Abstract
In the past few decades, major initiatives have been launched around the world to address chemical safety testing. These efforts aim to innovate and improve the efficacy of existing methods with the long-term goal of developing new risk assessment paradigms. The transcriptomic and toxicological profiling of mammalian cells has resulted in the creation of multiple toxicogenomic datasets and corresponding tools for analysis. To enable easy access and analysis of these valuable toxicogenomic data, we have developed ToxicoDB (toxicodb.ca), a free and open cloud-based platform integrating data from large in vitro toxicogenomic studies, including gene expression profiles of primary human and rat hepatocytes treated with 231 potential toxicants. To efficiently mine these complex toxicogenomic data, ToxicoDB provides users with harmonized chemical annotations, time- and dose-dependent plots of compounds across datasets, as well as the toxicity-related pathway analysis. The data in ToxicoDB have been generated using our open-source R package, ToxicoGx (github.com/bhklab/ToxicoGx). Altogether, ToxicoDB provides a streamlined process for mining highly organized, curated, and accessible toxicogenomic data that can be ultimately applied to preclinical toxicity studies and further our understanding of adverse outcomes.
INTRODUCTION
Compound toxicity and its effect on human health has been a major focus of toxicological research for the past few decades. From the traditional way of assessing toxicity using a single-compound approach, the direction is gradually moving towards high-throughput screens and alternative models. In pharmaceutical research, drug toxicity is one of the most significant reasons for high attrition rates in the drug discovery pipeline. Collectively, preclinical toxicity and adverse outcomes in humans contribute to approximately one-third of the drug failures in pipeline (1). In recent years, there have been massive developments in creating platforms to profile and understand mechanisms of toxicity, thereby reducing the drug attrition rates and the use of animal testing. Modern toxicology emphasizes the three Rs (replacement, reduction, and refinement) of animals in toxicology testing and great efforts have been made towards the advancement of these principles across the world. The integration of profiles from different domains (i.e. toxicology and genomics) provides a more powerful systematic approach to uncovering the effect of toxicants in the population. Several high-profile programs such as the Framework Programme 7 (FP7), Horizon 2020, Tox21, ToxCast and initiatives by major government agencies have been actively promoting in silico and toxicogenomic approaches for the past few decades (2). Toxicogenomics is a sub-discipline of toxicology that involves generation and interpretation of several ‘omics’ platforms such as transcriptomics, proteomics, and metabolomics, to understand chemical-induced toxicity. Recently, toxicogenomics has been considered to play a significant role in both predictive and mechanism-based toxicology in an effort to identify candidate chemical compounds with specific toxicological profiles, in a more efficient and economical way. In the past decades, the dearth of toxicogenomic data has been compensated by large-scale, systematic initiatives such as Open TG-GATEs (Toxicogenomics Project-Genomics Assisted Toxicity Evaluation System) (3) and DrugMatrix (4). Open TG-GATEs was developed to incorporate the efforts of The Toxicogenomics Project (TGP), a joint government-private sector project organized by the National Institute of Biomedical Innovation, National Institute of Health Sciences and 15 pharmaceutical companies. The project has been completed in two phases, TGP1 and TGP2, collectively profiling over 170 compounds in primary human hepatocytes (in vitro) and rat kidney and liver organs (in vivo). DrugMatrix was originally introduced as a commercial database in 2006 and transferred into the public domain in 2011, with more than 200 compounds tested in vivo in rat tissues such as liver and 125 compounds in the in vitro rat hepatocytes (5). In addition to these large-scale datasets, the Comparative Toxicogenomics Database (6), a public resource for vast toxicogenomic information, provides a triad of chemical-gene, chemical–disease, and gene–disease relationship information helpful in assessing how environmental toxins affect human health.
Many studies have demonstrated that the changes in gene expression patterns perturbed with toxicants are associated with the toxic endpoints and aid in the prediction of expression-based biomarkers. In vitro assay systems, derived from animal or human tissues, have been proposed as alternative methodologies from animal testing. Examples of widely-used in vitro systems are primary human or rat hepatocytes, immortalized cell lines (e.g. HepaRG and HepG2), 3D culture systems, and embryonic stem cells (7). Expression profiling of these systems has been employed to classify compounds and predict their mode of action, for toxicity assessment. Genomic analysis of carcinogens unveiled pathways associated with genotoxicity, such as the immune response, apoptosis, and cell cycle, whereas signaling transduction and protein phosphorylation pathways were modulated mainly for non-genotoxic carcinogens (8). The lack of reproducibility of results in toxicology has long been a concern in the drug discovery process (9,10). Several platforms have been developed to address this issue in a data-driven manner (11). Inter-laboratory comparisons investigating the variability induced by the heterogeneity of experimental and data analysis protocols have been reported (12). Inter- and intra-laboratory reproducibility study of in vitro toxicogenomics show promising results with consistent replication of the results (12). Reproducibility of key pathways involved in response to chemical stress across species has also been studied (13). Even though there has been a spike in the use of genomic data for toxicity prediction, data availability by itself is not promising. The disparate nature of these datasets and high-dimensional gene expression profiles hinder integrative and reproducible analysis of the data. Several roadblocks such as differences in experimental design, use of ambiguous compound and gene annotations, lack of unified statistical methods limit fitness of the data. Considerable efforts are therefore required for homogenous pre-processing of the data before meaningful analysis could commence.
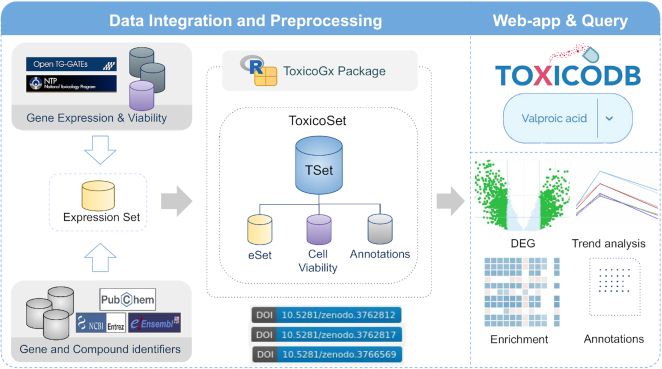
To address these issues, we have developed ToxicoDB, a web-application integrating three large in vitro toxicogenomic datasets. ToxicoDB offers an intuitive interface to explore these datasets by providing curated compound annotations, human and rat gene identifiers, and visualization of time- and dose-dependent chemical effects on genes. These datasets contain 6597 experiments for 231 chemical compounds. ToxicoDB users can view, analyze, and download differential gene expression and visualize enriched pathways for compounds of interest. Additionally, users can download normalized gene expression values along with experimental metadata for user-defined analysis. Here, we describe the content, pre-processing and web-interface (Figure 1).
Figure 1.
Schematic overview of ToxicoDB. Molecular profiles, viability assays and metadata have been integrated into a ToxicoSet (TSet) via the R ToxicoGx package and the ToxicoSet is subsequently used as the data source for ToxicoDB. As an example, users can query ‘valproic acid’ and ToxicoDB provides detailed information including compound annotations and analysis results such as differential expressed genes (DEGs), compound-gene trends over time, and associated pathways.
METHODS
Toxicogenomic datasets and statistics
ToxicoDB incorporates large in vitro toxicogenomic datasets such as Open TG-GATEs and DrugMatrix. To date, we have curated three datasets: (i) Primary human hepatocyte from Open TG-GATEs (TGH); (ii) rat hepatocyte from Open TG-GATEs (TGR); (iii) DrugMatrix (DM). The microarray data in TGH and TGR were downloaded from the Life Science Database Archive (dbarchive.biosciencedbc.jp/en/open-tggates/download.html) along with metadata and viability measurements. Biological molecules and compounds for which there was an ambiguity in concentration were excluded from the curation. Currently, TGH and TGR datasets contain 146 and 140 compounds, respectively. For DM, raw files and metadata for 125 chemical compounds have been obtained from the diXa Data Warehouse (wwwdev.ebi.ac.uk/fg/dixa/), study ID DIXA-033, an initiative developing a single resource to collect data produced by toxicogenomics studies (14). Probes were mapped to unique genes using the latest Brainarray CDF (version 24) for human and rat. Robust Multi-array Average (RMA) from the R affy package (version 1.62.0) has been used for batch normalization (15). Genes were mapped to Ensembl gene IDs using the latest version (Ensembl Release 99). Entrez gene ID and other attributes were mapped using the R biomaRt package (version 2.40.5) for both human and rat (16). Differential gene expression for experiments at all concentrations and time points have been performed using the R Limma package (version 3.40.6) (17). The normalized data was integrated into ToxicoSets (TSet) using our R package ToxicoGx, and can also be downloaded directly from the ToxicoDB webserver along with metadata.
Integration of the toxicogenomics datasets in ToxicoGx
To provide a unified framework for easy downloading and analysis of toxicogenomic datasets, we developed an R ToxicoGx package (github.com/bhklab/ToxicoGx) wherein raw data was accessed and extensively curated in-house and integrated as a new R object called ToxicoSet (TSet; Supplementary Figure S1). A TSet efficiently stores molecular profiles (gene expression upon compound exposure), viability assays (cytotoxicity), and metadata. Cell viability values from Open TG-GATEs include two types of assays to evaluate cytotoxicity: lactate dehydrogenase assay and DNA content of cells. A TSet can be downloaded to access the molecular profiles of all experimental conditions, detailed curation objects for compounds, and cell viability measurements pertaining to a dataset. ToxicoGx provides a suite of functions for summarizing complex experiments and computing compound-gene associations that allow users to analyze the data conveniently.
Semi-automated curation and annotation of chemical compound identifiers
To maintain consistency and to maximize the overlap across datasets, we developed a semi-automated curation of compound names. Firstly, compound names have been checked for exact case-insensitive matches against already curated unique compound names from PubChem. Names that did not match in the first step were subjected to partial matching with compound synonyms obtained from PubChem (18) or DrugBank (19). For the remaining unmapped compounds, SMILES, InChIKeys, or PubChem identifiers were used for mapping to human-readable names. Annotations of 58 compounds based on in vitro and in vivo genotoxic (GTX) and non-genotoxic (NGTX) carcinogenicity have been obtained from OpenRiskNet (20) (Supplementary Table S1).
Gene-set enrichment analysis and gene-sets
To analyze pathways associated with compound-induced gene expression changes, Gene-Set Enrichment Analysis (GSEA) was performed using the runGSA function of the R Piano package (version 2.0.2) (21). Biological Process (BP) Gene Ontology (GO) terms and Reactome (C2) pathways were downloaded using the R msigdbr package (version 7.0.1) to maintain consistency between the human and rat GMT files (22). Pathways associated with toxicity have been downloaded from Comparative Toxicogenomics Database (CTD) for Reactome and KEGG (April 2020 release).
Cross-dataset dose level selection
The cross-dataset correlation for 45 common compounds between TGR and DM were analyzed using the ToxicoGx package (version 1.0.0) in R (version 3.6.1). The drugPerturbationSig function was used to model compound signatures. This function returns the estimated linear model coefficient for the effect of compound concentration on gene expression and associated statistics (t-statistics and significance). Both TGR and DM provided gene expression measurements at three dose levels (low, middle, high), and reported the associated concentrations used. Compound signatures were estimated for all three dose levels. The Spearman correlations for the signatures of the same compounds across the two datasets were computed for all possible pairs of dose levels computed, and are reported in Supplementary Table S2. For subsequent analysis, the dose level for each compound was chosen such that it minimized the absolute concentration difference between the two studies. Compound signatures were filtered for genes with a significant false discovery rate (FDR < 0.05) in at least one compound in one or both datasets, and Spearman correlations between all compound signatures in the two datasets were computed. The effect of the absolute value of the concentration difference on the strength of compound signature correlation between the two datasets was assessed in two different ways: the Spearman rank correlation test was done between exact absolute values of concentration differences and the correlation of the same compounds across the two studies; and the absolute values of concentration differences were categorized into three categories: x ≤ 20 μM, 20 μM < x ≤ 1000 μM, x > 1000 μM, with a Kruskal–Wallis rank sum test applied to detect differences in location of the same compound correlation distributions across the three categories (Supplementary Figure S3).
WEB DEVELOPMENT AND INFRASTRUCTURE
Web implementation and API
The application consists of three layers: the front-end/client layer, the backend/server layer and the database layer. The client layer implementation in React (version 16.11.0) guarantees fast rendering and high performance, while data is being organized and visualized in the form of plots and tables. ToxicoDB tables allow users to download data in CSV format as spreadsheets. Rendering of the plots is done with d3.js (version 5) and ReactPlotlyJS (version 2.4.0), which are both JavaScript libraries for interactive and dynamic visualization built on HTML, CSS, and SVG. The backend/server layer is built using Node (version 10.16.0) and Express (version 4.17.1) with Representational State Transfer (RESTful) architecture. Knex.js (version 0.20.1) has been used as a SQL query builder to interact with the database layer of the app. The database layer is implemented as a relational database in MySQL with InnoDB storage engine (version 5.7). The schema for the ToxicoDB database is provided in Supplementary Figure S2.
All components of the web application are hosted on Microsoft Azure cloud infrastructure that provides PaaS (Platform as a Service) solutions to simplify application management and ensures an increased level of security, performance and flexibility. The web application leverages two Azure PaaS server resources. The Node server is deployed under Azure Web App Service, while the database server is using Azure Database for MySQL Server Service solution to support large batch queries against multiple tables, ACID (atomicity, consistency, isolation, durability) compliance and transactional support.
ToxicoDB provides a RESTful API (Application Programming Interface) that users may use directly to query the database and receive data in JSON format without using a web app interface (Supplementary Data). This provides a lot of flexibility for other developers to programmatically retrieve and use the most recent version of the data available in ToxicoDB and integrate it into their own software or automated solutions.
Toxicogenomic data availability and data access
The curated datasets are available as ToxicoSet (TSet) objects on Zenodo: Open TG-GATEs human (DOI: 10.5281/zenodo.3762812), Open TG-GATEs rat (DOI: 10.5281/zenodo.3762817), and DrugMatrix (DOI: 10.5281/zenodo.3766569).
Documentation and open-source code
The ToxicoDB code is open-source and publicly available on GitHub (github.com/bhklab/ToxicoDB-web). The ToxicoDB web application is documented with examples of use cases and synonym-based search, chemical compounds, genes, datasets, and pathways summary pages available. It also details the annotation and analysis pages for individual chemical compounds, genes, and datasets, as well as gene expression visualization pages with explanations of how to interpret the data. The roadmap for additional datasets in the pipeline is listed in the documentation. The documentation can be found at http://toxicodb.ca/documentation/.
WEB-INTERFACE AND ANALYSIS
ToxicoDB search
The main way to interact with ToxicoDB is through its search interface. The search bar found on the homepage of the website, allows users to query the data contained in the database, and functions as the main navigation tool around the web app. All chemical compounds, genes, datasets and pathway analysis can be accessed by clicking on the respective links on the top right corner of the front page. For simplicity, we implemented an intuitive search interface to query a compound or gene of interest. A plain search of a gene or compound name (or synonyms) shows both human and rat data. A pairwise compound-gene query can be used when users seek to visualize the dataset-specific effect of a compound on the gene implicated in a toxic response. The search bar is augmented with auto-completion which lets users know of the existence of that entity in the database. The pathways enriched by compounds of interest can be queried separately in the Pathways page of the web-application.
Analysis of differential gene expression induced by acetaminophen in primary human hepatocytes
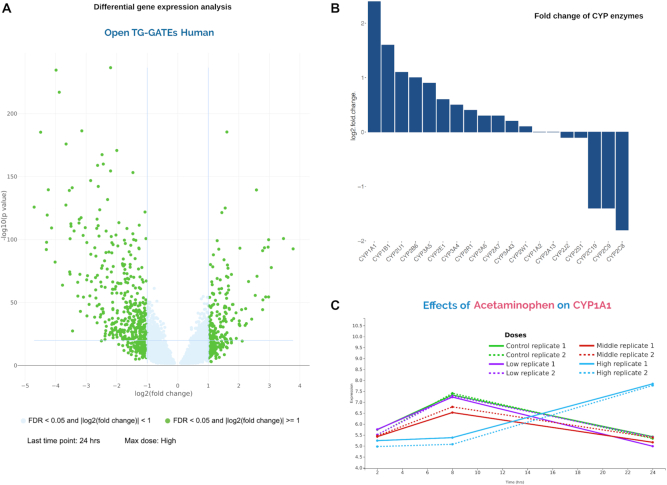
For this study, acetaminophen, a widely used analgesic antipyretic agent, which is also classified as ‘Most-DILI (Drug-Induced-Liver-Injury)-Concern’ drug by FDA, was queried in the search bar or selected from the list of all chemical compounds which directs to the compound page. This page starts with annotations of the compound such as PubChem CID linking to the database, SMILES, InChIKeys, carcinogenicity classification, and synonyms used across datasets. The top differentially expressed genes computed using limma are presented using a volcano plot for all datasets (Figure 2A). The significant genes are highlighted in green where the absolute value of the log2-fold-change is >1 and the FDR is <0.05. The name of the significant genes is visible upon hovering over the clickable dots. The same data is presented in a tabular, downloadable format for user-defined analysis. For this study, we sought to analyze the effect of acetaminophen (data downloaded for the highest dose and 24 h time point) on the Cytochrome P450 (CYP) enzymes in the TGH dataset. All other doses and time combinations are available for the user for visualization as well as analysis. CYP is a superfamily of heme-proteins that carry out oxidative metabolism of many endogenous and foreign compounds (23). Studies have implicated the effect of CYP enzymes on bioactivation of acetaminophen (24,25) We observed that acetaminophen downregulates the expression of certain CYP enzymes such as CYP2C8, CYP2C19 compared to CYP1A1 and CYP1B1 (Figure 2B). The Search interface also provides a line plot to visualize the time- and dose-dependent effect of the acetaminophen on genes of interest (Figure 2C).
Figure 2.
Query for ‘acetaminophen’. (A) Volcano plot displays differentially expressed genes for acetaminophen where significant genes are highlighted in green in TGH. (B) Bar plot shows a log2-fold-change in CYP 450 genes for acetaminophen. (C) The line plot shows the effect of time and dose of acetaminophen on CYP1A1 in TGH.
Effect of DNA synthesis inhibitors on toxicity-related pathways
The pathways section of ToxicoDB provides access to pathway enrichment analysis of the toxicity signatures for each compound, computed as described in the corresponding methods section. This page has four subquery sections enabling the selection of a dataset, chemical compounds of choice, ontologies such as GO and Reactome (MSigDB) as well as Comparative Toxicogenomics Database (CTD), and finally a list of pathway names. In order to analyze the effect of certain DNA synthesis inhibitors on compound metabolism, cellular stress, and cell cycle, compounds from TGH were selected to query in the Pathways section of ToxicoDB. Azathioprine, Colchicine, Cyclophosphamide, Doxorubicin, Etoposide and N-methyl-N-nitrosourea were queried for MSigDB Reactome pathways (Figure 3). The pathways related to cell cycle and DNA replication were downregulated whereas pathways associated with cytochrome P450, cellular response to stress, and metabolism of xenobiotic compounds were upregulated with exceptions to Cyclophosphamide. This trend aligns with the studies discussing xenobiotic-metabolizing enzymes and their role in activation and detoxification of chemicals (26) as well as oxidative stress induced by chemical compounds (27,28).
Figure 3.
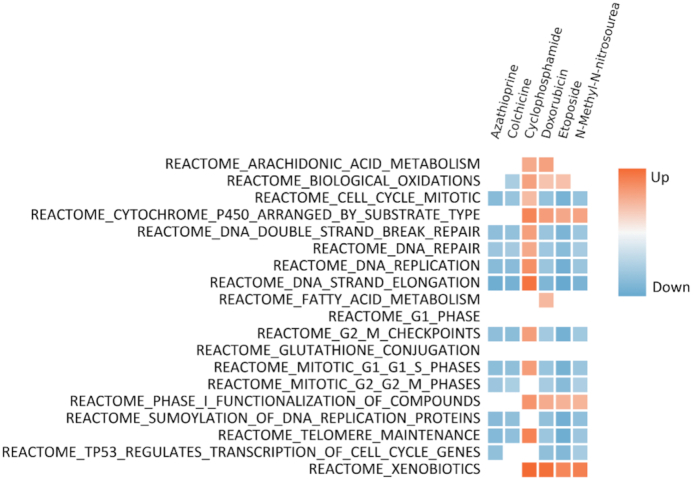
Heatmap of pathways associated with DNA synthesis inhibitors. Pathways are shown as rows and chemical compounds as columns. FDR significant (<0.05) are highlighted in colors, white indicates no significant enrichment. Upregulated (red) blocks indicate chemical compound metabolism and downregulated (blue) indicate cell cycle-related pathways.
Cross-dataset correlation analysis of TGR and DM using ToxicoGx
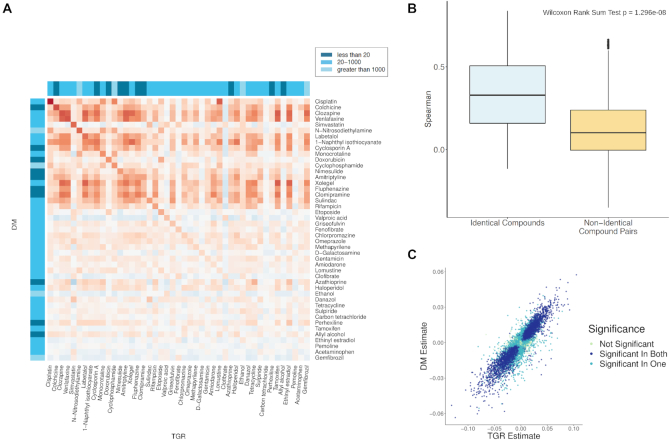
To check the consistency between TGR and DM rat primary hepatocyte data, we compared chemical-induced gene expression changes in both datasets (see METHODS). The union of genes significant in both studies was selected to avoid bias and was used for further analysis. To check if there are consistent signatures between the two datasets, we computed the rank-based correlation of the differentially expressed genes (coefficients estimated using ToxicoGx; Figure 4A) for the closest compound-specific doses between datasets. Among the top chemical compounds that were found to be correlated between the datasets, Cisplatin, a widely used chemotherapeutic agent (29), showed the highest correlation (Figure 4C) whereas Gemfibrozil, a peroxisome proliferator-activated receptor alpha (PPARA) inhibitor (30) was the least correlated. We further compared the correlation of chemically induced transcriptomic signatures between identical (well-correlated) and different compounds in both datasets. As expected, we found that identical compounds yield significantly higher correlations than pairs of different compounds (Wilcoxon rank-sum test, one-sided P-value < 1.296e–08; Figure 4B). We further assessed whether the low correlations of some identical compound signatures across the two datasets could be explained by differences in the compound concentrations used. We looked at the association between the absolute differences in concentration and the cross-dataset signature correlation in two ways. We first computed a rank-based correlation, finding a weak trend towards lower cross-dataset correlation at higher concentration differences (Spearman rho = −0.23, P = 0.13). We also categorized the concentration differences into ≤20 μM, 20–1000 μM and >1000 μM, but found no significant difference in cross-dataset correlations between these three groups (Kruskal−Wallis rank-sum test, P = 0.62). The results suggest that although differences in concentrations of compounds have a weak effect on the consistency of compound signatures between the two studies, further investigation is necessary to explain the variability observed. ToxicoDB allows researchers to directly access and explore data across datasets to answer such questions.
Figure 4.
Cross-dataset analysis between TGR and DM using Spearman correlation. (A) The compound-gene signature correlations for common compounds are shown in the heatmap. Red indicates the compound pairs that are similar (positive correlation), blue indicates a negative correlation, and intensity indicates strength. The three dose difference ranges are shown with labels (B) A comparison of the correlation between identical compounds in TGR and DM (light blue) vs non-identical pairs of compounds, with Spearman correlation between compound-gene signatures shown. This corresponds to the values on the diagonal and off-diagonal in (A) respectively. (C) An example of the compound-gene associations plotted for Cisplatin, the most correlated compound between the two datasets. The X- and Y-axis show linear model estimates from the drugPerturbationSig function in ToxicoGx, and points are colored by the significance of the association (FDR adjusted P-value < 0.05) in none, one or both of the datasets.
DISCUSSION AND FUTURE DIRECTIONS
The growing availability of large datasets, combining both genomic and toxicological profiles of mammalian cells, offers new opportunities to identify toxic compounds and the biological pathways associated with their toxicity. However, the disparate nature of these data hinders joint analysis of multiple toxicogenomic datasets. Collectively analyzing the gene expression changes exposed to chemical compounds at multiple time points and doses in replicates often present challenges. To address this issue, we have developed ToxicoDB, a web-application allowing users to easily mine large, highly curated toxicogenomic datasets. By using a unified nomenclature for annotations of chemical compounds and genomic features, ToxicoDB provides tidy, well-annotated data and analysis results to users through a convenient web-interface. Users who wish to conduct a more complex analysis of the toxicogenomic data can also use the companion ToxicoGx R package as a command-line tool.
Noteworthy efforts have been made by various groups to integrate and analyze toxicogenomic data. MoAviz (31) allows visualization of perturbed pathways using the data from Open TG-GATEs and DrugMatrix. It uses a metric Modified Jaccard Index (MJI) for the quantitative description of pathway similarity to evaluate the extent of association of gene expression changes with mode of action. The interface allows search of datasets and compounds and provides a quantitative description of genomic pathway similarity. Toxygates (32) was originally released as an interface to increase the accessibility of Open TG-GATEs. Currently, it provides an orthologous mode for data comparison among different species, interactive clustering, enrichment analysis, and user data uploading. Collaborative Toxicogenomics (CTox) (33) is an integrated web portal for gene expression analysis in safety studies wherein the results can be compared to Open TG-GATEs and DrugMatrix. Users can describe their experiments, upload the corresponding samples, evaluate their results using a variety of established and emerging systems biology analysis methods. LTmap (34) compares signatures of query compounds against pre-generated signatures from Open TG-GATEs.
While MoAviz, Toxygates, CTox and LTmap provide different platforms for dataset integration, ToxicoDB provides consistent standardized identifiers for compounds and genes, unlike the existing databases. The source package ToxicoGx coupled with ToxicoDB provides an excellent platform for power users who wish to further analyze the data. The feature comparison between databases is shown in Table 1.
Table 1.
Comparison of main functionalities between ToxicoDB and existing web-applications focusing on the query, visualization and analysis of toxicogenomic datasets.
| ToxicoDB | MoAviZ | Toxygates | LTmap | CTox | |
|---|---|---|---|---|---|
| Open source | ✓ | NA | ✓ | NA | ✓ |
| Model system (in vitro) | ✓ | ✓ | ✓ | ✓ | ✗ |
| Model system (in vivo) | ✗ | ✓ | ✓ | ✓ | ✓ |
| Use without login | ✓ | ✓ | ✓ | ✗ | ✗ |
| API | ✓ | ✗ | ✗ | ✗ | ✗ |
| Annotation of compounds and genes | ✓ | ✗ | ✗ | ✗ | ✗ |
ToxicoDB has potential limitations in terms of inclusion of model systems. It currently supports in vitro datasets for human and rat hepatocytes, but we plan to extend our data compendium to include in vivo toxicogenomic data to apply the current functionalities for integrative analyses and cross-study comparisons. We expect ToxicoDB to be of particular interest to machine learning researchers, as it provides normalized gene expression values that can be made use in modelling tasks such as toxicity signature identification. To our knowledge, ToxicoDB is unique in the way that it harmonizes heterogeneous data across in vitro toxicogenomic datasets, allowing users to easily query and summarize the associations between gene expression induced by potential toxicants.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Open TG-GATEs and DrugMatrix for the generation of valuable toxicogenomic data. We would like to thank diXa data warehouse for the hassle-free access of the data to the scientific community. Finally, we would like to thank OpenRiskNet for the support.
Contributor Information
Sisira Kadambat Nair, Princess Margaret Cancer Centre, University Health Network, Toronto, ON M5G 0A3, Canada.
Christopher Eeles, Princess Margaret Cancer Centre, University Health Network, Toronto, ON M5G 0A3, Canada.
Chantal Ho, Princess Margaret Cancer Centre, University Health Network, Toronto, ON M5G 0A3, Canada.
Gangesh Beri, Princess Margaret Cancer Centre, University Health Network, Toronto, ON M5G 0A3, Canada.
Esther Yoo, Princess Margaret Cancer Centre, University Health Network, Toronto, ON M5G 0A3, Canada.
Denis Tkachuk, Princess Margaret Cancer Centre, University Health Network, Toronto, ON M5G 0A3, Canada.
Amy Tang, Princess Margaret Cancer Centre, University Health Network, Toronto, ON M5G 0A3, Canada.
Parwaiz Nijrabi, Princess Margaret Cancer Centre, University Health Network, Toronto, ON M5G 0A3, Canada; Department of Medical Biophysics, University of Toronto, Toronto, ON M5G 1L7, Canada.
Petr Smirnov, Princess Margaret Cancer Centre, University Health Network, Toronto, ON M5G 0A3, Canada; Department of Medical Biophysics, University of Toronto, Toronto, ON M5G 1L7, Canada.
Heewon Seo, Princess Margaret Cancer Centre, University Health Network, Toronto, ON M5G 0A3, Canada.
Danyel Jennen, Department of Toxicogenomics, GROW School of Oncology and Development Biology, Maastricht University, Maastricht, The Netherlands.
Benjamin Haibe-Kains, Princess Margaret Cancer Centre, University Health Network, Toronto, ON M5G 0A3, Canada; Department of Medical Biophysics, University of Toronto, Toronto, ON M5G 1L7, Canada; Department of Computer Science, University of Toronto, Toronto, ON M5T 3A1, Canada; Ontario Institute for Cancer Research, Toronto, ON M5G 1L7, Canada; Vector Institute for Artificial Intelligence, Toronto, ON M5G 1L7, Canada.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Genome Canada [15414]; Edelweiss Connect [731075]. Funding for open access charges: Genome Canada [15414]; OpenRiskNet project funded by the European Commission within the Horizon 2020 EINFRA-22–2016 Programme [731075].
Conflict of interest statement. None declared.
REFERENCES
- 1. Guengerich FP. Mechanisms of drug toxicity and relevance to pharmaceutical development. Drug Metab. Pharmacokinet. 2011; 26:3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Liu Z., Huang R., Roberts R., Tong W.. Toxicogenomics: a 2020 Vision. Trends Pharmacol. Sci. 2019; 40:92–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Igarashi Y., Nakatsu N., Yamashita T., Ono A., Ohno Y., Urushidani T., Yamada H.. Open TG-GATEs: a large-scale toxicogenomics database. Nucleic Acids Res. 2015; 43:D921–D927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ganter B., Snyder R.D., Halbert D.N., Lee M.D.. Toxicogenomics in drug discovery and development: mechanistic analysis of compound/class-dependent effects using the DrugMatrix database. Pharmacogenomics. 2006; 7:1025–1044. [DOI] [PubMed] [Google Scholar]
- 5. Alexander-Dann B., Pruteanu L.L., Oerton E., Sharma N., Berindan-Neagoe I., Módos D., Bender A.. Developments in toxicogenomics: understanding and predicting compound-induced toxicity from gene expression data. Mol. Omics. 2018; 14:218–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mattingly C.J., Rosenstein M.C., Davis A.P., Colby G.T., Jr F.J.N., Boyer J.L.. The comparative toxicogenomics database: a cross-species resource for building chemical-gene interaction networks. Toxicol. Sci. 2006; 92:587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Doktorova T.Y., Yildirimman R., Vinken M., Vilardell M., Vanhaecke T., Gmuender H., Bort R., Brolen G., Holmgren G., Li R. et al.. Transcriptomic responses generated by hepatocarcinogens in a battery of liver-based in vitro models. Carcinogenesis. 2013; 34:1393–1402. [DOI] [PubMed] [Google Scholar]
- 8. Hochstenbach K., van Leeuwen D.M., Gottschalk R.W., Gmuender H., Stølevik S.B., Nygaard U.C., Løvik M., Granum B., Namork E., van Loveren H. et al.. Transcriptomic fingerprints in human peripheral blood mononuclear cells indicative of genotoxic and non-genotoxic carcinogenic exposure. Mutat. Res. 2012; 746:124–134. [DOI] [PubMed] [Google Scholar]
- 9. Poland C.A., Miller M.R., Duffin R., Cassee F.. The elephant in the room: reproducibility in toxicology. Part. Fibre Toxicol. 2014; 11:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Miller G.W. Improving reproducibility in toxicology. Toxicol. Sci. 2014; 139:1–3. [DOI] [PubMed] [Google Scholar]
- 11. Darde T.A., Gaudriault P., Beranger R., Lancien C., Caillarec-Joly A., Sallou O., Bonvallot N., Chevrier C., Mazaud-Guittot S., Jégou B. et al.. TOXsIgN: a cross-species repository for toxicogenomic signatures. Bioinformatics. 2018; 34:2116–2122. [DOI] [PubMed] [Google Scholar]
- 12. Herwig R., Gmuender H., Corvi R., Bloch K.M., Brandenburg A., Castell J., Ceelen L., Chesne C., Doktorova T.Y., Jennen D. et al.. Inter-laboratory study of human in vitro toxicogenomics-based tests as alternative methods for evaluating chemical carcinogenicity: a bioinformatics perspective. Arch. Toxicol. 2016; 90:2215–2229. [DOI] [PubMed] [Google Scholar]
- 13. El-Hachem N., Grossmann P., Blanchet-Cohen A., Bateman A.R., Bouchard N., Archambault J., Aerts H.J.W.L., Haibe-Kains B.. Characterization of Conserved Toxicogenomic Responses in Chemically Exposed Hepatocytes across Species and Platforms. Environ. Health Perspect. 2016; 124:313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hendrickx D.M., Aerts H.J.W.L., Caiment F., Clark D., Ebbels T.M.D., Evelo C.T., Gmuender H., Hebels D.G.A.J., Herwig R., Hescheler J. et al.. diXa: a data infrastructure for chemical safety assessment. Bioinformatics. 2015; 31:1505–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P.. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003; 4:249–264. [DOI] [PubMed] [Google Scholar]
- 16. Durinck S., Spellman P.T., Birney E., Huber W.. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009; 4:1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K.. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., Li Q., Shoemaker B.A., Thiessen P.A., Yu B. et al.. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 2019; 47:D1102–D1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wishart D.S., Knox C., Guo A.C., Cheng D., Shrivastava S., Tzur D., Gautam B., Hassanali M.. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008; 36:D901–D906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. OpenRiskNet - Risk Assessment e-Infrastructure https://openrisknet.org/.
- 21. Väremo L., Nielsen J., Nookaew I.. Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res. 2013; 41:4378–4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dolgalev I. MSigDB Gene Sets for Multiple Organisms in a Tidy Data Format [R package msigdbr version 7.0.1]. 2020; Comprehensive R Archive Network (CRAN)https://CRAN.R-project.org/package=msigdbr. [Google Scholar]
- 23. Danielson P.B. The cytochrome P450 superfamily: biochemistry, evolution and drug metabolism in humans. Curr. Drug Metab. 2002; 3:561–597. [DOI] [PubMed] [Google Scholar]
- 24. Laine J.E., Auriola S., Pasanen M., Juvonen R.O.. Acetaminophen bioactivation by human cytochrome P450 enzymes and animal microsomes. Xenobiotica. 2009; 39:11–21. [DOI] [PubMed] [Google Scholar]
- 25. Woolbright B.L., Jaeschke H.. Xenobiotic and endobiotic mediated interactions between the cytochrome P450 system and the inflammatory response in the liver. Adv. Pharmacol. 2015; 74:131–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Reed L., Arlt V.M., Phillips D.H.. The role of cytochrome P450 enzymes in carcinogen activation and detoxication: an in vivo-in vitro paradox. Carcinogenesis. 2018; 39:851–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yoon C.S., Kim H.K., Mishchenko N.P., Vasileva E.A., Fedoreyev S.A., Stonik V.A., Han J.. Spinochrome D attenuates doxorubicin-induced cardiomyocyte death via improving glutathione metabolism and attenuating oxidative stress. Mar. Drugs. 2018; 17:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sheweita S.A., El-Hosseiny L.S., Nashashibi M.A.. Protective effects of essential oils as natural antioxidants against hepatotoxicity induced by cyclophosphamide in mice. PLoS One. 2016; 11:e0165667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dasari S., Tchounwou P.B.. Cisplatin in cancer therapy: molecular mechanisms of action. Eur. J. Pharmacol. 2014; 740:364–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bossé Y., Pascot A., Dumont M., Brochu M., Prud’homme D., Bergeron J., Després J.-P., Vohl M.-C.. Influences of the PPARα-L162V polymorphism on plasma HDL2-cholesterol response of abdominally obese men treated with gemfibrozil. Genet Med. 2002; 4:311–315. [DOI] [PubMed] [Google Scholar]
- 31. McMullen P.D., Pendse S.N., Black M.B., Mansouri K., Haider S., Andersen M.E., Clewell R.A.. Addressing systematic inconsistencies between in vitro and in vivo transcriptomic mode of action signatures. Toxicol. In Vitro. 2019; 58:1–12. [DOI] [PubMed] [Google Scholar]
- 32. Nyström-Persson J., Natsume-Kitatani Y., Igarashi Y., Satoh D., Mizuguchi K.. Interactive toxicogenomics: gene set discovery, clustering and analysis in toxygates. Sci. Rep. 2017; 7:1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sutherland J.J., Stevens J.L., Johnson K., Elango N., Webster Y.W., Mills B.J., Robertson D.H.. A novel open access web portal for integrating mechanistic and toxicogenomic study results. Toxicol. Sci. 2019; 170:296–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xing L., Wu L., Liu Y., Ai N., Lu X., Fan X.. LTMap: a web server for assessing the potential liver toxicity by genome-wide transcriptional expression data. J. Appl. Toxicol. 2014; 34:805–809. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.