Abstract
Among the diverse location of the breakpoints (BPs) of structural variants (SVs), the breakpoints of fusion genes (FGs) are located in the gene bodies. This broken gene context provided the aberrant functional clues to study disease genesis. Many tumorigenic fusion genes have retained or lost functional or regulatory domains and these features impacted tumorigenesis. Full annotation of fusion genes aided by the visualization tool based on two gene bodies will be helpful to study the functional aspect of fusion genes. To date, a specialized tool with effective visualization of the functional features of fusion genes is not available. In this study, we built FGviewer, a tool for visualizing functional features of human fusion genes, which is available at https://ccsmweb.uth.edu/FGviewer. FGviewer gets the input of fusion gene symbols, breakpoint information, or structural variants from whole-genome sequence (WGS) data. For any combination of gene pairs/breakpoints to be involved in fusion genes, the users can search the functional/regulatory aspect of the fusion gene in the three bio-molecular levels (DNA-, RNA-, and protein-levels) and one clinical level (pathogenic-level). FGviewer will be a unique online tool in disease research communities.
INTRODUCTION
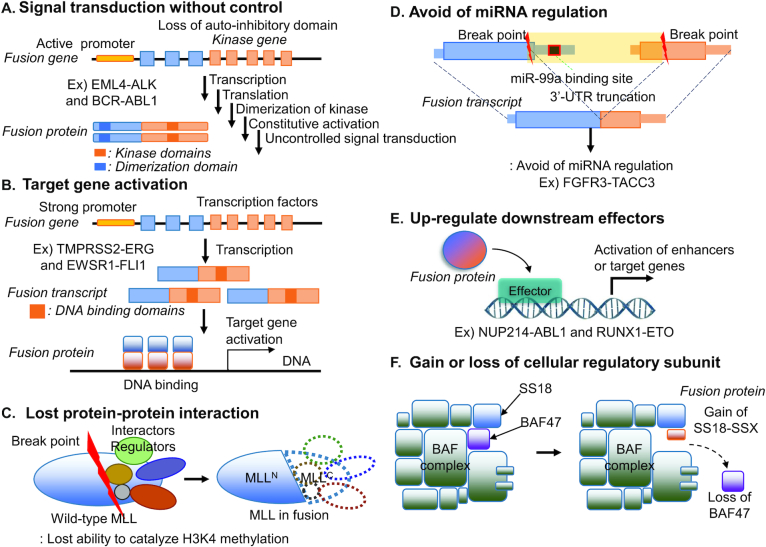
Accumulated WGS data sets from diverse disease consortiums/studies have analyzed structural variants (SVs). Among the diverse location of breakpoints (BPs) of structural variants in the human genome such as gene bodies, intergenic regions, and repeats, the breakpoints of fusion genes (FGs) are located in gene bodies. This broken gene context provided the aberrant functional clues to study disease genesis and there are many fusion genes that have been recognized as biomarkers and therapeutic targets in cancer (1). Many tumorigenic FGs have retained or lost oncogenic protein-domains or regulatory features (2). The maintenance or loss of the functional features directly impacts on tumor initiation, progression, and evolution. To date, there are multiple representative functional mechanisms of fusion genes studied as shown in Figure 1. If a partner gene, with the active promoter and dimerization functional domains, is fused with a kinase domain, it will constitutively drive the activated expression of the kinase, leading to uncontrolled cell proliferation. For example, ALK is not normally transcribed in adult lung, but the EML4-ALK fusion gene is transcribed under the control of the EML4 promoter. The dimerization domain of EML4 permits unregulated dimerization of the tyrosine kinase domain, constitutively activating downstream pathways (3). The BCR-ABL1 fusion gene is formed between BCR and the non-receptor tyrosine kinase, ABL1. The dimerization function of the coiled-coil domain in BCR and lacking auto-inhibitory N-terminal myristoylation in ABL1 and contribute to the constitutive activation of kinase function (4,5). TMPRSS2-ERG fusion gene has over 50% of frequency in the prostate cancer patients. The strong promoter of TMPRSS2, which is prostate tissue-specific and androgen-response gene, enhanced the function of proto-oncogene (ERG). Similarly, if a transactivation domain of EWSR1 is fused with a DNA-binding domain of FLI1, this fusion protein can recruit transcriptional coregulatory elements, resulting in target gene activation (6). Through fusion, one protein can also lose its interactions with important cellular regulators (i.e. MLL fusion genes) (7). If the 3′-UTR region of a fusion gene is truncated during fusion, such fusion transcript can avoid the regulation by its targeting miRNAs (i.e., FGFR3-TACC3) (8). Other FG mechanisms include the loss of function of tumor suppressors and DNA damage repairs through the deletion or frameshift open reading frame (ORF) of fusion transcripts (9,10). Recently, several studies were done for identifying downstream effectors of FGs (i.e. NUP214-ABL1, RUNX1-ETO and SS18-SSX) (11–13). SS18, which encodes the BAF complex subunit, fused with SSX and affected the chromatin accessibility (14). As here, there are multiple types of mechanisms of fusion genes in human diseases. Therefore, deep functional annotation of fusion genes is an urgent need for identifying novel driver fusion genes, understanding their roles in disease development, and developing new therapeutic strategies against diseases.
Figure 1.
Diverse tumorigenic mechanisms of FGs in cancer. (A) Signal transduction with control. The BCR with active promoter and dimerization domain is fused with a kinase domain of ABL1 and it leads to uncontrolled cell proliferation. (B) Target gene activation. If a transactivation domain of EWSR1 or tissue-specific androgen-responsive TMPRSS2 fused with a DNA-binding domain of FLI1 or ERG, it results in target gene activation. (C) Lost protein-protein interaction. MLL fusion proteins lose their interactions with important cellular. (D) Avoid of miRNA regulation. The 3′-UTR region of FGFR3 is truncated due to fusion gene, which can avoid the miRNA regulation. (E) Up-regulation of downstream effectors. Inhibition of STAT5, the downstream factor of NUP214-ABL1 led to leukemia cell death. (F) Gain/loss of cellular regulatory subunit. Due to SS18-SSX fusion gene in the BAF complex subunit, it affected the chromatin accessibility.
Predicting the functional classes of fusion genes is still a challenge. This is partly due to the dearth of experimental data and biased studies on several driver fusion genes. Current approaches to fusion gene annotation have mainly focused on ORF investigation of fusion transcript sequence and pointing the location of major protein domain in the fusion protein sequence (i.e. FusionHub, Pegasus) (15,16). Oncofuse introduced the gene expression level, length of 3′-UTR and the number of protein-protein interaction (PPIs), and GO terms on fusion partners (17). However, these approaches are not sufficiently capable of capturing the aberrant function of non-linear transcripts. Well-organized and arranged protein functional features, such as the protein annotation features of UniProt, are needed for the successful annotations (18). Furthermore, in order to better assist fusion gene studies, a unique visualization tool is an urgent need based on two different gene bodies. Current FG visualization tools, like INTEGRATE-Vis (19) and AGFusion (20), only provide the loci information of main protein domains in the fusion protein sequence. iFUSE shows the structures of genes involving in the structural variants vertically and provides FG sequences (21). The chimeraviz provides a visualization of the RNA-seq read depth with isoform structures (22). However, these visualizations are not helpful to give an intuitive understanding of the fusion gene structure on genomic-, transcriptomic-, protein- and pathogenic-levels. To do this, it needs a unique visualization tool or browser from the non-collinear genomic context between two different genes. However, there are no specialized tools with effective visualization of the functional features of human fusion genes for better prioritization and therapeutic target selection so far.
In this study, we developed FGviewer, a tool for visualizing functional features of human fusion genes, available at https://ccsmweb.uth.edu/FGviewer. FGviewer gets the input of fusion gene symbols, breakpoint information, or structural variants of the VCF (v4.3) file from WGS data. For any combination of gene pairs/breakpoints to be involved in gene fusion, the users can search the functional aspect of fusion genes at the three bio-molecular levels (DNA-, mRNA-, and protein-levels) and one clinical level. According to the breakpoint line defined/dragged by the user, FGviewer provides the automatically transcribed and translated breakpoint loci information with ORF annotation, retention status of functional/regulatory feature information, and fusion transcript/amino acid sequences. All of this information with the whole feature alignment image across four levels are downloadable. FGviewer will be useful for a better understanding of disease genesis, progression, and identification of drug-targetable features of fusion genes. We believe FGviewer will be a unique online tool in disease research communities.
MATERIALS AND METHODS
Human gene and transcript structures, and protein sequences
Since we have focused on the functional fusion genes, we have downloaded sequences of 18 963 protein accessions of reviewed from UniProt. For the transcript level, we have downloaded transcript and gene structures corresponding to these protein accessions from GENCODE (v19 of GRCh37 and v28 of GRCh38) (23).
Protein sequence annotation features and making the same coordinate map across four different levels
We downloaded the protein functional feature information in general feature format (GFF) of 18 963 accessions of UniProt corresponding to 18 963 genes (18). UniProt provides the locus information of 39 protein features, including six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and three secondary structure features. Based on the canonical protein sequence, we have all of these feature loci information. To have the same width among genomic-, transcript- and protein-levels, we calculated the individual coordinates following the central dogma. The coordinates of transcript start (txt-start) and end (txt-end) in DNA-level correspond to the start and end of the mRNA-level. To have consistent coordinates between DNA- and mRNA-level, we excluded the intron sequences at DNA-level. For the minus strand genes, we show the genomic coordinates reversely, then DNA-level annotation can keep its direction from left to right as all other annotation levels. In this way, FGviewer provides the truncated part, which is the non-retained zone in the fusion gene with gray shade, always in the middle of two gene structures. The coordinates of coding start (CDS-start) and end (CDS-end) in mRNA-level correspond the start and end of protein-level. Here, we multiplied three to each amino acid length so that all three different level's coordinates can be movable simultaneously with the same width at the browser. To map the individual protein functional features in this same coordinate map, we also multiplied three to the start and end positions of individual protein features. The loci of ClinVar variants on the canonical amino acid sequence also were multiplied three.
Transcription factors targeting individual human gene
TF-target relationship information was downloaded from TRRUST (version 2) (24), which is a manually curated database of human transcriptional regulatory networks. From TRRUST, we obtained 8427 pairs between 795 TFs and 2492 target genes.
miRNA targeting individual human gene
We downloaded the conserved human miRNA–target gene interaction information from TargetScan (release 7.2) (25). For more reliable interactions, we have filtered out the miRNA−target pairs when the context++ score and percent identity were bigger than −0.4 and <97%, respectively.
Pathogenic variant and disease information
The pathogenic variant of individual human genes was extracted from the public archive of interpretations of clinically relevant variants (ClinVar, December 2018) (26). Disease−gene information was extracted from DisGeNet (December 2018, version 5.0) (27), a database of gene-disease.
ORF annotation of fusion sequence and translation into the amino acid sequence
For each fusion gene BP pair, based on both GENCODE v19 (GRCh37) and v28 (GRCh38), we examined ORF of major isoform transcript sequences, which encodes the canonical protein amino acid sequence. When both BPs of two genes are located inside of the coding region (CDS) and the number of the nucleotides of CDS in fusion transcript is a multiple of three, then we reported such fusion gene transcript as ‘in-frame’. When one or two nucleotide insertions are present, we reported such transcripts as ‘frame-shift’. We translated the fusion transcript sequence to the fusion amino acid sequence using the dictionary of three nucleotides and amino acid codon pairs.
Web server architecture
The FGviewer system is designed based on a three-tier architecture: client, server, and database. It includes a user-friendly web interface, Python's Tornado web server (Tornado 5.1.1 with Python3.6), and MySQL database. This database was developed on MySQL 3.23 with the MyISAM storage engine.
OVERVIEW OF WEBSERVER
FGviewer aims to show the functional impacts of breakage of two genes across four interpretable levels according to the individual breakpoint loci defined by the users. The same breakpoint line across four tiers classifies between fusion gene involved (retained region) and non-involved zones (truncated region) with multiple types of functional features. Those features include swapped gene expression regulatory (i.e. transcription factor or miRNA binding sites), protein functional features (i.e. protein domains, protein-protein interactions, binding sites of all molecules, secondary structure level feature etc.), clinically relevant variants, fusion mRNA and amino acid sequences, and ORF assignment based on the user's breakpoint coordinates.
WEB INTERFACE AND ANALYSIS RESULTS
Search & browse fusion genes
FGviewer provides multiple fusion gene searching ways, such as inputting gene symbols or genomic BP coordinates and uploading multiple cases with a file. The users can also search fusion genes from the structural variants (SVs) with the VCF file from the analysis of WGS data. FGviewer annotates diverse types of SVs including deletion (DEL), insertion (INS), duplication (DUP), inversion (INV), copy number variation (CNV) and break ends (BND), and identifies fusion genes located in those breakpoints of structural variants according to VCF v4.3. Using this function, we analyzed dbVar data (a version of 1 November 2019) (28), which is the NCBI’s database of human genomic structural variation, and identified 40K fusion genes from 6 million of structural variant regions in 116 studies out of all 180 studies (Table 1). On the page of FGviewer, users can click their interested genomic studies gathered in dbVar and directly can go to the FGviewer analysis page for individual fusion genes among 40K fusion genes from 6 million of structural variants (Supplementary Table S1). Specifically, we found 1037 fusion genes from the structural variants detected from the Genome Aggregation Database (genomAD v2.1) of 15K population WGS data. Furthermore, one study (dbVar study ID: nstd33) in dbVar previously identified a recurrent 15q24 microdeletion from multiple individuals with mental retardation (29). From the SVs in 15q24, detected in this study, we found one novel fusion gene, CELF6-TMEM266. CELF6, CUGBP Elav-like family member 6, encodes CELF RNA binding proteins, which play roles in early embryonic development, heart, and skeletal muscle functions (30). From the analysis result of FGviewer on this fusion gene, we found that the binding site of miR-133a-3p.1 was lost in the 3′-UTR of CELF6 (Supplementary Figure S1). miR-133a-3p.1 is known as a motor neuron regulator enriched in amyotrophic lateral sclerosis (31) and differentially expressed in major depressive disorder (32). In this way, users can use FGviewer to predict the functional aspect of novel FGs and can identify driver FGs in human diseases.
Table 1.
The number of identified FGs from SV regions of 116 studies in dbVar. It is sorted by the number of identified FGs of each study
| Study ID | # variant regions | # FGs | Study ID | # variant regions | # FGs | Study ID | # variant regions | # FGs |
|---|---|---|---|---|---|---|---|---|
| nstd151 | 49 339 | 6140 | nstd4 | 2949 | 92 | nstd14 | 368 | 9 |
| nstd102 | 60 993 | 6076 | nstd79 | 64 | 92 | nstd149 | 36 | 8 |
| nstd100 | 70 319 | 3717 | estd229 | 16 676 | 90 | nstd99 | 11 116 | 8 |
| nstd37 | 20 391 | 3097 | estd20 | 20 206 | 85 | nstd141 | 123 | 7 |
| nstd54 | 81 345 | 2392 | nstd158 | 1621 | 85 | nstd51 | 229 | 6 |
| nstd71 | 45 084 | 2313 | estd24 | 1773 | 80 | nstd91 | 148 | 6 |
| nstd113 | 9010 | 1645 | nstd53 | 900 | 77 | estd22 | 780 358 | 5 |
| nstd68 | 5132 | 1103 | estd218 | 5513 | 76 | estd228 | 12 | 5 |
| nstd130 | 39 657 | 1043 | nstd111 | 2233 | 76 | nstd161 | 4 | 5 |
| nstd166 | 373 378 | 1037 | nstd157 | 1907 | 66 | nstd46 | 1183 | 5 |
| nstd122 | 94 923 | 798 | nstd29 | 388 | 66 | estd215 | 28 083 | 4 |
| nstd101 | 3105 | 783 | nstd75 | 927 | 66 | estd232 | 8 | 4 |
| nstd132 | 6173 | 725 | nstd145 | 8237 | 57 | nstd17 | 935 | 4 |
| nstd173 | 23 548 | 583 | nstd41 | 255 | 53 | nstd31 | 226 | 4 |
| nstd86 | 1030 | 528 | nstd73 | 9109 | 51 | nstd43 | 1318 | 4 |
| nstd174 | 38 176 | 455 | nstd97 | 2558 | 50 | nstd49 | 4197 | 4 |
| estd214 | 61 678 | 389 | nstd67 | 7950 | 44 | nstd65 | 1299 | 4 |
| nstd21 | 13 566 | 382 | nstd112 | 15 012 | 36 | nstd143 | 18 | 3 |
| estd1 | 3340 | 370 | nstd156 | 963 | 34 | nstd162 | 99 810 | 3 |
| estd55 | 6862 | 366 | estd206 | 513 | 31 | nstd177 | 22 | 3 |
| nstd85 | 1416 | 358 | estd211 | 100 | 31 | nstd36 | 395 | 3 |
| nstd27 | 13 843 | 357 | estd221 | 699 | 30 | nstd47 | 1167 | 3 |
| estd219 | 68 825 | 345 | estd231 | 7063 | 30 | nstd153 | 8 | 2 |
| nstd106 | 39 678 | 337 | estd49 | 225 | 30 | nstd172 | 4659 | 2 |
| estd59 | 228 871 | 323 | nstd45 | 364 | 30 | nstd28 | 6 | 2 |
| estd224 | 3576 | 245 | nstd82 | 28 214 | 30 | nstd50 | 2637 | 2 |
| nstd84 | 10 376 | 245 | estd208 | 81 | 27 | nstd58 | 7 | 2 |
| estd203 | 4192 | 244 | nstd32 | 119 | 24 | nstd83 | 25 | 2 |
| nstd8 | 791 | 209 | nstd64 | 1383 | 23 | estd210 | 4 | 1 |
| nstd107 | 8440 | 204 | estd225 | 1249 | 22 | estd236 | 7 | 1 |
| nstd12 | 2290 | 183 | estd216 | 879 | 20 | estd3 | 2682 | 1 |
| estd212 | 16 575 | 164 | estd213 | 497 | 17 | nstd1 | 297 | 1 |
| nstd30 | 1428 | 161 | nstd152 | 103 985 | 14 | nstd103 | 130 | 1 |
| estd220 | 3620 | 146 | nstd168 | 16 424 | 14 | nstd125 | 34 | 1 |
| nstd80 | 1512 | 144 | nstd2 | 7458 | 14 | nstd164 | 10 | 1 |
| nstd40 | 689 | 129 | nstd16 | 1290 | 12 | nstd20 | 539 | 1 |
| nstd133 | 246 | 123 | nstd22 | 1319 | 11 | nstd33 | 4 | 1 |
| estd233 | 1026 | 116 | nstd66 | 81 | 10 | nstd42 | 1 | 1 |
| estd201 | 36 558 | 105 | nstd92 | 245 | 10 |
FGviewer analysis result
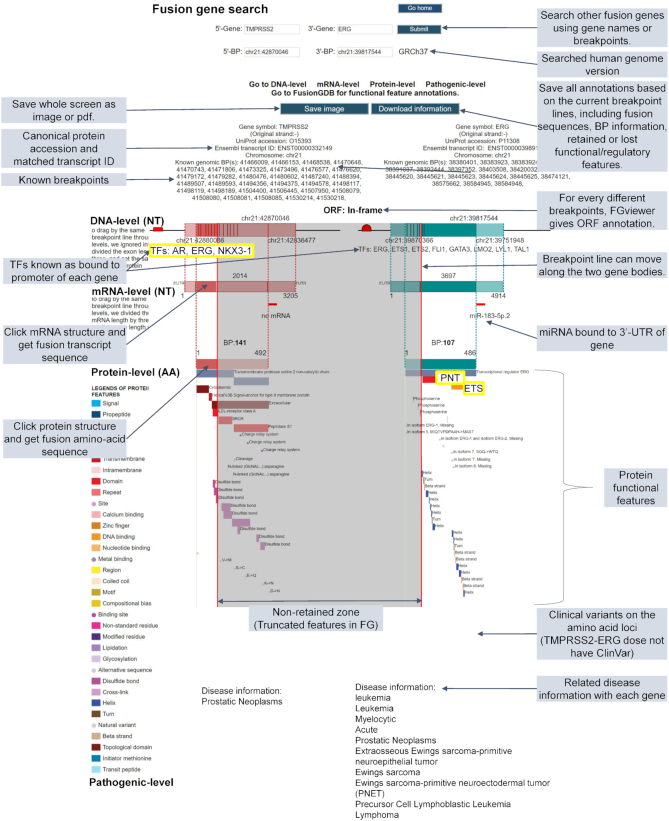
For a fusion gene searching, FGviewer displays four levels of annotations including DNA-, mRNA-, protein- and pathogenic-levels. The DNA-level annotation provides the genomic coordinates of breakpoints and the transcription factors known as binding to the promoter region of individual partner genes. Users can infer the effect when the 3′-partner gene lost the promoter region that has the TF binding sites, due to the formation of the fusion gene. mRNA-level annotation provides the transcript sequence-based breakpoint coordinates, fusion transcript sequence, ORF assignment to fusion transcript, and potential miRNAs predicted as binding at the 3′-UTR of individual partner genes. From this, users can infer the effect when the 5′-partner gene lost the 3′-UTR region that has the miRNA binding sites, due to the creation of a fusion gene. Protein-level annotation provides the amino-acid sequence-based breakpoint coordinates, fusion amino-acid sequence and 39-protein functional features across two gene bodies. From the gray color-shaded background presenting the non-fusion gene involved zone, users can recognize which protein functional features were lost or retained. Pathogenic-level annotation provides the variants, which is important in the clinical aspect, and the disease information, which is associated with individual partner genes. By integrating information from these four types of annotations, users can learn and understand about the potential roles of FGs in their diseases.
FGviewer provides a user-friendly interface and several unique browse utilities for FG functional annotation study. The main features of the FGviewer are summarized here. (i) Users can search fusion genes by gene symbols, genomic coordinates, or batch searching by input file of multiple gene symbols or genomic breakpoints for both of GRCh37 and GRCh38 human genomes. Users also can search fusion genes by inputting the structural variants of the VCF file from WGS data. (ii) Users can drag the breakpoint lines and these lines move simultaneously on the four different levels including DNA-, RNA-, protein- and pathogenic-levels. Accordingly, users can get the automatically transcribed and translated breakpoint loci information with ORF annotation and fusion sequences of transcript and amino-acid. (iii) To help users with more information, FGviewer provides the known human genomic BP information gathered from our previous studies (1,2). (iv) According to the user-defined breakpoint loci, the user can separate the functional features between retained and not-retained. (v) In the mRNA- and protein-levels, users can have the broken sequence of transcript and amino acids for both of FG partner genes based on the user-defined breakpoints. (vi) For individual functional features across four levels, users can see the detailed information by clicking it. (vii) Users can save the whole feature information with the retention/non-retention information according to the user-defined breakpoints and the whole feature alignment status across four levels into image or PDF file.
Figure 2 shows the example result of FGviewer using TMPRSS2-ERG as the input query. TMPRSS2 is a prostate-specific, androgen-responsive, transmembrane serine protease. ERG is the oncogenic transcription factor containing a highly conserved Ets DNA binding region and an N-terminal regulatory domain. The Ets domain serves as a DNA binding recognition site, as well as a protein–protein interaction site commonly used for interactions with other transcription factors. Therefore, the fusion between these genes enhances the production of the ERG transcription factor under the control of the androgen-sensitive promoter elements of TMPRSS2. Then, this enhanced ERG can induce their target gene expression (33). FGviewer provided these functional contexts as shown in Figure 2. The androgen receptor (AR) transcription factor binds to the promoter of TMPRSS2 and this binding is retained in the fusion gene since it is joined as the 5′-partner gene. In the fusion gene, ERG retained pointed (PNT) and Ets DNA-binding domains. Conserved PNT/SAM domain and an ETS domain are the common features of members of ETS related proteins. These domains play key roles in regulating downstream target genes that are crucial for several biological processes such as cellular proliferation, differentiation, development, transformation, and apoptosis (34). In the pathogenic-level of FGviewer, there was no ClinVar mutation in TMPRSS2 and ERG. However, the disease annotation shows the tissue-specificity of TMPRSS2 compared to ERG. This might be related to many ETS fusion genes in many cancers including leukemia and bone cancers. Supplementary Figure S2 shows the example result of FGviewer using EWSR1-FLI1 as the input query. About 95% of patients of the Ewing tumors, the second most frequent bone tumors in teenagers and young adults (35), have this FG. The chimeric protein consists of the transcription activation domain of EWSR1 and the DNA-binding domain of the transcription factor FLI1. The prion-like domains (PrLDs), which possess an unusual amino-acid composition enriched in glycine and uncharged polar amino acids, including glutamine, asparagine, tyrosine and serine, are found in human RNA-binding proteins (RBPs) (36). Aberrant regulation of RBP expression or activity can contribute to disease onset and progression (37). In Figure 2, we can see the retention of the EWS activation domain (EAD, Gln/Pro/Thr-rich) and 31 X approximate tandem repeats in the 5′-partner gene (EWSR1). The 3′-partner gene (FLI1) retained the ETS domain (PROSITE ID: PRU00237). In summary, from the result of FGviewer, EWSR1-FLI1 is anticipated to contribute to the aberrant regulation of target genes. Similarly, users can apply FGviewer to predict the functional features of interested FGs.
Figure 2.
Overview of the FGviewer result page of the TMPRSS2-ERG query search. FGviewer is composed of four levels including DNA-, mRNA-, protein- and pathogenic-levels. Details on individual functions are described with shaded text.
DISCUSSION
FGviewer is the first web server that visualizes the functional features across multi-levels in non-collinear gene structures. FGviewer provides a unique and efficient visualization tool for the functional annotation of FGs. To serve broad biomedical research communities, we will continuously update the user interface and gene annotation information such as more reliable TF and miRNA information. FGviewer can handle the diverse breakpoints of fusion genes. However, the current version of FGviewer does not cover the multiple gene isoform structures. The different gene isoforms affect the retention of functional/regulatory features covering different transcription factor binding sites, miRNA binding sites and functional domain or features due to the different exon composition. In the near future, we hope we can find an efficient way to visualize the alternative splicing isoforms in FGviewer. If possible, providing some general scores to show the promoter activity relating bound TFs, it would be helpful to infer the expressional activity of fusion genes. For the functional feature categorization, we arranged multiple potentially important functional features of known fusion genes in FusionGDB (https://ccsm.uth.edu/FusionGDB). With the help of FGviewer and FusionGDB, we hope users can identify abnormal functional features in the novel or known fusion gene structures for their study. Furthermore, even not for fusion genes, but for any single coding genes, users can see the landscape of the functional/regulatory features at a glance. FGviewer will be a useful web tool for many investigators in pathology, cancer genomics and precision medicine, drug, and therapeutic research, among others.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the members of the Center for Computational Systems Medicine (CCSM) for valuable discussion. We also thank the members of the Information Technology Team of SBMI, especially Mr David Ha and Mr Michael Phan.
Contributor Information
Pora Kim, School of Biomedical Informatics, The University of Texas Health Science Center at Houston, Houston, TX 77030, USA.
Ke Yiya, College of Electronic and Information Engineering, Tongji University, Shanghai, China.
Xiaobo Zhou, School of Biomedical Informatics, The University of Texas Health Science Center at Houston, Houston, TX 77030, USA; McGovern Medical School, The University of Texas Health Science Center at Houston, Houston, TX 77030, USA; School of Dentistry, The University of Texas Health Science Center at Houston, Houston, TX 77030, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [R01GM123037, U01AR069395, R01CA241930 to X.Z., in part]; the funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript. Funding for open access charge: Dr & Mrs Carl V. Vartian Chair Professorship Funds to Dr Zhou from the University of Texas Health Science Center at Houston.
Conflict of interest statement. None declared.
REFERENCES
- 1. Lee M., Lee K., Yu N., Jang I., Choi I., Kim P., Jang Y.E., Kim B., Kim S., Lee B. et al.. ChimerDB 3.0: an enhanced database for fusion genes from cancer transcriptome and literature data mining. Nucleic Acids Res. 2017; 45:D784–D789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kim P., Zhou X.. FusionGDB: fusion gene annotation DataBase. Nucleic Acids Res. 2019; 47:D994–D1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rosenbaum J.N., Bloom R., Forys J.T., Hiken J., Armstrong J.R., Branson J., McNulty S., Velu P.D., Pepin K., Abel H. et al.. Genomic heterogeneity of ALK fusion breakpoints in non-small-cell lung cancer. Mod. Pathol. 2018; 31:791–808. [DOI] [PubMed] [Google Scholar]
- 4. Hantschel O. Structure, regulation, signaling, and targeting of abl kinases in cancer. Genes Cancer. 2012; 3:436–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. O’Hare T., Deininger M.W., Eide C.A., Clackson T., Druker B.J.. Targeting the BCR-ABL signaling pathway in therapy-resistant Philadelphia chromosome-positive leukemia. Clin. Cancer Res. 2011; 17:212–221. [DOI] [PubMed] [Google Scholar]
- 6. Boulay G., Sandoval G.J., Riggi N., Iyer S., Buisson R., Naigles B., Awad M.E., Rengarajan S., Volorio A., McBride M.J. et al.. Cancer-specific retargeting of BAF complexes by a prion-like domain. Cell. 2017; 171:163–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang Y., Chen A., Yan X.M., Huang G.. Disordered epigenetic regulation in MLL-related leukemia. Int. J. Hematol. 2012; 96:428–437. [DOI] [PubMed] [Google Scholar]
- 8. Parker B.C., Annala M.J., Cogdell D.E., Granberg K.J., Sun Y., Ji P., Li X., Gumin J., Zheng H., Hu L. et al.. The tumorigenic FGFR3-TACC3 gene fusion escapes miR-99a regulation in glioblastoma. J. Clin. Invest. 2013; 123:855–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jin Y., Mertens F., Kullendorff C.M., Panagopoulos I.. Fusion of the tumor-suppressor gene CHEK2 and the gene for the regulatory subunit B of protein phosphatase 2 PPP2R2A in childhood teratoma. Neoplasia. 2006; 8:413–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Knijnenburg T.A., Wang L., Zimmermann M.T., Chambwe N., Gao G.F., Cherniack A.D., Fan H., Shen H., Way G.P., Greene C.S. et al.. Genomic and molecular landscape of DNA damage repair deficiency across the cancer genome atlas. Cell Rep. 2018; 23:239–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vanden Bempt M., Demeyer S., Broux M., De Bie J., Bornschein S., Mentens N., Vandepoel R., Geerdens E., Radaelli E., Bornhauser B.C. et al.. Cooperative enhancer activation by TLX1 and STAT5 drives development of NUP214-ABL1/TLX1-Positive T cell acute lymphoblastic leukemia. Cancer Cell. 2018; 34:271–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Martinez-Soria N., McKenzie L., Draper J., Ptasinska A., Issa H., Potluri S., Blair H.J., Pickin A., Isa A., Chin P.S. et al.. The oncogenic transcription factor RUNX1/ETO corrupts cell cycle regulation to drive leukemic transformation. Cancer Cell. 2018; 34:626–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Banito A., Li X., Laporte A.N., Roe J.S., Sanchez-Vega F., Huang C.H., Dancsok A.R., Hatzi K., Chen C.C., Tschaharganeh D.F. et al.. The SS18-SSX oncoprotein hijacks KDM2B-PRC1.1 to drive synovial sarcoma. Cancer Cell. 2018; 33:527–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McBride M.J., Pulice J.L., Beird H.C., Ingram D.R., D’Avino A.R., Shern J.F., Charville G.W., Hornick J.L., Nakayama R.T., Garcia-Rivera E.M. et al.. The SS18-SSX fusion oncoprotein hijacks BAF complex targeting and function to drive synovial sarcoma. Cancer Cell. 2018; 33:1128–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Panigrahi P., Jere A., Anamika K.. FusionHub: A unified web platform for annotation and visualization of gene fusion events in human cancer. PLoS One. 2018; 13:e0196588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Abate F., Zairis S., Ficarra E., Acquaviva A., Wiggins C.H., Frattini V., Lasorella A., Iavarone A., Inghirami G., Rabadan R.. Pegasus: a comprehensive annotation and prediction tool for detection of driver gene fusions in cancer. BMC Syst. Biol. 2014; 8:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shugay M., Ortiz de Mendibil I., Vizmanos J.L., Novo F.J.. Oncofuse: a computational framework for the prediction of the oncogenic potential of gene fusions. Bioinformatics. 2013; 29:2539–2546. [DOI] [PubMed] [Google Scholar]
- 18. UniProt Consortium, T UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2018; 46:2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang J., Gao T., Maher C.A.. INTEGRATE-Vis: a tool for comprehensive gene fusion visualization. Sci. Rep. 2017; 7:17808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Murphy C., Elemento O.. AGFusion: annotate and visualize gene fusions. 2016; 14 October 2016, preprint: not peer reviewed 10.1101/080903. [DOI]
- 21. Hiltemann S., McClellan E.A., van Nijnatten J., Horsman S., Palli I., Teles Alves I., Hartjes T., Trapman J., van der Spek P., Jenster G. et al.. iFUSE: integrated fusion gene explorer. Bioinformatics. 2013; 29:1700–1701. [DOI] [PubMed] [Google Scholar]
- 22. Lagstad S., Zhao S., Hoff A.M., Johannessen B., Lingjaerde O.C., Skotheim R.I.. chimeraviz: a tool for visualizing chimeric RNA. Bioinformatics. 2017; 33:2954–2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Frankish A., Diekhans M., Ferreira A.M., Johnson R., Jungreis I., Loveland J., Mudge J.M., Sisu C., Wright J., Armstrong J. et al.. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019; 47:D766–D773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Han H., Cho J.W., Lee S., Yun A., Kim H., Bae D., Yang S., Kim C.Y., Lee M., Kim E. et al.. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018; 46:D380–D386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Agarwal V., Bell G.W., Nam J.W., Bartel D.P.. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015; 4:e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Landrum M.J., Chitipiralla S., Brown G.R., Chen C., Gu B., Hart J., Hoffman D., Jang W., Kaur K., Liu C. et al.. ClinVar: improvements to accessing data. Nucleic Acids Res. 2020; 48:D835–D844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pinero J., Bravo A., Queralt-Rosinach N., Gutierrez-Sacristan A., Deu-Pons J., Centeno E., Garcia-Garcia J., Sanz F., Furlong L.I.. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017; 45:D833–D839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lappalainen I., Lopez J., Skipper L., Hefferon T., Spalding J.D., Garner J., Chen C., Maguire M., Corbett M., Zhou G. et al.. DbVar and DGVa: public archives for genomic structural variation. Nucleic Acids Res. 2013; 41:D936–D941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sharp A.J., Selzer R.R., Veltman J.A., Gimelli S., Gimelli G., Striano P., Coppola A., Regan R., Price S.M., Knoers N.V. et al.. Characterization of a recurrent 15q24 microdeletion syndrome. Hum. Mol. Genet. 2007; 16:567–572. [DOI] [PubMed] [Google Scholar]
- 30. Dasgupta T., Ladd A.N.. The importance of CELF control: molecular and biological roles of the CUG-BP, Elav-like family of RNA-binding proteins. Wiley Interdiscipl. Rev. RNA. 2012; 3:104–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hoye M.L., Koval E.D., Wegener A.J., Hyman T.S., Yang C., O’Brien D.R., Miller R.L., Cole T., Schoch K.M., Shen T. et al.. MicroRNA profiling reveals marker of motor neuron disease in ALS models. J. Neurosci. 2017; 37:5574–5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dwivedi Y. Emerging role of microRNAs in major depressive disorder: diagnosis and therapeutic implications. Dialog. Clin. Neurosci. 2014; 16:43–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. St John J., Powell K., Conley-Lacomb M.K., Chinni S.R.. TMPRSS2-ERG fusion gene expression in prostate tumor cells and its clinical and biological significance in prostate cancer progression. J. Cancer Sci. Ther. 2012; 4:94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sreenath T.L., Dobi A., Petrovics G., Srivastava S.. Oncogenic activation of ERG: a predominant mechanism in prostate cancer. J. Carcinog. 2011; 10:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Arndt C.A., Rose P.S., Folpe A.L., Laack N.N.. Common musculoskeletal tumors of childhood and adolescence. Mayo Clin. Proc. 2012; 87:475–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shorter J. Prion-like domains program Ewing's sarcoma. Cell. 2017; 171:30–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Svetoni F., Frisone P., Paronetto M.P.. Role of FET proteins in neurodegenerative disorders. RNA Biol. 2016; 13:1089–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.