Abstract
The current pandemic of 2019 novel coronavirus disease (COVID-19) caused by a novel virus strain, 2019-nCoV/SARS-CoV-2 have posed a serious threat to global public health and economy. It is largely unknown how the human immune system responds to this infection. A better understanding of the immune response to SARS-CoV-2 will be important to develop therapeutics against COVID-19. Here, we have used transcriptomic profile of human alveolar adenocarcinoma cells (A549) infected with SARS-CoV-2 and employed a network biology approach to generate human-virus interactome. Network topological analysis discovers 15 SARS-CoV-2 targets, which belongs to a subset of interferon (IFN) stimulated genes (ISGs). These ISGs (IFIT1, IFITM1, IRF7, ISG15, MX1, and OAS2) can be considered as potential candidates for drug targets in the treatments of COVID-19. We have identified significant interaction between ISGs and TLR3 agonists, like poly I: C, and imiquimod, and suggests that TLR3 agonists can be considered as a potential drug for drug repurposing in COVID-19. Our network centric analysis suggests that moderating the innate immune response is a valuable approach to target COVID-19.
Keywords: COVID-19, Network biology, Immune response, Interferon stimulating genes, Drug repurposing
Highlights
-
•
Differential gene expression analysis of SARS-CoV-2 infected transcriptome
-
•
Network based Human-SRAS-CoV-2 interactome analysis
-
•
Interferon (IFN) stimulated genes (ISGs) are the most important targets.
-
•
TLR3 agonists, like poly I:C, and imiquimod are identified as potential drugs.
-
•
Targeting the innate immune response is a valuable approach against COVID-19.
1. Introduction
The current pandemic of Coronavirus Disease-2019 (COVID-19) has led to over 5 million confirmed cases and almost 3.5 lakhs fatalities worldwide, since its emergence in late 2019. COVID-19 is caused by a novel virus coronavirus strain, SARS-CoV-2, an enveloped, positive-sense, single-stranded RNA, β-coronavirus of the family Coronaviridae [1].
Nearly 80% of COVID-19 patients show mild symptoms, such as cough and fever, and do not require hospitalization, but the remaining 20% do require [2]. However, of those 20%, almost half of them develop severe respiratory failure in the form of fatal acute respiratory distress syndrome (ARDS) [3]. Along with this fatal form, dysregulated immune response also mediated the severe COVID-19 pathogenesis. The immune dysregulation, called hypercytokinemia or “cytokine storm,” is frequently associated with ARDS [4]. Till date, it remains unclear how SARS-CoV-2 disrupt the host innate immune response. Several recent studies have reported the dysregulated secretion of proinflammatory cytokines in COVID-19 [[5], [6], [7]]. Recently, Melo et al. [5] reported a moderate interferon (IFN) response to SARS-CoV-2 infection in primary cells and showed that IFN can reduce SARS-CoV-2 replication in vitro. Zhou et al. [7] and Xiong et al. [6] recently examined the innate immune response to SARS-CoV-2 infection in bronchoalveolar lavage fluid (BALF). The differentially expressed genes were enriched in inflammatory pathways including Chemokine Signaling [7]. Whereas, in studies by Xiong et al. [6], the upregulated genes were largely related to viral infection with the most enriched biological processes being protein targeting to membrane and ER. These studies reported the significant upregulation of a subset of interferon-stimulated genes (ISGs) which are directly related to antiviral activity, such as ISG15, IFIH1, MX1, OAS1-3, IFITMs etc. These studies along with others revealed prominent role of innate immune response to COVID-19 infection [[8], [9], [10], [11]]. The two important themes are that COVID-19 consistently results in the upregulation of chemokines, and increased levels of proinflammatory cytokines such as IL-1β, IL-2, and IL-6, thus leads to tissue damage. The second is that SARS-CoV-2 triggered a robust IFN response, marked by the upregulation of several ISGs in the lungs.
Like other viruses, novel SARS-CoV-2 also utilizes host machinery for their growth and survival during infection. Systematic investigation of virus–host protein–protein interactions (PPIs) offers an effective way toward elucidating the mechanisms of viral infection and drug repurposing [12]. It has also been reported that SARS-CoV-2 can establish a higher rate of infectivity if it acquires combinatorial mutations at the S-protein/ACE2 interfacial residues [13]. Several studies have shown that targeting cellular antiviral targets could be considered as a novel strategy for the development of effective treatments for viral infections, including SARS-CoV [14], MERS-CoV [14], Ebola virus [15], Zika virus [16], and more recently in SARS-CoV-2 [17].
There are no vaccines or drugs approved for the novel COVID-19 infection yet, but more than 80 clinical trials have been launched to test coronavirus treatments. The current COVID-19 pandemic leads the scientific community to focus seriously on drug repurposing to tackle the COVID-19 infection [[18], [19], [20], [21], [22]]. Toward this goal, in this study, we have generated a human-SARS-CoV-2 interactome based on recently published RNA-Seq analysis of human adenocarcinomic alveolar basal epithelial (A549) cells infected with SARS-CoV-2, and identified disease-related functional genes that will provide the insights into the patho-mechanisms of COVID-19. Moreover, drug-protein interactions of these hub proteins, investigated in this study will leads to the identification of potential candidate drugs for repurposing.
2. Materials and methods
2.1. Data collection
We have downloaded the differential gene expression data of human alveolar adenocarcinoma cells infected with COVID-19 from Melo et.al study [23]. We have also downloaded all the possible drug-gene interactions data from Drug-bank database and Comparative Toxicogenomic Database (CTD), along with drug-disease interactions as well [24,25].
2.2. Identification of significantly dysregulated genes
Melo et.al [26] previously described 120 differentially expressed genes (DEGs) based on p-value cut-off (p-val < 0.05). Identification of significantly dysregulated genes on the basis of P-value is not quite precise. The amount of change in the expression of genes (log2fold change) should also be considered. We reanalysed the data with q-value cut-off (FDR < 0.05), as well as applied log2fold change cut-off (log2fold ≥ 2) for identifying significantly dysregulated genes from the data of 120 genes.
2.3. Gene ontology and pathway enrichment analysis
For comprehensively analyzing the biological function of DEGs, we used database for annotation, visualization and integrated discovery (DAVID) version 6.7 [27]. This database uses the GO and the Kyoto encyclopedia of genes and genomes (KEGG) analysis for analyzing the DEGs. The gene ontology analysis included the annotation at biological level, cellular level and molecular level. The pathways and functions with FDR < 0.05 were considered significant.
2.4. Protein-protein interaction study
Using the DEGs list of Melo et al. [26], protein-protein interaction network was constructed using STRING plugin of cytoscape tool [28]. STRING plugin uses co-expression, text-mining, gene fusion, neighbourhood, and experimental data for preparing PPI network. Cluego tool was used for identifying the biological roles of DEGs and DAVID tool was used for gene ontology (GO) analysis.
2.5. Hub genes identification
In this PPI network, each node indicates a protein and an edge indicates an interaction between proteins. We have also calculated the network's topological parameters such as degree centrality (k), betweenness centrality (C b), closeness connectivity and eccentricity using network analyzer plugin of the cytoscape tool. Hub genes are the highly connected genes having high correlation with other genes in the network. Any changes in the expression of hub genes have the potential to influence the major part of the network. Genes having high connectivity can transfer information rapidly in the network as compare to other genes. Top genes which have the higher degree of connectivity (k) and betweenness centrality value were considered as hub genes.
Degree centrality (k) is value assign to each node purely based on number of links held by it. Degree indicates the number of interactions held by a node with other nodes in the network and helps in measuring the node significance in controlling the network.
| (1) |
where, K u is the node set containing all the neighbours of node u, and w(u,v) is the edge weight connecting node u with node v.
Betweenness centrality (C b) measures how many times a node falls on the shortest path with other neighbouring nodes. It characterizes a nodes ability to control the signal processing and information flow in the network.
| (2) |
where p(k,u,f) is the number of interactions from k to f that passes through u, and p(k,f) represents total number of shortest interactions between node k and f.
Biological process enrichment analysis and GO analysis of the hub genes were done using Cluego and DAVID tool respectively.
2.6. Drug-gene interaction analysis
Drug–target interaction information for the hub genes was collected from the DrugBank database [24], and Comparative Toxicogenomics Database (CTD) [25]. The predicted drugs for hub genes through the gene-drug interaction databases were used for constructing drug-protein network using STITCH database [29]. STITCH is a database known to predict functional and physical interaction between chemicals/drugs and genes. The interactions in STITCH database is derived from five main sources, mainly by automated text-mining, high-throughput lab experiments, co-expression interaction data, interaction prediction by genomic context and by previous knowledges from databases. For each interaction between drug and gene, a combined score was calculated by STITCH database. The combined score was calculated by combining the probabilities of interaction from different evidence channels and corrected probability of randomly observing an interaction. The drug-gene interactions having network score more then 0.9 was consider significant and positively hit.
3. Results
3.1. Generation of human- SARS-CoV-2 Interactome by using transcriptome study
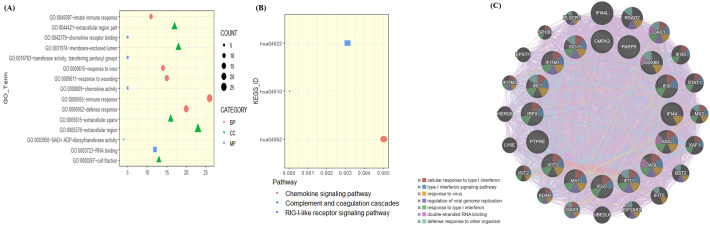
It is likely that the outcome of SARS-CoV-2 infection can largely be determined by the interaction between the host proteins and virus proteins. To build the human-SARS-CoV-2 interactome, we have used the transcriptome data of human alveolar adenocarcinoma (A549) cells infected with COVID-19 from Melo et al. [23]. Melo et al. identified 120 dysregulated genes with p-adj < 0.05, in which majority of genes were getting upregulated. Gene Ontology (GO) analysis of dysregulated genes using DAVID [30] revealed that the dysregulated genes were enriched in regulation of defence response to virus, regulation of protein export from nucleus, response to interferons, and regulation of response to cytokine stimulus (Fig. 1A). KEGG pathway enrichment analysis reveals the role of dysregulated genes in chemokine signaling pathways, complement and coagulation pathways and RIG-I-like receptor signaling pathways (Fig. 1B).
Fig. 1.
Enrichment analysis of significantly upregulated genes. (A) GO pathway showed that upregulated DEGs were particularly enriched in which Biological process, Molecular functions, and Cell component. (B) The most significantly enriched KEGG pathway of the upregulated DEGs. (C) Gene network of 16 upregulated genes derived from GeneMANIA along with functional enrichment.
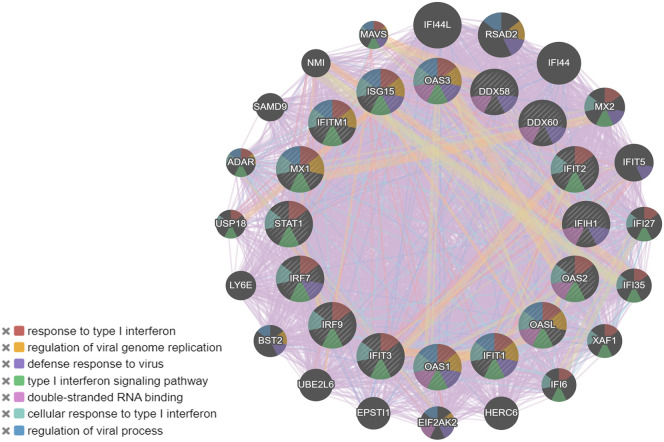
We reanalysed the transcriptomic gene expression signature based on log2fold change >2 and adj-p value < 0.05. We have identified 16 significantly dysregulated genes and of these genes, IFIT1, IFITM1, IRF7, ISG15, MX1, and OAS2 were highly upregulated. The GeneMANIA webserver was used to predict interactions between these DEGs genes in the network using the GO term “biological process” and source organism Homo sapiens as additional parameters. The gene enrichment of theses DEGs highlighted GO terms including response to virus, ribonucleotide binding and IFN-related signaling pathway (Fig. 1C). Overall, the analysis demonstrated that the upregulated genes are mainly linked to the host response to SARS-CoV-2 infection, type I interferon signaling and the cytokine-mediated signaling pathway.
We used the PPI network analysis to construct the interactome of the DEGs for COVID-19. For this, we used the STRING plugin of cytoscape tool [28], and obtained a PPI network with 117 nodes and 905 edges from the 120 dysregulated genes (Fig. 2A). The proteins are ranked based on their degree connectivity and betweenness centrality scores (Supporting information Table 1). Among these, STAT1, IRF7, IFIH1, MX1, ISG15, IFIT3, OAS2, and DDX58 has high degree and betweenness centrality values.
Fig. 2.
Human-SARS-CoV-2 interactome. (A) The human PPI network analysis of the 120 upregulated genes, visualized by Cytoscape tool. Nodes with bigger size are considered as hubs in this network and are represented in diamond shape with red colour. (B) Functional enrichment analysis of the upregulated 120 genes that characterized the host response of COVID-19.
To obtain a more in-depth understanding of the interactome, GO function and KEGG pathway analysis were applied using DAVID (Fig. 2B and Supporting information Table 2). GO analysis results showed that in the biological process, the interacting genes were mainly enrichment in the regulation of defence response to virus, innate immune response, inflammatory response, and also played an important role in complement activation, and response to nutrient and extracellular stimulus (Fig. 2B). The cellular components are significantly located in the extracellular region and membrane fraction, etc. Molecular functions were mainly enriched in chemokines or cytokine activity, RNA binding, transferase activity, and calcium ion binding. KEGG pathway enrichment analysis revealed the role of interacting genes in complement and coagulation pathways, RIG-1-like receptor signaling pathways, and chemokine signaling pathways (Supporting information Table 2).
Overall, the network analysis indicates that SARS-CoV-2 targets the proteins in the IFN signaling pathway to evade the immune system. This highlights the key role of the IFN-mediated antiviral responses.
3.2. Network topology-based functional analyses reveal that SARS-CoV-2 targets core signaling pathways of the host network
Several virus-human interactome have shown that viral proteins significantly interacts with hub proteins of a network, and thus the topological features of a network can be used to predict viral targets [[31], [32], [33], [34]]. In this regard, two topological features, degree (number of connections) and betweenness (the fraction of all shortest paths that include a node within a network), were calculated to identify candidate hub nodes using the Cytoscape's Network analyzer tool.
Collectively, we have identified top 15 nodes with high degree of connectivity and Betweenness value and was subsequently considered as hub genes in the network (Table 1 ).
Table 1.
Top 15 proteins based on the topological properties in PPI network of 120 dysregulated genes, and arranged on the basis of the decreasing degree of connectivity value.
| Hub genes | Degree | Betweenness centrality |
|---|---|---|
| STAT1 | 55 | 0.247 |
| IRF7 | 48 | 0.041 |
| IFIH1 | 45 | 0.044 |
| MX1 | 45 | 0.013 |
| ISG15 | 44 | 0.051 |
| IFIT3 | 44 | 0.028 |
| OAS2 | 44 | 0.012 |
| DDX58 | 43 | 0.015 |
| IRF9 | 42 | 0.019 |
| IFIT1 | 42 | 0.007 |
| OAS1 | 42 | 0.006 |
| OAS3 | 42 | 0.006 |
| DDX60 | 41 | 0.027 |
| OASL | 41 | 0.006 |
| IFIT2 | 41 | 0.003 |
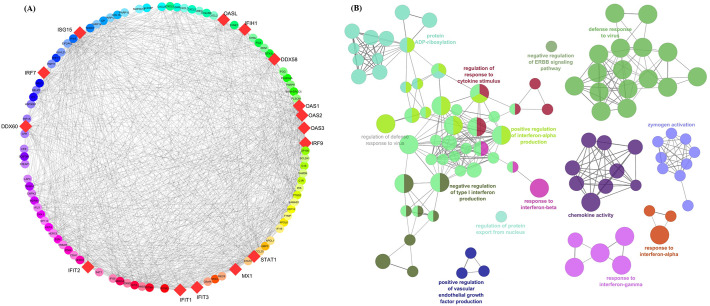
Biological process enrichment analysis of 15 hub genes using GeneMania webserver and GO analysis revealed their role in defence mechanism to viral response, cellular response to type I interferon, regulation of viral genome replication, double-stranded RNA binding, type I interferon signaling pathway, and regulation of innate immune response (Fig. 3 and Supporting information Table 3).
Fig. 3.
The PPIs network and gene enrichment analysis of 15 highly interactive Interferon stimulating genes (ISGs) along with functional enrichment.
These identified hub genes belong to the family of Interferon stimulating genes (ISGs). ISGs including IFIT and IFITM, ISG15, IFIH1, MX1, IRF7, OAS 1-3 and STAT1are known to potentiate IFN signaling and thus exert antiviral activity. These ISGs fall into two categories, one that is direct effectors of the innate immune response including: MX1, IFITM3, ISG15, IFIH1, and IRF-7, and the other includes the induction of viral RNA sensors such as DDX58 and the OAS1-3 genes. In addition to antiviral activity, ISGs may exert diverse functions including RNA and nucleotide binding, IFN regulation and inflammation regulation.
MX1, OAS1-3, IFIH1, IFITM1-3, ISG15 etc. are highly expressed in respiratory airways [35,36] and are associated with viral entry-associated ACE2 gene, as shown in single-cell RNA-sequencing data from various tissues from human [37,38]. Given the high expression of the viral entry-associated genes, it is reasonable that these nasal epithelial cells are trained to express these immune-associated genes to decrease viral susceptibility.
Further, we used Enrichr [39], a web-based server for gene-set enrichment analysis and provides different summaries of collective functions of gene lists. Interestingly, disease enrichment analysis demonstrated that the signature was highly associated with other viral diseases like West nile encephalitis, Tick-borne encephalitis, Dengue fever, Chikungunya, and SARS. Also, these hub genes are involved in the mammalian phenotype including increased susceptibility to viral infection induced morbidity/mortality, decreased interferon secretion, increased IgG level, abnormal T cell activation, and decreased interleukin-6 secretion.
Overall, these results clearly showed that the signatures are largely involved in innate immune response following SARS-CoV-2 infection (Supporting information Table 4).
3.3. Network based drug repurposing suggests toll like receptor 3 (TLR3) agonist as potential drugs to target COVID-19
Further using Drugbank database [24] and Comparative Toxicogenomics Database (CTD) [25], we identified the possible drugs which are known to have possible interaction with the hub genes (Supporting information Table 5). In total, we have identified 185 drug molecules which are known to interact with the hub genes (Supporting information Fig. 1). Apart from protein-drug interaction, we also looked for drug-disease interaction, i.e. which drug is used in which disease.
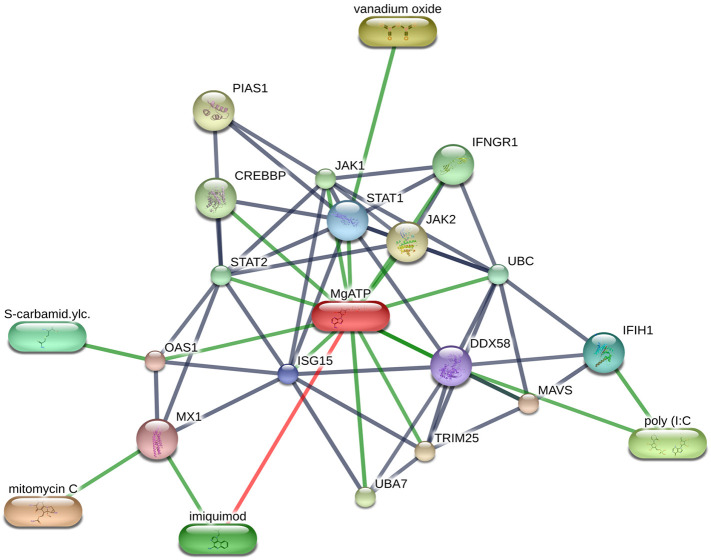
Based on the drug-gene interaction results, we used STITCH database [29] for final categorization of drug-gene interaction network based on interaction score which is ≥0.9. Interestingly, we found five proteins MX1, OAS1, STAT1, DDX58, and ISG15 were highly correlated to rare disease phenotype as well as to SARS in Enrichr database. These five proteins were also the most common influential proteins according to GO analysis. The STITCH drug-protein interaction network analysis of these five proteins has shown significant interaction with drugs/compound (Fig. 4 ). As shown in Fig. 4, MX1 gene interacts with Mitomycin-C, an antitumor and imiquimod, an immune response modifier. IFIH1 interacts with Polyinosinic:polycytidylic acid (poly I:C), which is an immunostimulant. OAS1 showed significant interaction with S-carbamidomethylcysteine (Cysteine-S-acetamide) and with MgATP. STAT1 interacts with Vanadium oxide and MgATP whereas, DDX58 and ISG15 interact with MgATP. Some of these identified drugs are used for induction of IFNs and thus play a key role in the body defence against SARS-CoV-2s infection.
Fig. 4.
Drug-gene interaction network of five important ISGs and their related drugs using STITCH database. Green colour interactions between drug and genes were significant interaction with combined score > 0.9. The network shows poly (I:C), Imiquimod, Mitomycin C and other related drugs to ISGs network.
4. Discussion
Here, we generated a comprehensive human-SARS-CoV-2 interactome from transcriptome studies of a lung cell infected with CoVID-19. The PPI network analysis indicates that the pathways are enriched in host response to virus infection, type I interferons signaling, and cytokine activation. Network topology analyses identified 15 high-value targets of SARS CoV-2, which belongs to a subset of canonical ISGs. These ISGs are largely involved in regulation of defence response to virus, innate immune response, inflammatory response, and RNA binding [40,41].
An interferon-inducible protein, MX Dynamin Like GTPase (MX1) is associated with influenza and viral encephalitis infection [42,43]. Also notable is the role of the Interferon Induced Protein with Tetratricopeptide Repeats 1 (IFIT1) and DExD/H-Box Helicase 58 (DDX58) as an antiviral activity. In Hepatitis E virus infection, polymerase binds to IFIT1 protecting the viral RNA from translation inhibition mediated by IFIT1 that boosts the interferon response in murine macrophage-like cells [44]. Recently, the significant upregulation of IFIT1-3, and DDX58 gene expression under COVID-19 viral infection has been reported [26]. In response to viral infections, several genes of host such as OAS1-3, IRF7, IRF9, STAT1 and IFIH1 are highly expressed and are highly correlated with host response to viral infections [[45], [46], [47]].
Studies show that CoVs are equipped with strategies to antagonize the IFN signaling pathway that facilitates the virus to escape host immune response. SARS-CoV escapes the host IFN signaling as its ORF6 protein blocks the expression of STAT1-activated genes [48]. Similarly, in MERS-CoV ORF4b inhibits IRF3 and IRF7 to antagonize the antiviral IFN-β response [49]. Additionally, papain-like proteases (PLPs) are expressed in both SARS-CoV and MERS-CoV that enables to delay the host immune response. Coronaviruses engages in interactions with IFN stimulated gene 15 (ISG15) and antagonizing the IFN-mediated antiviral response [50,51]. The antiviral activity of ISG15 has been shown in several viruses including human cytomegalovirus [52], HIV [53], West Nile virus [54], swine fever virus [55], MERS-CoV [56], and SARS-CoV [57].
IFNs play a key role in the body defence against viral infections, and in this regard, we here showed some candidate drugs for repurposing. One of these drugs is poly (I:C) (polyinosinic:polycytidylic acid), a synthetic double-stranded RNA immune-stimulant, which is used as adjuvant in vaccine production [58]. It is agonist for toll like receptor 3 (TLR3) which induces the expression of IFNs. Many studies demonstrated that the TLR3 agonists, poly ICLC and poly (I:C) increases the production of IFN-α, -β, and -γ, which inhibited CoV replication and minimized the inhibitory effects of CoV on IFN signaling pathways [[59], [60], [61]]. Interestingly, chloroquine, a recently proposed drugs for COVID-19 [62], inhibits poly (I:C)-mediated IFN-β induction [63]. Therefore, TLR3 agonists can be a considered as potential drugs for repurposing in COVID-19. In addition to TLR-3 agonists TLR7 agonists such as imiquimod [64], can induce IFN production in the human body as well. Imiquimod is a strong inducer of IFN-α and several proinflammatory mediators, including TNF-α, IL-12, and chemokines [65].
The other drug predicted to bind MX1 and OAS, Mitomycin C is a cancer drug that is used in the treatment of bladder, colon, and breast cancers. It functions as an alkylating agent that causes cross-linking of DNA and inhibits RNA as well as protein synthesis [66,67]. At high concentration, Mitomycin C inhibits the replication of influenza virus by blocking the RNA synthesis [68]. Mitomycin C has also been shown to inhibit B cell, T cell, and macrophage proliferation in vitro and impair antigen presentation, as well as the secretion of IFN-γ, TNF-α, and IL-2. Vanadium oxide, an activator of STAT-1, involved in immune-regulating mechanisms, including immune suppression and inflammation downregulation by stimulating and activating B/T cells.3 [69,70]
5. Conclusion
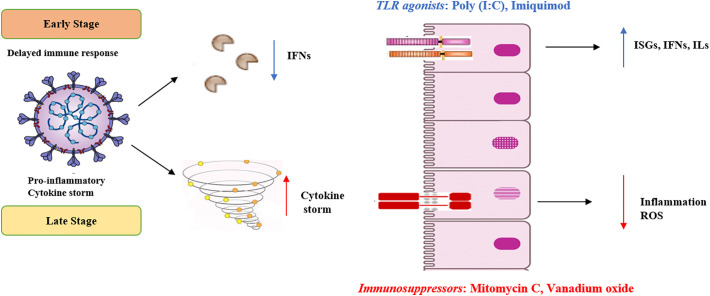
In summary, our integrative interactome and network topology analyses showed that SARS-CoV-2 induced a strong IFN response, marked by the increased expression of several ISGs, including MX1, OAS1-3, IFIH1, ISG15, IRFs, and IFITMs etc. These ISGs exert antiviral activity and could protect the host cells from the infection. This protective action of ISGs might account for the lesser percentage of severe cases and the lower fatality rate in COVID-19. However, the extent of protection or damage to the host cell depends on stage of infection, types of cells, SARS clade [[71], [72], [73]] and other factors like co-infection, age, and co-morbidities. Recently, Zou et al. [74] reported high SARS-CoV-2 loads very early during infection, suggesting that the virus may have developed arsenals that is able to delay the IFN response by inhibiting innate immune signaling. Thus, IFN induction in the incubation period and at the very early stages of the infection could be the key to prevent COVID-19 associated mortalities. Administration of interferon-inducing agents, such as Poly (I:C) and Imiquimod could reduce the mortality of SARS at the very early stages of the disease (Fig. 5 ). On the other hand, at the later stages of the disease, the balance of the immune system becomes impaired, leading to inflammatory over-reactions, cytokine storm, and possible autoimmune responses. In such circumstances, administration of mitomycin C and other immunosuppressants to reduce possible lung inflammations may be needed (Fig. 5).
Fig. 5.
Innate immune response as a protector to SARS-CoV-2. In the early stage of infection, SARS-CoV-2 inhibits host type I IFN antiviral immune defence. We propose to counteract this by utilizing toll like receptors (TLRs) agonists, poly (I:C) and Imiquimod to induce the production of IFNs and ISGs, which will reinstate the impaired immune responses. In the late stage of infection, there is a proinflammatory cytokine storm, which can be theoretically targeted by immunosuppressors, like Mitomycin C, and Vanadium oxide.
The following are the supplementary data related to this article.
The human PPI network analysis of the 120 dysregulated genes.
Functional enrichment analysis of 120 dysregulated genes using DAVID tool and with p-value ≤ 0.05 filter.
Functional enrichment analysis of 15 hub genes.
Gene set enrichment analysis for Molecular phenotype predictions in 15 Hub genes using Enrichr.
Drug-gene interactions for 15 hub genes.
Network analysis of drug-genes of 15 hub genes along with the disease-drug interactions. Green triangles represent hub genes, Red square represents drugs and Blue circles represent diseases.
CRediT authorship contribution statement
Kartikay Prasad:Conceptualization, Methodology, Data curation, Software.Fatima Khatoon:Data curation.Summya Rashid:Writing - original draft.Nemat Ali:Visualization, Investigation.Abdullah F. AlAsmari:Supervision.Mohammad Z. Ahmed:Software, Validation.Ali S. Alqahtani:Software, Validation.Mohammed S. Alqahtani:Writing - review & editing.Vijay Kumar:Conceptualization, Supervision, Writing - review & editing.
Acknowledgments
Acknowledgments
Authors sincerely thank to the Department of Science and Technology, Government of India. The authors extend their appreciation to the Deanship of Scientific Research at King Saud University for funding this work through research group no. RG-1441-461.
Declaration of competing interest
No potential conflict of interest was reported by the authors.
References
- 1.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., Du B., Li L.J., Zeng G., Yuen K.Y., Chen R.C., Tang C.L., Wang T., Chen P.Y., Xiang J., Li S.Y., Wang J.L., Liang Z.J., Peng Y.X., Wei L., Liu Y., Hu Y.H., Peng P., Wang J.M., Liu J.Y., Chen Z., Li G., Zheng Z.J., Qiu S.Q., Luo J., Ye C.J., Zhu S.Y., Zhong N.S. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jose R.J., Manuel A. COVID-19 cytokine storm: the interplay between inflammation and coagulation. Lancet Respir. Med. 2020;8:46–47. doi: 10.1016/S2213-2600(20)30216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Blanco-Melo D., Nilsson-Payant B.E., Liu W.C. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181(5):1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiong Y., Liu Y., Cao L., Wang D., Guo M., Jiang A., Guo D., Hu W., Yang J., Tang Z., Wu H., Lin Y., Zhang M., Zhang Q., Shi M., Zhou Y., Lan K., Chen Y. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microbes Infect. 2020;9(1):761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Z., Ren L., Zhang L., Zhong J., Xiao Y., Jia Z., Guo L., Yang J., Wang C., Jiang S., Yang D., Zhang G., Li H., Chen F., Xu Y., Chen M., Gao Z., Dong J., Liu B., Zhang X., Wang W., He K., Jin Q., Li M., Wang J., Zhou Z., Ren L., Zhang L., Zhong J., Xiao Y., Jia Z., Guo L., Yang J., Wang C., Jiang S., Yang D., Zhang G., Li H., Chen F., Xu Y., Chen M., Gao Z., Dong J., Liu B., Zhang X., Wang W., He K., Jin Q., Li M., Wang J. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020;27(6):883–890. doi: 10.1016/j.chom.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Golonka R.M., Saha P., Yeoh B.S., Chattopadhyay S., Gewirtz A.T., Joe B., Vijay-Kumar M. Harnessing innate immunity to eliminate SARS-CoV-2 and ameliorate COVID-19 disease. Physiol. Genomics. 2020;52(5):217–221. doi: 10.1152/physiolgenomics.00033.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi Y., Wang Y., Shao C., Huang J., Gan J., Huang X., Bucci E., Piacentini M., Ippolito G., Melino G. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 2020;27(5):1451–1454. doi: 10.1038/s41418-020-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fung T.S., Liu D.X. Human coronavirus: host-pathogen interaction. Annu. Rev. Microbiol. 2019;73:529–557. doi: 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- 11.McKechnie J.L., Blish C.A. The innate immune system: fighting on the front lines or fanning the flames of COVID-19? Cell Host Microbe. 2020;27(6):863–869. doi: 10.1016/j.chom.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang S., Fu C., Lian X., Dong X., Zhang Z. Understanding human-virus protein-protein interactions using a human protein complex-based analysis framework. mSystems. 2019;4(2) doi: 10.1128/mSystems.00303-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Padhi A.K., Kalita P., Zhang K.Y.J., Tripathi T. High throughput designing and mutational mapping of RBD-ACE2 interface guide non-conventional therapeutic strategies for COVID-19. BioRxiv. 2020 [Google Scholar]
- 14.Dyall J., Coleman C.M., Hart B.J., Venkataraman T., Holbrook M.R., Kindrachuk J., Johnson R.F., Olinger G.G., Jr., Jahrling P.B., Laidlaw M., Johansen L.M., Lear-Rooney C.M., Glass P.J., Hensley L.E., Frieman M.B. Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection. Antimicrob. Agents Chemother. 2014;58(8):4885–4893. doi: 10.1128/AAC.03036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johansen L.M., Brannan J.M., Delos S.E., Shoemaker C.J., Stossel A., Lear C., Hoffstrom B.G., Dewald L.E., Schornberg K.L., Scully C., Lehar J., Hensley L.E., White J.M., Olinger G.G. FDA-approved selective estrogen receptor modulators inhibit Ebola virus infection. Sci. Transl. Med. 2013;5(190):190ra79. doi: 10.1126/scitranslmed.3005471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrows N.J., Campos R.K., Powell S.T., Prasanth K.R., Schott-Lerner G., Soto-Acosta R., Galarza-Munoz G., McGrath E.L., Urrabaz-Garza R., Gao J., Wu P., Menon R., Saade G., Fernandez-Salas I., Rossi S.L., Vasilakis N., Routh A., Bradrick S.S., Garcia-Blanco M.A. A screen of FDA-approved drugs for inhibitors of Zika virus infection. Cell Host Microbe. 2016;20(2):259–270. doi: 10.1016/j.chom.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Huettenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.P., Liu Y., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O'Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu D., Wang H.Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d'Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., Garcia-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020:1–13. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:14. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ciliberto G., Mancini R., Paggi M.G. Drug repurposing against COVID-19: focus on anticancer agents. J. Exp. Clin. Cancer Res. 2020;39(1):86. doi: 10.1186/s13046-020-01590-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J. Fast identification of possible drug treatment of coronavirus disease-19 (COVID-19) through computational drug repurposing study. J. Chem. Inf. Model. 2020;60(6):3277–3286. doi: 10.1021/acs.jcim.0c00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ciliberto G., Cardone L. Boosting the arsenal against COVID-19 through computational drug repurposing. Drug Discov. Today. 2020;25(6):946–948. doi: 10.1016/j.drudis.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia-Cremades M., Solans B.P., Hughes E., Ernest J.P., Wallender E., Aweeka F., Luetkemeyer A.F., Savic R.M. Optimizing hydroxychloroquine dosing for patients with COVID-19: an integrative modeling approach for effective drug repurposing. Clin. Pharmacol. Ther. 2020 doi: 10.1002/cpt.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blanco-Melo D., Nilsson-Payant B., Liu W.-C., Møller R., Panis M., Sachs D., Albrecht R. SARS-CoV-2 launches a unique transcriptional signature from in vitro, ex vivo, and in vivo systems. BioRxiv. 2020 [Google Scholar]
- 24.Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis A.P., Grondin C.J., Johnson R.J., Sciaky D., McMorran R., Wiegers J., Wiegers T.C., Mattingly C.J. The comparative toxicogenomics database: update 2019. Nucleic Acids Res. 2019;47(D1):D948–D954. doi: 10.1093/nar/gky868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blanco-Melo Daniel, Nilsson-Payant Benjamin E., Liu Wen-Chun, Møller Rasmus, Panis Maryline, Sachs David, Albrecht Randy A., tenOever B.R. 2020. SARS-CoV-2 Launches a Unique Transcriptional Signature From In Vitro, Ex Vivo, and In Vivo Systems. [Google Scholar]
- 27.Huang D.W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuhn M., Mering C. von, Campillos M., Jensen L.J., Bork P. STITCH: interaction networks of chemicals and proteins. Nucleic Acids Res. 2007;36(suppl_1):D684–D688. doi: 10.1093/nar/gkm795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4(1):44. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 31.Pfefferle S., Schopf J., Kogl M., Friedel C.C., Muller M.A., Carbajo-Lozoya J., Stellberger T., von Dall’Armi E., Herzog P., Kallies S., Niemeyer D., Ditt V., Kuri T., Zust R., Pumpor K., Hilgenfeld R., Schwarz F., Zimmer R., Steffen I., Weber F., Thiel V., Herrler G., Thiel H.J., Schwegmann-Wessels C., Pohlmann S., Haas J., Drosten C., von Brunn A. The SARS-coronavirus-host interactome: identification of cyclophilins as target for pan-coronavirus inhibitors. PLoS Pathog. 2011;7(10) doi: 10.1371/journal.ppat.1002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shapira S.D., Gat-Viks I., Shum B.O., Dricot A., de Grace M.M., Wu L., Gupta P.B., Hao T., Silver S.J., Root D.E., Hill D.E., Regev A., Hacohen N. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell. 2009;139(7):1255–1267. doi: 10.1016/j.cell.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ackerman E.E., Kawakami E., Katoh M., Watanabe T., Watanabe S., Tomita Y., Lopes T.J., Matsuoka Y., Kitano H., Shoemaker J.E., Kawaoka Y. Network-guided discovery of influenza virus replication host factors. mBio. 2018;9(6) doi: 10.1128/mBio.02002-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bosl K., Ianevski A., Than T.T., Andersen P.I., Kuivanen S., Teppor M., Zusinaite E., Dumpis U., Vitkauskiene A., Cox R.J., Kallio-Kokko H., Bergqvist A., Tenson T., Merits A., Oksenych V., Bjoras M., Anthonsen M.W., Shum D., Kaarbo M., Vapalahti O., Windisch M.P., Superti-Furga G., Snijder B., Kainov D., Kandasamy R.K. Common nodes of virus-host interaction revealed through an integrated network analysis. Front. Immunol. 2019;10:2186. doi: 10.3389/fimmu.2019.02186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vieira Braga F.A., Kar G., Berg M., Carpaij O.A., Polanski K., Simon L.M., Brouwer S., Gomes T., Hesse L., Jiang J., Fasouli E.S., Efremova M., Vento-Tormo R., Talavera-Lopez C., Jonker M.R., Affleck K., Palit S., Strzelecka P.M., Firth H.V., Mahbubani K.T., Cvejic A., Meyer K.B., Saeb-Parsy K., Luinge M., Brandsma C.A., Timens W., Angelidis I., Strunz M., Koppelman G.H., van Oosterhout A.J., Schiller H.B., Theis F.J., van den Berge M., Nawijn M.C., Teichmann S.A. A cellular census of human lungs identifies novel cell states in health and in asthma. Nat. Med. 2019;25(7):1153–1163. doi: 10.1038/s41591-019-0468-5. [DOI] [PubMed] [Google Scholar]
- 36.Arguel Marie-Jeanne, Lebrigand Kevin, Paquet Agnès, Arguel Marie-Jeanne, Lebrigand Kevin, Paquet Agnès, Pee’r Dana, Marquette Charles-Hugo, Leroy Sylvie, Barbry P. A single-cell atlas of the human healthy airways. BioRxiv. 2019 [Google Scholar]
- 37.Sungnak W., Huang N., Becavin C., Berg M., Queen R., Litvinukova M., Talavera-Lopez C., Maatz H., Reichart D., Sampaziotis F., Worlock K.B., Yoshida M., Barnes J.L. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26(5):681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., Cao Y., Yousif A.S., Bals J., Hauser B.M., Feldman J., Muus C., Wadsworth M.H., II, Kazer S.W., Hughes T.K., Doran B., Gatter G.J., Vukovic M., Taliaferro F., Mead B.E., Guo Z., Wang J.P., Gras D., Plaisant M., Ansari M., Angelidis I., Adler H., Sucre J.M.S., Taylor C.J., Lin B., Waghray A., Mitsialis V., Dwyer D.F., Buchheit K.M., Boyce J.A., Barrett N.A., Laidlaw T.M., Carroll S.L., Colonna L., Tkachev V., Peterson C.W., Yu A., Zheng H.B., Gideon H.P., Winchell C.G., Lin P.L., Bingle C.D., Snapper S.B., Kropski J.A., Theis F.J., Schiller H.B., Zaragosi L.E., Barbry P., Leslie A., Kiem H.P., Flynn J.L., Fortune S.M., Berger B., Finberg R.W., Kean L.S., Garber M., Schmidt A.G., Lingwood D., Shalek A.K., Ordovas-Montanes J. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181(5):1016–1035. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., McDermott M.G., Monteiro C.D., Gundersen G.W., Ma’ayan A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneider W.M., Chevillotte M.D., Rice C.M. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong M.T., Chen S.S. Emerging roles of interferon-stimulated genes in the innate immune response to hepatitis C virus infection. Cell. Mol. Immunol. 2016;13(1):11–35. doi: 10.1038/cmi.2014.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verhelst J., Parthoens E., Schepens B., Fiers W., Saelens X. Interferon-inducible protein Mx1 inhibits influenza virus by interfering with functional viral ribonucleoprotein complex assembly. J. Virol. 2012;86(24):13445–13455. doi: 10.1128/JVI.01682-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ciancanelli M.J., Abel L., Zhang S.Y., Casanova J.L. Host genetics of severe influenza: from mouse Mx1 to human IRF7. Curr. Opin. Immunol. 2016;38:109–120. doi: 10.1016/j.coi.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pingale K.D., Kanade G.D., Karpe Y.A. Hepatitis E virus polymerase binds to IFIT1 to protect the viral RNA from IFIT1-mediated translation inhibition. J. Gen. Virol. 2019;100(3):471–483. doi: 10.1099/jgv.0.001229. [DOI] [PubMed] [Google Scholar]
- 45.Hamano E., Hijikata M., Itoyama S., Quy T., Phi N.C., Long H.T., Ha L.D., Ban V.V., Matsushita I., Yanai H., Kirikae F., Kirikae T., Kuratsuji T., Sasazuki T., Keicho N. Polymorphisms of interferon-inducible genes OAS-1 and MxA associated with SARS in the Vietnamese population. Biochem. Biophys. Res. Commun. 2005;329(4):1234–1239. doi: 10.1016/j.bbrc.2005.02.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kindler E., Gil-Cruz C., Spanier J., Li Y., Wilhelm J., Rabouw H.H., Zust R., Hwang M., V’Kovski P., Stalder H., Marti S., Habjan M., Cervantes-Barragan L., Elliot R., Karl N., Gaughan C., van Kuppeveld F.J., Silverman R.H., Keller M., Ludewig B., Bergmann C.C., Ziebuhr J., Weiss S.R., Kalinke U., Thiel V. Early endonuclease-mediated evasion of RNA sensing ensures efficient coronavirus replication. PLoS Pathog. 2017;13(2) doi: 10.1371/journal.ppat.1006195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Melchjorsen J., Kristiansen H., Christiansen R., Rintahaka J., Matikainen S., Paludan S.R., Hartmann R. Differential regulation of the OASL and OAS1 genes in response to viral infections. J. Interf. Cytokine Res. 2009;29(4):199–207. doi: 10.1089/jir.2008.0050. [DOI] [PubMed] [Google Scholar]
- 48.Frieman M., Yount B., Heise M., Kopecky-Bromberg S.A., Palese P., Baric R.S. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 2007;81(18):9812–9824. doi: 10.1128/JVI.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang Y., Ye F., Zhu N., Wang W., Deng Y., Zhao Z., Tan W. Middle East respiratory syndrome coronavirus ORF4b protein inhibits type I interferon production through both cytoplasmic and nuclear targets. Sci. Rep. 2015;5 doi: 10.1038/srep17554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Daczkowski C.M., Dzimianski J.V., Clasman J.R., Goodwin O., Mesecar A.D., Pegan S.D. Structural insights into the interaction of coronavirus papain-like proteases and interferon-stimulated gene product 15 from different species. J. Mol. Biol. 2017;429(11):1661–1683. doi: 10.1016/j.jmb.2017.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dzimianski J.V., Scholte F.E.M., Bergeron E., Pegan S.D. ISG15: it’s complicated. J. Mol. Biol. 2019;431(21):4203–4216. doi: 10.1016/j.jmb.2019.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim Y.J., Kim E.T., Kim Y.E., Lee M.K., Kwon K.M., Kim K.I., Stamminger T., Ahn J.H. Consecutive inhibition of ISG15 expression and ISGylation by cytomegalovirus regulators. PLoS Pathog. 2016;12(8) doi: 10.1371/journal.ppat.1005850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Okumura A., Lu G., Pitha-Rowe I., Pitha P.M. Innate antiviral response targets HIV-1 release by the induction of ubiquitin-like protein ISG15. Proc. Natl. Acad. Sci. U. S. A. 2006;103(5):1440–1445. doi: 10.1073/pnas.0510518103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dai J., Pan W., Wang P. ISG15 facilitates cellular antiviral response to dengue and west nile virus infection in vitro. Virol. J. 2011;8:468. doi: 10.1186/1743-422X-8-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li C., Wang Y., Zheng H., Dong W., Lv H., Lin J., Guo K., Zhang Y. Antiviral activity of ISG15 against classical swine fever virus replication in porcine alveolar macrophages via inhibition of autophagy by ISGylating BECN1. Vet. Res. 2020;51(1):22. doi: 10.1186/s13567-020-00753-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mielech A.M., Kilianski A., Baez-Santos Y.M., Mesecar A.D., Baker S.C. MERS-CoV papain-like protease has deISGylating and deubiquitinating activities. Virology. 2014;450–451:64–70. doi: 10.1016/j.virol.2013.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ratia K., Kilianski A., Baez-Santos Y.M., Baker S.C., Mesecar A. Structural basis for the ubiquitin-linkage specificity and deISGylating activity of SARS-CoV papain-like protease. PLoS Pathog. 2014;10(5) doi: 10.1371/journal.ppat.1004113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ngoi S.M., Tovey M.G., Vella A.T. Targeting poly(I:C) to the TLR3-independent pathway boosts effector CD8 T cell differentiation through IFN-alpha/beta. J. Immunol. 2008;181(11):7670–7680. doi: 10.4049/jimmunol.181.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao J., Wohlford-Lenane C., Fleming E., Lane T.E., McCray P.B., Jr., Perlman S. Intranasal treatment with poly(I*C) protects aged mice from lethal respiratory virus infections. J. Virol. 2012;86(21):11416–11424. doi: 10.1128/JVI.01410-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kumaki Y., Salazar A.M., Wandersee M.K., Barnard D.L. Prophylactic and therapeutic intranasal administration with an immunomodulator, Hiltonol((R)) (poly IC:LC), in a lethal SARS-CoV-infected BALB/c mouse model. Antivir. Res. 2017;139:1–12. doi: 10.1016/j.antiviral.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barnard D.L., Day C.W., Bailey K., Heiner M., Montgomery R., Lauridsen L., Chan P.K., Sidwell R.W. Evaluation of immunomodulators, interferons and known in vitro SARS-coV inhibitors for inhibition of SARS-coV replication in BALB/c mice. Antivir. Chem. Chemother. 2006;17(5):275–284. doi: 10.1177/095632020601700505. [DOI] [PubMed] [Google Scholar]
- 62.Gao J., Tian Z., Yang X. Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci. Trends. 2020;14(1):72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- 63.Kumar A., Zhang J., Yu F.S. Toll-like receptor 3 agonist poly(I:C)-induced antiviral response in human corneal epithelial cells. Immunology. 2006;117(1):11–21. doi: 10.1111/j.1365-2567.2005.02258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nerurkar L., McColl A., Graham G., Cavanagh J. The systemic response to topical Aldara treatment is mediated through direct TLR7 stimulation as Imiquimod enters the circulation. Sci. Rep. 2017;7(1) doi: 10.1038/s41598-017-16707-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gibson S.J., Imbertson L.M., Wagner T.L., Testerman T.L., Reiter M.J., Miller R.L., Tomai M.A. Cellular requirements for cytokine production in response to the immunomodulators imiquimod and S-27609. J. Interf. Cytokine Res. 1995;15(6):537–545. doi: 10.1089/jir.1995.15.537. [DOI] [PubMed] [Google Scholar]
- 66.Verweij J., Pinedo H.M. Mitomycin C: mechanism of action, usefulness and limitations. Anti-Cancer Drugs. 1990;1(1):5–13. [PubMed] [Google Scholar]
- 67.Snodgrass R.G., Collier A.C., Coon A.E., Pritsos C.A. Mitomycin C inhibits ribosomal RNA: a novel cytotoxic mechanism for bioreductive drugs. J. Biol. Chem. 2010;285(25):19068–19075. doi: 10.1074/jbc.M109.040477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nayak D.P., Rasmussen A.F., Jr. Influence of mitomycin C on the replication of influenza viruses. Virology. 1966;30(4):673–683. doi: 10.1016/0042-6822(66)90172-3. [DOI] [PubMed] [Google Scholar]
- 69.Tsave O., Petanidis S., Kioseoglou E., Yavropoulou M.P., Yovos J.G., Anestakis D., Tsepa A., Salifoglou A. Role of vanadium in cellular and molecular immunology: association with immune-related inflammation and pharmacotoxicology mechanisms. Oxidative Med. Cell. Longev. 2016;2016 doi: 10.1155/2016/4013639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zwolak I. Vanadium carcinogenic, immunotoxic and neurotoxic effects: a review of in vitro studies. Toxicol. Mech. Methods. 2014;24(1):1–12. doi: 10.3109/15376516.2013.843110. [DOI] [PubMed] [Google Scholar]
- 71.Channappanavar R., Fehr A.R., Vijay R., Mack M., Zhao J., Meyerholz D.K., Perlman S. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe. 2016;19(2):181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Channappanavar R., Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin. Immunopathol. 2017;39(5):529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Channappanavar R., Fehr A.R., Zheng J., Wohlford-Lenane C., Abrahante J.E., Mack M., Sompallae R., McCray P.B., Jr., Meyerholz D.K., Perlman S. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J. Clin. Invest. 2019;130:3625–3639. doi: 10.1172/JCI126363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J., Guo Q., Song T., He J., Yen H.L., Peiris M., Wu J. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382(12):1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The human PPI network analysis of the 120 dysregulated genes.
Functional enrichment analysis of 120 dysregulated genes using DAVID tool and with p-value ≤ 0.05 filter.
Functional enrichment analysis of 15 hub genes.
Gene set enrichment analysis for Molecular phenotype predictions in 15 Hub genes using Enrichr.
Drug-gene interactions for 15 hub genes.
Network analysis of drug-genes of 15 hub genes along with the disease-drug interactions. Green triangles represent hub genes, Red square represents drugs and Blue circles represent diseases.