Abstract
Millions of protein sequences are being discovered at an incredible pace, representing an inexhaustible source of biocatalysts. Despite genomic databases growing exponentially, classical biochemical characterization techniques are time-demanding, cost-ineffective and low-throughput. Therefore, computational methods are being developed to explore the unmapped sequence space efficiently. Selection of putative enzymes for biochemical characterization based on rational and robust analysis of all available sequences remains an unsolved problem. To address this challenge, we have developed EnzymeMiner—a web server for automated screening and annotation of diverse family members that enables selection of hits for wet-lab experiments. EnzymeMiner prioritizes sequences that are more likely to preserve the catalytic activity and are heterologously expressible in a soluble form in Escherichia coli. The solubility prediction employs the in-house SoluProt predictor developed using machine learning. EnzymeMiner reduces the time devoted to data gathering, multi-step analysis, sequence prioritization and selection from days to hours. The successful use case for the haloalkane dehalogenase family is described in a comprehensive tutorial available on the EnzymeMiner web page. EnzymeMiner is a universal tool applicable to any enzyme family that provides an interactive and easy-to-use web interface freely available at https://loschmidt.chemi.muni.cz/enzymeminer/.
INTRODUCTION
There are currently >259 million non-redundant protein sequences in the NCBI nr database (release 2020-02-10) (1). Despite their enormous promise for biological and biotechnological discovery, experimental characterization has been performed on only a small fraction of the available sequences. Currently, there are about 560 000 protein sequences reliably curated in the UniProtKB/Swiss-Prot database (release 2020_01) (2).
The low ratio of characterized to uncharacterized sequences reflects the sharp contrast in time-demanding/low-throughput biochemical techniques versus fast/high-throughput next-generation sequencing technology. Although more efficient biochemical techniques employing miniaturization and automation have been developed (3–5), the most widely used experimental methods do not provide sufficient capacity for biochemical characterization of proteins spanning the ever-increasing sequence space. Therefore, computational methods are currently the only way to explore the immense protein diversity available among the millions of uncharacterized sequence entries.
Two different computational strategies are generally used for exploration of the unknown sequence space. The first strategy takes a novel uncharacterized sequence as input and predicts functional annotations. The method involves annotating the unknown input sequences by predicting protein domains (6), Enzyme Commission (EC) number (7) or Gene Ontology terms that are a subject of the initiative named the Critical Assessment of Functional Annotation (8). These methods are often universal and applicable to any protein sequence. However, they often lack specificity as the automatic annotation rules or statistical models need to be substantially general. A significant advantage of these methods is their seamless integration into available databases. Submission of a query sequence to a database is sufficient, with no need for running computation- and memory-intensive bioinformatics pipelines locally. A model example of this approach is the automatic annotation workflow of the UniProtKB/TrEMBL database (2).
The second strategy takes a well-known characterized sequence as an input and applies a computational workflow, typically based on a homology search, to identify novel uncharacterized entries in genomic databases that are related to the input query sequence (5,9). The homology search is often followed by a filtration step, which checks the essential sequence properties, e.g. domain structure or presence of catalytic residues. The main advantage of these methods is the higher specificity of the analysis. A disadvantage is that it may be complicated to apply the developed workflow to protein families other than those for which it was designed. Moreover, these workflows typically require running complex bioinformatics pipelines and are usually not available through a web interface.
The fundamental unsolved problem is how to deal with the overwhelming number of sequence entries identified by these methods and select a small number of relevant hits for in-depth experimental characterization. For example, a database search for members of the haloalkane dehalogenase model family using the UniProt web interface yields 3598 sequences (UniProtKB release 2020_01). It is impossible to rationally select several tens of targets for experimental testing without additional bioinformatics analyses to help prioritize such a large pool of sequences.
To address the challenge of exploring the unmapped enzyme sequence space and rational selection of attractive targets, we have developed the EnzymeMiner web server. EnzymeMiner identifies novel enzyme family members, comprehensively annotates the targets and facilitates efficient prioritization and selection of representative hits for experimental characterization. To the best of our knowledge, there is currently no other tool available that allows such a comprehensive analysis in a single easy-to-run integrated workflow on the web.
MATERIALS AND METHODS
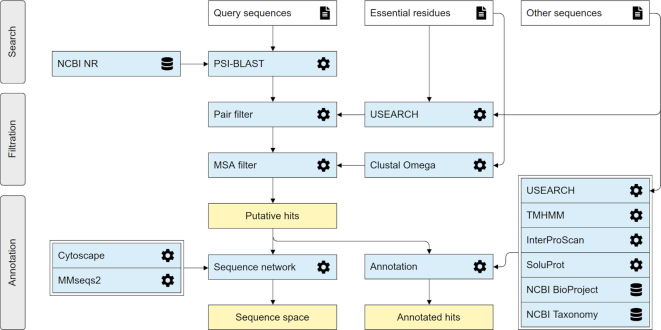
EnzymeMiner implements a three-step workflow: (i) homology search, (ii) essential residue based filtering and (iii) hits annotation (Figure 1). To execute these tasks, the server requires two different types of input information: (i) query sequences and (ii) essential residue templates. The query sequences serve as seeds for the initial homology search. The essential residue templates, defined as pairs of a protein sequence and a set of essential residues in that sequence, allow the server to prioritize hits that are more likely to display the enzyme function. Therefore, the essential residues may be the catalytic and ligand- or cofactor-binding residues that are indispensable for proper catalytic function. Each essential residue is defined by its name, position and a set of allowed amino acids for that position.
Figure 1.
The EnzymeMiner workflow. The workflow consists of three distinct steps: (i) sequence homology search, (ii) filtration of functional sequences, and (iii) annotation of hits. These steps are executed consecutively and automatically. EnzymeMiner has only two required inputs: (i) query sequences, and (ii) essential residue templates. The Other sequences are optional inputs that allow EnzymeMiner to calculate the sequence identity between these sequences and all the hits. Input files are highlighted by a white background, tools and databases have a light blue background, outputs are highlighted by a yellow background.
In the first homology search step, a query sequence is used as a query for a PSI-BLAST (10) two-iteration search in the NCBI nr database (1). If more than one query sequence is provided, a search is conducted for each sequence separately. Besides a minimum E-value threshold 10−20, the PSI-BLAST hits must share a minimum of 25% global sequence identity with at least one of the query sequences. Artificial protein sequences, i.e. sequences described by the term artificial, synthetic construct, vector, vaccinia virus, plasmid, halotag or replicon, are removed. EnzymeMiner sorts the PSI-BLAST hits by E-value and passes a maximum of 10,000 best hits to the next steps in the workflow. The default parameters for the homology search step, as well as the other steps, can be modified using advanced options in the web server.
In the second essential residue based filtering step, the homology search hits are filtered using the essential residue templates. First, the hits are divided into template clusters. Each cluster contains all hits matching essential residues of a particular template. Essential residues are checked using global pairwise alignment with the template calculated by USEARCH (11). When multiple essential residue templates match, the hit is assigned to the template with the highest global sequence identity. Second, for each cluster, an initial multiple sequence alignment (MSA) is constructed using Clustal Omega (12). The MSA is used to revalidate the essential residues of identified hits by checking the corresponding column in the MSA. Sequences not matching essential residues of the template are removed from the cluster. Third, the MSA is constructed again for each template cluster and the essential residues are checked for the last time. The final set of identified sequences reported by EnzymeMiner contains all sequences left in the template clusters.
In the third annotation step, the identified sequences are annotated using several databases and predictors: (i) transmembrane regions are predicted by TMHMM (13), (ii) Pfam domains are predicted by InterProScan (14), (iii) source organism annotation is extracted from the NCBI Taxonomy (15) and the NCBI BioProject database (16), (iv) protein solubility is predicted by the in-house tool SoluProt for prediction of soluble protein expression in Escherichia coli and (v) sequence identities to queries, hits or other optional sequences are calculated by USEARCH (11). SoluProt is based on a random forest regression model that employs 36 sequence-based features (https://loschmidt.chemi.muni.cz/soluprot/). It has been shown to achieve an accuracy of 58%, specificity of 73% and sensitivity of 44% on a balanced independent test set of 3788 sequences (Hon et al., manuscript in preparation). Alternative solubility prediction tools are summarised in a recently published review (17). It is not advised to use the solubility score for other expression systems because it was trained solely on E. coli data. We expect further intensive development of protein solubility predictors in coming years and will ensure that the solubility score in the EnzymeMiner stays at the cutting-edge in terms of its accuracy and reproducibility.
The sequence space of the identified hits is visualized using representative sequence similarity networks (SSNs) generated at various clustering thresholds using MMseqs2 (18) and Cytoscape (19). SSNs provide a clean visual approach to identify clusters of highly similar sequences and rapidly spot sequence outliers. SSNs proved to facilitate identification of previously unexplored sequence and function space (20). The SSN generation method used in EnzymeMiner is inspired by the EFI-EST tool (21). The minimum alignment score to include an edge between two representative sequences in an SSN is 40.
DESCRIPTION OF THE WEB SERVER
Job submission
New jobs can be submitted from the EnzymeMiner homepage. EnzymeMiner provides two conceptually different ways to define the input of the workflow: (i) using curated sequences from the UniProtKB/Swiss-Prot database and (ii) using custom sequences. We recommend the UniProtKB/Swiss-Prot option for users who do not have in-depth knowledge of the enzyme family. In contrast, the Custom sequences tab gives full control over the EnzymeMiner input—query sequences and essential residue templates are specified manually by the user. This is recommended for users who have good knowledge about the enzyme family and want to provide additional starting information to obtain refined results. The last option is a combination of both approaches, where Swiss-Prot sequences can be pre-selected first and then the input can be modified in the Custom sequences tab.
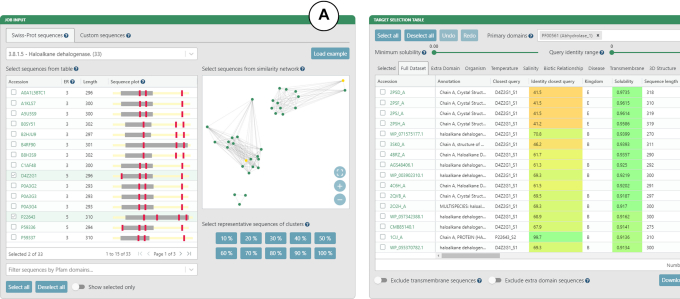
In the Swiss-Prot sequences tab (Figure 2A), sequences from the Swiss-Prot database can be queried by Enzyme Commission (EC) number. As a result, a table of all sequences annotated by the EC number and corresponding SSN is generated. The table has four columns: (i) sequence accessions hyperlinked to the UniProt database, (ii) number of essential residues, (iii) sequence length and (iv) sequence plot. The sequence plot summarizes two important features of the sequence – positions of essential residues and identified Pfam domains. The positions of essential residues are obtained from the Swiss-Prot database. The SSN visualizes the sequence space of all the sequences in the current EC group. Nodes represent Swiss-Prot sequences, whereas edge lengths are proportional to the pairwise sequence identities. Similar sequences are close to each other, whereas more distant sequences are not connected at all.
Figure 2.
The EnzymeMiner graphical user interface showing example inputs and results for the haloalkane dehalogenase family (EC 3.8.1.5). (A) Inputs based on curated sequences from the UniProtKB/Swiss-Prot database. The input sequences can be selected using: (i) the sequence table, (ii) the SSN or (iii) the sequence identity threshold. (B) Target selection table. The table is organized into eleven sheets that summarize the results from different perspectives. The table can be filtered using solubility and identity sliders, and transmembrane and extra domain exclusion switches.
There are three strategies possible for selecting Swiss-Prot sequences as the EnzymeMiner query: (i) select a row from the sequence table, (ii) select a node in the SSN and (iii) select cluster representatives by defining a sequence identity threshold. The sequence identity threshold buttons select cluster representatives at the given percentage threshold. Using this feature, the user can automatically select a small set of sequences that cover the whole known sequence space of the current EC group. All selected Swiss-Prot sequences are used as a query in the homology search step and also as essential residue templates for the filtration step. To modify the selected sets of queries and essential residue templates, the user can switch to the Custom sequences tab and refine the selection manually.
EnzymeMiner results
The results page is organized into four sections: (i) job information box, (ii) download results box, (iii) target selection table and (iv) sequence similarity network.
In the job information box, the user can find the job ID, title, start time and status of the job. There is also a rerun button for rerunning the same analysis without the need for re-entering the same input. This feature is handy for periodically mining new sequences as the sequence databases grow. For example, there are hundreds of new hits for the haloalkane dehalogenase family every year. In the download results box, the user can download the results table in XLSX format or tab-separated text format. A ZIP archive containing all output files from the EnzymeMiner workflow can also be downloaded.
The target selection table is the most important component of the EnzymeMiner results (Figure 2B). It presents all the putative enzyme sequences identified by EnzymeMiner and helps to select targets for experimental characterization. The table is organized into eleven sheets summarizing the results from different perspectives. (i) The Selected sheet shows all the sequences selected from individual sheets. It contains an extra column to track the argument used for the selection. By default, it is prefilled by the name of the sheet from which the sequence was selected, but it can be freely changed. (ii) The Full Dataset sheet shows all identified sequences. (iii) The Extra domain sheet shows sequences with extra Pfam domains found in the sequence but not listed in the Primary domains selection box. (iv) The Organism sheet shows sequences with known source organisms. (v) The Temperature sheet shows sequences from organisms having extreme optimum temperature annotation in the NCBI BioProject database, including sequences from thermophilic or cryophilic organisms. (vi) The Salinity sheet shows sequences from organisms having extreme salinity annotation in the NCBI BioProject database. (vii) The Biotic Relationship sheet shows sequences from organisms having biotic relationship annotation in the NCBI BioProject database. (viii) The Disease sheet shows sequences from organisms having disease annotation in the NCBI BioProject database. (ix) The Transmembrane sheet shows sequences with transmembrane regions predicted by the TMHMM tool. (x) The 3D Structure sheet shows sequences with an available 3D structure in the Protein Data Bank (22). (xi) The Network sheet shows sequences clustered into a selected sequence similarity network node.
There are four options for filtering the identified sequences displayed in the target selection table. The first option is the minimum solubility slider. Sequences with lower predicted solubility will be hidden. We recommend setting the solubility threshold to >0.5 to increase the probability of finding soluble protein expression in E. coli. We do not recommend to set the solubility threshold too high because of possible trade-off between enzyme solubility and activity (23). The second option is the identity range bar. Only sequences with identity to query sequences in the specified range will be visible. The third option is to exclude transmembrane proteins. We recommend removing these sequences as they are usually difficult to produce and tend to have lower predicted solubility. The fourth option is to exclude proteins with an extra domain. Extra domains are defined as domains found in the sequence but not listed in the Primary domains selection box. We recommend avoiding sequences with extra domains, but these sequences may also show interesting and unusual biological properties. The selection table can be sorted by clicking on a column header. Holding ‘Shift’ while clicking on the column headers allows sorting by multiple columns.
The SSN visualizes the sequence space of all identified sequences. Both clusters of similar sequences and sequence outliers can be easily identified. As there might be thousands of sequences, the sequences are clustered at the identity threshold and only an SSN of the representative sequences is shown for performance reasons. Sequences having greater sequence identity are consolidated into a single metanode. Edges indicate high sequence identity between representative sequences of the connected metanodes. Clicking on a metanode displays the Network sheet showing which sequences are represented by a particular metanode. The SSN can be downloaded as a Cytoscape session file for further analysis and custom visualization. Networks clustered at different identities are available. The numbers of nodes and edges are indicated for each identity threshold. The SSN is interactively linked to the target selection table. All nodes representing selected sequences are automatically highlighted in the SSN.
Target selection
The target selection table and SSN facilitate the selection of a diverse set of soluble putative enzyme sequences for experimental validation. First, we recommend setting the maximum sequence identity to queries to 90%. This will remove all hits that are very similar to already known proteins. Second, we recommend selecting a few sequences from individual sheets to cover different phyla from the domains Archea, Bacteria and Eukarya. The most exciting enzymes might be from extremophilic organisms. Third, the SSN can be used to check that the selection covers all sequence clusters. Fourth, users can select sequences from all subfamilies of the enzyme family of interest. The members of different subfamilies can be easily recognized by the Closest query or Closest known column in the selection table (note: requires representative sequences of subfamilies as job input). Fifth, the available filtering options can be used to (i) prioritize sequences with the highest predicted solubility, (ii) prioritize sequences with known tertiary structures, (iii) eliminate proteins with predicted transmembrane regions and (iv) eliminate sequences with extra domains.
EXPERIMENTAL VALIDATION OF THE EnzymeMiner WORKFLOW
The EnzymeMiner workflow has been thoroughly experimentally validated using the model enzymes haloalkane dehalogenases (5). The sequence-based search identified 658 putative dehalogenases. The subsequent analysis prioritized and selected 20 candidate genes for exploration of their protein structural and functional diversity. The selected enzymes originated from genetically unrelated Bacteria, Eukarya and, for the first time, also Archaea and showed novel catalytic properties and stabilities. The workflow helped to identify novel haloalkane dehalogenases, including (i) the most catalytically efficient enzyme (kcat/K0.5 = 96.8 mM−1 s−1), (ii) the most thermostable enzyme showing a melting temperature of 71°C, (iii) three different cold-adapted enzymes active at near to 0°C, (iv) highly enantioselective enzymes, (v) enzymes with a wide range of optimal operational temperature from 20 to 70°C and an unusually broad pH range from 5.7–10 and (vi) biocatalysts degrading the warfare chemical yperite and various environmental pollutants. The sequence mining, annotation, and visualization steps from the workflow published by Vanacek and co-workers (5) were fully automated in the EnzymeMiner web server. The successful use case for the haloalkane dehalogenase family is described in an easy-to-follow tutorial available on the EnzymeMiner web page. Additional extensive validation of the fully automated version of EnzymeMiner, experimentally testing the properties of another 45 genes of the haloalkane dehalogenases, is currently ongoing in our laboratory.
CONCLUSIONS AND OUTLOOK
The EnzymeMiner web server identifies putative members of enzyme families and facilitates their prioritization and well-informed manual selection for experimental characterization to reveal novel biocatalysts. Such a task is difficult using the web interfaces of the available protein databases, e.g. UniProtKB/TrEMBL and NCBI Protein, since additional analyses are often required. The major advantage of EnzymeMiner over existing protein sources is the flexibility of input and concise annotation-rich interactive presentation of results. The user can input custom queries and a custom description of essential residues to focus the search on specific protein families or subfamilies. The output of EnzymeMiner is an interactive selection table containing the annotated sequences divided into sheets based on various criteria. The table helps to select a diverse set of sequences for experimental characterization. Two key prioritization criteria are (i) the predicted solubility score, which can be used to prioritize the identified sequences and increase the chance of finding enzymes with soluble protein expression, and (ii) the sequence identity to query sequences complemented with an interactive SSN displayed directly on the web, which can be used to find diverse sequences. Additionally, source organism and domain annotations help to select sequences with diverse properties. EnzymeMiner is a universal tool applicable to any enzyme family. It reduces the time needed for data gathering, multi-step analysis and sequence prioritization from days to hours. All the EnzymeMiner features are implemented directly on the web server and no external tools are required. The web server was optimized for modern browsers including Chrome, Firefox and Safari. An EnzymeMiner job can take a few hours or days to compute, depending on the current load of the server. In the next EnzymeMiner version, we plan three major improvements. First, we will implement automated tertiary structure prediction based on homology modeling and threading for all identified sequences. The structural predictions will allow subsequent analysis of active site pockets/cavities and access tunnels. Structural features will significantly enrich the set of annotations and help to identify additional attractive targets for experimental characterization. Second, we will implement automated periodical mining. When enabled, EnzymeMiner will rerun the analysis periodically and inform the user about novel sequences found since the last search. Finally, we will implement a wizard for automated selection of hits based on input criteria provided by a user.
ACKNOWLEDGEMENTS
We thank the participants of the 1st Hands-on Computational Enzyme Design Course (Brno, Czech Republic) for giving valuable feedback on the EnzymeMiner user interface. Their comments inspired us to make the sequence similarity network visualization more interactive.
Contributor Information
Jiri Hon, Loschmidt Laboratories, Department of Experimental Biology and Research Center for Toxic Compounds in the Environment RECETOX, Faculty of Science, Masaryk University, Brno, Czech Republic; IT4Innovations Centre of Excellence, Faculty of Information Technology, Brno University of Technology, Bozetechova 2, Brno, Czech Republic; International Clinical Research Center, St. Anne's University Hospital Brno, Brno, Czech Republic.
Simeon Borko, Loschmidt Laboratories, Department of Experimental Biology and Research Center for Toxic Compounds in the Environment RECETOX, Faculty of Science, Masaryk University, Brno, Czech Republic; IT4Innovations Centre of Excellence, Faculty of Information Technology, Brno University of Technology, Bozetechova 2, Brno, Czech Republic.
Jan Stourac, Loschmidt Laboratories, Department of Experimental Biology and Research Center for Toxic Compounds in the Environment RECETOX, Faculty of Science, Masaryk University, Brno, Czech Republic; International Clinical Research Center, St. Anne's University Hospital Brno, Brno, Czech Republic.
Zbynek Prokop, Loschmidt Laboratories, Department of Experimental Biology and Research Center for Toxic Compounds in the Environment RECETOX, Faculty of Science, Masaryk University, Brno, Czech Republic; International Clinical Research Center, St. Anne's University Hospital Brno, Brno, Czech Republic.
Jaroslav Zendulka, IT4Innovations Centre of Excellence, Faculty of Information Technology, Brno University of Technology, Bozetechova 2, Brno, Czech Republic.
David Bednar, Loschmidt Laboratories, Department of Experimental Biology and Research Center for Toxic Compounds in the Environment RECETOX, Faculty of Science, Masaryk University, Brno, Czech Republic; International Clinical Research Center, St. Anne's University Hospital Brno, Brno, Czech Republic.
Tomas Martinek, IT4Innovations Centre of Excellence, Faculty of Information Technology, Brno University of Technology, Bozetechova 2, Brno, Czech Republic.
Jiri Damborsky, Loschmidt Laboratories, Department of Experimental Biology and Research Center for Toxic Compounds in the Environment RECETOX, Faculty of Science, Masaryk University, Brno, Czech Republic; International Clinical Research Center, St. Anne's University Hospital Brno, Brno, Czech Republic.
FUNDING
Czech Ministry of Education [857560, 02.1.01/0.0/0.0/18_046/0015975, CZ.02.1.01/0.0/0.0/16_026/0008451, CZ.02.1.01/0.0/0.0/16_019/0000868, LQ1602]; European Commission [720776, 814418]; AI Methods for Cybersecurity and Control Systems project of the Brno University of Technology [FIT-S-20-6293]; Computational resources were supplied by the project ‘e-Infrastruktura CZ’ [e-INFRA LM2018140] provided within the program Projects of Large Research, Development and Innovations Infrastructures, and by the ELIXIR-CZ project [LM2015047], part of the international ELIXIR infrastructure. Funding for open access charge: Czech Ministry of Education.
Conflict of interest statement. None declared.
REFERENCES
- 1. Sayers E.W., Agarwala R., Bolton E.E., Brister J.R., Canese K., Clark K., Connor R., Fiorini N., Funk K., Hefferon T. et al.. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2019; 47:D23–D28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. UniProt Consortium UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019; 47:D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Colin P.-Y., Kintses B., Gielen F., Miton C.M., Fischer G., Mohamed M.F., Hyvönen M., Morgavi D.P., Janssen D.B., Hollfelder F.. Ultrahigh-throughput discovery of promiscuous enzymes by picodroplet functional metagenomics. Nat. Commun. 2015; 6:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beneyton T., Thomas S., Griffiths A.D., Nicaud J.-M., Drevelle A., Rossignol T.. Droplet-based microfluidic high-throughput screening of heterologous enzymes secreted by the yeast Yarrowia lipolytica. Microb. Cell Fact. 2017; 16:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vanacek P., Sebestova E., Babkova P., Bidmanova S., Daniel L., Dvorak P., Stepankova V., Chaloupkova R., Brezovsky J., Prokop Z. et al.. Exploration of enzyme diversity by integrating bioinformatics with expression analysis and biochemical characterization. ACS Catal. 2018; 8:2402–2412. [Google Scholar]
- 6. El-Gebali S., Mistry J., Bateman A., Eddy S.R., Luciani A., Potter S.C., Qureshi M., Richardson L.J., Salazar G.A., Smart A. et al.. The Pfam protein families database in 2019. Nucleic Acids Res. 2019; 47:D427–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li Y., Wang S., Umarov R., Xie B., Fan M., Li L., Gao X.. DEEPre: sequence-based enzyme EC number prediction by deep learning. Bioinformatics. 2018; 34:760–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou N., Jiang Y., Bergquist T.R., Lee A.J., Kacsoh B.Z., Crocker A.W., Lewis K.A., Georghiou G., Nguyen H.N., Hamid M.N. et al.. The CAFA challenge reports improved protein function prediction and new functional annotations for hundreds of genes through experimental screens. Genome Biol. 2019; 20:244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mak W.S., Tran S., Marcheschi R., Bertolani S., Thompson J., Baker D., Liao J.C., Siegel J.B.. Integrative genomic mining for enzyme function to enable engineering of a non-natural biosynthetic pathway. Nat. Commun. 2015; 6:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Altschul S. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997; 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010; 26:2460–2461. [DOI] [PubMed] [Google Scholar]
- 12. Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J. et al.. Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011; 7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Krogh A., Larsson B., von Heijne G., Sonnhammer E.L.. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001; 305:567–580. [DOI] [PubMed] [Google Scholar]
- 14. Quevillon E., Silventoinen V., Pillai S., Harte N., Mulder N., Apweiler R., Lopez R.. InterProScan: protein domains identifier. Nucleic Acids Res. 2005; 33:W116–W120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Federhen S. The NCBI Taxonomy database. Nucleic Acids Res. 2012; 40:D136–D143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Barrett T., Clark K., Gevorgyan R., Gorelenkov V., Gribov E., Karsch-Mizrachi I., Kimelman M., Pruitt K.D., Resenchuk S., Tatusova T. et al.. BioProject and BioSample databases at NCBI: facilitating capture and organization of metadata. Nucleic Acids Res. 2012; 40:D57–D63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Musil M., Konegger H., Hon J., Bednar D., Damborsky J.. Computational design of Stable and Soluble Biocatalysts. ACS Catal. 2019; 9:1033–1054. [Google Scholar]
- 18. Steinegger M., Söding J.. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 2017; 35:1026–1028. [DOI] [PubMed] [Google Scholar]
- 19. Shannon P. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003; 13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Copp J.N., Akiva E., Babbitt P.C., Tokuriki N.. Revealing unexplored sequence-function space using sequence similarity networks. Biochemistry. 2018; 57:4651–4662. [DOI] [PubMed] [Google Scholar]
- 21. Gerlt J.A., Bouvier J.T., Davidson D.B., Imker H.J., Sadkhin B., Slater D.R., Whalen K.L.. Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): A web tool for generating protein sequence similarity networks. Biochim. Biophys. Acta (BBA) - Proteins Proteomics. 2015; 1854:1019–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E.. The Protein Data Bank. Nucleic Acids Res. 2000; 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Klesmith J.R., Bacik J.-P., Wrenbeck E.E., Michalczyk R., Whitehead T.A.. Trade-offs between enzyme fitness and solubility illuminated by deep mutational scanning. Proc. Natl Acad. Sci. U.S.A. 2017; 114:2265–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]