Abstract
Background
Recently, radiomics has emerged as a non-invasive, imaging-based tissue characterization method in multiple cancer types. One limitation for robust and reproducible analysis lies in the inter-reader variability of the tumor annotations, which can potentially cause differences in the extracted feature sets and results. In this study, the diagnostic potential of a rapid and clinically feasible VOI (Volume of Interest)-based approach to radiomics is investigated to assess MR-derived parameters for predicting molecular subtype, hormonal receptor status, Ki67- and HER2-Expression, metastasis of lymph nodes and lymph vessel involvement as well as grading in patients with breast cancer.
Methods
A total of 98 treatment-naïve patients (mean 59.7 years, range 28.0–89.4) with BI-RADS 5 and 6 lesions who underwent a dedicated breast MRI prior to therapy were retrospectively included in this study. The imaging protocol comprised dynamic contrast-enhanced T1-weighted imaging and T2-weighted imaging. Tumor annotations were obtained by drawing VOIs around the primary tumor lesions followed by thresholding. From each segmentation, 13.118 quantitative imaging features were extracted and analyzed with machine learning methods. Validation was performed by 5-fold cross-validation with 25 repeats.
Results
Predictions for molecular subtypes obtained AUCs of 0.75 (HER2-enriched), 0.73 (triple-negative), 0.65 (luminal A) and 0.69 (luminal B). Differentiating subtypes from one another was highest for HER2-enriched vs triple-negative (AUC 0.97), followed by luminal B vs triple-negative (0.86). Receptor status predictions for Estrogen Receptor (ER), Progesterone Receptor (PR) and Hormone receptor positivity yielded AUCs of 0.67, 0.69 and 0.69, while Ki67 and HER2 Expressions achieved 0.81 and 0.62. Involvement of the lymph vessels could be predicted with an AUC of 0.8, while lymph node metastasis yielded an AUC of 0.71. Models for grading performed similar with an AUC of 0.71 for Elston-Ellis grading and 0.74 for the histological grading.
Conclusion
Our preliminary results of a rapid approach to VOI-based tumor-annotations for radiomics provides comparable results to current publications with the perks of clinical suitability, enabling a comprehensive non-invasive platform for breast tumor decoding and phenotyping.
Background
Breast cancer is one of the most common causes of cancer-related death in women worldwide. The prognosis depends on the time of first diagnosis and treatment, which in turn strongly depends on the molecular and hormonal features of the tumor. In clinical routine, these features are assessed by invasive tissue sampling and histopathological analysis. Over the past few years, ‘radiomics’ or ‘radiogenomics’ has demonstrated its potential for non-invasive, imaging-based tissue characterization in multiple cancer types [1–4]. As a rapidly emerging field, radiomics is based on the extraction of numerous quantitative features from medical images to assess relationships between features and the underlying pathophysiology using machine learning methods for analysis [5]. While radiomics-based analysis for prediction of the molecular status has been shown highly successful for brain cancers and prostate cancer [6–8], results from previously published studies on breast cancer show promising, yet divergent results [9,10]. One limitation for robust and reproducible radiomics analysis lies in the impact of widely-divergent inter-reader variability in annotating the tumors [11]. To date most radiomics studies require an either fully manual or semi-automatic tumor segmentation by a radiologist to enable the feature extraction process [10,12–16], resulting in potentially strongly divergent differences of the tumor annotations. These differences have been shown to cause significant variations in the feature sets extracted from the annotated tumor images and the consequential results thereof.
Therefore, the aim of this study was to investigate whether the utilization of a rapid and reproducible approach of VOI-based semi-automatic radiomics can facilitate the prediction of the molecular subtype, hormonal receptor status, Ki67- and HER2-Expression, involvement of lymph nodes and lymph vessel involvement as well as grading in patients with breast cancer.
Materials and methods
Patients
Ethical approval for this retrospective study was granted by the local ethics committee. Informed consent was waived. All analyses were performed based on anonymized data. After a prospective database research a total of 114 patients with histopathological confirmation of breast cancer (BIRADS 5 or 6) between August 2014 and March 2018, who fulfilled the following criteria were included in this study: (1) dedicated breast MRI prior treatment, (2) breast MRI performed exclusively on one dedicated 1.5 Tesla MR scanner to reduce potential influence of varying field strengths or imaging parameters on texture analysis, (3) lesions >/ = 1cm on MRI to decrease potential partial volume effects on radiomic analysis [17], (4) 18 years or older, (5) neither pregnant nor breastfeeding. 16 of these 114 patients with incomplete MR sequences were excluded from this study, resulting in a total of 98 patients who were included in the analysis.
Data acquisition
Breast MR scans were performed on a 1.5 Tesla MR system (Aera, Siemens Healthcare, Erlangen) in prone position utilizing a dedicated 16-channel breast radiofrequency (RF) coil (Siemens Healthcare, Erlangen).
The study protocol comprised the following sequences:
A transversal 2-dimensional turbo-spin echo sequence (0.7 x 0.7 mm voxel size, 2.0 mm slice thickness, gap 0 mm, TR 6100 ms, TE 52 ms, FOV 340 x 340 mm2, flip angle 150 degrees, matrix size 256x256, 88 slices)
Six repetitions of a non-fatsaturated transversal 3-dimensional fast low-angle shot (FLASH) sequence for dynamic imaging (0.3 x 0.3 mm voxel size, 2.0 mm slice thickness, gap 0 mm, TR 8.7 ms, TE 4.76 ms, FOV 400 x 400 mm2, flip angle 25 degrees, matrix size 256x256, 88 slices). A dose of 0.1 mmol/kg bodyweight gadobutrol (Bayer Healthcare, Berlin, Germany) was injected intravenously after the first FLASH sequence with a flow of 2 mL/s using an automated injector (Spectris Solaris, MR Injection System; Medrad, Pittsburg, PA).
Subtraction imaging was acquired automatically after completion of the dynamic series.
Standard of reference
Histopathological analysis served as the reference standard and was based on either core needle biopsy (n = 79) or surgical resection (n = 19). Histological and molecular subtype classification was determined according to the 2013 St. Gallen guidelines [18]. IHC (ImmunoHistoChemistry) status analysis comprised estrogen receptor, progesterone receptor and HER2 status. Positivity for ER and PR was defined as the presence of 1% or more of positively stained nuclei. Molecular subtypes were classified accordingly: (1) luminal A for hormone receptor positive and HER2 negative, (2) luminal B for hormone receptor positive and HER2 positive, (3) triple-negative for hormone receptor and HER2 negative and (4) HER2-enriched for hormone receptor negative and HER2 positive [19–21]. Histological grading was determined in accordance with Elston-Ellis [22]. Elston-Ellis gradings 5, 6 and 7 were classified into a low-grade group, gradings 8 and 9 into a high-grade group. Ki67 was considered to be low, if the value was below 14% and high if otherwise [18]. A list of all outcome variables is displayed in Table 1.
Table 1. Display of the distribution of the outcome variables.
N denotes the sample size used for each outcome, while Positive and Negative denote the balance of the data.
| Outcome Variable | N | Positive | Negative |
|---|---|---|---|
| Molecular breast cancer subtypes | |||
| Luminal A | 95 | 58 (61%) | 37 (39%) |
| Luminal B | 95 | 16 (17%) | 79 (83%) |
| HER2-enriched | 95 | 6 (6%) | 89 (94%) |
| Triple-Negative | 95 | 15 (16%) | 80 (84%) |
| Differentiation of Molecular breast cancer subtypes | |||
| Luminal A vs Luminal B | 74 | 58 (78%) | 16 (22%) |
| Luminal A vs HER2-enriched | 64 | 58 (91%) | 6 (9%) |
| Luminal A vs Triple- Negative | 73 | 58 (79%) | 15 (21%) |
| Luminal B vs HER2-enriched | 22 | 16 (73%) | 6 (27%) |
| Luminal B vs Triple- Negative | 31 | 16 (52%) | 15 (48%) |
| HER2-enriched vs Triple-Negative | 21 | 6 (29%) | 15 (71%) |
| Receptor status | |||
| Estrogen Receptor (ER) | 95 | 73 (77%) | 22 (23%) |
| Progesterone Receptor (PR) | 95 | 66 (69%) | 29 (31%) |
| Hormone receptor positivity | 95 | 74 (78%) | 21 (22%) |
| Ki67- and HER2-Expression | |||
| Ki67 | 80 | 20 (25%) | 60 (75%) |
| Human epidermal growth factor receptor 2 (HER2) | 95 | 22 (23%) | 73 (77%) |
| Involvement of the lymphatic system | |||
| Lymph Vessel Involvement | 51 | 8 (16%) | 43 (84%) |
| Lymph Node Metastasis | 84 | 34 (40%) | 50 (60%) |
| Grading | |||
| Elston-Ellis Grading (EE) | 57 | 44 (77%) | 13 (23%) |
| Histological Grading | 90 | 18 (20%) | 72 (80%) |
Radiomics and statistical analysis
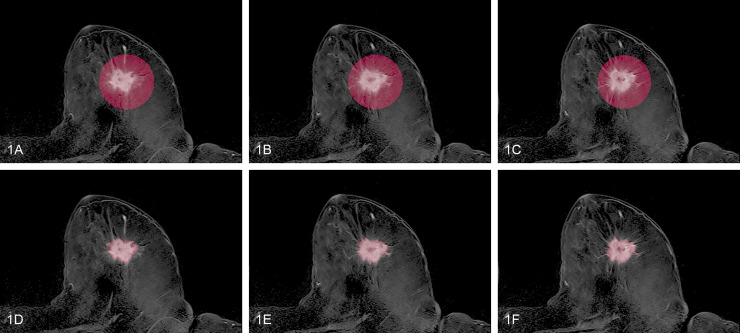
All MRI datasets were imported into the open source image processing software 3D-Slicer [23]. To ensure best conspicuity of the cancerous lesions the second subtraction series of the dynamic set was utilized for radiomic analysis. Tumor volumes were encircled utilizing a VOI (volume of interest) by an experienced breast radiologist with 15 years of experience (Fig 1A). A simple threshold of using 30% of the highest intensity inside the volume, followed with a morphological closing was used to obtain an automatic fine segmentation of the tumor mass (Fig 1B). Since this method may generate several segments, only the largest was retained. All segmentations were then reviewed by the above mentioned experienced breast radiologist and exported as DICOM-SEG. (http://dicom.nema.org/medical/dicom/current/output/chtml/part03/sect_A.51.html).
Fig 1. Imaging example of the VOI-encirclement of a tumor lesion.
Fig 1A–1C display three consecutive axial slices of a tumor lesion in the second subtraction series annotated via VOI-encirclement. Fig 1D–1F show the corresponding semi-automatic fine tumor segmentation obtained by thresholding.
Image preprocessing was performed in accordance with previous publications [1,5,24], e.g. a gradient filter was used to make the T1-weighted DCE-MRI series and the T2-weighted series more comparable. Overall, 13.118 generic features comprising shape, first order and higher order features were generated (a detailed list is displayed in S1 File). Five different feature selection methods (randomized logistic regression, chi-square, f-score, t-score, mutual information) were used to reduce the set of features. For machine learning analysis, three different classifiers (Naive Bayes, random forests and logistic regression) were employed. Classification performance was measured using a Receiver Operating Characteristic (ROC) analysis and reported as Area under the Curve (AUC) with a 95% confidence interval. A two-sided DeLong test was employed to compare if the AUC of ROC curves is different to that of a constant predictor. More details are listed in S3 File.
All analyses were performed using Python 3.6. Pyradiomics 2.0.1 [24] was used for the extraction of the radiomics features. Implementations of the feature selection methods and classifiers were taken from the scikit-learn 0.21.0 package.
Validation
For internal validation, a stratified 5-fold cross-validation (CV) was applied, selecting 80% of the samples randomly in each round for training, with the remaining 20% being reserved exclusively for testing. Feature selection was applied only to the training folds. The CV was repeated 25 times to get a more accurate estimation of the final performance. As some of the outcome variables showed imbalanced positive and negative classes (Table 1), stratified sampling was used.
The prediction capacity of all models was measured by averaging over the corresponding cross-validation test folds. Subsequently, AUC as well as sensitivity and specificity were computed.
Results
A total of 98 patients (mean age 59.7, range 28.0–89.4) were included in the study. Mean lesion size was 4.9 +/- 2.9 cm (range: 1.2–16.1 cm).
Due to low occurrence, patients with an Elston-Ellis grading below 5 (n = 3) or with a histopathological grading of 1 (n = 2) were removed from the corresponding subanalyses. The sample sizes of all subanalyses ranged from 21 to 95. The distributions of the outcome variables are listed in Table 1.
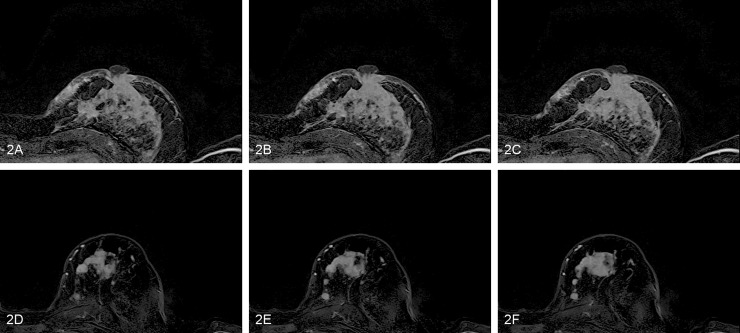
Trained models produced prediction scores that were used for the ROC analysis (Fig 2 and violin plots in S4 File). Predictions showed varying results, ranging from AUC 0.62 to 0.97. Detailed results of classification accuracies are displayed in Table 2 and visualized in ROC plots in S5 File.
Fig 2. Example of two patients with low and high prediction scores using the model trained to predict low vs high Ki67.
(A-C) Three consecutive axial slices of a tumor lesion in the second subtraction series for the patient with the highest prediction score. Histological reference revealed a Ki67 count of 80. (D-F) The corresponding axial slices for the patient with the lowest prediction score. Histological reference revealed a Ki67 count of 10.
Table 2. Detailed results of the classification accuracies.
Results for all classifiers. P-Values were computed using a DeLong test to compare ROC curves.
| Outcome | Classifier | Feature Selection Method | Number of Features | AUC [95% CI], p-Value | Sensitivity | Specificity | Accuracy |
|---|---|---|---|---|---|---|---|
| Molecular breast cancer subtypes | |||||||
| Luminal A | Random forest | Mutual Information | 32 | 0.65 [0.54, 0.77], p = 0.009 | 0.53 | 0.78 | 0.62 |
| Luminal B | Random forest | t-Score | 32 | 0.69 [0.55, 0.84], p = 0.008 | 0.56 | 0.76 | 0.72 |
| HER2-enriched | Logistic Regression | Randomized Logistic Regression | 2 | 0.75 [0.58, 0.91], p = 0.003 | 0.67 | 0.82 | 0.80 |
| Triple- Negative | Random forest | Chi Square | 32 | 0.73 [0.58, 0.87], p = 0.002 | 0.8 | 0.6 | 0.59 |
| Differentiation of Molecular breast cancer subtypes | |||||||
| Luminal A vs Luminal B | Random forest | Randomized Logistic Regression | 4 | 0.64 [0.47, 0.81], p = 0.115 | 0.78 | 0.56 | 0.72 |
| Luminal A vs HER2-enriched | Random forest | Mutual Information | 16 | 0.79 [0.58, 0.99], p = 0.003 | 0.79 | 0.83 | 0.78 |
| Luminal A vs Triple- Negative | Random forest | t-Score | 8 | 0.74 [0.61, 0.87], p < 0.001 | 0.66 | 0.8 | 0.66 |
| Luminal B vs HER2-enriched | Logistic Regression | F-Score | 2 | 0.78 [0.59, 0.97], p = 0.003 | 0.69 | 0.83 | 0.68 |
| Luminal B vs Triple- Negative | Naive Bayes | Randomized Logistic Regression | 1 | 0.86 [0.71, 1.0], p < 0.001 | 0.88 | 0.87 | 0.84 |
| HER2-enriched vs Triple Negative | Naive Bayes | Mutual Information | 2 | 0.97 [0.89, 1.0], p < 0.001 | 0.83 | 1.0 | 0.90 |
| Receptor status | |||||||
| Estrogen Receptor (ER) | Naive Bayes | Mutual Information | 32 | 0.67 [0.53, 0.8], p = 0.014 | 0.68 | 0.68 | 0.63 |
| Progesterone Receptor (PR) | Naive Bayes | t-Score | 32 | 0.69 [0.57, 0.8], p = 0.002 | 0.71 | 0.62 | 0.69 |
| Hormone receptor positivity | Logistic Regression | Chi Square | 1 | 0.69 [0.57, 0.81], p = 0.002 | 0.65 | 0.71 | 0.61 |
| Ki67- and HER2-Expression | |||||||
| Ki67 | Logistic Regression | Randomized Logistic Regression | 8 | 0.81 [0.7, 0.92], p < 0.001 | 0.75 | 0.68 | 0.84 |
| Human epidermal growth factor receptor 2 (HER2) | Random forest | Randomized Logistic Regression | 16 | 0.62 [0.48, 0.75], p = 0.079 | 0.64 | 0.58 | 0.51 |
| Involvement of the lymphatic system | |||||||
| Lymph Vessel Involvement | Random forest | Mutual Information | 8 | 0.8 [0.65, 0.95], p < 0.001 | 0.88 | 0.67 | 0.68 |
| Lymph Node Metastasis | Logistic Regression | Mutual Information | 1 | 0.71 [0.6, 0.83], p < 0.001 | 0.71 | 0.74 | 0.71 |
| Grading | |||||||
| Elston-Ellis Grading (EE) | Logistic Regression | Chi Square | 32 | 0.71 [0.55, 0.88], p = 0.009 | 0.64 | 0.85 | 0.67 |
| Histological Grading | Naive Bayes | Randomized Logistic Regression | 16 | 0.74 [0.62, 0.87], p < 0.001 | 0.72 | 0.72 | 0.62 |
Molecular breast cancer subtypes
Highest accuracies for correct classification were achieved for the differentiation of HER2-enriched from triple-negative with an AUC of 0.97, followed by luminal B from triple-negative (AUC 0.86) and luminal A and B from HER2-enriched (AUC 0.79 and 0.78 respectively). Classification of the individual molecular subtype against all other subtypes showed moderate to low accuracies, with best accuracies achieved for prediction of HER2-enriched (AUC 0.75) and lowest for luminal A (AUC 0.65).
Receptor status
Classification of the hormonal receptor status was highest for the pairwise discrimination of the overall hormone receptor positivity and Progesterone receptor positivity with AUCs of 0.69. Comparable, yet slightly lower AUCs were achieved for Estrogen receptor positivity with an AUC of 0.67.
Ki67- and HER2-expression
Prediction of Ki67 Expression was achieved with an AUC of 0.81. Classification of HER2-Expression was rather low with an AUC of 0.62.
Involvement of the lymphatic system
Prediction of lymph vessel involvement achieved high accuracies (AUC 0.80), while classification of involvement of lymph nodes was moderate with an AUC of 0.72.
Grading
Both gradings (histological grading and Elston-Ellis grading) showed comparably moderate performance, with AUC 0.71 for the prediction of the Elston-Elliis grading and 0.75 for histological grading.
Features and feature selection methods
Regarding the number of features, no general trend could be observed (a detailed list of the selected features is displayed in S2 File while a correlation matrix of all extracted features with the outcomes is visualized in S6 File). The number of selected features of the best models differed for each outcome variable, e.g. selecting 1 feature yielded the best model for lymph node metastasis, while the model using 32 features showed the best performance for prediction of the Elston-Ellis grading. Similarly, the most often selected features differed: Considering only those 114 features that have been selected more than 50% in all training repeats across all sub-analyses, most often features from the DCE-MRI sequence were selected (70%, N = 80) with lowest features derived from T2 imaging (11%, N = 12). Considering the preprocessing, wavelet features were most often used (41%, N = 47), followed by local binary pattern features (24%, N = 27). No shape features were selected by any of the feature selection methods. While most often first order features (36%, N = 41) and GLSZM as well as GLCM features (22%, N = 25 and 21%, N = 24 resp.) were used, no clear pattern can be seen in the features themselves. Here, the most often selected feature was LargeAreaHighGrayLevelEmphasis (8%, N = 9), while variants of the emphasis feature was most often used (32%, N = 36).
No clear pattern was observed regarding the feature selection method and the classifier. Models using randomized logistic regression (N = 6) were equally successful in producing best performing models as mutual information (N = 6). Similarly, no superiority of any of the classifiers was seen: Though random forests were used slightly more often than logistic regression (N = 8 and N = 6), both were not outperforming Naive Bayes (N = 5).
Discussion
Inter- and intratumoral heterogeneity is one of the hallmarks of breast cancer [25] having induced refinements of the previously mainly pathology-driven classification to molecular classifications and subtyping [26]. Each of these subtypes is associated to different risk factors for incidence, response to treatment, risk of disease progression, and preferential organ sites of metastases. Luminal tumors are positive for estrogen and progesterone receptors, and the majority respond well to hormonal interventions, whereas HER2+ tumors have amplification and overexpression of the ERBB2 oncogene and can be effectively controlled with a diverse array of anti-HER2 therapies. Basal-like tumors in general lack hormone receptors and HER2; thus, the majority of these tumors are also called triple-negative breast cancer (TNBC). While the “personalization” of treatment, by means of adaptation in accordance with molecular subtyping, has significantly changed and improved breast cancer treatment, invasive tissue sampling may be restricted in its assessment of whole-tumor heterogeneity, as particularly in patients undergoing neoadjuvant chemotherapy only a minor part of the tumor is histopathologically analyzed prior to treatment.
Over the past few years, Radiomics has been demonstrated to facilitate a promising platform for non-invasive whole-tumor tissue characterization for a number of cancer types [6,7,27]. An increasing number of research studies have found radiomic signatures to be predictive markers of underlying gene-expression patterns, therapy response, relapse, patient survival and other clinical and histopathological outcomes, building a bridge between imaging and genomics, also known as radiogenomics [4,6,28]. In one of the largest studies on molecular subtyping, Saha et al. [9] extracted 529 features from DCE-MRI imaging of 922 patients and predicted luminal A, triple-negative, ER and PR status with AUCs of 0.70, 0.66, 0.66 and 0.62, respectively. These results coincide well with our study results, with the biggest difference lying in the prediction of triple-negative cancers (AUC 0.66 vs 0.73). Similarly, in a smaller study, Guo et al. [10] predict Stage I vs III, ER, PR, HER2 as well as lymph node metastases (LNM) from radiological and genetic data. The AUCs of the radiomics only models were 0.87, 0.79, 0.69, 0.65 and 0.69 for Stage, ER, PR, HER2 and LNM respectively. Again, these numbers lie within the 95% CI of our models, differing strongest in prediction of ER (AUC 0.79 vs 0.67). Our results in differentiation of molecular subtypes also go in line with a previous publication by Leithner et al. [29], revealing comparable results for the differentiation of luminal A from HER2-enriched, luminal B from triple-negative and superior results for distinguishing HER2-enriched from triple-negative cancers.
Similar results have also been obtained using computer-assisted diagnostics (CAD), where instead of a broad set of automatically generated features only a few manual ones are extracted and analyzed via statistical or machine learning models. Baltzer et al. [30] demonstrated that volume enhancement characteristics are significantly associated with LNM, ER, PR and HER2. Dietzel et al. [31] used a neural network to predict lymph node status and obtained an AUC of 0.74, which is comparable to our result.
Nevertheless, our study differs from a number of previous trials in three important factors: Firstly, we favored a rather simple and robust imaging protocol for analysis, comprising dynamic T1-weighted imaging and T2-weighted imaging. While DWI is known to be susceptible to artifacts and suffer from an overall lack of standardization, it is considered as an important source of functional data and has been shown to improve specificity [32]. Nonetheless DWI was not used in this study as it was not part of our standard imaging protocol. As it was not consistently available for all patients, it was excluded from our analysis. Secondly, our approach to non-invasive tissue characterization was composed broader than most previous studies in comprising the molecular subtype as well as parameters like Ki67, grading and involvement of lymph tissue. While the prediction of grading and involved lymph nodes may be considered rather moderate with AUCs of 0.71 and 0.75, respectively, prediction of Ki67 achieved clinically relevant and high AUCs of 0.81. Ki67 expression and Ki67 index have been shown to play considerable roles as predictive factors for disease free survival, overall survival and may be used for treatment and follow-up assessment [33]. The third and most distinctive difference is our approach towards tumor annotation. Most previous radiomics studies were based on labor-intensive manual or semi-automatic full-tumor annotations performed by the radiologists, resulting in potential inter-reader variabilities depending on each radiologists´ appreciation of the tumor shape and size as well as expertise. Our approach was simplified in restricting the radiologists´ influence to recognition and a VOI-based encirclement of the tumor and a subsequent sub-automatic segmentation by the machine learning algorithm, using a simple threshold of 30% of the highest intensity inside the volume to obtain an automatic fine segmentation of the tumor mass. A similar segmentation approach was adopted recently by Dietzel et al. [34]. By thresholding voxels in the initial enhancement in the dynamics having more than 30% difference they identify the active tumor tissue and extract vascularization patterns.
This simplified approach endorses two important factors: first, the time for tumor annotation is significantly reduced from a dedicated whole-tumor manual / semi-automatic annotation to a simple VOI demarcation as commonly used in hybrid imaging for assessment of tracer uptake, making this approach feasible in clinical routine. Secondly, these VOI and intensity-based segmentations reduce potential inter-reader variabilities based on manual / semi-automatic segmentation by radiologist. This simplified approach to radiomics goes in line with the recent recognition of the potential impact of inter-reader variability in radiomics. Saha et al. [11] recently published an overview of algorithmic features and the potential impact of inter-reader variability in annotating tumors. They conclude that breast MRI radiomics features widely vary in terms of their stability, ranging from an average inter-reader stability for all features of 0.85 to as low as 0.63 for tumor-based features.
Compared to previous publications [35,36] our study results do not facilitate clear recommendations regarding the feature selection method or the applied classifiers. This underlines that in high dimensional spaces no method generally outperforms others. This also holds true for the feature selection, where similarly no pattern could be clearly detected. Overall, dynamic T1-weighted imaging seems to facilitate the extraction of more relevant features than T2 weighted imaging, which goes in line with previous publications on this matter [32,37].
While the present study offers several strong points, including a rather broad scope and simplified approach to tumor-annotation, some limitations apply: The small sample size might have introduced a bias in form of overfitting and impeded the splitting of the data into strict train and test sets. This stems from the fact that the lesions of many patients were clip-marked at the time of examination and thus were excluded from the study. Furthermore, a total of 24 patients with BIRADS 6 were included. As a biopsy was performed in these patients prior to imaging, an effect on the radiomics analysis cannot be ruled out. A meaningful subgroup analysis would require a higher sample size and was thus not carried out. Another bias may be caused by the fact, that all examinations in this study were performed with a scanner from a single provider. Finally, due to the “proof-of-concept” nature of this study, another restriction may be elicited by the lack of an external validation data set, as the primary study goal was to establish a simplified approach to radiomics. Hence, all of these weak points should be addressed in future trials, comprising larger patient cohorts of multi-center, multi-vendor studies.
Conclusions
Overall, our preliminary study results demonstrate the usability of a simplified and rapid approach to tumor for MRI-based tumor decoding and phenotyping of breast cancer.
Supporting information
This file contains a list of the 13,118 extracted features as well as the applied preprocessing methods.
(DOCX)
This file contains a list of features that were selected by the best performing methods.
(DOCX)
This file contains a detailed description of the performed analysis.
(DOCX)
This file contains all violin plots for the prediction scores.
(DOCX)
This file contains all ROC curves.
(DOCX)
This file contains a plot depicting the correlation of all extracted features with the outcomes.
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.”
Funding Statement
The authors received no specific funding for this work.
References
- 1.Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RGPM, Granton P, et al. Radiomics: Extracting more information from medical images using advanced feature analysis. European Journal of Cancer. 2012;48: 441–446. 10.1016/j.ejca.2011.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coroller TP, Agrawal V, Narayan V, Hou Y, Grossmann P, Lee SW, et al. Radiomic phenotype features predict pathological response in non-small cell lung cancer. Radiotherapy and Oncology. 2016;119: 480–486. 10.1016/j.radonc.2016.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bodalal Z, Trebeschi S, Nguyen-Kim TDL, Schats W, Beets-Tan R. Radiogenomics: bridging imaging and genomics. Abdom Radiol. 2019;44: 1960–1984. 10.1007/s00261-019-02028-w [DOI] [PubMed] [Google Scholar]
- 4.Pinker K, Shitano F, Sala E, Do RK, Young RJ, Wibmer AG, et al. Background, Current Role and Potential Applications of Radiogenomics. J Magn Reson Imaging. 2018;47: 604–620. 10.1002/jmri.25870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aerts HJWL, Velazquez ER, Leijenaar RTH, Parmar C, Grossmann P, Carvalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nature Communications. 2014;5 10.1038/ncomms5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith CP, Czarniecki M, Mehralivand S, Stoyanova R, Choyke PL, Harmon S, et al. Radiomics and radiogenomics of prostate cancer. Abdom Radiol. 2019;44: 2021–2029. 10.1007/s00261-018-1660-7 [DOI] [PubMed] [Google Scholar]
- 7.Lotan E, Jain R, Razavian N, Fatterpekar GM, Lui YW. State of the Art: Machine Learning Applications in Glioma Imaging. American Journal of Roentgenology. 2019;212: 26–37. 10.2214/AJR.18.20218 [DOI] [PubMed] [Google Scholar]
- 8.Haubold J, Demircioglu A, Gratz M, Glas M, Wrede K, Sure U, et al. Non-invasive tumor decoding and phenotyping of cerebral gliomas utilizing multiparametric 18F-FET PET-MRI and MR Fingerprinting. Eur J Nucl Med Mol Imaging. 2019. [cited 6 Jan 2020]. 10.1007/s00259-019-04602-2 [DOI] [PubMed] [Google Scholar]
- 9.Saha A, Harowicz MR, Grimm LJ, Kim CE, Ghate SV, Walsh R, et al. A machine learning approach to radiogenomics of breast cancer: a study of 922 subjects and 529 DCE-MRI features. Br J Cancer. 2018;119: 508–516. 10.1038/s41416-018-0185-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo W, Li H, Zhu Y, Lan L, Yang S, Drukker K, et al. Prediction of clinical phenotypes in invasive breast carcinomas from the integration of radiomics and genomics data. J Med Imag. 2015;2: 041007 10.1117/1.JMI.2.4.041007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saha A, Harowicz MR, Mazurowski MA. Breast cancer MRI radiomics: An overview of algorithmic features and impact of inter-reader variability in annotating tumors. Med Phys. 2018;45: 3076–3085. 10.1002/mp.12925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mazurowski MA, Grimm LJ, Zhang J, Marcom PK, Yoon SC, Kim C, et al. Recurrence-free survival in breast cancer is associated with MRI tumor enhancement dynamics quantified using computer algorithms. European Journal of Radiology. 2015;84: 2117–2122. 10.1016/j.ejrad.2015.07.012 [DOI] [PubMed] [Google Scholar]
- 13.Chen W, Giger ML, Bick U. A Fuzzy C-Means (FCM)-Based Approach for Computerized Segmentation of Breast Lesions in Dynamic Contrast-Enhanced MR Images1. Academic Radiology. 2006;13: 63–72. 10.1016/j.acra.2005.08.035 [DOI] [PubMed] [Google Scholar]
- 14.Nie K, Chen J-H, Yu HJ, Chu Y, Nalcioglu O, Su M-Y. Quantitative Analysis of Lesion Morphology and Texture Features for Diagnostic Prediction in Breast MRI. Academic Radiology. 2008;15: 1513–1525. 10.1016/j.acra.2008.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho N, Im S-A, Park I-A, Lee K-H, Li M, Han W, et al. Breast Cancer: Early Prediction of Response to Neoadjuvant Chemotherapy Using Parametric Response Maps for MR Imaging. Radiology. 2014;272: 385–396. 10.1148/radiol.14131332 [DOI] [PubMed] [Google Scholar]
- 16.Ashraf AB, Daye D, Gavenonis S, Mies C, Feldman M, Rosen M, et al. Identification of Intrinsic Imaging Phenotypes for Breast Cancer Tumors: Preliminary Associations with Gene Expression Profiles. Radiology. 2014;272: 374–384. 10.1148/radiol.14131375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayerhoefer ME, Schima W, Trattnig S, Pinker K, Berger‐Kulemann V, Ba‐Ssalamah A. Texture-based classification of focal liver lesions on MRI at 3.0 Tesla: A feasibility study in cysts and hemangiomas. Journal of Magnetic Resonance Imaging. 2010;32: 352–359. 10.1002/jmri.22268 [DOI] [PubMed] [Google Scholar]
- 18.Harbeck N, Thomssen C, Gnant M. St. Gallen 2013: Brief Preliminary Summary of the Consensus Discussion. Breast Care. 2013;8: 102–109. 10.1159/000351193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hon JDC, Singh B, Sahin A, Du G, Wang J, Wang VY, et al. Breast cancer molecular subtypes: from TNBC to QNBC. Am J Cancer Res. 6: 1864–1872. [PMC free article] [PubMed] [Google Scholar]
- 20.Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, et al. Race, Breast Cancer Subtypes, and Survival in the Carolina Breast Cancer Study. JAMA. 2006;295: 2492–2502. 10.1001/jama.295.21.2492 [DOI] [PubMed] [Google Scholar]
- 21.Mazurowski MA, Zhang J, Grimm LJ, Yoon SC, Silber JI. Radiogenomic Analysis of Breast Cancer: Luminal B Molecular Subtype Is Associated with Enhancement Dynamics at MR Imaging. Radiology. 2014;273: 365–372. 10.1148/radiol.14132641 [DOI] [PubMed] [Google Scholar]
- 22.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. C. W. Elston & I. O. Ellis. Histopathology 1991; 19; 403–410. Histopathology. 2002;41: 151–151. 10.1111/j.1365-2559.1991.tb00229.x [DOI] [PubMed] [Google Scholar]
- 23.Kikinis R, Pieper SD, Vosburgh KG. 3D Slicer: A Platform for Subject-Specific Image Analysis, Visualization, and Clinical Support In: Jolesz FA, editor. Intraoperative Imaging and Image-Guided Therapy. New York, NY: Springer; 2014. pp. 277–289. 10.1007/978-1-4614-7657-3_19 [DOI] [Google Scholar]
- 24.van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, et al. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017;77: e104–e107. 10.1158/0008-5472.CAN-17-0339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Polyak K. Breast cancer: origins and evolution. J Clin Invest. 2007;117: 3155–3163. 10.1172/JCI33295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perez EA. Breast Cancer Management: Opportunities and Barriers to an Individualized Approach. The Oncologist. 2011;16: 20–22. 10.1634/theoncologist.2011-S1-20 [DOI] [PubMed] [Google Scholar]
- 27.Yip SSF, Aerts HJWL. Applications and limitations of radiomics. Phys Med Biol. 2016;61: R150–R166. 10.1088/0031-9155/61/13/R150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grove O, Berglund AE, Schabath MB, Aerts HJWL, Dekker A, Wang H, et al. Quantitative Computed Tomographic Descriptors Associate Tumor Shape Complexity and Intratumor Heterogeneity with Prognosis in Lung Adenocarcinoma. PLoS One. 2015;10 10.1371/journal.pone.0118261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leithner D, Horvat JV, Marino MA, Bernard-Davila B, Jochelson MS, Ochoa-Albiztegui RE, et al. Radiomic signatures with contrast-enhanced magnetic resonance imaging for the assessment of breast cancer receptor status and molecular subtypes: initial results. Breast Cancer Res. 2019;21: 106 10.1186/s13058-019-1187-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baltzer PAT, Vag T, Dietzel M, Beger S, Freiberg C, Gajda M, et al. Computer-aided interpretation of dynamic magnetic resonance imaging reflects histopathology of invasive breast cancer. Eur Radiol. 2010;20: 1563–1571. 10.1007/s00330-010-1722-x [DOI] [PubMed] [Google Scholar]
- 31.Dietzel M, Baltzer PAT, Dietzel A, Vag T, Gröschel T, Gajda M, et al. Application of artificial neural networks for the prediction of lymph node metastases to the ipsilateral axilla–initial experience in 194 patients using magnetic resonance mammography. Acta Radiol. 2010;51: 851–858. 10.3109/02841851.2010.498444 [DOI] [PubMed] [Google Scholar]
- 32.Maforo N, Li H, Weiss W, Lan L, Giger M. SU-D-BRA-02: Radiomics of Multi-Parametric Breast MRI in Breast Cancer Diagnosis: A Quantitative Investigation of Diffusion Weighted Imaging, Dynamic Contrast-Enhanced, and T2-Weighted Magnetic Resonance Imaging. Medical Physics. 2015;42: 3213–3213. 10.1118/1.4923882 [DOI] [Google Scholar]
- 33.Soliman NA, Yussif SM. Ki-67 as a prognostic marker according to breast cancer molecular subtype. Cancer Biol Med. 2016;13: 496–504. 10.20892/j.issn.2095-3941.2016.0066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dietzel M, Schulz-Wendtland R, Ellmann S, Zoubi R, Wenkel E, Hammon M, et al. Automated volumetric radiomic analysis of breast cancer vascularization improves survival prediction in primary breast cancer. Sci Rep. 2020;10: 3664 10.1038/s41598-020-60393-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang B, He X, Ouyang F, Gu D, Dong Y, Zhang L, et al. Radiomic machine-learning classifiers for prognostic biomarkers of advanced nasopharyngeal carcinoma. Cancer Letters. 2017;403: 21–27. 10.1016/j.canlet.2017.06.004 [DOI] [PubMed] [Google Scholar]
- 36.Parmar C, Grossmann P, Rietveld D, Rietbergen MM, Lambin P, Aerts HJWL. Radiomic Machine-Learning Classifiers for Prognostic Biomarkers of Head and Neck Cancer. Front Oncol. 2015;5 10.3389/fonc.2015.00272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhooshan N, Giger M, Lan L, Li H, Marquez A, Shimauchi A, et al. Combined use of T2-weighted MRI and T1-weighted dynamic contrast–enhanced MRI in the automated analysis of breast lesions. Magnetic Resonance in Medicine. 2011;66: 555–564. 10.1002/mrm.22800 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This file contains a list of the 13,118 extracted features as well as the applied preprocessing methods.
(DOCX)
This file contains a list of features that were selected by the best performing methods.
(DOCX)
This file contains a detailed description of the performed analysis.
(DOCX)
This file contains all violin plots for the prediction scores.
(DOCX)
This file contains all ROC curves.
(DOCX)
This file contains a plot depicting the correlation of all extracted features with the outcomes.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.”