Abstract
VarFish is a user-friendly web application for the quality control, filtering, prioritization, analysis, and user-based annotation of DNA variant data with a focus on rare disease genetics. It is capable of processing variant call files with single or multiple samples. The variants are automatically annotated with population frequencies, molecular impact, and presence in databases such as ClinVar. Further, it provides support for pathogenicity scores including CADD, MutationTaster, and phenotypic similarity scores. Users can filter variants based on these annotations and presumed inheritance pattern and sort the results by these scores. Variants passing the filter are listed with their annotations and many useful link-outs to genome browsers, other gene/variant data portals, and external tools for variant assessment. VarFish allows users to create their own annotations including support for variant assessment following ACMG-AMP guidelines. In close collaboration with medical practitioners, VarFish was designed for variant analysis and prioritization in diagnostic and research settings as described in the software's extensive manual. The user interface has been optimized for supporting these protocols. Users can install VarFish on their own in-house servers where it provides additional lab notebook features for collaborative analysis and allows re-analysis of cases, e.g. after update of genotype or phenotype databases.
INTRODUCTION
Targeted sequencing (1) such as gene panel or whole exome sequencing (WES) has become common in clinical genetics research and diagnostic applications. Whole genome sequencing (WGS) is an emerging approach for such applications, yet interpretation of small non-coding variants remains challenging, and there currently appears to be no evidence for WGS being superior to WES in the clinic in relation its the higher costs; WES is still considered the most cost-efficient (2,3). Of course, the exome-like variants from WGS data can be analyzed in the same fashion as WES data. The interest in this area is shown by the large number of tools available for the scoring, filtering, and prioritization of exome-wide variants. We limit our discussion to freely accessible, web-based tools allowing for exome-wide variant analysis which focus on rare diseases; these include Exomiser (4), eXtasy (5), GeneTalk (6), MutationDistiller (7), OVA (8), Phen-Gen (9), Variant Ranker (10), VCF-Server (11), VEP (12) and wANNOVAR (13). These prior works cover different feature sets (shown in Table 1) and differ in stability and availability of their source code. The latter is particularly important when authors discontinue their service. Common features include variant pathogenicity and gene-phenotype similarity scores, annotations and filtering by population frequencies. In the light of rapid growth and frequent updates of public databases, we view certain topics to be important emerging themes in the field of variant filtering and prioritizing. These include joint filtering of multiple cases, user-based annotation of variants and sharing thereof, building databases of analysed cases with variant assessments, and the re-evaluation of cases. Approaching these topics commonly requires duplicate work or advanced bioinformatics skills. Here, we report on our web-based application VarFish developed to tackle these challenges. VarFish is freely available without login at https://varfish-kiosk.bihealth.org and a demo version showcasing additional features available on in-house installations is available at https://varfish-demo.bihealth.org. Its source code is available for free at https://github.com/bihealth/varfish-server.
Table 1.
Feature comparison of state-of-the-art web-based tools for variant filtering and prioritization. The tools are ordered by the date of the most recent update, ties are broken by number of features
| Latest update | HPO | OMIM | Gene lists/ regions | No registration | Gene info | Demo/ Tutorial | Quality filter | Interactivity | Tabular download | Multi-sample VCF | Bring your server | User annotations | ACMG-AMP class | Quality control | Sanity checks | Multiple VCFs | In-house DB | Collaboration | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| eXtasy | 2013 | ✓ | ✓ | ✓ | ✓ | ||||||||||||||
| Phen-Gen | 2014 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |||||||||||
| OVA | 2015 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |||||||||||
| Variant Ranker | 2017 | ✓ | ✓ | ✓ | ✓ | ||||||||||||||
| wANNOVAR | 2017 | ✓ | ✓ | ✓ | ✓ | ✓ | (✓) | ||||||||||||
| GeneTalk | 2018 | (✓) | (✓) | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||||||
| VCF- Server | 2019 | ✓ | ✓ | ✓ | ✓ | ||||||||||||||
| Exomiser | 2020 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | (✓) | ✓ | ||||||||||
| VEP | 2020 | ✓ | ✓ | ✓ | ✓ | ||||||||||||||
| Mutation Distiller | 2020 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |||||||||
| VarFish | 2020 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓* | ✓* | ✓* |
A tick mark (✓) indicates that the feature is present. ‘Interactivity’ allows users to interactively change filter and sorting options. ‘Tabular downloads’ allows users to download a spreadsheet (e.g. Excel) file with their results. ‘User annotations’ allow users to leave flag, color coded, or free-text comments, ‘ACMG-AMPs support’ assists users in creating variant assessments following the ACMG guidelines. ‘Quality control’ allows users to perform visual quality control, e.g. for depth of coverage, while ‘sanity checks’ allow to compare the relatedness or sex inferred from the data with user-provided meta data. ‘Multi-sample VCFs’ supports cases with more than one sample while ‘multiple VCFs’ supports querying multiple cases at the same time. ‘Bring your server’ allows users to create their own installations on their own server. ‘In-house DB’ allows to build a database of variants identified at the user's institution. An asterisk (*) indicates that the feature is only available on installations on the user's own server. Parentheses around the tick mark in ‘multi-sample VCF’ row indicates that filtering is restricted to predefined models of inheritance. Parentheses for GeneTalk indicate that the feature is only available when using the PEDIA tool.
RESULTS
Feature comparison with state-of-the-art tools
Table 1 shows the features implemented in VarFish in comparison to state-of-the art web tools (based on Figure 1 from (7)). The year of the latest update is important as databases on variants and genes are growing quickly. The support of the Human Phenotype Ontology (HPO) and Online Mendelian Inheritance in Man (OMIM) is relevant for prioritizing single exomes. Filtering based on affecting genes and regions is of interest when characterizing patient cohorts for variants in known disease-causing genes. Furthermore, support for multi-sample files or even multiple VCF files at once allows more advanced analyses. Implementing quality control features is important to gauge the quality of data in an integrated platform. Dynamic variant reports greatly improve usability over repeated submission of queries to batch systems. Tabular downloads (e.g. as spreadsheet files) make it possible to archive cases on the user's computer and store them in clinical or laboratory information management system. Supporting users in annotating variants with flags, colour codes extends such systems in the manner of a laboratory notebook. Supporting users in the ACMG-AMP (14) classification of variants is useful for creating diagnostic reports. The possibility to organize cases in projects with project-based access control further fosters collaboration in data analysis and allows for using the in-house data in the variant analysis (as further explained in Section S1 in the Supplemental Material). Custom installations on the user's own server can address data privacy issues. Finally, the availability of extensive documentation, tutorials and providing the tools without requiring user registration lowers the entry barrier.
Figure 1.
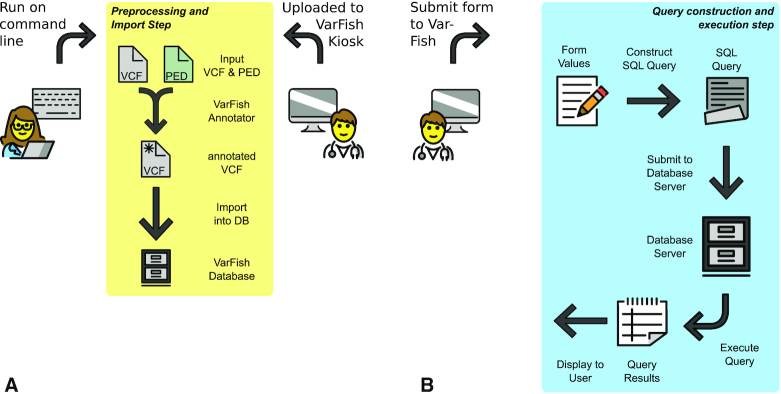
Illustration of the VarFish workflow. Sub figure (A) shows the preprocessing and import step that can be either triggered by computational on the command line (e.g. in parallel on a compute cluster) or by non-computational staff by upload to VarFish Kiosk. The annotated files are then imported into the VarFish database. Sub figure (B) shows the query construction and execution step. The form values are converted into a SQL query that is sent to the database server for execution. After execution, the results are reported to the user.
The VarFish workflow
There are two major parts in the VarFish data processing workflow: the data preprocessing and import and the query construction and execution step, shown in Figure 1.
The input to the preprocessing and import step is a file in VCF (Variant Call Format) format (15) and optionally a PLINK (16) pedigree file. Each variant record is read from the file and annotated with molecular impact using the Jannovar (17) library for both RefSeq (18) and ENSEMBL (19) transcripts, with the distance to the closest exon in either database, with its population frequencies in the ExAC (20), gnomAD (21), and Thousand Genomes Project (22) databases, and its presence in ClinVar (23). The variants are assigned a case identifier and are then imported together with properties such as quality scores from the VCF file into a PostgreSQL database table following the star schema pattern common in data warehouse applications.
The input to the query construction and execution step consists of the identifier of the case and the query settings from the user (see below). It constructs a SQL database query for selecting the variants based on the input criteria and joins the central variant table to further metadata tables (e.g. providing information about genes, variants, or conservation information). This query is then submitted to the database system for execution. VarFish was developed for use in genetics of rare diseases where users desire to create short lists of variants (say <200) for further analysis based on population frequency, genotype/segregation in families, molecular impact, and other criteria (24). In particular, the first three criteria can be used to greatly reduce the number of resulting variants. By employing the star schema pattern, database indices can be created for the most common queries and the (small) number of rows returned from the query on the central variant table can be obtained fast. The extensive metadata acquisition can then be limited to this small number of rows. As a periodic background job, VarFish tabulates the number of samples that each variant occurs in heterozygous, hemizygous and homozygous state. This allows for removing variants seen in many cases as is the case for local polymorphisms or artifacts not seen in the population databases because of differences in variant calling. This feature is not available in the public ‘kiosk’ mode where users cannot be trusted and could run reidentification attacks (25). Further aspects of the query generation and execution are explained in Section S3 in the Supplemental Material.
Quality control functionality
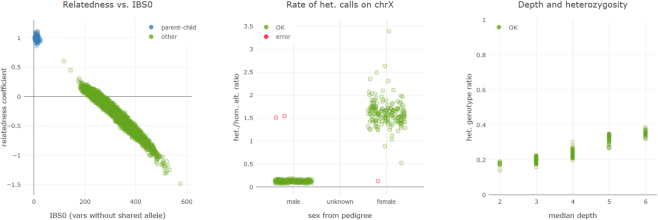
Another feature in VarFish is a global quality control function. Figure 2 shows an example of the quality control (QC) plots available. The three plots follow the Peddy methodology (26) and allow samples be examined for (un)expected relatedness, the sex derived from X-chromosomal variants, and depth-of-coverage vs. fraction of heterozygous variants. A detailed description and interpretation guide is available in the online VarFish user manual. Further, VarFish allows users to provide quality control information for each sample that cannot be derived from VCF files, such as coverage information in JSON format (https://json.org). This allows an integrated display of QC information suitable for clinicians as suggested by Shyr, cf. (27). Finally, the user can consider this information for all samples in a project to evaluate a sample in comparison to similarly processed ones or for a whole cohort.
Figure 2.
Quality control plots following the Peddy (26) approach. The plots are described in the main text.
Database- and user-based annotation
VarFish integrates a growing list of databases with Table 2 showing a list of the most important ones (a full list of all databases is given in the online manual). Databases such as gnomAD provide information on the variant frequencies within (sub)populations, dbSNP provides identifiers for registered variants, ClinVar provides variant pathogenicity and PubMed identifiers. Protein-level conservation information is derived from UCSC genome browser (28) data, the NCBI gene database provides gene summaries and gene reference into function (RIF) information, and the HPO (29) provides phenotype information. The background databases need to be imported when installing VarFish locally. An archive file with this data is provided for download. We provide the full Snakemake (30) workflow for downloading the data from open and free sources for reproducibility.
Table 2.
A selection of the most important databases whose data is integrated into VarFish or that VarFish links out to. A full list can be found in the online manual available at https://varfish-server.rtfd.io contains an updated version
| Category | Database | |
|---|---|---|
| Integrated | Frequency | gnomAD |
| Clinical | ClinVar | |
| Phenotype | HPO | |
| Gene description | NCBI Gene & GeneRIF | |
| Constraint scores | gnomAD pLI/LOEUF | |
| Conservation | UCSC 100 Vertebrates | |
| Link-Out | Gene database | NCBI Entrez, GeneCards, PubMed, PanelApp |
| Variant score/tool | MutationTaster, varSEAKSplicing, VariantValidator | |
| Variant database | Beacon Network, VarSome | |
| Genome browser | Locus in local IGV, Public UCSC, DGV, ENSEMBL |
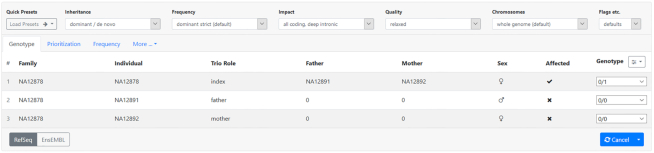
The VarFish result display lists extensive links to databases and data portals providing additional information. Furthermore, functionality for remote control of the integrative genome viewer (IGV) (31) and assessment of variants by tools such as MutationTaster (32) are provided. Resulting variant lists can be directly uploaded into MutationDistiller (7) for a complementary analysis. Further, users can annotate variants with flags, color codes and free-form text as shown in Figure 3. This figure also shows the support for computing and storing using the ACMG-AMP guidelines (11). Most criteria in ACMG-AMP require expert knowledge and some even critical reading of the literature. We thus decided that VarFish (at least in its current version) should not perform an automatic assessment but rather allow the user to select the points of evidence that they think the variant agrees with.
Figure 3.
User annotation of variants. Users can apply flags and color codes to variants and leave free-text annotations. Flags include ‘bookmark’, ‘reported as candidate’ and ‘final causative variant’ as well as ‘no phenotype linked to gene’. Color codes can be assigned in categories ‘raw data visual inspection’, ‘gene clinical/phenotype match’ and ‘validation results’ as well as an overall summary color. Also see Section S2 in the Supplemental Material.
The user-based annotations of variants and cases are visible to all users with access to the project. This is meant to foster collaboration within the center who have access to the cases in a project by providing a central place for storing the variant assessment. Also, this helps in the case of working a case over time where an analyst might choose to further scrutinize a case that was screened earlier by another person. Section S2 from the Supplemental Material illustrates the variant annotation further.
Filtering interface
Figure 4 illustrates the filtering interface and workflow from the user's perspective. The aim is to provide an easy access for inexperienced users yet offer high flexibility for experts. This is realized by providing two levels of presets. With no preset, VarFish provides a high degree of configurability for genotype, population frequency, variant quality measures (including variant call quality and variant allelic balance) and ClinVar annotation. On the first preset level, it provides default settings for several categories. For example, there are separate ‘super strict’, ‘strict’ and ‘relaxed’ population frequency settings under the assumption of dominant mode of inheritance, separate ones for assumed recessive mode of inheritance, and ‘strict’ and ‘relaxed’ settings for variant quality measures. On the second preset level, VarFish allows the user to select between settings such as ‘de novo’, ‘dominant inheritance’ and ‘recessive inheritance.’ For example, the second-level preset ‘recessive inheritance’ will set the genotype filter to return homozygous variants and compound heterozygous variants in the index (enforcing appropriate genotypes for the parents if present) with appropriate (yet strict) population frequencies and strict quality thresholds. The rationale is that users prefer to start reviewing a few promising variants first and then relax the filters while the total number of variants remains manageable. The user browses each variant in the raw data using IGV as well as the gene summary information and gene phenotype links. The results of this research can then be documented for each variant and flags are generated for the different categories (see above). In contrast, the second-level preset ‘de novo’ sets the genotype filter to ‘heterozygous’ for the index, and ‘reference’ for all other members in the pedigree, the population frequencies are set to very restrictive values, while the quality thresholds are relaxed and deeply intronic variants with a distance of up to 100 bp are included. The rationale is that de novo variants are very rare in trios and that even non-coding and low-covered variants are worth getting inspected, the latter also under the aspect of possible mosaic disease causing variants (33).
Figure 4.
The filtering interface. The ‘Quick Presets’ control allows for the coarsest (yet easiest to use) update of filter criteria. The other fields in the top row allow presets for each category while the tabs in the form below allow to fine-tune filter and priorization options where necessary.
Joint filtering of multiple cases
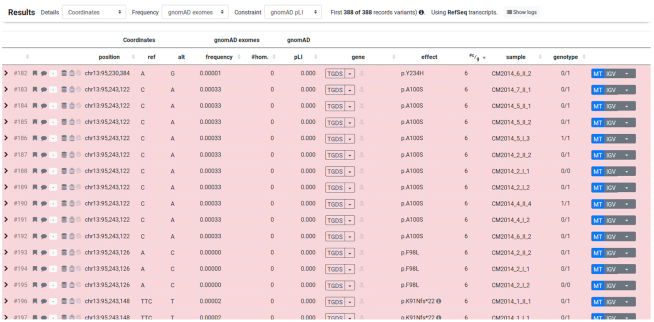
VarFish is capable of filtering the variants of multiple cases at once. The resulting variants are annotated with the number of cases that have at least one variant identified in a given gene. The result can then be sorted by that number to identify genes that carry rare variants of interesting impact and mode of inheritance. To demonstrate this, we performed a re-analysis of the original cohort used for identifying TGDS as a disease gene for Catel-Manzke syndrome published earlier (34). For this analysis, the second preset ‘recessive inheritance’ was applied first, followed by relaxing the quality thresholds as the data was generated in the early days of WES sequencing. Figure 5 shows the top results. Of the resulting 388 variant records the only gene carrying variants in all six sequenced cases was TGDS, the next gene listed matched only two cases.
Figure 5.
Catel-Manzke cohort filtering results (first 15 variants shown) to reproduce the finding that TGDS is the most likely candidate for being the disease gene.
DISCUSSION
We here present VarFish, a flexible platform for the automated annotation of small variants, their filtering and prioritization. We demonstrated its use and effectiveness in a practical use case. The system aims at empowering biomedical and clinician researchers to perform complex, customizable variant prioritization and filtering in a maximally flexible way. Instead of implementing new custom scores, VarFish builds on and combines best-in-class scoring algorithms such as CADD and MutationTaster for variant pathogenicity prediction and gene-phenotype similarity computation.
Annotation, filtering, and prioritization are shown to the user, allowing swift interactive processing by sorting variants according to different criteria. Comprehensive link-outs to external gene and variant databases are provided, and the integration of the IGV genome browser enables raw data inspection. Allowing the user to leave annotation flags, color code and free-text comments has proven highly useful for our in-house users. Data can be analyzed in an integrated platform up to ACMG-AMP variant assessment, followed by downloading a spreadsheet file for documentation in external systems.
We remark that the flexibility of VarFish comes at the prices of many filter settings that introduce a high degree of complexity when compared to Exomiser or MutationDistiller that work more similar to a ‘single click’ fashion. Both approaches (high vs. low flexibility/complexity) have their advantages and disadvantages and we plan to implement a ‘beginner mode’ in VarFish in future versions to lower the entry barrier for new users.
Further advanced features such as collecting cases in projects and performing joint queries on multiple cases at once allow answering research questions that previously required bioinformatics expertise. For example, this allows an integrated characterization of disease cohorts by screening for pathogenic variants in known disease-associated genes followed by a joint analysis of the remaining cases, e.g. to identify jointly mutated genes.
Finally, the pipeline to generate the background database files from publicly available sources and all software is free to use in academic and commercial settings. This ensures full transparency, allows for setting up a fully reproducible variant analysis pipeline, and provides users with the advantage of open source systems in that there is no vendor lock-in (in concordance with the FAIR (35) data management principles) and users are less dependent of the original software author. Both open and closed source software have their advantages and disadvantages, including (in)dependency on commercial vendor's decisions regarding pricing and discontinuing products and how easy it is to adjust the software to one's needs. VarFish is used in the authors’ daily work and actively maintained. We welcome questions, comments, and suggestions via email or the Github project's issue tracker.
CONCLUSION
VarFish is a flexible and powerful platform for variant filtering, prioritization, and user-based annotation. With its laboratory notebook features, it promotes collaborative analysis of cases within a center and the re-analysis of variants at multiple points in time. Future development will focus on the extension of re-analysis features, cross-center collaboration and supporting the analysis of structural variants, and support for whole genome data.
WEB SERVER IMPLEMENTATION
VarFish is implemented in Python 3 with the Django web framework based on SODAR-core (https://github.com/bihealth/sodar_core) using PostgreSQL 11 for data storage and querying and VCFPy (36) for file parsing. In our installation, it runs on a Linux container server with 128GB of RAM, 16 cores and 1TB of disk.
DATA AVAILABILITY
VarFish is available for public usage at https://varfish-kiosk.bihealth.org. A demonstration instance with additional collaboration features has been setup at https://varfish-demo.bihealth.org. The source code is available from https://github.com/bihealth/varfish-server.
No new data was generated for this study. For demonstration purposes we used data from a previous publication (34).
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Hannah Kemmer, Jirko Kühnisch, and Elisa Schäfer for their feedback on VarFish. Further, the authors thank Murim Choi for his help with the UCSC vertebrate conservation data. The icons used in Figure 1 and the visual abstract were taken from openmoji.org.
Contributor Information
Manuel Holtgrewe, CUBI – Core Unit Bioinformatics, Berlin Institute of Health, Berlin 10117, Germany; Charité – Universitätsmedizin Berlin, corporate member of Freie Universität Berlin, Humboldt-Universität zu Berlin, and Berlin Institute of Health, Berlin 10117, Germany.
Oliver Stolpe, CUBI – Core Unit Bioinformatics, Berlin Institute of Health, Berlin 10117, Germany; Charité – Universitätsmedizin Berlin, corporate member of Freie Universität Berlin, Humboldt-Universität zu Berlin, and Berlin Institute of Health, Berlin 10117, Germany.
Mikko Nieminen, CUBI – Core Unit Bioinformatics, Berlin Institute of Health, Berlin 10117, Germany; Max Delbrück Center for Molecular Medicine, Berlin 13125, Germany.
Stefan Mundlos, Institute of Medical Genetics and Human Genetics, Charité – Universitätsmedizin Berlin, corporate member of Freie Universität Berlin, Humboldt-Universität zu Berlin, and Berlin Institute of Health, Berlin 13353, Germany; Development and Disease Group, Max Planck Institute for Medical Genetics, Berlin 14195, Germany.
Alexej Knaus, Institute for Genomic Statistics and Bioinformatics, University Hospital Bonn, Bonn 53127, Germany.
Uwe Kornak, Institute of Medical Genetics and Human Genetics, Charité – Universitätsmedizin Berlin, corporate member of Freie Universität Berlin, Humboldt-Universität zu Berlin, and Berlin Institute of Health, Berlin 13353, Germany; Development and Disease Group, Max Planck Institute for Medical Genetics, Berlin 14195, Germany.
Dominik Seelow, Institute of Medical Genetics and Human Genetics, Charité – Universitätsmedizin Berlin, corporate member of Freie Universität Berlin, Humboldt-Universität zu Berlin, and Berlin Institute of Health, Berlin 13353, Germany; Berlin Institute of Health (BIH), Anna-Louisa-Karsch-Str. 2, 10178 Berlin, Germany.
Lara Segebrecht, Institute of Medical Genetics and Human Genetics, Charité – Universitätsmedizin Berlin, corporate member of Freie Universität Berlin, Humboldt-Universität zu Berlin, and Berlin Institute of Health, Berlin 13353, Germany.
Malte Spielmann, Development and Disease Group, Max Planck Institute for Medical Genetics, Berlin 14195, Germany; Institut für Humangenetik Lübeck, Universität zu Lübeck, 23538 Lübeck, Germany.
Björn Fischer-Zirnsak, Institute of Medical Genetics and Human Genetics, Charité – Universitätsmedizin Berlin, corporate member of Freie Universität Berlin, Humboldt-Universität zu Berlin, and Berlin Institute of Health, Berlin 13353, Germany; Development and Disease Group, Max Planck Institute for Medical Genetics, Berlin 14195, Germany.
Felix Boschann, Institute of Medical Genetics and Human Genetics, Charité – Universitätsmedizin Berlin, corporate member of Freie Universität Berlin, Humboldt-Universität zu Berlin, and Berlin Institute of Health, Berlin 13353, Germany.
Ute Scholl, Berlin Institute of Health (BIH), Anna-Louisa-Karsch-Str. 2, 10178 Berlin, Germany; Charité – Universitätsmedizin Berlin, corporate member of Freie Universität Berlin, Humboldt-Universität zu Berlin, and Berlin Institute of Health, Department of Nephrology and Medical Intensive Care, BCRT – Berlin Institute of Health Center for Regenerative Therapies, 13353 Berlin, Germany.
Nadja Ehmke, Institute of Medical Genetics and Human Genetics, Charité – Universitätsmedizin Berlin, corporate member of Freie Universität Berlin, Humboldt-Universität zu Berlin, and Berlin Institute of Health, Berlin 13353, Germany.
Dieter Beule, CUBI – Core Unit Bioinformatics, Berlin Institute of Health, Berlin 10117, Germany; Max Delbrück Center for Molecular Medicine, Berlin 13125, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Stiftung Charité (to U.S.); Charité Rahel-Hirsch-Stipendium (to N.E.); Funding for open access charge: Berlin Institute of Health/Charité funds.
Conflict of interest statement. None declared.
REFERENCES
- 1. Shendure J., Ji H.. Next-generation DNA sequencing. Nat. Biotechnol. 2008; 26:1135–1145. [DOI] [PubMed] [Google Scholar]
- 2. Sun Y., Ruivenkamp C.A.L., Hoffer M.J.V., Vrijenhoek T., Kriek M., van Asperen C.J., den Dunnen J.T., Santen G.W.E.. Next-generation diagnostics: gene panel, exome, or whole genome. Hum. Mutat. 2015; 36:648–655. [DOI] [PubMed] [Google Scholar]
- 3. Alfares A., Aloraini T., Subaie L.A., Alissa A., Qudsi A.A., Alahmad A., Mutairi F.A., Alswaid A., Alothaim A., Eyaid W. et al.. Whole-genome sequencing offers additional but limited clinical utility compared with reanalysis of whole-exome sequencing. Genet. Med. 2018; 20:1328–1333. [DOI] [PubMed] [Google Scholar]
- 4. Smedley D., Jacobsen J.O.B., Jäger M., Köhler S., Holtgrewe M., Schubach M., Siragusa E., Zemojtel T., Buske O.J., Washington N.L. et al.. Next-generation diagnostics and disease-gene discovery with the Exomiser. Nat. Protoc. 2015; 10:2004–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sifrim A., Popovic D., Tranchevent L.-C., Ardeshirdavani A., Sakai R., Konings P., Vermeesch J.R., Aerts J., De Moor B., Moreau Y.. eXtasy: variant prioritization by genomic data fusion. Nat. Methods. 2013; 10:1083–1084. [DOI] [PubMed] [Google Scholar]
- 6. Kamphans T., Krawitz P.M.. GeneTalk: an expert exchange platform for assessing rare sequence variants in personal genomes. Bioinformatics. 2012; 28:2515–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hombach D., Schuelke M., Knierim E., Ehmke N., Schwarz J.M., Fischer-Zirnsak B., Seelow D.. MutationDistiller: user-driven identification of pathogenic DNA variants. Nucleic Acids Res. 2019; 47:W114–W120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Antanaviciute A., Watson C.M., Harrison S.M., Lascelles C., Crinnion L., Markham A.F., Bonthron D.T., Carr I.M.. OVA: integrating molecular and physical phenotype data from multiple biomedical domain ontologies with variant filtering for enhanced variant prioritization. Bioinformatics. 2015; 31:3822–3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Javed A., Agrawal S., Ng P.C.. Phen-Gen: combining phenotype and genotype to analyze rare disorders. Nat. Methods. 2014; 11:935–937. [DOI] [PubMed] [Google Scholar]
- 10. Alexander J., Mantzaris D., Georgitsi M., Drineas P., Paschou P.. Variant ranker: a web-tool to rank genomic data according to functional significance. BMC Bioinformatics. 2017; 18:341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jiang J., Gu J., Zhao T., Lu H.. VCF-Server: a web-based visualization tool for high-throughput variant data mining and management. Mol Genet Genomic Med. 2019; 7:e00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R.S., Thormann A., Flicek P., Cunningham F.. The Ensembl variant effect predictor. Genome Biol. 2016; 17:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chang X., Wang K.. wANNOVAR: annotating genetic variants for personal genomes via the web. J. Med. Genet. 2012; 49:433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E. et al.. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015; 17:405–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T. et al.. The variant call format and VCFtools. Bioinformatics. 2011; 27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J. et al.. PLINK: A tool set for Whole-Genome association and Population-Based linkage analyses. Am. J. Hum. Genet. 2007; 81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jäger M., Wang K., Bauer S., Smedley D., Krawitz P., Robinson P.N.. Jannovar: a java library for exome annotation. Hum. Mutat. 2014; 35:548–555. [DOI] [PubMed] [Google Scholar]
- 18. O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D. et al.. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016; 44:D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zerbino D.R., Achuthan P., Akanni W., Amode M.R., Barrell D., Bhai J., Billis K., Cummins C., Gall A., Girón C.G. et al.. Ensembl 2018. Nucleic Acids Res. 2018; 46:D754–D761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B. et al.. Analysis of protein-coding genetic variation in 60, 706 humans. Nature. 2016; 536:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P.. The mutational constraint spectrum quantified from variation in 141,456 humans. 2020; bioRxiv doi:08 April 2020, preprint: not peer reviewed Genomics 10.1101/531210. [DOI] [PMC free article] [PubMed]
- 22. 1000 Genomes Project Consortium Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A. et al.. A global reference for human genetic variation. Nature. 2015; 526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Jang W. et al.. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018; 46:D1062–D1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Robinson P.N., Piro R.M., Jäger M.. Computational Exome and Genome Analysis. 2018; Boca Raton: CRC Press. [Google Scholar]
- 25. Shringarpure S.S., Bustamante C.D.. Privacy risks from genomic data-sharing beacons. Am. J. Hum. Genet. 2015; 97:631–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pedersen B.S., Quinlan A.R.. Who's who? Detecting and resolving sample anomalies in human DNA sequencing studies with peddy. Am. J. Hum. Genet. 2017; 100:406–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shyr C., Kushniruk A., van Karnebeek C.D.M., Wasserman W.W.. Dynamic software design for clinical exome and genome analyses: insights from bioinformaticians, clinical geneticists, and genetic counselors. J. Am. Med. Inform. Assoc. 2016; 23:257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D.. The human genome browser at UCSC. Genome Res. 2002; 12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Robinson P.N., Köhler S., Bauer S., Seelow D., Horn D., Mundlos S.. The human phenotype ontology: a tool for annotating and analyzing human hereditary disease. Am. J. Hum. Genet. 2008; 83:610–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Köster J., Rahmann S.. Snakemake–a scalable bioinformatics workflow engine. Bioinformatics. 2012; 28:2520–2522. [DOI] [PubMed] [Google Scholar]
- 31. Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P.. Integrative genomics viewer. Nat. Biotechnol. 2011; 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schwarz J.M., Cooper D.N., Schuelke M., Seelow D.. MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods. 2014; 11:361–362. [DOI] [PubMed] [Google Scholar]
- 33. Cao Y., Tokita M.J., Chen E.S., Ghosh R., Chen T., Feng Y., Gorman E., Gibellini F., Ward P.A., Braxton A. et al.. A clinical survey of mosaic single nucleotide variants in disease-causing genes detected by exome sequencing. Genome Med. 2019; 11:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ehmke N., Caliebe A., Koenig R., Kant S.G., Stark Z., Cormier-Daire V., Wieczorek D., Gillessen-Kaesbach G., Hoff K., Kawalia A. et al.. Homozygous and Compound-heterozygous mutations in TGDS cause Catel-Manzke syndrome. Am. J. Hum. Genet. 2014; 95:763–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wilkinson M.D., Dumontier M., Aalbersberg I.J., Appleton G., Axton M., Baak A., Blomberg N., Boiten J.-W., da Silva Santos L.B., Bourne P.E. et al.. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data. 2016; 3:160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Holtgrewe M., Beule D.. VCFPy: a Python 3 library with good support for both reading and writing VCF. JOSS. 2016; 1:85. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
VarFish is available for public usage at https://varfish-kiosk.bihealth.org. A demonstration instance with additional collaboration features has been setup at https://varfish-demo.bihealth.org. The source code is available from https://github.com/bihealth/varfish-server.
No new data was generated for this study. For demonstration purposes we used data from a previous publication (34).