Abstract
The amount of biological data, generated with (single cell) omics technologies, is rapidly increasing, thereby exacerbating bottlenecks in the data analysis and interpretation of omics experiments. Data mining platforms that facilitate non-bioinformatician experimental scientists to analyze a wide range of experimental designs and data types can alleviate such bottlenecks, aiding in the exploration of (newly generated or publicly available) omics datasets. Here, we present BIOMEX, a browser-based software, designed to facilitate the Biological Interpretation Of Multi-omics EXperiments by bench scientists. BIOMEX integrates state-of-the-art statistical tools and field-tested algorithms into a flexible but well-defined workflow that accommodates metabolomics, transcriptomics, proteomics, mass cytometry and single cell data from different platforms and organisms. The BIOMEX workflow is accompanied by a manual and video tutorials that provide the necessary background to navigate the interface and get acquainted with the employed methods. BIOMEX guides the user through omics-tailored analyses, such as data pretreatment and normalization, dimensionality reduction, differential and enrichment analysis, pathway mapping, clustering, marker analysis, trajectory inference, meta-analysis and others. BIOMEX is fully interactive, allowing users to easily change parameters and generate customized plots exportable as high-quality publication-ready figures. BIOMEX is open source and freely available at https://www.vibcancer.be/software-tools/biomex.
INTRODUCTION
The recent growth of unbiased high-throughput sequencing and profiling technologies has revolutionized the generation and analysis of biological data (1). The commoditization, exponential growth and increased throughput of these technologies (2) has helped the community to develop breakthroughs in bioanalytical research. For example, using next generation sequencing technologies, it is possible to analyze whole genome and transcriptome sequences within an hour (3). In addition, using mass spectrometry, thousands of proteins and metabolites can be measured simultaneously (4,5). Until recently, traditional profiling methods could be applied only to ‘bulk’ samples homogenized from whole tissue or organ extracts. With the advent of single cell genomics, transcriptomics and proteomics profiling technologies, individual cell contents can now be measured (6) allowing the characteristics of individual cells to be studied (7,8), further increasing the volume of data to analyze.
Availability of such large and complex datasets introduces multiple challenges. Computational challenges relate to the handling, processing and analysis of the data; new bioinformatics tools are continuously developed in tandem with technological advances. Biological challenges stem from the need to understand the biological significance of the information in the data and require in-depth knowledge of the biological question. Consequently, detailed biological interpretation of omics data requires a synthesis of domain-knowledge and computational skills, which continues to inspire interdisciplinary projects between researchers with complementary expertise. Various tools exist to meet the challenges related to analyzing omics data, and can be loosely defined as workflow tools (e.g. Galaxy (9), Taverna (10)), pre-processing tools (e.g. XCMS Online (11), MaxQuant (12)), specialized tools and more broad data analysis platforms. Easy-to-use data analysis platforms that allow experimental scientists to autonomously analyze omics data have been highly successful as solutions to bridge the gap between data generation and interpretation (e.g. Perseus (13), EXPANDER (14), InstantClue (15), MetaboAnalyst (16)). However, currently available data analysis platforms mostly focus on analyses of bulk omics data types, and in many cases they are not tailored to support the pretreatment and analysis of a wide range of omics experiments within the same interactive framework (e.g. RNA-sequencing, gene expression microarrays, metabolomics, proteomics), making the unified analysis of all these varieties of omics data challenging. Moreover, these tools do not scale, nor provide a structured data mining workflow to explore single cell datasets (e.g. single cell RNA-sequencing and mass cytometry).
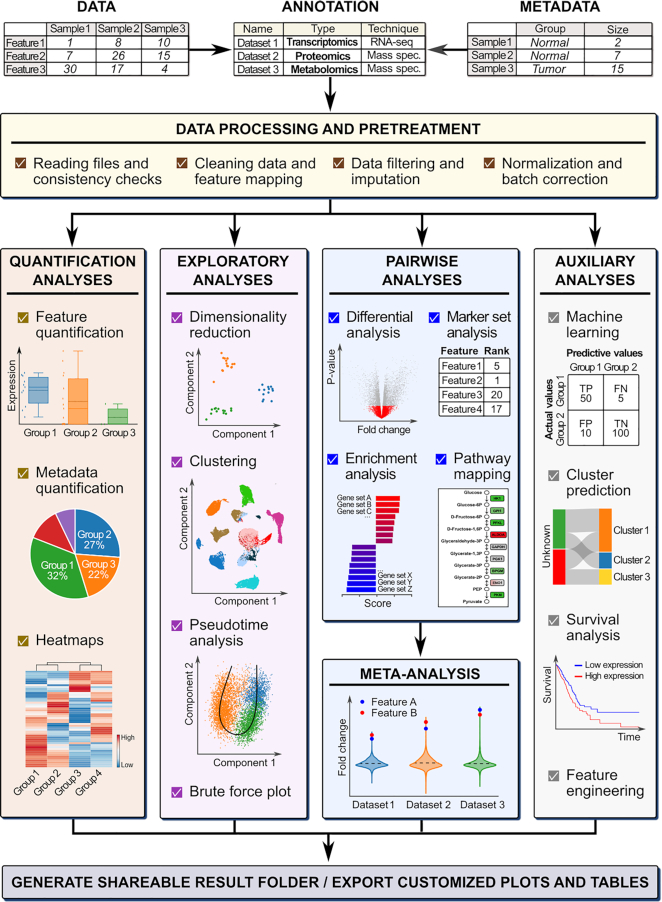
Here we present BIOMEX, a data mining software developed for the Biological Interpretation Of Multi-omics EXperiments. BIOMEX integrates a range of publicly available algorithms and field-tested data analysis approaches into a well-defined and guided stepwise workflow that accommodates a wide variety of experimental designs and multi-omics data, including metabolomics, transcriptomics, proteomics, single cell RNA-sequencing and mass cytometry experiments (Figure 1). The software is capable of handling large-scale data such as single cell omics experiments, scaling from tens to hundreds of thousands of cells.
Figure 1.
The BIOMEX workflow. The workflow guides the user through distinct analysis steps. The data (and metadata) need to be uploaded, and each uploaded dataset must be annotated with the relevant information (i.e. the omics data type, technology, feature identifier, etc.). In the processing step, the data is cleaned and consistency checks are performed to verify that the data was uploaded in the correct format. After the feature identifiers are mapped to feature names, the data is filtered and normalized in the pretreatment step. Depending on the omics data type, the data can also be imputed or batch corrected. Once the data is pretreated, it is ready for downstream analysis. The different analyses available in BIOMEX can be divided into five categories: (i) quantification analyses quantify the abundance level of the features in the data (and metadata); (ii) exploratory analyses assist in understanding the underlying structure of the data; (iii) pairwise analyses reveal the functional differences between groups; (iv) meta-analysis combines results from different studies in a singular, unique and robust result and (v) auxiliary analyses (e.g. machine learning and survival analysis). Ultimately, all the analyses can be saved in a self-contained folder that can be shared between scientists and results can be customized and exported either as tables or high quality publication-ready figures. Abbreviations: TP, true positive, FN, false negative, TN, true negative, FP, false positive. Note: The term ‘features’ is used to indicate genes, metabolites and proteins.
BIOMEX aims to alleviate the bottlenecks in the biological data-analysis-to-interpretation pipeline of omics experiments (13,17): these bottlenecks have become prominent in light of the increased need to understand the underlying biological phenomena that are now measureable at a much higher resolution.
OVERVIEW
Functional requirements and design rationale
BIOMEX is designed to allow non-bioinformatician experimental scientists to perform interactive data mining of bulk and single cell omics datasets. We therefore defined the following functional requirements for the software:
To accommodate multi-omics data across select biological species and computational platforms.
To allow users to interactively analyze complex experimental designs using state-of-the-art and field-tested algorithms.
To facilitate the re-use of publicly available data.
To provide a flexible, well-defined data analysis workflow.
To provide self-contained, non-technical background information to aid the meaningful use of each analysis module.
To enable the generation of highly-customizable publication-ready plots and figures.
BIOMEX is implemented in the open source R programming language (https://cran.r-project.org/); the majority of algorithms required for biological data mining are available through open source R packages. BIOMEX integrates these algorithms and packages into a workflow, in which parameters can be interactively tuned using the Shiny web framework (https://shiny.rstudio.com/, the full list of packages used in BIOMEX is available in Supplementary Table 1) (18). Together, BIOMEX creates a workflow that allows users to iteratively fine-tune complex analyses in order to facilitate detailed biological interpretation through interactive visualizations.
Manual and video tutorials
A comprehensive web manual that describes all functionalities, data formats, parameters and analyses related to the workflow is provided within the BIOMEX software. The manual guides the user through the step-by-step procedure required to execute the workflow and is complemented with video tutorials that introduce users to all interface elements and software functionalities.
DATA IMPORT: DATA AND METADATA MATRIX
BIOMEX requires two files for each experiment, the data and metadata matrix.
Data matrix
The data matrix contains typical omics (i.e. transcriptomics, metabolomics, proteomics, mass cytometry) output in a text (.txt) or comma separated values (.csv) format. The data file is organized such that the first column contains feature identifiers (i.e. genes, metabolites, proteins), while the first row contains descriptors (i.e. sample or cell IDs). The data matrix can be uploaded as unprocessed gene expression values (raw read counts, unique molecular identifier counts for single cell RNA-sequencing, non-log transformed intensities for microarrays) or absolute abundances for metabolomics and proteomics data. Alternatively, BIOMEX accepts preprocessed data (e.g. filtered and normalized, batch corrected, etc.).
BIOMEX automatically checks the uniformity and compatibility of the data, while also dealing with irrelevant (empty) observations and features. For transcriptomics and proteomics, the feature (gene or protein) identifiers are mapped to feature names to allow downstream interpretation of the results and further analyses.
Metadata matrix
The second required file is the metadata file (.txt or .csv format) that contains all the auxiliary information about the experimental design (variables). The metadata file is organized such that the first column contains descriptors matching with the data file, while the first row contains the variables (e.g. factors, numeric, etc.). This file can be modified interactively within the software.
Example datasets
We provide example datasets (the data and metadata matrices) that are readily available to be uploaded into the software. These example datasets encompass all the omics data types supported by BIOMEX, and they include case studies (described below) available directly from the software via the ‘Case studies’ section.
DATA ANALYSIS: ANALYSIS MODULES
The BIOMEX workflow contains 10 data analysis modules, which are briefly described below.
Module 1: Data pretreatment
After data upload, the data is filtered to remove low quality features, normalized, and, if necessary, corrected for unwanted technical variation (e.g. batch effects) (19–22). Algorithms for quality filtering, normalization and regression are often omics-type specific: BIOMEX automatically suggests applicable algorithms and parameter settings depending on the type of data being analyzed. For example, single cell RNA-sequencing data can be corrected for batch effects by using the mutual nearest neighbor (MNN) method (22).
The output of this module is a clean/corrected data matrix that can be used for downstream analysis.
Module 2: Feature engineering
Complementing unbiased and automated methods, domain knowledge can be used to craft new features from the existing features in the data to estimate biological variation (e.g. from gene expression to pathway activity). During this (optional) step, BIOMEX employs gene set variation analysis (23) (GSVA) to convert the features-by-observations data matrix (output of module 1) into an engineered sets-by-observations data matrix. The newly created engineered data can then be used to perform downstream analysis, including differential analysis. BIOMEX includes the KEGG sets by default, but users can also upload custom sets. Alternatively, feature engineered data, created with independent methods, can be directly uploaded to BIOMEX and subsequently used in downstream analysis.
Module 3: Visualization of trends
Feature magnitudes and trends are visualized in bar plots, box plots, violin plots and density kernel estimation plots. These plots are grouped based on the information present in the metadata. The uploaded metadata can also be explored through pie charts and horizontal bar plots.
Module 4: Unsupervised analyses
Unsupervised analysis aims to unbiasedly detect patterns in the data. For dimensionality reduction and visualization, BIOMEX includes Principal Component Analysis (24) (PCA, flashPCA package (25)), t-distributed Stochastic Neighbor Embedding (26) (t-SNE, Rtsne package) and Uniform Manifold Approximation and Projection (27) (UMAP, umap package). Also, BIOMEX supports K-means, hierarchical and graph-based clustering (Seurat (21) and FlowSOM (28) packages). The output of hierarchical clustering can be visualized via dendrograms, and the associated uncertainty can be assessed using multi-scale bootstrap resampling (pvclust package (29)). BIOMEX provides interactive heatmaps (heatmaply package (30)) to visualize inherent associations between groups or clusters.
Module 5: Supervised analyses
Supervised pairwise analyses are used to explore quantitative differences in expression or abundance levels between groups (differential analysis). BIOMEX uses linear models (limma and MAST packages (31,32)) to describe the relationship between expression levels of features between two groups. This enables handling of complex experimental designs, and allows including covariates in the modeling process. The magnitude of differential expression (log fold change) and the P-values are provided for each feature, together with the false discovery rate adjusted P-values calculated with the Benjamini-Hochberg method (33). Volcano plots are used to visually represent the differential analysis results.
As an extension of pair-wise differential analysis, BIOMEX includes marker analysis that can be used to detect key discriminating features between multiple groups (or clusters in single cell data). This analysis consists of a two-step intra-dataset meta-analysis approach. First, BIOMEX performs a differential analysis for each group against all the other groups separately and filters out features that are not consistently differentially expressed (34). Subsequently, marker features are ranked using a product-based meta-analysis (median-, sum- or P-value-based meta-analysis can be used to rank features) (35).
Functional analysis of omics data can be performed in BIOMEX using several tools. These include Gene Set Enrichment Analysis (36) (GSEA, clusterProfiler (37) package) used to perform the competitive set enrichment analysis, and rotation gene set tests (ROAST) (38) to perform self-contained set enrichment analysis. Results of such analyses can be either displayed as a waterfall plot or a horizontal bar plot. These analyses provide functional information regarding, for example, pathways or biological processes that may be deregulated in a select set of conditions. The default setting in the software is to use the (metabolic) KEGG pathway sets (39), but this can be changed by the user to include other pathways or biological processes. BIOMEX uses the KEGG pathways to map features in the data (genes, proteins, metabolites) to well defined and constructed pathways using the pathview (40) package. The pathway visualizations are interactive and can be customized by the user to incorporate a priori biological insight (e.g. irrelevant isoforms can be manually excluded).
Module 6: Single cell specific algorithms
Single cell data can be used to infer differentiation trajectories using computational methods. BIOMEX includes Monocle (41) and SCORPIUS (42) to infer branched and linear cell trajectories, respectively. BIOMEX also uses locally estimated scatterplot smoothing (LOESS) regression to subsequently model the dynamic behavior of features in pseudotime. As a second single cell-specific approach, BIOMEX includes scmap (43) to project cluster identities from a reference dataset to another non-clustered dataset by calculating the similarities between cells of the non-clustered dataset and the cluster centroids in the reference dataset.
Module 7: Survival analysis
Survival analysis is implemented in BIOMEX in order to link omics data to a disease outcome. For example, using The Cancer Genome Atlas (TCGA) (44) and other resources, it is possible to infer the effect of deregulation of a given gene to a treatment outcome or patient survival. BIOMEX uses the Kaplan–Meier (45) test to generate the survival functions, and the logrank test (46) to assess the significance of those survival functions (survival package).
Module 8: Machine learning
Machine learning is a set of approaches that can model the relationship between a set of variables (features) and instances (observations) based on a given training dataset. BIOMEX includes the ranger (47) implementation of the random forest model to perform classification and regression tasks (48). Recursive feature elimination (RFE) (49) is used as the feature selection method of choice to select the most predictive features. The machine learning pipeline is based on the caret package and includes cross-validation strategies to assess the predictive performance of the model (50).
Module 9: Meta-analysis of bulk omics data
Integrative data analysis approaches have been successfully used to analyze multiple datasets simultaneously to compare the results of independent experiments (51,52). With the availability of added-value databases, publicly available preprocessed data can be easily accessed by scientists and used to perform meta-analyses. In a meta-analysis, (i) a pair-wise differential analysis is performed for each dataset independently; (ii) we rank the features in each dataset by a metric (e.g. fold change) and (iii) we combine the rank numbers for all features using a product-based (or median-, sum- and P-value-based) meta-analysis approach. As a result, we obtain a ranked list of features, which are consistently differentially expressed across all the selected comparisons (i.e. differential analyses) in different datasets. The results can be visually explored through violin plots.
Module 10: Single cell meta-analysis
Meta-analysis can also be performed by measuring the similarity between clusters (53). This analysis, developed specifically for single cell omics data, assesses the conservation of cell phenotypes between different tissues, organs, studies, conditions, etc. BIOMEX performs the cluster similarity analysis by combining the results obtained during the marker set analysis. Similarity between the clusters present in the marker set results are calculated using the pairwise Jaccard similarity coefficients (54) for all clusters against all other clusters. The output of this analysis is a similarity score matrix, which describes quantitatively how each cluster is similar to other clusters. PCA is applied to the pairwise Jaccard similarity coefficient matrix to visually represent the similarity between clusters.
DATA EXPORT: PLOTS AND TABLES
All the plots and tables (plotly, ggplot2 (55), DT packages) can be fully customized and exported in a variety of high quality formats (e.g. vectorized image format). BIOMEX saves all parameters and results in a self-contained folder, which can be shared between users and loaded into BIOMEX, improving the reproducibility of analyses.
CASE STUDIES
To showcase the analysis modules implemented in BIOMEX, we provide two case studies. A step-by-step tutorial on how to reproduce the results obtained in both case studies is available in the manual, which includes all the parameters used to perform the analyses and to generate the plots.
Bulk data: exploration of the TCGA cholangiocarcinoma dataset
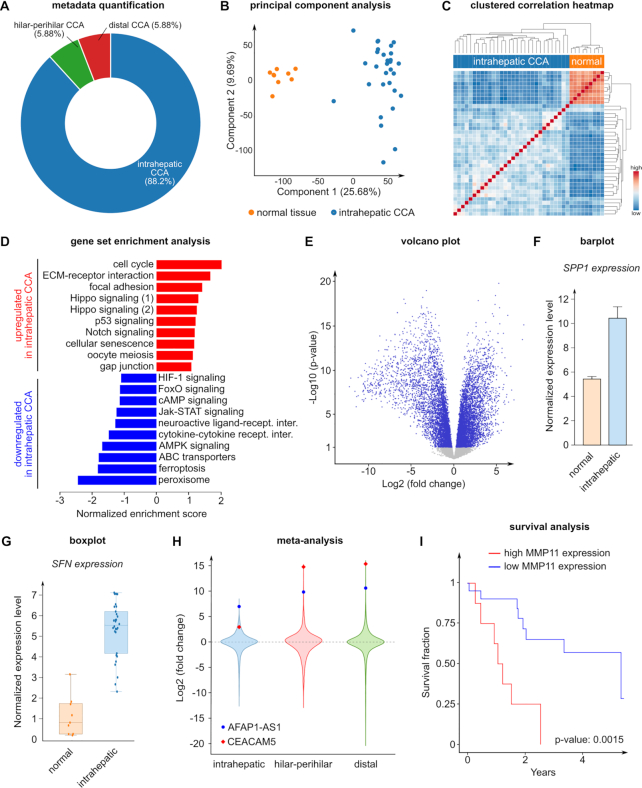
To provide an illustrative example on how bulk data can be analyzed, we explored a publicly available TCGA dataset (TCGA-CHOL) on cholangiocarcinoma (CCA), a cancer from the bile duct that represents the second most commonly diagnosed primary liver tumor (56). Even when diagnosed at an early stage, CCA is a very aggressive malignancy with poor patient outcome and limited treatment opportunities (56). According to their anatomical location, CCAs are classified as intrahepatic, hilar-perihilar and distal, which represent respectively 88.2%, 5.9% and 5.9% of the patients in this analysis (Figure 2A). Dimensionality reduction (PCA) and correlation heatmap analyses showed that biopsies from normal tissue have a clearly distinct transcriptomic signature compared to intrahepatic CCA resections (Figure 2B, C). Although there is a strong inter tumor sample heterogeneity between patients (Figure 2C), we aimed at determining transcriptomic similarities that could be involved in overall CCA pathogenesis. Enrichment analysis of normal versus intrahepatic CCA samples indicated that cell cycle and extracellular matrix (ECM)-receptor interaction gene sets were the most upregulated (Figure 2D). Consistently, differential gene expression analysis showed that several key mitotic checkpoints (e.g. CDK1, E2F1, CDC45, SFN) and mitotic spindle assembly/control genes (e.g. CDC20, CDC25, TUBB3), as well as numerous genes encoding laminin, integrin, collagen and ECM-secreted proteins (e.g. SPP1, COMP, TNC) were upregulated in the tumor samples (Figure 2E–G and not shown). To explore whether this signature was conserved across the other classes of CCA, we performed a meta-analysis of normal versus tumor samples from intrahepatic, hilar-perihilar and distal CCAs. We identified two genes, namely CEACAM5 and AFAP1-AS1, ranking in the top 2 in a product-rank meta-analysis (Figure 2H). Interestingly, CEACAM5 (carcinoembryonic antigen, CEA) is a well-established prognostic marker in CCA (57), while the long non-coding RNA AFAP1-AS1 has been linked to metastasis (a process requiring complex ECM remodeling) and cancer cell proliferation in CCA. Hence, the meta-analysis results further supported the importance of cell proliferation and ECM-cell adhesion in CCA.
Figure 2.
Cholangiocarcinoma TCGA data analysis results. (A) Overall histological type percentage of the TCGA cholangiocarcinoma dataset. (B) PCA of normal and intrahepatic tumor samples. (C) Clustered heatmap based on the correlation of normal and intrahepatic tumor samples. (D) Competitive enrichment analysis of normal versus intrahepatic tumor samples using the gene sets related to the ‘Environmental Information Processing’ and ‘Cellular Process’ KEGG pathway maps. The upregulated gene sets are shown in red, the downregulated gene sets are shown in blue. Note: There are two enriched KEGG gene sets related to Hippo signaling, indicated separately in the figure. Hippo signaling (1): KEGG Hippo signaling pathway; Hippo signaling (2): KEGG Hippo signaling pathway—multiple species. (E) Differential analysis of normal versus intrahepatic tumor samples shown in a volcano plot. The significantly different genes (P < 0.05) are shown in blue, the non-significant genes are shown in grey. (F) Barplot visualization of SPP1 expression in normal and intrahepatic tumor samples. The error bar represents the standard error. (G) Boxplot visualization of SFN expression in normal and intrahepatic tumor samples. The box represents the range between the first quartile (Q1) and the third quartile (Q3), the horizontal line represents the median, the whiskers represent the interquartile ranges (IQR, 1.5 × IQR below Q1 and 1.5 × IQR above Q3). (H) Meta-analysis of intrahepatic, hilar-perihilar and distal tumor types. Each violin plot represents the differential analysis of normal versus the corresponding tumor type. The top 2 most consistently upregulated genes (CEACAM5 and AFAP1-AS1) are highlighted. (I) Survival analysis based on MMP11 gene expression of intrahepatic tumor samples. All the results shown in the figure can be directly explored in the BIOMEX ‘Case studies’ section. The parameters used to generate these plots can be found in the manual.
A crucial step during invasion and metastasis is the remodeling of the ECM by proteolytic degradation, involving matrix metalloproteinases (MMPs) as pivotal actors. Interestingly, MMP11 has been correlated to poor survival in several cancer types including CCA (58), breast and pancreatic cancers. Consistently, we found that high MMP11 expression was correlated to poor survival in patients with intrahepatic CCA (P-value = 0.0015) (Figure 2I). Together, these findings show that cell proliferation and ECM adhesion/remodeling are conserved features across the CCA classes. Hence, (novel) insights can be derived by exploring publicly available data, showcasing the potential and value of BIOMEX.
Single cell data: re-analysis of the endothelial cell atlas dataset
To provide an illustrative example on how single cell data can be analyzed, we selected heart, liver and lung endothelial cells (ECs) from the recently published murine EC atlas scRNA-seq dataset (59). We intended to showcase a logical sequence of analyses that a BIOMEX user can employ to (re-)analyze single cell data.
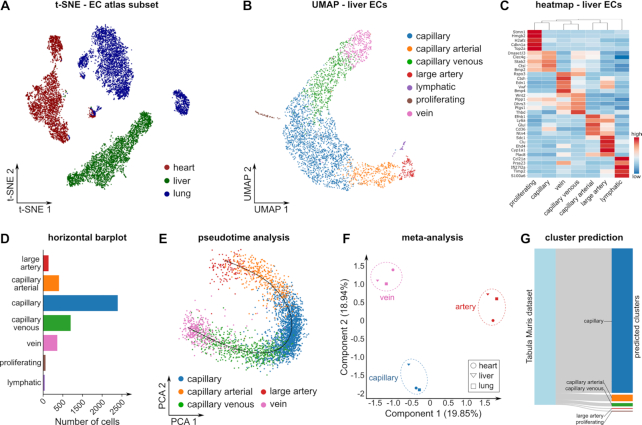
ECs line the lumen of blood vessels and are known to be heterogeneous along the vascular tree. Consistently, dimensionality reduction and visualization using PCA and t-SNE indicated that ECs from the lung, heart and liver vascular beds have a distinct transcriptional profile (Figure 3A). To explore heterogeneity of ECs within a single vascular bed, we performed dimensionality reduction using UMAP to visualize the subclusters as they were detected in the EC atlas (59) (Figure 3B). Next, we performed rank-product based marker set analysis, and visualized the top 5 marker genes for each cluster using a heatmap (Figure 3C). Marker genes were consistent with previously described markers of (sublineages of) arterial, capillary, venous, lymphatic and proliferating ECs (59). Quantification of the number of cells per cluster showed that capillary ECs constitute the majority of the liver single cell population (Figure 3D). Further, unbiased linear trajectory inference reconstructed a phenotypic continuum of arterial, capillary and venous phenotypes, consistent with the known anatomical topography of liver ECs (Figure 3E). To explore whether the cluster signatures are conserved across tissues, we performed a similar analysis for the lung and heart ECs (not shown), and subsequently performed a Jaccard similarity analysis. This analysis revealed that marker genes of arterial, capillary and venous phenotypes are conserved across vascular beds (Figure 3F). Finally, we used scmap to project liver ECs from an independent reference dataset (Tabula Muris dataset (8)), onto the cluster identified in the EC atlas liver ECs (Figure 3G).
Figure 3.
Endothelial cell atlas data analysis results. (A) t-SNE plot of ECs from three murine tissues (heart, liver, lung). (B) UMAP of liver tissue showing the endothelial cell clusters as described in the EC atlas. (C) Clustered heatmap showing the top 5 marker genes for each cluster. Colors represent row-wise scaled gene expression with a mean of 0 and a standard deviation of 1 (Z scores). (D) Number of cells for each cluster in liver ECs. (E) Differentiation trajectory of the classic EC phenotypes (arteries, capillaries, veins) in liver. (F) PCA on the pairwise Jaccard similarity coefficients between the top 50 marker genes of the classic EC phenotypes (arteries, capillaries and veins) in heart, lung and liver. (G) Sankey diagram showing the scmap cluster projection of the EC atlas liver data on the Tabula Muris EC liver data. All the results shown in the figure can be directly explored in the BIOMEX ‘Case studies’ section. The parameters used to generate these plots can be found in the manual.
Together, this sequence of relatively simple analysis steps shows the power of the BIOMEX workflow to easily explore single cell datasets in detail.
CONCLUSION
To facilitate bench scientists in solving the computational problems arising from omics experiments, we designed and developed BIOMEX, a data mining software for the Biological Interpretation Of Multi-omics EXperiments. BIOMEX aims to alleviate the data-analysis-to-interpretation bottlenecks, lowering the barriers needed to extract the biological information embedded in omics measurements. With its user-friendly, highly interactive web-like interface, users can address complex biological questions by using advanced computational tools and fine-tune the analyses in real time. In addition, its design is unconstrained and allows multi-omics data to be simultaneously uploaded and analyzed into one unified framework, providing a well-defined workflow to analyze, interpret and visualize large-scale data such as single cell measurements. BIOMEX also aids the exploration of datasets generated from publicly available profiling efforts (e.g. Tabula Muris (8), Human Cell Atlas (60), The Cancer Genome Atlas (44)), repositories (e.g. ArrayExpress (61), Gene Expression Omnibus (62)) and added-value databases (e.g. EndoDB (63)). Furthermore, it facilitates the shareability of results, reproducibility of analyses and execution of meta-analyses between different experiments. Due to its convenient user interface and comprehensive manual, BIOMEX could also be used as a didactical tool to introduce researchers to the field of biological data science.
To further promote detailed data mining of (single cell) omics datasets accessible to non-bioinformatician experimental scientists, we made BIOMEX freely available for Windows and Linux at https://www.vibcancer.be/software-tools/biomex. The source code is deposited at https://bitbucket.org/ftaverna/biomex.
Supplementary Material
ACKNOWLEDGEMENTS
Author contributions: P.C. & J.G. conceptualized the BIOMEX design; F.T. developed the software; F.T., J.G., S.K., K.R., L.T., A.S. evaluated the software; F.T., T.K., J.G., L.T, P.C. wrote the manuscript; L.T., F.T. made the figures; K.R., A.S., S.K., G.E, M.D., L.S. provided advice and helped improve the manuscript. All authors commented on the manuscript.
Notes
Present address: Jermaine Goveia, Department of Laboratory Medicine, University Hospitals Leuven, Leuven, Belgium.
Present address: Tobias K. Karakach, Bioinformatics Core Laboratory, Children's Hospital Research Institute of Manitoba and Rady Faculty of Health Sciences, Department of Pediatrics and Child Health, Health Sciences Centre, Winnipeg, Manitoba, Canada.
Present address: Shawez Khan, Center for Cancer Immune Therapy, Herlev Hospital, Department of Oncology, University of Copenhagen, Herlev, Denmark.
Contributor Information
Federico Taverna, Laboratory of Angiogenesis and Vascular Metabolism, Department of Oncology and Leuven Cancer Institute (LKI), KU Leuven, VIB Center for Cancer Biology, Leuven 3000, Belgium.
Jermaine Goveia, Laboratory of Angiogenesis and Vascular Metabolism, Department of Oncology and Leuven Cancer Institute (LKI), KU Leuven, VIB Center for Cancer Biology, Leuven 3000, Belgium.
Tobias K Karakach, Laboratory of Angiogenesis and Vascular Metabolism, Department of Oncology and Leuven Cancer Institute (LKI), KU Leuven, VIB Center for Cancer Biology, Leuven 3000, Belgium.
Shawez Khan, Laboratory of Angiogenesis and Vascular Metabolism, Department of Oncology and Leuven Cancer Institute (LKI), KU Leuven, VIB Center for Cancer Biology, Leuven 3000, Belgium.
Katerina Rohlenova, Laboratory of Angiogenesis and Vascular Metabolism, Department of Oncology and Leuven Cancer Institute (LKI), KU Leuven, VIB Center for Cancer Biology, Leuven 3000, Belgium.
Lucas Treps, Laboratory of Angiogenesis and Vascular Metabolism, Department of Oncology and Leuven Cancer Institute (LKI), KU Leuven, VIB Center for Cancer Biology, Leuven 3000, Belgium.
Abhishek Subramanian, Laboratory of Angiogenesis and Vascular Metabolism, Department of Oncology and Leuven Cancer Institute (LKI), KU Leuven, VIB Center for Cancer Biology, Leuven 3000, Belgium.
Luc Schoonjans, Laboratory of Angiogenesis and Vascular Metabolism, Department of Oncology and Leuven Cancer Institute (LKI), KU Leuven, VIB Center for Cancer Biology, Leuven 3000, Belgium.
Mieke Dewerchin, Laboratory of Angiogenesis and Vascular Metabolism, Department of Oncology and Leuven Cancer Institute (LKI), KU Leuven, VIB Center for Cancer Biology, Leuven 3000, Belgium.
Guy Eelen, Laboratory of Angiogenesis and Vascular Metabolism, Department of Oncology and Leuven Cancer Institute (LKI), KU Leuven, VIB Center for Cancer Biology, Leuven 3000, Belgium.
Peter Carmeliet, Laboratory of Angiogenesis and Vascular Metabolism, Department of Oncology and Leuven Cancer Institute (LKI), KU Leuven, VIB Center for Cancer Biology, Leuven 3000, Belgium.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Fonds voor Wetenschappelijk Onderzoek (FWO-Vlaanderen) (to J.G., K.R., L.T., P.C.); Kom op Tegen Kanker (Stand up to Cancer, Flemish Cancer Society) (to S.K., P.C.); Devotion (Developmental Origins of Chronic Diseases in Children) Network (to T.K.); Methusalem funding (to P.C.); Foundation against Cancer [2016-078 to P.C.]; European Research Council Proof of Concept [ERC-713758 to P.C.]; Advanced European Research Council Grant [EU-ERC743074 to P.C.]. Funding for open access charge: VIB.
Conflict of interest statement. None declared.
REFERENCES
- 1. Manzoni C., Kia D.A., Vandrovcova J., Hardy J., Wood N.W., Lewis P.A., Ferrari R.. Genome, transcriptome and proteome: the rise of omics data and their integration in biomedical sciences. Brief. Bioinform. 2018; 19:286–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stephens Z.D., Lee S.Y., Faghri F., Campbell R.H., Zhai C., Efron M.J., Iyer R., Schatz M.C., Sinha S., Robinson G.E.. Big data: astronomical or genomical. PLoS Biol. 2015; 13:e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lightbody G., Haberland V., Browne F., Taggart L., Zheng H., Parkes E., Blayney J.K.. Review of applications of high-throughput sequencing in personalized medicine: barriers and facilitators of future progress in research and clinical application. Brief. Bioinform. 2019; 20:1795–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aebersold R., Mann M.. Mass spectrometry-based proteomics. Nature. 2003; 422:198–207. [DOI] [PubMed] [Google Scholar]
- 5. Dettmer K., Aronov P.A., Hammock B.D.. Mass spectrometry-based metabolomics. Mass Spectrom. Rev. 2007; 26:51–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hwang B., Lee J.H., Bang D.. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp. Mol. Med. 2018; 50:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bhaduri A., Nowakowski T.J., Pollen A.A., Kriegstein A.R.. Identification of cell types in a mouse brain single-cell atlas using low sampling coverage. BMC Biol. 2018; 16:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schaum N., Karkanias J., Neff N.F., May A.P., Quake S.R., Wyss-Coray T., Darmanis S., Batson J., Botvinnik O., Chen M.B. et al.. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018; 562:367–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Blankenberg D., Von Kuster G., Coraor N., Ananda G., Lazarus R., Mangan M., Nekrutenko A., Taylor J.. Galaxy: a web-based genome analysis tool for experimentalists. Curr. Protoc. Mol. Biol. 2010; doi:10.1002/0471142727.mb1910s89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wolstencroft K., Haines R., Fellows D., Williams A., Withers D., Owen S., Soiland-Reyes S., Dunlop I., Nenadic A., Fisher P. et al.. The Taverna workflow suite: designing and executing workflows of Web Services on the desktop, web or in the cloud. Nucleic Acids Res. 2013; 41:W557–W561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tautenhahn R., Patti G.J., Rinehart D., Siuzdak G.. XCMS Online: a web-based platform to process untargeted metabolomic data. Anal. Chem. 2012; 84:5035–5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cox J., Mann M.. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008; 26:1367–1372. [DOI] [PubMed] [Google Scholar]
- 13. Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J.. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016; 13:731–740. [DOI] [PubMed] [Google Scholar]
- 14. Hait T.A., Maron-Katz A., Sagir D., Amar D., Ulitsky I., Linhart C., Tanay A., Sharan R., Shiloh Y., Elkon R. et al.. The EXPANDER integrated platform for transcriptome analysis. J. Mol. Biol. 2019; 431:2398–2406. [DOI] [PubMed] [Google Scholar]
- 15. Nolte H., MacVicar T.D., Tellkamp F., Kruger M.. Instant Clue: a software suite for interactive data visualization and analysis. Sci. Rep. 2018; 8:12648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xia J., Psychogios N., Young N., Wishart D.S.. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009; 37:W652–W660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alyass A., Turcotte M., Meyre D.. From big data analysis to personalized medicine for all: challenges and opportunities. BMC Med. Genomics. 2015; 8:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mattmann C.A. Computing: A vision for data science. Nature. 2013; 493:473–475. [DOI] [PubMed] [Google Scholar]
- 19. Li B., Tang J., Yang Q., Li S., Cui X., Li Y., Chen Y., Xue W., Li X., Zhu F.. NOREVA: normalization and evaluation of MS-based metabolomics data. Nucleic Acids Res. 2017; 45:W162–W170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Robinson M.D., McCarthy D.J., Smyth G.K.. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010; 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Satija R., Farrell J.A., Gennert D., Schier A.F., Regev A.. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015; 33:495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Haghverdi L., Lun A.T.L., Morgan M.D., Marioni J.C.. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol. 2018; 36:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hanzelmann S., Castelo R., Guinney J.. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013; 14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jolliffe I.T., Cadima J.. Principal component analysis: a review and recent developments. Philos. Trans. A Math. Phys. Eng. Sci. 2016; 374:20150202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Abraham G., Inouye M.. Fast principal component analysis of large-scale genome-wide data. PLoS One. 2014; 9:e93766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van der Maaten L.J.P., Hinton G.E.. Visualizing high-dimensional data using t-SNE. J. Mach. Learn. Res. 2008; 9:2579–2605. [Google Scholar]
- 27. McInnes L., Healy J., Melville J.. Umap: uniform manifold approximation and projection for dimension reduction. 2018; arXiv doi:06 December 2018, preprint: not peer reviewedhttps://arxiv.org/abs/1802.03426.
- 28. Van Gassen S., Callebaut B., Van Helden M.J., Lambrecht B.N., Demeester P., Dhaene T., Saeys Y.. FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A. 2015; 87:636–645. [DOI] [PubMed] [Google Scholar]
- 29. Suzuki R., Shimodaira H.. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006; 22:1540–1542. [DOI] [PubMed] [Google Scholar]
- 30. Galili T., O’Callaghan A., Sidi J., Sievert C.. heatmaply: an R package for creating interactive cluster heatmaps for online publishing. Bioinformatics. 2018; 34:1600–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K.. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Finak G., McDavid A., Yajima M., Deng J., Gersuk V., Shalek A.K., Slichter C.K., Miller H.W., McElrath M.J., Prlic M. et al.. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015; 16:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Benjamini Y., Drai D., Elmer G., Kafkafi N., Golani I.. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001; 125:279–284. [DOI] [PubMed] [Google Scholar]
- 34. Bruning U., Morales-Rodriguez F., Kalucka J., Goveia J., Taverna F., Queiroz K.C.S., Dubois C., Cantelmo A.R., Chen R., Loroch S. et al.. Impairment of angiogenesis by fatty acid synthase inhibition involves mTOR Malonylation. Cell Metab. 2018; 28:866–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hong F., Breitling R., McEntee C.W., Wittner B.S., Nemhauser J.L., Chory J.. RankProd: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics. 2006; 22:2825–2827. [DOI] [PubMed] [Google Scholar]
- 36. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. et al.. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. PNAS. 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yu G., Wang L.G., Han Y., He Q.Y.. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012; 16:284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu D., Lim E., Vaillant F., Asselin-Labat M.L., Visvader J.E., Smyth G.K.. ROAST: rotation gene set tests for complex microarray experiments. Bioinformatics. 2010; 26:2176–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kanehisa M., Goto S.. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000; 28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Luo W., Brouwer C.. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics. 2013; 29:1830–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., Rinn J.L.. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014; 32:381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cannoodt R., Saelens W., Sichien D., Tavernier S., Janssens S., Guilliams M., Lambrecht B., Preter K.D., Saeys Y.. SCORPIUS improves trajectory inference and identifies novel modules in dendritic cell development. 2016; bioRxiv doi:07 October 2016, preprint: not peer reviewed 10.1101/079509. [DOI]
- 43. Kiselev V.Y., Yiu A., Hemberg M.. scmap: projection of single-cell RNA-seq data across data sets. Nat. Methods. 2018; 15:359–362. [DOI] [PubMed] [Google Scholar]
- 44. Tomczak K., Czerwinska P., Wiznerowicz M.. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp. Oncol. (Poznan, Poland). 2015; 19:A68–A77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Goel M.K., Khanna P., Kishore J.. Understanding survival analysis: Kaplan-Meier estimate. Int. J. Ayurveda Res. 2010; 1:274–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bland J.M., Altman D.G.. The logrank test. BMJ. 2004; 328:1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wright M.N., Ziegler A.. ranger: a fast implementation of random forests for high dimensional data in C++ and R. J.Stat. Softw. 2017; 77:1–17. [Google Scholar]
- 48. Breiman L. Random Forests. Mach. Learn. 2001; 45:5–32. [Google Scholar]
- 49. Guyon I., Weston J., Barnhill S., Vapnik V.. Gene selection for cancer classification using support vector machines. Mach. Learn. 2002; 46:389–422. [Google Scholar]
- 50. Refaeilzadeh P., Tang L., Liu H.. Liu L, ÖZsu MT. Encyclopedia of Database Systems. 2009; Boston: Springer US; 532–538. [Google Scholar]
- 51. Chen J., Qian Z., Li F., Li J., Lu Y.. Integrative analysis of microarray data to reveal regulation patterns in the pathogenesis of hepatocellular carcinoma. Gut Liver. 2017; 11:112–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cahan P., Rovegno F., Mooney D., Newman J.C., Laurent St, 3rd G., McCaffrey T.A.. Meta-analysis of microarray results: challenges, opportunities, and recommendations for standardization. Gene. 2007; 401:12–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Goveia J., Rohlenova K., Taverna F., Treps L., Conradi L.C., Pircher A., Geldhof V., de Rooij L., Kalucka J., Sokol L. et al.. An integrated gene expression landscape profiling approach to identify lung tumor endothelial cell heterogeneity and angiogenic candidates. Cancer Cell. 2020; 37:21–36. [DOI] [PubMed] [Google Scholar]
- 54. Levandowsky M., Winter D.. Distance between Sets. Nature. 1971; 234:34–35. [Google Scholar]
- 55. Wickham H. ggplot2: Elegant Graphics for Data Analysis. 2009; Incorporated: Springer Publishing Company. [Google Scholar]
- 56. Rizvi S., Khan S.A., Hallemeier C.L., Kelley R.K., Gores G.J.. Cholangiocarcinoma - evolving concepts and therapeutic strategies. Nat. Rev. Clin. Oncol. 2018; 15:95–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Loosen S.H., Roderburg C., Kauertz K.L., Koch A., Vucur M., Schneider A.T., Binnebösel M., Ulmer T.F., Lurje G., Schoening W. et al.. CEA but not CA19-9 is an independent prognostic factor in patients undergoing resection of cholangiocarcinoma. Sci. Rep. 2017; 7:16975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhong W., Dai L., Liu J., Zhou S.. Cholangiocarcinomaassociated genes identified by integrative analysis of gene expression data. Mol Med Rep. 2018; 17:5744–5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kalucka J., de Rooij L.P.M.H., Goveia J., Rohlenova K., Dumas S.J., Meta E., Conchinha N.V., Taverna F., Teuwen L.-A., Veys K. et al.. Single-Cell transcriptome atlas of murine endothelial cells. Cell. 2020; 180:764–779. [DOI] [PubMed] [Google Scholar]
- 60. Regev A., Teichmann S.A., Lander E.S., Amit I., Benoist C., Birney E., Bodenmiller B., Campbell P., Carninci P., Clatworthy M. et al.. The human cell atlas. eLife. 2017; 6:e27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kolesnikov N., Hastings E., Keays M., Melnichuk O., Tang Y.A., Williams E., Dylag M., Kurbatova N., Brandizi M., Burdett T. et al.. ArrayExpress update–simplifying data submissions. Nucleic Acids Res. 2015; 43:D1113–D1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Barrett T., Troup D.B., Wilhite S.E., Ledoux P., Rudnev D., Evangelista C., Kim I.F., Soboleva A., Tomashevsky M., Edgar R.. NCBI GEO: mining tens of millions of expression profiles–database and tools update. Nucleic Acids Res. 2007; 35:D760–D765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Khan S., Taverna F., Rohlenova K., Treps L., Geldhof V., de Rooij L., Sokol L., Pircher A., Conradi L.C., Kalucka J. et al.. EndoDB: a database of endothelial cell transcriptomics data. Nucleic Acids Res. 2019; 47:D736–D744. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.