Abstract
Major histocompatibility complex (MHC) molecules are expressed on the cell surface, where they present peptides to T cells, which gives them a key role in the development of T-cell immune responses. MHC molecules come in two main variants: MHC Class I (MHC-I) and MHC Class II (MHC-II). MHC-I predominantly present peptides derived from intracellular proteins, whereas MHC-II predominantly presents peptides from extracellular proteins. In both cases, the binding between MHC and antigenic peptides is the most selective step in the antigen presentation pathway. Therefore, the prediction of peptide binding to MHC is a powerful utility to predict the possible specificity of a T-cell immune response. Commonly MHC binding prediction tools are trained on binding affinity or mass spectrometry-eluted ligands. Recent studies have however demonstrated how the integration of both data types can boost predictive performances. Inspired by this, we here present NetMHCpan-4.1 and NetMHCIIpan-4.0, two web servers created to predict binding between peptides and MHC-I and MHC-II, respectively. Both methods exploit tailored machine learning strategies to integrate different training data types, resulting in state-of-the-art performance and outperforming their competitors. The servers are available at http://www.cbs.dtu.dk/services/NetMHCpan-4.1/ and http://www.cbs.dtu.dk/services/NetMHCIIpan-4.0/.
INTRODUCTION
The Major histocompatibility complex (MHC) is a fundamental cell surface protein of the cellular immune system of vertebrates. The primary function of MHC is to bind to peptides (small protein fragments) derived from the digestion of intracellular or extracellular proteins and display them to the intercellular space. If T cells recognize and bind to a peptide–MHC complex, an immune response can be triggered and the compromised cell will undergo lysis. Given this, the binding of antigenic peptides to MHC molecules represents a necessary step for cellular immunity, and understanding the rules of this event has large and valuable potential in human health applications.
MHC comes in two main variants: MHC Class I (MHC-I) and MHC Class II (MHC-II). MHC-I binds peptides from intracellular proteins after these undergo proteasomal degradation, and serves as a control mechanism for antigenic variations in the self-peptidome repertoire. On the other hand, the MHC-II binds peptides generated by protease-digestion of extracellular proteins; with this, both MHC systems can exert control over foreign organisms via the presentation of non-self proteins to T cells (1). In view of this fact, important efforts have been committed to developing computational methods capable of accurately predicting peptide binding to both MHC-I and MHC-II (reviewed in (2)).
Different types of experimental data have been used to train these methods. According to the nature of such training data, we can classify peptide-MHC binding predictors in three main categories. The first category corresponds to predictors trained on binding affinity (BA) data (3–6). This type of data imposes a substantial limitation on prediction performances, since it only models the single event of peptide-MHC binding, and neglects any other biological feature involved in the process. The second category covers methods that are either trained with data retrieved from mass spectrometry (MS) experiments, known as eluted ligands (EL) (7–11), or trained integrating both BA and EL data (5,12–15). This latter data type incorporates information not only related to the peptide-MHC binding event, but also information about prior steps in the biological antigen presentation pathway processes. However, except for genetically engineered cells, cellular MHC expression profile is very diverse due to the multiple MHC allelic variants. Also, antibodies employed to purify peptide–MHC complexes in MS EL pipelines are mostly pan- or locus-specific, leading to inherently poly-specific (or Multi Allelic, MA) data (i.e., the data contains peptides matching multiple cognate MHC binding motifs). Thus, a prior, user biased peptide-MHC annotation criteria are, in general, needed in order to interpret such EL MA data, transform them to Single Allelic (EL SA, or single peptide-MHC annotations) and employ them for the training of MHC-specific binding predictors (16).
The third and last category of algorithms seeks to resolve this limitation of the second type of models, and incorporates, together with the training of a prediction algorithm, the capability of annotating EL MA sequences to single MHC restrictions (17,18). One such method is termed NNAlign_MA (17), which during the training process can cluster EL sequences with ambiguous cognate MHCs into single MHC specificities, using a strategy called pseudo-labeling. This enables not only the possibility of novel motif discovery, but also a considerable expansion of the training set size, and therefore an overall improvement of the method's predictive power.
In this work, we deploy NNAlign_MA to update NetMHCpan and NetMHCIIpan, augmenting their training capabilities and also increasing their predictive performance. We do this by incorporating NNAlign_MA to the core of the new models, allowing us to expand their training sets greatly. Moving further, we perform a full independent epitope evaluation on both models and show how the updated methods outperform other current state-of-the-art algorithms.
The NNAlign_MA machine learning framework
The updated versions of NetMHCpan and NetMHCIIpan differ from their predecessors in two critical aspects: the training data and the machine-learning modeling framework. The training data have been vastly extended by accumulating MHC BA and EL data from the public domain. In particular, EL data were extended to include MA data. The combined dataset used for training of NetMHCpan-4.1 consists of 13 245 212 data points covering 250 distinct MHC class I molecules, and the combined dataset used for training of NetMHCIIpan-4.0 consists of 4 086 230 data points covering a total of 116 distinct MHC class II molecules. For specific details on the training sets and data partitioning refer to Supplementary Materials. The machine learning framework was updated from NNAlign to NNAlign_MA to allow for effective handling of these MA data. In short, the NNAlign framework is a single-allele framework permitting the integration of mixed data types (BA and EL) in the model training, which allows information to be leveraged across the different data types, resulting in a boosted predictive power (12,13). NNAlign_MA extends this training framework to allow for the incorporation of EL MA data. This is achieved by iteratively annotating the best single-allele to the MA data during the model training, effectively deconvoluting the MA binding motifs (17). For specific details on the model hyper-parameters and cross-validation training performance, please refer to Supplementary Material.
WEB INTERFACE
Submission page
Input data
Both servers accept two different types of input; FASTA and PEPTIDE. The input data can be directly pasted into a submission box or uploaded from the user's local disk. For FASTA input, the user can specify the peptide length(s) to be included in the predictions (for class I, the length range goes from 8 to 14 amino acids, default is 8–11; for class II only one length is admitted with 15 being the default value).
Also, for Class II, one can specify if CONTEXT encoding (13) is to be used. This context consists of amino acids spanning the source protein N and C terminal parts of the ligand.
The submission page includes examples of input data for all accepted formats and provides buttons to upload sample data automatically.
MHC selection
Next, the servers provide a drop-down menu in order to select which MHC family and molecule(s) to be used. NetMHCpan-4.1 covers more than 11 000 MHC molecules, spanning human (HLA-A, HLA-B, HLA-C, HLA-E, HLA-G), mouse (H-2), cattle (BoLA), primates (Patr, Mamu, Gogo), swine (SLA), equine (EQCA) and dog (DLA), and NetMHCIIpan-4.0 covers a total of close to 1000 human (HLA-DR, HLA-DQ, HLA-DP) and mouse (H-2) MHC alleles. For DQ and DP, the user can make combinations of the covered alpha and beta protein chains. Furthermore, given the pan-specific nature of both methods, predictions can be run for any MHC molecule of known sequence by uploading a full-length MHC protein sequence in FASTA format.
Additional configuration
Both NetMHCpan methods inform if a sequence is a strong MHC binder (SB) or a weak MHC binder (WB) based on a %Rank score. Briefly, %Rank is a transformation that normalizes prediction scores across different MHC molecules and enables inter-specific MHC binding prediction comparisons. %Rank of a query sequence is computed by comparing its prediction score to a distribution of prediction scores for the MHC in question, estimated from a set of random natural peptides. Given this, a %Rank value of 1% means that a queried sequence obtained a prediction score that corresponds to the top 1% scores obtained from random natural peptides. The %Rank values for detecting SBs and WBs can be modified by specifying the corresponding thresholds (by default, %Rank < 0.5% and %Rank < 2% thresholds are considered for detecting SBs and WBs for class I and %Rank < 2% and %Rank < 10%, for SBs and WBs for class II). In addition, an option is available to only report sequences with a lower than a defined %Rank threshold, and for class II to print only the strongest binding peptide overlapping a given binding core if FASTA was selected as the input format.
Additionally, the user may opt to get the BA prediction scores of input sequences together with the EL likelihood, and to sort the output according to the corresponding EL predicted values (from high to low). In addition, and for user convenience, the possibility to save the output as a *. XLS file (readable to most spreadsheet software) is also provided.
Output page
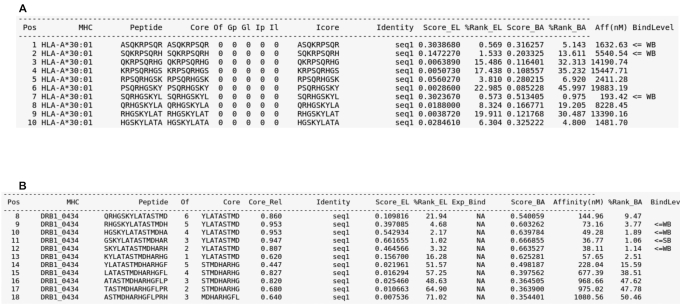
The output from both servers details the binding prediction values of the provided input sequence(s) for the selected MHC molecule(s), together with additional information to guide the interpretation of results. As seen in Figure 1, NetMHCpan and NetMHCIIpan output consist of several plain text columns, which exhibit different pieces of information regarding the prediction outcome.
Figure 1.
Example outputs for the NetMHCpan-4.1 and NetMHCIIpan-4.0 tools. (A) Example output for NetMHCpan-4.1, using as input the web server's FASTA sample data and the HLA-A*30:01 allele, with a peptide length of nine and other options set to default. (B) Example output for NetMHCIIpan-4.0, using as input the web server's FASTA sample data and the DRB1*04:34 allele, with all other options set to default. By default, prediction scores are for both methods displayed in terms of a Score_EL (the likelihood of a peptide being an MHC ligand) column and a ‘%Rank_EL’ column (the EL percentile Rank score); if the user selects to include BA predictions, such values are reported as well. The ‘BindLevel’ column displays the presence of Strong Binders (SB) or Weak Binders (WB) amongst the queried peptides. ‘Peptide’ informs the list of peptides that have been interrogated against the selected MHC molecule(s) (exhibited in the ‘MHC’ column). The ‘Pos’ entry refers to the queried peptide's position in the selected FASTA input, and ‘Core’ refers to such peptide's identified binding core. ‘Identity’ is an automatically generated ID that is assigned to the input. Other columns refer to specific properties that depend on the MHC class being employed. For additional details on the interpretation of the different columns of the output, refer to the ‘output format’ page on both web servers homepages.
EVALUATION AND EXAMPLES
As independent validations, the models were benchmarked on sets of T-cell epitope data and for class I also EL SA data. For MHC class I the epitope dataset was taken from Jurtz et. al (12) combined with a comprehensive set of MHC multimer validated epitopes obtained from the IEDB and for MHC class II from Reynisson et al. (19). The EL SA data were obtained from (20). In all cases, the data were filtered to ensure no overlap with the training data (for further details on the data sets refer to Supplementary Material). For the epitope data, the predictive performance was estimated in terms of FRANK (12). That is, for each epitope-HLA pair, binding to the HLA was predicted for all overlapping peptides of the source protein using the eluted ligand likelihood prediction score and the FRANK value was reported as the proportion of peptides with a prediction score higher than that of the epitope. Using this measure, a value of 0 corresponds to a perfect prediction (the known epitope is identified with the highest predicted binding value among all peptides found within the source protein), while a value of 0.5 corresponds to a random prediction. Further, was the corresponding AUC for each epitope reported, again assigning all overlapping peptides in the source protein except the epitope as negatives. For further details on the CD8 epitope benchmark, refer to Supplementary Table 7. For the EL SA dataset, negative decoy peptides were added as described in the “Training and Test data” section of the Supplementary Material in ‘Materials and Methods’ and the performance evaluated in terms of AUC, AUC0.1 and PPV. Here, PPV was estimated from the fraction of positive peptides within the top N predictions, where N is equal to the total number of ligands times 0.95 (to account for potential MS contaminants). For additional information on the EL SA benchmark, refer to Supplementary Table 8.
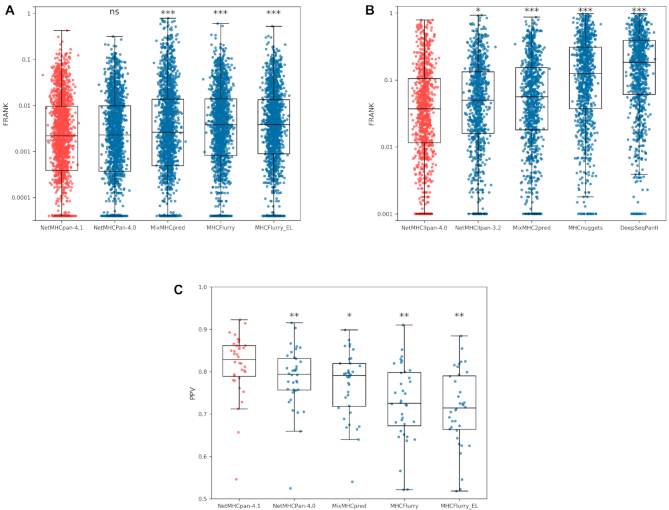
The results of these benchmarks are shown in Figure 2. Here, NetMHCpan-4.1 was compared to NetMHCpan-4.0 (12), MixMHCpred (18,21), MHCFlurry (5) and MHCFlurry_EL (an unpublished version of MHCFLurry trained with EL SA data, available at GitHub (22)). For this benchmark, because MixMHCpred cannot make predictions for peptides containing ‘X’ (wildcard amino acid symbol), such peptides were removed from the benchmark dataset. NetMHCIIpan-4.0 was compared in a similar manner to NetMHCIIpan-3.2 (23), MixMHC2pred (11), MHCnuggets (24) and DeepSeqPanII (25).
Figure 2.
Epitope benchmark results for the NetMHCpan-4.1 and NetMHCIIpan-4.0 web servers. (A) Performance results for the CD8+ epitope benchmark. Median FRANK values for the different methods are: NetMHCpan-4.1, 0.00220; NetMHCpan-4.0, 0.00230; MixMHCpred, 0.00264; MHCFlurry, 0.00383; and MHCFlurry_EL, 0.00386. (B) FRANK performance results for the CD4+ epitope benchmark. The median FRANK for the different methods are: NetMHCIIpan-4.0, 0.0351; NetMHCIIpan-3.2, 0.04825; MixMH2Cpred, 0.0513; MHCnuggets, 0.1219; and DeepSeqPanII, 0.1767. (C) PPV performance results for the MS MHC class I eluted ligand benchmark. Median PPV values for the different methods are: NetMHCpan-4.1, 0.8291; NetMHCpan-4.0, 0.7940; MixMHCpred, 0.7911; MHCFlurry, 0.7256; and MHCFlurry_EL, 0.7144. P-values are shown as * P < 0.05, ** P < 10−6 and *** P < 10−9. All p-values were calculatated using a two-tailed binomial test. The plotted boxes extend from the lower to upper quartile values of the data (25th to 75th percentile), with a line at the median; whiskers extend from the box to show the range of the data to the most extreme, non-outlier data points.
With the exception of NetMHCpan-4.1 and NetMHCpan-4.0 when tested on the epitope benchmark, all three benchmarks confirmed a significantly superior performance of NetMHCpan-4.1 and NetMHCIIpan-4.0 over all other methods included in the respective benchmarks. For the class I epitope benchmark, NetMHCpan-4.1 and NetMHCpan-4.0 were found to share comparable predictive performance. For NetMHCpan-4.1 a consistent improvement was found for HLA-B and HLA-C molecules for both the epitopes and ligand benchmarks when compared to NetMHCpan-4.0 (consistent with the very large increased coverage of these loci by the EL dataset used for the training of NetMHCpan-4.1). Note, also that in contrast to what was observed when evaluating the performance on eluted ligand data (19), but in line with earlier works (13,19,26), a drop in the performance of NetMHCIIpan-4.0 was observed when including context information (Supplementary Figure S3).
DISCUSSION
Over the last years, large amounts of novel MS-eluted MHC ligand data have become available, enabling a highly enriched characterization of the MHC-presented ligandome. Here, we have benefitted from this data, and combining it with an extensive set of MHC peptide-binding data available in the IEDB, have developed updated versions of the NetMHCpan and NetMHCIIpan tools. Both methods are capable of predicting a peptide's likelihood of antigen presentation (and BA) to MHC class I and class II molecules. Both tools were trained using the NNAlign_MA machine learning framework, which enables the integration of MS ligand datasets obtained from cell lines expressing multiple MHC alleles. The benchmarking of these methods against other available state-of-the-art algorithms exhibited a significantly improved predictive power for the prediction of MHC ligands and T-cell epitopes.
For both NetMHCpan-4.1 and NetMHCIIpan-4.0, the performance gain was found most pronounced for prediction of MS identified MHC ligands. This in particular for class I, where the NetMHCpan-4.1 method on the epitope benchmark was found to perform at par with its most recent ancestor NetMHCpan-4.0. Many possible reasons for this limited impact on the performance for epitope prediction exists, including biases in the epitope data currently available toward past prediction methods and in-vitro experimental validation techniques, and biases in the MS EL data not shared with T-cell epitopes. Future work will resolve the impact and importance of these biases, and allow us to access to what degree the improved power for prediction of MS MHC ligands translates into an improved power also for prediction of T-cell epitopes.
Benchmark evaluation of the tools demonstrated an overall robust power of the NNAlign_MA machine learning framework to perform motif deconvolution across all MHC molecules included in the training data. However, results also pointed to a lower performance for MHC molecules characterized by limited ligand datasets such as HLA-C and HLA-DQ. While this low number of ligands annotated to MHC from these two loci in part can be explained from their relative low protein expression, other causes could include differences in the HLA-loci specificities of the antibodies used for immunoprecipitation (IP) when purifying MHC molecules prior to running MS experiments. Future work may tell if working with antibodies with improved HLA-DQ specificities or using engineered cell lines with, for instance, tagged HLA molecules as suggested by (8) can help resolve this.
Even though one of the main contributions to the improved performance of the prediction methods proposed here (and other recently published methods) is the integration of MS derived EL data, MS data itself contains an inherent bias imposed resulting in for instance overrepresentation of ‘flyable’ (27) and neglecting cysteine-containing peptides (7). These biases impose limitations on the set of ligands detectable in MS and hence subsequent limitations on the learned binding motifs. Given this, further complementary technological platforms for high throughput detection of MHC peptide interactions might be warranted to complete our understanding of HLA antigen presentation.
Both NetMHCpan and NetMHCIIpan have an easy to use user interface, allowing for simple uploads of query sequence data, and a selection of MHC alleles to be interrogated for binding. As the only current publicly available tools, both methods demonstrate a truly pan-specific capability, allowing users to make predictions for all MHC molecules, including those not previously characterized by binding data. The output from the tools is provided in simple text format with guided information, aiding the user to select relevant epitope/MHC-ligand candidates.
Given the demonstrated high performances and their ease of use, we expect the updated web servers to become relevant tools to guide future rational epitope discovery projects.
DATA AVAILABILITY
The two web servers described in this work are hosted at http://www.cbs.dtu.dk/services/NetMHCpan-4.1 and http://www.cbs.dtu.dk/services/NetMHCIIpan-4.0. The servers will likewise be made available from the IEDB analysis resource.
Supplementary Material
Contributor Information
Birkir Reynisson, Department of Bio and Health Informatics, Technical University of Denmark, Kgs. Lyngby, DK 28002, Denmark.
Bruno Alvarez, Instituto de Investigaciones Biotecnológicas, Universidad Nacional de San Martín, Buenos Aires, BA 16503, Argentina.
Sinu Paul, La Jolla Institute for Immunology, La Jolla, CA 92037, USA.
Bjoern Peters, La Jolla Institute for Immunology, La Jolla, CA 92037, USA; Department of Medicine, University of California, San Diego, CA 92093, USA.
Morten Nielsen, Department of Bio and Health Informatics, Technical University of Denmark, Kgs. Lyngby, DK 28002, Denmark; Instituto de Investigaciones Biotecnológicas, Universidad Nacional de San Martín, Buenos Aires, BA 16503, Argentina.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [75N93019C00001]; EIT Health through the grant No. [19638]. EIT Health is supported by the EIT, a body of the European Union. Funding for open access charge: National Institutes of Health [75N93019C00001].
Conflict of interest statement. None declared.
REFERENCES
- 1. Duan L., Mukherjee E.. Janeway’s Immunobiology, Ninth Edition. Yale Journal of Biology and Medicine. 2016; 89:424–425. [Google Scholar]
- 2. Peters B., Nielsen M., Sette A.. T cell epitope predictions. Annu. Rev. Immunol. 2020; 38:123–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nielsen M., Andreatta M.. NetMHCpan-3.0; improved prediction of binding to MHC class I molecules integrating information from multiple receptor and peptide length datasets. Genome Med. 2016; 8:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Karosiene E., Rasmussen M., Blicher T., Lund O., Buus S., Nielsen M.. NetMHCIIpan-3.0, a common pan-specific MHC class II prediction method including all three human MHC class II isotypes, HLA-DR, HLA-DP and HLA-DQ. Immunogenetics. 2013; 65:711–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. O’Donnell T.J., Rubinsteyn A., Bonsack M., Riemer A.B., Laserson U., Hammerbacher J.. MHCflurry: open-source Class I MHC binding affinity prediction. Cell Syst. 2018; 7:129–132. [DOI] [PubMed] [Google Scholar]
- 6. Kim Y., Sidney J., Pinilla C., Sette A., Peters B.. Derivation of an amino acid similarity matrix for peptide: MHC binding and its application as a Bayesian prior. BMC Bioinformatics. 2009; 10:394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Abelin J.G., Keskin D.B., Sarkizova S., Hartigan C.R., Zhang W., Sidney J., Stevens J., Lane W., Zhang G.L., Eisenhaure T.M. et al.. Mass spectrometry profiling of HLA-associated peptidomes in mono-allelic cells enables more accurate epitope prediction. Immunity. 2017; 46:315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Abelin J.G., Harjanto D., Malloy M., Suri P., Colson T., Goulding S.P., Creech A.L., Serrano L.R., Nasir G., Nasrullah Y. et al.. Defining HLA-II ligand processing and binding rules with mass spectrometry enhances cancer epitope prediction. Immunity. 2019; 51:766–779. [DOI] [PubMed] [Google Scholar]
- 9. Bassani-Sternberg M., Gfeller D.. Unsupervised HLA peptidome deconvolution improves ligand prediction accuracy and predicts cooperative effects in peptide-HLA interactions. J. Immunol. 2016; 197:2492–2499. [DOI] [PubMed] [Google Scholar]
- 10. Bulik-Sullivan B., Busby J., Palmer C.D., Davis M.J., Murphy T., Clark A., Busby M., Duke F., Yang A., Young L. et al.. Deep learning using tumor HLA peptide mass spectrometry datasets improves neoantigen identification. Nat. Biotechnol. 2018; 37:55–63. [DOI] [PubMed] [Google Scholar]
- 11. Racle J., Michaux J., Rockinger G.A., Arnaud M., Bobisse S., Chong C., Guillaume P., Coukos G., Harari A., Jandus C. et al.. Robust prediction of HLA class II epitopes by deep motif deconvolution of immunopeptidomes. Nat. Biotechnol. 2019; 37:1283–1286. [DOI] [PubMed] [Google Scholar]
- 12. Jurtz V., Paul S., Andreatta M., Marcatili P., Peters B., Nielsen M.. NetMHCpan-4.0: improved peptide-MHC class i interaction predictions integrating eluted ligand and peptide binding affinity data. J. Immunol. 2017; 199:3360–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Barra C., Alvarez B., Paul S., Sette A., Peters B., Andreatta M., Buus S., Nielsen M.. Footprints of antigen processing boost MHC class II natural ligand predictions. Genome Med. 2018; 10:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Alvarez B., Barra C., Nielsen M., Andreatta M.. Computational tools for the identification and interpretation of sequence motifs in immunopeptidomes. Proteomics. 2018; 18:e1700252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Garde C., Ramarathinam S.H., Jappe E.C., Nielsen M., Kringelum J.V., Trolle T., Purcell A.W.. Improved peptide-MHC class II interaction prediction through integration of eluted ligand and peptide affinity data. Immunogenetics. 2019; 71:445–454. [DOI] [PubMed] [Google Scholar]
- 16. Nielsen M., Connelley T., Ternette N.. Improved prediction of bovine leucocyte antigens (BoLA) presented ligands by use of mass-spectrometry-determined ligand and in vitro binding data. J. Proteome Res. 2018; 17:559–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alvarez B., Reynisson B., Barra C., Buus S., Ternette N., Connelley T., Andreatta M., Nielsen M.. NNAlign_MA; MHC peptidome deconvolution for accurate mhc binding motif characterization and improved t-cell epitope predictions. Mol. Cell Proteomics. 2019; 18:2459–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bassani-Sternberg M., Chong C., Guillaume P., Solleder M., Pak H., Gannon P.O., Kandalaft L.E., Coukos G., Gfeller D.. Deciphering HLA-I motifs across HLA peptidomes improves neo-antigen predictions and identifies allostery regulating HLA specificity. PLoS Comput. Biol. 2017; 13:e1005725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Reynisson B., Barra C., Kaabinejadian S., Hildebrand W.H., Peters B., Nielsen M.. Improved prediction of MHC II antigen presentation through integration and motif deconvolution of mass spectrometry MHC eluted ligand data. 2020; bioRxiv doi:19 February 2020, preprint: not peer reviewed 10.1101/799882. [DOI] [PubMed]
- 20. Sarkizova S., Klaeger S., Le P.M., Li L.W., Oliveira G., Keshishian H., Hartigan C.R., Zhang W., Braun D.A., Ligon K.L. et al.. A large peptidome dataset improves HLA class I epitope prediction across most of the human population. Nat. Biotechnol. 2020; 38:199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gfeller D., Guillaume P., Michaux J., Pak H.-S., Daniel R.T., Racle J., Coukos G., Bassani-Sternberg M.. The length distribution and multiple specificity of naturally presented HLA-I ligands. J. Immunol. 2018; 201:3705–3716. [DOI] [PubMed] [Google Scholar]
- 22. O’Donnell T.J., Rubinsteyn A., Bonsack M., Riemer A.B., Laserson U., Hammerbacher J.. 2020; MHCFlurryhttps://github.com/openvax/mhcflurry. [DOI] [PubMed]
- 23. Jensen K.K., Andreatta M., Marcatili P., Buus S., Greenbaum J.A., Yan Z., Sette A., Peters B., Nielsen M.. Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology. 2018; 154:394–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shao X.M., Bhattacharya R., Huang J., Sivakumar I.K.A., Tokheim C., Zheng L., Hirsch D., Kaminow B., Omdahl A., Bonsack M. et al.. High-throughput prediction of MHC class i and ii neoantigens with MHCnuggets. Cancer Immunol. Res. 2020; 8:396–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu Z., Jin J., Cui Y., Xiong Z., Nasiri A., Zhao Y., Hu J.. DeepSeqPanII: an interpretable recurrent neural network model with attention mechanism for peptide-HLA class II binding prediction. 2019; bioRxiv doi:24 October 2019, preprint: not peer reviewed 10.1101/817502. [DOI] [PubMed]
- 26. Paul S., Karosiene E., Dhanda S.K., Jurtz V., Edwards L., Nielsen M., Sette A., Peters B.. Determination of a predictive cleavage motif for eluted major histocompatibility complex class II ligands. Front. Immunol. 2018; 9:1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jarnuczak A.F., Lee D.C.H., Lawless C., Holman S.W., Eyers C.E., Hubbard S.J.. Analysis of intrinsic peptide detectability via integrated label-free and SRM-based absolute quantitative proteomics. J. Proteome Res. 2016; 15:2945–2959. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The two web servers described in this work are hosted at http://www.cbs.dtu.dk/services/NetMHCpan-4.1 and http://www.cbs.dtu.dk/services/NetMHCIIpan-4.0. The servers will likewise be made available from the IEDB analysis resource.