Abstract
Recent innovations in translational research have ushered an exponential increase in the discovery of novel biomarkers, thereby elevating the hope for deeper insights into “personalized” medicine approaches to disease phenotyping and care. However, a critical gap exists between the fast pace of biomarker discovery and the successful translation to clinical use. This gap underscores the fundamental biomarker conundrum across various acute and chronic disorders: how does a biomarker address a specific unmet need? Additionally, the gap highlights the need to shift the paradigm from a focus on biomarker discovery to greater translational impact and the need for a more streamlined drug approval process. The unmet need for biomarkers in acute respiratory distress syndrome (ARDS) is for reliable and validated biomarkers that minimize heterogeneity and allow for stratification of subject selection for enrollment in clinical trials of tailored therapies. This unmet need is particularly highlighted by the ongoing SARS-CoV-2/COVID-19 pandemic. The unprecedented numbers of COVID-19-induced ARDS cases has strained health care systems across the world and exposed the need for biomarkers that would accelerate drug development and the successful phenotyping of COVID-19-infected patients at risk for development of ARDS and ARDS mortality. Accordingly, this review discusses the current state of ARDS biomarkers in the context of the drug development pipeline and highlight gaps between biomarker discovery and clinical implementation while proposing potential paths forward. We discuss potential ARDS biomarkers by category and by context of use, highlighting progress in the development continuum. We conclude by discussing challenges to successful translation of biomarker candidates to clinical impact and proposing possible novel strategies.
INTRODUCTION
Innovations in laboratory biochemistries, molecular biology, and “omics” medicine have ushered in an era with the potential to unravel the Gordian knot of identifying validated molecular markers of disease.1 , 2 The emergence of precision medicine and high throughput precision technologies elevated aspirations for defining novel biomarkers that would accelerate improved treatment of diverse adverse health conditions by facilitating the identification of responders to promising novel or repurposed therapeutic strategies.3 , 4 A cursory review of the medical literature5, 6, 7 over the past 3 decades revealed the emergence of an increasing number of biomarker candidates. However, the exponential rate of initial discovery has now completely outpaced the ability of the biomedical community to successfully develop and validate the clinical utility of prospective biomarkers.7 , 8 In fact, only ∼0.1% of potentially clinically relevant biomarkers described in the literature have progressed to utility as a meaningful and routinely utilized clinical readout.9 The reasons for this massive disconnect are multifactorial including the stark reality that the majority of biomarkers identified are by investigators in government-funded university laboratories that are ill-resourced to complete the biomarker development and validation continuum.5 This realization led the U.S. Congress under the 21st Century Cures Act of 2016, to encourage the U.S. Food and Drug Administration (FDA) to create the biomarker qualification program as part of the drug development toolkit, an effort to guide researchers and accelerate the development of promising biomarkers.10, 11, 12, 13
Prior reviews of biomarkers in acute respiratory distress syndrome (ARDS), a serious critical care illness in dire need of validated and clinically useful biomarkers, have largely served as diligent but descriptive approaches outlining new technologies or summarizing the pathobiology of current biomarkers.14, 15, 16, 17, 18, 19, 20 In contrast, this current review is highly divergent from prior reports and seeks to discuss the current state of ARDS biomarkers in the context of the drug development pipeline and to highlight the gaps between discovery and clinical implementation while proposing potential paths forward. Our intent is to shift the paradigm from a focus on biomarker discovery that is currently relegated to demonstrating a correlation between a specific biomarker and either the development of ARDS or ARDS severity, to a focus on the clinical utility and implementation of the biomarker within well-defined contexts of use including subject stratification in clinical trials.4 , 5 The need for such a translational focus is particularly highlighted by the ongoing SARS-CoV-2/COVID-19 pandemic. COVID-19-induced ARDS has strained health care systems across the world and exposed the need for biomarkers that would accelerate disease phenotyping and drug development.
The clinical definition of the highly heterogeneous ARDS includes acute arterial hypoxemia and a ratio of partial pressure of arterial oxygen [PaO2] to fraction of inspired oxygen [FiO2] that is less than 300, bilateral pulmonary opacities, and the exclusion of cardiac failure or other reversible primary causes.21 Since lung biopsies are not routinely obtained in ARDS, this clinical definition aims to identify patients with noncardiogenic pulmonary edema, a process characterized by increased protein permeability of the alveolar-capillary membrane.22 , 23 Diagnostic uncertainty in ARDS further exacerbates disease heterogeneity and is a potential source of bias in conducting clinical trials.23 There is a compelling unmet medical need to identify clinical and/or disease-specific biochemical parameters that risk-stratify patients for both accurate prognostication and clinical trial purposes. Stratification of ARDS patients with reliable biomarkers that are predictive of mortality would optimize participant selection for clinical trial enrollment by focusing on those subjects most likely to benefit from novel clinical interventions.24 , 25 More than 45 promising candidate biomarkers in ARDS have been described in the medical literature, however, to date no biomarker has been successfully developed as an accepted point of care surrogate marker of disease.14 , 15 The heterogeneity of the ARDS phenotype, the variability of candidate biomarkers, and the focus on biomarker discovery without consideration of biomarker development, are all serious contributors to the abysmal record for ARDS biomarker development and the poor performance record of ARDS clinical trials.
Biomarkers, objectively measured as characteristic of clinical, pathologic, or physiologic processes, can be bioanalytical (proteins, metabolites, DNA genetic variants, RNA types), histologic, or radiographic.12 , 26 The ideal bioanalytical biomarker is easily measured in blood or in other bodily fluids, has an excellent analytical sensitivity, high statistical sensitivity and specificity, varies rapidly in response to impactful therapies, aids in subject stratification, and exhibits biologic plausibility.26 Although traditional clinical or laboratory variables such as blood pressure readings, PaO2, hemoglobin A1C, and glomerular filtration rates are examples of “biomarkers,” in the context of translational research, the term often refers to a marker used to provide insight into a “personalized medicine” approach to phenotyping and care.27
In this review, we have attempted to summarize the current state of ARDS biomarkers based upon FDA-proposed categories with assessment of the advancement of each ARDS biomarker in the drug development continuum. Finally, we have sought to identify potential challenges to the successful translation of candidates through the pipeline of biomarker development. Of note, the biomarkers covered in this review are not an exhaustive list of possible ARDS biomarkers.
ARDS BIOMARKERS BY CATEGORY
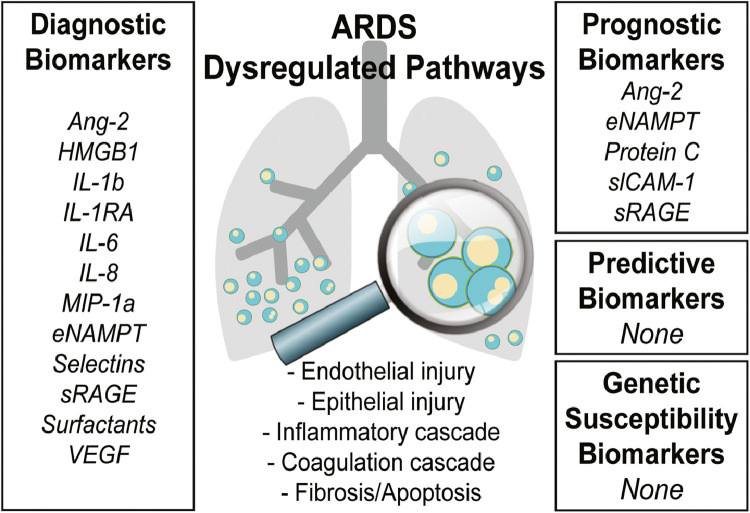
The pathogenesis of ARDS includes a combination of endothelial injury, epithelial injury, an intense inflammatory cascade, dysregulated coagulation, fibrosis, and apoptosis in response to diverse stimuli14 (Fig 1 ). This acute dysregulation of various biochemical and cellular pathways allows for the generation of numerous potential biomarkers associated with risk or severity of ARDS.14 , 15 However, to address the specific unmet medical needs in ARDS, biomarkers must provide specific contexts such as early diagnosis, pathobiologic disease classification, or guide successful development of novel therapies (Fig 1). For example, a diagnostic biomarker could lead to the early diagnosis of ARDS, promote improved patient selection for clinical trial enrollment and provide useful and reliable implications for clinical care. A predictive biomarker in the disease-causal pathway could enhance our understanding of the pathophysiology and, by extension, identify likely novel therapeutic targets and strategies. A prognostic biomarker would optimally stratify ARDS patients by likelihood of treatment response, again enriching future clinical trials for detection of a treatment effect. Whereas previous efforts have focused on discovery by demonstrating an association between a specific biomarker and ARDS risk and mortality, Table 1 presents a summary of ARDS biomarkers classified by general biomarker categories and potential examples of the corresponding context for biomarker use as recommended by the FDA.11
Fig 1.
ARDS biomarkers by category.
The majority of candidate ARDS biomarkers involve dysregulation of the following pathways: endothelial injury, epithelial injury, inflammatory cascade, coagulation cascade, or fibrosis and apoptosis. Studies have mostly assessed the diagnostic and prognostic performance of these candidate biomarkers. Ang-2, angiopoietin 2; HMGB1, high mobility group box nuclear protein 1; IL-1β, interleukin 1 beta; IL-1RA, interleukin 1 receptor antagonist; IL-6, interleukin 6; IL-8, interleukin 8; MIP-1α, macrophage inflammatory protein-1α; eNAMPT, extracellular nicotinamide phosphoribosyltransferase; sRAGE, soluble receptor for advanced glycation end products; VEGF, vascular endothelial growth factor.
Table 1.
Biomarker categories and examples of corresponding context of use
| Biomarker category | Context of use example |
|---|---|
| Diagnostic biomarkers |
|
| Predictive biomarkers |
|
| Prognostic biomarkers |
|
| Biomarkers for susceptibility or risk |
|
| Biomarkers of treatment responses and pharmacodynamics |
|
| Biomarkers of safety |
|
Diagnostic biomarkers in ARDS
In the context of ARDS, an ideal diagnostic biomarker is a surrogate that identifies the early stages of the syndrome, minimizes heterogeneity, reflects the natural history, and is a potential target for a clinical trial. Currently, the most promising diagnostic biomarkers do correlate with ARDS susceptibility, however, they do not meet the full criteria for surrogacy. Specific examples include:
Receptor for advanced glycation end products (RAGE)
RAGE, a transmembrane pattern recognition receptor of the immunoglobulin superfamily is abundantly expressed in the lung and primarily located on the basal surface of alveolar type 1 epithelial cells (AT1).28 , 29 The soluble form of RAGE (sRAGE) comprising the extracellular domain is produced through cleavage by matrix metalloproteinases. Soluble RAGE is a marker of AT1 cell injury and a key mediator of alveolar inflammation30, 31, 32 and sRAGE expression is enhanced in the early stage of ARDS.33 Plasma and bronchoalveolar lavage (BAL) fluid levels of sRAGE are elevated during ARDS and correlate with disease severity by lung CT. Although a very promising diagnostic biomarker, sRAGE measurements have not progressed to clinical utility and validation.
Angiopoeitin-2 (Ang-2)
Ang-2 is an endothelial-derived protein that increases the junction instability of the endothelial junction thereby enhancing vascular leak.34 As a potential diagnostic biomarker, higher plasma levels of Ang-2 were associated with development of ARDS in a cohort of patients admitted to the ICU.35 Similarly, among surgical ICU patients, plasma levels of Ang-2 were higher in ARDS patients compared to those without ARDS.36 Ang-2 remains a very promising diagnostic biomarker but without validation of clinical utility in ARDS.
Vascular endothelial growth factor (VEGF)
VEGF belongs to the platelet-derived growth factor superfamily which play central roles in the regulation of angiogenesis and lymphangiogenesis.37 VEGF is released from various alveolar type 2 epithelial (AT2) cells, neutrophils, alveolar macrophages, and activated T cells and depending on the degree of epithelial or endothelial damage, VEGF expression in ARDS varies.14 Studies suggesting a correlation of plasma or BAL VEGF levels with the diagnosis of ARDS have not been consistently replicated.14
Surfactant proteins
Because of the vital role in maintaining the integrity of the alveolar-capillary interface, the surfactant-associated proteins (SP-A, SP-B, SP-C, and SP-D) were considered early on as natural candidate diagnostic biomarkers in ARDS.38 , 39 However, initial reports correlating high plasma levels of SP-A and SP-B or reduced BAL levels of SP-D with a diagnosis of ARDS have not been confirmed.35 , 40
Selectins
These membrane-associated glycoproteins mediate the adhesion of leukocytes and platelets to the vascular endothelium.41 Plasma levels of P-selectin and E-selectin are elevated in patients with acute lung injury compared to those with sepsis without lung injury.42 , 43 However, these initial reports have not been robustly replicated.
Proinflammatory cytokines
The intense inflammatory cascade characteristic of ARDS is associated with increased plasma and alveolar levels of a number of inflammatory cytokines such as IL-1β and TNF-α that are both secreted by activated macrophages in the early inflammatory phase of ARDS and drive the release of other proinflammatory chemokines including monocyte chemotactic protein-1, macrophage inflammatory protein-1α, IL-6, and IL-8.44 These proinflammatory chemokines propagate the inflammatory cascade by further damage of the endothelial-alveolar barrier, recruitment of inflammatory cells into the airspaces, and impairment of fluid transport.44 Naturally, these proinflammatory cytokines have been extensively studied as possible diagnostic candidate biomarkers in ARDS.14 , 15 However, to date, no individual proinflammatory cytokine has been clinically validated as a robust diagnostic biomarker.
Anti-inflammatory cytokines
The innate immune system responds to the acute inflammatory cascade with specific (IL-1 receptor antagonist or IL-1RA) and nonspecific anti-inflammatory systems (IL-10).44 IL-1RA45 but not IL-1046 has emerged as a promising diagnostic ARDS biomarker but has yet to be validated.
Cytozymes
Cytozymes are a class of proteins that retain an intracellular function as an enzyme but are highly proinflammatory when secreted into the extracellular space or circulation. High mobility group box nuclear protein 1 (HMGB1) dually serves an intracellular function as a DNA nuclear binding protein and is an inflammatory cytokine when secreted by monocytes and macrophages.47 Plasma and alveolar levels of HMGB1 increase early after severe trauma and correlate with development of ARDS.47 However, critical significant associations between HMGB1 levels at ICU admission and clinical outcomes in critically ill patients has yet to be demonstrated.48 Macrophage migration inhibitory factor (MIF) is an intracellular tautomerase that is a potent inflammatory mediator when secreted. MIF is postulated to play a crucial pathological role in the development of alveolar inflammation in ARDS. MIF, and its close structural D-dopachrome homolog (MIF-2 or DDT), are potential diagnostic biomarkers in sepsis, trauma and ARDS that require further clinical validation.49 , 50 Similarly, the nicotinamide phosphoribosyltransferase known as NAMPT (also known as pre-B-cell colony-enhancing factor or visfatin) regulates intracellular NAD metabolism, whereas extracellular secreted eNAMPT is an innate immunity regulator which binds Toll-like receptor 4.51 eNAMPT levels are increased in ARDS with trending toward ARDS severity, but requires robust clinical validation as a diagnostic biomarker.52, 53, 54
Plasminogen activator inhibitor (PAI-I)
Plasminogen activator (PA) and PAI-I regulate fibrinolysis through the conversion of plasminogen to plasmin.55 During lung injury, alveolar epithelial cells and activated macrophages overexpress PAI-1, thus contributing to decreased alveolar fibrinolytic activity.55 Initial reports suggesting PAI-1 as a diagnostic biomarker were not confirmed in a large prospective cohort.56
Predictive biomarkers in ARDS
Many promising therapies for ARDS have failed in phase 3 studies due to phenotypic heterogeneity, a challenge potentially addressed by validated predictive biomarkers that identify individuals more likely to respond to a treatment type. Predictive biomarkers that reside in the causal pathway of the disease are obvious potential therapeutic targets. By identifying patients in whom a larger treatment effect can be obtained, biomarkers may significantly impact clinical trial design and sample size considerations. The best example of a causal pathway biomarker is low-density lipoprotein which has been implicated in the development of atherosclerotic cardiovascular disease.57 Casual inference of low-density lipoprotein and atherosclerotic cardiovascular disease is supported by evidence from genetic risk studies on inherited disorders of metabolism, prospective epidemiologic studies, Mendelian randomization studies, and randomized controlled trials.57 In ARDS, no single biomarker has been shown to reliably predict clinical outcomes.
Prognostic biomarkers in ARDS
Prognostic biomarkers provide information addressing the overall disease outcome and are potentially useful in stratifying patients for enrollment in clinical trials. In ARDS, enrichment of study subjects most likely to have a poor outcome can aid the design of clinical trials and enhance the ability to detect beneficial effects from potential therapies.
RAGE
Monitoring of sRAGE levels have been used to identify the subgroup of ARDS patients more likely to respond to alveolar recruitment maneuvers.58 Unfortunately, however, initial reports of an association between plasma levels of sRAGE and 28-day or 90-day mortality in patients with ARDS58 , 59 have not been consistently replicated.60
Angiopoeitin-2 (Ang-2)
Elevated levels of Ang-2 have been associated with increased risk of mortality among patients with infection-related ARDS.61 , 62 Higher levels of Ang-2, measured as part of a panel of 6 biomarkers was associated with increased risk of mortality.63 , 64
Soluble intercellular adhesion molecule-1 (sICAM-1)
sICAM-1 is an inducible glycoprotein that is expressed on the endothelial cell surface.65 Levels of sICAM-1 are upregulated during inflammation in response to stimulation by interferon-γ or IL-1.66 In multiple studies, elevated baseline levels of sICAM have been associated with increased mortality from ARDS.67, 68, 69
Protein C
Protein C is synthesized in the liver as an inactive form and transformed on cell surface to its active form by the thrombomodulin-thrombin complex.70 , 71 Activated protein C is an important endogenous regulator of coagulation and fibrinolysis with anti-inflammatory properties that can improve endothelial permeability,72 and exert antiapoptotic effects.73 Activated protein C also inactivates PAI-1 thus promoting fibrinolysis.74 Low plasma levels of protein C have been associated with higher ARDS mortality.75
Proinflammatory cytokines
High plasma levels of IL-1β, TNF-α, IL-6, IL-1, and IL-18 have been associated with increased mortality from ARDS.15 , 44 However, none of these proinflammatory biomarkers have sufficient specificity to serve as a stand-alone prognostic biomarker. Higher plasma levels of IL-6, IL-8, IL-1RA, measured as part of a panel of 6 biomarkers were associated with increased risk of mortality.63 High eNAMPT levels at the time of admission to the intensive care unit correlate with disease severity and may predict mortality in patients with sepsis and ARDS.76 , 77 Unfortunately, no single biomarker has been shown to reliably provide information about the patient's overall disease outcome and thus stratify patients for enrollment in clinical trials. However, recent efforts to combine biomarkers demonstrate that prognostic ability can be greatly enhanced.24 , 25 , 63
Biomarkers of ARDS genetic susceptibility
Despite the challenges in studying ARDS phenotypes, genomic/genetic methodologies have generated novel insights into the pathogenesis of ARDS78 and elucidated previously unknown mechanistic pathways in the pathogenesis of ARDS.79 , 80 For example, the association between NAMPT and development of ARDS and ventilator-induced lung injury (VILI)-driven pathobiology was discovered utilizing high throughput functional genomic approaches.52 Extensive microarray-based lung gene expression profiling in canine, murine, and human acute lung injury models identified increased expression of NAMPT 52 whose genetic variants are now confirmed to be associated with ARDS susceptibility and severity.52 , 53 , 81 Single nucleotide polymorphisms (SNP)-driven ARDS approaches, including genome-wide association studies for ARDS susceptibility, are historically limited by low statistical power and the daunting heterogeneity of the ARDS phenotype. However, these strategies have identified a S1PR382 , 83 and the polypeptide-interacting protein alpha-1 (PPFIA1) as risk factors for developing acute lung injury including after major trauma.84 Recently, an association between variants in the selectin P ligand gene (SELPLG) and the development of ARDS in African-American patients with sepsis85 was described with the potential as a viable ARDS biomarker. Despite herculean efforts, no genotype-driven biomarker of genetic susceptibility to ARDS has reached the level of clinical utility. Mendelian randomization analysis with genetic variation as an instrumental variable linked to plasma levels of a biomarker can infer causal inference under certain assumptions.78 , 86 Plasma levels of Ang-2, sRAGE, and S1P3 have been identified as potential casual biomarkers in sepsis-associated ARDS using these techniques.87 , 88
Combining biomarkers to improve diagnostic, predictive, and prognostic value
Thus far, no individual ARDS biomarker candidate has demonstrated acceptable statistical sensitivity and specificity to serve as an ideal predictive, diagnostic, or prognostic biomarker. Recently, researchers have combined biological markers to improve the sensitivity and sensitivity. For example, a panel of 7 biomarkers (sRAGE, procollagen peptide III, brain natriuretic peptide, Ang-2, IL-10, TNF-α, and IL-8) were recently found to exhibit a high diagnostic accuracy for differentiating acute lung injury among trauma patients from controls as reflected by an area under the receiver operating characteristic curve (AUC -- 0.86).89 Similarly, a combination of biomarkers of epithelial injury (CC16, SP-D, sRAGE) and inflammation (IL-6, IL-8) was demonstrated to exhibit reasonable diagnostic value for ARDS in patients with severe sepsis (AUC -- 0.75).90 Recently, a combination of 6 biomarkers (Ang-2, MIF, IL-8, IL-1RA, IL-6, and eNAMPT) showed promising prognostic value.63 A subphenotype of ARDS patients with high plasma levels of these cytokines exhibited significantly higher mortality when compared to those with lower levels.63 Additional attempts to combine clinical and biological markers to enhance the diagnostic accuracy or stratify ARDS patients by prognosis have met with less success.24 , 25 , 91
ARDS BIOMARKERS IN THE DEVELOPMENT CONTINUUM
The biomarker development pipeline can be divided into the following phases: biomarker discovery, analytical validation, validation for clinical utility, regulatory qualification, and approval (Fig 2 ). Biomarker discovery is the initial preclinical description of an association with a disease process. Analytical validation involves the assessment of the performance of the assay in specific samples. How reliably does the test measure the analytes of interest in the patient specimen? Clinical validation assesses how robustly and reliably is the test result correlated with the clinical phenotype or outcome of interest. Regulatory qualification and approval is required prior to clinical implementation of a biomarker. Many candidate biomarkers in ARDS, such as TNF-α, IL-6, and IL-8, are based on preclinical investigations of dysregulated cellular pathways characteristic of acute lung injury14 , 15 , 44 and have proven to share an element of analytical validity.44 Ideally, novel biomarker assays would typically undergo optimization and confirmation of analytical validity.92 Unfortunately, without standardization of platform techniques and robust external analytical validation, it is difficult to rule out analytical flaws and laboratory errors in many studies claiming an association between blood biomarker candidates and ARDS. Over the past 60 years, immunoassays used for qualitative and quantitative detection of analytes have evolved from uniplex/conventional enzyme-linked immunosorbent assay formats that rely on colorimetric enzymatic substrates for detection to multiplex enzyme-linked immunosorbent assay systems that adopt chemiluminescent/fluorescent reporter systems.92 Contemporary multiplex immunoassay systems include platforms such as Luminex, Cytometric Bead Arrays, and Bio-PlexPro that employ a suspension format or platforms that rely on a planar format such as the Mesoscale Discovery Technology Platform (MSD) and the Q-Plex array (Quansys Biosciences).92 While these multiplex platforms present a theoretical benefit in terms of comprehensive profiling in complex phenotypes such as ARDS, challenges such as cross-reactivity of capture and/or detection antibodies may compromise readout viability.93 , 94 Fig. 3 summarizes the state of ARDS biomarkers in the development continuum and a majority of the current candidates have commercially available assays that have undergone a degree of analytical validation. However, details regarding the quality of samples and reproducibility of the platform are not always reported.92
Fig 2.
Steps in the biomarker development pipeline. There are 4 phases in the biomarker development pipeline. Biomarker discovery is the initial preclinical description of an association with a disease process. Analytical validation involves the assessment of the performance of the assay in specific samples. How reliably does the test measure the analytes of interest in the patient specimen? Clinical validation assesses how robustly and reliably is the test result correlated with the clinical phenotype or outcome of interest. Regulatory qualification and approval is required prior to clinical implementation of a biomarker. The FDA regulates initial biomarker qualification in the USA. IVD, in vitro diagnostic products are classified by the FDA to determine the appropriate premarket process. The FDA does not enforce premarket review of LDT – laboratory developed test – in vitro diagnostic test designed, manufactured and used within a single laboratory. CLIA, Clinical Laboratory Improvement Amendments, regulate laboratory testing and require clinical laboratories to be certified by the Center for Medicare and Medicaid Services (CMS) before they can accept human samples for diagnostic testing. CPT, Common Procedural Terminology.
Fig 3.
ARDS biomarkers in the development continuum. The majority of the current candidate biomarkers have laboratory developed or commercially available assays that have undergone some form of analytical validation. No biomarker has cleared the threshold for robust and reliable clinical validation. Biomarkers are listed by type of dysregulated pathway. Ang-2, angiopoietin 2; CC16, Clara cell secretary protein; HGF, hepatocyte growth factor; HTI56, human alveolar type I cell protein; KL-6, Krebs von den Lungen-6; HMGB1, high mobility group box nuclear protein 1; iCAM-1, intracellular adhesion molecule 1; IL-1β, interleukin 1 beta; IL-1RA, interleukin 1 receptor antagonist; IL-6, interleukin 6; IL-8, interleukin 8; KGF*, keratinocyte growth factor; MMP, matrix metalloproteinase; MIF, macrophage migration inhibitory factor; MIP-1α, macrophage inflammatory protein-1α; eNAMPT, extracellular nicotinamide phosphoribosyltransferase; PAI-1, plasminogen activator inhibitor-1; S1PR3, sphingosine-1phosphate receptor 3; SP-D, surfactant protein D; sRAGE, soluble receptor for advanced glycation end products; TNF-α, tumor necrosis factor alpha; VEGF, vascular endothelial growth factor; vWF, von Willebrand factor. Elastin*, Laminin*. *No evidence of analytical validation.
Validation for clinical utility should distinguish between at-risk ICU controls and ARDS patients and between ARDS survivors and nonsurvivors. Ideally, validation for clinical utility should include an assessment of performance against clinically meaningful outcomes in multiple prospective cohort studies. For ARDS, mortality is the universal endpoint, however, the context of use determines the endpoint chosen. As noted above, the historical focus in the ARDS biomarker research community has been on discovery. No ARDS biomarker candidate has cleared regulatory qualification and approval.
CHALLENGES TO SUCCESSFUL TRANSLATION OF ARDS BIOMARKERS
As noted above, general biomarker development, including specific utility in ARDS, remains a limited priority and pales in comparison to efforts in government-funded university laboratories where novel biomarker discovery is the focus and centers on demonstrating disease associations5 but do not address issues of clinical validation. The focus of these laboratories is entirely understandable given the lack of capacity and resources to undertake the stringent biomarker profiling required to attract investment in clinical trials.5 Unidentified flaws at the time of biomarker discovery can hinder the subsequent progression along the development pipeline. Such flaws include, but are not limited to, the poor quality of biospecimens (frequent freeze-thawing, etc.), insufficient sample numbers (leading to inadequate statistical power), and incomplete phenotypic clinical data linked to the assayed samples.5 Another challenge is the variability in test accuracy and reproducibility of analytical platforms, resulting in inconsistent performance of various measurement assays and inability to replicate the original claims. Publication bias in favor of positive results is another challenge to proper clinical validation. There are currently no standards, best practices, or guidelines to guide investigators in the ARDS biomarker research and development. A final concern is the relative absence of multidisciplinary coordinated efforts in the ARDS research community to address the unmet need for ARDS biomarkers which requires integrative and collaborative approaches.
THE WAY FORWARD
In order to address the major barriers of moving forward beyond association and toward causation, mechanism, and predicting response, researchers need to veer from working in “silos” and instead move to the forging of new collaborative, integrative approaches to biomarker development.5 This is important because specific failures of biomarkers begin at the discovery and analytical validation phases. The need for broadly accepted standards to inform every module and decision point of biomarker development cannot be overemphasized.5 In terms of clinical validation, beyond strategies such as increasing statistical power, deeper phenotyping to minimize heterogeneity, and more robust replication, there is a need for innovative study designs such as cell based screening of candidate biomarkers,95 single patient (N-of-1) designs based on biomarker profiles,96 adaptive signature designs97 and the use of mediation analysis98 or Mendelian randomization.99 Another emerging approach is to leverage the ready availability of rich and expansive datasets and advances in multi-omic technologies and computational platforms to identify novel biomarkers.100 , 101 Machine learning unsupervised algorithms capitalize on the vast amount of human genetic information in large populations, comparing transcriptomic, proteomic, and metabolomics profiling of patients with disease vs healthy individuals to identify novel biomarkers.102 Strategies to improve the rates of ARDS biomarker validation should include new collaborative research networks that include all stakeholder communities (researchers, funding agencies, and pharmaceutical companies) mobilizing resources and diverse expertise. For example, the national biomarker development alliance has proposed standards-based, end-to-end systems approach to facilitate the seamless flow of meritorious biomarker candidates within and across the modules of the discovery and development pipeline.5 The ongoing SARS-CoV-2/COVID-19 pandemic presents a unique opportunity given the dramatic increase in number of COVID-19-associated ARDS cases worldwide. Therefore, a national federal-funded ARDS biomarker consortium in partnership with industry would appear to serve as an excellent starting point.
CONCLUSIONS
A critical gap exists between the fast pace of biomarker discovery in ARDS and successful translation to clinical use. This gap underscores the fundamental biomarker conundrum across various acute and chronic disorders: how does a biomarker address a specific unmet need? In addition, this gap highlights the need to shift the paradigm from a focus on biomarker discovery to greater translational impact. In ARDS, the unmet need is for reliable validated ARDS biomarkers that minimize heterogeneity and allow for stratification of subject selection for enrollment in clinical trials, tailored therapies for specific endotypes as suggested by biomarkers, and a more streamlined drug approval process. This will require multilateral collaboration and, while challenging, has never before been as within reach as it is today.
Acknowledgments
Conflicts of Interest: All authors declare no relevant conflict of interest.
Funding: This work was supported by K08 HL141623 (C.B.); P01 HL126609 (J.G.N.G.); R41 HL147769 (J.G.N.G.); R42 HL145930 (J.G.N.G.).
All authors have read the journal's authorship agreement and the manuscript has been reviewed by and approved by all named authors. The authors did not have any editorial support for preparation of this manuscript.
References
- 1.Ginsburg G.S. Genomics-inspired biomarkers and diagnostics—where are they? Clin Chem. 2017;63:255–257. doi: 10.1373/clinchem.2016.266114. [DOI] [PubMed] [Google Scholar]
- 2.Sandhu C, Qureshi A, Emili A. Panomics for precision medicine. Trends Mol Med. 2018;24:85–101. doi: 10.1016/j.molmed.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mayeux R. Biomarkers: potential uses and limitations. NeuroRx. 2004;1:182–188. doi: 10.1602/neurorx.1.2.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amur S, LaVange L, Zineh I, Buckman-Garner S, Woodcock J. Biomarker qualification: toward a multiple stakeholder framework for biomarker development, regulatory acceptance, and utilization. Clin Pharmacol Ther. 2015;98:34–46. doi: 10.1002/cpt.136. [DOI] [PubMed] [Google Scholar]
- 5.Barker AD, Compton CC, Poste G. The National Biomarker Development Alliance accelerating the translation of biomarkers to the clinic. Biomark Med. 2014;8:873–876. doi: 10.2217/bmm.14.52. [DOI] [PubMed] [Google Scholar]
- 6.Wickstrom K, Moseley J. Biomarkers and surrogate endpoints in drug development: a European regulatory view. Invest Ophthalmol Visual Sci. 2017;58 doi: 10.1167/iovs.17-21778. Bio27-bio33. [DOI] [PubMed] [Google Scholar]
- 7.Burke H.B. Predicting clinical outcomes using molecular biomarkers. Biomark Cancer. 2016;8 doi: 10.4137/BIC.S33380. BIC.S33380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poste G. Bring on the biomarkers. Nature. 2011;469:156–157. doi: 10.1038/469156a. [DOI] [PubMed] [Google Scholar]
- 9.Kern SE. Why your new cancer biomarker may never work: recurrent patterns and remarkable diversity in biomarker failures. Cancer Res. 2012;72:6097–6101. doi: 10.1158/0008-5472.CAN-12-3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sauer JM, Porter AC. Preclinical biomarker qualification. Exp Biol Med (Maywood, NJ). 2018;243:222–227. doi: 10.1177/1535370217743949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mattes WB, Goodsaid F. Regulatory landscapes for biomarkers and diagnostic tests: qualification, approval, and role in clinical practice. Exp Biol Med (Maywood, NJ) 2018;243:256–261. doi: 10.1177/1535370217739629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Group F-NBW. Food and Drug Administration (US); Silver Spring, MD: 2016. BEST (Biomarkers, EndpointS, and other Tools) resource. [PubMed] [Google Scholar]
- 13.H.R. 6. 21st Century Cures Act . Government Printing Office; Washington, DC: 2016. 114th Congress. U.S. House. [Google Scholar]
- 14.Blondonnet R, Constantin JM, Sapin V, Jabaudon M. A pathophysiologic approach to biomarkers in acute respiratory distress syndrome. Dis Mark. 2016;2016 doi: 10.1155/2016/3501373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spadaro S, Park M, Turrini C. Biomarkers for acute respiratory distress syndrome and prospects for personalised medicine. J Inflamm. 2019;16:1. doi: 10.1186/s12950-018-0202-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Laorden MI, Lorente JA, Flores C, Slutsky AS, Villar J. Biomarkers for the acute respiratory distress syndrome: how to make the diagnosis more precise. Ann Transl Med. 2017;5:283. doi: 10.21037/atm.2017.06.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu AC, Kiley JP, Noel PJ. Current status and future opportunities in lung precision medicine research with a focus on biomarkers. An American Thoracic Society/National Heart, Lung, and Blood Institute Research statement. Am. J Respir Crit Care Med. 2018;198:e116–e136. doi: 10.1164/rccm.201810-1895ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyer NJ, Calfee CS. Novel translational approaches to the search for precision therapies for acute respiratory distress syndrome. Lancet Respir Med. 2017;5:512–523. doi: 10.1016/S2213-2600(17)30187-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walter JM, Wilson J, Ware LB. Biomarkers in acute respiratory distress syndrome: from pathobiology to improving patient care. Expert Rev Respir Med. 2014;8:573–586. doi: 10.1586/17476348.2014.924073. [DOI] [PubMed] [Google Scholar]
- 20.Capelozzi VL, Allen TC, Beasley MB. Molecular and immune biomarkers in acute respiratory distress syndrome: a perspective from Members of the Pulmonary Pathology Society. Arch Pathol Lab Med. 2017;141:1719–1727. doi: 10.5858/arpa.2017-0115-SA. [DOI] [PubMed] [Google Scholar]
- 21.Ranieri VM, Rubenfeld GD, Thompson BT. Acute respiratory distress syndrome: the Berlin Definition. JAMA. 2012;307:2526–2533. doi: 10.1001/jama.2012.5669. [DOI] [PubMed] [Google Scholar]
- 22.Slutsky AS, Ranieri VM. Ventilator-induced lung injury. New Engl J Med. 2013;369:2126–2136. doi: 10.1056/NEJMra1208707. [DOI] [PubMed] [Google Scholar]
- 23.Bellani G, Laffey JG, Pham T. Epidemiology, patterns of care, and mortality for patients with acute respiratory distress syndrome in intensive care units in 50 countries trends in acute respiratory distress syndrome in 50 countries. JAMA. 2016;315:788–800. doi: 10.1001/jama.2016.0291. [DOI] [PubMed] [Google Scholar]
- 24.Calfee CS, Delucchi K, Parsons PE, Thompson BT, Ware LB, Matthay MA. Subphenotypes in acute respiratory distress syndrome: latent class analysis of data from two randomised controlled trials. Lancet Respir Med. 2014;2:611–620. doi: 10.1016/S2213-2600(14)70097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calfee CS, Delucchi KL, Sinha P. Acute respiratory distress syndrome subphenotypes and differential response to simvastatin: secondary analysis of a randomised controlled trial. Lancet Respir Med. 2018;6:691–698. doi: 10.1016/S2213-2600(18)30177-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Califf RM. Biomarker definitions and their applications. Exp Biol Med (Maywood, NJ). 2018;243:213–221. doi: 10.1177/1535370217750088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Selleck MJ, Senthil M, Wall NR. Making meaningful clinical use of biomarkers. Biomark Insights. 2017;12 doi: 10.1177/1177271917715236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shirasawa M, Fujiwara N, Hirabayashi S. Receptor for advanced glycation end-products is a marker of type I lung alveolar cells. Genes Cells. 2004;9:165–174. doi: 10.1111/j.1356-9597.2004.00712.x. [DOI] [PubMed] [Google Scholar]
- 29.Neeper M, Schmidt AM, Brett J. Cloning and expression of a cell surface receptor for advanced glycosylation end products of proteins. J Biol Chem. 1992;267:14998–15004. [PubMed] [Google Scholar]
- 30.Jabaudon M, Berthelin P, Pranal T. Receptor for advanced glycation end-products and ARDS prediction: a multicentre observational study. Sci Rep. 2018;8:2603. doi: 10.1038/s41598-018-20994-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson MD, Widdicombe JH, Allen L, Barbry P, Dobbs LG. Alveolar epithelial type I cells contain transport proteins and transport sodium, supporting an active role for type I cells in regulation of lung liquid homeostasis. Proc Natl Acad Sci. 2002;99:1966–1971. doi: 10.1073/pnas.042689399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanford LE, Enghild JJ, Valnickova Z. Purification and characterization of mouse soluble receptor for advanced glycation end products (sRAGE) J Biol Chem. 2004;279:50019–50024. doi: 10.1074/jbc.M409782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jabaudon M, Blondonnet R, Roszyk L. Soluble receptor for advanced glycation end-products predicts impaired alveolar fluid clearance in acute respiratory distress syndrome. Am J Respir Crit Care Med. 2015;192:191–199. doi: 10.1164/rccm.201501-0020OC. [DOI] [PubMed] [Google Scholar]
- 34.Lucas R, Verin AD, Black SM, Catravas JD. Regulators of endothelial and epithelial barrier integrity and function in acute lung injury. Biochem Pharmacol. 2009;77:1763–1772. doi: 10.1016/j.bcp.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agrawal A, Matthay MA, Kangelaris KN. Plasma angiopoietin-2 predicts the onset of acute lung injury in critically ill patients. Am J Respir Crit Care Med. 2013;187:736–742. doi: 10.1164/rccm.201208-1460OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wada T, Jesmin S, Gando S. The role of angiogenic factors and their soluble receptors in acute lung injury (ALI)/ acute respiratory distress syndrome (ARDS) associated with critical illness. J Inflamm. 2013;10:6. doi: 10.1186/1476-9255-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shibuya M. Vascular endothelial growth factor and its receptor system: physiological functions in angiogenesis and pathological roles in various diseases. J Biochem. 2012;153:13–19. doi: 10.1093/jb/mvs136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ashbaugh D, Boyd Bigelow D, Petty T, Levine B. Acute respiratory distress in adults. Lancet. 1967;290:319–323. doi: 10.1016/s0140-6736(67)90168-7. [DOI] [PubMed] [Google Scholar]
- 39.Frerking I, Günther A, Seeger W, Pison U. Pulmonary surfactant: functions, abnormalities and therapeutic options. Intensive Care Med. 2001;27:1699–1717. doi: 10.1007/s00134-001-1121-5. [DOI] [PubMed] [Google Scholar]
- 40.Eisner MD, Parsons P, Matthay MA, Ware L, Greene K. Plasma surfactant protein levels and clinical outcomes in patients with acute lung injury. Thorax. 2003;58:983–988. doi: 10.1136/thorax.58.11.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McEver RP. Selectins: lectins that initiate cell adhesion under flow. Curr Opin Cell Biol. 2002;14:581–586. doi: 10.1016/s0955-0674(02)00367-8. [DOI] [PubMed] [Google Scholar]
- 42.Sakamaki F, Ishizaka A, Handa M. Soluble form of P-selectin in plasma is elevated in acute lung injury. Am J Respir Crit Care Med. 1995;151:1821–1826. doi: 10.1164/ajrccm.151.6.7539327. [DOI] [PubMed] [Google Scholar]
- 43.Boldt J, Wollbrück M, Kuhn D, Linke LC, Hempelmann G. Do plasma levels of circulating soluble adhesion molecules differ between surviving and nonsurviving critically iii patients. Chest. 1995;107:787–792. doi: 10.1378/chest.107.3.787. [DOI] [PubMed] [Google Scholar]
- 44.Cross LJ, Matthay MA. Biomarkers in acute lung injury: insights into the pathogenesis of acute lung injury. Crit Care Clin. 2011;27:355–377. doi: 10.1016/j.ccc.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kovach MA, Stringer KA, Bunting R. Microarray analysis identifies IL-1 receptor type 2 as a novel candidate biomarker in patients with acute respiratory distress syndrome. Respir Res. 2015;16:29. doi: 10.1186/s12931-015-0190-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Armstrong L, Millar AB. Relative production of tumour necrosis factor alpha and interleukin 10 in adult respiratory distress syndrome. Thorax. 1997;52:442–446. doi: 10.1136/thx.52.5.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cohen MJ, Brohi K, Calfee CS. Early release of high mobility group box nuclear protein 1 after severe trauma in humans: role of injury severity and tissue hypoperfusion. Crit Care. 2009;13:R174. doi: 10.1186/cc8152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yagmur E, Buendgens L, Herbers U. High mobility group box 1 as a biomarker in critically ill patients. J Clin Lab Anal. 2018;32:e22584. doi: 10.1002/jcla.22584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lai KN, Leung JC, Metz CN, Lai FM, Bucala R, Lan HY. Role for macrophage migration inhibitory factor in acute respiratory distress syndrome. J Pathol. 2003;199:496–508. doi: 10.1002/path.1291. [DOI] [PubMed] [Google Scholar]
- 50.Pohl J, Hendgen-Cotta UB, Stock P, Luedike P, Rassaf T. Elevated MIF-2 levels predict mortality in critically ill patients. J Crit Care. 2017;40:52–57. doi: 10.1016/j.jcrc.2017.03.012. [DOI] [PubMed] [Google Scholar]
- 51.Camp SM, Ceco E, Evenoski CL. Unique toll-like receptor 4 activation by NAMPT/PBEF induces NFkappaB signaling and inflammatory lung injury. Sci Rep. 2015;5:13135. doi: 10.1038/srep13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ye SQ, Simon BA, Maloney JP. Pre-B-cell colony-enhancing factor as a potential novel biomarker in acute lung injury. Am J Respir Crit Care Med. 2005;171:361–370. doi: 10.1164/rccm.200404-563OC. [DOI] [PubMed] [Google Scholar]
- 53.Bajwa EK, Yu CL, Gong MN, Thompson BT, Christiani DC. Pre-B-cell colony-enhancing factor gene polymorphisms and risk of acute respiratory distress syndrome. Crit Care Med. 2007;35:1290–1295. doi: 10.1097/01.CCM.0000260243.22758.4F. [DOI] [PubMed] [Google Scholar]
- 54.Hong SB, Huang Y, Moreno-Vinasco L. Essential role of pre-B-cell colony enhancing factor in ventilator-induced lung injury. Am J Respir Crit Care Med. 2008;178:605–617. doi: 10.1164/rccm.200712-1822OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ware LB, Bastarache JA, Wang L. Coagulation and fibrinolysis in human acute lung injury—new therapeutic targets. Keio J Med. 2005;54:142–149. doi: 10.2302/kjm.54.142. [DOI] [PubMed] [Google Scholar]
- 56.Prabhakaran P, Ware LB, White KE, Cross MT, Matthay MA, Olman MA. Elevated levels of plasminogen activator inhibitor-1 in pulmonary edema fluid are associated with mortality in acute lung injury. Am J Physiol-Lung Cell Mol Physiol. 2003;285:L20–L28. doi: 10.1152/ajplung.00312.2002. [DOI] [PubMed] [Google Scholar]
- 57.Ference BA, Ginsberg HN, Graham I. Low-density lipoproteins cause atherosclerotic cardiovascular disease. 1. Evidence from genetic, epidemiologic, and clinical studies. A consensus statement from the European Atherosclerosis Society Consensus Panel. Eur Heart J. 2017;38:2459–2472. doi: 10.1093/eurheartj/ehx144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Calfee CS, Ware LB, Eisner MD. Plasma receptor for advanced glycation end products and clinical outcomes in acute lung injury. Thorax. 2008;63:1083–1089. doi: 10.1136/thx.2008.095588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nakamura T, Sato E, Fujiwara N, Kawagoe Y, Maeda S, Yamagishi S. Increased levels of soluble receptor for advanced glycation end products (sRAGE) and high mobility group box 1 (HMGB1) are associated with death in patients with acute respiratory distress syndrome. Clin Biochem. 2011;44:601–604. doi: 10.1016/j.clinbiochem.2010.12.014. [DOI] [PubMed] [Google Scholar]
- 60.Terpstra ML, Aman J, van Nieuw Amerongen GP, Groeneveld AB. Plasma biomarkers for acute respiratory distress syndrome: a systematic review and meta-analysis. Crit Care Med. 2014;42:691–700. doi: 10.1097/01.ccm.0000435669.60811.24. [DOI] [PubMed] [Google Scholar]
- 61.Calfee CS, Gallagher D, Abbott J, Thompson BT, Matthay MA. Plasma angiopoietin-2 in clinical acute lung injury: prognostic and pathogenetic significance. Crit Care Med. 2012;40:1731–1737. doi: 10.1097/CCM.0b013e3182451c87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Calfee CS, Janz DR, Bernard GR. Distinct molecular phenotypes of direct vs indirect ARDS in single-center and multicenter studies. Chest. 2015;147:1539–1548. doi: 10.1378/chest.14-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bime C, Casanova N, Oita RC. Development of a biomarker mortality risk model in acute respiratory distress syndrome. Crit Care. 2019;23:410. doi: 10.1186/s13054-019-2697-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pregernig A, Müller M, Held U, Beck-Schimmer B. Prediction of mortality in adult patients with sepsis using six biomarkers: a systematic review and meta-analysis. Ann Intensive Care. 2019;9:125. doi: 10.1186/s13613-019-0600-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dustin ML, Rothlein R, Bhan AK, Dinarello CA, Springer TA. Induction by IL 1 and interferon-gamma: tissue distribution, biochemistry, and function of a natural adherence molecule (ICAM-1) J Immunol. 1986;137:245–254. [PubMed] [Google Scholar]
- 66.Roebuck KA, Finnegan A. Regulation of intercellular adhesion molecule-1 (CD54) gene expression. J Leukocyte Biol. 1999;66:876–888. doi: 10.1002/jlb.66.6.876. [DOI] [PubMed] [Google Scholar]
- 67.Calfee CS, Eisner MD, Parsons PE. Soluble intercellular adhesion molecule-1 and clinical outcomes in patients with acute lung injury. Intensive Care Med. 2009;35:248–257. doi: 10.1007/s00134-008-1235-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Agouridakis P, Kyriakou D, Alexandrakis MG. The predictive role of serum and bronchoalveolar lavage cytokines and adhesion molecules for acute respiratory distress syndrome development and outcome. Respir Res. 2002;3:27. doi: 10.1186/rr193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Flori HR, Ware LB, Glidden D, Matthay MA. Early elevation of plasma soluble intercellular adhesion molecule-1 in pediatric acute lung injury identifies patients at increased risk of death and prolonged mechanical ventilation. Pediatric Crit Care Med. 2003;4:315–321. doi: 10.1097/01.PCC.0000074583.27727.8E. [DOI] [PubMed] [Google Scholar]
- 70.Conway EM. Thrombomodulin and its role in inflammation. Semin Immunopathol. 2012;34:107–125. doi: 10.1007/s00281-011-0282-8. [DOI] [PubMed] [Google Scholar]
- 71.Koutsi A, Papapanagiotou A, Papavassiliou AG. Thrombomodulin: from haemostasis to inflammation and tumourigenesis. Int J Biochem Cell Biol. 2008;40:1669–1673. doi: 10.1016/j.biocel.2007.06.024. [DOI] [PubMed] [Google Scholar]
- 72.Finigan JH, Dudek SM, Singleton PA. Activated protein C mediates novel lung endothelial barrier enhancement: role of sphingosine 1-phosphate receptor transactivation. J Biol Chem. 2005;280:17286–17293. doi: 10.1074/jbc.M412427200. [DOI] [PubMed] [Google Scholar]
- 73.Nold MF, Nold-Petry CA, Fischer D. Activated protein C downregulates p38 mitogen-activated protein kinase and improves clinical parameters in an in-vivo model of septic shock. Thromb Haemostasis. 2007;98:1118–1126. doi: 10.1160/th07-01-0052. [DOI] [PubMed] [Google Scholar]
- 74.Christiaans SC, Wagener BM, Esmon CT, Pittet JF. Protein C and acute inflammation: a clinical and biological perspective. Am J Physiol-Lung Cell Mol Physiol. 2013;305:L455–L466. doi: 10.1152/ajplung.00093.2013. [DOI] [PubMed] [Google Scholar]
- 75.Ware LB, Matthay MA, Parsons PE, Thompson BT, Januzzi JL, Eisner MD. Pathogenetic and prognostic significance of altered coagulation and fibrinolysis in acute lung injury/acute respiratory distress syndrome. Crit Care Med. 2007;35:1821–1828. doi: 10.1097/01.CCM.0000221922.08878.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Koch A, Weiskirchen R, Krusch A. Visfatin serum levels predict mortality in critically ill patients. Dis Mark. 2018;2018 doi: 10.1155/2018/7315356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee KA, Gong MN. Pre-B-cell colony-enhancing factor and its clinical correlates with acute lung injury and sepsis. Chest. 2011;140:382–390. doi: 10.1378/chest.10-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reilly JP, Christie JD, Meyer NJ. Fifty years of research in ARDS. Genomic contributions and opportunities. Am J Respir Crit Care Med. 2017;196:1113–1121. doi: 10.1164/rccm.201702-0405CP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Simon BA, Easley RB, Grigoryev DN. Microarray analysis of regional cellular responses to local mechanical stress in acute lung injury. Am J Physiol Lung Cell Mol Physiol. 2006;291:L851–L861. doi: 10.1152/ajplung.00463.2005. [DOI] [PubMed] [Google Scholar]
- 80.Grigoryev DN, Ma SF, Irizarry RA, Ye SQ, Quackenbush J, Garcia JG. Orthologous gene-expression profiling in multi-species models: search for candidate genes. Genome Biol. 2004;5:R34. doi: 10.1186/gb-2004-5-5-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wurfel MM. Microarray-based analysis of ventilator-induced lung injury. Proc Am Thoracic Soc. 2007;4:77–84. doi: 10.1513/pats.200608-149JG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sun X, Singleton PA, Letsiou E. Sphingosine-1-phosphate receptor-3 is a novel biomarker in acute lung injury. Am J Respir Cell Mol Biol. 2012;47:628–636. doi: 10.1165/rcmb.2012-0048OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sun X, Ma SF, Wade MS. Functional promoter variants in sphingosine 1-phosphate receptor 3 associate with susceptibility to sepsis-associated acute respiratory distress syndrome. Am J Physiol. Lung Cell Mol Physiol. 2013;305:L467–L477. doi: 10.1152/ajplung.00010.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Christie JD, Ma SF, Aplenc R. Variation in the myosin light chain kinase gene is associated with development of acute lung injury after major trauma. Crit Care Med. 2008;36:2794–2800. doi: 10.1097/ccm.0b013e318186b843. [DOI] [PubMed] [Google Scholar]
- 85.Bime C, Pouladi N, Sammani S. Genome-wide association study in African Americans with acute respiratory distress syndrome identifies the selectin P ligand gene as a risk factor. Am J Respir Crit Care Med. 2018;197:1421–1432. doi: 10.1164/rccm.201705-0961OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lawlor DA, Harbord RM, Sterne JAC, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27:1133–1163. doi: 10.1002/sim.3034. [DOI] [PubMed] [Google Scholar]
- 87.Reilly JP, Wang F, Jones TK. Plasma angiopoietin-2 as a potential causal marker in sepsis-associated ARDS development: evidence from Mendelian randomization and mediation analysis. Intensive Care Med. 2018;44:1849–1858. doi: 10.1007/s00134-018-5328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jones TK, Feng R, Kerchberger VE. Plasma sRAGE acts as genetically regulated causal intermediate in sepsis-associated ARDS. Am J Respir Crit Care Med. 2019;201(1):47–56. doi: 10.1164/rccm.201810-2033OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fremont RD, Koyama T, Calfee CS. Acute lung injury in patients with traumatic injuries: utility of a panel of biomarkers for diagnosis and pathogenesis. J Trauma. 2010;68:1121–1127. doi: 10.1097/TA.0b013e3181c40728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ware LB, Koyama T, Zhao Z. Biomarkers of lung epithelial injury and inflammation distinguish severe sepsis patients with acute respiratory distress syndrome. Crit Care. 2013;17:R253. doi: 10.1186/cc13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ware LB, Koyama T, Billheimer DD. Prognostic and pathogenetic value of combining clinical and biochemical indices in patients with acute lung injury. Chest. 2010;137:288–296. doi: 10.1378/chest.09-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tighe PJ, Ryder RR, Todd I, Fairclough LC. ELISA in the multiplex era: potentials and pitfalls. Proteomics – Clin Applic. 2015;9:406–422. doi: 10.1002/prca.201400130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pla-Roca M, Leulmi RF, Tourekhanova S. Antibody colocalization microarray: a scalable technology for multiplex protein analysis in complex samples. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.011460. M111.011460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Juncker D, Bergeron S, Laforte V, Li H. Cross-reactivity in antibody microarrays and multiplexed sandwich assays: shedding light on the dark side of multiplexing. Curr Opin Chem Biol. 2014;18:29–37. doi: 10.1016/j.cbpa.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 95.Balis FM. Evolution of anticancer drug discovery and the role of cell-based screening. JNCI: J Natl Cancer Inst. 2002;94:78–79. doi: 10.1093/jnci/94.2.78. [DOI] [PubMed] [Google Scholar]
- 96.Demeyin WA, Frost J, Ukoumunne OC, Briscoe S, Britten N. N of 1 trials and the optimal individualisation of drug treatments: a systematic review protocol. Syst Rev. 2017;6:90. doi: 10.1186/s13643-017-0479-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Freidlin B, Simon R. Adaptive signature design: an adaptive clinical trial design for generating and prospectively testing a gene expression signature for sensitive patients. Clin Cancer Res. 2005;11:7872–7878. doi: 10.1158/1078-0432.CCR-05-0605. [DOI] [PubMed] [Google Scholar]
- 98.Richmond RC, Hemani G, Tilling K, Davey Smith G, Relton CL. Challenges and novel approaches for investigating molecular mediation. Hum Mol Genet. 2016;25(R2):R149–r156. doi: 10.1093/hmg/ddw197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Burgess S, Daniel RM, Butterworth AS, Thompson SG. Network Mendelian randomization: using genetic variants as instrumental variables to investigate mediation in causal pathways. Int J Epidemiol. 2015;44:484–495. doi: 10.1093/ije/dyu176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sodek KL, Evangelou AI, Ignatchenko A. Identification of pathways associated with invasive behavior by ovarian cancer cells using multidimensional protein identification technology (MudPIT) Mol bioSyst. 2008;4:762–773. doi: 10.1039/b717542f. [DOI] [PubMed] [Google Scholar]
- 101.Cohen Freue GV, Meredith A, Smith D. Computational biomarker pipeline from discovery to clinical implementation: plasma proteomic biomarkers for cardiac transplantation. PLOS Comput Biol. 2013;9 doi: 10.1371/journal.pcbi.1002963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vamathevan J, Clark D, Czodrowski P. Applications of machine learning in drug discovery and development. Nat Rev Drug Discov. 2019;18:463–477. doi: 10.1038/s41573-019-0024-5. [DOI] [PMC free article] [PubMed] [Google Scholar]