Abstract
Many common illnesses differentially affect men and women for unknown reasons. The autoimmune diseases lupus and Sjögren’s syndrome affect nine times more women than men1, whereas schizophrenia affects men more frequently and severely2. All three illnesses have their strongest common genetic associations in the Major Histocompatibility Complex (MHC) locus, an association that in lupus and Sjögren’s syndrome has long been thought to arise from alleles of the human leukocyte antigen (HLA) genes at that locus3–6. Here we show that the complement component 4 (C4) genes, which are also in the MHC locus and were recently found to increase risk for schizophrenia7, generate 7-fold variation in risk for lupus (95% CI: 5.88–8.61; p < 10−117 in total) and 16-fold variation in risk for Sjögren’s syndrome (95% CI: 8.59–30.89; p < 10−23 in total) among individuals with common C4 genotypes, with C4A protecting more strongly than C4B in both illnesses. The same alleles that increase risk for schizophrenia greatly reduced risk for lupus and Sjögren’s syndrome. In all three illnesses, C4 alleles acted more strongly in men than in women: common combinations of C4A and C4B generated 14-fold variation in risk for lupus, 31-fold variation in risk for Sjögren’s syndrome, and 1.7-fold variation in schizophrenia risk among men (vs. 6-fold, 15-fold, and 1.26-fold among women respectively). At a protein level, both C4 and its effector C3 were present at greater levels in men than women in cerebrospinal fluid (p < 10−5 for both C4 and C3) and plasma8,9 among adults ages 20–50, corresponding to the ages of differential disease vulnerability. Sex differences in complement protein levels may help explain the larger effects of C4 alleles in men, women’s greater risk of SLE and Sjögren’s, and men’s greater vulnerability in schizophrenia. These results implicate the complement system as a source of sexual dimorphism in vulnerability to diverse illnesses.
Systemic lupus erythematosus (SLE, or “lupus”) is a systemic autoimmune disease of unknown cause. Risk of SLE is heritable (66%10), although SLE may have environmental triggers, as its onset often follows events that damage cells, such as infections and severe sunburns11. Most SLE patients produce autoantibodies against nucleic acid complexes, including ribonucleoproteins and DNA12.
In genetic studies, SLE associates most strongly with variation across the major histocompatibility complex (MHC) locus, which contains the human leukocyte antigen (HLA) genes3. However, conclusive attribution of this association to specific genes and alleles has been difficult; the identities of the most likely genetic sources have been frequently revised as genetic studies have grown in size4,5. In several other autoimmune diseases, including type 1 diabetes, celiac disease, and rheumatoid arthritis, strong effects of the MHC locus arise from HLA alleles that cause the peptide binding groove of HLA proteins to present a disease-critical autoantigen13,14. In SLE, by contrast, genetic variants in the MHC locus (including SNPs and HLA alleles) associate broadly with the presence of diverse autoantibodies15.
The complement component 4 (C4A and C4B) genes are also present in the MHC genomic region, between the class I and class II HLA genes. Classical complement proteins help eliminate debris from dead and damaged cells, attenuating the visibility of diverse intracellular proteins to the adaptive immune system. C4A and C4B commonly vary in genomic copy number16 and encode complement proteins with distinct affinities for molecular targets17,18. SLE frequently presents with hypocomplementemia that worsens during flares, possibly reflecting increased active consumption of complement19. Rare cases of severe, early-onset SLE can involve complete deficiency of a complement component (C4, C2, or C1Q)20,21, and one of the strongest common-variant associations in SLE maps to ITGAM, which encodes a receptor for C3, the effector of C422. Although total C4 gene copy number associates with SLE risk23,24, this association is thought to arise from linkage disequilibrium (LD) with alleles of nearby HLA genes25, which have been the focus of fine-mapping analyses3,4.
The complex genetic variation at C4 – arising from many alleles with different numbers of C4A and C4B genes – has been challenging to analyze in large cohorts. A recently feasible approach to this problem is based on imputation: people share long haplotypes with the same combinations of SNP and C4 alleles, such that C4A and C4B gene copy numbers can be imputed from SNP data7. To analyze C4 in large cohorts, we developed a way to identify C4 alleles from whole-genome sequence (WGS) data (Extended Data Fig. 1a, b), then analyzed WGS data from 1,265 individuals (from the Genomic Psychiatry Cohort26,27) to create a large multi-ancestry panel of 2,530 reference haplotypes of MHC-region SNPs and C4 alleles (Extended Data Fig. 1c) – ten times more than in earlier work7. We then analyzed SNP data from the largest SLE genetic association study3 (ImmunoChip 6,748 SLE cases and 11,516 controls of European ancestry) (Extended Data Fig. 2a, b), imputing C4 alleles to estimate the SLE risk associated with common combinations of C4A and C4B gene copy numbers (Fig. 1a).
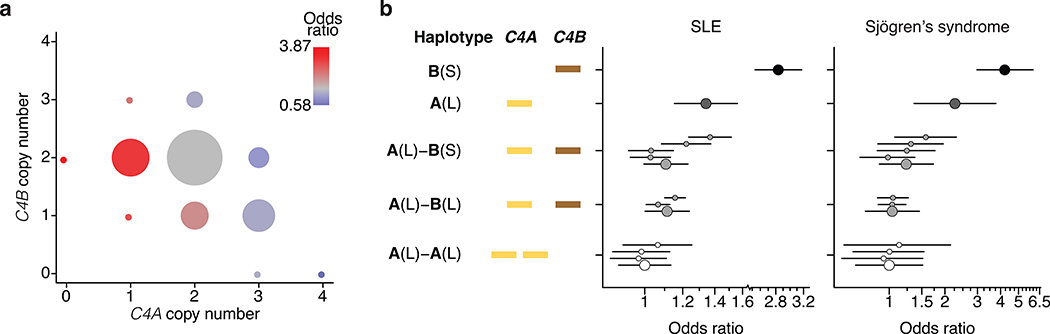
Figure 1. Association of SLE and Sjögren’s syndrome (SjS) with C4 alleles.
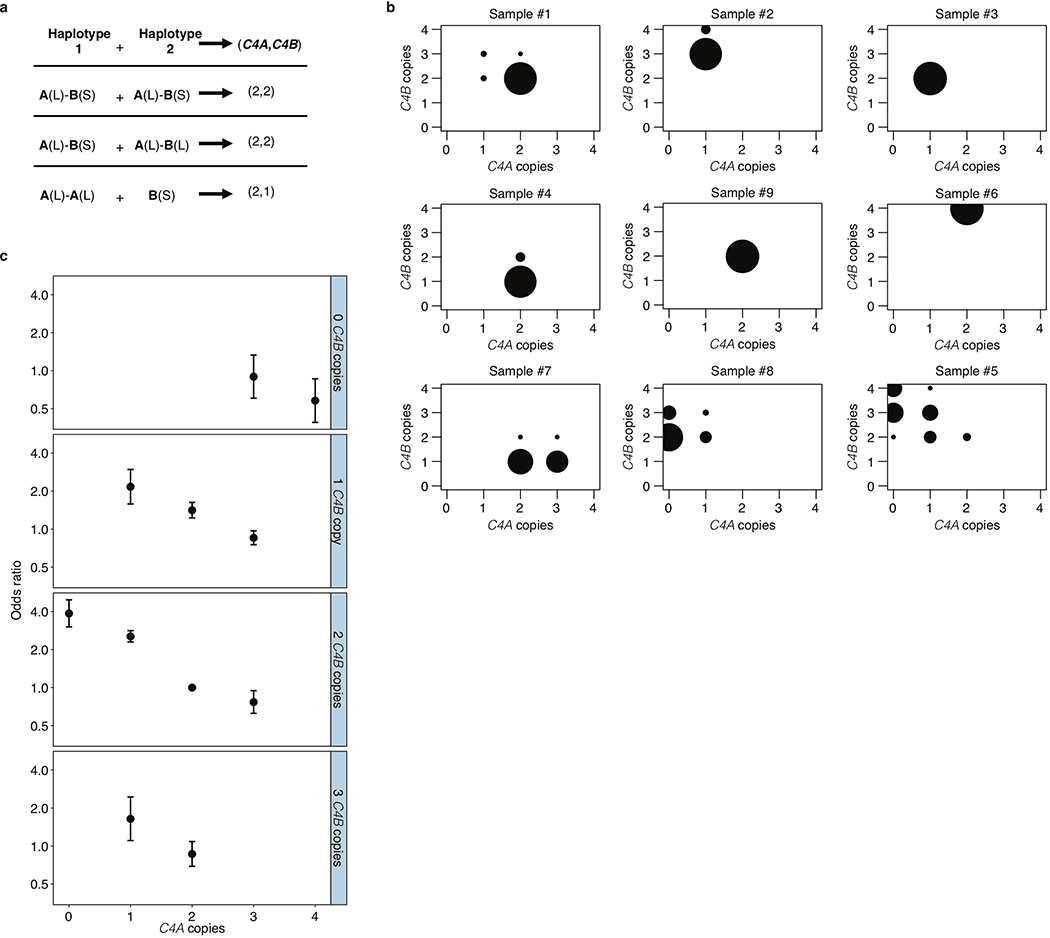
(a) Levels of SLE risk associated with 11 common combinations of C4A and C4B gene copy number. The color of each circle reflects the level of SLE risk (odds ratio) associated with a specific combination of C4A and C4B gene copy numbers relative to the most common combination (two copies of C4A and two copies of C4B) in gray. The area of each circle is proportional to the number of individuals with that number of C4A and C4B genes. Paths from left to right on the plot reflect the effect of increasing C4A gene copy number (greatly reduced risk); paths from bottom to top reflect the effect of increasing C4B gene copy number (modestly reduced risk); and diagonal paths from upper left to lower right reflect the effect of exchanging C4B for C4A copies (modestly reduced risk). Data are from analysis of 6,748 SLE cases and 11,516 controls of European ancestry. The odds ratios are reported with confidence intervals in Extended Data Fig. 2c.
(b) SLE and SjS risk associated with common combinations of C4 structural allele and MHC SNP haplotype. For each C4 locus structure, separate odds ratios are reported for each “haplogroup,” i.e., the MHC SNP haplotype background on which the C4 structure segregates. Data are from analyses of 6,748 SLE cases and 11,516 controls for the left plot and 673 SjS cases and 1,153 controls for the right plot. Error bars represent 95% confidence intervals around the effect size estimate for each allele.
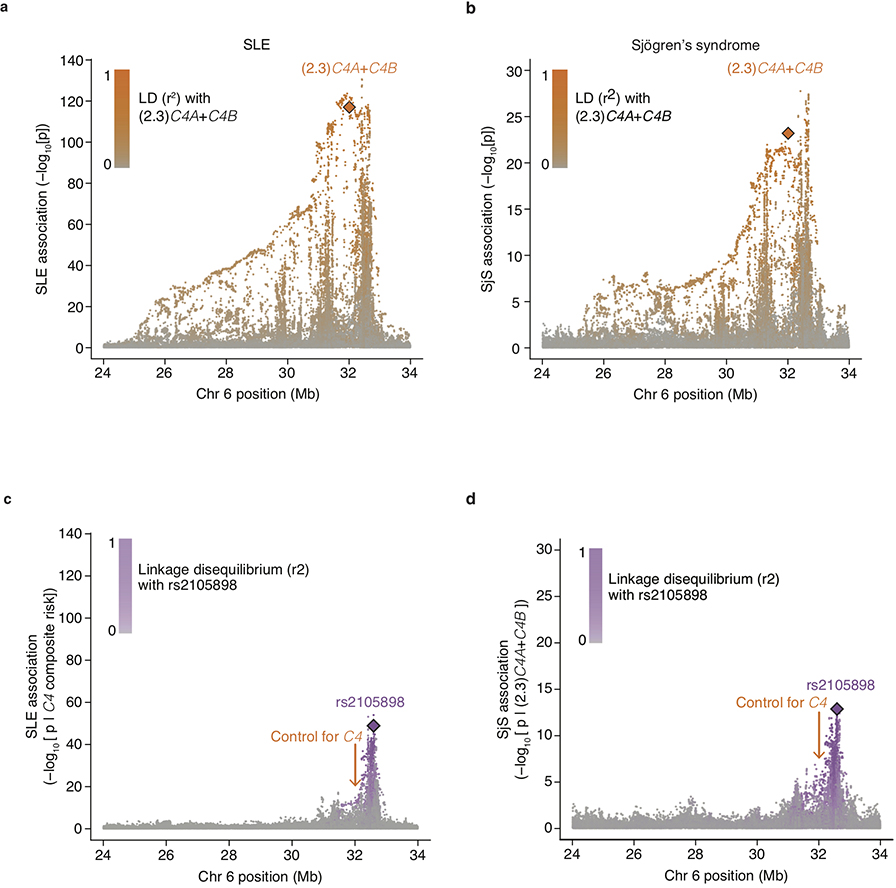
Groups of research participants with the eleven most common combinations of C4A and C4B gene copy number exhibited 7-fold variation in their relative risk of SLE (Fig. 1a, Extended Data Fig. 2c). The relationship between SLE risk and C4 gene copy number exhibited consistent, logical patterns across the 11 genotype groups. For each C4B copy number, greater C4A copy number associated with reduced SLE risk (Fig. 1a, Extended Data Fig. 2c). For each C4A copy number, greater C4B copy number associated with more modestly reduced risk (Fig. 1a). Logistic-regression analysis estimated that the protection afforded by each copy of C4A (OR: 0.54; 95% CI: [0.51, 0.57]) was equivalent to that of 2.3 copies of C4B (OR: 0.77; 95% CI: [0.71,0.82]). We calculated an initial C4-derived risk score as 2.3 times the number of C4A genes, plus the number of C4B genes, in an individual’s genome. Despite clear limitations of this risk score – it is imperfectly imputed from flanking SNP haplotypes (r2 = 0.77, Extended Data Table 1) and only approximates C4-derived risk by using a simple, linear model (to avoid over-fitting the genetic data) – SNPs across the MHC genomic region tended to associate with SLE in proportion to their level of LD with this risk score (Extended Data Fig. 3a).
Combinations of many different C4 alleles generate the observed variation in C4A and C4B gene copy number; particular C4A and C4B gene copy numbers have also arisen recurrently on multiple SNP haplotypes7 (Extended Data Fig. 1c). Analysis of SLE risk in relation to each of these C4 alleles and SNP haplotypes reinforced the conclusion that C4A contributes strong protection, and C4B more modest protection, from SLE, and that C4 genes (rather than nearby variants) are the principal drivers of this variation in risk levels (Fig. 1b).
These results prompted us to consider whether other autoimmune disorders with similar patterns of genetic association at the MHC genomic region might also be driven in part by C4 variation. Primary Sjögren’s syndrome (SjS) is a heritable (54%28) systemic autoimmune disorder of exocrine glands, characterized primarily by dry eyes and mouth with other systemic effects. At a protein level, SjS is (like SLE) characterized by diverse autoantibodies, including antinuclear antibodies targeting ribonucleoproteins29, and hypocomplementemia30. The largest source of common genetic risk for SjS lies in the MHC genomic locus31, with associations to the same haplotype(s) as in SLE6 and with heterogeneous HLA associations in different ancestries32. We imputed C4 alleles into existing SNP data from a European-ancestry SjS case-control cohort (673 cases and 1153 controls). As in SLE, logistic-regression analyses found both C4A copy number (OR: 0.41; 95% CI: [0.34, 0.49]) and C4B copy number (OR: 0.67; 95% CI: [0.53, 0.86]) to be protective against SjS. The risk-equivalent ratio of C4B to C4A gene copies was similar in SjS and SLE (about 2.3 to 1); also, as with SLE, nearby SNPs associated with SjS in proportion to their LD with a C4-derived risk score ((2.3)C4A+C4B ) (Extended Data Fig. 3b). The distribution of SjS risk across the individual C4 alleles and haplotypes revealed a pattern that, as in SLE, supported greater protective effect from C4A than C4B, and little effect of flanking SNP haplotypes (Fig. 1b).
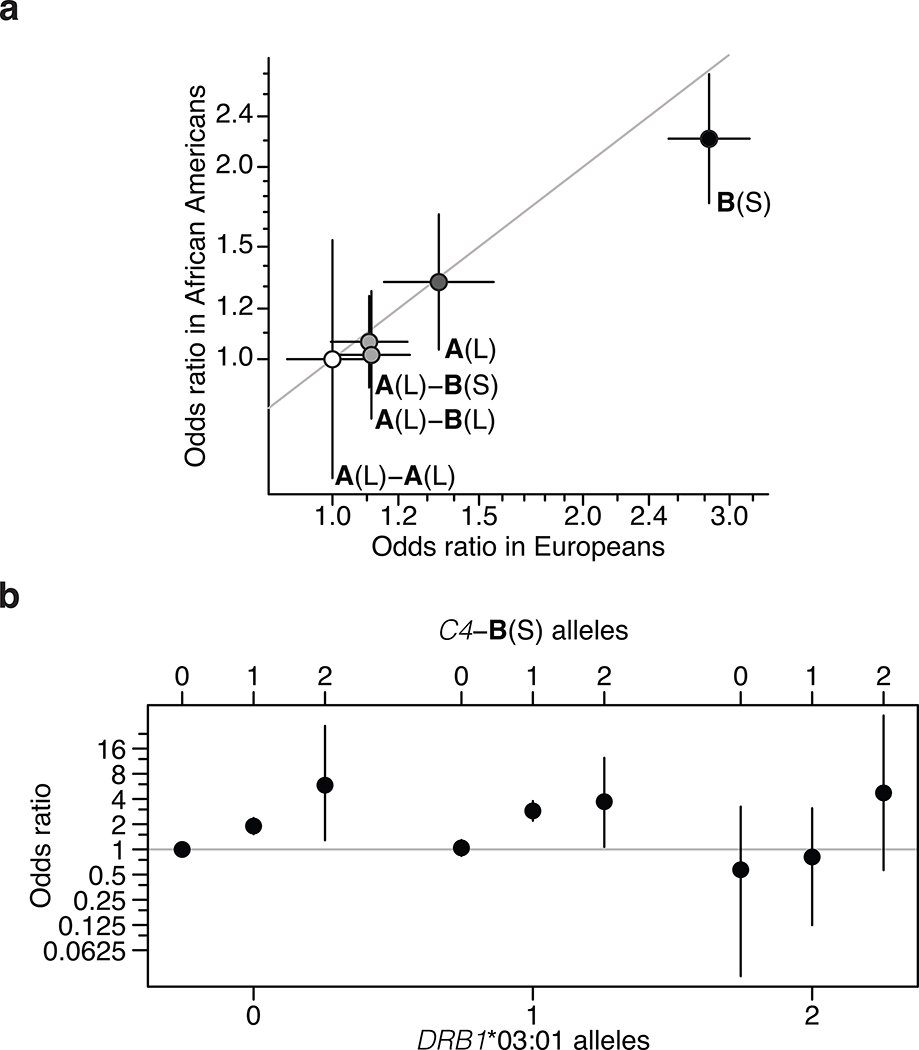
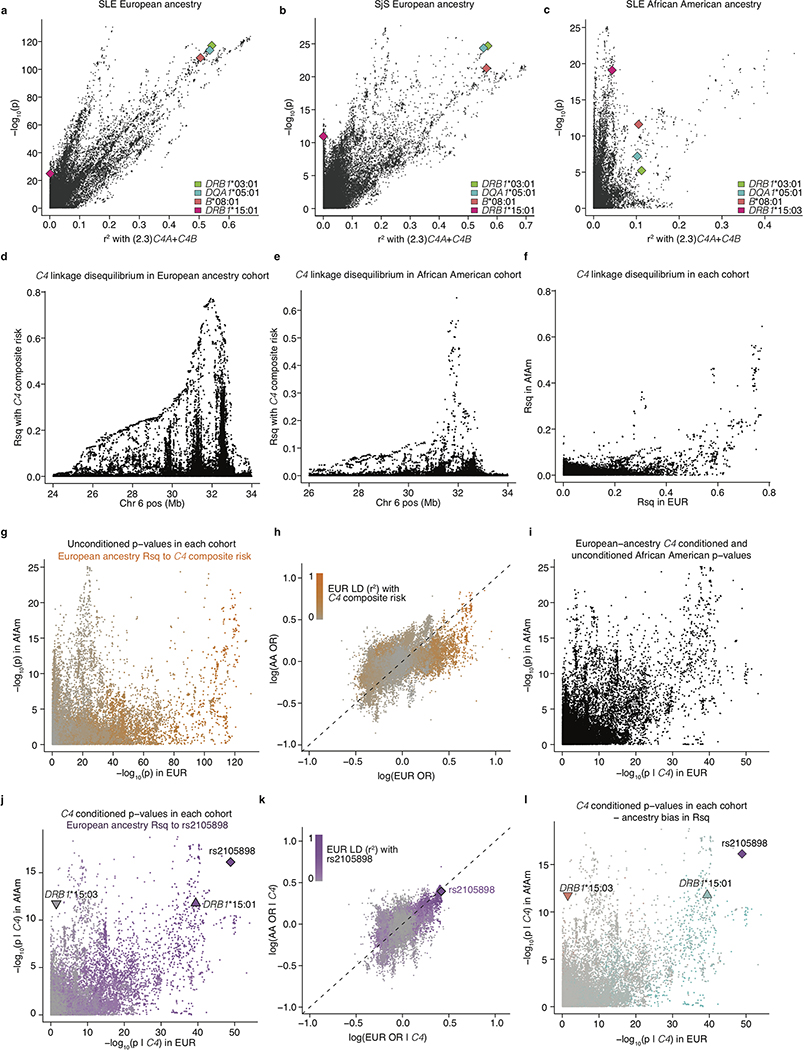
The association of SLE and SjS with C4 gene copy number has long been attributed to the HLA-DRB1*03:01 allele. In European populations, DRB1*03:01 is in strong LD (r2 = 0.71) with the common C4-B(S) allele, which lacks any C4A gene and is the highest-risk C4 allele in our analysis (Fig. 1b); many MHC-region SNPs associated with SLE and SjS in proportion to their LD correlations with both C4 and DRB1*03:01 (Extended Data Fig. 4a, b). Cohorts with other ancestries can have recombinant haplotypes that disambiguate the contributions of alleles that are in LD in Europeans. Among African Americans, we found that common C4 alleles exhibited far less LD with HLA alleles; in particular, the LD between C4-B(S) and DRB1*03:01 was low (r2 = 0.10) (Extended Data Table 2). Thus, genetic data from an African American SLE cohort (1,494 cases, 5,908 controls) made it possible to distinguish between these potential genetic effects. Joint association analysis of C4A, C4B, and DRB1*0301 implicated C4A (p < 10−14) and C4B (p < 10−5) but not DRB1*0301 (p = 0.29) (Extended Data Table 3). Each C4 allele associated with effect sizes of similar magnitude on SLE risk in Europeans and African Americans (Fig. 2a). An analysis specifically of combinations of C4-B(S) and DRB1*03:01 allele dosages in African Americans showed that C4-B(S) alleles consistently increased SLE risk regardless of DRB1*03:01 status, whereas DRB1*03:01 had no consistent effect when controlling for C4-B(S) (Fig. 2b). Although C4 alleles had less LD with nearby variants on African American than on European haplotypes, SNPs across the genomic region associated with SLE in proportion to LD correlations with C4 in African Americans as well (Extended Data Fig. 4c).
Figure 2. C4 and trans-ancestral analysis of the MHC association signal in SLE.
(a) Common C4 alleles exhibit similar strengths of association (odds ratios) in European-ancestry and African American (1,494 SLE cases; 5,908 controls) cohorts. Error bars represent 95% confidence intervals around the effect size estimate for each sex.
(b) Analysis of SLE risk across combinations of C4-B(S) and DRB1*03:01 genotypes in an African American SLE case–control cohort, in which the two alleles exhibit very little LD (r2 = 0.10). On each DRB1*03:01 genotype background, additional C4-B(S) alleles increase risk (i.e. within each grouping). Whereas on each C4-B(S) background, DRB1*03:01 alleles have no appreciable relationship with risk (this can be seen by comparing, for example, the first of the three points from each group). Error bars represent 95% confidence intervals around the effect size estimate for each combination of C4-B(S) and DRB1*03:01.
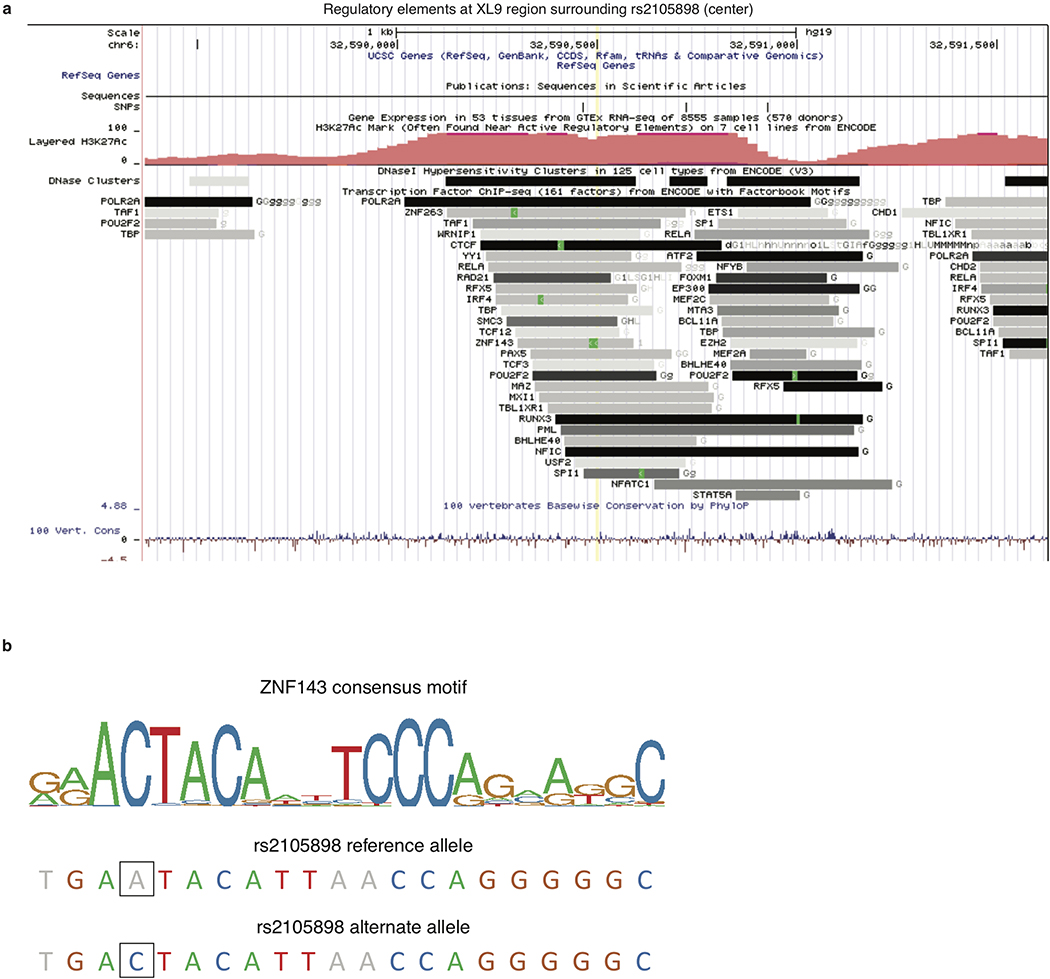
Accounting for C4 alleles in jointly analyzing the SLE association data from African American and European ancestry cohorts also enabled the mapping of an additional, more-modest genetic effect independent of C4; this effect (tagged by rs2105898 and rs9271513) appeared to involve noncoding variation in the HLA class II XL9 region that associates most strongly with expression levels (rather than the coding sequence) of many HLA class II genes (Extended Data Figs. 3c, d, 4d–l, 5, and Supplementary Note 1).
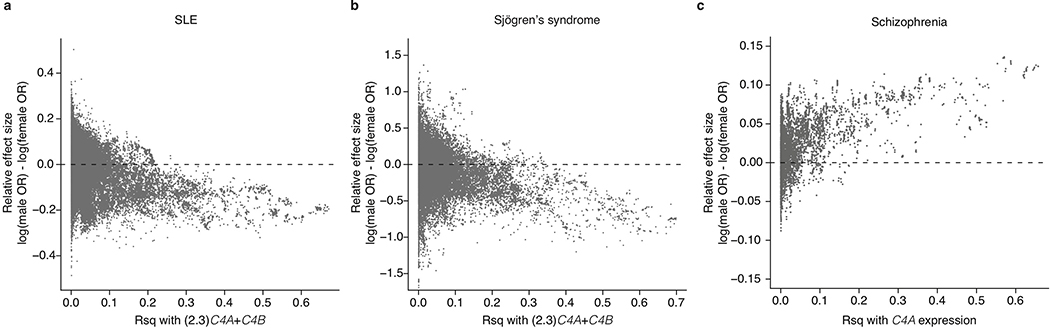
Alleles at C4 that increase dosage of C4A (and to a more modest extent C4B) appear to protect strongly against SLE and SjS (Fig. 1a, b); by contrast, alleles that increase expression of C4A in the brain are more common among research participants with schizophrenia6. These same illnesses exhibit striking, and opposite, sex differences: SLE and SjS are nine times more common among women of childbearing age than among men of a similar age1, whereas in schizophrenia, women exhibit less severe symptoms, more frequent remission of symptoms, lower relapse rates, and lower overall incidence2. Though the vast majority of genetic associations in complex diseases are shared between men and women33, the SNPs that most strongly associate with SLE risk within the MHC region associate to larger potential effect sizes in men34. Hence, we sought to evaluate the possibility that the effects of C4 alleles on risk in SLE, SjS, and schizophrenia might differ between men and women.
Analysis indicated that the effects of C4 alleles were stronger in men. When a sex-by-C4 interaction term was included in association analyses, this term was significant for both SLE (p = 0.002) and schizophrenia (p = 0.0024), with larger C4 effects in men for both disorders. (Analysis of SjS had limited power due to the small number of men affected by SjS.) For both SLE and schizophrenia, the individual C4 alleles consistently associated with stronger effects in men than women (Fig. 3a, b). SNPs across the MHC genomic region exhibited sex-biased association to SLE, SjS, and schizophrenia to the extent of their LD with C4 (Extended Data Fig. 6a–c).
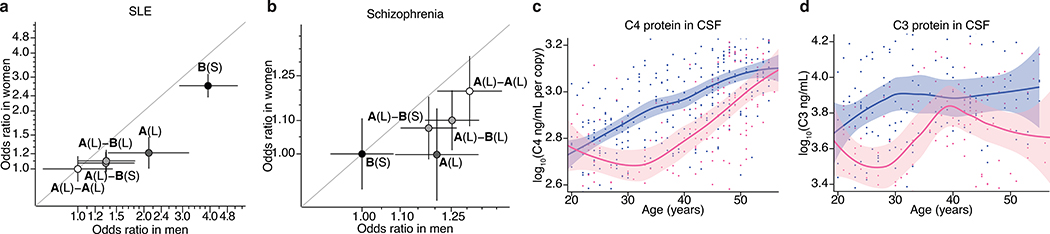
Figure 3. Sex differences in the magnitude of C4 genetic effects and complement protein concentrations.
(a) SLE risk (odds ratios) associated with the four most common C4 alleles in men (x-axis) and women (y-axis) among 6,748 affected and 11,516 unaffected individuals of European ancestry. For each sex, the lowest-risk allele (C4-A(L)-A(L)) is used as a reference (odds ratio of 1.0). Shading of each point reflects the relative level of SLE risk (darker = greater risk) conferred by C4A and C4B copy numbers as in Fig. 2b. Error bars represent 95% confidence intervals around the effect size estimate for each sex.
(b) Schizophrenia risk (odds ratios) associated with the four most common C4 alleles in men (x-axis) and women (y-axis) among 28,799 affected and 35,986 unaffected individuals of European ancestry, aggregated by the Psychiatric Genomics Consortium43. For each sex, the lowest-risk allele (C4-B(S)) is used as a reference (odds ratio of 1.0). For visual comparison with a, shading of each allele reflects the relative level of SLE risk. Error bars represent 95% confidence intervals around the effect size estimate for each sex.
(c) Concentrations of C4 protein in cerebrospinal fluid sampled from 340 adult men (blue) and 167 adult women (pink) as a function of age with local polynomial regression (LOESS) smoothing. Concentrations are normalized to the number of C4 gene copies in an individual’s genome (a strong independent source of variance, Extended Data Fig. 7a) and shown on a log10 scale as a LOESS curve. Shaded regions represent 95% confidence intervals derived during LOESS.
(d) Levels of C3 protein in cerebrospinal fluid from 179 adult men and 125 adult women as a function of age. Concentrations are shown on a log10 scale as a LOESS curve. Shaded regions represent 95% confidence intervals derived during LOESS.
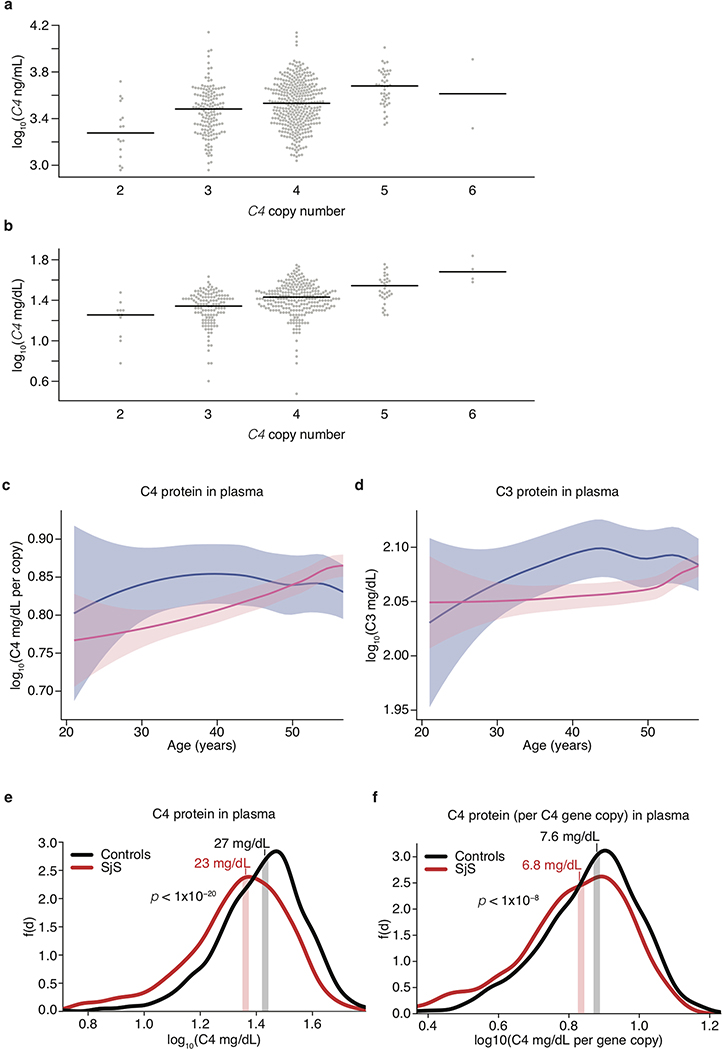
The stronger effects of C4 alleles on male relative to female risk could arise from sex differences in C4 RNA expression, C4 protein levels, or downstream responses to C4. Analysis of RNA expression in human tissues, using data from GTEx35, identified no sex differences in C4 RNA expression in brain, blood, liver, or lymphoblastoid cells (a more-detailed description of this analysis can be found in Supplementary Note 2). We then analyzed C4 protein in cerebrospinal fluid (CSF) from two panels of adult research participants (n = 589 total) in whom we had also measured C4 gene copy number (by direct genotyping or imputation). CSF C4 protein levels correlated strongly with C4 gene copy number (p < 10−10, Extended Data Fig. 7a), so we normalized C4 protein measurements to the number of C4 gene copies. CSF from adult men contained on average 27% more C4 protein per C4 gene copy than CSF from women (meta-analysis p = 9.9 × 10−6, Fig. 3c). C4 acts by activating the complement component 3 (C3) protein, promoting C3 deposition onto targets in tissues. CSF levels of C3 protein were also on average 42% higher among men than women (meta-analysis p = 7.5 × 10−7, Fig. 3d).
The elevated concentrations of C3 and C4 proteins in CSF of men parallel earlier findings that, in plasma, C3 and C4 are also present at higher levels in men than women8,9. The large sample size (n > 50,000) of the plasma studies allows sex differences to be further analyzed as a function of age. Both men and women undergo age-dependent elevation of C4 and C3 levels in plasma, but this occurs early in adulthood (age 20–30) in men and closer to menopause (age 40–50) in women, with the result that male–female differences in complement protein levels are observed primarily during the reproductive years (ages 20–50)8,9. We replicated these findings using measurements of C3 and gene copy number-corrected C4 protein in plasma from adults, finding (as in the earlier plasma studies8,9 and in CSF, Fig. 3c, d) that these differences are most pronounced during the reproductively active years of adulthood (ages 20–50) (Extended Data Fig. 7b–d). We also observed that SjS patients have lower C4 serum levels than controls (p < 1×10−20, Extended Data Fig. 7e) even after correcting for C4 gene copy number (p < 1×10−8, Extended Data Fig. 7f), suggesting that hypocomplementemia in SjS is not simply due to C4 genetics but also reflects disease effects on ambient complement levels, for example due to complement consumption. The ages of pronounced sex difference in complement levels corresponded to the ages at which men and women differ in disease incidence: in schizophrenia, men outnumber women among cases incident in early adulthood, but not among cases incident after age 402; in SLE, women greatly outnumber men among cases incident during the child-bearing years, but not among cases incident after age 50 or during childhood36; in SjS, the large relative vulnerability of women declines in magnitude after age 5037.
Our results indicate that the MHC genomic region shapes vulnerability in lupus and SjS – two of the three most common rheumatic autoimmune diseases – in a very different way than in type I diabetes, rheumatoid arthritis, and celiac disease. In the latter diseases, precise interactions between specific HLA protein variants and specific autoantigens determine risk 13,14. In SLE and SjS, however, the genetic variation implicated here points instead to the continuous, chronic interaction of the immune system with very many potential autoantigens. Because complement facilitates the rapid clearance of debris from dead and injured cells, elevated levels of C4 protein likely attenuate interactions between the adaptive immune system and ribonuclear self-antigens at sites of cell injury, pre-empting the development of autoimmunity. The additional C4-independent genetic risk effect described here (associated with rs2105898) may also affect autoimmunity broadly, rather than antigen-specifically, by regulating expression of many HLA class II genes (including DRB1, DQA1, and DQB1). Mouse models of SLE indicate that once tolerance is broken for one self-antigen, autoreactive germinal centers generate B cells targeting other self-antigens38; such “epitope spreading” could lead to autoreactivity against many related autoantigens, regardless of which antigen(s) are involved in the earliest interactions with immune cells. Further supporting such a model, higher copy number of C4 associates with lower risk of AQP4-IgG-seropositive neuromyelitis optica (NMO-IgG+)39, in which seropositive patients have increased incidence of other non-organ-specific autoantibodies such as those seen in SLE and SjS40. B-cells also express the complement receptors CR1 and CR241, providing an additional candidate mechanism for regulation by C4 and C3.
We note that the role of complement proteins in preventing the emergence of autoimmunity may be very different than their (potentially disease-exacerbating) role once autoimmunity has been established. Also, our genetic findings address the development of SLE and SjS rather than complications that arise in any specific organ. A few percent of SLE patients develop neurological complications that can include psychosis42; though psychosis is also a symptom of schizophrenia, neurological complications of SLE do not resemble schizophrenia more broadly, and likely have a different etiology.
The same C4 alleles that increase vulnerability to schizophrenia appeared to protect strongly against SLE and SjS. This pleiotropy will be important to consider in efforts to engage the complement system therapeutically. The complement system contributed to these pleiotropic effects more strongly in men than in women. Moreover, though the natural allelic series at C4 allowed human-genetic analysis to establish dose-risk relationships for C4 in men and women, sexual dimorphism in the complement-protein levels also included complement component 3 (C3). Why and how biology has come to create this sexual dimorphism in the complement system in humans presents interesting questions for immune and evolutionary biology.
Methods
Creation of a C4 reference panel from whole-genome sequence data
We constructed a reference panel for imputation of C4 structural haplotypes using whole-genome sequencing data for 1265 individuals from the Genomic Psychiatry Cohort26. The reference panel included individuals of diverse ancestry, including 765 Europeans, 250 African Americans, and 250 people of reported Latino ancestry.
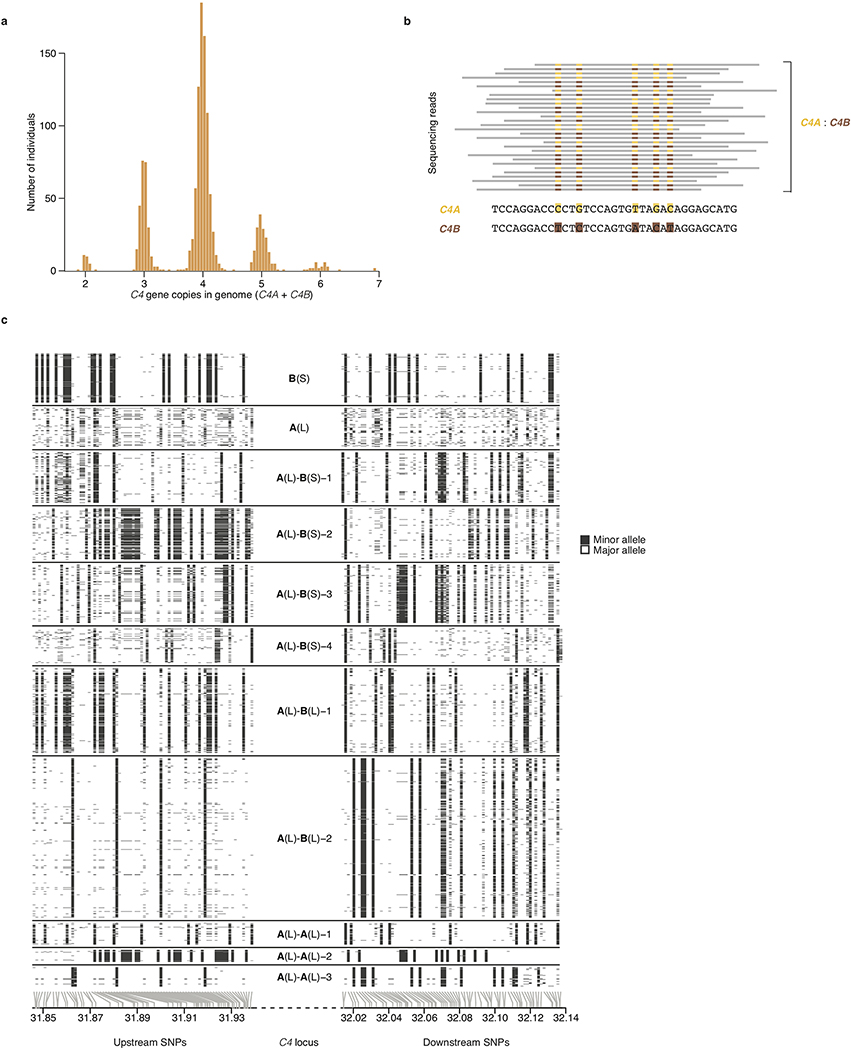
We estimated the diploid C4 copy number, and separately the diploid copy number of the contained HERV segment, using Genome STRiP44. Briefly, Genome STRiP carefully calibrates measurements of read depth across specific genomic segments of interest by estimating and normalizing away sample-specific technical effects such as the effect of GC content on read depth (estimated from the genome-wide data). To estimate C4 copy number, we genotyped the segments 6:31948358–31981050 and 6:31981096–32013904 (hg19) for total copy number, but masked the intronic HERV segments that distinguish short (S) from long (L) C4 gene isotypes. For the HERV region, we genotyped segments 6:31952461–31958829 and 6:31985199–31991567 (hg19) for total copy number. Across the 1,265 individuals, the resultant locus-specific copy-number estimates exhibited a strongly multi-modal distribution (Extended Data Fig. 1a) from which individuals’ total C4 copy numbers could be readily inferred.
We then estimated the ratio of C4A to C4B genes in each individual genome. To do this, we extracted reads mapping to the paralogous sequence variants that distinguish C4A from C4B (hg19 coordinates 6:31963859–31963876 and 6:31996597–31996614) in each individual, combining reads across the two sites. We included only reads that aligned to one of these segments in its entirety. We then counted the number of reads matching the canonical active site sequences for C4A (CCC TGT CCA GTG TTA GAC) and C4B (CTC TCT CCA GTG ATA CAT). We combined these counts with the likelihood estimates of diploid C4 copy number (from Genome STRiP) to determine the maximum likelihood combination of C4A and C4B in each individual (Extended Data Fig. 1b). We estimated the genotype quality of the C4A and C4B estimate from the likelihood ratio between the most likely and second most likely combinations.
To phase the C4 haplotypes, we first used the GenerateHaploidCNVGenotypes utility in Genome STRiP to estimate haplotype-specific copy-number likelihoods for C4 (total C4 gene copy number), C4A, C4B, and HERV using the diploid likelihoods from the prior step as input. Default parameters for GenerateHaploidCNVGenotypes were used, plus -genotypeLikelihoodThreshold 0.0001. The output was then processed by the GenerateCNVHaplotypes utility in Genome STRiP to combine the multiple estimates into likelihood estimates for a set of unified structural alleles. GenerateCNVHaplotypes was run with default parameters, plus -defaultLogLikelihood −50, -unknownHaplotypeLikelihood −50, and -sampleHaplotypePriorLikelihood 2.0. The resultant VCF was phased using Beagle 4.1 (beagle_4.1_27Jul16.86a) in two steps: first, performing genotype refinement from the genotype likelihoods using the Beagle gtgl= and maxlr=1000000 parameters, and then running Beagle again on the output file using gt= to complete the phasing.
Our previous work suggested that several C4 structures segregate on different haplotypes, and probably arose by recurrent mutation on different haplotype backgrounds7. The GenerateCNVHaplotypes utility requires as input an enumerated set of structural alleles to assign to the samples in the reference cohort, including any structurally equivalent alleles, with distinct labels to mark them as independent, plus a list of samples to assign (with high likelihood) to specific labeled input alleles to disambiguate among these recurrent alleles. The selection of the set of structural alleles to be modeled, along with the labeling strategy, is important to our methodology and the performance of the reference panel. In the reference panel, each input allele represents a specific copy number structure and optionally includes a label that differentiates the allele from other independent alleles with equivalent structure. We use the notation <H_n_n_n_n_L> to identify each allele, where the four integers following the H are, respectively, the (redundant) haploid count of the total number of C4 copies, C4A copies, C4B copies and HERV copies on the haplotype. For example, <H_2_1_1_1> was used to represent the “AL-BS” haplotype. The optional final label L is used to distinguish potentially recurrent haplotypes with otherwise equivalent structures (under the model) that should be treated as independent alleles for phasing and imputation.
To build the reference panel, we experimentally evaluated a large number of potential sets of structural alleles and methods for assigning labels to potentially recurrent alleles. For each evaluation, we built a reference panel using the 1265 reference samples, and then evaluated the performance of the panel via cross-validation, leaving out 10 different samples in each trial (5 samples in the last trial) and imputing the missing samples from the remaining samples in the panel. The imputed results for all 1265 samples were then compared to the original diploid copy number estimates to evaluate the performance of each candidate reference panel (Extended Data Table 1).
Using this procedure, we selected a final panel for downstream analysis that used a set of 29 structural alleles representing 16 distinct allelic structures (as listed in the reference panel VCF file). Each allele contained from one to three copies of C4. Three allelic structures (AL-BS, AL-BL, and AL-AL) were represented as a set of independently labeled alleles with 9, 3, and 4 labels, respectively.
To identify the number of labels to use on the different alleles and the samples to “seed” the alleles, we generated “spider plots” of the C4 locus based on initial phasing experiments run without labeled alleles, and then clustered the resulting haplotypes in two dimensions based on the Y-coordinate distance between the haplotypes on the left and right sides of the spider plot. Clustering was based on visualizing the clusters (Extended Data Fig. 1c) and then manually choosing both the number of clusters (labels) to assign and a set of confidently assigned haplotypes to use to “seed” the clusters in GenerateCNVHaplotypes. This procedure was iterated multiple times using cross-validation, as described above, to evaluate the imputation performance of each candidate labeling strategy.
Within the data set used to build the reference panel, there is evidence for individuals carrying seven or more diploid copies of C4, which implies the existence of (rare) alleles with four or more copies of C4. In our experiments, attempting to add additional haplotypes to model these rare four-copy alleles reduced overall imputation performance. Consequently, we conducted all downstream analyses using a reference panel that models only alleles with up to three copies of C4. In the future, larger reference panels might benefit from modeling these rare four-copy alleles.
The reference panel will be available in dbGaP (accession # pending) with broad permission for research use.
Genetic data for SLE
For analysis of systemic lupus erythematosus (SLE), collection and genotyping of the European-ancestry cohort (6,748 cases, 11,516 controls, genotyped by ImmunoChip) as previously described3. Collection and genotyping of the African-American cohort (1,494 cases, 5,908 controls, genotyped by OmniExpress) as previously described5.
Genetic data for SjS
For analysis of Sjögren’s syndrome (SjS), collection and genotyping of the European-ancestry cohort (673 cases, 1,153 controls, genotyped by Omni2.5) as previously described32 and available in dbGaP under study accession number phs000672.v1.p1.
Genetic data for schizophrenia
The schizophrenia analysis made use of genotype data from 40 cohorts of European ancestry (28,799 cases, 35,986 controls) made available by the Psychiatric Genetics Consortium (PGC) as previously described43. Genotyping chips used for each cohort are listed in Supplementary Table 3 of that study.
Imputation of C4 alleles
The reference haplotypes described above were used to extend the SLE, SjS, or schizophrenia cohort SNP genotypes by imputation. SNP data in VCF format were used as input for Beagle v4.145,46 for imputation of C4 as a multi-allelic variant. Within the Beagle pipeline, the reference panel was first converted to bref format. From the cohort SNP genotypes, we used only those SNPs from the MHC region (chr6:24–34 Mb on hg19) that were also in the haplotype reference panel. We used the conform-gt tool to perform strand-flipping and filtering of specific SNPs for which strand remained ambiguous. Beagle was run using default parameters with two key exceptions: we used the GRCh37 PLINK recombination map, and we set the output to include genotype probability (i.e., GP field in VCF) for correct downstream probabilistic estimation of C4A and C4B joint dosages.
Imputation of HLA alleles
For HLA allele imputation, sample genotypes were used as input for the R package HIBAG47. For both European ancestry and African American cohorts, publicly available multi-ethnic reference panels generated for the most appropriate genotyping chip (i.e. Immunochip for European ancestry SLE cohort, Omni 2.5 for European ancestry SjS cohort, and OmniExpress for African American SLE cohort) were used48. Default parameters were used for all settings. All class I and class II HLA genes were imputed. Output haplotype posterior probabilities were summed per allele to yield diploid dosages for each individual.
Associating single and joint C4 structural allele dosages to SLE and SjS in European ancestry individuals
The analysis described above yields dosage estimates for each of the common C4 structural haplotypes (e.g., AL-BS, AL-AL, etc.) for each genome in each cohort. In addition to performing association analysis on these structures (Fig 1b), we also performed association analysis on the dosages of each underlying C4 gene isotype (i.e. C4A, C4B, C4L, and C4S). These dosages were computed from the allelic dosage (DS) field of the imputation output VCF simply by multiplying the dosage of a C4 structural haplotype by the number of copies of each C4 isotype that haplotype contains (e.g., AL-BL contains one C4A gene and one C4B gene).
C4 isotype dosages were then tested for disease association by logistic regression, with the inclusion of four available ancestry covariates derived from genome-wide principal component analysis (PCA) as additional independent variables, PCc,
| (1) |
where θ=E[SLE|X]. For SjS, the model instead included two available multiethnic ancestry covariates from dbGaP that correlated strongly with European-specific ancestry covariates (specifically, PC5 and PC7) and smoking status as independent variables. Coefficients for relative weighting of C4A and C4B dosages were obtained from a joint logistic regression,
| (2) |
The values per individual of β1C4A + β2C4B were used as a combined C4 risk term for estimating both association strength (Extended Data Fig. 3a, b) as well as evaluating the relationship between the strength of nearby variants’ association with SLE or SjS and linkage with C4 variation (Extended Data Fig. 4a–c).
Joint dosages of C4A and C4B for each individual in the same cohort were estimated by summing across their genotype probabilities of paired structural alleles that encode for the same diploid copy numbers of both C4A and C4B (Extended Data Fig. 2a, b). For each individual/genome, this yields a joint dosage distribution of C4A and C4B gene copy number, reflecting any possible imputed haplotype-level dosages with nonzero probability. Joint dosages for C4A and C4B diploid copy numbers were tested for association with SLE in a joint model with the same ancestry covariates (Fig. 1a),
| (3) |
Calculation of composite C4 risk for SLE
Because SLE risk strongly associated with C4A and C4B copy numbers (Fig. 1a) in a manner that can be approximated as – but is not necessarily linear or independent – a composite C4 risk score was derived by taking the weighted sum of joint C4A and C4B dosages multiplied by the corresponding effect sizes from the aforementioned model of the joint C4A and C4B diploid copy numbers. The weights for calculating this composite C4 risk term were computed from the data from the European ancestry cohort, and then applied unchanged to analysis of the African American cohort.
Associations of variants across the MHC region to SLE and SjS
Genotypes for non-array SNPs were imputed with IMPUTE2 using the 1000 Genomes reference panel; separate analyses were performed for the European-ancestry and African American cohorts. Unless otherwise stated, all subsequent SLE analyses were performed identically for both European ancestry and African American cohorts. Dosage of each variant, vi, was tested for association with SLE or SjS in a logistic regression including available ancestry covariates (and smoking status for SjS) first alone (Extended Data Fig. 3a, b),
| (4) |
then with C4 composite risk (Extended Data Fig. 3c),
| (5) |
where θ=E[SLE|X]. For SjS, the simpler weighted (2.3)C4A+C4B model was used instead of composite risk term, as the cohort’s size gave poor precision to estimates of risk for many joint (C4A, C4B) copy numbers (Extended Data Fig. 3d). The Pearson correlation between the C4 composite risk term and each other variant was computed and squared (r2) to yield a measure of linkage disequilibrium between C4 composite risk and that variant in that cohort.
Association analyses for specific C4 structural alleles
The C4 structural haplotypes were tested for association with disease (Fig. 1b, 2a) in a joint logistic regression that included (i) terms for dosages of the five most common C4 structural haplotypes (AL-BS, AL-BL, AL-AL, BS, and AL), (ii) (for SLE and SjS) rs2105898 genotype, and (iii) ancestry covariates and (for SjS) smoking status,
| (6) |
where θ=E[SLE|X]. Several of these common C4 structural alleles arose multiple times on distinct haplotypes; we term the set of haplotypes in which such a common allele appeared as “haplogroups”. The haplogroups can be further tested in a logistic regression model in which the structural allele appearing in all member haplotypes is instead encoded as dosages for each of the SNP haplotypes in which it appears. These association analyses (Fig. 1b, 2a) were performed as in (6), with structural allele dosages for ALBS, ALBL, and ALAL replaced by multiple terms for each distinct haplotype.
To delineate the relationship between C4-BS and DRB1*03:01 alleles – which are highly linked in European ancestry haplotypes – allelic dosages per individual in the African American SLE cohort were rounded to yield the most likely integer dosage for each. Although genotype dosages for each are reported by BEAGLE and HIBAG respectively, probabilities per haplotype are not linked and multiplying possible diploid dosages could yield incorrect non-zero joint dosages. Joint genotypes were tested as individual terms in a logistic regression model (Fig. 2b),
| (7) |
Sex-stratified associations of C4 structural alleles and other variants with SLE, SjS, and schizophrenia
Determination of an effect from sex on the contribution of overall C4 variation to risk for each disorder was done by including an interaction term between sex and C4; ie. (2.3)C4A+C4B for SLE and SjS and estimated C4A expression for schizophrenia:
| (8) |
Each variant in the MHC region was tested for association with among European ancestry cases and cohorts in a logistic regression as in models (4)–(6) using only male cases and controls, and then separately using only female cases and controls (Extended Data Fig. 6a–c). Likewise, allelic series analyses were performed as in (7), but in separate models for men and women (Fig. 3a, b).
To assess the relationship between sex bias in the risk associated with a variant and linkage to C4 composite risk (as non-negative r2), male and female log-odds were multiplied by the sign of the Pearson correlation between that variant and C4 composite risk before taking the difference.
Analyses of cerebrospinal fluid
Cerebrospinal fluid (CSF) from healthy individuals was obtained from two research panels. The first panel, consisting of 533 donors (327 male, 126 female) from hospitals around Utrecht, Netherlands, was described previously49,50. The donors were generally healthy research participants undergoing spinal anesthesia for minor elective surgery. The same donors were previously genotyped using the Illumina Omni SNP array. To estimate C4 copy numbers, we used SNPs from the MHC region (chr6:24–34 Mb on hg19) as input for C4 allele imputation with Beagle, as described above in Imputation of C4 alleles.
The second CSF panel sampled specimens from 56 donors (14 male, 42 female) from Brigham and Women’s Hospital (BWH; Boston, MA, USA) under a protocol approved by the institutional review board at BWH (IRB protocol ID no. 1999P010911) with informed consent. These samples were originally obtained to exclude the possibility of infection, and clinical analyses had revealed no evidence of infection. Donors ranged in age from 18 to 64 years old. Blood samples from the same individuals were used for extraction of genomic DNA, and C4 gene copy number was measured by droplet digital PCR (ddPCR) as previously described7. Samples were excluded from measurements if they lacked C4 genotypes, sex information, or contained visible blood contamination.
C4 measurements were performed by sandwich ELISA of 1:400 dilutions of the original CSF sample using goat anti-sera against human C4 as the capture antibody (Quidel, A305, used at 1:1000 dilution), FITC-conjugated polyclonal rabbit anti-human C4c as the detection antibody (Dako, F016902–2, used at 1:3000 dilution), and alkaline phosphatase–conjugated polyclonal goat anti-rabbit IgG as the secondary antibody (Abcam, ab97048, used at 1:5000 dilution). C3 measurements were performed using the human complement C3 ELISA kit (Abcam, ab108823).
Because C4 gene copy number had a large and proportional effect on C4 protein concentration in these CSF samples (Extended Data Fig. 7a), we corrected for C4 gene copy number in our analysis of relationship between sex and C4 protein concentration, by normalizing the ratio of C4 protein (in CSF) to C4 gene copies (in genome). Therefore, these analyses included only samples for which DNA was available or C4 was successfully imputed. In total, 495 (332 male, 163 female) C4 and 304 (179 male, 125 female) C3 concentrations were obtained across both cohorts. Log-concentrations of C3 (ng/mL) and C4 (ng/[mL, per C4 gene copy number]) protein were then used separately in linear regression models to estimate a sex-unbiased cohort-specific offset for each protein,
| (9) |
to be applied to all concentrations for that protein. Estimation of average measurements by age for each sex was done by local polynomial regression smoothing (LOESS) (Fig. 3c, d). To evaluate the significance of sex effects, we used these cohort-corrected concentrations estimates and analyzed them with the non-parametric unsigned Mann-Whitney rank–sum test comparing concentration distributions for males and females.
Analyses of blood plasma
Blood plasma was collected and immunoturbidimetric measurements of C3 and C4 protein in 1,844 individuals (182 men, 1662 women) by Sjögren’s International Collaborative Clinical Alliance (SICCA) from individuals with and without SjS as previously described51. C4 copy numbers for these individuals were previously imputed for use in logistic regression of SjS risk. As C4 copy number has an effect on measured C4 protein similar to CSF (Extended Data Fig. 7b), we normalized C4 levels to them in all following analyses. Estimation of average measurements by age for each sex was done by local polynomial regression smoothing (LOESS) on log-concentrations of C3 (mg/dL) and C4 (mg/[dL, per C4 gene copy number]) protein (Extended Data Fig. 7c, d). To evaluate the significance of sex bias within age ranges displaying the greatest difference (informed by LOESS), we analyzed individuals in these bins with the non-parametric unsigned Mann-Whitney rank–sum test comparing concentration distributions for males and females.
Difference in C4 protein levels between individual with and without SjS was done by performing a non-parametric unsigned Mann-Whitney rank–sum test on C4 protein levels with and without normalization to C4 genomic copy number (Extended Data Fig. 7e, f).
Data Availability Statement
Individual genotype data for Sjögren’s syndrome cases and controls and individual plasma concentrations for C4 and C3 are available in dbGaP under accession number phs000672.v1.p1. Individual genotype data for schizophrenia cases and controls are available by application to the Psychiatric Genomics Consortium (PGC). Questions regarding individual genotype data for SLE cases and controls of European and/or African American ancestry can be directed to Timothy J. Vyse (timothy.vyse@kcl.ac.uk). Data resources (reference haplotypes), software scripts and instructions for imputing C4 alleles into SNP data sets are available on the McCarroll lab web site at http://mccarrolllab.org/resources/resources-for-c4/. Genotype and protein concentration data for CSF samples are available upon request.
Extended Data
Extended Data Figure 1. A panel of 2,530 reference haplotypes (created from whole-genome sequence data) containing C4 alleles and SNPs across the MHC genomic region enables imputation of C4 alleles into large-scale SNP data.
(a) Distributions (across 1,265 individuals) of total C4 gene copy number (C4A + C4B), as measured from read depth of coverage across the C4 locus, in whole-genome sequencing data.
(b) The relative numbers of reads that overlap sequences specific to C4A or C4B (together with the total C4 gene copy number as in a) are used to infer the underlying copy numbers of the C4A and C4B genes. For example, in an individual with four C4 genes, the presence of equal numbers of reads specific to C4A or C4B suggests the presence of two copies each of C4A and C4B. Precise statistical approaches (including inference of probabilistic dosages), and further approaches for phasing C4 allelic states with nearby SNPs to create reference haplotypes, are described in Methods.
(c) The SNP haplotypes flanking each C4 allele are shown as rows (SNPs as columns), with white and black representing the major and minor allele of each SNP. Gray lines at the bottom indicate the physical location of each SNP along chromosome 6. The differences among the haplotypes are most pronounced closest to C4 (toward the center of the plot), as historical recombination events in the flanking megabases will have caused the haplotypes to be less consistently distinct at greater genomic distances from C4. The patterns indicate that many combinations of C4A and C4B gene copy numbers have arisen recurrently on more than one SNP haplotype, a relationship that can be used in association analyses (Fig. 1b).
Extended Data Figure 2. Aggregation of joint C4A and C4B genotype probabilities per individual across imputed C4 structural alleles for estimation of SLE risk for each combination.
(a) An individual’s joint C4A and C4B gene copy number can be calculated by summing the C4A and C4B gene contents for each possible pair of two inherited alleles. Many pairings of possible inherited alleles result in the same joint C4A and C4B gene copy number.
(b) Each individual’s C4A and C4B gene copy number was imputed from their SNP data, using the reference haplotypes summarized in Extended Data Fig. 1c. For >95% of individuals (exemplified by samples 1–6 in the figure), this inference can be made with >90% certainty/confidence (the areas of the circles represent the posterior probability distribution over possible C4A/C4B gene copy numbers). For the remaining individuals (exemplified by samples 7–9 in the figure), greater statistical uncertainty persists about C4 genotype. To account for this uncertainty, in downstream association analysis, all C4 genotype assignments are handled as probabilistic gene dosages – analogous to the genotype dosages that are routinely used in large-scale genetic association studies that use imputation.
(c) Odds ratios and 95% confidence intervals underlying each of the C4-genotype risk estimates in Fig. 1a presented as a series of panels for each observed copy number of C4B, with increasing copy number of C4A for that C4B dosage (x-axis). Data are from analysis of 6,748 SLE cases and 11,516 controls of European ancestry.
Extended Data Figure 3. Conditional association analyses for genetic markers across the extended MHC genomic region within the European-ancestry SLE and Sjögren’s syndrome (SjS) cohort.
(a) Association of SLE with genetic markers (SNPs and imputed HLA alleles) across the extended MHC locus within the European-ancestry SLE cohort (6,748 cases and 11,516 controls). Orange diamond: an initial estimate of C4-related genetic risk, calculated as a weighted sum of the number of C4A and C4B gene copies: (2.3)C4A+C4B, with the weights derived from the relative coefficients estimated from logistic regression of SLE risk vs. C4A and C4B gene dosages. This risk score is imputed with an accuracy (r2) of 0.77. Points representing all other genetic variants in the MHC locus are shaded orange according to their level of linkage disequilibrium–based correlation to this C4-derived risk score.
(b) As in a, but for a European-ancestry Sjögren’s syndrome (SjS) cohort (673 cases and 1,153 controls). The orange diamond here also represents (2.3)C4A+C4B, with this weighting derived from the relative coefficients estimated from logistic regression of SjS risk vs. C4A and C4B gene dosages
(c) Association of SLE with genetic markers (SNPs and imputed HLA alleles) across the extended MHC locus within the European-ancestry SLE cohort controlling for C4 composite risk (weighted sum of risk associated with various combinations of C4A and C4B). Variants are shaded in purple by their LD with rs2105898, an independent association identified from trans-ancestral analyses.
(d) As in c, but in association with a European-ancestry SjS cohort. Here a simpler linear model of risk contributed by C4A and C4B was used instead of a weighted sum across all possible combinations.
Extended Data Figure 4. Using C4 gene variation to understand the appearance of trans-ancestral disparity in MHC association signals, and to fine-map an additional genetic effect.
All panels show association signals (for SLE and SjS) for variants in a multi-megabase region of human chromosome 6 containing the MHC region including the HLA and C4 genes.
(a) Relationship between SLE association [-log10(p), y-axis] and LD to the weighted C4 risk score (x-axis) for genetic markers and imputed HLA alleles across the extended MHC locus. In this European-ancestry cohort, it is unclear (from this analysis alone) whether the association with the markers in the predominant ray of points (at a ~45° angle from the x-axis) is driven by variation at C4 or by the long haplotype containing DRB1*03:01 (green), DQA1*05:01 (blue), and B*08:01 (red). In addition, at least one independent association signal (a ray of points at a higher angle in the plot, with strong association signals and only weak LD-based correlation to C4 and DRB1*0301) with some LD to DRB1*15:01 (maroon) is also present.
(b) Analysis as in a, but for associations to SjS in a cohort of European ancestry. As in SLE, it is initially unclear whether the genetic association signal is driven by variation at C4 or by linked HLA alleles, DRB1*03:01 (green), DQA1*05:01 (blue), and B*08:01 (red). There is also an independent association signal with LD to DRB1*15:01 (maroon).
(c) Analysis as in a, but of an African American SLE case–control cohort (in which LD in the MHC region is more limited). Many MHC-region SNPs associate with SLE in proportion to their LD with the weighted C4 risk score inferred from the earlier analysis of the European-ancestry cohort; this C4-derived risk score itself associates with SLE at p = 4.3×10−19 in a logistic regression on 1,494 SLE cases and 5,908 controls. No similarly strong association is observed for DRB1*03:01, DQA1*05:01, or B*08:01, HLA alleles which are in strong LD with C4 risk on European-ancestry (but not African American) haplotypes. An independent association signal is also present in this cohort, more clearly in LD with the DRB1*15:03 allele (maroon).
(d) LD in the European-ancestry SLE cohort between the composite C4 risk term (weighted sum of risk associated with various combinations of C4A and C4B from Fig. 2a) and variants in the MHC region as r2 (y-axis).
(e) As in d, but for the African American SLE cohort.
(f) LD (to C4 composite risk) for the same variants in European-ancestry individuals (x-axis) and African Americans (y-axis). Note the abundance of variants that have greater LD with C4 risk among European-ancestry individuals than among African Americans. Also, several groups of variants have equivalent LD (to C4 risk) in European ancestry individuals but exhibit a range of LD to C4 risk among African Americans.
(g) Associations with SLE (-log10 p-values) for the same variants in European ancestry (x-axis) and African American (y-axis) case-control cohorts. Orange shading represents the extent of LD with C4 risk in European ancestry individuals. Variants with strong European-specific association to SLE are generally in strong LD with C4 risk among Europeans-ancestry individuals.
(h) Comparison of the inferred effect size from association of genetic markers with SLE (unconditioned log-odds ratios) among European-ancestry (x-axis) and African American (y-axis) research participants. As also seen in g, variants with discordant associations to SLE (across populations) tend also to be in strong LD to C4 risk among European-ancestry individuals.
(i) As in g, but now controlling for the effect of C4 variation in analysis of the European-ancestry cohort (x-axis). Note that controlling for C4 risk in European-ancestry individuals alone greatly aligns (relative to g) the patterns of association between European ancestry and African American cohorts.
(j) As in i, but now also controlling for the effect of C4 in associations of the African American cohort. Note that due to the lack of strong LD relationships between C4 and variants in the MHC region in African Americans (e), this further adjustment does not change results strongly (relative to i). The independent signal, rs2105898, and HLA alleles, DRB1*15:01 and DRB1*15:03, are also highlighted. LD with rs2105898 in European-ancestry individuals is indicated by purple shading.
(k) Comparison of the inferred effect sizes from association of genetic markers with SLE (log-odds ratios) controlling for C4-derived risk among European-ancestry (x-axis) and African American (y-axis) research participants. Two SNPs (rs2105898 and rs9271513) that form a short haplotype common to both ancestry groups are among the strongest associations in both cohorts. (Their association to SLE in the European-ancestry cohort was initially much less remarkable than that of other SNPs that are in strong LD with C4.) LD with rs2105898 in European-ancestry individuals is indicated by purple shading.
(l) As in i, but with variants shaded by whether they exhibit greater LD to rs2105898 in Europeans (blue) or African Americans (red).
Extended Data Figure 5. Relationship of rs2105898 alleles to a known ZNF143 binding motif in the XL9 region of the MHC class II locus.
(a) Location of rs2105898 (yellow line at center) within the XL9 region, with relevant tracks showing overlapping histone marks and transcription factor binding peaks (from ENCODE52), visualized with the UCSC genome browser53.
(b) ZNF143 consensus binding motif as a sequence logo, with the letters colored if the base is present in >5% of observed instances. The alleles of rs2105898 are indicated by outlined box surrounding the base.
Extended Data Figure 6. Relationships between sex bias of disease associations and LD to C4 risk for variants in the MHC region.
(e) Relationship between male bias in SLE risk (difference between male and female log–odds ratios) and LD with C4 risk for common (minor allele frequency [MAF] > 0.1) genetic markers across the extended MHC region. For each SNP, the allele for which sex risk bias is plotted is the allele that is positively correlated (via LD) with C4-derived risk score.
(f) Relationship between male bias in SjS risk (log-odds ratios) and LD with C4 risk for common (minor allele frequency [MAF] > 0.1) genetic markers across the extended MHC region. For each SNP, the allele for which sex risk bias is plotted is the allele that is positively correlated (via LD) with C4-derived risk score.
(g) Relationship of male bias in schizophrenia risk (log–odds ratios) and LD to C4A expression for common (MAF > 0.1) genetic markers across the extended MHC region. For each SNP, the allele for which sex risk bias is plotted is the allele that is positively correlated (via LD) with imputed C4A expression, as previously described7.
Extended Data Figure 7. Correlation of C4 protein measurements in cerebrospinal fluid and blood plasma with imputed C4 gene copy number and relationship of plasma complement to sex and SjS status.
(a) Measurements of C4 protein in CSF obtained by ELISA are presented as log10(ng/mL) (y-axis) for each observed or imputed copy number of total C4 (x-axis, here showing most likely copy number from imputation). Because C4 gene copy number affects C4 protein levels so strongly, we normalized C4 protein measurements to each donor’s C4 gene copy number in subsequent analyses (Fig. 3c). Bars indicate median values for each C4 copy number.
(b) Measurements of C4 protein in blood plasma obtained by immunoturbidimetric assays are presented as log10(mg/dL) (y-axis) for each imputed most-likely copy number of C4 genes (x-axis). Because C4 gene copy number affects C4 protein levels so strongly, we normalized C4 protein measurements by C4 gene copy number in subsequent analyses as in c. Due to the number of observations (n = 1,844 total), the plot is downsampled to 500 points; the median bars shown are for all individuals (before downsampling).
(c) Levels of C4 protein in blood plasma from 182 adult men and 1662 adult women as a function of age. Concentrations are normalized to the number of C4 gene copies in an individual’s genome (a strong independent source of variance) and shown on a log10 scale as a LOESS curve. Shaded regions represent 95% confidence intervals derived during LOESS.
(d) Levels of C3 protein in blood plasma as a function of age from the same individuals in panel c. Concentrations are shown on a log10 scale as a LOESS curve. Shaded regions represent 95% confidence intervals derived during LOESS.
(e) C4 protein in blood plasma was measured in 670 individuals with SjS (red) and 1,151 individuals without SjS (black) and is shown on a log10 scale (x-axis). Vertical stripes represent median levels for cases and controls separately. Comparison of the two sets was done with a non-parametric two-sided Mann-Whitney rank–sum test (p = 4.8×10−21).
(f) As in e, but concentrations are normalized to the number of C4 gene copies in an individual’s genome and this per-copy amount is shown on a log10 scale (x-axis). Comparison of the two sets was done with a non-parametric two-sided Mann-Whitney rank–sum test (p = 7.6×10−9).
Extended Data Table 1. Imputation accuracy for C4 copy numbers in European ancestry and African American haplotypes.
Imputation accuracy was evaluated by correlation of imputation results to C4 gene copy numbers directly inferred from WGS data. Aggregated copy numbers imputed from each round of leaving 10 individuals out were correlated with the directly-typed measurements and are reported as r2 for each feature of C4 structural variation for European ancestry and African American members of the reference panel separately.
| Imputation accuracy (r2) |
||
|---|---|---|
| Gene copy number | European ancestry | African Americans |
| C4 | 0.80 | 0.58 |
| C4A | 0.78 | 0.65 |
| C4B | 0.74 | 0.61 |
| C4-HERV | 0.91 | 0.76 |
| 2.3(C4A)+C4B | 0.77 | 0.64 |
Extended Data Table 2. Frequency of common C4 alleles and their LD-based correlation with HLA alleles in European ancestry and African American cohorts.
For each common C4 allele and HLA gene, the allele with strongest LD (r2) is listed if present on more than half of the haplotypes with that C4 allele (exact fraction in %). r2 values greater than 0.4 are highlighted to point out particularly strong C4-HLA allele correlations, such as for several HLA alleles with the C4-B(S) allele in European ancestry individuals. Some common C4 alleles are further subdivided into distinct haplotypes used in imputation (and in Fig. 1b), as defined by shared alleles from variants flanking C4. Note that some alleles such as C4-A(L)-A(L)-3 are present at a low frequency in African Americans that might reflect their presence on admixed European-origin haplotypes spanning this region, whereas others such as C4-B(S) are likely to also exist on African haplotypes – these differences between C4 alleles are also reflected in the similarity of LD with HLA alleles to the corresponding row of the European ancestry section.
| European ancestry | |||||||||||||||||||
|
A |
B |
C |
C4 allele | Allele Frequency |
DRB1 |
DQA1 |
DQB1 |
||||||||||||
| allele | % | r2 | allele | % | r2 | allele | % | r2 | allele | % | r2 | allele | % | r2 | allele | % | r2 | ||
| 01:01 | 69 | 0.27 | 08:01 | 93 | 0.75 | 07:01 | 93 | 0.57 | B(S) | 13.7% | 03:01 | 94 | 0.71 | 05:01 | 94 | 0.7 | 02:01 | 94 | 0.7 |
| A(L) | 4.8% | ||||||||||||||||||
| 06:02 | 69 | 0.31 | A(L)-B(S)-1 | 6.1% | 07:01 | 74 | 0.25 | 02:01 | 74 | 0.25 | |||||||||
| 44:03 | 54 | 0.28 | 16:01 | 53 | 0.39 | A(L)-B(S)-2 | 4.5% | 07:01 | 57 | 0.1 | 02:01 | 57 | 0.1 | 02:02 | 55 | 0.14 | |||
| A(L)-B(S)-3 | 3.8% | ||||||||||||||||||
| A(L)-B(S)-4 | 4.5% | ||||||||||||||||||
| 07:02 | 64 | 0.42 | 07:02 | 63 | 0.35 | A(L)-B(L)-1 | 15.5% | 15:01 | 73 | 0.49 | 01:02 | 74 | 0.32 | 06:02 | 70 | 0.47 | |||
| A(L)-B(L)-2 | 23.1% | ||||||||||||||||||
| 35:01 | 55 | 0.2 | 04:01 | 57 | 0.09 | A(L)-A(L)-1 | 3.2% | 01:01 | 65 | 0.14 | 01:01 | 65 | 0.11 | 05:01 | 64 | 0.1 | |||
| A(L)-A(L)-2 | 2.1% | 13:01 | 67 | 0.16 | 01:03 | 65 | 0.13 | 06:03 | 67 | 0.15 | |||||||||
| 02:01 | 65 | 0.03 | 44:02 | 74 | 0.24 | 05:01 | 72 | 0.23 | A(L)-A(L)-3 | 4.5% | 04:01 | 80 | 0.29 | 03:03 | 79 | 0.37 | 03:01 | 82 | 0.15 |
| African American | |||||||||||||||||||
|
A |
B |
C |
C4 allele | Allele Frequency |
DRB1 |
DQA1 |
DQB1 |
||||||||||||
| allele | % | r2 | allele | % | r2 | allele | % | r2 | allele | % | r2 | allele | % | r2 | allele | % | r2 | ||
| B(S) | 5.0% | 01:02 | 51 | 0.01 | |||||||||||||||
| A(L) | 7.5% | ||||||||||||||||||
| A(L)-B(S)-1 | 14.1% | ||||||||||||||||||
| A(L)-B(S)-2 | 18.1% | ||||||||||||||||||
| A(L)-B(S)-3 | 17.7% | ||||||||||||||||||
| A(L)-B(S)-4 | 6.5% | ||||||||||||||||||
| A(L)-B(L)-1 | 4.4% | 15:01 | 67 | 0.2 | 01:02 | 72 | 0.04 | 06:02 | 59 | 0.06 | |||||||||
| A(L)-B(L)-2 | 4.5% | ||||||||||||||||||
| A(L)-A(L)-1 | 0.7% | 01:01 | 57 | 0.07 | 01:01 | 53 | 0.01 | ||||||||||||
| A(L)-A(L)-2 | 0.8% | ||||||||||||||||||
| 02:01 | 72 | 0.03 | 44:02 | 86 | 0.31 | 05:01 | 78 | 0.17 | A(L)-A(L)-3 | 0.8% | 04:01 | 93 | 0.27 | 03:03 | 86 | 0.14 | 03:01 | 87 | 0.03 |
Extended Data Table 3. Results of association analyses of SLE risk against C4 variation, HLA alleles, and/or rs2105898 in European ancestry and African American cohorts.
Coefficients (beta, standard error) and p-values (as −log10(p)) for individual terms composing several relevant logistic regression models for predicting SLE risk in a European ancestry cohort of 6,748 SLE cases and 11,516 controls and an African American cohort of 1,494 SLE cases and 5,908 controls. Each analysis also included ancestry-specific covariates. For each model, the Akaike information criterion (AIC) and overall p-value (as determined by Chi-squared likelihood-ratio test) are given at the right to indicate the relative strengths of similar models for each ancestry cohort.
| European ancestry | ||||||||||||||||||||
| C4 | C4A | C4B | DRB1*03:01 | B*08:01 | rs2105898 | |||||||||||||||
| Model | beta | se | −log10(p) | beta | se | −log10(p) | beta | se | −log10(p) | beta | se | −log10(p) | beta | se | −log10(p) | beta | se | −log10(p) | AIC | LRT −log10(p) |
| C4 | −0.55 | 0.027 | 92.7 | 22855.26 | 260.2 | |||||||||||||||
| C4A | −0.53 | 0.024 | 105.3 | 22790.05 | 274.3 | |||||||||||||||
| C4A+C4B | −0.62 | 0.028 | 112 | −0.27 | 0.037 | 12.3 | 22739.8 | 284.4 | ||||||||||||
| DRB1*03:01 | 0.7 | 0.03 | 117.1 | 22748.33 | 283.3 | |||||||||||||||
| B*08:01 | 0.69 | 0.031 | 108.4 | 22790.65 | 274.2 | |||||||||||||||
| rs2105898 | −0.32 | 0.027 | 30.7 | 23153.86 | 195.5 | |||||||||||||||
| C4A + C4B + DRB1*03:01 | −0.35 | 0.041 | 17.2 | −0.11 | 0.041 | 2.3 | 0.4 | 0.046 | 17.5 | 22666.1 | 299.6 | |||||||||
| C4A + C4B + B*08:01 | −0.41 | 0.039 | 24.6 | −0.17 | 0.039 | 4.7 | 0.35 | 0.044 | 14.4 | 22680.53 | 296.4 | |||||||||
| C4A + C4B + rs2105898 | −0.67 | 0.028 | 122.8 | −0.32 | 0.038 | 16.4 | −0.38 | 0.028 | 41.1 | 22558.42 | 322.8 | |||||||||
| African American | ||||||||||||||||||||
| C4 | C4A | C4B | DRB1*03:01 | B*08:01 | rs2105898 | |||||||||||||||
| Model | beta | se | −log10(p) | beta | se | −log10(p) | beta | se | −log10(p) | beta | se | −log10(p) | beta | se | −log10(p) | beta | se | −log10(p) | AIC | LRT −log10(p) |
| C4 | −0.51 | 0.059 | 17.3 | 7358.65 | 19.7 | |||||||||||||||
| C4A | −0.43 | 0.062 | 11.2 | 7385.17 | 14 | |||||||||||||||
| C4A+C4B | −0.62 | 0.068 | 18.7 | −0.41 | 0.068 | 8.6 | 7351.45 | 20.9 | ||||||||||||
| DRB1*03:01 | 0.41 | 0.091 | 5.2 | 7413.36 | 8 | |||||||||||||||
| B*08:01 | 0.78 | 0.11 | 11.6 | 7387.33 | 13.6 | |||||||||||||||
| rs2105898 | −0.46 | 0.047 | 21.9 | 7339.35 | 23.9 | |||||||||||||||
| C4A + C4B + DRB1*03:01 | −0.59 | 0.073 | 15 | −0.38 | 0.071 | 7.1 | 0.1 | 0.099 | 0.5 | 7352.34 | 20.4 | |||||||||
| C4A + C4B + B*08:01 | −0.51 | 0.073 | 11.7 | −0.37 | 0.069 | 7.2 | 0.49 | 0.12 | 4.4 | 7337.24 | 23.6 | |||||||||
| C4A + C4B + rs2105898 | −0.52 | 0.07 | 13.2 | −0.43 | 0.069 | 9.4 | −0.42 | 0.048 | 17.8 | 7277.78 | 36.2 | |||||||||
Supplementary Material
Acknowledgements
This work was supported by the National Human Genome Research Institute (HG006855), the National Institute of Mental Health (MH112491, MH105641, MH105653), the Stanley Center for Psychiatric Research, and the National Institute for Health Research Biomedical Research Centre (NIHR BRC) at Guy’s and St Thomas’ NHS Foundation and King’s College London. We thank Christina Usher and Christopher Patil for contributions to the figures and manuscript text. We thank Marta Florio for suggestions regarding figure display.
Schizophrenia Working Group of the Psychiatric Genomics Consortium
Stephan Ripke16,17, Benjamin M. Neale16,17,18,19, Aiden Corvin20, James T. R. Walters21, Kai-How Farh16, Peter A. Holmans21,22, Phil Lee16,17,19, Brendan Bulik-Sullivan16,17, David A. Collier23,24, Hailiang Huang16,18, Tune H. Pers18,25,26, Ingrid Agartz27,28,29, Esben Agerbo30,31,32, Margot Albus33, Madeline Alexander34, Farooq Amin35,36, Silviu A. Bacanu37, Martin Begemann38, Richard A Belliveau Jr17, Judit Bene39,40, Sarah E. Bergen 17,41, Elizabeth Bevilacqua17, Tim B Bigdeli 37, Donald W. Black42, Richard Bruggeman43, Nancy G. Buccola44, Randy L. Buckner45,46,47, William Byerley48, Wiepke Cahn49, Guiqing Cai50,51, Murray J. Cairns54,135,185, Dominique Campion52, Rita M. Cantor53, Vaughan J. Carr54,55, Noa Carrera21, Stanley V. Catts54,56, Kimberly D. Chambert17, Raymond C. K. Chan57, Ronald Y. L. Chen58, Eric Y. H. Chen58,59, Wei Cheng60, Eric F. C. Cheung61, Siow Ann Chong62, C. Robert Cloninger63, David Cohen64, Nadine Cohen65, Paul Cormican20, Nick Craddock21,22, Benedicto Crespo-Facorro225, James J. Crowley66, David Curtis67,68, Michael Davidson69, Kenneth L. Davis51, Franziska Degenhardt70,71, Jurgen Del Favero72, Lynn E. DeLisi143,144, Ditte Demontis32,73,74, Dimitris Dikeos75, Timothy Dinan76, Srdjan Djurovic29,77, Gary Donohoe20,78, Elodie Drapeau51, Jubao Duan79,80, Frank Dudbridge81, Naser Durmishi82, Peter Eichhammer83, Johan Eriksson84,85,86, Valentina Escott-Price21, Laurent Essioux87, Ayman H. Fanous88,89,90,91, Martilias S. Farrell66, Josef Frank92, Lude Franke93, Robert Freedman94, Nelson B. Freimer95, Marion Friedl96, Joseph I. Friedman51, Menachem Fromer16,17,19,97, Giulio Genovese17, Lyudmila Georgieva21, Elliot S. Gershon224, Ina Giegling96,98, Paola Giusti-Rodríguez66, Stephanie Godard99, Jacqueline I. Goldstein16,18, Vera Golimbet100, Srihari Gopal101, Jacob Gratten102, Lieuwe de Haan103, Marina Mitjans38, Marian L. Hamshere21, Mark Hansen104, Thomas Hansen32,105, Vahram Haroutunian51,106,107, Annette M. Hartmann96, Frans A. Henskens54,108,109, Stefan Herms70,71,110, Joel N. Hirschhorn18,26,111, Per Hoffmann70,71,110, Andrea Hofman70,71, Mads V. Hollegaard112, David M. Hougaard112, Masashi Ikeda113, Inge Joa114, Antonio Julià115, René S. Kahn49, Luba Kalaydjieva116,117, Sena Karachanak-Yankova118, Juha Karjalainen93, David Kavanagh21, Matthew C. Keller119, Brian J. Kelly135, James L. Kennedy120,121,122, Andrey Khrunin123, Yunjung Kim66, Janis Klovins124, James A. Knowles125, Bettina Konte96, Vaidutis Kucinskas126, Zita Ausrele Kucinskiene126, Hana Kuzelova- Ptackova127, Anna K. Kähler41, Claudine Laurent34,128, Jimmy Lee Chee Keong62,129, S. Hong Lee102, Sophie E. Legge21, Bernard Lerer130, Miaoxin Li58,59,131 Tao Li132, Kung-Yee Liang133, Jeffrey Lieberman134, Svetlana Limborska123, Carmel M. Loughland54,135, Jan Lubinski136, Jouko Lönnqvist137, Milan Macek Jr127, Patrik K. E. Magnusson41, Brion S. Maher138, Wolfgang Maier139, Jacques Mallet140, Sara Marsal115, Manuel Mattheisen32,73,74,141, Morten Mattingsdal29,142, Robert W. McCarley143,144, Colm McDonald145, Andrew M. McIntosh146,147, Sandra Meier92, Carin J. Meijer103, Bela Melegh39,40, Ingrid Melle29,148, Raquelle I. Mesholam-Gately143,149, Andres Metspalu150, Patricia T. Michie54,151, Lili Milani150, Vihra Milanova152, Younes Mokrab23, Derek W. Morris20,78, Ole Mors32,73,153, Kieran C. Murphy154, Robin M. Murray155, Inez Myin-Germeys156, Bertram Müller- Myhsok157,158,159, Mari Nelis150, Igor Nenadic160, Deborah A. Nertney161, Gerald Nestadt162, Kristin K. Nicodemus163, Liene Nikitina-Zake124, Laura Nisenbaum164, Annelie Nordin165, Eadbhard O’Callaghan166, Colm O’Dushlaine17, F. Anthony O’Neill167, Sang-Yun Oh168, Ann Olincy94, Line Olsen32,105, Jim Van Os156,169, Psychosis Endophenotypes International Consortium170, Christos Pantelis54,171, George N. Papadimitriou75, Agnes A. Steixner38, Elena Parkhomenko51, Michele T. Pato125, Tiina Paunio172,173, Milica Pejovic-Milovancevic174, Diana O. Perkins175, Olli Pietiläinen173,176, Jonathan Pimm68, Andrew J. Pocklington21, John Powell155, Alkes Price18,177, Ann E. Pulver162, Shaun M. Purcell97, Digby Quested178, Henrik B. Rasmussen32,105, Abraham Reichenberg51, Mark A. Reimers179, Alexander L. Richards21, Joshua L. Roffman45,47, Panos Roussos97,180, Douglas M. Ruderfer21,97, Veikko Salomaa86, Alan R. Sanders79,80, Ulrich Schall54,135, Christian R. Schubert181, Thomas G. Schulze92,182, Sibylle G. Schwab183, Edward M. Scolnick17, Rodney J. Scott54,184,185, Larry J. Seidman143,149, Jianxin Shi186, Engilbert Sigurdsson187, Teimuraz Silagadze188, Jeremy M. Silverman51,189, Kang Sim62, Petr Slominsky123, Jordan W. Smoller17,19, Hon-Cheong So58, Chris C. A. Spencer190, Eli A. Stahl18,97, Hreinn Stefansson191, Stacy Steinberg191, Elisabeth Stogmann192, Richard E. Straub193, Eric Strengman194,49, Jana Strohmaier92, T. Scott Stroup134, Mythily Subramaniam62, Jaana Suvisaari137, Dragan M. Svrakic63, Jin P. Szatkiewicz66, Erik Söderman27, Srinivas Thirumalai195, Draga Toncheva118, Paul A. Tooney54,135,185, Sarah Tosato196, Juha Veijola197,198, John Waddington199, Dermot Walsh200, Dai Wang101, Qiang Wang132, Bradley T. Webb37, Mark Weiser69, Dieter B. Wildenauer201, Nigel M. Williams21, Stephanie Williams66, Stephanie H. Witt92, Aaron R. Wolen179, Emily H. M. Wong58, Brandon K. Wormley37, Jing Qin Wu54,185, Hualin Simon Xi202, Clement C. Zai120,121, Xuebin Zheng203, Fritz Zimprich192, Naomi R. Wray102, Kari Stefansson191, Peter M. Visscher102, Wellcome Trust Case-Control Consortium 2204, Rolf Adolfsson165, Ole A. Andreassen29,148, Douglas H. R. Blackwood147, Elvira Bramon205, Joseph D. Buxbaum50,51,106,206, Anders D. Børglum32,73,74,153, Sven Cichon70,71,110,207, Ariel Darvasi208, Enrico Domenici209, Hannelore Ehrenreich38, Tõnu Esko18,26,111,150, Pablo V. Gejman79,80, Michael Gill20, Hugh Gurling68, Christina M. Hultman41, Nakao Iwata113, Assen V. Jablensky54,117,201,210, Erik G. Jönsson27,29, Kenneth S. Kendler211, George Kirov21, Jo Knight120,121,122, Todd Lencz212,213,214, Douglas F. Levinson34, Qingqin S. Li101, Jianjun Liu203,215, Anil K. Malhotra212,213,214, Steven A. McCarroll17,111, Andrew McQuillin68, Jennifer L. Moran17, Preben B. Mortensen30,31,32, Bryan J. Mowry102,216, Markus M. Nöthen70,71, Roel A. Ophoff53,95,49, Michael J. Owen21,22, Aarno Palotie17,19,176, Carlos N. Pato125, Tracey L. Petryshen17,143,217, Danielle Posthuma218,219,220, Marcella Rietschel92, Brien P. Riley211, Dan Rujescu96,98, Pak C. Sham58,59,131 Pamela Sklar97,106,180, David St Clair221, Daniel R. Weinberger193,222, Jens R. Wendland181, Thomas Werge32,105,223, Mark J. Daly16,17,18, Patrick F. Sullivan41,66,175 & Michael C. O’Donovan21,22
16Analytic and Translational Genetics Unit, Massachusetts General Hospital, Boston, Massachusetts 02114, USA.
17Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, Massachusetts 02142, USA.
18Medical and Population Genetics Program, Broad Institute of MIT and Harvard, Cambridge, Massachusetts 02142, USA.
19Psychiatric and Neurodevelopmental Genetics Unit, Massachusetts General Hospital, Boston, Massachusetts 02114, USA.
20Neuropsychiatric Genetics Research Group, Department of Psychiatry, Trinity College Dublin, Dublin 8, Ireland.
21MRC Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, School of Medicine, Cardiff University, Cardiff, CF24 4HQ, UK.
22National Centre for Mental Health, Cardiff University, Cardiff, CF24 4HQ, UK.
23Eli Lilly and Company Limited, Erl Wood Manor, Sunninghill Road, Windlesham, Surrey, GU20 6PH, UK.
24Social, Genetic and Developmental Psychiatry Centre, Institute of Psychiatry, King’s College London, London, SE5 8AF, UK.
25Center for Biological Sequence Analysis, Department of Systems Biology, Technical University of Denmark, DK-2800, Denmark.
26Division of Endocrinology and Center for Basic and Translational Obesity Research, Boston Children’s Hospital, Boston, Massachusetts, 02115 USA.
27Department of Clinical Neuroscience, Psychiatry Section, Karolinska Institutet, SE-17176 Stockholm, Sweden.
28Department of Psychiatry, Diakonhjemmet Hospital, 0319 Oslo, Norway.
29NORMENT, KG Jebsen Centre for Psychosis Research, Institute of Clinical Medicine, University of Oslo, 0424 Oslo, Norway.
30Centre for Integrative Register-based Research, CIRRAU, Aarhus University, DK-8210 Aarhus, Denmark.
31National Centre for Register-based Research, Aarhus University, DK-8210 Aarhus, Denmark.
32The Lundbeck Foundation Initiative for Integrative Psychiatric Research, iPSYCH, Denmark.
33State Mental Hospital, 85540 Haar, Germany.
34Department of Psychiatry and Behavioral Sciences, Stanford University, Stanford, California 94305, USA.
35Department of Psychiatry and Behavioral Sciences, Atlanta Veterans Affairs Medical Center, Atlanta, Georgia 30033, USA.
36Department of Psychiatry and Behavioral Sciences, Emory University, Atlanta Georgia 30322, USA.
37Virginia Institute for Psychiatric and Behavioral Genetics, Department of Psychiatry, Virginia Commonwealth University, Richmond, Virginia 23298, USA.
38Clinical Neuroscience, Max Planck Institute of Experimental Medicine, Göttingen 37075, Germany.
39Department of Medical Genetics, University of Pécs, Pécs H-7624, Hungary.
40Szentagothai Research Center, University of Pécs, Pécs H-7624, Hungary.
41Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Stockholm SE-17177, Sweden.
42Department of Psychiatry, University of Iowa Carver College of Medicine, Iowa City, Iowa 52242, USA.
43University Medical Center Groningen, Department of Psychiatry, University of Groningen NL-9700 RB, The Netherlands.
44School of Nursing, Louisiana State University Health Sciences Center, New Orleans, Louisiana 70112, USA.
45Athinoula A. Martinos Center, Massachusetts General Hospital, Boston, Massachusetts 02129, USA.
46Center for Brain Science, Harvard University, Cambridge, Massachusetts, 02138 USA.
47Department of Psychiatry, Massachusetts General Hospital, Boston, Massachusetts, 02114 USA.
48Department of Psychiatry, University of California at San Francisco, San Francisco, California, 94143 USA.
49University Medical Center Utrecht, Department of Psychiatry, Rudolf Magnus Institute of Neuroscience, 3584 Utrecht, The Netherlands.
50Department of Human Genetics, Icahn School of Medicine at Mount Sinai, New York, New York 10029 USA.
51Department of Psychiatry, Icahn School of Medicine at Mount Sinai, New York, New York 10029 USA.
52Centre Hospitalier du Rouvray and INSERM U1079 Faculty of Medicine, 76301 Rouen, France.
53Department of Human Genetics, David Geffen School of Medicine, University of California, Los Angeles, California 90095, USA.
54Schizophrenia Research Institute, Sydney NSW 2010, Australia.
55School of Psychiatry, University of New South Wales, Sydney NSW 2031, Australia.
56Royal Brisbane and Women’s Hospital, University of Queensland, Brisbane, St Lucia QLD 4072, Australia.
57Institute of Psychology, Chinese Academy of Science, Beijing 100101, China.
58Department of Psychiatry, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong, China.
59State Key Laboratory for Brain and Cognitive Sciences, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong, China.
60Department of Computer Science, University of North Carolina, Chapel Hill, North Carolina 27514, USA.
61Castle Peak Hospital, Hong Kong, China.
62Institute of Mental Health, Singapore 539747, Singapore.
63Department of Psychiatry, Washington University, St. Louis, Missouri 63110, USA.
64Department of Child and Adolescent Psychiatry, Assistance Publique Hopitaux de Paris, Pierre and Marie Curie Faculty of Medicine and Institute for Intelligent Systems and Robotics, Paris, 75013, France.
65 Blue Note Biosciences, Princeton, New Jersey 08540, USA
66Department of Genetics, University of North Carolina, Chapel Hill, North Carolina 27599–7264, USA.
67Department of Psychological Medicine, Queen Mary University of London, London E1 1BB, UK.
68Molecular Psychiatry Laboratory, Division of Psychiatry, University College London, London WC1E 6JJ, UK.
69Sheba Medical Center, Tel Hashomer 52621, Israel.
70Department of Genomics, Life and Brain Center, D-53127 Bonn, Germany.
71Institute of Human Genetics, University of Bonn, D-53127 Bonn, Germany.
72Applied Molecular Genomics Unit, VIB Department of Molecular Genetics, University of Antwerp, B-2610 Antwerp, Belgium.
73Centre for Integrative Sequencing, iSEQ, Aarhus University, DK-8000 Aarhus C, Denmark.
74Department of Biomedicine, Aarhus University, DK-8000 Aarhus C, Denmark.
75First Department of Psychiatry, University of Athens Medical School, Athens 11528, Greece.
76Department of Psychiatry, University College Cork, Co. Cork, Ireland.
77Department of Medical Genetics, Oslo University Hospital, 0424 Oslo, Norway.
78Cognitive Genetics and Therapy Group, School of Psychology and Discipline of Biochemistry, National University of Ireland Galway, Co. Galway, Ireland.
79Department of Psychiatry and Behavioral Neuroscience, University of Chicago, Chicago, Illinois 60637, USA.
80Department of Psychiatry and Behavioral Sciences, NorthShore University HealthSystem, Evanston, Illinois 60201, USA.
81Department of Non-Communicable Disease Epidemiology, London School of Hygiene and Tropical Medicine, London WC1E 7HT, UK.
82Department of Child and Adolescent Psychiatry, University Clinic of Psychiatry, Skopje 1000, Republic of Macedonia.
83Department of Psychiatry, University of Regensburg, 93053 Regensburg, Germany.
84Department of General Practice, Helsinki University Central Hospital, University of Helsinki P.O. Box 20, Tukholmankatu 8 B, FI-00014, Helsinki, Finland
85Folkhälsan Research Center, Helsinki, Finland, Biomedicum Helsinki 1, Haartmaninkatu 8, FI-00290, Helsinki, Finland.
86National Institute for Health and Welfare, P.O. BOX 30, FI-00271 Helsinki, Finland.
87Translational Technologies and Bioinformatics, Pharma Research and Early Development, F. Hoffman-La Roche, CH-4070 Basel, Switzerland.
88Department of Psychiatry, Georgetown University School of Medicine, Washington DC 20057, USA.
89Department of Psychiatry, Keck School of Medicine of the University of Southern California, Los Angeles, California 90033, USA.
90Department of Psychiatry, Virginia Commonwealth University School of Medicine, Richmond, Virginia 23298, USA.
91Mental Health Service Line, Washington VA Medical Center, Washington DC 20422, USA.
92Department of Genetic Epidemiology in Psychiatry, Central Institute of Mental Health, Medical Faculty Mannheim, University of Heidelberg, Heidelberg, D-68159 Mannheim, Germany.
93Department of Genetics, University of Groningen, University Medical Centre Groningen, 9700 RB Groningen, The Netherlands.
94Department of Psychiatry, University of Colorado Denver, Aurora, Colorado 80045, USA.
95Center for Neurobehavioral Genetics, Semel Institute for Neuroscience and Human Behavior, University of California, Los Angeles, California 90095, USA.
96Department of Psychiatry, University of Halle, 06112 Halle, Germany.
97Division of Psychiatric Genomics, Department of Psychiatry, Icahn School of Medicine at Mount Sinai, New York, New York 10029, USA.
98Department of Psychiatry, University of Munich, 80336, Munich, Germany.
99Departments of Psychiatry and Human and Molecular Genetics, INSERM, Institut de Myologie, Hôpital de la Pitiè-Salpêtrière, Paris, 75013, France.
100Mental Health Research Centre, Russian Academy of Medical Sciences, 115522 Moscow, Russia.
101Neuroscience Therapeutic Area, Janssen Research and Development, Raritan, New Jersey 08869, USA.
102Queensland Brain Institute, The University of Queensland, Brisbane, Queensland, QLD 4072, Australia.
103Academic Medical Centre University of Amsterdam, Department of Psychiatry, 1105 AZ Amsterdam, The Netherlands.
104Illumina, La Jolla, California, California 92122, USA.
105Institute of Biological Psychiatry, Mental Health Centre Sct. Hans, Mental Health Services Copenhagen, DK-4000, Denmark.
106Friedman Brain Institute, Icahn School of Medicine at Mount Sinai, New York, New York 10029, USA.
107J. J. Peters VA Medical Center, Bronx, New York, New York 10468, USA.
108Priority Research Centre for Health Behaviour, University of Newcastle, Newcastle NSW 2308, Australia.
109School of Electrical Engineering and Computer Science, University of Newcastle, Newcastle NSW 2308, Australia.
110Division of Medical Genetics, Department of Biomedicine, University of Basel, Basel, CH-4058, Switzerland.
111Department of Genetics, Harvard Medical School, Boston, Massachusetts 02115, USA.
112Section of Neonatal Screening and Hormones, Department of Clinical Biochemistry, Immunology and Genetics, Statens Serum Institut, Copenhagen, DK-2300, Denmark.
113Department of Psychiatry, Fujita Health University School of Medicine, Toyoake, Aichi, 470–1192, Japan.
114Regional Centre for Clinical Research in Psychosis, Department of Psychiatry, Stavanger University Hospital, 4011 Stavanger, Norway.
115Rheumatology Research Group, Vall d’Hebron Research Institute, Barcelona, 08035, Spain.
116Centre for Medical Research, The University of Western Australia, Perth, WA 6009, Australia.
117The Perkins Institute for Medical Research, The University of Western Australia, Perth, WA 6009, Australia.
118Department of Medical Genetics, Medical University, Sofia1431, Bulgaria.
119Department of Psychology, University of Colorado Boulder, Boulder, Colorado 80309, USA.
120Campbell Family Mental Health Research Institute, Centre for Addiction and Mental Health, Toronto, Ontario, M5T 1R8, Canada.
121Department of Psychiatry, University of Toronto, Toronto, Ontario, M5T 1R8, Canada.
122Institute of Medical Science, University of Toronto, Toronto, Ontario, M5S 1A8, Canada.
123Institute of Molecular Genetics, Russian Academy of Sciences, Moscow123182, Russia.
124Latvian Biomedical Research and Study Centre, Riga, LV-1067, Latvia.
125Department of Psychiatry and Zilkha Neurogenetics Institute, Keck School of Medicine at University of Southern California, Los Angeles, California 90089, USA.
126Faculty of Medicine, Vilnius University, LT-01513 Vilnius, Lithuania.
127 Department of Biology and Medical Genetics, 2nd Faculty of Medicine and University Hospital Motol, 150 06 Prague, Czech Republic.
128 Department of Child and Adolescent Psychiatry, Pierre and Marie Curie Faculty of Medicine, Paris 75013, France.
129Duke-NUS Graduate Medical School, Singapore 169857, Singapore.
130Department of Psychiatry, Hadassah-Hebrew University Medical Center, Jerusalem 91120, Israel.
131Centre for Genomic Sciences, The University of Hong Kong, Hong Kong, China.
132Mental Health Centre and Psychiatric Laboratory, West China Hospital, Sichuan University, Chengdu, 610041, Sichuan, China.
133Department of Biostatistics, Johns Hopkins University Bloomberg School of Public Health, Baltimore, Maryland 21205, USA.
134Department of Psychiatry, Columbia University, New York, New York 10032, USA.
135Priority Centre for Translational Neuroscience and Mental Health, University of Newcastle, Newcastle NSW 2300, Australia.
136Department of Genetics and Pathology, International Hereditary Cancer Center, Pomeranian Medical University in Szczecin, 70–453 Szczecin, Poland.
137Department of Mental Health and Substance Abuse Services; National Institute for Health and Welfare, P.O. BOX 30, FI-00271 Helsinki, Finland
138Department of Mental Health, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland 21205, USA.
139Department of Psychiatry, University of Bonn, D-53127 Bonn, Germany.
140Centre National de la Recherche Scientifique, Laboratoire de Génétique Moléculaire de la Neurotransmission et des Processus Neurodégénératifs, Hôpital de la Pitié Salpêtrière, 75013, Paris, France.
141Department of Genomics Mathematics, University of Bonn, D-53127 Bonn, Germany.
142Research Unit, Sørlandet Hospital, 4604 Kristiansand, Norway.
143Department of Psychiatry, Harvard Medical School, Boston, Massachusetts 02115, USA.
144VA Boston Health Care System, Brockton, Massachusetts 02301, USA.
145Department of Psychiatry, National University of Ireland Galway, Co. Galway, Ireland.
146Centre for Cognitive Ageing and Cognitive Epidemiology, University of Edinburgh, Edinburgh EH16 4SB, UK.
147Division of Psychiatry, University of Edinburgh, Edinburgh EH16 4SB, UK.
148Division of Mental Health and Addiction, Oslo University Hospital, 0424 Oslo, Norway.
149Massachusetts Mental Health Center Public Psychiatry Division of the Beth Israel Deaconess Medical Center, Boston, Massachusetts 02114, USA.
150Estonian Genome Center, University of Tartu, Tartu 50090, Estonia.
151School of Psychology, University of Newcastle, Newcastle NSW 2308, Australia.
152First Psychiatric Clinic, Medical University, Sofia 1431, Bulgaria.
153Department P, Aarhus University Hospital, DK-8240 Risskov, Denmark.
154Department of Psychiatry, Royal College of Surgeons in Ireland, Dublin 2, Ireland.
155King’s College London, London SE5 8AF, UK.
156Maastricht University Medical Centre, South Limburg Mental Health Research and Teaching Network, EURON, 6229 HX Maastricht, The Netherlands.
157Institute of Translational Medicine, University of Liverpool, Liverpool L69 3BX, UK.
158Max Planck Institute of Psychiatry, 80336 Munich, Germany.
159Munich Cluster for Systems Neurology (SyNergy), 80336 Munich, Germany.
160Department of Psychiatry and Psychotherapy, Jena University Hospital, 07743 Jena, Germany.
161Department of Psychiatry, Queensland Brain Institute and Queensland Centre for Mental Health Research, University of Queensland, Brisbane, Queensland, St Lucia QLD 4072, Australia.
162Department of Psychiatry and Behavioral Sciences, Johns Hopkins University School of Medicine, Baltimore, Maryland 21205, USA.
163Department of Psychiatry, Trinity College Dublin, Dublin 2, Ireland.
164Eli Lilly and Company, Lilly Corporate Center, Indianapolis, 46285 Indiana, USA.
165Department of Clinical Sciences, Psychiatry, Umeå University, SE-901 87 Umeå, Sweden.
166DETECT Early Intervention Service for Psychosis, Blackrock, Co. Dublin, Ireland.
167Centre for Public Health, Institute of Clinical Sciences, Queen’s University Belfast, Belfast BT12 6AB, UK.
168Lawrence Berkeley National Laboratory, University of California at Berkeley, Berkeley, California 94720, USA.
169Institute of Psychiatry, King’s College London, London SE5 8AF, UK.
170A list of authors and affiliations appear in the Supplementary Information.
171Melbourne Neuropsychiatry Centre, University of Melbourne & Melbourne Health, Melbourne, Vic 3053, Australia.
172Department of Psychiatry, University of Helsinki, P.O. Box 590, FI-00029 HUS, Helsinki, Finland.
173Public Health Genomics Unit, National Institute for Health and Welfare, P.O. BOX 30, FI-00271 Helsinki, Finland.
174Medical Faculty, University of Belgrade, 11000 Belgrade, Serbia.
175Department of Psychiatry, University of North Carolina, Chapel Hill, North Carolina 27599–7160, USA.
176Institute for Molecular Medicine Finland, FIMM, University of Helsinki, P.O. Box 20 FI-00014, Helsinki, Finland.
177Department of Epidemiology, Harvard School of Public Health, Boston, Massachusetts 02115, USA.
178Department of Psychiatry, University of Oxford, Oxford, OX3 7JX, UK.
179Virginia Institute for Psychiatric and Behavioral Genetics, Virginia Commonwealth University, Richmond, Virginia 23298, USA.
180Institute for Multiscale Biology, Icahn School of Medicine at Mount Sinai, New York, New York 10029, USA.
181PharmaTherapeutics Clinical Research, Pfizer Worldwide Research and Development, Cambridge, Massachusetts 02139, USA.
182Department of Psychiatry and Psychotherapy, University of Gottingen, 37073 Göttingen, Germany.
183Psychiatry and Psychotherapy Clinic, University of Erlangen, 91054 Erlangen, Germany.
184Hunter New England Health Service, Newcastle NSW 2308, Australia.
185School of Biomedical Sciences and Pharmacy, University of Newcastle, Callaghan NSW 2308, Australia.
186Division of Cancer Epidemiology and Genetics, National Cancer Institute, Bethesda, Maryland 20892, USA.
187University of Iceland, Landspitali, National University Hospital, 101 Reykjavik, Iceland.
188Department of Psychiatry and Drug Addiction, Tbilisi State Medical University (TSMU), N33, 0177 Tbilisi, Georgia.
189Research and Development, Bronx Veterans Affairs Medical Center, New York, New York 10468, USA.
190Wellcome Trust Centre for Human Genetics, Oxford, OX3 7BN, UK.
191deCODE Genetics, 101 Reykjavik, Iceland.
192Department of Clinical Neurology, Medical University of Vienna, 1090 Wien, Austria.
193Lieber Institute for Brain Development, Baltimore, Maryland 21205, USA.
194Department of Medical Genetics, University Medical Centre Utrecht, Universiteitsweg 100, 3584 CG, Utrecht, The Netherlands.
195Berkshire Healthcare NHS Foundation Trust, Bracknell RG12 1BQ, UK.
196Section of Psychiatry, University of Verona, 37134 Verona, Italy.
197Department of Psychiatry, University of Oulu, P.O. BOX 5000, 90014, Finland
198University Hospital of Oulu, P.O.BOX 20, 90029 OYS, Finland.
199Molecular and Cellular Therapeutics, Royal College of Surgeons in Ireland, Dublin 2, Ireland.
200Health Research Board, Dublin 2, Ireland.
201School of Psychiatry and Clinical Neurosciences, The University of Western Australia, Perth WA6009, Australia.
202Computational Sciences CoE, Pfizer Worldwide Research and Development, Cambridge, Massachusetts 02139, USA.
203Human Genetics, Genome Institute of Singapore, A*STAR, Singapore 138672, Singapore.
205University College London, London WC1E 6BT, UK.
206Department of Neuroscience, Icahn School of Medicine at Mount Sinai, New York, New York 10029, USA.
207Institute of Neuroscience and Medicine (INM-1), Research Center Juelich, 52428 Juelich, Germany.
208Department of Genetics, The Hebrew University of Jerusalem, 91905 Jerusalem, Israel.
209Neuroscience Discovery and Translational Area, Pharma Research and Early Development, F. Hoffman-La Roche, CH-4070 Basel, Switzerland.
210Centre for Clinical Research in Neuropsychiatry, School of Psychiatry and Clinical Neurosciences, The University of Western Australia, Medical Research Foundation Building, Perth WA 6000, Australia.
211Virginia Institute for Psychiatric and Behavioral Genetics, Departments of Psychiatry and Human and Molecular Genetics, Virginia Commonwealth University, Richmond, Virginia 23298, USA.
212The Feinstein Institute for Medical Research, Manhasset, New York, 11030 USA.
213The Hofstra NS-LIJ School of Medicine, Hempstead, New York, 11549 USA.
214The Zucker Hillside Hospital, Glen Oaks, New York,11004 USA.
215Saw Swee Hock School of Public Health, National University of Singapore, Singapore 117597, Singapore.
216Queensland Centre for Mental Health Research, University of Queensland, Brisbane 4076, Queensland, Australia.
217Center for Human Genetic Research and Department of Psychiatry, Massachusetts General Hospital, Boston, Massachusetts 02114, USA.
218Department of Child and Adolescent Psychiatry, Erasmus University Medical Centre, Rotterdam 3000, The Netherlands.
219Department of Complex Trait Genetics, Neuroscience Campus Amsterdam, VU University Medical Center Amsterdam, Amsterdam 1081, The Netherlands.
220Department of Functional Genomics, Center for Neurogenomics and Cognitive Research, Neuroscience Campus Amsterdam, VU University, Amsterdam 1081, The Netherlands.
221University of Aberdeen, Institute of Medical Sciences, Aberdeen, AB25 2ZD, UK.
222Departments of Psychiatry, Neurology, Neuroscience and Institute of Genetic Medicine, Johns Hopkins School of Medicine, Baltimore, Maryland 21205, USA.
223Department of Clinical Medicine, University of Copenhagen, Copenhagen 2200, Denmark.
224Departments of Psychiatry and Human Genetics, University of Chicago, Chicago, Illinois 60637, USA.
225University Hospital Marqués de Valdecilla, Instituto de Formación e Investigación Marqués de Valdecilla, University of Cantabria, ED39008 Santander, Spain.
Footnotes
Competing interests
The authors declare no competing interests.
Supplementary Information is available for this paper.
Reprints and permissions information is available at www.nature.com/reprints
References
- 1.Ngo ST, Steyn FJ & McCombe PA Gender differences in autoimmune disease. Front Neuroendocrinol 35, 347–369, doi: 10.1016/j.yfrne.2014.04.004 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Abel KM, Drake R & Goldstein JM Sex differences in schizophrenia. Int Rev Psychiatry 22, 417–428, doi: 10.3109/09540261.2010.515205 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Langefeld CD et al. Transancestral mapping and genetic load in systemic lupus erythematosus. Nature Communications 8, 16021, doi: 10.1038/ncomms16021 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.International MHC et al. Mapping of multiple susceptibility variants within the MHC region for 7 immune-mediated diseases. Proc Natl Acad Sci U S A 106, 18680–18685, doi: 10.1073/pnas.0909307106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanscombe KB et al. Genetic fine mapping of systemic lupus erythematosus MHC associations in Europeans and African Americans. Hum Mol Genet 27, 3813–3824, doi: 10.1093/hmg/ddy280 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cruz-Tapias P, Rojas-Villarraga A, Maier-Moore S & Anaya JM HLA and Sjogren’s syndrome susceptibility. A meta-analysis of worldwide studies. Autoimmun Rev 11, 281–287, doi: 10.1016/j.autrev.2011.10.002 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Sekar A et al. Schizophrenia risk from complex variation of complement component 4. Nature 530, 177–183 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaya da Costa M et al. Age and Sex-Associated Changes of Complement Activity and Complement Levels in a Healthy Caucasian Population. Front Immunol 9, 2664, doi: 10.3389/fimmu.2018.02664 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ritchie RF et al. Reference distributions for complement proteins C3 and C4: a practical, simple and clinically relevant approach in a large cohort. Journal of clinical laboratory analysis 18, 1–8, doi: 10.1002/jcla.10100 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lawrence JS, Martins CL & Drake GL A family survey of lupus erythematosus. 1. Heritability. J Rheumatol 14, 913–921 (1987). [PubMed] [Google Scholar]
- 11.Lipsky PE Systemic lupus erythematosus: an autoimmune disease of B cell hyperactivity. Nat Immunol 2, 764–766, doi: 10.1038/ni0901-764 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Ippolito A et al. Autoantibodies in systemic lupus erythematosus: comparison of historical and current assessment of seropositivity. Lupus 20, 250–255, doi: 10.1177/0961203310385738 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Lee KH, Wucherpfennig KW & Wiley DC Structure of a human insulin peptide-HLA-DQ8 complex and susceptibility to type 1 diabetes. Nat Immunol 2, 501–507, doi: 10.1038/88694 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Raychaudhuri S et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat Genet 44, 291–296, doi: 10.1038/ng.1076 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morris DL et al. MHC associations with clinical and autoantibody manifestations in European SLE. Genes Immun 15, 210–217, doi: 10.1038/gene.2014.6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Banlaki Z, Doleschall M, Rajczy K, Fust G & Szilagyi A Fine-tuned characterization of RCCX copy number variants and their relationship with extended MHC haplotypes. Genes Immun 13, 530–535, doi: 10.1038/gene.2012.29 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Isenman DE & Young JR The molecular basis for the difference in immune hemolysis activity of the Chido and Rodgers isotypes of human complement component C4. J Immunol 132, 3019–3027 (1984). [PubMed] [Google Scholar]
- 18.Law SK, Dodds AW & Porter RR A comparison of the properties of two classes, C4A and C4B, of the human complement component C4. EMBO J 3, 1819–1823 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Birmingham DJ et al. The complex nature of serum C3 and C4 as biomarkers of lupus renal flare. Lupus 19, 1272–1280, doi: 10.1177/0961203310371154 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ross SC & Densen P Complement deficiency states and infection: epidemiology, pathogenesis and consequences of neisserial and other infections in an immune deficiency. Medicine (Baltimore) 63, 243–273 (1984). [PubMed] [Google Scholar]
- 21.Wu YL, Hauptmann G, Viguier M & Yu CY Molecular basis of complete complement C4 deficiency in two North-African families with systemic lupus erythematosus. Genes Immun 10, 433–445, doi: 10.1038/gene.2009.10 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.International Consortium for Systemic Lupus Erythematosus, G. et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet 40, 204–210, doi: 10.1038/ng.81 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Y et al. Gene copy-number variation and associated polymorphisms of complement component C4 in human systemic lupus erythematosus (SLE): low copy number is a risk factor for and high copy number is a protective factor against SLE susceptibility in European Americans. Am J Hum Genet 80, 1037–1054, doi: 10.1086/518257 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juptner M et al. Low copy numbers of complement C4 and homozygous deficiency of C4A may predispose to severe disease and earlier disease onset in patients with systemic lupus erythematosus. Lupus 27, 600–609, doi: 10.1177/0961203317735187 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boteva L et al. Genetically determined partial complement C4 deficiency states are not independent risk factors for SLE in UK and Spanish populations. Am J Hum Genet 90, 445–456, doi: 10.1016/j.ajhg.2012.01.012 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pato MT et al. The genomic psychiatry cohort: partners in discovery. Am J Med Genet B Neuropsychiatr Genet 162B, 306–312, doi: 10.1002/ajmg.b.32160 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanders SJ et al. Whole genome sequencing in psychiatric disorders: the WGSPD consortium. Nat Neurosci 20, 1661–1668, doi: 10.1038/s41593-017-0017-9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuo CF et al. Familial Risk of Sjogren’s Syndrome and Co-aggregation of Autoimmune Diseases in Affected Families: A Nationwide Population Study. Arthritis Rheumatol 67, 1904–1912, doi: 10.1002/art.39127 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fayyaz A, Kurien BT & Scofield RH Autoantibodies in Sjogren’s Syndrome. Rheum Dis Clin North Am 42, 419–434, doi: 10.1016/j.rdc.2016.03.002 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramos-Casals M et al. Hypocomplementaemia as an immunological marker of morbidity and mortality in patients with primary Sjogren’s syndrome. Rheumatology (Oxford) 44, 89–94, doi: 10.1093/rheumatology/keh407 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Chused TM, Kassan SS, Opelz G, Moutsopoulos HM & Terasaki PI Sjogren’s syndrome association with HLA-Dw3. N Engl J Med 296, 895–897, doi: 10.1056/NEJM197704212961602 (1977). [DOI] [PubMed] [Google Scholar]
- 32.Taylor KE et al. Genome-Wide Association Analysis Reveals Genetic Heterogeneity of Sjogren’s Syndrome According to Ancestry. Arthritis Rheumatol 69, 1294–1305, doi: 10.1002/art.40040 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khramtsova EA, Davis LK & Stranger BE The role of sex in the genomics of human complex traits. Nat Rev Genet 20, 173–190, doi: 10.1038/s41576-018-0083-1 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Hughes T et al. Analysis of autosomal genes reveals gene-sex interactions and higher total genetic risk in men with systemic lupus erythematosus. Ann Rheum Dis 71, 694–699, doi: 10.1136/annrheumdis-2011-200385 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.GTEx Consortium et al. Genetic effects on gene expression across human tissues. Nature 550, 204–213, doi: 10.1038/nature24277 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brinks R et al. Age-specific and sex-specific incidence of systemic lupus erythematosus: an estimate from cross-sectional claims data of 2.3 million people in the German statutory health insurance 2002. Lupus Sci Med 3, e000181, doi: 10.1136/lupus-2016-000181 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim HJ et al. Incidence, mortality, and causes of death in physician-diagnosed primary Sjogren’s syndrome in Korea: A nationwide, population-based study. Semin Arthritis Rheum 47, 222–227, doi: 10.1016/j.semarthrit.2017.03.004 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Degn SE et al. Clonal Evolution of Autoreactive Germinal Centers. Cell 170, 913–926 e919, doi: 10.1016/j.cell.2017.07.026 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Estrada K et al. A whole-genome sequence study identifies genetic risk factors for neuromyelitis optica. Nat Commun 9, 1929, doi: 10.1038/s41467-018-04332-3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pittock SJ et al. Neuromyelitis optica and non organ-specific autoimmunity. Arch Neurol 65, 78–83, doi: 10.1001/archneurol.2007.17 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Erdei A et al. Expression and role of CR1 and CR2 on B and T lymphocytes under physiological and autoimmune conditions. Mol Immunol 46, 2767–2773, doi: 10.1016/j.molimm.2009.05.181 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Unterman A et al. Neuropsychiatric syndromes in systemic lupus erythematosus: a meta-analysis. Semin Arthritis Rheum 41, 1–11, doi: 10.1016/j.semarthrit.2010.08.001 (2011). [DOI] [PubMed] [Google Scholar]
References
- 43.Schizophrenia Working Group of the Psychiatric Genomics, C. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427, doi: 10.1038/nature13595 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Handsaker RE et al. Large multiallelic copy number variations in humans. Nat Genet 47, 296–303, doi: 10.1038/ng.3200 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Browning SR & Browning BL Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet 81, 1084–1097, doi: 10.1086/521987 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Browning BL & Browning SR Genotype Imputation with Millions of Reference Samples. Am J Hum Genet 98, 116–126, doi: 10.1016/j.ajhg.2015.11.020 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng X et al. HIBAG--HLA genotype imputation with attribute bagging. Pharmacogenomics J 14, 192–200, doi: 10.1038/tpj.2013.18 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng X Imputation-Based HLA Typing with SNPs in GWAS Studies. Methods Mol Biol 1802, 163–176, doi: 10.1007/978-1-4939-8546-3_11 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Luykx JJ et al. A common variant in ERBB4 regulates GABA concentrations in human cerebrospinal fluid. Neuropsychopharmacology 37, 2088–2092, doi: 10.1038/npp.2012.57 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Albersen M et al. Vitamin B-6 vitamers in human plasma and cerebrospinal fluid. Am J Clin Nutr 100, 587–592, doi: 10.3945/ajcn.113.082008 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Malladi AS et al. Primary Sjogren’s syndrome as a systemic disease: a study of participants enrolled in an international Sjogren’s syndrome registry. Arthritis Care Res (Hoboken) 64, 911–918, doi: 10.1002/acr.21610 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Consortium, E. P. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74, doi: 10.1038/nature11247 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kent WJ et al. The human genome browser at UCSC. Genome Res 12, 996–1006, doi: 10.1101/gr.229102 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Individual genotype data for Sjögren’s syndrome cases and controls and individual plasma concentrations for C4 and C3 are available in dbGaP under accession number phs000672.v1.p1. Individual genotype data for schizophrenia cases and controls are available by application to the Psychiatric Genomics Consortium (PGC). Questions regarding individual genotype data for SLE cases and controls of European and/or African American ancestry can be directed to Timothy J. Vyse (timothy.vyse@kcl.ac.uk). Data resources (reference haplotypes), software scripts and instructions for imputing C4 alleles into SNP data sets are available on the McCarroll lab web site at http://mccarrolllab.org/resources/resources-for-c4/. Genotype and protein concentration data for CSF samples are available upon request.