Abstract
The gut microbiota synthesize hundreds of molecules, many of which are known to impact host physiology. Among the most abundant metabolites are the secondary bile acids deoxycholic acid (DCA) and lithocholic acid (LCA), which accumulate at ~500 μM and are known to block C. difficile growth1, promote hepatocellular carcinoma2, and modulate host metabolism via the GPCR TGR53. More broadly, DCA, LCA and their derivatives are a major component of the recirculating bile acid pool4; the size and composition of this pool are a target of therapies for primary biliary cholangitis and nonalcoholic steatohepatitis. Despite the clear impact of DCA and LCA on host physiology, incomplete knowledge of their biosynthetic genes and a lack of genetic tools in their native producer limit our ability to modulate secondary bile acid levels in the host. Here, we complete the pathway to DCA/LCA by assigning and characterizing enzymes for each of the steps in its reductive arm, revealing a strategy in which the A-B rings of the steroid core are transiently converted into an electron acceptor for two reductive steps carried out by Fe-S flavoenzymes. Using anaerobic in vitro reconstitution, we establish that a set of six enzymes is necessary and sufficient for the 8-step conversion of cholic acid to DCA. We then engineer the pathway into Clostridium sporogenes, conferring production of DCA and LCA on a non-producing commensal and demonstrating that a microbiome-derived pathway can be expressed and controlled heterologously. These data establish a complete pathway to two central components of the bile acid pool.
The human gut microbiota harbor hundreds of pathways, most of which are encoded by genes that have not yet been identified5–8. Their small molecule products are of interest for three reasons: (i) Most derive predominantly or exclusively from the microbiota (i.e., there is no host source), and many enter the circulation, where they can have effects on peripheral tissues and organ systems. (ii) Their concentrations are similar to or exceed that of a typical drug; for example, indoxyl sulfate can accumulate in the human host at 130 mg/day9. Moreover, their concentration ranges are large, typically >10-fold10, which could help explain microbiome-mediated biological differences among people. (iii) Of the few high-abundance molecules whose biological functions are well understood, most are ligands for a key host receptor; for example, short-chain fatty acids modulate host immune function via GPR41/GPR4311–13. Thus, high-abundance, microbiota-derived molecules are responsible for a remarkably broad range of phenotypes conferred on the host by bacteria.
Among these pathways, 7α-dehydroxylation of the primary bile acids cholic acid (CA) and chenodeoxycholic acid (CDCA) is particularly notable because the organisms that carry it out are present at very low abundance—an estimated 1:106 in a typical gut community14—yet they fully process a pool of primary bile acids that is ~1 mM in concentration15. Therefore, the flux through this pathway must be very high in the small subset of cells in which it operates, and the low-abundance organisms in the microbiome that perform this transformation have an unusually large impact on the pool of metabolites that enters the host. This pathway’s products—deoxycholic acid (DCA) and lithocholic acid (LCA)—are the most abundant secondary bile acids in humans (up to 450–700 μM in cecal contents)16, and are known to be important in three biological contexts: prevention of Clostridium difficile outgrowth1, induction of hepatocellular carcinogenesis2, and modulation of the host metabolic and immune responses17–19. More broadly, DCA, LCA, and their derivatives are a major component of the recirculating bile acid pool, representing >90% of the pool in the intestine and >25% in the gallbladder15. These microbiome-derived bile acids are therefore central to understanding the efficacy of therapeutics that target the bile acid pool and are approved or in clinical trials for primary biliary cholangitis and nonalcoholic steatohepatitis4.
In pioneering work, Hylemon and coworkers showed that the gut bacterium Clostridium scindens VPI 12708 carries out the 7α-dehydroxylation of CA to produce DCA20. CA serves as an inducer of 7α-dehydroxylation, leading to the discovery of a bile-acid-induced operon (termed bai) containing eight genes (Fig. 1 and Extended Data Fig. 1)21. Through heterologous expression and characterization of individual bai gene products, enzymes have been attributed to each step of the oxidative arm of the pathway22–27, but the reductive arm of the pathway remains poorly characterized28. A complete understanding of the pathway would enable efforts to control the composition of the bile acid pool by engineering the microbiome.
Fig. 1. Schematic of the bai operon and 7α-dehydroxylation.
(A) The bai operon consists of eight genes: seven encode enzymes and the eighth, baiG, encodes a transporter. It is conserved in every bacterial species known to 7α-dehydroxylate primary bile acids, and its gene products have been linked to specific steps in the pathway. (B) A simplified schematic showing the dehydroxylation of CA to DCA.
Here, by purifying and assaying pathway enzymes under anaerobic conditions, we reconstituted 7α-dehydroxylation in vitro. We then transferred the pathway from its genetically intractable producer Clostridium scindens into Clostridium sporogenes, conferring production of DCA and LCA on a non-producing commensal bacterial species. These data establish a complete pathway for two central components of the bile acid pool, and they provide a genetic basis for controlling the bile acid output of the microbiome.
Reconstitution of bile acid 7α-dehydroxylation
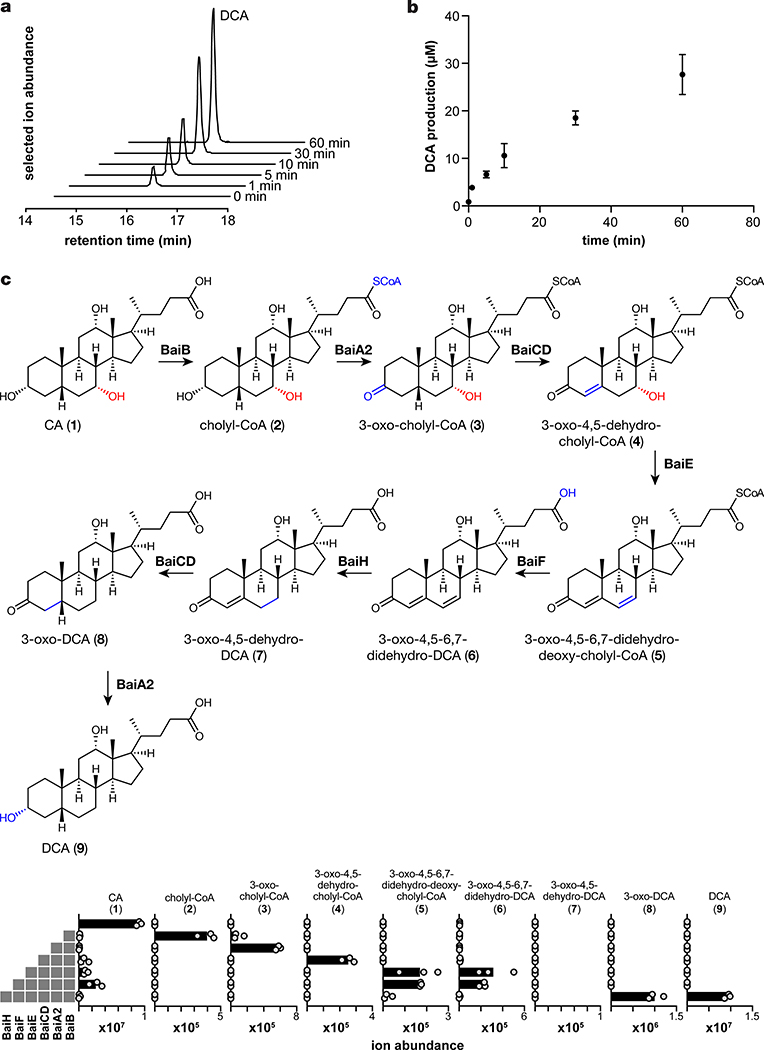
We first set out to de-orphan the remaining steps in the 7α-dehydroxylation pathway. Since previous studies of the bai enzymes involved expressing them individually in E. coli, we reasoned that an alternative approach in which enzymes were purified, mixed, and assayed in vitro could help delineate the set of enzymes necessary and sufficient for 7α-dehydroxylation. Given that the eight-gene bai operon is shared among all known 7α-dehydroxylating strains, we focused our efforts on the enzymes encoded by the operon. We cloned three orthologs of each enzyme, expressed them individually in E. coli under micro-aerobic conditions, and purified them anaerobically as N-terminal His6 fusions. Using this strategy, we obtained at least one soluble, purified ortholog of each Bai enzyme (Extended Data Fig. 2). When we incubated a mixture of the purified Bai enzymes with CA, NAD+, coenzyme A, and ATP under anaerobic conditions and monitored the reaction by LC-MS, we observed the time-dependent conversion of CA to DCA, indicating that the combination of BaiB, BaiCD, BaiA2, BaiE, BaiF, and BaiH is sufficient for 7α-dehydroxylation; no additional enzymes are required (Fig. 2a, b).
Fig. 2. Establishing the complete 7α-dehydroxylation pathway in vitro.
(A) EICs showing time-dependent production of DCA by six purified Bai enzymes. BaiB, BaiCD, BaiA2, BaiE, BaiF, and BaiH were purified and assayed anaerobically in the presence of NAD+, CoA, and ATP. Reactions were initiated by the addition of CA, and aliquots were analyzed by LC-MS at the indicated timepoints. The experiment was repeated twice independently with similar results. (B) Time course of DCA production by a mixture of BaiB, BaiCD, BaiA2, BaiE, BaiF, and BaiH. Data points indicate the mean level of DCA ± SD (three biological replicates). (C) Complete proposed pathway for the 7α-dehydroxylation of CA to DCA. LC-MS ion abundance for DCA and pathway intermediates produced by a step-wise reconstitution assay in which the indicated enzymes were co-incubated as described in (A). Bars indicate mean of three independent biological replicates.
To test our hypotheses regarding the order of steps in the pathway, we performed stepwise reconstitutions in which enzymes were added one at a time and intermediates were allowed to build up at each step in the pathway (Fig. 2c). From these data, we draw two conclusions: First, the six enzymes used in the reconstitution are not just sufficient but also necessary, and the pathway proceeds according to the scheme shown in Fig. 4. We directly observed mass ions consistent with each of the proposed intermediates, providing direct evidence for the previously proposed portion of the biosynthetic route. See Supplementary Table 1 and Extended Data Fig. 3 for data supporting our provisional structural assignments; two important limitations are that we do not have authentic standards for all intermediates, and the ability to distinguish bile acid isomers by LC-MS can be limited. In spite of its conservation in all known dehydroxylating species, BaiI is dispensable for CA dehydroxylation in vitro. Since BaiI is a predicted Δ5-ketosteroid isomerase, it may process a substrate other than CA, likely one with a 4,5- or 5,6-olefin.
Fig. 4. In vivo activity of the 7α-dehydroxylation pathway.
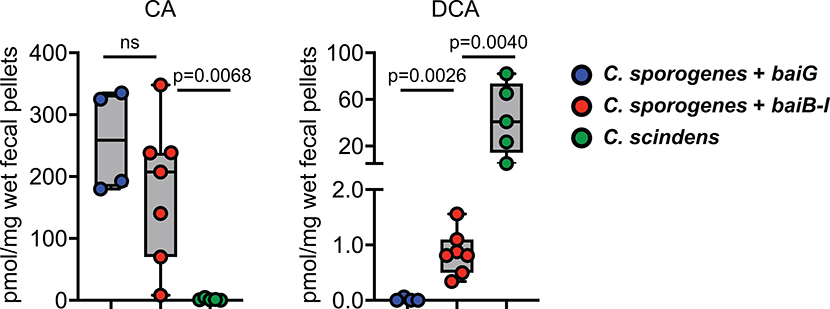
Germ-free mice were mono-colonized with C. sporogenes harboring the transporter baiG, C. sporogenes harboring baiB-I, MF001, or the native 7α-dehydroxylating strain C. scindens. Fecal pellets were obtained on day 6 and analyzed by LC-MS to determine the quantity of the host-derived substrate CA and the pathway product DCA. P values were determined by a two-tailed t-test; ns = not significant. Box and whisker plots show median values, 25th–75th percentile, and range for n = 4–7 independent biological replicates.
Second, to our surprise, the absence of BaiH caused the pathway to stall at the highly oxidized intermediate 3-oxo-4,5–6,7-didehydro-DCA, and its addition resulted in two successive 2e- reductions to form 3-oxo-DCA. BaiH had previously been proposed to oxidize an alternative substrate, 3-oxo-4,5-dehydro-UDCA25, so a potential role in the reductive arm of the pathway was unexpected. To explore this finding further, we incubated purified BaiH with synthetic 3-oxo-4,5–6,7-didehydro-DCA; we observed that the enzyme catalyzes a 2e- reduction to 3-oxo-4,5-dehydro-DCA, but does not reduce this intermediate further (Fig. 3e). Notably, 3-oxo-4,5-dehydro-DCA does not build up in the reconstitution reaction containing BaiH, suggesting that another enzyme present in the mixture catalyzes the second reductive step. Hypothesizing that the BaiH homolog BaiCD catalyzes the second reductive step, we incubated it with synthetic 3-oxo-4,5-dehydro-DCA, revealing that it reduces this substrate to 3-oxo-DCA (Extended Data Fig. 4). Together, these data show that the pathway employs an unusual redox strategy in which the A and B rings of the steroid core are converted into a highly oxidized intermediate, 3-oxo-4,5–6,7-didehydro-DCA; and that the two key reductive steps are catalyzed by two homologous enzymes in the Fe-S flavoenzyme superfamily, BaiH and BaiCD.
Fig. 3. Transferring the 7α-dehydroxylation pathway in Clostridium sporogenes.
(A) The bai operon was divided among three plasmids: baiB-baiF in pMTL83153 (pMF01), baiG in pMTL83353 (pMF02), and baiH-baiI in pMTL83253 (pMF03). pMF01, pMF02, and pMF03 were conjugated successively into Clostridium sporogenes ATCC 15579 to create MF001. (B) Combined extracted ion chromatograms (EICs) showing the conversion of CA to DCA by MF001 versus a control strain of C. sporogenes harboring the transporter baiG (MF012). The strains were grown with 1 μM CA for 72 hr, extracted with acetone, and analyzed by LC-MS. The asterisk indicates isoDCA. The experiment was repeated three times independently with similar results. (C) Combined EICs showing time-dependent conversion of CA to DCA by MF001. The strain was grown with 1 μM CA and aliquots from the indicated timepoints were analyzed as described in (B). The experiment was repeated twice independently with similar results. (D) LC-MS ion abundances are shown for DCA, pathway intermediates, and derivatives produced by C. sporogenes strains with single gene deletions within the bai operon. Bars indicate mean of three independent biological replicates. (E) Combined EICs showing the conversion of 3-oxo-4,5–6,7-didehydro-DCA to 3-oxo-4,5-dehydro-DCA by C. sporogenes + baiG/baiH (left), and the conversion of 3-oxo-4,5-dehydro-DCA to 3-oxo-DCA by C. sporogenes + baiG/baiCD (right). Each strain was cultivated with synthetic 3-oxo-4,5,6,7-didehydro-DCA (left) or 3-oxo-4,5-dehydro-DCA (right) for 72 hr and culture extracts were analyzed as in (B). The portion of the traces in the gray box has been scaled up 10x to make it easier to visualize the peaks corresponding to 3-oxo-DCA and DCA. The experiment was repeated twice independently with similar results.
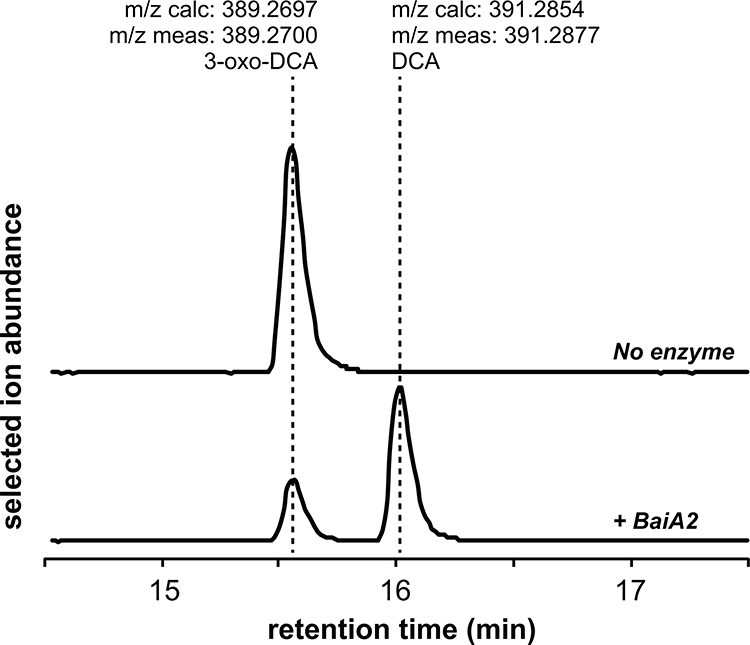
Finally, the last step in the pathway—reduction of 3-oxo-DCA to DCA—is carried out by BaiA2, as confirmed by assaying purified BaiA2 alone (Extended Data Fig. 4). Thus, BaiA2 and BaiCD both act twice in the pathway, catalyzing its first two and last two redox steps.
Engineering the pathway into C. sporogenes
Having determined the set of enzymes that are necessary and sufficient for the pathway, we sought to gain genetic control over the pathway as a first step toward engineering the bile acid output of the gut community. We began by attempting to construct a mutation in the baiCD gene of the native producer, C. scindens, using the ClosTron group II intron system; however, we were unsuccessful due to an inability to introduce DNA constructs into C. scindens by conjugation. As an alternative approach, we considered expressing the bai pathway in a gut commensal that is unable to carry out 7α-dehydroxylation; however, with notable exceptions29–32, methods for transferring pathways in Clostridium are underdeveloped. To our knowledge, no pathway from the human microbiome has been mobilized from one Clostridium species to another.
We selected Clostridium sporogenes ATCC 15579 as the recipient for two reasons: it is related to C. scindens, making it likely that ancillary metabolic requirements for the pathway (e.g., cofactor biogenesis) would be met; and genetic tools have been developed that enable plasmids to be transformed into C. sporogenes33. Our initial attempts to clone the entire 8-gene bai operon (baiB-baiI) into an E. coli-C. sporogenes shuttle vector failed to yield clones harboring the complete operon. Reasoning that there might be a gene in the cluster that is toxic to E. coli, we cloned various fragments of the cluster under the control of different promoters (detailed in Supplementary Table 2), eventually managing to split the cluster into three pieces, each in its own E. coli-C. sporogenes shuttle vector: baiB-baiF in pMTL83153 (pMF01), baiG in pMTL83353 (pMF02), and baiH-baiI in pMTL83253 (pMF03) (Fig. 3a, Extended Data Fig. 5). Genes in pMF01 and pMF03 were placed under the control of the spoIIE promoter from C. sporogenes ATCC 15579, which is expressed during the late stages of Clostridium growth34, while baiG in pMF02 was driven by the strong fdx promoter. We conjugated these plasmids sequentially into C. sporogenes to yield strain MF001.
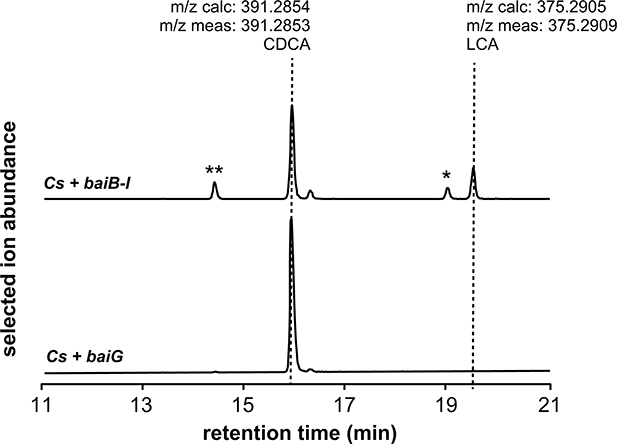
When incubated with CA, MF001 produces DCA in a time-dependent manner, in contrast to a control strain that harbors only the transporter (baiG) (Fig. 3b, c), which does not. Additionally, MF001 converts CDCA to LCA (Extended Data Fig. 7). These data show that the eight genes in the core bai cluster (Fig. 1) are sufficient to confer bile acid 7α-dehydroxylation on C. sporogenes, although they do not rule out the participation of one or more genes endogenous to C. sporogenes.
Identifying branch points in the pathway
To uncover potential branch points for engineering the biosynthesis of non-native pathway products, we constructed a set of strains in which each of the eight genes were individually deleted (Extended Data Fig. 6). We grew these strains with CA and assayed their culture supernatant for the build-up of intermediates (Fig. 3d). Deletion of genes in the oxidative arm of the pathway resulted in the buildup of early pathway intermediates, as expected. Two exceptions were the baiE mutant, which produced only cholyl-CoA; and the baiF-deficient strain, which generated a small quantity of the final product DCA, suggesting there might be a compensatory CoA hydrolase or that non-enzymatic hydrolysis of the CoA thioester happens to some extent in vivo.
Intriguingly, the baiH mutant accumulates a key intermediate in the reductive arm of the pathway, 3-oxo-4,5–6,7-didehydro-DCA (Fig. 3d), supporting our finding that BaiH catalyzes the first reductive step in the pathway. Moreover, strains of C. sporogenes expressing BaiG/BaiH and BaiG/BaiCD convert, respectively, 3-oxo-4,5–6,7-didehydro-DCA to 3-oxo-4,5-dehydro-DCA and 3-oxo-4,5-dehydro-DCA to 3-oxo-DCA (Fig. 3e), providing access to intermediates that do not accumulate in a culture of C. scindens. Notably, the fully oxidized and partially reduced intermediates are branch points for the production of allo (5α) bile acids, which have important biological activities including the induction of regulatory T cells35. Thus, gaining genetic control over the pathway by expressing it in an alternative gut provides opportunities for rational and deliberate control of bile acid metabolism and the production of alternative molecules with distinct biological properties.
Colonizing mice with engineered C. sporogenes
Finally, we colonized germ-free mice with MF001 to see whether it would confer the production of pathway products on the host. We included two other experimental groups: As a negative control, germ-free mice mono-colonized by C. sporogenes + baiG (the bile acid transporter); and as a positive control, germ-free mice mono-colonized by wild-type Clostridium scindens, a native bai operon-containing (7α-dehydroxylating) strain. As shown in Fig. 4, the engineered C. sporogenes + baiB-I strain conferred production of DCA on the host. The level of production was substantially lower than that observed from C. scindens; we suspect this is because of the need to include additional genes, not yet known, that couple the pathway to a pool of reduced cofactor from membrane-bound components of the electron transport chain, increasing flux. Nonetheless, our data provide an important starting point for subsequent efforts to study and engineer the bile acid pool.
Engineering pathways from the microbiome
These results establish the complete bile acid 7α-dehydroxylation pathway, bringing this pathway closer to the level of knowledge we have about endogenous human metablic pathways. Key features of the pathway might serve as a model for other pathways that produce high-abundance metabolites in the gut (see SI Discussion and Extended Data Fig. 8).
The gut microbiome harbors hundreds of pathways, many of which likely modulate host biology, but to date only a few of these pathways have been a target of engineering39,40. This stands in contrast to natural product pathways from terrestrial and marine microorganisms and plants, which are commonly expressed in heterologous hosts41,42 and engineered to generate non-native products43. Two technology gaps need to be overcome in order to make microbiome-derived pathways amenable to engineering: (i) efficient strategies for identifying pathways for known metabolites and small molecule products of orphan gene clusters, and (ii) tools for transferring pathways into bacterial hosts native to the gut and manipulating them to produce novel molecules. The work described here is a starting point for these efforts. If it can be generalized to other species of Clostridia, it could lead to a set of tools for deorphaning, heterologously expressing, and engineering pathways from the microbiome.
METHODS
Bacterial strains, culture conditions, and bile acids
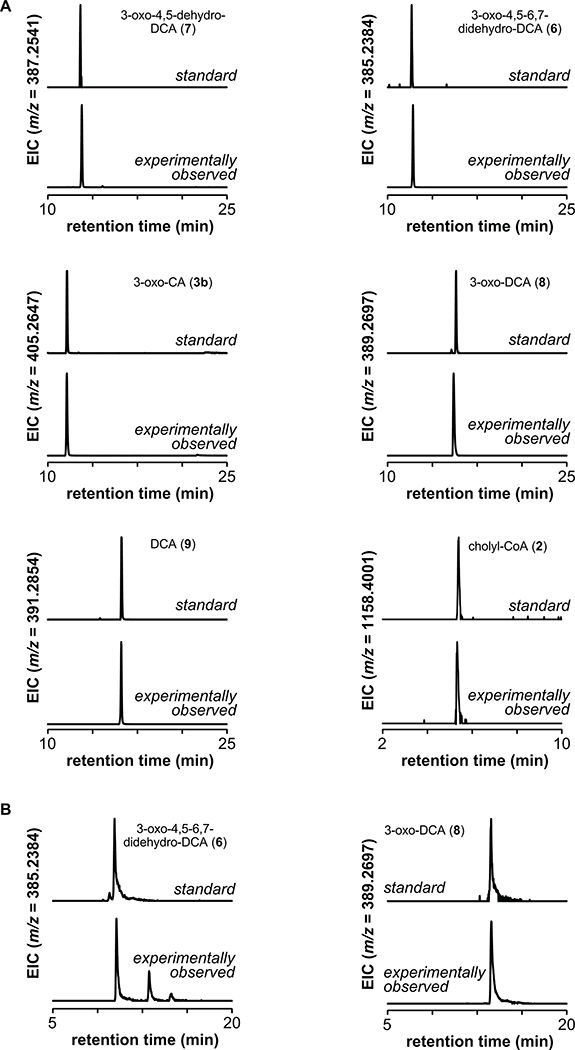
Clostridium scindens VPI 12708 and Clostridium sporogenes ATCC 15579 were obtained from the Japan Collection of Microorganisms (JCM) and the American Type Culture Collection (ATCC), respectively. Engineered C. sporogenes strains used in this study are shown in Supplementary Table 3. They were cultured in TYG (3% w/v tryptone, 2% w/v yeast extract, 0.1% w/v sodium thioglycolate) broth at 37 ˚C in an anaerobic chamber from Coy Laboratories. Escherichia coli CA434 (HB101/pRK24) was cultured at 37 ˚C in LB broth supplemented with 12 μg/mL tetracycline and 100 μg/mL carbenicillin. In addition, 20 μg/mL chloramphenicol, 100 μg/mL spectinomycin or 250 μg/mL erythromycin was used for the selection of series of plasmids of pMTL83153, pMTL83353 or pMTL83253 respectively. Plasmids used in this study are shown in Supplementary Table 2. Cholic acid (1), chenodeoxycholic acid, deoxycholic acid (9) and lithocholic acid were purchased from Sigma-Aldrich. 3-oxo-cholic acid (3b) and 3-oxo-deoxycholic acid (8) were purchased from Steraloids. 3-oxo-4,5–6,7-didehydro-DCA (6) and 3-oxo-4,5-dehydro-DCA (7) were synthesized using previously reported procedures44. Structural assignments for the remaining pathway intermediates and derivatives shown in Fig. 2 and Fig. 3 are provisional, and were made on the basis of mass spectra, retention times, and comparison to chemically related standards.
Cloning of the bai operon
All PCR amplification was conducted using PrimeSTAR Max DNA polymerase (Takara Bio) according to the manufacturer’s instructions. Sequences of primers for target genes and cloning vectors were shown in Supplementary Table 4. For the heterologous expression of bai genes under fdx promoter, pMTL vectors were amplified with primers 1 and 2. For the expression of bai genes under spoIIE promoter, pMTL vectors harboring spoIIE promoter was constructed at first. pMTL vectors were amplified with primers 1 and 3 to remove the fdx promoter and spoIIE promoter region, which is the 277 bp sequence upstream of CLOSPO_01065, was amplified with primers 4 and 5. Then these two PCR fragments were assembled by overlap PCR. The target gene sequences were amplified with the primers pair shown in Supplementary Table 4. PCR fragments were assembled with the amplified fragments of vectors using Gibson assembly kit (New England Bio Labs). E. coli Stbl4 competent cells (Invitrogen) were transformed with the assembled plasmids by electroporation and transformants were confirmed by PCR. Positive clones harboring assembled plasmids were cultivated, and the plasmid was obtained by miniprep and verified by sequencing.
Heterologous expression in C. sporogenes
Bacterial cultures were incubated in a Coy anaerobic chamber under an atmosphere consisting of 10% CO2, 5% H2, 85% N2. Growth media were pre-reduced by overnight pre-incubation in the anaerobic chamber. For the heterologous expression experiments, plasmids were transferred into C. sporogenes by conjugation using E. coli CA434. E. coli CA434 was electroporated with the individual plasmids and recovered overnight in selective media. 1 mL of overnight culture of the resultant transformants was harvested. The cell pellet was washed with PBS to remove residual antibiotics and re-suspended with 200 μL of an overnight culture of C. sporogenes in anaerobic chamber. Eight drops of 25 μL of the suspension were pipetted on TYG agar plate without antibiotics and the plate was incubated anaerobically at 37 ˚C for 2 days. The bacterial biomass was scraped up and resuspended in 300 μL of PBS. The whole cell suspension was then plated on TYG agar plates supplemented with 250 μg/mL d-cycloserine and appropriate antibiotics (15 μg/mL thiamphenicol for pMTL83153, 500 μg/mL spectinomycin for pMTL83353 or 5 μg/mL erythromycin for pMTL83253). After a few days, antibiotic resistant colonies were picked and re-streaked on agar containing the same antibiotic. The resulting clones were confirmed by PCR amplification using appropriate primers (Supplementary Table 4). Multiple plasmids were introduced sequentially, using the same procedure.
Extraction of metabolites
Engineered strains were cultured anaerobically in TYG medium supplemented with appropriate antibiotics from frozen glycerol stocks. 10 μL of the overnight culture was inoculated in 1 mL of TYG medium supplemented with appropriate antibiotics and 1 μM substrate. After 72 hr, unless otherwise noted, the culture was extracted with 20% acetone and centrifuged. The supernatant was analyzed by LC/MS.
LC/MS analysis of metabolite extracts
Metabolite extracts were analyzed using an Agilent 1290 LC system coupled to an Agilent 6530 QTOF with a 1.7 μm, 2.1 × 100 mm Kinetex C18 column (Phenomenex). Water with 0.05% formic acid (A) and acetone with 0.05% formic acid (B) was used as the mobile phase at a flow rate of 0.35 mL/min over a 32 min gradient: 0–1 min, 25% B; 1–25 min, 25–75% B; 25–26 min, 75–100% B; 26–30 min, 100% B; 30–32 min 75–25% B. All data were collected in negative ion mode.
For detection of CoA conjugates and flavin cofactors, a 1.8 μm, 2.1 × 50 mm ZORBAX SB-C18 column (Agilent Technologies) and water with 10 mM ammonium acetate pH 9.0 (A) and acetonitrile (B) was used. A flow rate of 0.3 mL/min was used over the 17 min gradient: 0–2 min, 15% B; 2–14 min, 15–50% B; 14–14.1 min 50–95% B, 14.1–17 min, 85% B. All data were collected in positive ion mode.
Cloning of bai operon genes
To increase the probability of assembling a complete bai operon, we cloned the genes encoding baiB, baiA2, baiCD, baiE, baiF, and baiH from Clostridium scindens VPI12708, Clostridium hylemonae, and Clostridium hiranonis using the primers in Supplementary Table 5 and the KOD Xtreme™ Hot Start PCR kit (Millipore) following the manufacturer’s protocol. Each PCR-amplified gene contains ligation independent cloning (LIC) sites that are complimentary to the pSGC vector. PCR products were purified with the Agencourt Ampure XP PCR clean-up kit (Beckman Coulter) according to the manufacturer’s protocol. The pSGC vector was prepared for LIC by linearization with the restriction enzyme BsaI. LIC sites were installed by adding T4 DNA polymerase (NEB) to 10 μg of linearized plasmid in a 50 μL reaction containing 2.5 mM GTP, 1x NEB Buffer 2, and 1x BSA for 1 hr at 22 °C. T4 DNA polymerase was heat-inactivated by incubation at 75 °C for 20 min. PCR products (in a volume of 2 μL) were treated with T4 DNA polymerase in a 10 μL reaction containing 2.5 mM CTP, 1x NEB Buffer 2, and 1x BSA for 1 hr at 22 °C. T4 DNA polymerase was heat inactivated by incubation at 75 °C for 20 min . The LIC reaction was assembled by mixing 15 ng of digested vector DNA with ~40 ng of digested PCR product; the reaction mixture was then incubated at 22 °C for 10 min. A 30 μL aliquot of DH10B cells (NEB) were transformed with 2 μL of the LIC reaction mixture using standard bacterial transformation protocols. This cloning procedure adds a His6 tag to the N-terminus of each protein with the following sequence: MHHHHHHSSGVDLGTENLYFQS. All final constructs were sequence verified (Genescript).
Expression and purification of BaiH and BaiCD
BL-21(DE3) cells containing the pPH151 plasmid were transformed with the pSGC plasmid containing either BaiCD or BaiH. The transformants were selected on an LB/agar plate containing 50 μg/mL kanamycin and 34 μg/mL chloramphenicol. A single colony was used to inoculate 20 mL of LB overnight culture containing the above antibiotics. The overnight culture was used to inoculate 2 L of Studier’s auto induction media (ZYP-5052 supplemented with 1 mM flavin mononucleotide and 200 μM FeCl3) housed in a 2 L PYREX® media bottle. Cultures were grown with constant aeration using a sparging stone attached to a pressurized, 0.22 μm filtered air source all in a water bath maintained at 37 °C. After 5 hr, aeration was stopped and the culture was placed in an ice bath for 1 hr. The culture was returned to a 22 °C water bath and light aeration was resumed. After 5 min, cysteine was added to a final concentration of 600 μM. The culture was grown at 22 °C for ~ 20 hr before being harvested by centrifugation at 10,000 × g. Cell pellets were flash frozen and stored in liquid N2 until purification. All subsequent steps were carried out in an MBraun anaerobic chamber maintained at < 0.1 ppm oxygen (MBraun, Stratham, NH). Plastics were brought into the chamber and allowed to sit for two weeks before use. All solvents and buffer stocks were degassed by sparging with argon gas for 4 hr before being taken into the chamber. In a typical purification, ~30 grams of BaiCD or BaiH cell paste was resuspended in 30 mL of lysis buffer containing 50 mM HEPES, pH 7.5, 300 mM KCl, 4 mM imidazole, 10 mM 2-mercaptoethanol (BME), 10% glycerol, 1 mM FMN, 1 mM FAD, and 1% Triton-X305. The resuspension was subjected to 50 rounds of sonic disruption (80% output, 3 s pulse on, 12 s pulse of) at 4 °C. The lysate was cleared by centrifugation at 4 °C for 1 hr at 15,000 × g. The supernatant was loaded with an ÄKTA express FPLC system onto a 5 mL fast-flow HisTrap™ column (GE Healthcare Life Sciences) equilibrated in lysis buffer lacking FMA, FAD, and Triton-X305. The column was washed with 10 column volumes of lysis buffer before elution with 5 mL of buffer containing 50 mM HEPES, pH 7.5, 300 mM KCl, 300 mM imidazole, 10 mM BME, and 10% glycerol. The fractions containing protein, based on absorbance at 280 nm, were pooled and reconstituted with Fe and sulfur as previously described. The reconstituted proteins were then passed over a HiPrep 16/60 Sephacryl S-200 HR column equilibrated in 20 mM HEPES, pH 7.5, 300 mM KCl, 5 mM DTT, and 10% glycerol. The proteins were concentrated to ~ 1 mL with a vivaspin 20 concentrator (Sartorius Stedium Biotech). The protein concentration was estimated by A280 using the extinction coefficient calculated based on its corresponding amino acid sequence. The presence of FAD and FMN were confirmed by precipitating purified enzyme with sulfuric acid and analyzing the resulting supernatant by LC-MS, comparing analytes to authentic standards of FAD and FMN.
Expression and purification of BaiB, BaiA2, BaiE, and BaiF
BL-21(DE3) cells containing the pRIL plasmid were transformed with the plasmid containing BaiB, BaiA2, BaiE, or BaiF. Each transformant was selected on an LB/agar plate containing 50 μg/mL kanamycin and 34 μg/mL chloramphenicol. A single colony was used to inoculate 20 mL of LB overnight culture containing the above antibiotics. The overnight culture was used to inoculate 2 L of Studier’s auto induction media (ZYP-5052) housed in a 2 L PYREX® media bottle. Cultures were grown with constant aeration using a sparging stone attached to a pressurized, 0.22 μm filtered air source in a water bath at 37 °C. After 5 hr, aeration was stopped and the culture was placed in an ice bath for 1 hr. The culture was returned to a 22 °C water bath and light aeration was resumed. The culture was grown at 22 °C for ~ 20 hr before being harvested by centrifugation at 10,000 × g. Cell pellets were flash frozen and stored in liquid N2 until purification. All subsequent steps were carried out in an MBraun anaerobic chamber maintained at < 0.1 ppm oxygen as above with minor modifications. Briefly, a typical purification, ~ 30 – 40 grams of cell paste was resuspended in 30 – 40 mL of lysis buffer containing 50 mM HEPES, pH 7.5, 300 mM KCl, 4 mM imidazole, 10 mM 2-mercaptoethanol (BME), 10% glycerol, and 1% Triton-X305. The resuspension was subjected to 50 rounds of sonic disruption (80% output, 3 s pulse on, 12 s pulse of) at 4 °C. The lysate was cleared by centrifugation at 4 °C for 1 hr at 15,000 × g. The supernatant was loaded with an ÄKTA express FPLC system onto a 5 mL fast-flow HisTrap™ column (GE Healthcare Life Sciences) equilibrated in lysis buffer lacking Triton-X305. The column was washed with 10 column volumes of lysis buffer before elution with 5 mL of buffer containing 50 mM HEPES, pH 7.5, 300 mM KCl, 300 mM imidazole, 10 mM BME, and 10% glycerol. The fractions containing protein, based on absorbance at 280 nm, were pooled and immediately passed over a HiPrep 16/60 Sephacryl S-200 HR column equilibrated in 20 mM HEPES, pH 7.5, 300 mM KCl, 5 mM DTT, and 10% glycerol. The proteins were concentrated to ~ 1 mL with a vivaspin 20 concentrator (Sartorius Stedium Biotech). The protein concentration was estimated by A280 using the extinction coefficient calculated based on its corresponding amino acid sequence.
Bile acid pathway in vitro reconstitution
Six assays each contained 50 mM HEPES pH 7.5, 50 mM KCl, 200 μM NAD, 100 μM CoA, and 200 μM ATP. In addition, each assay contained 0.1 mM of 1–6 of the following enzymes: BaiB from C. scindens, BaiA2 from C. scindens, BaiCD from C. hiranonis, BaiE from C. hiranonis, BaiF from C. hylemonae, and BaiH from C. scindens. All reactions were initiated with the addition of cholic acid and incubated at 22 °C for 30 min before being quenched by the additions of an equal volume of 100 % acetone. Each assay was performed in triplicate. Product formation was monitored by LC/MS described above.
Bile acid pathway in vitro reconstitution kinetics
To determine the rate of DCA production by the in vitro pathway, assays were performed with 50 mM HEPES pH 7.5, 50 mM KCl, 200 μM NAD, 100 μM CoA, and 200 μM ATP, 0.1 mM of BaiB from C. scindens, BaiA2 from C. scindens, BaiCD from C. hiranonis, BaiE from C. hiranonis, BaiF from C. hylemonae, and BaiH from C. scindens. Reactions were initiated with the addition of cholic acid and incubated at 22 °C. Samples of the reaction were removed and mixed with an equal volume (100 mM H2SO4 at designated times. Each assay was performed in triplicate. Product formation was monitored by LC/MS described above.
KM assay for BaiCD
Kinetic parameters for BaiCD from C. hiranonis were determined in assays that contained 0.45 μM enzyme, 50 mM HEPES pH 7.5, 50 mM KCl, and 500 μM NADH. Reactions mixtures were incubated for 5 min at 22 °C before being initiated with 3-oxo-4,5-dehydro-deoxycholic acid. Concentrations of substrate were varied between 3.91 μM and 500 μM. 20 μL samples were removed and mixed with an equal volume of 100 mM H2SO4 to stop the reaction. Product formation was determined by LC/MS described above. Reactions were performed in triplicate and the data were fit to the Michaelis-Menten equation by the least squares method.
KM assay for BaiH
Kinetic parameters for BaiH from C. scindens were determined in assays that contained 0.45 μM enzyme, 50 mM HEPES pH 7.5, 50 mM KCl, and 500 μM NADH. Reactions mixtures were incubated for 5 min at 22 °C before being initiated with 3-oxo-4,5,6,7-didehydro-deoxycholic acid. Concentrations of substrate were varied between 0.78 μM and 100 μM. 20 μL samples were removed and mixed with an equal volume 100 mM H2SO4 to stop the reaction. Product formation was determined by LC/MS described above. Reactions were performed in triplicate and the data were fit to the Michaelis-Menten equation by the least squares method.
Colonization of germ-free mice with engineered Clostridium strains
Engineered C. sporogenes strains or C. scindens ATCC 15579 were cultured anaerobically in TYG medium (supplemented with appropriate antibiotics) or BHI medium from frozen glycerol stocks. 50 μL of the overnight culture was used to inoculate 5 mL of fresh growth medium containing 50 μM CA. After 48 hr, bacterial cells were pelleted by centrifugation, washed twice with PBS, resuspended in 25% glycerol solution, and stored at −80 °C. Germ free C57BL/6 mice (male, ~8 weeks of age, n = 6 or 7 per group) were obtained from Taconic Biosciences (Hudson, NY) and colonies were maintained in gnotobiotic isolators in accordance with A-PLAC, the Stanford IACUC. The mice were maintained on a Teklad Custom Diet (TD.180755, w/0.5% Cholic Acid). Mice were gavaged 1x daily for three successive days using the following procedure: Glycerol stocks of engineered strains of C. sporogenes or C. scindens ATCC 15579 were thawed to room temperature and mice were inoculated by oral gavage with ~200 μl of the thawed glycerol stock (~1 × 107 CFU). Six days post colonization, fecal pellets were collected and then mice were euthanized humanely by CO2 asphyxiation. Cecal contents were also collected, snap-frozen in liquid nitrogen, and stored at −80 °C.
Murine fecal samples (~30 mg) or intestinal contents (~100 mg) were pre-weighed into a 2 ml screw top tube containing six 6 mm ceramic beads (Precellys® CK28 Lysing Kit). 300 μL or 1 ml of a mixture of ice-cold acetonitrile, methanol, and water (4/4/2, v/v/v) was then added to each tube, and samples were homogenized by vigorous shaking using a QIAGEN Tissue Lyser II at 25/s for 10 min. The resulting homogenates were centrifuged for 15 min at 14,000 g at 4 °C. 100 μl of the supernatant was then combined with 100 μL of an internal standard solution (2 μM d4-cholic acid in H2O). The resulting mixtures were filtered through a Durapore PVDF 0.22 μm membrane using Ultrafree centrifugal filters (Millipore, UFC30GV00) and 5 μL was analyzed by LC-MS as described in LC/MS analysis of metabolite extracts.
Extended Data
Extended Data Fig. 1. Previously proposed pathway for 7α-dehydroxylation of CA in C. scindens VPI 12708.
See main text for details and a summary of the previous literature.
Extended Data Fig. 2. Purification of recombinant Bai proteins.
(A) SDS-PAGE analysis of purified Bai proteins after Ni-affinity and size exclusion purification visualized by Coomassie Brilliant Blue staining. The image was generated using a Bio-Rad Gel Doc Universal Hood II Molecular Imager. MWM, molecular weight marker; 1, BaiB from C. scindens; 2, BaiB from C. hylemonae; 3, BaiCD from C. hiranonis; 4, BaiE from C. scindens; 5, BaiE from C. hiranonis; 6, BaiA2 from C. scindens; 7, BaiF from C. hylemonae; 8, BaiH from C. scindens; 9, BaiI from C. scindens; 10, BaiI from C. hiranonis. B) UV-visible spectra of BaiCD from C. hiranonis (24 μM, left) and BaiH from C. scindens (13 μM, right). Features at 370 nm and 450 nm are indicative of flavin bound to BaiCD and BaiH, which are partially obscured by the presence of a [4Fe-4S] cluster, which have broad absorbance between 300 nm and 700 nm. C) The presence of FMN and FAD were confirmed by mass spectrometry. Experiments in A-C were reported twice independently with similar results.
Extended Data Fig. 3. Bile acid standards.
(A) For each compound in the study for which we have an authentic standard, we show an extracted ion chromatogram of the authentic standard and the experimentally observed compound. Since the data shown here were collected from samples run at different times, retention time drift may be responsible for the peak pairs that do not have identical retention times. (B) We observed a retention time drift in the LC-MS data collected for the experiment shown in Figure 2C. For two representative compounds from that data set, we show an extracted ion chromatogram of the experimentally observed compound and an authentic standard run contemporaneously, showing that the retention times remain consistent with our peak assignments.
Extended Data Fig. 4. Kinetic parameters for BaiCD and BaiH.
(A) Michaelis-Menten analysis of the conversion of 3-oxo-4,5-dehydro-DCA to 3-oxo-DCA by BaiCD. Reaction mixtures contained 0.45 μM BaiCD, 1 mM NADH, with the substrate concentration varied between 15 μM and 500 μM. B) Michaelis-Menten analysis of the conversion of 3-oxo-4,5,6,7-didehydro-DCA to 3-oxo-4,5-dehydro-DCA by BaiH. Reaction mixtures contained 0.45 μM BaiH, 1 mM NADH, with the substrate concentration varied between 3 μM and 100 μM. Data indicate the average product level ± one SD (three biological replicates).
Extended Data Fig. 5. Biochemical analysis of 3-oxo-DCA reduction by BaiA2.
Combined extracted ion chromatograms showing the conversion of 3-oxo-DCA to DCA by recombinant BaiA2. This experiment was performed once.
Extended Data Fig. 6. Constructs for expressing the bai operon and portions thereof in C. sporogenes.
Each of the plasmids has replication origins for E. coli and Clostridium, traJ to enjable conjugal plasmid transfer, and an antibiotic resistance gene. bai genes were introduced into these plasmids under the control of the fdx or spoIIE promoter. For the genetic analysis of baiCD and baiH function, pMTL83153-based plasmids were used.
Extended Data Fig. 7. 7α-dehydroxylation of CDCA in vivo.
Combined extracted ion chromatograms showing the conversion of CDCA to LCA by a C. sporogenes strain harboring the complete bai operon on three plasmids (MF001) versus a control strain of C. sporogenes harboring the transporter baiG (MF012). The strains were cultivated with 1 μM CA for 72 hr; an acetone extract of the culture supernatant was analyzed by HPLC-MS. The single asterisk indicates isoLCA, and the double asterisk is provisionally assigned as isoCDCA. This experiment was performed once.
Extended Data Fig. 8. Metabolic logic of the 7α-dehydroxylation pathway.
Highly oxidized metabolic intermediates as anaerobic electron acceptors. In the first half of the 7α-dehydroxylation pathway, two successive two-electron oxidations set up a vinylogous dehydration of the 7-hydroxyl, yielding the highly oxidized intermediate 3-oxo-4,5–6,7-didehydro-DCA. In the second half of the pathway, three successive two-electron reductions reduce this molecule to DCA, resulting in a net 2e- reduction. The first two of these reductions are carried out by Fe-S flavoenzymes, which harbor a suite of four cofactors that enable them to convert two-electron inputs to a one-electron manifold. A schematic of of the previously proposed pathway is shown in Extended Data Fig. 1A.
Supplementary Material
ACKNOWLEDGMENTS
We are deeply indebted to Christopher T. Walsh, Dylan Dodd, Colleen O’Loughlin, and members of the Fischbach and Almo laboratories for helpful comments on the manuscript. This work was supported by NIH grants DP1 DK113598 (M.A.F.), R01 DK110174 (M.A.F.), P01 GM118303-01 (S.C.A.), U54 GM093342 (S.C.A.), and U54 GM094662 (S.C.A.), the Chan-Zuckerberg Biohub (M.A.F.), an HHMI-Simons Faculty Scholars Award (M.A.F.), an Investigators in the Pathogenesis of Infectious Disease award from the Burroughs Wellcome Foundation (M.A.F.), and the Price Family Foundation (S.C.A.).
Footnotes
DATA AVAILABILITY STATEMENT
Mass spectrometry data that support the findings of this study have been deposited in MassIVE with the accession code MSV000085048. The source data for the animal experiments are available as Supplementary Table 1.
SUPPLEMENTARY INFORMATION
Supplementary Table 1 is available in the online version of the paper.
MAIN TEXT REFERENCES
- 1.Buffie CG et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoshimoto S et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 499, 97–101 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Duboc H, Taché Y & Hofmann AF The bile acid TGR5 membrane receptor: from basic research to clinical application. Dig. Liver Dis. 46, 302–312 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arab JP, Karpen SJ, Dawson PA, Arrese M & Trauner M Bile acids and nonalcoholic fatty liver disease: Molecular insights and therapeutic perspectives. Hepatology 65, 350–362 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicholson JK et al. Host-gut microbiota metabolic interactions. Science 336, 1262–1267 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Lee W-J & Hase K Gut microbiota-generated metabolites in animal health and disease. Nat. Chem. Biol. 10, 416–424 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Koppel N, Maini Rekdal V & Balskus EP Chemical transformation of xenobiotics by the human gut microbiota. Science 356, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donia MS & Fischbach MA HUMAN MICROBIOTA. Small molecules from the human microbiota. Science 349, 1254766 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel KP, Luo FJ-G, Plummer NS, Hostetter TH & Meyer TW The production of p-cresol sulfate and indoxyl sulfate in vegetarians versus omnivores. Clin. J. Am. Soc. Nephrol. 7, 982–988 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bouatra S et al. The human urine metabolome. PLoS One 8, e73076 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furusawa Y et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Maslowski KM et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 461, 1282–1286 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith PM et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wells JE, Berr F, Thomas LA, Dowling RH & Hylemon PB Isolation and characterization of cholic acid 7α-dehydroxylating fecal bacteria from cholesterol gallstone patients. J. Hepatol. 32, 4–10 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Ridlon JM, Kang D-J & Hylemon PB Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 47, 241–259 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Hamilton JP et al. Human cecal bile acids: concentration and spectrum. Am. J. Physiol. Gastrointest. Liver Physiol. 293, G256–63 (2007). [DOI] [PubMed] [Google Scholar]
- 17.de Aguiar Vallim TQ, Tarling EJ & Edwards PA Pleiotropic roles of bile acids in metabolism. Cell Metab. 17, 657–669 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wahlström A, Sayin SI, Marschall H-U & Bäckhed F Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metab. 24, 41–50 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Brestoff JR & Artis D Commensal bacteria at the interface of host metabolism and the immune system. Nat. Immunol. 14, 676–684 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lipsky WRL, Fricke RJ & Hylemon PB Bile acid induction specificity of 7α-dehydroxylase activity in an intestinal Eubacterium species. Steroids (1980). [DOI] [PubMed] [Google Scholar]
- 21.Ridlon JM, Harris SC, Bhowmik S, Kang D-J & Hylemon PB Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 7, 22–39 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mallonee DH, Lijewski MA & Hylemon PB Expression in Escherichia coli and characterization of a bile acid-inducible 3α-hydroxysteroid dehydrogenase from Eubacterium sp. strain VPI 12708. Curr Microbiol (1995). [DOI] [PubMed] [Google Scholar]
- 23.Mallonee DH, Adams JL & Hylemon PB The bile acid-inducible baiB gene from Eubacterium sp. strain VPI 12708 encodes a bile acid-coenzyme A ligase. J. Bacteriol. 174, 2065–2071 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhowmik S et al. Structural and functional characterization of BaiA, an enzyme involved in secondary bile acid synthesis in human gut microbe. Proteins: Structure, Function, and Bioinformatics 82, 216–229 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang D-J, Ridlon JM, Moore DR, Barnes S & Hylemon PB Clostridium scindens baiCD and baiH genes encode stereo-specific 7alpha/7beta-hydroxy-3-oxo-delta4-cholenoic acid oxidoreductases. Biochim. Biophys. Acta 1781, 16–25 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dawson JA, Mallonee DH, Björkhem I & Hylemon PB Expression and characterization of a C24 bile acid 7 alpha-dehydratase from Eubacterium sp. strain VPI 12708 in Escherichia coli. J. Lipid Res. 37, 1258–1267 (1996). [PubMed] [Google Scholar]
- 27.Ye HQ, Mallonee DH, Wells JE, Björkhem I & Hylemon PB The bile acid-inducible baiF gene from Eubacterium sp. strain VPI 12708 encodes a bile acid-coenzyme A hydrolase. J. Lipid Res. 40, 17–23 (1999). [PubMed] [Google Scholar]
- 28.Harris SC et al. Identification of a gene encoding a flavoprotein involved in bile acid metabolism by the human gut bacterium Clostridium scindens ATCC 35704. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1863, 276–283 (2018). [DOI] [PubMed] [Google Scholar]
- 29.González-Pajuelo M et al. Metabolic engineering of Clostridium acetobutylicum for the industrial production of 1,3-propanediol from glycerol. Metab. Eng. 7, 329–336 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Higashide W, Li Y, Yang Y & Liao JC Metabolic engineering of Clostridium cellulolyticum for production of isobutanol from cellulose. Appl. Environ. Microbiol. 77, 2727–2733 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kovács K et al. Secretion and assembly of functional mini-cellulosomes from synthetic chromosomal operons in Clostridium acetobutylicum ATCC 824. Biotechnol Biofuels 6, 117 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mingardon F, Chanal A, Tardif C & Fierobe H-P The issue of secretion in heterologous expression of Clostridium cellulolyticum cellulase-encoding genes in Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 77, 2831–2838 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heap JT, Pennington OJ, Cartman ST & Minton NP A modular system for Clostridium shuttle plasmids. J. Microbiol. Methods 78, 79–85 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Wang Y et al. Bacterial Genome Editing with CRISPR-Cas9: Deletion, Integration, Single Nucleotide Modification, and Desirable “Clean” Mutant Selection in Clostridium beijerinckiias an Example. ACS Synthetic Biology 5, 721–732 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Hang S et al. Bile acid metabolites control Th17 and Treg cell differentiation. BioRxiv (2018). doi: 10.1101/465344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caldeira J, Feicht R, White H & Teixeira M EPR and Moessbauer spectroscopic studies on enoate reductase. Journal of Biological … (1996). [DOI] [PubMed] [Google Scholar]
- 37.Rohdich F, Wiese A, Feicht R, Simon H & Bacher A Enoate reductases of Clostridia. Cloning, sequencing, and expression. J. Biol. Chem. 276, 5779–5787 (2001). [DOI] [PubMed] [Google Scholar]
- 38.Lu Z & Imlay JA The Fumarate Reductase of Bacteroides thetaiotaomicron, unlike That of Escherichia coli, Is Configured so that It Does Not Generate Reactive Oxygen Species. MBio 8, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheridan PO et al. Heterologous gene expression in the human gut bacteria Eubacterium rectale and Roseburia inulinivorans by means of conjugative plasmids. Anaerobe 59, 131–140 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Hao T et al. An anaerobic bacterium host system for heterologous expression of natural product biosynthetic gene clusters. Nat. Commun. 10, 3665 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huo L et al. Heterologous expression of bacterial natural product biosynthetic pathways. Nat Prod Rep (2019). doi: 10.1039/c8np00091c [DOI] [PubMed] [Google Scholar]
- 42.Keasling JD Manufacturing molecules through metabolic engineering. Science 330, 1355–1358 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Pickens LB, Tang Y & Chooi Y-H Metabolic engineering for the production of natural products. Annu. Rev. Chem. Biomol. Eng. 2, 211–236 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
ADDITIONAL REFERENCES
- 44.Leppik RA Improved synthesis of 3-keto, 4-ene-3-keto, and 4, 6-diene-3-keto bile acids. Steroids (1983). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.