Abstract
The rMAPS2 (RNA Map Analysis and Plotting Server 2) web server, freely available at http://rmaps.cecsresearch.org/, has provided the high-throughput sequencing data research community with curated tools for the identification of RNA binding protein sites. rMAPS2 analyzes differential alternative splicing or CLIP peak data obtained from high-throughput sequencing data analysis tools like MISO, rMATS, Piranha, PIPE-CLIP and PARalyzer, and then, graphically displays enriched RNA-binding protein target sites. The initial release of rMAPS focused only on the most common alternative splicing event, skipped exon or exon skipping. However, there was a high demand for the analysis of other major types of alternative splicing events, especially for retained intron events since this is the most common type of alternative splicing in plants, such as Arabidopsis thaliana. Here, we expanded the implementation of rMAPS2 to facilitate analyses for all five major types of alternative splicing events: skipped exon, mutually exclusive exons, alternative 5′ splice site, alternative 3′ splice site and retained intron. In addition, by employing multi-threading, rMAPS2 has vastly improved the user experience with significant reductions in running time, ∼3.5 min for the analysis of all five major alternative splicing types at once.
INTRODUCTION
RNA-binding proteins (RBPs) play important roles in regulation of transcriptional and post-transcriptional gene expression in eukaryotic cells (1). Alternative splicing (AS) is a prevalent mechanism for generating transcriptomic and proteomic complexity and diversity in eukaryotic cells (2). The regulation of AS, one of several roles of RBPs, is an important post-transcriptional regulatory mechanism. It is known that RBP regulation of AS is dependent on the binding site positions of RBPs with respect to the regulated AS events (3–7). Thus, spatial analyses of RBP binding sites and motifs around regulated AS events can help determine whether AS events in specific biological processes are regulated by specific RBPs.
Interactions between RBPs and RNA can be studied using high-throughput sequencing data generated from RNA sequencing (RNA-seq) (8) and cross-linking immunoprecipitation sequencing (CLIP-seq). Antibodies are attached to RBPs of interest, and the RNA sequences bound to these RBPs are then extracted through immunoprecipitation (9). With the advancement of high-throughput sequencing depth and quality, more reliable differentially regulated AS studies have become available. Studying differentially expressed AS events in two different conditions can provide profound insights on gene regulation related to RBP activities.
In 2016, we introduced rMAPS, a web server to perform RBP motif and binding site analyses with data on alternatively spliced exons (10), in order to fulfil the need for a user-friendly way to integrate RNA-seq analysis of AS with information about RBP consensus motifs and CLIP-seq peaks. rMAPS tries to find global patterns of motif enrichment found across similar alternative skipped exons by scanning for motifs near the exon boundaries. rMAPS searches for a global pattern of certain motifs, specifically located near the alternatively spliced exon boundaries, giving rise to the upregulation (enhancer) or downregulation (silencer) of the alternative skipped exon as compared to background (unregulated exons). Since its initial release, rMAPS has received more than 120 000 visits from over 35 countries, generating 15 RNA maps per week. During the past 12 months, usage has increased from 11 to 19 RNA maps per week, and the number of citations has doubled during the same time period. Clearly, rMAPS is getting more attention from the high-throughput sequencing and RBP research communities, and there is every indication that these numbers will continue to rise as knowledge of rMAPS’s capability spreads.
Existing RBP analysis web servers include RBPmap (11), which predicts RBP binding sites on RNA sequences, PNImodeler (12), which infers protein-binding NTs from sequence data, RBPmotif (13), which performs de novo motif finding and secondary structure analysis, and HotSPRing (14), which predicts the hot spots in RBPs. However, none of these tools are known to perform RBP motif or CLIP peak search in a binding-site-position dependent manner.
Although the initial release of rMAPS provided researchers with much functionality, allowing interpretation of differential AS events or CLIP peaks obtained from massive sequencing data analysis tools such as MISO (15), rMATS (16–18), Piranha (19), PIPE-CLIP (20) and PARalyzer (21), it was restricted to RBP motif analysis of skipped exon (SE) events only. The initial release of rMAPS focused on SE events because these are the most common alternative splicing events in higher eukaryotes. However, the demand for RBP motif analysis on other types of AS events was high, especially for retained intron (RI) events, which are a prominent type of AS event in plants. Here we implement and release a new version, rMAPS2, with several improvements. In addition to a contemporary graphical user interface (GUI) (see new features and updates section), rMAPS2 provides RBP motif enrichment analysis and CLIP peak enrichment analysis for all five major types of AS events: skipped exon (SE), mutually exclusive exon (MXE), alternative 5′ splice site (A5SS), alternative 3′ splice site (A3SS) and retained intron (RI). To broaden its use across the scientific community, rMAPS2 is now capable of analyzing more than 13 different species including the most widely researched plant species, Arabidopsis thaliana, with plans to continue adding more species in the future (see Supplementary Table S1 for the list of supported species). To improve running time, major parts of the code have been restructured to employ multi-threading, and consequently, rMAPS2 can run more than 10 times faster, dropping from up to 8 min for a single SE event type to about 3.5 min for all five AS types together.
rMAPS2 WEB SERVER
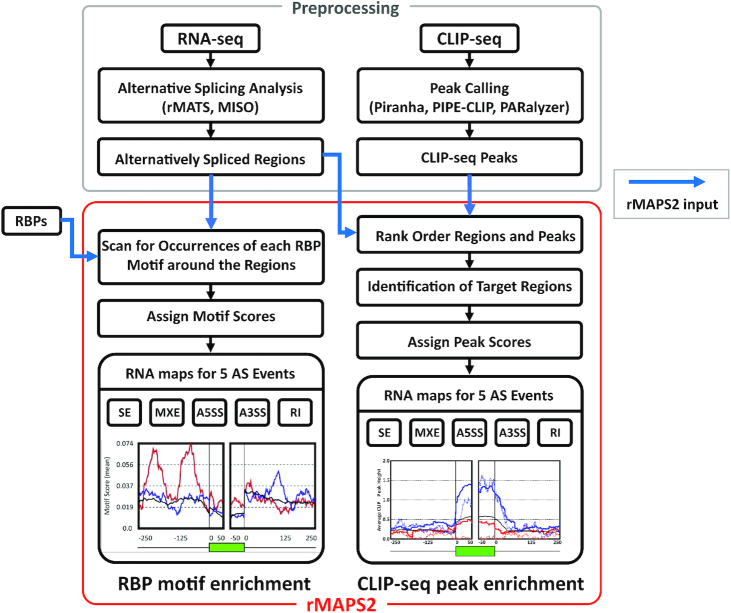
rMAPS2, RNA Map Analysis and Plotting Server 2, provides two major types of analyses: RBP motif enrichment analysis and CLIP-seq binding sites (or peak) enrichment analysis. As shown in Figure 1, rMAPS2 systematically performs binding site sequence enrichment analyses of RBPs around AS event sites, or within the exonic and flanking intronic regions. Then, rMAPS2 generates plots that show the spatial distributions of RBP motif density (RNA maps) for a set of known RBP motifs collected from the previous studies (22–24) (see Supplementary Table S2 for the list of RBPs) or from user input motifs. rMAPS2 can also examine RBP motifs within endogenous RNAs identified by CLIP-seq and create an RNA map, visualizing the spatial distribution of binding sites for the RBP of interest. Each plot generated can provide insights on position-dependent functions of an RBP. Users can easily upload their differential alternative splicing event data from two sample groups to search RBP motif enrichment for over 100 known RBPs motifs or for their own binding motif sequences of interest.
Figure 1.
The overall workflow of rMAPS2. The RBP motif analysis takes the AS analysis results from RNA-seq data, then scans for the occurrences of the known RBP motifs around the differentially regulated AS regions to generate an RNA map for each RBP. Similarly, the CLIP-seq binding site analysis generates an RNA map using the signals of CLIP-seq binding sites around the differentially regulated AS regions. Blue arrows indicate rMAPS2 input files.
RBP motif analysis
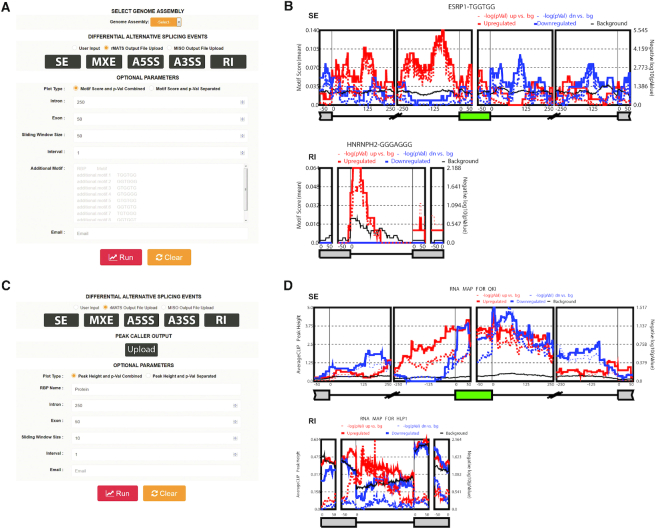
As the one of the main analyses of rMAPS2, RBP motif enrichment analysis searches significantly enriched RBP motifs from differentially regulated AS events between two sample groups as compared to non-regulated background AS events. rMAPS2 generates RNA maps to visualize spatial distributions of RBP motif density around the associated exonic and flanking intronic regions. RBP motif enrichment analysis takes AS events identified from RNA-seq data and consensus motifs of RBPs as input data. For differentially regulated AS events data from RNA-seq, users can simply upload output files obtained from rMATS and MISO, or user defined files following rMAPS2 input file format. By default, the rMAPS2 server provides a number of known consensus motifs including numerous well-characterized splicing factors from previous studies. Users can also upload additional motifs of interest. Once a user specifies all required inputs (genome assembly and AS events) and optional parameters such as plot type, intron and exon size, sliding window size, interval, email address and additional motifs (Figure 2A), RBP motif enrichment analysis evaluates the occurrences of each RBP motif in the upstream exon, upstream flanking intron, target exon, downstream flanking intron and downstream exon separately (having additional exon and flanking intron regions for MXE events). For intronic sequences, RBP motif enrichment analysis excludes the 20 bp sequence within the 3′ splice site and the 6 bp sequence within the 5′ splice site, which are strongly constrained by the consensus splice site signals. For each motif, motif scores representing density are calculated within a given sliding window (default 50 bp) as the overall percentage of nucleotides covered by the motif. After computing motif scores separately for the set of upregulated, downregulated and background (non-regulated) exons, rMAPS2 plots the motif scores for these three sets of exons in red, blue and black solid lines, respectively. In addition, to identify specific positions with a significant difference in motif score between different sets of exons, rMAPS2 employs Wilcoxon's rank sum test, as described in the previous version (10), for each sliding window between upregulated versus background or downregulated versus background exons, and plots negative log10P-values with red and blue dotted lines for upregulated versus background and downregulated versus background respectively. Note that the solid lines and dashed lines can be displayed on a separate plot using the optional parameter ‘Plot Type’. Plots with separate dashed lines are shown in Supplementary Figure S1. RBP motif enrichment analysis provides URL links for the entire set of results, or partial results, for future use (see Figure 2B for example output). Researchers can visually examine the output of RBP motif enrichment analysis (i.e. RNA maps) to identify RBPs that show high motif scores with low P-values which, in turn, suggests that given RBPs have an important role in AS regulation. Further downstream studies such as CLIP-seq analysis can be performed for such RBPs.
Figure 2.
A view of rMAPS2 input and output pages. (A) Input page for the RBP motif analysis tool. After providing all required inputs, clicking on the ‘Run’ button will start the analysis. (B) An example output page of the motif analysis tool for SE and RI events (see Supplementary Figure S2 for the A5/3SS and MXE cases). It generates RNA maps for known RBP motifs for each AS type along with a URL link for the entire results for future use. (C) Input page for the CLIP-seq binding site analysis tool. It requires AS events from RNA-seq data and peaks from CLIP-seq data. (D) An example output page of the CLIP-seq binding site analysis tool for SE and RI events (see Supplementary Figure S3 for the A5/3SS and MXE cases). The RI case is generated from Arabidopsis thaliana datasets to illustrate the importance of RI events in plants. It generates an RNA map for the RBP of interest for each AS type and a URL link for the entire results for future use.
CLIP-seq binding site (peak) analysis
CLIP-seq peak analysis combines differentially regulated AS events data obtained from RNA-seq with peaks called from CLIP-seq. CLIP-seq provides insight into how an RBP interacts with its target RNA by revealing their location of binding. The CLIP-seq peak, which represents the location where an RBP interacts with RNA, provides clues to how the RBP functions in the regulation of gene expression, and the significance of RBP location can be confrimed by looking at the enrichment of the peaks related to various AS events. As discussed in Yee's RBP-Maps study (25), CLIP-seq peak analysis can be affected by the use of peak versus read density. Since rMAPS2 uses the output of the peak callers, it utilizes peaks information only to generate an RNA map of peaks near the AS events. As input, CLIP-seq peak analysis requires AS events data from RNA-seq, peaks called from CLIP-seq peak callers such as Piranha, PIPE_CLIP and PARalyzer, the RBP name used in the CLIP-seq, and optional parameters similar to those used in RBP motif analysis (Figure 2C).
CLIP-seq peak analysis examines the CLIP-seq peaks in the upstream exon, upstream flanking intron, target exon, downstream flanking intron and downstream exon separately (having additional exon and flanking intron regions for MXE events) following specified parameters. The resulting RNA map visualizes the spatial distributions of average CLIP-seq signals in the sliding window (default 50bp) for upregulated, downregulated and background exons using the same colors as those in the RBP motif enrichment analysis (i.e. red for upregulated, blue for downregulated and black for background). Output files can be downloaded via provided URL links for future use (see Figure 2D for an example output).
NEW FEATURES AND UPDATES
In addition to more insightful user interface improvements including an animated step-by-step user guide, the following key features and updates are implemented in rMAPS2 to support five major types of AS events for a wide range of species, and to reduce the running time.
Five major types of AS events
Unlike its predecessor that can run with only one type of AS event (SE), rMAPS2 can run with five major types of AS events: SE, MXE, A5SS, A3SS and RI. The initial release of rMAPS only focused on one type of AS event, SE, because it is the most common type of AS events in higher eukaryotes. There has been increasing demand for the systematic analysis of additional major types of AS events: MXE, A5SS, A3SS and RI. In particular, there was a high demand for the analysis related to RI events since RI events are known to be the most common type of AS events in plants. Since the regulatory role of RBPs is not limited to the SE event, examining all five major types of AS events for the enrichment of RBP motifs would help uncover more regulatory roles of RBPs. The code was restructured to take different types of AS events as input and to generate RNA maps accordingly.
Wide range of species
Since the initial release, there has been a high demand for supporting more species. We actively expanded the list of supporting species by compiling a genome assembly, constructing a search index, and intensively testing with real high-throughput sequencing data. Up to date, rMAPS2 supports 17 genome assemblies for 13 species: human (hg19, hg38), mouse (mm10), rat (rn6), fruit fly (dm3, dm6), roundworm (ce11), zebrafish (danRer10, danRer11), African clawed frog (xenLae2), western clawed frog (xenTro9), pig (susScr11), chicken (galGal5, galGal6), cow (bosTau9), thale cress (araTha1), and rice (oSa7). Some species have more than one genome assembly because multiple genome assemblies are in use in the research field. For example, both hg19 and hg38 assemblies are actively used for human studies. A few notable genomes are added to meet users’ requests. Besides the addition of the most frequently studied plant genome, araTha1, two frog genomes (xenLae2 and xenTro9) have been recently added. As the CRISPR/Cas9 system is getting more popular, there has been a high demand for these genomes as well. rMAPS2 will continuously add more species to facilitate RBP studies across organisms.
Code restructuring
Although basic algorithms are still inherited from the initial version which was released in 2016, the code for rMAPS2 has been restructured (i) to support more types of AS events without changing external behavior, (ii) to provide more insightful user experience and (iii) to achieve a significantly reduced running time. To support additional types of AS events, the existing algorithm for SE event was revised to precisely construct the boundaries of exons and introns associated with other types of AS events. Especially for RI events, since the length of introns are relatively long (>3000 bp on average for human introns in CDS regions (26)), the intron of interest was binned to 100 bins and the motif scores were computed separately for each bin. To provide a more insightful and seamless user experience, we rewrote the front-end of the server from HTML and PHP to ASP.NET Model-View-Controller (MVC) framework which is known to perform better than PHP and updated rMAPS2 user guide with step-by-step animations. To make significant reductions in running time, we employed multi-threading to generate RNA maps for multiple AS types at the same time. Previously, a single SE event required 8 min to run. However, currently the RBP motif enrichment analysis for all five types of AS events can be completed in about 3.5 min.
Technical implementation
The server consists of a front-end web user interface, based on ASP .NET MVC 4, and a back-end application component written in Python that performs the actual analysis and plotting. The web front-end runs on IBM System x3850 × 5 server, equipped with Intel Xeon E7–4820 CPUs on Microsoft Windows Server 2016 Datacenter. To ensure a reasonable run time for each analysis, the actual analysis is performed on a cluster that runs on HPE Proliant DL360p Gen8 server, equipped with Intel Xeon E5–2687W V2 CPUs and 128 GB of memory on CentOS 7.6. The typical run time for an RBP motif enrichment analysis is about 3.5 min and CLIP-seq peak enrichment analysis take less than 1 min. rMAPS2 was tested on multiple browsers including Google Chrome 80.0.3987.87, Microsoft Edge 44.18362.449.0, Microsoft IE 11.0.9600.18618 and Mozilla Firefox 73.0.1.
USE CASE
To illustrate the new features and updates introduced with rMAPS2, we used RNA-seq data published by Bebee et al. (3) for RBP motif enrichment analysis and used closely related RNA-seq and QKI CLIP-seq data published by Bebee et al. (27) and Pillman et al. (28) for CLIP-seq peak enrichment analysis. Both datasets used for CLIP-seq peak analysis are from a human epithelial-mesenchymal transition (EMT) study. We chose these datasets to show that rMAPS2 can work not only on datasets from the same experiment but also on datasets from similar or closely related experiments. At last, the importance of the RI events in plants was illustrated using RNA-seq and CLIP-seq data from A. thaliana.
To conduct RBP motif analysis, we ran the rMATS (16–18) framework on the first RNA-seq data (CTRL versus ESRP double knockout). We then ran rMAPS2 RBP motif enrichment analysis on all five types of AS events reported from rMATS, and it took less than 3 min. As we can see from the SE case in Figure 2B, ESRP motifs (G-rich) are enriched in the upstream intron and within the target exon of the upregulated exons (solid red line is significantly higher than solid black line) while ESRP motifs are enriched in the downstream intron of the downregulated exons (solid blue line is significantly higher than solid black line). ESRP is known to promote inclusion of the exons when it binds to the upstream intron of the target exon or target exon body and suppress inclusion of the exons when it binds to the downstream intron of the target exon (4). Since the RNA-seq was performed on ESRP knockdown data, Figure 2B, the SE case clearly shows that rMAPS2 accurately depicts the expected pattern. The RI case in Figure 2B shows that HNRNPH2 promotes intron retention when it binds to the intron (solid red line), which is in agreement with a previous splicing modulator detection study (29).
For the CLIP-seq peak enrichment analysis, we ran the rMATS framework on the second RNA-seq data (day 0 versus day 7) and ran Piranha (19) on CLIP-seq data. Using the output from rMATS and Piranha, we ran the rMAPS2 CLIP-seq peak enrichment analysis which took about 30 s. The SE case in Figure 2D shows the enrichment of a CLIP-seq peak in the upstream intron of upregulated exons (solid red line and small P-values indicated by dotted red line) and a CLIP-seq peak in the downstream intron of downregulated exons (solid blue line with small P-values indicated by dotted blue line). This agrees with the Pillman's study (28) which was done only for the SE case, and it illustrates that CLIP-seq peak enrichment studies can be performed on separate datasets as long as both datasets are from closely related experiments. To illustrate the importance of the RI events in plants, we generated the RI case in Figure 2D using additional RNA-seq data from NCBI GEO (accession number: GSE146189) and HLP1 CLIP-seq data from Zhang's study (30). The RI case in Figure 2D clearly shows the enrichment of HLP1 CLIP peaks in the upstream exons and within the intron of upregulated introns (solid red line with small P-values) which suggests HLP1 could promote retention of the intron when it binds to the upstream exon or intron itself.
This use case clearly illustrates that rMAPS2 now works with all major types of AS events (see Supplementary Figures S2 and 3 for A5/3SS and MXE cases) with significantly reduced running time. The resulting plots from both analyses agree with known binding patterns and these plots can provide insightful visualization of regulatory roles of RBPs in major types of AS event.
CONCLUSION AND FUTURE PERSPECTIVES
The new version of rMAPS2, a web server originally designed to integrate RNA-seq results of differential alternative splicing with information about RBP motif occurrences and CLIP-seq binding sites across the transcriptome, has large improvements as a result of continued development. rMAPS2 now supports all five major types of AS events, has a significantly reduced running time, supports an extended list of species and provides a more insightful user interface. rMAPS2 offers over 110 known RBP motif sequences obtained from the literature, and users can provide RBP motif sequences specifically related to their research. This large set of RBPs and their motifs provides a good basis for the study of RBP regulation of splicing. To respond to the high demand from users, rMAPS2 now supports the most widely used plant species, A. thaliana, and two frog genome assemblies (African clawed frog and Western clawed frog), commonly used with the CRISPR/Cas9 system. We will continue to add more species and RBP motifs. One future improvement includes generating and displaying plots on the fly. With the right development technique, we will be able to generate and display plots in real time so that users do not have to wait until all plots are generated. Overall, the new release of rMAPS2 is a powerful web server to explore binding patterns of RBPs in splicing and is certain to improve our understanding of the regulatory role of RBPs in various types of AS events.
DATA AVAILABILITY
rMAPS2 is available from https://rmaps.cecsresearch.org/. The website is free and open to all users and there is no login requirement.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Ron Lile for extensive testing and constructive comments. We would like to thank Julia Chariker for insightful discussions.
Contributor Information
Jae Y Hwang, Department of Computer Science and Engineering, University of Louisville, Louisville, KY 40292, USA.
Sungbo Jung, Department of Computer Science and Engineering, University of Louisville, Louisville, KY 40292, USA.
Tae L Kook, Department of Computer Science and Engineering, University of Louisville, Louisville, KY 40292, USA.
Eric C Rouchka, Department of Computer Science and Engineering, University of Louisville, Louisville, KY 40292, USA; KBRIN Bioinformatics Core, University of Louisville, Louisville, KY 40292, USA.
Jinwoong Bok, Department of Anatomy, Yonsei University College of Medicine, Seoul 03722, Republic of Korea; Department of Otorhinolaryngology, Yonsei University College of Medicine, Seoul 03722, Republic of Korea; BK21 PLUS project for Medical Science, Yonsei University College of Medicine, Seoul 03722, Republic of Korea.
Juw W Park, Department of Computer Science and Engineering, University of Louisville, Louisville, KY 40292, USA; KBRIN Bioinformatics Core, University of Louisville, Louisville, KY 40292, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R15GM126446, P20GM103436 to J.P.]; National Research Foundation of Korea [2016R1A5A2008630, 2017R1A2B3009133 to J.B.]; Funding for open access charge: National Institutes of Health [R15GM126446].
Conflict of interest statement. None declared.
REFERENCES
- 1. Li X., Kazan H., Lipshitz H.D., Morris Q.D.. Finding the target sites of RNA-binding proteins. Wiley Interdiscip. Rev. RNA. 2014; 5:111–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen M., Manley J.L.. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 2009; 10:741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bebee T.W., Park J.W., Sheridan K.I., Warzecha C.C., Cieply B.W., Rohacek A.M., Xing Y., Carstens R.P.. The splicing regulators Esrp1 and Esrp2 direct an epithelial splicing program essential for mammalian development. Elife. 2015; 4:e08954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Warzecha C.C., Jiang P., Amirikian K., Dittmar K.A., Lu H., Shen S., Guo W., Xing Y., Carstens R.P.. An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition. EMBO J. 2010; 29:3286–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Martinez-Contreras R., Fisette J.F., Nasim F.U., Madden R., Cordeau M., Chabot B.. Intronic binding sites for hnRNP A/B and hnRNP F/H proteins stimulate pre-mRNA splicing. PLoS Biol. 2006; 4:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ule J., Stefani G., Mele A., Ruggiu M., Wang X., Taneri B., Gaasterland T., Blencowe B.J., Darnell R.B.. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006; 444:580–586. [DOI] [PubMed] [Google Scholar]
- 7. Yeo G.W., Coufal N.G., Liang T.Y., Peng G.E., Fu X.D., Gage F.H.. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat. Struct. Mol. Biol. 2009; 16:130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lu Z.X., Huang Q., Park J.W., Shen S., Lin L., Tokheim C.J., Henry M.D., Xing Y.. Transcriptome-wide landscape of pre-mRNA alternative splicing associated with metastatic colonization. Mol. Cancer Res. 2015; 13:305–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shapiro I.M., Cheng A.W., Flytzanis N.C., Balsamo M., Condeelis J.S., Oktay M.H., Burge C.B., Gertler F.B.. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLos Genet. 2011; 7:e1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Park J.W., Jung S., Rouchka E.C., Tseng Y.T., Xing Y.. rMAPS: RNA map analysis and plotting server for alternative exon regulation. Nucleic Acids Res. 2016; 44:W333–W338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Paz I., Kosti I., Ares M. Jr, Cline M., Mandel-Gutfreund Y.. RBPmap: a web server for mapping binding sites of RNA-binding proteins. Nucleic Acids Res. 2014; 42:W361–W367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Im J., Tuvshinjargal N., Park B., Lee W., Huang D.S., Han K.. PNImodeler: web server for inferring protein-binding nucleotides from sequence data. BMC Genomics. 2015; 16(Suppl. 3):S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kazan H., Morris Q.. RBPmotif: a web server for the discovery of sequence and structure preferences of RNA-binding proteins. Nucleic Acids Res. 2013; 41:W180–W186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Barik A., Nithin C., Karampudi N.B., Mukherjee S., Bahadur R.P.. Probing binding hot spots at protein-RNA recognition sites. Nucleic Acids Res. 2016; 44:e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Katz Y., Wang E.T., Airoldi E.M., Burge C.B.. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods. 2010; 7:1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shen S., Park J.W., Lu Z.X., Lin L., Henry M.D., Wu Y.N., Zhou Q., Xing Y.. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl Acad. Sci. U.S.A. 2014; 111:E5593–E5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Park J.W., Tokheim C., Shen S., Xing Y.. Identifying differential alternative splicing events from RNA sequencing data using RNASeq-MATS. Methods Mol. Biol. 2013; 1038:171–179. [DOI] [PubMed] [Google Scholar]
- 18. Shen S., Park J.W., Huang J., Dittmar K.A., Lu Z.X., Zhou Q., Carstens R.P., Xing Y.. MATS: a Bayesian framework for flexible detection of differential alternative splicing from RNA-Seq data. Nucleic Acids Res. 2012; 40:e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Uren P.J., Bahrami-Samani E., Burns S.C., Qiao M., Karginov F.V., Hodges E., Hannon G.J., Sanford J.R., Penalva L.O., Smith A.D.. Site identification in high-throughput RNA-protein interaction data. Bioinformatics. 2012; 28:3013–3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen B., Yun J., Kim M.S., Mendell J.T., Xie Y.. PIPE-CLIP: a comprehensive online tool for CLIP-seq data analysis. Genome Biol. 2014; 15:R18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Corcoran D.L., Georgiev S., Mukherjee N., Gottwein E., Skalsky R.L., Keene J.D., Ohler U.. PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol. 2011; 12:R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Anderson E.S., Lin C.H., Xiao X., Stoilov P., Burge C.B., Black D.L.. The cardiotonic steroid digitoxin regulates alternative splicing through depletion of the splicing factors SRSF3 and TRA2B. RNA. 2012; 18:1041–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dittmar K.A., Jiang P., Park J.W., Amirikian K., Wan J., Shen S., Xing Y., Carstens R.P.. Genome-wide determination of a broad ESRP-regulated posttranscriptional network by high-throughput sequencing. Mol. Cell. Biol. 2012; 32:1468–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ray D., Kazan H., Cook K.B., Weirauch M.T., Najafabadi H.S., Li X., Gueroussov S., Albu M., Zheng H., Yang A. et al.. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013; 499:172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yee B.A., Pratt G.A., Graveley B.R., Van Nostrand E.L., Yeo G.W.. RBP-Maps enables robust generation of splicing regulatory maps. RNA. 2019; 25:193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hong X., Scofield D.G., Lynch M.. Intron size, abundance, and distribution within untranslated regions of genes. Mol. Biol. Evol. 2006; 23:2392–2404. [DOI] [PubMed] [Google Scholar]
- 27. Yang Y., Park J.W., Bebee T.W., Warzecha C.C., Guo Y., Shang X., Xing Y., Carstens R.P.. Determination of a comprehensive alternative splicing regulatory network and combinatorial regulation by key factors during the Epithelial-to-Mesenchymal transition. Mol. Cell. Biol. 2016; 36:1704–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pillman K.A., Phillips C.A., Roslan S., Toubia J., Dredge B.K., Bert A.G., Lumb R., Neumann D.P., Li X., Conn S.J. et al.. miR-200/375 control epithelial plasticity-associated alternative splicing by repressing the RNA-binding protein Quaking. EMBO J. 2018; 37:e99016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mudvari P., Movassagh M., Kowsari K., Seyfi A., Kokkinaki M., Edwards N.J., Golestaneh N., Horvath A.. SNPlice: variants that modulate Intron retention from RNA-sequencing data. Bioinformatics. 2015; 31:1191–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang Y., Gu L., Hou Y., Wang L., Deng X., Hang R., Chen D., Zhang X., Zhang Y., Liu C. et al.. Integrative genome-wide analysis reveals HLP1, a novel RNA-binding protein, regulates plant flowering by targeting alternative polyadenylation. Cell Res. 2015; 25:864–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
rMAPS2 is available from https://rmaps.cecsresearch.org/. The website is free and open to all users and there is no login requirement.