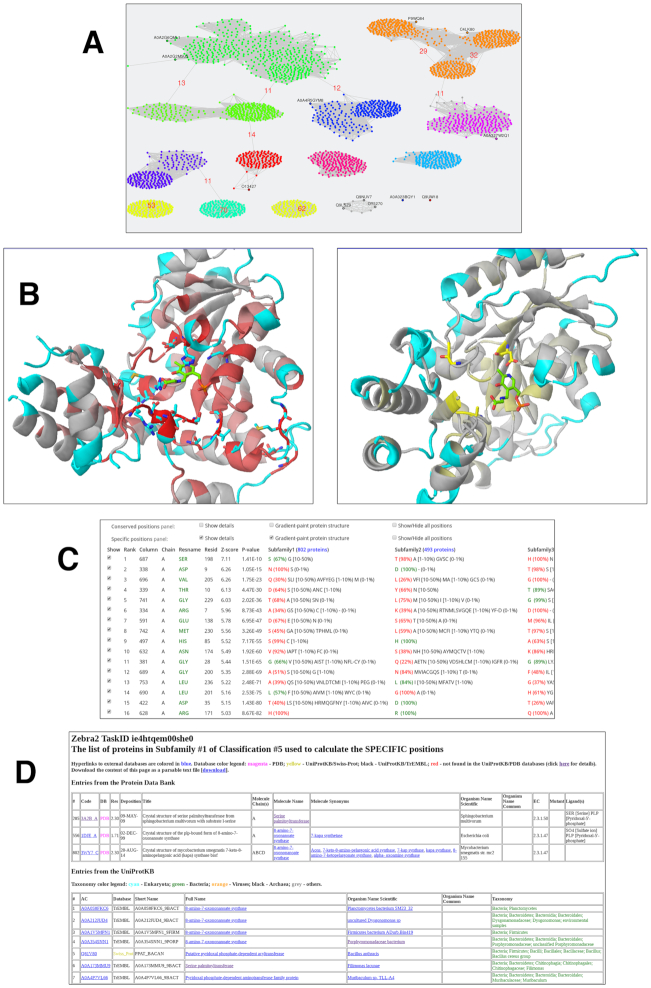
The web-server provides a detailed on-line log of all its activities, available immediately after the task is accepted, and interactively updated while processing the user data. Users are advised to always check this on-line log for warnings and errors. In particular, if a task fails, this on-line log will contain a description of the problem. If successful, the following output will be created: the first output is a list of automatically proposed classifications of proteins into specificity groups; the second output is a list of conserved and specific positions for each proposed classification. The identified conserved and specific positions are automatically prioritized (i.e., ranked) according to the statistical significance as previously discussed (9). The automatically proposed classifications are ranked in declining order of statistical significance of the SSPs/SDPs they produce. The most significant positions/classifications are ranked first. The Zebra2 output is primarily web-based and can be studied on-site. In the Mode 1 (i.e. when multiple alignment and a representative 3D-structure are submitted to the web-server), the results are presented at four information levels: sequence similarity network can be used to evaluate the automatically proposed classifications and study functional trends across protein superfamilies from the context of sequence similarity within and between the functional families/subfamilies (35) (Figure 1A); interfaces to 3D-structure-based and sequence-based analysis can be used to highlight the most conserved and specific parts of the common fold, as well as to visualize the selected conserved and subfamily-specific positions in the structure of the query protein (Figure 1B and C); a detailed annotation of proteins featuring links to the external databases (i.e., PDB (29), UniProt (32), BRENDA (33) and BacDive (34)) can help to interpret the results of the bioinformatic analysis (Figure 1D), as further explained in the Results section. In the Mode 2 (i.e. when multiple alignment is provided as the sole input to the web-server) only the sequence similarity network analysis is available on-line. The Zebra2 results are also available for download as text files (in both Modes 1 and 2) and binary 3D-annotations for PyMOL Molecular Graphics System (only in the Mode 1). The online interactivity is implemented in HTML5 using JSmol (36) and Cytoscape (37) plug-ins. The layout of the sequence similarity network is defined by the Compound Spring Embedder algorithm of the Cytoscape.js library using pairwise sequence identities of at least 40%. Illustrated user manual describing the Zebra2 output is available at https://biokinet.belozersky.msu.ru/zebra-output.
Figure 1.
The on-line interactive analysis toolkit is the hallmark of Zebra2. The results are provided at four information levels: (A) as a sequence similarity network; interfaces to (B) 3D-structure-based and (C) sequence-based analysis of conservation and variability; and (D) accompanied by the detailed annotation of proteins accumulated from the integrated databases with links to the external resources (i.e. PDB, UniProt, BacDive, BRENDA). In (B), the gradient paint indicates the statistical significance of the subfamily-specific (red-to-cyan) and conserved (yellow-to-grey) positions. To operate this example on-line use the ‘Demo mode (PLP)’.
RESULTS
The Zebra2+Mustguseal is a new highly automated web-tool featuring an intuitive and easy-to-use interface to identify the subfamily-specific and conserved positions in diverse protein superfamilies. The input and output can be created, processed and studied on-site via the web-interface. To alleviate the burden on the end-user of identifying suitable homologs, the Zebra2 web-server connects with a companion Mustguseal web-server that handles the construction of large multiple alignments of protein families based on all available information about their 3D-structures and sequences in public databases (see ‘Input’ in Methods). It takes just a few clicks, starting from the Zebra2 landing page, to create an alignment of up to thousands of proteins and subject it to a comprehensive bioinformatic analysis. At the same time, separate interfaces to the two bioinformatic pipelines provide the opportunity to customize Mustguseal and Zebra2 for a particular research objective. The Zebra2 output can be studied on-site or downloaded to a local desktop station. Implementation of the on-line interactive analysis toolkit is the hallmark of Zebra2, i.e., the results are presented at four information levels in a way that should be the most convenient to a human expert (see ‘Output’ in Methods). The Zebra2 can help to study the patterns of local structure associated with a function/property specific to a particular family/subfamily or common among all homologs (4), and assist to select hot-spots for enzyme engineering (5,14–16): to improve stereo- and substrate specificity, increase stability or introduce novel catalytic activity into the query protein (e.g. 1, 2, 17, 20). The key limitations of the previously developed SSP/SDP-prediction tools were outlined in the Introduction. A detailed comparison of the new Zebra2 with the Zebra v.1 (10), as well as other available web-servers Multi-Harmony (12), SDPpred (3), SPEER-SERVER (11) and Spial (13) is provided as a Supplementary Data (Supplementary Table S1, Supplementary Figure S1). We further discuss a recent case-study where the positions selected by the Zebra bioinformatic analysis algorithm were studied experimentally. The Zebra2 examples are also available on-line at https://biokinet.belozersky.msu.ru/zebra-examples.
The fold type I pyridoxal-5′-phosphate (PLP)-dependent enzymes represent a prime example for the natural diversity of catalytic activities that evolved within one structural scaffold (38). Bioinformatic analysis of this superfamily was carried out using Zebra2+Mustguseal to demonstrate the potential of the new tool to explore the structure-function relationship in proteins and design novel enzymes. This example is qualitatively similar to a recent study of the determinants of reaction specificity and catalytic promiscuity of l-threonine aldolase from Aeromonas jandaei or LTAaj (2). To operate this example on-line you can request the ‘Demo mode (PLP)’ at the Zebra2 submission page. The PDB code 3WGB (chain A) of LTAaj was submitted to the Mustguseal to automatically construct the alignment of a representative set of 4185 homologs with high structure similarity but low sequence identity to the query protein. The LTAaj was selected as the query because it was available for the experimental site-directed mutagenesis in that study (2). The automatically constructed alignment was then subjected to analysis by the Zebra2. The proteins in input alignment were automatically assigned to 11 specificity groups by maximizing the statistical significance of the corresponding specific positions (Figure 1A). The analysis of the available functional annotation of members in the predicted specificity groups (e.g. Figure 1D) revealed that they correspond to families of the fold-type I PLP-dependent enzymes with different reaction specificities (e.g., Threonine aldolases, Cysteine desulfurases, Threonine-phosphate decarboxylases, Tyrosine phenol-lyases, Glutamine aminotransferases etc.). The identified conserved positions Gly143, Asp168 and Lys199 (Figure 1B and C) were previously shown to be catalytically important and involved in the cofactor-binding in the superfamily, and their mutation greatly reduced the enzyme catalytic activity (2). The roles of the identified specific positions (Figure 1B and C) in the catalytic mechanism and reaction specificity of LTAaj can be further studied by implementing the knowledge-driven expert analysis, molecular modelling, and experimental site-directed mutagenesis, as recently discussed (2). In that study, it was shown that specific positions conserved within functional families, but different between them, determine the reaction specificity of LTAaj by coordinating the PLP cofactor, thus affecting its activation and tautomeric equilibrium of the catalytic intermediates. Mutagenesis at the positions identified by the bioinformatic analysis in LTAaj reduced the native aldol activity and facilitated promiscuous reactions, i.e., substitutions at the selected family-specific positions increased the transaminase activity up to 6-fold and introduced a novel alanine racemase activity (2). We conclude that the specific positions identified by the bioinformatic analysis can be employed to study the structural basis of functional diversity and implemented as hot-spots to design enzymes with novel properties, while the conserved positions as ‘irreplaceable’ should probably be excluded from the list of potential mutation sites for protein engineering.
DISCUSSION
Bioinformatic analysis has a potential of becoming a particularly important tool in protein engineering for systematic analysis and study of the ever increasing volumes of protein sequences and 3D-structures attributed to functionally diverse superfamilies. The common problem of many bioinformatic programs/algorithms is that they were advertised as helpful tools to assist experimental studies, but usually required profound skills in computational biology, what impeded practical use by many investigators in a daily laboratory practice. The recent trend is to provide advanced computational methods directly to the wet-lab scientists by developing easy-to-use tools featuring content-rich interactive on-line output (23,39–41).
The Zebra2 is an advanced web-server to study the patterns of conservation and variability of amino acid residues in a functionally diverse superfamily using a web-browser as the only mandatory tool. The first novelty of the Zebra2 is integration with the recently introduced Mustguseal engine to automatically collect and align functionally diverse homologs with high structural, but low sequence similarity to the query protein of interest (24). The second novelty is implementation of the interactive analysis toolkit to present the results at four information levels and thereby facilitate the knowledge-driven expert selection of the most promising positions on-line. Mustguseal and Zebra2 can be used to collect and study the currently available sequence variants within a common 3D-structural fold of a superfamily, to identify conserved positions that play common functional and structural role shared by all homologs, and to reveal specific residues responsible for diversity. The value of positions selected by the bioinformatic analysis can be assessed by an expert and followed by experimental site-directed mutagenesis, or can be studied by molecular modelling to reveal the mechanisms of their involvement in a protein function (2,14,26,42). Generally speaking, conserved positions represent protein sites where no changes were allowed during the evolution, as only the particular amino acid residues were able to provide the necessary structure and function. Such invariant positions in a large superfamily should probably be excluded from the list of potential mutation sites for protein engineering, as their replacement is known to diminish the catalytic activity (e.g. 2). On the other side, the subfamily-specific positions are determinants of functional diversity within a common structural fold and present an experimentally testable hypothesis to engineer biocatalysts (4,5,8,14).
The integration of web-based bioinformatic tools Zebra2 and Mustguseal provides an out-of-the-box easy-to-use solution, first of its kind, to identify subfamily-specific and conserved positions in protein superfamilies—to study function-associated patterns of local structure, to assist at selecting hot-spots for rational design, and to prepare focused libraries for directed evolution. We hope the new Zebra2+Mustguseal integrated web-tool will further bridge the gap between methods of advanced computational biology and experimental studies, thus promoting the value of bioinformatic analysis in protein engineering.
Supplementary Material
ACKNOWLEDGEMENTS
The use of HPC computing resources at the Lomonosov Moscow State University supported by the project RFMEFI62117X0011 to prepare the PDB and UniProt databases in a format compatible with Mustguseal and Zebra2 is acknowledged (43).
Contributor Information
Dmitry Suplatov, Lomonosov Moscow State University, Belozersky Institute of Physicochemical Biology and Faculty of Bioengineering and Bioinformatics, Lenin Hills 1-73, Moscow 119234, Russia.
Yana Sharapova, Lomonosov Moscow State University, Belozersky Institute of Physicochemical Biology and Faculty of Bioengineering and Bioinformatics, Lenin Hills 1-73, Moscow 119234, Russia.
Elizaveta Geraseva, Lomonosov Moscow State University, Belozersky Institute of Physicochemical Biology and Faculty of Bioengineering and Bioinformatics, Lenin Hills 1-73, Moscow 119234, Russia.
Vytas Švedas, Lomonosov Moscow State University, Belozersky Institute of Physicochemical Biology and Faculty of Bioengineering and Bioinformatics, Lenin Hills 1-73, Moscow 119234, Russia.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Russian Foundation for Basic Research according to the research project [18-29-13060]. Funding for open access charge: Russian Foundation for Basic Research [18-29-13060].
Conflict of interest statement. None declared.
REFERENCES
- 1. Suplatov D.A., Besenmatter W., Švedas V.K., Svendsen A.. Bioinformatic analysis of alpha/beta-hydrolase fold enzymes reveals subfamily-specific positions responsible for discrimination of amidase and lipase activities. Protein Eng. Des. Sel. 2012; 25:689–697. [DOI] [PubMed] [Google Scholar]
- 2. Fesko K., Suplatov D., Švedas V.. Bioinformatic analysis of the fold type I PLP−dependent enzymes reveals determinants of reaction specificity in I−threonine aldolase from Aeromonas jandaei. FEBS Open Biol. 2018; 8:1013–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kalinina O.V., Novichkov P.S., Mironov A.A., Gelfand M.S., Rakhmaninova A.B.. SDPpred: a tool for prediction of amino acid residues that determine differences in functional specificity of homologous proteins. Nucleic Acids Res. 2004; 32:W424–W428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Suplatov D., Kirilin E., Švedas V.. Svendsen A. Bioinformatic analysis of protein families to select function-related variable positions. Understanding Enzymes: Function, Design, Engineering, and Analysis. 2016; Singapore, Pan Stanford Publishing; 351–385. [Google Scholar]
- 5. Pleiss J. Systematic analysis of large enzyme families: identification of specificity−and selectivity−determining hotspots. ChemCatChem. 2014; 6:944–950. [Google Scholar]
- 6. De Juan D., Pazos F., Valencia A.. Emerging methods in protein co-evolution. Nat. Rev. Genet. 2013; 14:249–261. [DOI] [PubMed] [Google Scholar]
- 7. Casari G., Sander C., Valencia A.. A method to predict functional residues in proteins. Nat. Struct. Biol. 1995; 2:171. [DOI] [PubMed] [Google Scholar]
- 8. Chagoyen M., García-Martín J.A., Pazos F.. Practical analysis of specificity-determining residues in protein families. Brief. Bioinform. 2016; 17:255–261. [DOI] [PubMed] [Google Scholar]
- 9. Suplatov D., Shalaeva D., Kirilin E., Arzhanik V., Švedas V.. Bioinformatic analysis of protein families for identification of variable amino acid residues responsible for functional diversity. J. Biomol. Struct. Dyn. 2014; 32:75–87. [DOI] [PubMed] [Google Scholar]
- 10. Suplatov D., Kirilin E., Takhaveev V., Švedas V.. Zebra: a web server for bioinformatic analysis of diverse protein families. J. Biomol. Struct. Dyn. 2014; 32:1752–1758. [DOI] [PubMed] [Google Scholar]
- 11. Chakraborty A., Mandloi S., Lanczycki C.J., Panchenko A.R., Chakrabarti S.. SPEER-SERVER: a web server for prediction of protein specificity determining sites. Nucleic Acids Res. 2012; 40:W242–W248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brandt B.W., Feenstra K.A., Heringa J.. Multi-harmony: detecting functional specificity from sequence alignment. Nucleic Acids Res. 2010; 38:W35–W40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wuster A., Venkatakrishnan A.J., Schertler G.F., Babu M.M.. Spial: analysis of subtype-specific features in multiple sequence alignments of proteins. Bioinformatics. 2010; 26:2906–2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Suplatov D., Voevodin V., Švedas V.. Robust enzyme design: bioinformatic tools for improved protein stability. Biotechnol. J. 2015; 10:344–355. [DOI] [PubMed] [Google Scholar]
- 15. Romero-Rivera A., Garcia-Borràs M., Osuna S.. Computational tools for the evaluation of laboratory-engineered biocatalysts. Chem. Commun. 2017; 53:284–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Damborsky J., Brezovsky J.. Computational tools for designing and engineering enzymes. Curr. Opin. Chem. Biol. 2014; 19:8–16. [DOI] [PubMed] [Google Scholar]
- 17. Demming R.M., Hammer S.C., Nestl B.M., Gergel S., Fademrecht S., Pleiss J., Hauer B.. Asymmetric enzymatic hydration of unactivated, aliphatic alkenes. Angew. Chem. Int. Ed. Engl. 2019; 58:173–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cao T.P., Choi J.M., Kim S.W., Lee S.H.. The crystal structure of methanol dehydrogenase, a quinoprotein from the marine methylotrophic bacterium methylophaga aminisulfidivorans MPT. J. Microbiol. 2018; 56:246–254. [DOI] [PubMed] [Google Scholar]
- 19. Popinako A., Antonov M., Tikhonov A., Tikhonova T., Popov V.. Structural adaptations of octaheme nitrite reductases from haloalkaliphilic Thioalkalivibrio bacteria to alkaline pH and high salinity. PLoS One. 2017; 12:e0177392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Suplatov D., Panin N., Kirilin E., Shcherbakova T., Kudryavtsev P., Švedas V.. Computational design of a pH stable enzyme: understanding molecular mechanism of penicillin acylase's adaptation to alkaline conditions. PLoS One. 2014; 9:e100643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Malhis N., Jones S.J., Gsponer J.. Improved measures for evolutionary conservation that exploit taxonomy distances. Nat. Commun. 2019; 10:1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang K.K., Wu Z., Arnold F.H.. Machine-learning-guided directed evolution for protein engineering. Nat. Methods. 2019; 16:687–694. [DOI] [PubMed] [Google Scholar]
- 23. Rozewicki J., Li S., Amada K.M., Standley D.M., Katoh K.. MAFFT-DASH: integrated protein sequence and structural alignment. Nucleic Acids Res. 2019; 47:W5–W10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Suplatov D.A., Kopylov K.E., Popova N.N., Voevodin V.V., Švedas V.K.. Mustguseal: a server for multiple structure-guided sequence alignment of protein families. Bioinformatics. 2018; 34:1583–1585. [DOI] [PubMed] [Google Scholar]
- 25. Suplatov D., Sharapova Y., Shegay M., Popova N., Fesko K., Voevodin V., Švedas V.. Voevodin V, Sobolev S. High-performance hybrid computing for bioinformatic analysis of protein superfamilies. Communications in Computer and Information Science. 2019; Basel: Springer Nature Switzerland AG; 249–264. [Google Scholar]
- 26. Maria-Solano M.A., Serrano-Hervás E., Romero-Rivera A., Iglesias-Fernández J., Osuna S.. Role of conformational dynamics in the evolution of novel enzyme function. Chem. Commun. 2018; 54:6622–6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Valdar W.S., Thornton J.M.. Protein–protein interfaces: analysis of amino acid conservation in homodimers. Proteins. 2001; 42:108–124. [PubMed] [Google Scholar]
- 28. Fu L., Niu B., Zhu Z., Wu S., Li W.. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012; 28:3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Burley S.K., Berman H.M., Bhikadiya C., Bi C., Chen L., Di Costanzo L., Christie C., Dalenberg K., Duarte J.M., Dutta S. et al.. RCSB protein data bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2019; 47:D464–D474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Krissinel E., Henrick K.. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 2004; 60:2256–2268. [DOI] [PubMed] [Google Scholar]
- 31. Sharapova Y., Suplatov D., Švedas V.. Neuraminidase a from Streptococcus pneumoniae has a modular organization of catalytic and lectin domains separated by a flexible linker. FEBS J. 2018; 285:2428–2445. [DOI] [PubMed] [Google Scholar]
- 32. UniProt Consortium UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019; 47:D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jeske L., Placzek S., Schomburg I., Chang A., Schomburg D.. BRENDA in 2019: a European ELIXIR core data resource. Nucleic Acids Res. 2019; 47:D542–D549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Reimer L.C., Vetcininova A., Carbasse J.S., Söhngen C., Gleim D., Ebeling C., Overmann J.. BacDive in 2019: bacterial phenotypic data for High-throughput biodiversity analysis. Nucleic Acids Res. 2019; 47:D631–D636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zallot R., Oberg N., Gerlt J.A.. The EFI web resource for genomic enzymology tools: leveraging protein, genome, and metagenome databases to discover novel enzymes and metabolic pathways. Biochemistry. 2019; 58:4169–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hanson R.M., Prilusky J., Renjian Z., Nakane T., Sussman J.L.. JSmol and the next-generation web−based representation of 3D molecular structure as applied to proteopedia. Isr. J. Chem. 2013; 53:207–216. [Google Scholar]
- 37. Otasek D., Morris J.H., Bouças J., Pico A.R., Demchak B.. Cytoscape automation: empowering workflow-based network analysis. Genome Biol. 2019; 20:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Voß M., Xiang C., Esque J., Nobili A., Menke M.J., André I., Höhne M., Bornscheuer U.T.. Creation of (R)-amine transaminase activity within an α-amino acid transaminase scaffold. ACS Chem. Biol. 2020; 15:416–424. [DOI] [PubMed] [Google Scholar]
- 39. Suplatov D., Timonina D., Sharapova Y., Švedas V.. Yosshi: a web-server for disulfide engineering by bioinformatic analysis of diverse protein families. Nucleic Acids Res. 2019; 47:W308–W314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sumbalova L., Stourac J., Martinek T., Bednar D., Damborsky J.. HotSpot Wizard 3.0: web server for automated design of mutations and smart libraries based on sequence input information. Nucleic Acids Res. 2018; 46:W356–W362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Buß O., Buchholz P.C., Gräff M., Klausmann P., Rudat J., Pleiss J.. The ω−transaminase engineering database (oTAED): a navigation tool in protein sequence and structure space. Proteins. 2018; 86:566–580. [DOI] [PubMed] [Google Scholar]
- 42. Pirhadi S., Sunseri J., Koes D.R.. Open source molecular modeling. J. Mol. Graph. Model. 2016; 69:127–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sadovnichy V., Tikhonravov A., Voevodin V., Opanasenko V., Vetter J.S.. Lomonosov: supercomputing at moscow state university. Contemporary High Performance Computing: From Petascale Toward Exascale (Chapman & Hall/CRC Computational Science). 2013; Boca Raton, CRC Press; 283–307. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.