Abstract
Inferring homology relationships across genes in different species is a central task in comparative genomics. Therefore, a large number of resources and methods have been developed over the years. Some public databases include phylogenetic trees of homologous gene families which can be used to further differentiate homology relationships into orthology and paralogy. MetaPhOrs is a web server that integrates phylogenetic information from different sources to provide orthology and paralogy relationships based on a common phylogeny-based predictive algorithm and associated with a consistency-based confidence score. Here we describe the latest version of the web server which includes major new implementations and provides orthology and paralogy relationships derived from ∼8.2 million gene family trees—from 13 different source repositories across ∼4000 species with sequenced genomes. MetaPhOrs server is freely available, without registration, at http://orthology.phylomedb.org/
INTRODUCTION
Accurate prediction of orthology is central to comparative genomics. Although many orthology prediction tools have been developed, many users rely on pre-computed relationships for model and non-model organisms that are provided by a growing number of orthology databases (1). Homologous genes can be orthologs or paralogs, depending on whether they diverged from their common ancestor through speciation or duplication, respectively, and this is best inferred through phylogenetic analysis (2,3). Hence phylogenetic trees derived from sets of homologous genes (i.e. gene trees) can be used to infer orthology and paralogy relationships and can help uncover relevant evolutionary events. In contrast to popular similarity network-based approaches that build clusters of orthologous genes comprising orthologs and in-paralogs, phylogeny-based inference of orthology and paralogy provides pair-wise homology relationships and is able to reconstruct complex patterns of co-orthology (i.e. one-to-many or many-to-many relationships (3).
Different databases provide access to thousands of gene trees across different taxa, but they often do not provide specific orthology or paralogy information. If they do, different parameters and methods are used which may result in incongruent results that are difficult to compare by the end-user. MetaPhOrs (Meta Phylogeny Based Orthologs) fills this gap by integrating phylogenetic information derived from different databases into a single framework for the inference of orthology and paralogy relationships (4). MetaPhOrs uses the species-overlap orthology prediction algorithm (2,5) over gene trees retrieved from heterogeneous sources and integrates the resulting orthology and paralogy pairwise relationships using a consistency-based approach (4). Hence, MetaPhOrs serves as a global repository of highly accurate, phylogeny-based orthology and paralogy predictions that is easily accessible for non-expert users. The accuracy of orthology predictions provided by MetaPhOrs has been benchmarked alongside other methods, showing the superiority of the integrative approach over the individual methods or databases (4,6). Since its first release a decade ago, MetaPhOrs, has been regularly updated and expanded and here we describe the features and characteristics of the last and major update to the server, which provides significantly increased functionality.
MATERIALS AND METHODS
Web server
The MetaPhOrs web server is currently hosted at the Barcelona Supercomputing Center (BSC-CNS) and runs on Ubuntu Linux with 16 GB memory. Other significant software packages used include: Apache (version 2.4.18, https://httpd.apache.org), PHP (version 7.0.15, http://www.php.net), MariaDB (version 10.2.22, htps://mariadb.org/), ETE (version 3.1.1, (7)) and Python (version 3.7.4, https://www.python.org/) on the backend. The client-side user interface was implemented using Drupal (version 7.69), SQLite (version 3.7.3) and JavaScript libraries, including jQuery (version 1.11.0). Page tracking is provided by Google Analytics (http://www.google.com/analytics/).
Overview of the consistency score
Combined orthology/paralogy assignment in MetaPhOrs is based on the consistency score (CS). CS defines the overall agreement of source gene trees about a given prediction and it ranges from 0 to 1, the closer the value of CS to 1, the more confident the prediction. Further details on how CS is calculated, can be found in the original MetaPhOrs' publication (4). In addition, MetaPhOrs assigns an Evidence Level (EL) to each prediction, defined as the number of independent sources used for inferring this prediction.
METAPHORS WEB SERVER DESCRIPTION
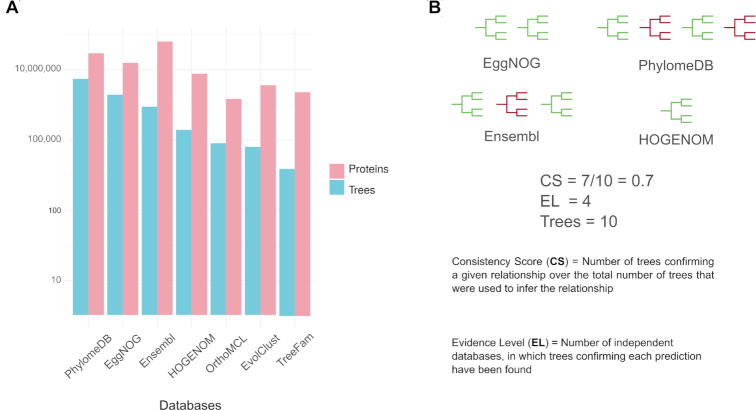
MetaPhOrs is a public repository of phylogeny-based orthologs and paralogs that were computed using phylogenetic trees available in several repositories. In addition, MetaPhOrs computes ∼77 000 gene trees for OrthoMCL repository using the PhylomeDB pipeline (8) (Figure 1A). The MetaPhOrs web server has undergone several upgrades since its first release in 2010. The current version represents a major upgrade of the web interface and a significant expansion of the taxonomic scope covered by the web server. In this new version, orthology and paralogy predictions were computed from data available in 13 different large-scale databases (Figure 1A). These orthology and paralogy predictions for ∼117 million proteins (Figure 1A) were inferred based on the analyses of ∼8.2 million maximum-likelihood (ML) trees covering ∼4000 different fully-sequenced species. For each prediction, MetaPhOrs provides a CS (defining overall agreement of source gene trees about a given prediction) and EL (informing about the number of repositories from which prediction is retrieved) describing its goodness, together with the number of trees and links to their source databases (Figure 1B).
Figure 1.
(A) Bar plot showing the contribution of the different source repositories for the total of ∼8.2 million gene family trees and for the total of ∼117 million proteins, included in the new MetaPhOrs release. Note the base-10 log scale used for the Y-axis. Ensembl databases including vertebrates, bacteria, fungi, Metazoa, Pan, plants and protists. (B) Overview of the CS, and EL used in MetaPhOrs for Orthology and Paralogy assignment. Trees in green are in agreement with gene trees about a given prediction, trees in red are not.
By dynamically interacting through a highly intuitive web interface, users can interrogate orthology and paralogy relationships for a given gene, set of genes or species, across a defined set of species or taxonomic range. The user can then visualize or explore evolutionary or functional information associated with the set of retrieved orthologs, as well as download homology tables, sequences or related information. Using the web interface, the orthology and paralogy information can be retrieved in different ways: (i) by searching for a particular gene (ii) by providing a sequence as input or (iii) by given pair of species and searching all predictions for this pair. Queries can be limited to a species, or a set of species, a source database, and the output can be filtered by levels of CS or EL values, depending on the level of stringency desired by the user.
IMPROVEMENTS IN THE NEW RELEASE
The latest update introduces major features that greatly improve the functions and usability of MetaPhOrs. Here we describe the most relevant that were implemented in this new release.
Redesign user interface
The original MetaPhOrs web server was developed using a Drupal content management system, and custom scripts in Python to process the data. But the incredible advances in the client-side languages have rendered this design obsolete, not very fast for the user and incompatible with mobile devices. Thus, we have redesigned the web interface for fast interactivity and full functionality with any of the popular browsers (e.g. Chrome, Edge, Firefox and Safari). The front-end software development uses Responsive Web Design technologies, supporting desktop/laptops computers, smartphones and tablet devices combined with a robust Drupal+JS application to manage the user interface. Furthermore, by incorporating modern software features like asynchronous javascript, we have made MetaPhOrs more secure in terms of errors, more intuitive to the users and we have improved the data retrieval performance, which will, in turn, be able to make more efficient use of the underlying database.
Multiple comparisons between species
The previous version of MetaPhOrs did not support whole-genome orthology comparisons in one search due to limitations in computational capacity and visualization methods. While the previous version of MetaPhOrs required that the user perform a different search for each gene, this new version allows retrieving genome-wide predictions of orthology or paralogy for a given pair of species.
New organisms and database update
The previous publication of MetaPhOrs (4) includes orthology and paralogy predictions across ∼800 organisms. In this main update of MetaPhOrs’ web server, we made a significant expansion of the taxonomic scope covered by the web server, which now covers ∼4000 different organisms with available genomes.
In this update of the database, we combine different large-scale databases of phylogenetic information, including the last releases of PhylomeDB (5), seven database from Ensembl Compara (including vertebrates, bacteria, fungi, Metazoa, Pan, plants and protists, release 98) (9), EggNOG (version 4.5.1) (10), TreeFam (Release 9) (11), Evolclust (version 1) (12) and Hogenom (release 6) (13). Additionally, we have reconstructed ML trees from protein families alignments stored in OrthoMCL (version 5) (14) (Figure 1A). Then, a score is assigned to each orthology and paralogy prediction based on its level of consistency across the different sources (Figure 1B).
The MetaPhOrs v2 server is running on MareNostrum 4 supercomputer at the Barcelona Supercomputing Center (www.bsc.es), which provides us with state of the art access to high storing and computing facilities. In addition, we have recently improved the pipeline to retrieve the information from the different sources and to compute the CS and EL, that will make it easier to schedule new releases.
Additional features
In addition to the main changes in this update, we have implemented other new features in order to serve the users with an easy-to-use and fast website. To explore orthologous relationships, the user can inspect the results through several interactive tables where they can filter the table (e.g. select only the results from a specific species, gene or CS) and re-order it. Furthermore, the results can be copied to the clipboard, or downloaded in several formats such as, PDF, comma-separated value, and, importantly, the OrthoXML standard format (15) which has been adopted by the Quest for Orthologs consortium to facilitate interoperability across orthology databases (1). Furthermore, users can download a multi-FASTA file with the orthologous (or paralogous) proteins in different species. Another feature added to MetaPhOrs is the addition of a History tab, that shows the last 10 searches performed by the user and the incorporation of error handling. Furthermore, we provided renewed help pages accessible through the header menu. At last, we added an FTP server that contains dumps of data that are served through the MetaPhOrs portal, which provides an archive of current and past releases, and facilitates reproducibility of studies based on MetaPhOrs data.
USE CASES
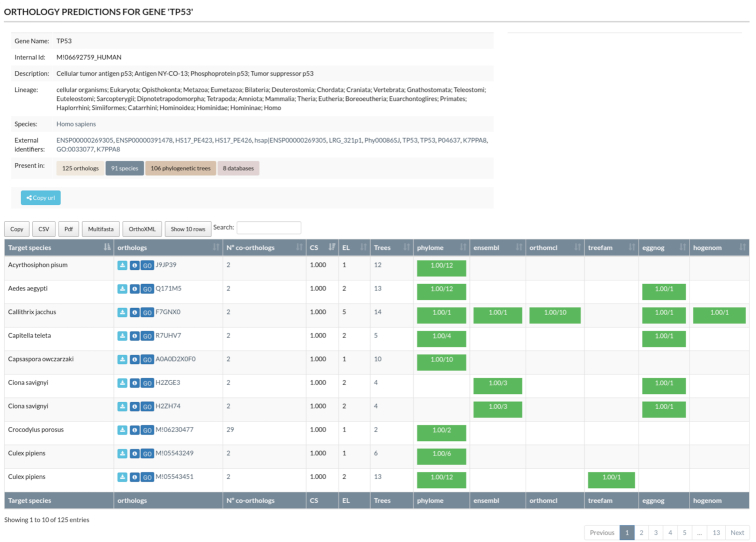
With MetaPhOrs, users can take advantage of multiple phylogenetic evidence to derive orthology and paralogy predictions. To illustrate the usage of MetaPhOrs, and some of the new features included in the latest release, we search for orthologs and paralogs for TP53 gene in human (Figure 2). At the top of the result page, MetaPhOrs’ website shows a description of the gene of interest. Thus, It shows that TP53 has 125 orthologs (CS > 0.5) in 91 species, as inferred from 106 phylogenetic trees retrieved from eight different sources (Figure 2). Below the description, there is a table of orthology and paralogy relationships for TP53 gene, one row for each of the 125 orthologs found (Figure 2). The homology table can be filtered by Confidence Score, EL values or species, and can be sorted by any column (Figure 2).
Figure 2.
Screenshot of MetaPhOrs web-server interface showing the orthologs and paralogs for TP53 gene in human. In the top, a description of the TP53 gene. In the bottom, a table of orthology and paralogy relationships for the TP53 gene.
Since its initial release in 2010, MetaPhOrs usage continues to increase with ∼290 visitors per month in 2019 based on usage data gathered using Google Analytics. MetaPhOrs server constitutes a resource of broad interest and applicability and it was used in several relevant projects. For example, it was used to determine the evolutionary ages of the germ layers in Caenorhabditis elegans (16) or to understand the differentiation and regulation of tissue-specific gene products in plants (17) and helped resolving early branches in the tree of life of modern birds (18).
BENCHMARKING
Orthology predictions have been benchmarked on the Quest for Orthologs reference proteomes. The quest for orthologs initiative aims to improve and standardize orthology predictions through collaboration and sharing knowledge among users and developers of algorithms and databases in this field (19,20). The accuracy of orthology predictions provided by MetaPhOrs has been benchmarked alongside other methods, showing the superiority of the integrative approach over the individual methods or databases (6), especially when looking at both precision and recall. Furthermore, MetaPhOrs has been benchmarked using alternative approaches and datasets in the original MetaPhOrs’ publication (4). Most benchmarks show a good compromise between accuracy and sensitivity in predictions provided by MetaPhOrs, comparable to the best scoring methods.
CONCLUSION
We describe the latest developments in MetaPhOrs, including new functions and a significant expansion of the database. The new MetaPhOrs web server offers a unique, modern interactive user interface providing phylogeny-based orthology and paralogy predictions.
ACKNOWLEDGEMENTS
We thank members of the Gabaldón group, and particularly Marina Marcet-Houben, for useful comments on the new release. We are thankful to all users that have approached us over the years with suggestions for improvements.
Contributor Information
Uciel Chorostecki, Barcelona Supercomputing Centre (BSC-CNS), 08034 Barcelona, Spain; Institute for Research in Biomedicine (IRB), The Barcelona Institute of Science and Technology, 08028 Barcelona, Spain.
Manuel Molina, Barcelona Supercomputing Centre (BSC-CNS), 08034 Barcelona, Spain; Institute for Research in Biomedicine (IRB), The Barcelona Institute of Science and Technology, 08028 Barcelona, Spain.
Leszek P Pryszcz, Centre for Genomic Regulation, 08003 Barcelona, Spain; International Institute of Molecular and Cell Biology, 4 Ks. Trojdena Street, 02-109 Warsaw, Poland.
Toni Gabaldón, Barcelona Supercomputing Centre (BSC-CNS), 08034 Barcelona, Spain; Institute for Research in Biomedicine (IRB), The Barcelona Institute of Science and Technology, 08028 Barcelona, Spain; ICREA, 08010 Barcelona, Spain.
FUNDING
H2020 Marie Skłodowska-Curie Actions [H2020-MSCA-IF-2017-793699 to U.C.]; Spanish Ministry of Economy, Industry, and Competitiveness (MEIC) [PGC2018-099921-B-I00]; CERCA Programme/Generalitat de Catalunya; Catalan Research Agency (AGAUR) SGR423; European Union's Horizon 2020 Research and Innovation Programme [ERC-2016-724173]; INB [PT17/0009/0023 - ISCIII-SGEFI/ERDF to T.G.]. Funding for open access charge: H2020Grant.
Conflict of interest statement. None declared.
REFERENCES
- 1. Glover N., Dessimoz C., Ebersberger I., Forslund S.K., Gabaldón T., Huerta-Cepas J., Martin M.-J., Muffato M., Patricio M., Pereira C. et al.. Advances and applications in the quest for orthologs. Mol. Biol. Evol. 2019; 36:2157–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gabaldón T. Large-scale assignment of orthology: back to phylogenetics. Genome Biol. 2008; 9:235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gabaldón T., Koonin E.V.. Functional and evolutionary implications of gene orthology. Nat. Rev. Genet. 2013; 14:360–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pryszcz L.P., Huerta-Cepas J., Gabaldón T.. MetaPhOrs: orthology and paralogy predictions from multiple phylogenetic evidence using a consistency-based confidence score. Nucleic Acids Res. 2011; 39:e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huerta-Cepas J., Bueno A., Dopazo J., Gabaldón T.. PhylomeDB: a database for genome-wide collections of gene phylogenies. Nucleic Acids Res. 2008; 36:D491–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Altenhoff A.M., Boeckmann B., Capella-Gutierrez S., Dalquen D.A., DeLuca T., Forslund K., Huerta-Cepas J., Linard B., Pereira C., Pryszcz L.P. et al.. Standardized benchmarking in the quest for orthologs. Nat. Methods. 2016; 13:425–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huerta-Cepas J., Serra F., Bork P.. ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 2016; 33:1635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huerta-Cepas J., Capella-Gutierrez S., Pryszcz L.P., Denisov I., Kormes D., Marcet-Houben M., Gabaldón T.. PhylomeDB v3.0: an expanding repository of genome-wide collections of trees, alignments and phylogeny-based orthology and paralogy predictions. Nucleic Acids Res. 2011; 39:D556–D560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vilella A.J., Severin J., Ureta-Vidal A., Heng L., Durbin R., Birney E.. Ensemblcompara genetrees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009; 19:327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huerta-Cepas J., Szklarczyk D., Forslund K., Cook H., Heller D., Walter M.C., Rattei T., Mende D.R., Sunagawa S., Kuhn M. et al.. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016; 44:D286–D293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schreiber F., Patricio M., Muffato M., Pignatelli M., Bateman A.. TreeFam v9: a new website, more species and orthology-on-the-fly. Nucleic Acids Res. 2014; 42:D922–D925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marcet-Houben M., Gabaldón T.. EvolClust: automated inference of evolutionary conserved gene clusters in eukaryotes. Bioinformatics. 2020; 36:1265–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Penel S., Arigon A.-M., Dufayard J.-F., Sertier A.-S., Daubin V., Duret L., Gouy M., Perrière G.. Databases of homologous gene families for comparative genomics. BMC Bioinformatics. 2009; 10(Suppl. 6):S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li L., Stoeckert C.J. Jr, Roos D.S.. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003; 13:2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schmitt T., Messina D.N., Schreiber F., Sonnhammer E.L.L.. Letter to the editor: SeqXML and OrthoXML: standards for sequence and orthology information. Brief. Bioinform. 2011; 12:485–488. [DOI] [PubMed] [Google Scholar]
- 16. Hashimshony T., Feder M., Levin M., Hall B.K., Yanai I.. Spatiotemporal transcriptomics reveals the evolutionary history of the endoderm germ layer. Nature. 2015; 519:219–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu Q. Mutational bias and translational selection shaping the codon usage pattern of tissue-specific genes in rice. PLoS One. 2012; 7:e48295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jarvis E.D., Mirarab S., Aberer A.J., Li B., Houde P., Li C., Ho S.Y.W., Faircloth B.C., Nabholz B., Howard J.T. et al.. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science. 2014; 346:1320–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dessimoz C., Gabaldón T., Roos D.S., Sonnhammer E.L.L., Herrero J. Quest for Orthologs Consortium . Toward community standards in the quest for orthologs. Bioinformatics. 2012; 28:900–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gabaldón T., Dessimoz C., Huxley-Jones J., Vilella A.J., Sonnhammer E.L., Lewis S.. Joining forces in the quest for orthologs. Genome Biol. 2009; 10:403. [DOI] [PMC free article] [PubMed] [Google Scholar]