Noninvasive detection of exosomal MCT1/CD147 can monitor the progression of glioma, a fatal brain tumor, as a liquid biopsy.
Abstract
Malignant glioma is a fatal brain tumor whose pathological progression is closely associated with glycolytic reprogramming, leading to the high expression of monocarboxylate transporter 1 (MCT1) and its ancillary protein, cluster of differentiation 147 (CD147) for enhancing lactate efflux. In particular, malignant glioma cells (GMs) release tremendous number of exosomes, nanovesicles of 30 to 200 nm in size, promoting tumor progression by the transport of pro-oncogenic molecules to neighboring cells. In the present study, we found that hypoxia-induced malignant GMs strongly enhanced MCT1 and CD147 expression, playing a crucial role in promoting calcium-dependent exosome release. Furthermore, it was first identified that hypoxic GMs-derived exosomes contained significantly high levels of MCT1 and CD147, which could be quantitatively detected by noninvasive localized surface plasmon resonance and atomic force microscopy biosensors, demonstrating that they could be precise surrogate biomarkers for tracking parent GMs’ metabolic reprogramming and malignant progression as liquid biopsies.
INTRODUCTION
Glioma is the most common type of brain cancer that predominantly originates from neuroglial stem cells (1). Accumulating evidence has revealed that a key hallmark during the malignant progression of glioma is metabolic reprogramming toward aerobic glycolysis, known as the Warburg effect (2). Consequently, malignant glioma cells (GMs) increase glucose consumption and lactate production through rapid glycolysis to meet the high demand of energy substrates, biosynthetic precursors, and signaling molecules, by which their growth and migration are promoted (3). Malignant GMs enhance the levels of monocarboxylate transporter 1 (MCT1) and cluster of differentiation 147 (CD147) as well as their localization at the plasma membrane to remove intracellular lactate out of cells for the maintenance of continuous glycolysis. This leads to the accumulation of lactate in the tumor microenvironment (TME) (4). This extracellular lactate can also be taken up by surrounding fasting GMs and stromal cells in the hypoxic TME to produce adenosine triphosphate (ATP), eventually establishing the metabolic coupling among heterogeneous neighboring cells (5). Recent reports have demonstrated that lactate in the TME can serve not only as an energy substrate and biosynthetic precursor but also as a signaling molecule in promoting tumor progression (6). However, the exact role of lactate as a signaling molecule in glioma progression remains largely elusive.
MCT1, a major MCT in the central nervous system (7), has been known to play a crucial role in the proton-linked transport of lactate and ketone bodies across the cell membrane by cooperative action with its binding protein, CD147 (8). In particular, MCT1 and CD147 in various tumors, including glioma, are significantly up-regulated during malignant progression of the tumor. Therefore, their levels and distribution in glioma tissues have been considered as crucial indicators to determine glioma malignancy, particularly that associated with metabolic adaptation (9). Blocking the function of MCT1 and CD147, genetically or chemically, has been shown to suppress the growth, metastasis, and invasion of GMs as well as angiogenesis in in vitro and in vivo experimental models (10). Such findings have led to the ongoing development of their inhibitors as anticancer agents via controlling tumor metabolism.
Increased glycolysis is an important survival mechanism of GMs in the metabolically stressful TME. GMs in the hypoxic TME enhance the expression of hypoxia-inducible factor 1 (HIF-1)–dependent glycolytic genes, including MCT1 and CD147, which produces high levels of ATP and lactate (9). A recent study has demonstrated that hypoxic cancer cells, including malignant GMs, also promote the release of a substantial number of exosomes, a major type of extracellular vesicles (EVs), which facilitates tumor progression (11). However, it remains largely unknown how up-regulated MCT1 and CD147 in malignant GMs are associated with the increased release of exosomes and the production of pro-oncogenic exosomes for glioma progression.
Most cells, including cancer cells, release exosomes, by which functional molecules can be delivered to neighboring or distal cells (12). Malignant GMs release a significantly high number of exosomes, partly by which their invasion, metastasis, and growth can be promoted (11, 13). GMs-derived exosomes contain tumor-associated proteins and microRNA (13). For example, they contain tumor-specific mRNA and microRNAs, including mRNA of c-Myc, mutant isocitrate dehydrogenase 1, mutant epidermal growth factor receptor variant III (EGFRvIII), and microRNA-21 (14, 15).
GMs-derived exosomes with a size ranging from 30 to 200 nm (16) can spread into systemic bio-fluids, such as cerebrospinal fluid (CSF) (17) and blood (18) by crossing the blood-CSF barrier (BCSFB) and the blood-brain barrier (BBB). Therefore, GMs-derived exosomes have been proposed as great platforms for the discovery of effective biomarkers to track glioma progression (19). Conventionally, the diagnosis and prognosis of glioma have been mainly dependent on magnetic resonance imaging (MRI) and computed tomography (CT) scans, as well as intracranial biopsies (20, 21). However, the detection of precise molecular signatures of glioma progression and metabolic adaptation has been difficult to ascertain. Therefore, the development of additional diagnostic tools with precise biomarkers has been in high demand to better monitor the metabolic reprogramming and malignant progression of glioma. While recent studies have suggested that highly sensitive detection of exosomes and exosomal components can improve the accuracy of diagnosis and prognosis of tumors such as glioma (11, 22, 23), the detailed characterization of GMs-derived exosomes requires additional investigation to provide a more thorough understanding of how to track glioma progression and metabolic adaptation. For example, differential biophysical properties, such as zeta potential, adhesiveness, stiffness, and roughness, as well as the release amount of daughter exosomes have been proposed as informative indicators to better understand and determine the malignant transition of parent GMs (24). In particular, the surface proteins of GM-derived exosomes can be reliable diagnostic biomarkers that can be measured by cost-effective, label-free, real-time, and highly sensitive detection tools such as localized surface plasmon resonance (LSPR) and atomic force microscopy (AFM) biosensors (25). LSPR biosensing is a powerful biocompatible technique with a high sensitivity, allowing it to detect single molecular interactions, such as antigen-antibody interactions. It also has high spatial resolution owing to the change of the dielectric property of surroundings in the functionalized sensing chip (26). AFM, on the other hand, is a versatile scanning probe microscope that can measure single molecular interactions with nanoscale spatial resolution achieved through the detection of the adhesive force between functionalized probe tips and the sample on the discs (27). The capability of imaging for soft samples in air and liquid without causing much damage makes AFM a powerful tool for the analysis of biological samples, including exosomes (28). Therefore, the quantitative detection of exosomal surface proteins by LSPR and AFM biosensors could provide the needed insights into the development of diagnostic and prognostic tools with precise biomarkers for glioma as a liquid biopsy.
In the present study, MCT1 and CD147, two major proteins associated with the glycolytic reprogramming and malignant progression of glioma, were first identified in the surface of GM-derived exosomes, and exosomal MCT1 and CD147 were quantitatively detected by label-free sensitive LSPR and AFM biosensors.
RESULTS
MCT1 and its ancillary protein, CD147, play an important role in the malignant progression of GMs
To determine whether hypoxia could promote cancer progression, U251 and U87 GMs were exposed to low oxygen tension (1% O2) in the hypoxic chamber, or they were treated with CoCl2 (100 μM) for 24 hours in the regular chamber. As reported previously (2, 3), the effect of hypoxia on the phenotypic change of GMs was significant in proliferation and migration assays. Transwell cell migration and Scratch assays revealed that hypoxia significantly promoted the migration of U251 (fig. S1, A to C and M to Q) and U87 (fig. S2, A to C) GMs. In addition, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) and 5-bromo-2′-deoxyuridine (BrdU) cell proliferation assays demonstrated that hypoxia enhanced their proliferation as well (figs. S1, J and L, and S2J).
GMs’ malignant progression is associated with metabolic reprogramming by the increased expression of glycolytic genes, such as MCT1 and CD147 (9, 10). To determine whether enhanced MCT1 and CD147 in GMs could induce their phenotypic change, the effect of gain of MCT1 or CD147 function on their migration and proliferation was tested by the expression of Lenti-MCT1cDNA–internal ribosome entry sequence (IRES)–enhanced green fluorescent protein (eGFP) or Lenti-CD147cDNA-IRES-eGFP in GMs (see Materials and Methods). Enhanced expression of MCT1 or CD147 in GMs (Fig. 1, D1 to S1, and fig. S3, S1 to T1, V1, and W1) promoted their migration and proliferation (figs. S1, D, E, G, I, and K, and S2, D, E, G, I, and K), mimicking the effect of hypoxia on GMs and indicating the crucial role of MCT1 and CD147 in the malignant progression of GMs. In addition, the effect of loss of MCT1 or CD147 function on GMs’ migration and proliferation was investigated by the expression of Lenti-H1-MCT1shRNA-CMV-eGFP for MCT1 or antisense locked nucleic acid (LNA) GapmeR for CD147 in GMs. Notably, MCT1 or CD147 knockdown (KD) in GMs (fig. S3, S1 to T1, V1, and W1) reduced their migration and proliferation (figs. S1, D, F, H, I, and K, and S2, D, F, H, I, and K), further demonstrating the crucial role of MCT1 and CD147 in tumor progression.
Fig. 1. Hypoxia promotes U251 GMs’ glycolytic reprogramming, including enhancing MCT1 and CD147 expression.
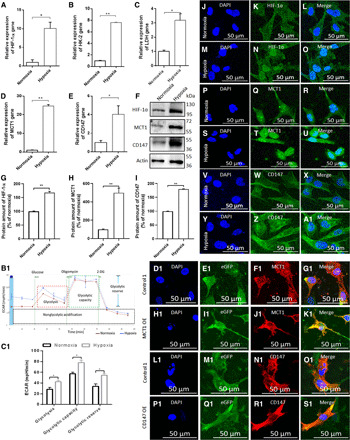
(A to E) Change in the mRNA expression of HIF-1ɑ, HK-2, LDH, MCT1, and CD147 in GMs in response to hypoxia (1% O2) (n = 3), as determined by quantitative real-time polymerase chain reaction (qRT-PCR). (F to I) Protein-level change of HIF-1ɑ, MCT1, and CD147 in GMs in response to hypoxia (1% O2) (n = 3), as determined by Western blot (WB). (J to Z and A1) Immunofluorescent staining for HIF-1ɑ, MCT1, and CD147 in GMs under normoxia and hypoxia. (B1) A representative graph of ECAR outputs from the XF24 analyzer for normoxic and hypoxic GMs and their response to glucose, oligomycin, and 2-deoxyglucose (2-DG) in the measurement of the status of glycolytic metabolism. (C1) Comparison of glycolysis, glycolytic capacity, and glycolytic reserve between normoxic and hypoxic GMs (n = 3). Immunofluorescent staining for MCT1 in GMs treated with (D1 to G1) empty backbone lentivirus (control 1) and (H1 to K1) MCT1 OE lentivirus for 24 hours. Immunofluorescent staining for CD147 in GMs treated with (L1 to O1) empty backbone lentivirus (control 1) and (P1 to S1) CD147 OE lentivirus for 24 hours. All data were shown as the means ± SD. Significance level: **P < 0.01, *P < 0.05, hypoxia versus normoxia.
Increased exosome release from hypoxic GMs is associated with enhanced MCT1 and CD147
To determine the effect of hypoxia on tumor progression, GMs were exposed to hypoxic chamber (1% O2), or CoCl2 (100 μM), by which the expression of HIF-1ɑ and its nuclear localization were significantly enhanced (Fig. 1, A, F, G, and J to O). Furthermore, glycolytic genes, including hexokinase-2 (HK-2), lactate dehydrogenase (LDH), MCT1, and CD147, were markedly up-regulated in hypoxic GMs (Fig. 1, B to F, H to I, P to Z, and A1, and fig. S3, A to R and A1 to R1). Enhanced glycolysis in hypoxic GMs was also observed in the measurement of the output of extracellular acidification rate (ECAR) from the Seahorse XF24 Extracellular Flux Analyzer (Fig. 1, B1 and C1).
Glycolytic reprogramming of GMs is crucial for their survival. A recent report demonstrated that hypoxic GMs released a large quantity of exosomes, supporting their survival through the autologous or heterologous interactions with GMs or surrounding cells in the TME (11). To investigate the correlation between the malignant transition of hypoxic GMs and their production and release of exosomes, a secretion assay for exosomes was conducted using nanoparticle tracking analysis (NTA). As compared with normoxic U251 GMs, hypoxic U251 GMs released a significantly higher number of exosomes (248.9% increase) (Fig. 2, A to C). Enhanced exosome release was also observed in hypoxic U87, U118, A172, C6, GL261 GMs, SF7761 glioma stem cells (GSCs), and adult GSCs (67.52, 163.61, 138.16, 80, 200, 270, and 226.07% increase compared to that of normoxic GMs, respectively) (figs. S4, A to C, Y, Z, and A1 to D1, and S5, A to C and M to O).
Fig. 2. Enhanced MCT1 and CD147 in hypoxic U251 GMs promote intracellular Ca2+-dependent exosome release.
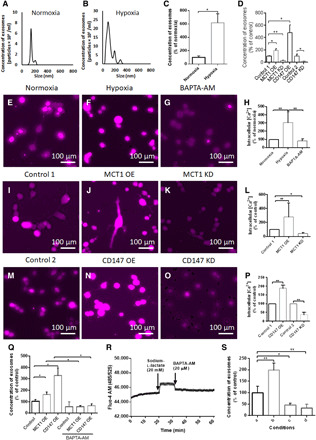
(A and B) Size distribution and quantity of exosomes released from normoxic and hypoxic GMs for 24 hours (NTA analysis). (C) Enhanced release of exosomes from hypoxic GMs (versus normoxic GMs). (D) Analysis of exosome release from GMs treated with control 1, MCT1 OE, MCT1 KD, CD147 OE (all lentivirus), and control 2 and CD147 KD (antisense oligonucleotides) constructs. (E to P) Representative images of Fura Red calcium dye- loaded- hypoxic (versus normoxic), MCT1 OE- or MCT1 KD- (versus control 1) induced, CD147 OE- or CD147 KD- (versus control 1 & 2) induced, and BAPTA-AM (20 μM)-treated GMs. (Q) Enhanced exosome release from MCT1 OE– and CD147 OE–induced (versus control 1) GMs, followed by a marked decline in exosome release by treatment with BAPTA-AM (20 μM, 100 μl). (R) Enhanced intracellular Ca2+ levels in GMs by treatment with sodium-l-lactate (20 mM, 100 μl), followed by distinctive decline in intracellular Ca2+ level by treatment with BAPTA-AM (20 μM, 100 μl). (S) NTA exosome release assay from GMs exposed to four different conditions for 10 min described in Materials and Methods. Briefly, a, Exo–fetal bovine serum (FBS) medium; b, sodium-l-lactate (20 mM), c, BAPTA-AM; d, BAPTA-AM with the pretreatment of sodium-l-lactate (20 mM). All chemicals were dissolved in the Exo-FBS medium containing 1% dimethyl sulfoxide. All data were shown as the means ± SD. Significance level: **P < 0.01, *P < 0.05, hypoxia versus normoxia, BAPTA-AM versus control, MCT1 KD lentivirus versus empty backbone lentivirus (control 1), and CD147 antisense oligonucleotides versus antisense control oligonucleotides (control 2).
To determine whether MCT1 and CD147 in GMs could be involved in regulating exosome release, the effect of gain or loss of MCT1 or CD147 functions in the release of exosomes from U251 GMs was investigated under normoxia and hypoxia. Under normoxic condition, MCT1 or CD147 overexpression (OE) in U251 GMs significantly increased exosome release (92.57 and 381.16%, respectively, compared to that of control). In contrast, MCT1 or CD147 KD in U251 GMs reduced exosome release (73.84 and 82.49%, respectively), indicating the essential role of MCT1 and CD147 in controlling exosome release from normoxic U251 GMs (Fig. 2D). MCT1 or CD147 KD in hypoxic U251 GMs significantly reduced hypoxia-induced exosome release; however, MCT1 or CD147 OE in hypoxic U251 GMs did not alter hypoxia-induced exosome release much (fig. S4V), suggesting that hypoxia in this condition enhanced the amount of MCT1 and CD147 for the maximum release of exosomes. In addition, to investigate the effect of mutual interaction between MCT1 and CD147 on exosome secretion, rescue experiments with MCT1 OE or CD147 OE in hypoxic U251 GMs were performed to reverse reduced exosome release by MCT1 KD or CD147 KD, respectively. NTA analysis revealed that both MCT1 OE and CD147 OE in hypoxic GMs reversed MCT1 or CD147 KD-dependent reduced exosome release, suggesting that the interaction between MCT1 and CD147 in GMs was important in the regulation of exosome release (fig. S4, W and X). Furthermore, the combinatorial additive rescue effect of MCT1 and CD147 OE in exosome release was also observed after MCT1 KD, but not CD147 KD in the condition (fig. S4, W and X), indicating the possible presence of an MCT1-independent CD147 pathway for exosome secretion.
GSCs in the TME were known to be crucial in glioma malignancy and recurrence. Therefore, it was wondered whether their response to hypoxia and exosome release was similar to that of other GMs lines, such as U251 and U87 GMs. NTA analysis indicated that hypoxic SF7761 GSCs and adult GSCs released 3 times and 3.26 times more exosomes, respectively (fig. S5, A to C and M to O). Furthermore, hypoxic SF7761 GSC– and adult GSC–derived exosomes contained significantly higher amounts of MCT1 and CD147 compared to normoxic cells (fig. S5, D to K and P to W). Moreover, MCT1 or CD147 KD in SF7761 GSCs and adult GSCs reduced exosome release, indicating their important role in controlling exosome release (fig. S5, L and X).
Hypoxia- and lactate-induced enhanced exosome release from GMs is controlled by MCT1 and CD147 in a calcium-dependent manner
To further investigate whether the change of intracellular Ca2+ levels could be associated with hypoxia-induced enhancement of exosome release, Fluo Red acetoxymethyl (AM) Ca2+ imaging and Fluo-4 AM Ca2+ assay were conducted with normoxic and hypoxic U251 GMs, as previously performed (29). Hypoxia increased both exosome release and intracellular Ca2+ levels in U251 GMs (Fig. 2, A to C, E, F, and H) and, furthermore, chelating intracellular Ca2+ with 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid (BAPTA)–AM blocked the enhanced release of exosomes from U251 GMs (Fig. 2, G and H), suggesting the regulatory role of intracellular Ca2+ in exosome release. In addition, to determine whether MCT1 and CD147 in GMs could influence levels of intracellular Ca2+, Fluo Red AM Ca2+ imaging was conducted with the MCT1- or CD147-enriched or -deficient U251 GMs by expressing Lenti-CMVP-MCT1cDNA-IRES-eGFP, Lenti-CMVP-CD147cDNA-IRES-eGFP, Lenti-H1-MCT1shRNA-CMV-eGFP, or CD147 antisense LNA GapmeR, respectively. The result revealed that MCT1 or CD147 OE in GMs enhanced intracellular Ca2+ levels and exosome release (Fig. 2, I, J, L to N, and P), whereas MCT1 or CD147 KD in GMs reduced both, demonstrating a strong positive correlation for each other (Fig. 2, I, K, L, M, O, and P). These findings indicated that MCT1 and CD147 in GMs could regulate exosome release in a calcium-dependent manner. The increase in exosome release by MCT1 or CD147 OE in GMs was reversed by treatment with BAPTA-AM (Fig. 2Q), further suggesting that the enhanced release of exosomes from GMs by MCT1 and CD147 is calcium dependent.
To recapitulate hypoxia-induced acidic TME (30), sodium-l-lactate (20 mM) was applied to the culture medium of GMs. High levels of extracellular lactate enhanced intracellular Ca2+ levels in GMs as determined by Fluo-4 AM Ca2+ assay (Fig. 2R). Increased intracellular Ca2+ levels further stimulated exosome release, which was blocked by BAPTA-AM (Fig. 2S), demonstrating that accumulated lactate in the TME could promote exosome release in a calcium-dependent manner.
Hypoxic GMs-derived exosomes contain significant amount of MCT1 and CD147
Exosomes from normoxic and hypoxic U251 GMs were further characterized by NTA and transmission electron microscopy (TEM) analysis. Both exosomes from normoxic and hypoxic GMs were mainly round-shaped nanovesicles ranging from 30 to 200 nm in size, as determined by NTA and TEM analysis (Figs. 2, A and B, and 3, A and B). Most GMs-derived exosomes were also positive for CD63, a major exosome marker, which was first revealed by immunogold EM, and exosomal CD63 levels were then further quantified by Western blot (WB) (Fig. 3, C, D, I, and J), ensuring the reliability of their isolation and characterization. MCT1 and CD147 in malignant GMs are enriched in the plasma membrane, thus incorporating them into the membrane of daughter exosomes. Therefore, to determine whether MCT1 and CD147 were present in the membrane of GMs-derived exosomes, immunogold EM was conducted. Both MCT1 and CD147 were present in the membrane of exosomes from all investigated GMs lines, including U251, U87, U118, and A172 GMs lines (Fig. 3, E to H, and fig. S4, D to U). In addition, quantitative analysis was conducted with normoxic and hypoxic GMs-derived exosomes to determine whether levels of exosomal MCT1 and CD147 could reflect their quantity in parent GMs. MCT1 and CD147 levels in parent GMs and their daughter exosomes were detected and measured by immunogold EM, WB, ICC, and enzyme-linked immunosorbent assay (ELISA) (Figs. 1, F, H, I, P to Z, and A1, and 3, E to L, and fig. S3, A to R, A1 to R1, and U1). MCT1 and CD147 levels in parent U251 GMs were positively correlated with those levels in daughter exosomes, revealing that exosomal MCT1 and CD147 could be reliable surrogate biomarkers to monitor their levels in parent GMs, which were related with malignant progression. Most notably, in the validation experiments, MCT1 and CD147 OE in parent U251 GMs showed increased levels in their daughter exosomes; in contrast, MCT1 and CD147 KD in parent U251 GMs displayed reduced levels in their daughter exosomes (fig. S3, S1, T1, and V1 to Y1).
Fig. 3. MCT1 and CD147 are significantly present in exosomes from both normoxic and hypoxic U251 GMs.
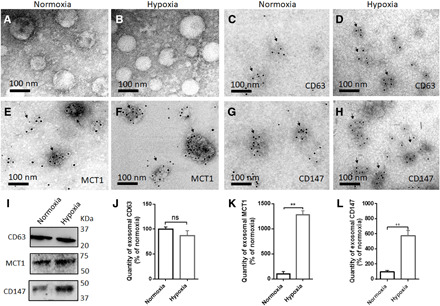
(A and B) TEM images of exosomes derived from normoxic and hypoxic GMs. (C to H) Representative immunogold EM images for CD63, MCT1, and CD147 in exosomes from normoxic and hypoxic GMs. (I) Determining the quantity of CD63, MCT1, and CD147 in exosomes from normoxic and hypoxic GMs by WB. (J to L) Bar graphs showing the relative quantity of CD63, MCT1, and CD147 in exosomes from normoxic and hypoxic GMs (n = 4) as detected by ELISA. All data were shown as the mean ± SD. Significance level: **P < 0.01; ns, not significant, hypoxia versus normoxia.
MCT1 and CD147 levels in parent GMs have an effect on the biophysical properties of daughter exosomes
As noted above, MCT1 and CD147 levels in GMs-derived exosomes were proportional to those of parent GMs. Therefore, enhanced levels of MCT1 and CD147 in hypoxic GMs might change the biophysical properties of daughter exosomes, suggesting that they could influence their uptake into recipient cells, such as endothelial cells (ECs). Therefore, daughter exosomes’ biophysical properties were investigated by measuring the zeta potential of hypoxic GMs-derived exosomes, as compared to normoxic GMs-derived exosomes, with Zetasizer Nano ZS. The zeta potential value of hypoxic GMs-derived exosomes was significantly lower than that of normoxic GMs-derived exosomes (fig. S6A), indicating the increased instability associated with the coagulation, membrane fusion, and uptake of exosomes into recipient cells. Next, to investigate whether MCT1 and CD147 levels in parent GMs could be directly associated with the zeta potential change in daughter exosomes, MCT1 and CD147 KD or OE in parent GMs were conducted as noted above, wherein results showed reduced or increased levels of MCT1 or CD147 in their corresponding daughter exosomes. Increased MCT1 or CD147 levels in exosomes made their zeta potential lower, recapitulating hypoxia-driven reduction of exosomal zeta potential. In contrast, decreased MCT1 or CD147 levels in exosomes made their zeta potential higher (fig. S6B), thereby presumably reducing their fusion into recipient cells. AFM analysis further revealed significant changes in biophysical properties, including roughness, Young’s modulus (elasticity and stiffness), and adhesion force, of hypoxic GMs-derived exosomes as compared with those of normoxic ones. The roughness, stiffness, and adhesion force in hypoxic GMs-derived exosomes were approximately 1.3 times bigger, 7 times smaller, and 3 times bigger, respectively (fig. S6, C, E, and G), demonstrating that their values could be informative to track GMs’ hypoxic and malignant status. Theoretically, enhanced adhesion force and increased zeta potential in hypoxic GMs-derived exosomes might facilitate their uptake into recipient cells. The uptake of hypoxic GMs-derived exosomes by ECs was much higher after incubation for 24 hours (fig. S7, A to U). This resulted in the promotion of their tube formation, which was determined by the quantification of number of branches, branching intervals, junctions, meshes, and segments (fig. S7, V to Z, A1, and B1), suggesting that the hypoxic GMs-derived exosomes have a crucial impact on angiogenesis. Increased or decreased MCT1 and CD147 levels in parent GMs by genetic modifications also produced altered roughness, stiffness, and adhesion force properties of daughter exosomes (fig. S6, D, F, and H), recapitulating hypoxia-induced biophysical alterations in exosomes, further supporting the potential role of MCT1 and CD147 in controlling the uptake of GMs-derived exosomes into recipient cells as well as controlling their release in the TME.
Exosomal MCT1 and CD147 are precisely detected by label-free LSPR and AFM biosensors
Recent reports demonstrated that exosomes could cross the BBB and BCSFB, supporting that their components, including membrane proteins and microRNAs, could be used as promising surrogate markers and systemic biomarkers for the diagnosis and prognosis of brain disorders, including glioma (17–19). Therefore, MCT1 and CD147 in GMs-derived exosomes could be potential biomarkers to monitor the metabolic and malignant status of parent GMs. As shown in the analysis of immunogold EM, MCT1 and CD147 were present mainly in the membrane of exosomes (Fig. 3, E to H, and fig. S4, F to I, L to O, and R to U). In addition, the analysis of SF7761 GSCs-derived exosomes and adult GSC-derived exosomes by immunogold EM and ELISA revealed the presence of exosomal MCT1 and CD147 as well (fig. S5, D to K and P to W). Thus, sensitive label-free LSPR and AFM biosensors were used to noninvasively detect exosomal MCT1 and CD147 with the functionalized self-assembly gold nanoislands (SAM-AuNIs) chip and silicon nitride cantilever tip with the antibody (AB) toward MCT1 or CD147. For the quantitative assessment of detection sensitivity and specificity for exosomal MCT1 and CD147 by two biosensors, reduced or increased levels of GMs-derived exosomes were first produced by genetic modifications such as OE or KD of MCT1 or CD147 in parent GMs (Fig. 1, D1 to S1, and fig. S3, S1, T1, V1, and W1). In summary, increased levels of MCT1 or CD147 in parent GMs enhanced MCT1 or CD147 levels in their daughter exosomes as well. In the same way, decreased levels of MCT1 or CD147 in parent GMs directly reduced MCT1 or CD147 levels in their daughter exosomes (fig. S3, X1 and Y1).
The noninvasive LSPR biosensor with the functionalized SAM-AuNIs sensing chip with anti-MCT1 AB (Fig. 5A) or anti-CD147 AB (Fig. 5B) was sensitive enough to quantitatively detect exosomal MCT1 or CD147 (Fig. 4, A, B, E, and F). The specificity of LSPR biosensing was demonstrated by the correlated LSPR phase response to the levels of exosomal MCT1 and CD147. For example, the higher their levels were, the bigger their LSPR response (Fig. 5, C and D). In particular, the LSPR biosensor precisely detected enhanced MCT1 or CD147 levels in exosomes from hypoxic GMs (Fig. 5, E and F). Furthermore, exosomal MCT1 and CD147 were accurately detected by a high-resolution AFM biosensor. To quantitatively measure them, the spring constant of silicon nitride cantilever of the AFM biosensor was calibrated to be 0.3744 N/m. It was first shown that the ScanAsyst-fluid mode of AFM imaging for exosomes captured on the functionalized SAM-AuNIs sample discs with anti-CD63 AB could produce great resolution for two-dimensional and three-dimensional AFM topographic images, facilitating better analysis of their biophysical properties (Fig. 5, I and J). Height profile analysis in the three-dimensional AFM topographic image also showed captured exosomes in the sample discs (Fig. 5, K and L). After the immobilization of exosomes on discs, the AFM biosensor was used to quantitatively detect exosomal MCT1 and CD147 by the functionalized cantilever tip with anti-MCT1 AB or anti-CD147 AB (fig. S8, A to D). This was the first “consecutive capture and sensing” method to detect exosomal surface proteins by AFM. Conclusively, a high degree of sensitivity and specificity of previously unreported AFM biosensing was established and validated by using MCT1- or CD147-deficient or -enriched exosomes (Fig. 4, C, D, G, and H). Last, the AFM biosensor precisely detected enhanced MCT1 or CD147 levels in exosomes from hypoxic GMs (Fig. 5, M to P).
Fig. 5. Noninvasive sensitive detection of increased MCT1 and CD147 in hypoxic GM-derived exosomes by LSPR and AFM biosensors.
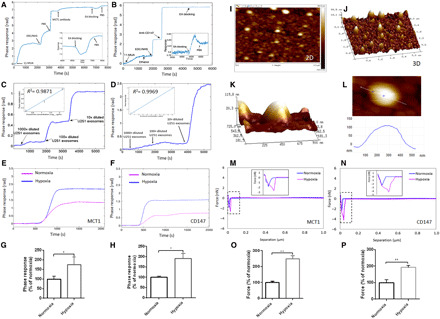
(A and B) Baseline phase response of the LSPR biosensor with the functionalized SAM-AuNIs sensing chip with anti-MCT1 AB or anti-CD147 AB after sequential treatment with 11-mercaptoundecanoic acid (MUA) and 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC)/N-hydroxysuccinimide (NHS). (C and D) Phase response of the LSPR biosensor toward three different concentrations (serial dilution of 1300 μg/ml exosome solution: 1000×, 100×, and 10×) of U251 GM-derived exosomes. Standard curve fitting for phase responses toward anti-MCT1 AB (R2 = 0.9871) or anti-CD147 AB (R2 = 0.9969). (E and F) Representative phase response of the LSPR biosensor toward equal amount of normoxic and hypoxic GM-derived exosomes (50 μg/ml). (G and H) Bar graph showing the relative strength of LSPR responses toward exosomal MCT1 (E) and CD147 (F) from normoxic or hypoxic GMs (n = 3). (I to K) Two-dimensional, three-dimensional, and high resolution of three-dimensional AFM topographic images for U251 GM-derived exosomes immobilized on the SAM-AuNIs sensing chip. (L) Height profile of single U251 GM-derived exosome by AFM scanning. (M and N) Representative separation force responses of the AFM biosensor with the functionalized cantilever sensing tip with anti-MCT1 AB, or anti-CD147 AB toward equal amount (50 μg/ml) of normoxic and hypoxic GM-derived exosomes captured on the SAM-AuNIs sample discs. (O and P) Bar graph showing the relative strength of AFM separation force responses toward exosomal MCT1 (M) and CD147 (N) from normoxic or hypoxic GMs (n = 12). All data were shown as the means ± SD. Significance level: **P < 0.01, *P < 0.05, hypoxia versus normoxia.
Fig. 4. Label-free quantitative detection of exosomal MCT1 and CD147 to monitor their expression levels in parent GMs, as novel surrogate biomarkers.
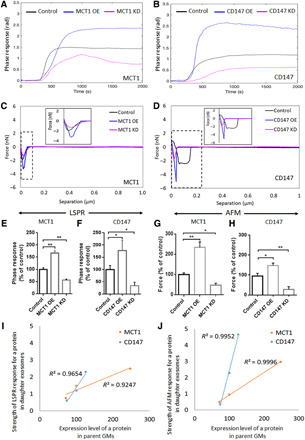
(A and B) Representative phase responses of the LSPR biosensor with the functionalized SAM-AuNIs sensing chip with anti-MCT1 AB or anti-CD147 AB and (C and D) separation force responses of the AFM biosensor with the functionalized silicon nitride tip with anti-MCT1 AB or anti-CD147 AB toward equal amount of daughter exosomes (50 μg/ml) from parent U251 GMs with no treatment (control), MCT1 OE, MCT1 KD, CD147 OE, and CD147 KD. (E to H) Bar graph showing the relative strength of LSPR responses (n = 3) or AFM forces (n = 12) toward exosomal MCT1 [from (A) and (C)] and CD147 [from (B) and (D)]. (I and J) Correlation curve between MCT1 or CD147 levels in parent GMs and the strength of LSPR responses toward exosomal MCT1 or CD147, respectively [for MCT1, coefficient of determination (R2) = 0.9247, and for CD147, R2 = 0.9654], or the strength of AFM forces toward exosomal MCT1 or CD147, respectively (for MCT1, R2 = 0.9996, and for CD147, R2 = 0.9952). The correlation analysis was performed based on the data obtained from (A) to (D). All data were shown as the means ± SD. Significance level: **P < 0.01, *P < 0.05, MCT1 OE and MCT1 KD group versus control. CD147 OE and CD147 KD group versus control.
Overall, a strong positive correlation between the levels of cellular MCT1 and CD147 and the response strength of LSPR [for MCT1, coefficient of determination (R2) = 0.9247, and for CD147, R2 = 0.9654] and AFM (for MCT1, R2 = 0.9996, and for CD147, R2 = 0.9952) for exosomal MCT1 and CD147 was observed (Fig. 4, I and J), strongly supporting the potential application of noninvasive LSPR- and AFM-based detection for exosomal MCT1 and CD147 to monitor GMs’ glycolytic metabolism associated with their malignant progression.
LSPR and AFM biosensors noninvasively detect MCT1 and CD147 in the blood serum–derived exosomes from a mouse model of glioma
MRI scans have been used as a major diagnostic method for glioma as well as an in vivo glioma study. In addition, MRI has also been applied to discover glioma pathologies in patients (20, 21, 31). However, new techniques have been demanded to help detect molecular and metabolic signatures of glioma even at its early stage to aid in a more precise diagnosis. Therefore, the noninvasive liquid biopsy for detecting metabolic biomarkers of glioma has been investigated. In this study, exosomal MCT1 and CD147 in blood serum were investigated in the course of glioma formation by using label-free LSPR and AFM biosensors. To do so, an in vivo mouse model of glioma was first established by the intracranial implantation of U251 or U87 GMs in immunodeficient mice as described in Materials and Methods. In the course of glioma formation, an MRI scan for each mouse was conducted, and blood from each mouse was then consecutively obtained for the isolation of serum-derived exosomes. Glioma formation was identified by an MRI scan at approximately 10 days after the implantation of U251 and U87 GMs into the brain (with a size range of 0.7 to 1.1 mm3) (Fig. 6, A to C). Characterization of isolated serum-derived exosomes from each mouse was conducted by NTA, TEM, ELISA, and immunogold EM (fig. S9, A to P). NTA demonstrated that the number of serum-derived exosomes from a mouse model of glioma was significantly higher (fig. S9, A to D), indicating the systemic impact of glioma formation in the body. TEM results showed the heterogeneous morphology and size of serum-derived exosomes (fig. S9, E to G). ELISA and immunogold EM revealed a higher amount of MCT1 and CD147 in serum-derived exosomes from a mouse model of glioma as compared to those of wild-type mice (fig. S9, H to P). Last, LSPR and AFM responses toward exosomal MCT1 and CD147 in serum-derived exosomes from a mouse model of glioma were significantly greater compared to those from control mice (Fig. 6, D to K). These data strongly suggested that, together with MRI images, label-free sensitive detection of exosomal MCT1 and CD147 in serum-derived exosomes could be supportive for the better diagnosis and prognosis of glioma.
Fig. 6. Label-free sensitive detection of MCT1 and CD147 in serum-derived exosomes from a mouse model of glioma.
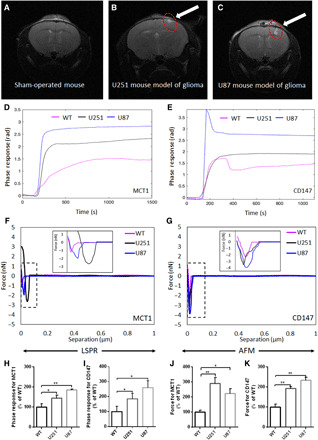
(A to C) Representative MRI images for the brain of sham-operated mice and U251 and U87 mouse models of glioma. (D and E) Representative phase responses of the LSPR biosensor with the functionalized SAM-AuNIs sensing chip with anti-MCT1 AB or anti-CD147 AB and (F and G) representative separation force curves of the AFM biosensor with the functionalized silicon nitride cantilever tip with anti-MCT1 AB or anti-CD147 AB toward serum-derived exosomes from sham-operated mice and U251 and U87 mouse models of glioma. (H to K) Bar graph summarizing the relative strength of LSPR responses (n = 3) or AFM forces (n = 3) toward exosomal MCT1 [e.g., (D) and (E)] and CD147 [e.g., (F) and (G)]. Detailed processes of LSPR and AFM biosensing were described in Materials and Methods. All data were expressed as the means ± SD. Significance level: **P < 0.01, *P < 0.05, U251 or U87 mouse model of glioma versus sham-operated severe combined immunodeficient mouse. WT, wild type.
DISCUSSION
It has been well known that cancer cells in the hypoxic TME can survive through their metabolic reprogramming, in which glycolysis-related genes, such as MCT1 and CD147, are up-regulated (32, 33). The present study demonstrated that hypoxia increased levels of MCT1 and CD147 in U251 (Fig. 1, A to Z and A1), U87 (fig. S3, A to F and A1 to F1), U118 (fig. S3, G to L and G1 to L1), and A172 (fig. S3, M to R and M1 to R1) GMs, which partly promoted their migration and proliferation as well, and further loss- and gain-of-function studies confirmed the essential role of MCT1 and CD147 in GMs’ survival and migration (figs. S1 and S2), supporting that they could be druggable targets for glioma therapy.
Malignant hypoxic GMs also release a tremendous number of exosomes, containing unique pro-oncogenic components such as mRNA of c-Myc and microRNA 221 and 128, which supports their survival by facilitating communication with neighboring cells (11, 13, 34). Therefore, increased exosome release has been proposed as a biomarker for determining tumor malignancy (11, 13). However, it has been difficult to ascertain whether metabolic reprogramming, such as up-regulation of MCT1 and CD147, in malignant GMs is directly associated with increased exosome release. To test this, loss- and gain-of-function studies were conducted with genetic modifications for MCT1 and CD147. The OE of MCT1 or CD147 in GMs recapitulated hypoxia-induced enhanced exosome release, whereas their KD in GMs reduced exosome release (Fig. 2D and fig. S3, S1, T1, V1, and W1). In particular, we discovered that up-regulation of MCT1 and CD147 enhanced exosome release from GMs, which was dependent on intracellular calcium levels. GMs with hypoxia exposure or lactate treatment enhanced exosome release by increased intracellular calcium, which was blocked by treatment with BAPTA-AM as well as MCT1 or CD147 KD (Fig. 2, A to C and E to P), implying that the effects of MCT1 and CD147 expression was dependent on intracellular calcium levels. High levels of extracellular lactate, common in the hypoxic TME (5), also increased intracellular Ca2+-dependent release of exosomes from GMs (Fig. 2, Q to S). One potential mechanism might be that extracellular lactate could be a signal for GMs to increase intracellular Ca2+ through interaction with a lactate receptor (35), facilitating exosome release. Another possibility might be that extracellular lactate can induce MCT1 expression (33), promoting exosome release.
The increased uptake of exosomes by surrounding cells or GMs in the TME is important for cancer survival; however, an in depth look at the underlying mechanisms has not been thoroughly investigated in GMs. Nonetheless, accumulated evidence has revealed that exosome release is promoted in malignant cancer and the uptake of tumor-derived exosomes into surrounding cells is significantly enhanced. In this study, we demonstrated that GM-derived exosomes had unique biophysical properties for the promotion of their uptake into surrounding cells. Exosomes from GMs with hypoxia exposure or OE for MCT1 or CD147 showed a much smaller zeta potential and stiffness, but displayed more roughness and a higher adhesion force (fig. S6, A to H). Presumably this is one of the means that drive their higher uptake into ECs, further enhancing tube formation (fig. S7). Our results additionally demonstrated that biophysical properties of exosomes could also be informative biomarkers to reflect the malignant status of GMs associated with MCT1 and CD147 expression, either directly or indirectly.
Precise detection of the malignant progression of glioma, which is predominantly associated with its metabolic reprogramming, is critical in the development of anti-glioma agents and glioma therapy, as well as in its diagnosis and prognosis. Malignant GMs release large amounts of exosomes within 30 to 200 nm in size, which can spread into the peripheral fluids, suggesting that they can be not only systemic functional mediators but also great platforms for the identification of biomarkers and fingerprints as a liquid biopsy for glioma. Therefore, it is important to find a link between exosome components and glioma malignancy. In the present study, two major proteins involved in the metabolic adaptation of GMs, MCT1 and a binding protein, CD147, were found mainly in the membrane of GM-derived exosomes and their exosomal levels recapitulated their levels in parent GMs (Fig. 3 and fig. S4, D to U). Thus, exosomal MCT1 and CD147 were studied to determine their feasibility as surrogate biomarkers for monitoring glioma progression and metabolism using label-free sensitive LSPR and AFM biosensors (26–28). Compared with general techniques used in the analysis of exosomes, such as flow cytometry, WB, immunogold EM, ELISA, as well as “omics”, label-free LSPR and AFM biosensing is simple, sensitive, and cost-effective. In our previous work, label-free LSPR and AFM biosensors were successfully used to characterize tumor-derived exosomes and MVs (16) and detect glioma-specific EGFRvIII in exosomes (36). In the present work, the functionalized SAM-AuNIs sensing chip with anti-MCT1 AB or anti-CD147 AB was used in LSPR biosensing, and it provided a great specificity and sensitivity of LSPR response toward exosomal MCT1 and CD147, whereas the functionalized AFM sensing tip with anti-MCT1 AB or anti-CD147 AB generated a magnificent response of AFM separation force with high-resolution image toward MCT1 and CD147 in the exosomes captured on the SAM-AuNIs sample discs coated with anti-CD63 AB (Fig. 5). The novel AFM biosensing with SAM-AuNIs sample disc-based consecutive capture and sensing of exosomes provided a potential great opportunity to characterize a specific population of disease-specific exosomes in a future study. In the test of specificity, LSPR and AFM biosensors quantitatively detected reduced or increased MCT1 and CD147 in daughter exosomes with genetic modifications, supporting their accurate detection for exosomal proteins as well as the strong quantitative correlation between their cellular and exosomal amount (Fig. 4), leading to the conclusion that LSPR and AFM biosensors had great capability to detect exosomal MCT1 and CD147, faithful biomarkers for monitoring GMs’ malignant progression and metabolic adaptation. In addition, LSPR and AFM biosensors precisely detected their increased amount in hypoxic GM-derived exosomes (quantity per same protein amount), even without consideration of a marked increase of exosomes by stimulation, proving its great capability in sensitive biosensing (Fig. 5).
As described above, one of challenges in glioma therapy is how to carry out affordable early detection of its molecular and metabolic changes as a liquid biopsy because MRI- and CT scan–based diagnosis primarily only determines later stages of glioma. Therefore, analysis of exosomes from blood and CSF of animal models and patients of glioma have become more and more important in the basic and clinical research of glioma. In the present study, we demonstrated that the quantity of serum-derived exosomes from a mouse model of glioma was much higher when the glioma became enlarged, as detected by MRI scanning (Fig. 6, A to C, and fig.S9, A to D). In terms of the morphology and size of serum-derived exosomes, their parent cells might be diverse, suggesting that glioma formation in the brain could have a systemic effect on the periphery and the quantity of exosomes in the peripheral circulation as well. Nonetheless, the origin of serum-derived exosomes was not clear; many of them had MCT1 and CD147 in their membranes (fig. S9, I to P). Using label-free sensitive LSPR and AFM biosensors, we precisely detected significantly higher levels of MCT1 and CD147 in serum-derived exosomes from the mouse model of glioma (Fig. 6, D to K). Notably, their LSPR and AFM response in the detection was positively correlated with glioma formation and progression, implying that MCT1 and CD147 from serum-derived exosomes could provide additive information to track glioma progression together with currently available MRI scans (Fig. 6 and fig. S9). However, it might be more informative to analyze pure GM-derived exosomes directly. Therefore, in the future, the development of isolation techniques and the enrichment of GM-derived exosomes from CSF of a mouse model of glioma and/or patients, along with precise detection of MCT1 and CD147 with those exosomes by LSPR and AFM biosensors, will serve as a requisite advancement in tracking glioma metabolism and progression.
In conclusion, we demonstrated that hypoxia promoted GMs’ malignant progression and that calcium-dependent exosome release was associated with enhanced MCT1 and CD147. Moreover, we revealed that hypoxic GM-derived exosomes had unique biophysical properties that promoted their uptake into ECs. In particular, we first found that GM-derived exosomes contained both MCT1 and CD147, the quantity of which was proportional to those of parent GMs, and exosomal MCT1 and CD147 could be precisely detected by noninvasive sensitive LSPR and AFM biosensors, demonstrating that they are likely to be promising surrogate biomarkers for tracking glioma metabolic malignancy. The present study supported the hypothesis that MCT1 and CD147 in GMs can also control the release, composition, and uptake of exosomes, providing great insights into the additional mechanism of MCT1 and CD147 inhibitors as anticancer agents in preventing glioma progression through exosome shuttling among neighboring cells (fig. S10).
MATERIALS AND METHODS
Animals
All animal experiments followed the Institutional Animal Care and Use Committee (IACUA) guidelines and were approved by the Animal Research Ethics Sub-Committee at City University of Hong Kong and Department of Health, Government of the Hong Kong Special Administrative Region. Implantation experiments with U257 and U87 GMs were performed using 6-week-old female severe combined immunodeficient (SCID) mice (Laboratory Animal Services Centre, The Chinese University of Hong Kong). Mice had free access to water and food ad libitum under a 12-hour light/12-hour dark cycle.
Cell culture
U251, U87, U118, A172, GL261, and C6 GMs and bEnd.3 ECs (Guangzhou Cellcook Biotech Co. Ltd., China) were cultured in Dulbecco’s modified Eagle’s medium with high glucose (DMEM-H) (Invitrogen, catalog no. 10569-010) supplemented with 10% fetal bovine serum (FBS) (Gibco, catalog no. 10270-106) and 1% penicillin-streptomycin (Pen-Strep) (Gibco, catalog no. 15140-122). Human embryonic kidney (HEK) 293T cells were cultured in DMEM supplemented with 10% FBS and 1% Pen-Strep. Human pediatric diffuse intrinsic pontine SF7761 GSCs were cultured in the specific medium, StemPro NSC SFM (catalog no. A10509-01, Invitrogen). Human adult GSCs (catalog no. 36104-41, Celprogen) were cultured in human GSC serum-free colony-forming unit media (catalog no. M36104-40CF). Cell cultures were maintained in a humidified incubator containing 5% CO2 at 37°C.
Hypoxia induction
For the induction of hypoxia with low oxygen, GMs were incubated in the chamber (Smartor 118 hypoxia chamber) flushed with a gas mixture, containing 1% O2, 5% CO2, and 94% N2 at 2 pounds per square inch, at 37°C for 24 hours. For the induction of hypoxia with a chemical, GMs were treated with 100 μM cobalt chloride (CoCl2) (Sigma-Aldrich, catalog no. 232696) at 37°C for 24 hours. In control experiments, GMs were exposed to normoxia (21% oxygen) at 37°C for 24 hours. When validating hypoxia, nuclear trafficking and localization of HIF-1α were used.
Cell migration assays
Migration of GMs was examined by both Transwell cell migration and Scratch assays (37). In brief, GMs (1 × 105 cells/100 μl of medium per well) were seeded in the upper chamber of Transwell Permeable Support chambers (Costar, catalog no. 3422) with a pore size of 8.0-μm mesh separating the upper and lower chambers, and 500 μl of complete GM culture medium was added to the lower chamber. GMs were allowed to migrate for 24 hours at 37°C to the lower chamber. Next, GMs in the lower chamber were fixed for 10 min by 4% paraformaldehyde (PFA) and then were stained with 0.1% Crystal Violet solution for 20 min. Last, pictures were taken by using an inverted microscope at 10× magnification. The number of GMs that migrated between chambers was counted using ImageJ software. Scratch assays were conducted following the standard protocol (37).
Cell proliferation assays
The proliferation of GMs was assessed by Vybrant MTT Cell Proliferation Assay Kit (Thermo Fisher Scientific, catalog no. V13154) and a BrdU Cell Proliferation Assay Kit (colorimetric) (ab126556) per the manufacturer’s guidelines.
RNA extraction and qRT-PCR
Total RNA was extracted from GMs using TRIzol reagent (Life Technologies, catalog no. 15596018), according to the manufacturer’s instructions, and its concentration was determined with a NanoDrop 2000 (Thermo Fisher Scientific). Reverse transcription (RT) was performed with total RNA using a SuperScript IV First-Strand cDNA Synthesis kit (Life Technologies). The mRNA expression of HIF-1ɑ, MCT1, CD147, LDH, HK, and β-actin was examined by quantitative real-time polymerase chain reaction (qRT-PCR) using KAPA SYBR FAST qPCR kit Master Mix (2×) Universal (catalog no. KK4650). The thermal cycling conditions involved denaturation at 95°C for 3 min and proceeded with 40 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 1 min, and extension at 72°C for 30 s. All reactions of qRT-PCR were performed in triplicate and Ct values of target genes were normalized to that of β-actin.
Western blotting
Identification and quantification of HIF-1α, CD-63, MCT1, and CD147 in both GMs and GM-derived exosomes were conducted by WB (36). Briefly, GM (20 μg) and exosome (50 to 100 μg) lysates were separated by 12 and 8% SDS-gel electrophoresis, respectively, and then transferred to polyvinylidene difluoride membrane. After its incubation with 5% skim milk in tris-buffered saline (TBS) (blocking buffer) for 1 hour, the membrane was further incubated with specific primary ABs (anti-MCT1 AB 1:200, anti-CD147 AB 1:1000 dilution for GMs; anti-MCT1 AB 1:100, anti-CD147 AB 1:200 dilution for exosomes) in blocking buffer (5% skim milk for GMs, 1% skim milk for exosomes) overnight at 4°C, followed by washing three times with TBST and further incubation with goat anti-rabbit immunoglobulin G (IgG) H&L [horseradish peroxidase (HRP)] secondary AB (1∶2000 dilution for GMs, 1:1000 dilution for exosomes) for 2 hours at room temperature. Immunoreactive bands were detected using enhanced chemiluminescence substrate (Bio-Rad) and imaged using the Azure Biosystems Gel Documentation system (C600).
Enzyme-linked immunosorbent assay
Following the manufacturer’s guidelines, 50 μl of GM lysates (200 μg/ml) and exosomes (400 μg/ml) were added to the wells of an ELISA micro-titer plate, incubated for 2 hours at 37°C, and further incubated for 1 hour at room temperature with primary ABs, anti-MCT1 AB and anti-CD147 AB (both 1:100 dilution) for GMs and exosomes, and anti-CD63 AB (1:100 dilution) only for exosomes. After washing three times, primary AB–treated samples were further incubated with anti-rabbit HRP-conjugated secondary AB (1:5000 dilution) for 1 hour at room temperature. After washing three times, tetramethyl-benzidine substrates were applied to secondary AB–treated samples and further incubated in a dark place for 30 min at room temperature. After washing three times, the absorbance at 450 nm, via a microplate reader, was recorded within 2 min after the addition of “Stop Solution” to the wells. Control was the absorbance value obtained from the well without any sample. The kits used are ExoELISA kit (System Biosciences, catalog no. EXOEL-CD63A-1), Human MCT1 ELISA Kit (LifeSpan Biosciences, catalog no. LS-F9108), and Human CD147 ELISA Kit (Abcam, catalog no. ab219631).
Immunocytochemistry
GMs were grown on poly-d-lysine (Merck, catalog no. A-003-E)–coated coverslips until 60 to 70% confluence before conducting the experiments with specific conditions, including exposure to normoxic and hypoxic condition; transduction of lenti-eGFP (control), MCT1 KD, MCT1 OE, and CD147 OE lentiviral particles; and transfection of antisense control and CD147 antisense LNA GapmeR. Next, the GMs were fixed with 4% PFA in phosphate-buffered saline (PBS) for 30 min on ice. After washing with 0.1% Triton X-100 in PBS (PBST) three times, the fixed GMs were further incubated in 5% bovine serum albumin (BSA) in PBST for 1 hour (for MCT1 staining, the fixed GMs were incubated in 1% BSA-PBST). After washing with PBS three times, the blocked GMs were incubated with primary ABs in 0.1% BSA-PBS (dilution: anti HIF-1ɑ AB at 1:200, anti-MCT1 AB at 1:50, and anti-CD147 AB at 1:100) overnight at 4°C in a dark humidified chamber, and followed by the incubation with goat anti-rabbit Alexa Fluor 488 secondary AB in 0.1% BSA-PBS (dilution at 1:500) for 2 hours at room temperature after washing with PBS. Last, immunostained GMs were counterstained and mounted by the medium with 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories, catalog no. H-1200). Images were taken on a Zeiss Laser Scanning Microscopy LSM 880 NLO with Airyscan. The ABs used in the experiment were rabbit polyclonal anti–HIF-1α AB (Abcam, catalog no. ab82832), rabbit polyclonal anti-MCT1 AB (Alomone Labs, catalog no. AMT-011), rabbit polyclonal anti-CD147 AB (Abcam, catalog no. ab64616), rabbit polyclonal anti-CD63 AB (Abcam, catalog no. ab68418), rabbit polyclonal anti–β-actin AB (Abcam, catalog no. ab16039), goat anti-rabbit IgG H&L (HRP) secondary AB (Abcam, catalog no. ab6721), and goat anti-rabbit IgG (H + L) Alexa Fluor 488 secondary AB (Thermo Fisher Scientific, catalog no. A11034).
Measurement of glycolysis in GMs
Glycolysis was measured by using the Agilent seahorse XF glycolysis stress test kit (Seahorse Biosciences; catalog no. 103020-100) according to the manufacturer’s guidelines. The value of basal glycolysis, glycolytic capacity, and glycolytic reserve from normoxic and hypoxic GMs was obtained by the analysis of ECAR after the sequential addition of glucose, oligomycin, and 2-deoxyglucose to the Agilent seahorse XF24 flux analyzer. Each experiment was conducted in triplicate.
Calcium imaging and assay
Calcium imaging for U251 GMs was performed by using Fura Red AM fluorescent indicator dye (catalog no. F3020, Invitrogen) with its detection by Nikon Eclipse Ti-S Calcium imaging system (38). In brief, cultured GMs in a 24-well plate was loaded with 10 μM Fura Red AM in Hanks’ balanced salt solution (HBSS) (Gibco, catalog no. 14175095) for 1 hour and kept free from light, followed by washing three times with HBSS buffer. The Fura Red AM–labeled GMs were maintained in HBSS buffer during calcium imaging. The intracellular Ca2+ levels in GMs were quantified using ImageJ software.
For calcium assay, U251 GMs were grown to 90% confluence in a 96-well plate. After washing them with HBSS buffer, GMs were loaded with the Ca2+ indicator, 50 μl of 4 μM Fluo-4 AM (Invitrogen, catalog no. F14201), in HBSS buffer. Next, GMs were incubated at room temperature for 1 hour and kept under dark conditions. After washing them with HBSS buffer, the continuous measurement of fluorescence kinetics was performed (excitation, 485 nm; emission, 525 nm) in a microplate reader. The results were plotted for an average reading over each kinetics cycle done in six replicates. BAPTA-AM (20 μM; Thermo Scientific, catalog no. B6769) was used to chelate intracellular Ca2+. Sodium-l-lactate (20 mM; Sigma-Aldrich, catalog no. 7022) was used to evaluate the effect of extracellular lactate on the intracellular Ca2+ levels in GMs.
OE and KD of MCT1 and CD147 in GMs
Lentiviral particles for MCT1 OE, MCT1 KD, and CD147 OE were produced in HEK 293T cells via the co-transfection of Lenti-Δ8.9 and Lenti-VSVG with Lenti-CMV promoter (CMVP)–MCT1 cDNA-IRES-eGFP, Lenti-H1promoter (H1P)–MCT1 shRNA-CMVP-eGFP, and Lenti-CMVP-CD147 cDNA-IRES-eGFP, respectively. Lentiviral particles that only expressed CMVP-eGFP were used as a negative control. A standard protocol for the transduction of lentiviral particles into GMs was used as follows:
1) Lenti-CMVP-MCT1 cDNA-IRES-eGFP and Lenti-CMVP-CD147 cDNA-IRES-eGFP: IRES oligonucleotides were first inserted into the lenti-FUGW-CMVP-eGFP backbone construct. Then, mMCT1 cDNA (1482 bp) and mCD147 cDNA (816 bp) were amplified by PCR and inserted into the lenti-FUGW-CMVP-IRES-eGFP backbone construct (7). All sequences and their expression were validated.
2) Lenti-H1P-MCT1 shRNA-CMVP-eGFP: Based on previous work (7), the following MCT1 shRNA sense oligonucleotides and antisense oligonucleotides were subcloned into the lenti-FUGW-backbone construct. All sequences, their expression, and their KD efficiency were validated.
5′-GATCCCCGTATCATGCTTTACGATTATTCAAGAGATAATCGTAAAGCATGATACTTTTTTC-3′
5′-TCGAGAAAAAAGTATCATGCTTTACGATTATCTCTTGAATAATCGTAAAGCATGATACGGG-3′
3) CD147 LNA GapmeR antisense oligonucleotides: Antisense oligonucleotides LNA GapmeR and antisense control oligonucleotides (Bio-stations Ltd.; positive control, catalog no. 300632-101; negative control, catalog no. 300610; and for CD147, catalog no. 300600) were transfected directly into GMs (“Gymnosis method”) as per the manufacturer’s guidelines. GMs were maintained in Opti-MEM medium (Gibco, catalog no. 31985070) with oligonucleotides for 24 hours before further analysis. Efficiency of CD147 KD in GMs with antisense oligonucleotides LNA GapmeR was validated by WB analysis.
Exosome isolation
Exosomes were isolated either by a modified differential ultracentrifugation method with filtration (16) or by a low-speed centrifugation method with a total exosome isolation reagent (Invitrogen, catalog no. 4478359) as per the manufacturer’s protocol. Briefly, in the ultracentrifugation method, extracellular medium from cultured GMs within exosome-isolation medium for 24 hours was first centrifuged at 300g for 10 min. The resultant supernatant was further centrifuged at 16,500g for 20 min, followed by the consecutive filtration of supernatant through a 0.22-μm filter (Jet Biofil, catalog no. FPE-204-030). The filtered solution was ultracentrifuged at 120,000g for 70 min. The resultant supernatant was aspirated and discarded to obtain exosome pellets.
In the low-speed centrifugation method with a total exosome isolation reagent, extracellular medium from cultured GMs within exosome-isolation medium for 24 hours was first centrifuged at 2000g for 30 min. Then, the resultant supernatant was mixed with 0.5 volumes of a total exosome isolation reagent. The mixture was incubated at 4°C overnight, followed by its centrifugation at 10,000g for 1 hour at 4°C. The resultant supernatant was aspirated and discarded and the remaining exosome pellet was diluted with 1× PBS for NTA or with 1× radioimmunoprecipitation assay buffer for WB, or fixed with PFA for TEM and immunogold EM.
Analysis of exosomes by NTA
The number and size distribution of GM-derived exosomes were characterized by NTA by using a Malvern NanoSight NS300 instrument (16). In brief, a monochromatic laser beam at 405 nm was applied to 500 ml of exosome solutions loaded into the sample chamber. Three video captures for exosome movements within a 30-s duration were recorded and further analyzed by NTA software (version 2.2, NanoSight) through the optimization for the identification and tracking of exosomes on a frame-by-frame basis. The released number of exosomes from GMs with various conditions was calculated by NTA analysis.
TEM and immunogold EM
The size and morphology of GM-derived exosomes were detected by TEM analysis (16). Briefly, exosomes were fixed with 2.5% PFA for 30 min, washed twice with PBS, dissolved in PBS/0.5% BSA, deposited onto formvar carbon-coated EM grids (catalog no. BZ1102XX, Beijing Zhongjingkeyi Technology Co., Ltd), and exposed for 10 min in a dry environment. Then, exosomes on the grids were washed five times (3 min each) with PBS/0.5% BSA. Afterward, fixed exosomes on the grid were washed twice with PBS/50 mM glycine (3 min each) and lastly with PBS/0.5% BSA (10 min), stained with 2% uranyl acetate for 5 min, and then viewed by an electron microscope (FEI/Philips Tecnai 12 BioTWIN at City University of Hong Kong EM core facility). For the immunogold labeling with ABs, fixed exosomes on the grid were incubated with 5% BSA for 30 min at room temperature, washed five times with PBS/0.5% BSA (3 min), transferred to a drop of the AB (1:50 dilution for anti-CD63 AB, anti-MCT1 AB, and anti-CD147 AB) in PBS/0.5% BSA, and incubated for 2 hours at room temperature. Afterward, exosomes in the grids were washed five times with PBS/0.5% BSA (3 min), incubated with goat anti-rabbit IgG H&L Gold (10 nm) preadsorbed (Abcam, catalog no. ab27234) in PBS/0.5% BSA for 1 hour at room temperature, and then washed five times (3 min) with PBS/0.5% BSA. Last, exosomes on the grids were stained with 2% uranyl acetate and then viewed under an electron microscope.
Uptake of U251 GM-derived exosomes by ECs
bEnd.3 ECs (approximately 30,000 cells per well) were cultured on the chamber slide (Lab-Tek, Thermo Scientific, USA) for 24 hours with normal growth medium at 37°C in a 5% CO2 incubator. On the next day, the cells were washed twice with PBS and replenished again with normal growth medium supplemented with normoxic or hypoxic GM-derived exosomes (250 μg), which were labeled with Exo-Green Exosome Protein Fluorescent Label (System Biosciences, catalog no. EXOG200A-1) (100 μl), and further maintained for 24 hours. Afterward, bEnd.3 ECs were washed three times with PBS and fixed with 4% PFA on ice for 30 min. Fixed bEnd.3 ECs on the slide were stained with rhodamine-conjugated phalloidin AB (1:200 dilutions) (catalog no. R415, Invitrogen) at room temperature for 1 hour to detect their actin cytoskeleton protein, followed by washing with PBS. Then, stained bEnd.3 ECs on the slide were mounted with Vectashield medium containing DAPI and observed under a laser scanning confocal microscope.
Tube formation assay
Twenty-four–well culture plates were coated with 125 μl of GeltrexTM (Gibco, no. A1413201) per well to induce the formation of a monolayer of bEND.3 ECs in the medium containing 0.2% FBS. When bEND.3 ECs reached 80 to 90% confluence after seeding (7 × 104 cells per well), the medium was replaced with one containing normoxic or hypoxic GM-derived exosomes (approximately 250 μg/250 μl medium plus 0.2% FBS). After leaving the culture for an additional 24 hours, tube formation of bEND.3 ECs was examined under a bright-field microscope and its representative pictures were taken at 10× magnification.
LSPR detection of exosomal MCT1 and CD147 with the functionalized SAM-AuNIs sensing chip
The functionalization of gold nanoparticles for LSPR biosensing has been well established in our laboratory and others (39). In brief, dry SAM-AuNIs sensing chips were sequentially rinsed with absolute ethanol (Sigma-Aldrich), incubated in 11-mercaptoundecanoic acid (MUA) solution (10 mM) for 30 min, and followed by rinsing off excess MUA molecules with absolute ethanol. Then, 2-(N-morpholino)ethane sulfonic acid (MES) was prepared by mixing equal volumes of 0.4 M 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) and 0.15 M N-hydroxysuccinimide, and then the freshly prepared MES solution was added to the SAM-AuNIs sensing chip for 20 min to activate the MUA carboxyl functional group. Afterward, 300 μl of polyclonal primary ABs in PBS (2 μg/ml; anti-MCT1 AB; dilution 1:100, and anti-CD147 AB; dilution 1:200) was applied to the SAM-AuNIs sensing chip for 40 min for the immobilization of ABs. Excessive ABs were rinsed away by PBS buffer, and nonspecific sites were further blocked by treatment with 1 M ethanolamine. The common-path interferometric sensing system and differential phase detection method were used to monitor the baseline phase responses during the functionalization process by adding each chemical and ABs to the chip, sequentially, and thereafter we performed the label-free detection of exosomal proteins with the LSPR biosensor with the functionalized chip as described in our previous work (36). For the detection of exosomal MCT1 and CD147 via LSPR biosensing, PBS was used as a basic running buffer. After rinsing SAM-AuNIs sensing chips with PBS, exosome solutions (50 μg/ml PBS) were introduced over the AB-functionalized surface of the sensing chip by using a peristaltic pump at a constant rate of 30 μl/min. The SAM-AuNIs sensing chip was subsequently flushed again by PBS to check the binding affinity and removal of the nonspecific binding of exosomes to ABs. LSPR experiments with exosomes in each experiment were performed three times independently.
AFM detection for MCT1 and CD147 in exosomes immobilized on SAM-AuNIs discs
Biosensing single molecular interaction between surface antigens of immobilized exosomes in SAM-AuNIs discs and anti-MCT1 or anti-CD147 AB functionalized in the sensing tip was conducted by using BioScope Catalyst AFM (Bruker). The spring constant of AFM silicon nitride cantilever was calibrated to be 0.3756 N/m in the detection of exosomal proteins. To capture exosomes, the surface of SAM-AuNIs sample discs of AFM was functionalized with anti-CD63 AB as described above (16). Two hundred microliters of exosome solution [PBS (50 μg/ml)] was first added to the sample discs, incubated for 10 min, and replaced with 1 ml of fresh PBS by mild decantation. Immunocaptured exosomes on the surface of the discs were further confirmed and analyzed by AFM scanning.
To determine exosomal MCT1 and CD147 levels by the measurement of intermolecular force between antigens and ABs, the silver nitride AFM tip (ScanAsyst-Fluid, TELTEC semiconductor pacific limited) was functionalized with either anti-MCT1 AB or anti-CD147 AB. In brief, primary ABs (anti-MCT1 AB; dilution 1:100, and anti-CD147 AB; dilution 1:200) were covalently attached to the Si3N4 tip of AFM via thiol ester linkage (Bruker). The probe tip was washed with PBS, incubated in blocking solution (1% BSA-PBS) for 1 hour, followed by a series of washing with PBS.
All measurements of exosomal proteins with AFM were recorded in PBS. Separation forces between MCT1 or CD147 in exosomes on SAM-AuNIs discs and anti-MCT1 or anti-CD147 AB on the sensing tips were measured by AFM ramp mode. Exosomal MCT1 and CD147 levels were determined and analyzed by the maximum peak of the AFM force-distance curve. Biophysical properties, including roughness, Young’s modulus, and adhesion force, were recorded for exosomes captured on the SAM-AuNIs discs by single ramping mode by using a bare AFM sensing tip with a spring constant of 0.3801 N/m (40). A bare SAM-AuNIs sample disc was used as a control in the experiment. Each AFM force curve was obtained from at least three independent experiments.
Zeta potential analysis of exosomes
Zeta potential of exosomes was measured and analyzed by Malvern Zetasizer Nano ZS using equally diluted samples prepared with equal amount of exosomes (50 μg/ml) within PBS for each group (16).
Orthotopic mouse models of glioma
Six-week-old female SCID mice were anesthetized by 1 to 2% isoflurane mixed in oxygen and fixed in a stereotactic frame. The injection coordinates for GM implantation into the brain were 0.2 mm anterior and 2.2 mm lateral from the bregma and 2.3 to 2.8 mm deep from the outer border of the cranium, respectively. In brief, a hole was drilled into the mouse skull in the cortex of the right frontal lobe. Then, 10 μl of 3 × 104 U251 or U87 GMs was injected through the hole by a Hamilton syringe with a 26-gauge needle at a flow rate of 0.5 μl/min using a microinjector.
Magnetic resonance imaging
All MRI images were acquired with a horizontal bore 3-T preclinical Bruker MRI system (Bruker, Ettlingen, Germany) with a 23-mm-diameter surface coil. Mice were anesthetized with 1 to 2% isoflurane carried in oxygen. After anesthesia induction, mice were placed on the animal bed with a warm pad to keep body temperature at 37°C. During an MRI scan, continuous monitoring of mouse respiration was conducted (SA Instrumentation). T2-weighted MRI for the brain was performed on days 5 and 10 after GM implantation to check glioma size, and the blood was then collected for the isolation of exosomes. The parameters of T2-weighted images were as follows: repetition time/echo time = 2146/16 ms, field of view = 16 × 16 mm, data acquisition matrix = 256 × 256, number of averages = 8, and rare acquisition with relaxation enhancement (RARE) factor = 10. The size of a tumor was calculated using ImageJ software.
Statistical analysis
All graphs were made, and statistical analyses were conducted using GraphPad Prism, or Microsoft Excel 2010. Statistical significance was analyzed by either an unpaired, two-tailed Student’s t test or one-way analysis of variance (ANOVA) with multiple comparisons by Dunnett’s test, or two-way ANOVA followed by post hoc analysis. A respective control for each experimental group was precisely chosen and used for all statistical comparisons. Statistical analyses indicated *P < 0.05 and **P < 0.01 as significance level. All quantitative data from multiple independent experiments were calculated and presented as the means ± SD as described in the legend of each figure.
Supplementary Material
Acknowledgments
We thank C. C. Fong (BMS, City University of Hong Kong) for technical support, K. M. Chan (BMS, City University of Hong Kong) for providing SF7761 GSCs, J. S. Yoo (HTI, The Hong Kong Polytechnic University) for providing the GL261 GMs, and R. G. Jesky (BMS, City University of Hong Kong) for editing the manuscript. Funding: This work was supported by City University of Hong Kong (grant nos. 9610340 and 7200472), Early Career Scheme (ECS)–UGC (grant no. 21102517), and General Research Fund (GRF)–UGC (grant no. 11103918) awarded to Y.L. at City University of Hong Kong. Author contributions: Y.L. designed the whole experiments. A.T. conducted most of the experiments. G.Q. and C.X. performed the LSPR-related experiments. G.Q., C.X., S.P.N., and C.M.L.W. analyzed the LSPR-related data. X.H. established the mouse model of glioma and performed MRI analysis. X.H. and K.W.Y.C. analyzed the MRI-related data. A.T. and Y.L. drafted the manuscript. All the authors revised and approved the content of the manuscript. Competing interests: A.T., G.Q., C.X., T.Y., S.P.N., C.M.L.W., and Y.L. are inventors on a patent to be filed with the USPTO related to this work for the LSPR- and AFM-based detection of exosomal MCT1 and CD147 for monitoring glioma progression. This work has also been submitted as part of the Ph.D. thesis of A.T. at City University of Hong Kong. All other authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/26/eaaz6119/DC1
REFERENCES AND NOTES
- 1.Sanai N., Alvarez-Buylla A., Berger M. S., Neural stem cells and the origin of gliomas. N. Engl. J. Med. 353, 811–822 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Koppenol W. H., Bounds P. L., Dang C. V., Otto Warburg’s contributions to current concepts of cancer metabolism. Nat. Rev. Cancer 11, 325–337 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Vander Heiden M. G., Cantley L. C., Thompson C. B., Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birsoy K., Wang T., Possemato R., Yilmaz O. H., Koch C. E., Chen W. W., Hutchins A. W., Gultekin Y., Peterson T. R., Carette J. E., Brummelkamp T. R., Clish C. B., Sabatini D. M., MCT1-mediated transport of a toxic molecule is an effective strategy for targeting glycolytic tumors. Nat. Genet. 45, 104–108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martinez-Outschoorn U. E., Peiris-Pagés M., Pestell R. G., Sotgia F., Lisanti M. P., Cancer metabolism: A therapeutic perspective. Nat. Rev. Clin. Oncol. 14, 11–31 (2017). [DOI] [PubMed] [Google Scholar]
- 6.San-Millán I., Brooks G. A., Reexamining cancer metabolism: Lactate production for carcinogenesis could be the purpose and explanation of the Warburg Effect. Carcinogenesis 38, 119–133 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee Y., Morrison B. M., Li Y., Lengacher S., Farah M. H., Hoffman P. N., Liu Y., Tsingalia A., Jin L., Zhang P.-W., Pellerin L., Magistretti P. J., Rothstein J. D., Oligodendroglia metabolically support axons and contribute to neurodegeneration. Nature 487, 443–448 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halestrap A. P., The monocarboxylate transporter family—Structure and functional characterization. IUBMB Life 64, 1–9 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Miranda-Gonçalves V., Granja S., Martinho O., Honavar M., Pojo M., Costa B. M., Pires M. M., Pinheiro C., Cordeiro M., Bebiano G., Costa P., Reis R. M., Baltazar F., Hypoxia-mediated upregulation of MCT1 expression supports the glycolytic phenotype of glioblastomas. Oncotarget 7, 46335–46353 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colen C. B., Shen Y., Ghoddoussi F., Yu P., Francis T. B., Koch B. J., Monterey M. D., Galloway M. P., Sloan A. E., Mathupala S. P., Metabolic targeting of lactate efflux by malignant glioma inhibits invasiveness and induces necrosis: An in vivo study. Neoplasia 13, 620–632 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kucharzewska P., Christianson H. C., Welch J. E., Svensson K. J., Fredlund E., Ringnér M., Mörgelin M., Bourseau-Guilmain E., Bengzon J., Belting M., Exosomes reflect the hypoxic status of glioma cells and mediate hypoxia-dependent activation of vascular cells during tumor development. Proc. Natl. Acad. Sci. U.S.A. 110, 7312–7317 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalluri R., The biology and function of exosomes in cancer. J. Clin. Invest. 126, 1208–1215 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skog J., Würdinger T., van Rijn S., Meijer D. H., Gainche L., Sena-Esteves M., Curry W. T. Jr., Carter B. S., Krichevsky A. M., Breakefield X. O., Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 10, 1470–1476 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D’Asti E., Chennakrishnaiah S., Lee T. H., Rak J., Extracellular vesicles in brain tumor progression. Cell. Mol. Neurobiol. 36, 383–407 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chistiakov D. A., Chekhonin V. P., Extracellular vesicles shed by glioma cells: Pathogenic role and clinical value. Tumor Biol. 35, 8425–8438 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Thakur A., Qiu G., Ng S.-P., Guan J., Yue J., Lee Y., Wu C.-M. L., Direct detection of two different tumor-derived extracellular vesicles by SAM-AuNIs LSPR biosensor. Biosens. Bioelectron. 94, 400–407 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Figueroa J. M., Skog J., Akers J., Li H., Komotar R., Jensen R., Ringel F., Yang I., Kalkanis S., Thompson R., LoGuidice L., Berghoff E., Parsa A., Liau L., Curry W., Cahill D., Bettegowda C., Lang F. F., Chiocca E. A., Henson J., Kim R., Breakefield X., Chen C., Messer K., Hochberg F., Carter B. S., Detection of wild-type EGFR amplification and EGFRvIII mutation in CSF-derived extracellular vesicles of glioblastoma patients. Neuro Oncol. 19, 1494–1502 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.García-Romero N., Carrión-Navarro J., Esteban-Rubio S., Lázaro-Ibáñez E., Peris-Celda M., Alonso M. M., Guzmán-De-Villoria J., Fernández-Carballal C., de Mendivil A. O., García-Duque S., Escobedo-Lucea C., Prat-Acín R., Belda-Iniesta C., Ayuso-Sacido A., DNA sequences within glioma-derived extracellular vesicles can cross the intact blood-brain barrier and be detected in peripheral blood of patients. Oncotarget 8, 1416–1428 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Indira Chandran V., Welinder C., Gonçalves de Oliveira K., Cerezo-Magaña M., Månsson A.-S., Johansson M. C., Marko-Varga G., Belting M., Global extracellular vesicle proteomic signature defines U87-MG glioma cell hypoxic status with potential implications for non-invasive diagnostics. J. Neurooncol 144, 477–488 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu X., Yadav N. N., Knutsson L., Hua J., Kalyani R., Hall E., Laterra J., Blakeley J., Strowd R., Pomper M., Barker P., Chan K., Liu G., McMahon M. T., Stevens R. D., van Zijl P. C. M., Dynamic Glucose-Enhanced (DGE) MRI: Translation to human scanning and first results in glioma patients. Tomography 1, 105–114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hochberg F. H., Pruitt A., Assumptions in the radiotherapy of glioblastoma. Neurology 30, 907–911 (1980). [DOI] [PubMed] [Google Scholar]
- 22.Shi R., Wang P.-Y., Li X.-Y., Chen J.-X., Li Y., Zhang X.-Z., Zhang C.-G., Jiang T., Li W.-B., Ding W., Cheng S.-J., Exosomal levels of miRNA-21 from cerebrospinal fluids associated with poor prognosis and tumor recurrence of glioma patients. Oncotarget 6, 26971–26781 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Indira Chandran V., Welinder C., Månsson A.-S., Offer S., Freyhult E., Pernemalm M., Lund S. M., Pedersen S., Lehtiö J., Marko-Varga G., Johansson M. C., Englund E., Sundgren P. C., Belting M., Ultrasensitive immunoprofiling of plasma extracellular vesicles identifies syndecan-1 as a potential tool for minimally invasive diagnosis of glioma. Clin. Cancer Res. 25, 3115–3127 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Im H., Shao H., Il Park Y., Peterson V. M., Castro C. M., Weissleder R., Lee H., Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat. Biotechnol. 32, 490–495 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rupert D. L. M., Claudio V., Lässer C., Bally M., Methods for the physical characterization and quantification of extracellular vesicles in biological samples. Biochim. Biophys. Acta Gen. Subj. 1861, 3164–3179 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Sepúlveda B., Angelomé P. C., Lechuga L. M., Liz-Marzán L. M., LSPR-based nanobiosensors. Nano Today 4, 244–251 (2009). [Google Scholar]
- 27.Hinterdorfer P., Baumgartner W., Gruber H. J., Schilcher K., Schindler H., Detection and localization of individual antibody-antigen recognition events by atomic force microscopy. Proc. Natl. Acad. Sci. U.S.A. 93, 3477–3481 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parisse P., Rago I., Ulloa Severino L., Perissinotto F., Ambrosetti E., Paoletti P., Ricci M., Beltrami A. P., Cesselli D., Casalis L., Atomic force microscopy analysis of extracellular vesicles. Eur. Biophys. J. 46, 813–820 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Savina A., Furlán M., Vidal M., Colombo M. I., Exosome release is regulated by a calcium-dependent mechanism in K562 cells. J. Biol. Chem. 278, 20083–20090 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Roland C. L., Arumugam T., Deng D., Liu S. H., Philip B., Gomez S., Burns W. R., Ramachandran V., Wang H., Cruz-Monserrate Z., Logsdon C. D., Cell surface lactate receptor gpr81 is crucial for cancer cell survival. Cancer Res. 74, 5301–5310 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coffey R. J., Lunsford D. L., Taylor F. H., Survival after stereotactic biopsy of malignant gliomas. Neurosurgery 22, 465–473 (1988). [DOI] [PubMed] [Google Scholar]
- 32.Kirk P., Wilson M. C., Heddle C., Brown M. H., Barclay A. N., Halestrap A. P., CD147 is tightly associated with lactate transporters MCT1 and MCT4 and facilitates their cell surface expression. EMBO J. 19, 3896–3904 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hashimoto T., Hussien R., Oommen S., Gohil K., Brooks G. A., Lactate sensitive transcription factor network in L6 cells: Activation of MCT1 and mitochondrial biogenesis. FASEB J. 21, 2602–2612 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Banelli B., Forlani A., Allemanni G., Morabito A., Pistillo M. P., Romani M., MicroRNA in glioblastoma: An overview. Int. J. Genomics. 2017, 7639084 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Romero-Garcia S., Moreno-Altamirano M. M. B., Prado-Garcia H., Sánchez-García F. J., Lactate contribution to the tumor microenvironment: Mechanisms, effects on immune cells and therapeutic relevance. Front. Immunol. 7, 52 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qiu G., Thakur A., Xu C., Ng S.-P., Lee Y., Wu C.-M. L., Detection of glioma-derived exosomes with the biotinylated antibody-functionalized titanium nitride plasmonic biosensor. Adv. Funct. Mater. 29, 1806761 (2019). [Google Scholar]
- 37.Justus C. R., Leffler N., Ruiz-Echevarria M., Yang L. V., In vitro cell migration and invasion assays. J. Vis. Exp. 2014, 51046 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barreto-Chang O. L., Dolmetsch R. E., Calcium imaging of cortical neurons using Fura-2 AM. J. Vis. Exp. 2009, 1067 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loo F.-C., Ng S.-P., Wu C.-M. L., Kong S. K., An aptasensor using DNA aptamer and white light common-path SPR spectral interferometry to detect cytochrome-c for anti-cancer drug screening. Sens. Actuators B 198, 416–423 (2014). [Google Scholar]
- 40.Bippes C. A., Muller D. J., High-resolution atomic force microscopy and spectroscopy of native membrane proteins. Reports Prog. Phys. 74, 086601 (2011). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/26/eaaz6119/DC1


