Abstract
To support rapid chemical toxicity assessment and mechanistic hypothesis generation, here we present an intuitive webtool allowing a user to identify target organs in the human body where a substance is estimated to be more likely to produce effects. This tool, called Tox21BodyMap, incorporates results of 9,270 chemicals tested in the United States federal Tox21 research consortium in 971 high-throughput screening (HTS) assays whose targets were mapped onto human organs using organ-specific gene expression data. Via Tox21BodyMap's interactive tools, users can visualize chemical target specificity by organ system, and implement different filtering criteria by changing gene expression thresholds and activity concentration parameters. Dynamic network representations, data tables, and plots with comprehensive activity summaries across all Tox21 HTS assay targets provide an overall picture of chemical bioactivity. Tox21BodyMap webserver is available at https://sandbox.ntp.niehs.nih.gov/bodymap/.
INTRODUCTION
Chemical hazard characterization in the twenty-first century has evolved to encompass large high-throughput screening (HTS) research programs, designed to produce quantitative data on the activity of thousands of chemicals across a multitude of biological targets and pathways, a strategy that has long been used in drug discovery. Such efforts, exemplified by the federal Tox21 research consortium (1,2) are intended to facilitate rapid chemical hazard screening, predictive computational toxicology using machine learning and artificial intelligence techniques, and human-relevant systems biology models that provide mechanistic insight into chemical toxicity (e.g. (3)). Currently the Tox21 program, including the U.S. EPA’s ToxCast component, has screened up to 10 000 chemicals across approximately a thousand assay targets, and testing is still ongoing.
Mapping data from biological assays onto organs in the human body is an important challenge in both targeted therapeutics, to specifically deliver a drug to an organ or a pathway, and in toxicology to understand and better predict chemical effects on specific organs. Different target organ mappings and knowledge organization systems have been developed for diverse sources of biological data; for example the human protein atlas has been used to map protein expression to cell types and organ pathology (4–6). As discussed in (7), with more than 37 trillion cells in the body, establishing the links between in vitro assay measurements at molecular and cellular resolution and the organ systems of the human body is a real challenge considering the diversity and cross-communication between individual body regions and cells.
To date, no efforts have been made to map the broad suite of Tox21/ToxCast assays to specific organs within the human body. It is made all the more complex when considering the heterogeneity of the assay platforms and technology types, i.e. some assays were designed to target expression of a specific gene, binding to a receptor, or activation of a particular molecular signalling pathway, while other assays have been developed to assess toxicological potential more broadly, for example liver cell viability or mitochondrial function. Such a mapping would allow for potentially more informative chemical toxicity prediction and mechanistic insight into potential target organ effects based on patterns of in vitro bioactivity.
In this study, we attempt to increase the utility of the Tox21/ToxCast dataset by creating a mapping of the molecular and cellular assay targets onto human organs and systems by combining organ-specific gene expression with an expert driven approach. Following this mapping, we built a webserver called Tox21BodyMap, providing an interactive and intuitive body mapping of the chemical-specific bioactivity patterns observed across the Tox21 dataset. Users input a chemical from the Tox21 library and adjust gene expression thresholds as well as AC50 (half-maximal activity concentrations) cutoffs to investigate organ specificity by chemical. Different data representations have been developed including interactive tables, activity summary plots, and dynamic networks, with downloadable outputs provided in table format.
MATERIALS AND METHODS
Tox21/ToxCast data
Tox21/ToxCast assay results were extracted from the EPA InVitroDB version 3.2 (10-2019) (https://www.epa.gov/chemical-research/exploring-toxcast-data-downloadable-data). The resource provides AC50s for ∼10 000 chemicals included in the Tox21 chemical library, tested in subsets of 1400 assays. All Tox21 chemicals were run in ∼100 assays in ultra HTS format, and out of these ∼2000 chemicals were tested in an additional ∼1200 assays as part of ToxCast, see (2) for details. Assays developed to measure technological interference and assays where it was not possible to establish a dominant human organ / gene mapping were removed, resulting in a final set of 970 assays for mapping. We excluded, for example, investigational assays and assays used to detect interference chemicals specific for one technology. The assay list is available in Supporting Information S1 with the associated organ mapping.
Gene expression by human system and organ
Gene expression data were extracted from the body atlas available in the platform Illumina Correlation Engine (version 12-2019) (http://www.nextbio.com/). For each gene, the expression level for each organ was extracted, as well as a median expression across all of the available organs. For each gene, we computed a ratio of organ-specific expression to that gene's median expression across all organs and mapped this information to each assay gene target. We defined a second ratio based on the organ-specific expression for a gene to the median expression of all genes in that organ. Supporting Information S2 contains the expression levels by gene for each human organ, as well as the list of organs used. Supporting S3 contains the median expression by organ.
Cell type and viability driven organ mapping
Some assays lacking specific gene targets were mapped using tissue/cell type based on input from experts familiar with each specific assay platform and technology type. Viability assays were mapped on immune system, liver, lung and stomach organs. The reason behind this choice is most chemicals that were active in these assays were reactive, and would therefore plausibly be cytotoxic at point of contact (e.g. stomach, lung) or first pass (liver), or if they made it into systemic circulation, they would target rapidly dividing cells (e.g. immune system/bone marrow).
Webserver development
The webserver was developed using Django in python 3.6.8 on a Centos7 virtual machine and used JavaScript for network representation and body mapping using agGrid (https://www.ag-grid.com/), vis.js (https://visjs.org/) and smiles-drawer (8) libraries. The webserver is connected to a PostgreSQL database including assay results by chemical and gene expression levels mapped onto organs and assays.
RESULTS
Assays mapping on human organ
971 assays have been mapped on 33 organs based on three types of mapping, i.e. gene target, viability assays, and cell/tissue type-based mapping. Most of the assays (521) were mapped using their specific gene target, an additional 329 were mapped using the cell/tissue-type mapping, and the remaining 121 assays were mapped using the viability target (stomach, liver, immune system and lung); see Supplemental Figure S1 for details. In Supplemental Tables S1–S3 we report the count of assays by technology, tissue and cell-type used. In term of technology platform, fluorescence and luminescence were the dominant technologies in the Tox21/ToxCast database. In terms of cell and tissue-type, liver was the most represented organ with many assays using HepG2 cells and hepatocytes (see Supplemental Figures S2, S3, and Supplemental Table S2).
Ratio of gene expression
We computed an organ-specific gene expression ratio based on two methods: the median expression by gene, i.e. dividing a gene's expression in one specific organ by the median expression level of that gene across all organs, or the median expression by organ, i.e. dividing a gene's expression in one specific organ by the median expression of all genes in that organ. Tox21 BodyMap webtool users can choose which ratio to use and apply different cutoffs on this ratio to examine the estimated chemical specificity by organ. Supporting information Figures S2 and S3 demonstrate the count of assays mapped for different expression cutoffs. First, we were able to identify organs with a lack of assays mapped, e.g. the pancreas, oesophagus, bladder or eye, and inversely organs which represent the majority of mapped assays, e.g. liver, immune system, stomach and lung. This overrepresentation was explained in part by the mapping schema used for viability assays, but also is a product of these assays being specifically designed for toxicity screening, hence the reliance on the liver. By comparing the two gene expression ratio methods, the mapping based on a ratio using the median expression by organ allowed for mapping more assays, especially when applying a higher cutoff of expression (indicating higher specificity for that gene target-organ combination).
Workflow
As an input, Tox21 BodyMap users choose a particular chemical from the Tox21 10k chemical library using a CASRN (Chemical Abstracts Service Registry Number) or a generic name. The chemical identifier search is indexed and the user selects by name or CASRN from a drop down list. The chemical name selection tool is alphabetically organized; the user can also begin typing the chemical name of interest and the tool will focus on the appropriate section of the chemical list. The ratio and associated cutoff applied by the user can be based on the median expression either by gene or by organ (see methods). Once a chemical is selected, the results page is divided into four sections: (i) chemical information, (ii) the body mapping, (iii) interactive results matrix and (iv) a network representation.
Chemical information section
In this section chemical structure is represented in 2-dimensional layout and links are provided to the chemical-specific page on the Comptox Chemicals Dashboard (https://comptox.epa.gov/dashboard) and chemmaps.com (https://sandbox.ntp.niehs.nih.gov/chemmaps/). Via these linked resources, users are provided with detailed information on the chemical and can visualize the structure within the chemical universe, respectively.
Body mapping section
The Tox21/ToxCast bioactivity profile of the selected chemical is mapped onto a picture of the human body containing 33 organs. The organs that are most likely to be affected based on the chemical bioactivity profile are highlighted. Using the dynamic sliders, users can change both the AC50 cutoff, range 1 nM–300 μM, and the gene expression threshold, range 2–50-fold of control expression (by gene or by organ, depending on ratio method chosen, see Materials and Methods). Adjusting the AC50 cutoff allows users to distinguish potent activity (low AC50 values) from activity occurring at higher testing concentrations. The gene expression threshold represents the ratio of organ-specific expression for a particular assay gene target vs expression of that target across all organs in the body (by gene) or vs expression of all genes within that particular organ (by organ), where a higher threshold equates to higher relative expression level of the gene in that organ.
Assay results section
Assay results are represented via a summary dotplot and an interactive table. The user can hover over each dot to see the assay name. Assays are divided into organ systems based on their target genes/organs or by their cell types (see Methods). The table contains information on all the assays where the chemical displayed activity, the assay gene targets (where applicable), the expression levels of those targets in each organ, and the mapping of each assay to the organ and organ system. Users can download the full table by chemical used to generate the representation.
Network representation
This section includes a dynamic network where all the mapping elements are projected, i.e. chemical, assays and AC50, gene mapping and gene expression as well as organ and system. Each node is colored based on the type of feature represented, red for chemical, pink for assay, green for gene, blue for organ and yellow for organ system. The fold of expression between assay target and organ is shown on edges between these node types. Edges between chemicals and assays are labeled with AC50 values. The network plots are automatically generated based on the activity of that particular chemical across all the assays tested in the Tox21 program, and their respective target-organ mapping, after applying user-selected cutoffs for potency (AC50) and for gene expression by organ (specificity ratio).
Case Study
Chemical bioactivity target mapping
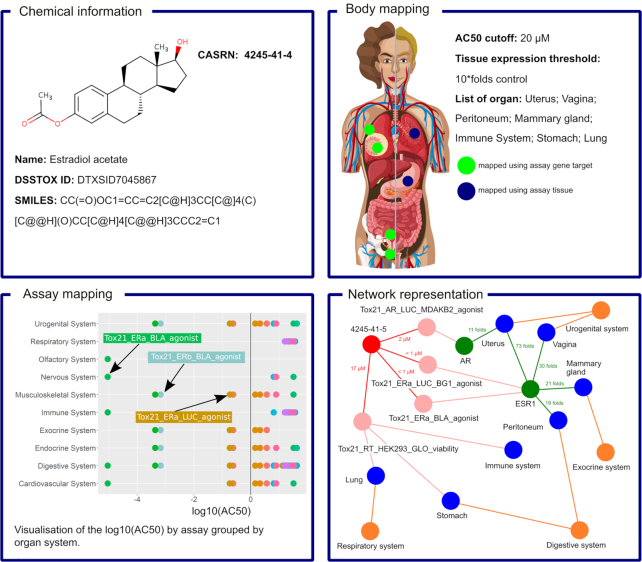
Estradiol acetate (CASRN: 4245–41-4) was chosen as an example to demonstrate the functionality of Tox21BodyMap, shown in Figure 1. It is an endogenous estrogen hormone produced in the human ovaries and generated in other tissues through aromatizing testosterone, with an important role in the regulation of the female reproductive cycle and a wide range of other hormone-driven biological functions, e.g. immune signalling and cardiovascular function and brain development (9,10). Exogenously, estradiol is used as a drug to treat menopause symptoms, and links between estradiol exposure and breast cancer, cardiovascular effects, and reproductive dysfunction are well established (11,12). It is also classified as a reference endocrine disrupting compound (13). As part of the Tox21 chemical library, estradiol acetate was tested across all ToxCast and Tox21 assays included in the HTS program.
Figure 1.
Tox21BodyMap visualisation of the activity of Estradiol acetate, generated by setting the in vitro activity concentration (AC50) cutoff at 20 μM and the organ-specific gene target expression threshold at 10 * fold above control. Three different mappings are shown: on the human body, as an activity barplot by organ system, and using a network that combines gene expression, assay results and organ mapping.
The chemical-specific results page of Tox21 BodyMap is divided into several sections, as described above. Under the chemical information section, the user can click on the DSSTOX ID or CASRN to visit the page of the U.S. EPA Chemicals Dashboard specific to estradiol acetate, to find detailed information and data on its physicochemical properties, presence in regulatory lists, environmental fate and transport, exposure, hazard characterization based on traditional animal toxicology studies, similar compounds, and much more. Within the body mapping section, the dynamic sliders allow the user to adjust the potency threshold or the threshold for gene target specificity within a particular organ. Applying an initial potency threshold of AC50 < 1 μM, without requiring significant specificity of gene expression within particular organs (organ expression threshold of 2-fold) demonstrates that estradiol is active at sub-micromolar concentrations in numerous endocrine related assays (e.g. ER, AR, RAR), whose targets are expressed across a wide range of organs. As the user increases the gene-target organ expression threshold to 20-fold, the uterus, vagina, and mammary gland are the only organs that are still identified as specific targets of estradiol, consistent with their role in endogenous estrogen-sensing and signaling. Figure 1 demonstrates the visualizations produced using cutoffs of AC50 < 20 μM and setting the organ-specific gene target expression ratio threshold at 10-fold above control. At these thresholds, we identified that most of estradiol's predicted effects were reported in the female urogenital system, vagina, uterus, and breast tissue, based on potent activity against multiple assays targeting ER and AR. Consistent with effects reported in the literature (9), we also identified potential effects in the lung, in the digestive system, and in the immune system, based on the most sensitive HTS assay targets and their organ-specific gene expression.
DISCUSSION
This work describes a unique scientific resource that facilitates the use of Tox21/ToxCast assay data for the identification and mechanistic linkage of chemicals to potential target organ effects in humans. Tox21BodyMap provides a variety of visualizations that help the user understand how target organs are linked to molecular/cellular targets. It is important to note that this work is biased by the coverage of the assays in the Tox21/ToxCast database. For example, due to their application in toxicity screening, more assays were developed to target the liver, a frequent target identified in regulatory toxicity studies, and less on other organs as identified in results. Second, in this initial work we relied upon the Illumina body atlas to develop assay target mapping onto only 33 organs which yielded the best match with the set of assays selected from the Tox21 database. Future iterations of the tool will explore alternative resources for target expression including resources that cover both protein (e.g, Human Protein Atlas) and RNA expression (e.g. GTEX). This work could be expanded by breaking down the organ classification into tissue type and taking into consideration key sex differences, outside of sex-specific organs, that would influence chemical effects. Finally, it is important to note that the mapping does not consider any ADME (absorption, distribution, metabolism, and excretion) chemical properties. The body mapping reflects the potential effect of the chemicals on a specific pathway in a specific organ which can be used, for example, to design cell-based assays but does not reflect distribution of the chemical in a whole organism. We envision diverse potential uses of the Tox21BodyMap, including hypothesis generation, chemical prioritization, mechanistic analyses, data gap identification, and in vivo study design. Ultimately, this webtool provides an easy and novel way to explore and visualize the Tox21/ToxCast data.
DATA AVAILABILITY
The Body Atlas is a part of Illumina Correlation Engine available on at http://www.nextbio.com/. The EPA InVitroDB version 3.2 is available at https://www.epa.gov/chemical-research/exploring-toxcast-data-downloadable-data. All of the information presented in this server can be downloaded in a table format. Scripts used to build this analysis are available in the GitHub platform (https://github.com/ABorrel/Tox21BodyMap). Tox21BodyMap (https://sandbox.ntp.niehs.nih.gov/bodymap/) is free and open to all users and there is no login requirement.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Paul Windsor from the NIEHS Office of Communications and Public Liaison for the human body picture conception and the NIEHS Office of Data Sciences for their help and support to set up the webserver. The work presented here does not represent the official position of any US government agency.
Contributor Information
Alexandre Borrel, NIH/NIEHS/DIR/BCBB, RTP, NC 27709, USA.
Scott S Auerbach, NIH/NIEHS/DNTP/BSB, RTP, NC 27709, USA.
Keith A Houck, EPA/ORD/CCTE, RTP, NC 27709, USA.
Nicole C Kleinstreuer, NIH/NIEHS/DIR/BCBB, RTP, NC 27709, USA; NIH/NIEHS/DNTP/NICEATM, RTP, NC 27709, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: National Institute of Environmental Health Science.
Conflict of interest statement. None declared.
REFERENCES
- 1. Collins F.S., Gray G.M., Bucher J.R.. Transforming environmental health protection. Science. 2008; 319:906–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Thomas R. The US Federal Tox21 Program: a strategic and operational plan for continued leadership. ALTEX. 2018; 35:163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kleinstreuer N.C., Yang J., Berg E.L., Knudsen T.B., Richard A.M., Martin M.T., Reif D.M., Judson R.S., Polokoff M., Dix D.J. et al.. Phenotypic screening of the ToxCast chemical library to classify toxic and therapeutic mechanisms. Nat. Biotechnol. 2014; 32:583–591. [DOI] [PubMed] [Google Scholar]
- 4. Thul P.J., Åkesson L., Wiking M., Mahdessian D., Geladaki A., Ait Blal H., Alm T., Asplund A., Björk L., Breckels L.M. et al.. A subcellular map of the human proteome. Science. 2017; 356:eaal3321. [DOI] [PubMed] [Google Scholar]
- 5. Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjostedt E., Asplund A. et al.. Tissue-based map of the human proteome. Science. 2015; 347:1260419. [DOI] [PubMed] [Google Scholar]
- 6. Uhlen M., Zhang C., Lee S., Sjöstedt E., Fagerberg L., Bidkhori G., Benfeitas R., Arif M., Liu Z., Edfors F. et al.. A pathology atlas of the human cancer transcriptome. Science. 2017; 357:eaan2507. [DOI] [PubMed] [Google Scholar]
- 7. Roy A.L., Conroy R.S.. Toward mapping the human body at a cellular resolution. Mol. Biol. Cell. 2018; 29:1779–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Probst D., Reymond J.-L.. SmilesDrawer: Parsing and drawing SMILES-Encoded molecular structures using Client-Side JavaScript. J. Chem. Inf. Model. 2018; 58:1–7. [DOI] [PubMed] [Google Scholar]
- 9. Hariri L., Rehman A.. Estradiol. 2020; http://www.ncbi.nlm.nih.gov/pubmed/31747204. [PubMed] [Google Scholar]
- 10. McCarthy M.M. Estradiol and the developing brain. Physiol. Rev. 2008; 88:91–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hill D.A., Crider M., Hill S.R.. Hormone therapy and other treatments for symptoms of menopause. Am. Fam. Physician. 2016; 94:884–889. [PubMed] [Google Scholar]
- 12. Samavat H., Kurzer M.S.. Estrogen metabolism and breast cancer. Cancer Lett. 2015; 356:231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mansouri K., Kleinstreuer N., Abdelaziz A.M., Alberga D., Alves V.M., Andersson P.L., Andrade C.H., Bai F., Balabin I., Ballabio D. et al.. CoMPARA: Collaborative modeling project for androgen receptor activity. Environ. Health Perspect. 2020; 128:27002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Body Atlas is a part of Illumina Correlation Engine available on at http://www.nextbio.com/. The EPA InVitroDB version 3.2 is available at https://www.epa.gov/chemical-research/exploring-toxcast-data-downloadable-data. All of the information presented in this server can be downloaded in a table format. Scripts used to build this analysis are available in the GitHub platform (https://github.com/ABorrel/Tox21BodyMap). Tox21BodyMap (https://sandbox.ntp.niehs.nih.gov/bodymap/) is free and open to all users and there is no login requirement.