Abstract
Objective:
Anemia and Proteobacteria-dominant intestinal dysbiosis in very low birth weight (VLBW) infants have been linked to necrotizing enterocolitis, a severe gut inflammatory disease. We hypothesize that anemia of prematurity is related to the development of intestinal dysbiosis.
Study Design:
342 weekly stool samples collected prospectively from 80 VLBW infants were analyzed for bacterial microbiomes (with 16S rRNA). Linear mixed effects model was used to determine the relationships between the onsets of anemia and intestinal dysbiosis.
Results:
Hematocrit was associated with intestinal microbiomes, with lower Hct occurring with increased Proteobacteria and decreased Firmicutes. Infants with a hematocrit <30% had intestinal microbiomes that diverged toward Proteobacteria dominance and low diversity after the first postnatal month. The microbiome changes were also related to the severity of anemia.
Conclusions:
This finding supports a potential microbiological explanation for anemia as a risk factor for intestinal dysbiosis in preterm infants.
Introduction:
Anemia is a common hematologic condition that requires treatment in preterm infants. In very low birth weight (VLBW) infants, anemia has been implicated in the development of necrotizing enterocolitis (NEC), a severe inflammatory and necrotic bowel disease of prematurity with high rates of morbidity and mortality (1, 2). More recent studies show that mucosal inflammation and injury, the hallmarks of NEC, are present in infants with anemia (3, 4) and neonatal mouse models demonstrate that anemia itself is pro-inflammatory (5, 6). In addition, one mouse model indicates that the severity of NEC correlates with the severity of anemia (6). NEC is a multifactorial disease. Additional evidence is needed to clarify the role of anemia in the development of NEC since the management of anemia is one risk factor under the control of the clinical care team (2, 7).
Proteobacteria dominant intestinal dysbiosis is a risk factor for NEC (8). In healthy guts, the growth of facultative anaerobes, such as a number of gram-negative pathogens from Proteobacteria phylum, are low and suppressed by the growth of obligate anaerobes from Bacteroidetes and Firmicutes phyla (9). Intestinal dysbiosis occurs when this healthy bacterial composition is disturbed, resulting in an increase in the proportion of facultative anaerobes with a concomitant decrease in the proportion of obligate anaerobes. This dysbiosis potentially negatively affects host well-being (9).The gut microbiomes in VLBW infants have very small percentages of Bacteroidetes and are dominated by Firmicutes and Proteobacteria at different ratios (10). Overwhelming evidence supports the occurrence of a Proteobacterial bloom prior to the onset of NEC (8, 11).
As anemia is emerging as an important and previously unknown risk factor in NEC development, examining the temporal relationship between the development of anemia and the composition of the intestinal microbiomes in VLBW infants is essential. We hypothesized that the development of anemia in VLBW infants is associated with intestinal microbial changes toward Proteobacteria dominant intestinal dysbiosis.
Methods
Infant selection, data and sample collection:
The University of South Florida Institutional Review Board approved this prospective study. This study was a part of an NIH-funded study investigating the gut microbiome in VLBW infants. Infants were recruited at a single tertiary NICU from May 2012 to December 2013. Inclusion criteria included birth weight <1500 g, no major chromosomal or intestinal anomalies, and parental informed consent. Infant stool samples were collected weekly from soiled diapers within three hours and stored immediately at −80°C until processing for DNA extraction for microbiome analysis. The stool collection started at enrollment until about 8 weeks postnatal age.
Perinatal information and infant clinical outcomes including sepsis, oxygen requirement, and chronic lung disease were retrieved from the Vermont Oxford Network (VON) database and from the electronic medical records. Each stool sample was matched with the closest hematocrit (Hct) (when available) obtained within 5 days of the stool collection with the preference given to the Hct taken prior to the stool collection. If there was a blood transfusion given within 72 hours of the stool collection, the Hct prior to the blood transfusion was recorded. Sepsis was defined as having a positive blood culture with contamination ruled out. Chronic lung disease was defined as oxygen requirement at 36 weeks corrected gestational age. We reported the total number of days on parenteral antibiotics during the stool collection period. We also computed the SNAPPE-II (Score for Neonatal Acute Physiology with Perinatal Extension-II) to represent infant illness severity (12). Microbiome was examined by postnatal age intervals based on potential clinical influence, ≤ 2 weeks (perinatal factors), 2–4 weeks (nutrition), and 4–8 weeks (feeding and growing period).
DNA Extraction, Amplification and Library Preparation:
The manufacturer’s instructions for MoBio PowerFecal DNA extraction kit (Qiagen, Carlsbad, CA) were followed exactly to extract total DNA from stool samples. The V4–5 region of the 16S rRNA gene was amplified by polymerase chain reaction with modified 515F and 806R primers, isolated, and sequenced using the Illumina Miseq platform (Illumina, San Diego, CA, USA) to generate 250 base-pair paired-end reads(13, 14).
Analyses of DNA data:
The Divisive Amplicon De-noising Algorithm 2 (DADA2) pipeline was used to analyze the V4–5 region of 16S rRNA gene amplicon (15). Taxonomy was assigned and the percentages were calculated using the Greengenes v13.8 database. Bacterial diversity within a sample, or alpha diversity, was represented by the number of species identified by exact sequence variants (ESVs) and Shannon index. Higher ESVs and Shannon index imply more richness and evenness within the samples. To examine the changes of one taxon relative to other taxa, we used Gneiss (balance tree) analysis (16).
Sample size and power analysis:
For the mixed model, a sample size of 49 anemic and 31 non-anemic infants, with an average of 4 samples per infant, can detect a 40% difference in mean bacterial percentages at 90% power and 0.050 significant level using intraclass correlation coefficient of 0.4 (17). For a multilevel regression model, a sample size of 64 infants can detect an effect size of 0.2 attributable to 1 independent variable and adjusted for an additional 11 covariates with 90% power and a significant level of 0.050 (18) (PASS2019, v19.0.3). The study was not powered to detect differences in NEC or feeding intolerance rates.
Statistical Analysis:
Continuous variables were described by mean and standard deviations (SD) or median and interquartile range (IQR) while categorical variables were described by percentages. The association between categorical variables was evaluated by the Fisher exact test. For continuous variables, independent samples t-tests and Mann Whitney U tests were applied for normally distributed and skewed data, respectively. To analyze repeated measurements over time with non-parametric variables, a series of linear mixed-effects models, using maximum likelihood estimation, were fitted to examine the relationships between Hct as a continuous variable or anemia as a ranked variable and the intestinal bacterial percentages. Anemia was defined as having a recorded Hct<30% (19, 20). We further stratified anemia into mild and severe anemia which were defined as having a recorded Hct 25–30% and Hct ≤25% respectively. The models adjusted for clinical factors that had potential effect on intestinal microbiome or anemia, including birth gestational age, postnatal age, days on antibiotics, history of sepsis, feeding types, respiratory distress syndrome, chorioamnionitis, vaginal birth, multiple birth, SNAPPE-II, and blood transfusion history as fixed effects (21, 22). Statistical interactions were tested when indicated. Significant correlations (coefficient >0.7) between the variables were excluded. The best-fitting models were determined by the lowest Akaike’s and Schwarz’s Bayesian criterion. Histograms was used to test normality of residuals (23) and homoskedasticity of the data was tested by the residual vs. predicted value plot. The 95% confidence intervals were used to describe the precision of the estimates. The two-tailed statistical tests were considered significant at p<0.05. All analyses were performed using IBM SPSS statistical software package (IBM Corp. released 2017, IBM SPSS Statistics, Version 25.0 Armonk, NY:IBM Corp).
Results
Infant and sample collection characteristics:
Eighty-three consecutive VLBW infants met the inclusion and exclusion criteria and eighty of them had at least one collected stool sample before death or completion of the study. A total of 342 stool samples from the 80 infants were included in the final analysis and 239 stool samples had matched Hct. The median number of stool samples per infant was 5 (IQR 3–5). Sixty-four infants had ≥2 Hct matched stool samples (median = 3 [IQR 2–4]) and only 3 infants without any Hct matched samples. The maternal and infant clinical characteristics are summarized in Table 1. Twenty-nine infants received at least one PRBC transfusions at the median postnatal age of 9 (IQR=4–24) days and none received probiotics.
Table 1:
Perinatal and Neonatal Clinical Characteristics
| Characteristic | All infants n=80 |
Anemic (Hct <30%, n=49) |
Non-anemic (Hct ≥30%, n=31) |
p-value |
|---|---|---|---|---|
| Gestational age, weeks, mean, (SD) | 28.1 (2.4) | 27.2 (2.0) | 29.5 (2.3) | <0.000 |
| Birth weight, grams, mean, (SD) | 1088 (224) | 1019 (216) | 1196 (194) | <0.000 |
| Maternal body mass index, mean, (SD) | 28.3 (7.4) | 29.0 (6.7) | 27.1 (8.3) | 0.312 |
| Male, n, (%) | 41 (51.2) | 28 (57.1) | 13 (41.9) | 0.252 |
| Hispanic ethnicity, n, (%) | 15 (18.8) | 9 (22.6) | 6 (19.4) | 1.000 |
| Race, n, (%) | ||||
| Black | 35 (43.8) | 21 (42.9) | 14 (45.2) | 0.729 |
| White | 43 (53.8) | 26 (53.1) | 17 (54.8) | 0.729 |
| Antenatal medications, n, (%) | ||||
| Steroids | 70 (87.5) | 41 (83.7) | 29 (93.5) | 0.301 |
| Magnesium | 60 (75) | 36 (73.5) | 24 (77.4) | 0.794 |
| Vaginal delivery, n, (%) | 15 (18.8) | 14 (28.6) | 1 (3.2) | 0.007 |
| Multiple birth, n, (%) | 18 (22.5) | 6 (12.2) | 12 (38.7) | 0.012 |
| Rupture of membrane (hrs) mean, (SD) | 24.1 (100.0) | 31.8 (118.8) | 11.8 (56.8) | 0.391 |
| Chorioamnionitis, n, (%) | 37 (46.3) | 26 (53.1) | 11 (35.5) | 0.023 |
| Maternal hypertension, n, (%) | 29 (36.3) | 14 (28.6) | 15 (48.4) | 0.096 |
| Small for gestational age, n, (%) | 10 (12.5) | 4 (8.2) | 6 (19.4) | 0.167 |
| Respiratory distress syndrome, n, (%) | 46 (57.5) | 37 (75.5) | 9 (29.0) | <0.000 |
| Oxygen on day 28, n, (%) | 23 (28.7) | 21 (42.9) | 2 (6.5) | 0.001 |
| Oxygen on day 36, n, (%) | 4 (5) | 4 (6.5) | 0 | 0.035 |
| Patent ductus arteriosus, n, (%) | 20 (25) | 16 (8.2) | 4 (12.9) | 0.064 |
| Indomethacin, n, (%) | 6 (7.5) | 6 (12.2) | 0 | 0.077 |
| Retinopathy of prematurity required treatment, n, (%) | 1 (1.3) | 1 (2) | 0 | 1.000 |
| Necrotizing enterocolitis, n, (%) | 3 (3.8) | 2 (4.1) | 1 (3.2) | 1.000 |
| Intraventricular Hemorrhage | 8 (10) | 7 (14.3) | 1 (3.2) | 0.124 |
| Days on antibiotics, days, mean, (SD) | 4.3 (4.6) | 5.3 (5.2) | 2.8 (3.0) | 0.010 |
| Positive blood culture, n, (%) | 8 (10) | 6 (12.2) | 2 (6.5) | 0.704 |
| Red blood cell transfusion, n, (%) | 29 (36.3) | 28 (57.1) | 1 (3.2) | <0.000 |
| Feeding type, n, (%) | ||||
| Maternal breast milk only | 47 (58.8) | 34 (69.4) | 13 (41.9) | 0.024 |
| Formula only | 2 (2.5) | 0 | 2 (6.5) | 0.024 |
| Mixed feeding types | 31 (38.8) | 16 (32.7) | 15 (48.4) | 0.024 |
| Discharge weight <10th percentile, n, (%) | 28 (35) | 17 (34.7) | 11 (35.5) | 1.000 |
| Length of stay, days, mean, (SD) | 68.8 (35.2) | 76.5 (33.9) | 56.7 (34.4) | 0.013 |
Independent t-test for continuous variables and Fisher exact test for categorical variables
Infants reached the lowest recorded Hct at the median age of 29 days (IQR=25–47). There were 49 infants in the anemic group which included 28 severely anemic and 21 mildly anemic infants, and 31 in the non-anemic group. The anemic infants had lower birth gestational age and birth weight, higher rates of respiratory support and oxygen requirement, more days on antibiotics, and received more exclusive mother’s own milk and red blood cell transfusions during the NICU course (Table 1). The difference in days on antibiotics between the two groups was significant during the first 2 weeks (p=0.015) but not after 2 weeks (p=0.203) (Mann Whitney U test).
Stool microbiomes of all infants:
There were 3,305,384 16S rRNA amplicon reads from 342 samples included in the final analysis with a range of 1,304 to 38,973 reads (mean = 12,962; median = 11,485) per sample. Sequence depth was rarified to 1,275 reads per sample and 1,221 ESVs were identified. We were able to assign all ESVs to known microbial taxa with the majority to the genus level.
We examined the correlations of the bacterial composition and diversity in relation to postnatal age. Over time, the percentage of the following bacterial taxonomies decreased: Bacteroidetes (Spearman’s rho = −0.118, p = 0.030), Firmicutes (Spearman’s rho = −0.138, p = 0.011), and Staphylococcus (Spearman’s rho = −0.475, p < 0.001); and the percentage of the following bacterial taxonomies increased: Actinobacteria (Spearman’s rho = 0.260, p < 0.001), Proteobacteria (Spearman’s rho = 0.124, p = 0.023), Bifidobacterium (Spearman’s rho = 0.241, p < 0.001), Enterococcus (Spearman’s rho = 0.262, p < 0.001) , Streptococcus (Spearman’s rho = 0.171, p = 0.002), Clostridium, (Spearman’s rho = 0.307, p < 0.001), Veillonella (Spearman’s rho = 0.237, p < 0.001), Escherichia (Spearman’s rho = 0.191, p < 0.001), and Proteus (Spearman’s rho = 0.1183, p < 0.001). Bacterial alpha diversity, represented by the number of ESVs and Shannon index, increased over time (Spearman’s rho = 0.361, p < 0.001 and Spearman’s rho = 0.317, p < 0.001 respectively).
Stool microbiome and Hct:
We examined the concurrent changes in Hct and microbiome composition with the mixed effects models. Over the 8 week enrollment period, Hct was associated positively with percentage of Firmicutes (B=1.09, F=10.53, p=0.001) and negatively with Proteobacteria (B=−1.18, F=10.10, p=0.002) (Figure 1). We then analyzed the changes in Hct and microbiome by postnatal age intervals. The mean Hct at ≤ 2 weeks (0–14 days), 2–4 weeks (15–28 days), and 4–8 weeks (29–60 days) were 38% (SD=7.5%), 32% (SD=5.4%), and 29% (SD=4.0%), respectively. Hct was associated positively with the percentage of Firmicutes (B=1.39, F=9.10, p=0.003) and negatively with Proteobacteria (B=−1.61, F=5.95, p=0.017) during 4–8 weeks postnatal age but not before 4 weeks (Table 2).
Figure 1: Hematocrit is associated with stool microbiome in VLBW infants.
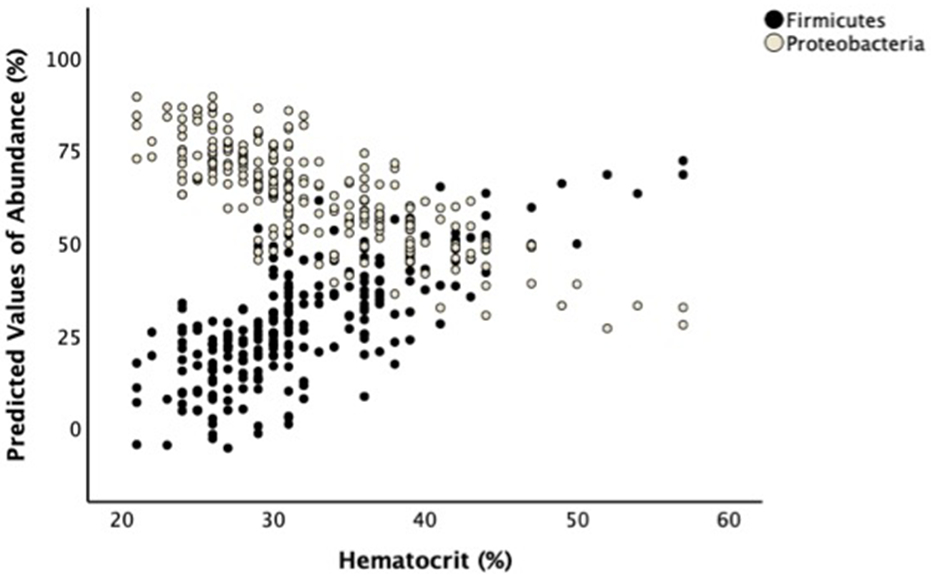
A linear mixed-effects model shows lower Hct is associated with higher Proteobacteria and lower Firmicutes abundances after adjusting for birth gestational age, postnatal age, days on antibiotics, history of sepsis, feeding types, respiratory distress syndrome, chorioamnionitis, vaginal birth, multiple birth, and blood transfusion history. The graph shows the scattered plots of the predicted values for bacterial percentages vs. Hct.
Table 2:
Linear mixed-effects model showing Hct is associated with Firmicutes and Proteobacteria at 4–8 weeks postnatal age
| Proteobacteria | ||||||
|---|---|---|---|---|---|---|
| Variables | Estimate | Std. Error | 95% Confidence Interval | F | P values | |
| Lower Bound | Upper Bound | |||||
| Intercept | 265.98 | 73.99 | 118.92 | 413.04 | 12.92 | 0.00 |
| SNAPPEII | −0.04 | 0.17 | −0.39 | 0.31 | 0.06 | 0.82 |
| Birtd gestational age (wks) | −4.63 | 2.25 | −9.09 | −0.16 | 4.23 | 0.04 |
| Postnatal age (d) | 0.13 | 0.41 | −0.69 | 0.95 | 0.11 | 0.75 |
| Days on antibiotics | −1.28 | 0.90 | −3.07 | 0.51 | 2.02 | 0.16 |
| Feeding type | −2.63 | 3.38 | −9.34 | 4.09 | 0.60 | 0.44 |
| Sepsis | 20.74 | 13.79 | −6.68 | 48.15 | 2.26 | 0.14 |
| Respiratory distress syndrome | −19.13 | 7.29 | −33.62 | −4.64 | 6.89 | 0.01 |
| Chorioamnionitis | −1.07 | 1.09 | −3.24 | 1.09 | 0.97 | 0.33 |
| Vaginal delivery | −13.88 | 7.29 | −28.37 | 0.60 | 3.63 | 0.06 |
| Multiple birtd | −4.59 | 7.51 | −19.52 | 10.34 | 0.37 | 0.54 |
| Blood transfusion | 0.15 | 6.47 | −12.71 | 13.02 | 0.00 | 0.98 |
| Hct (%) | −1.61 | 0.66 | −2.93 | −0.30 | 5.95 | 0.02 |
| Firmicutes | ||||||
| Variables | Estimate | Sth. Error | 95% Confidence Interval | F | P values | |
| Lower Bound | Upper Bound | |||||
| Intercept | −81.46 | 51.68 | −184.18 | 21.26 | 2.48 | 0.12 |
| SNAPPEII | 0.10 | 0.12 | −0.14 | 0.35 | 0.72 | 0.40 |
| Birtd gestational age (wks) | 2.20 | 1.57 | −0.92 | 5.32 | 1.96 | 0.17 |
| Postnatal age (d) | −0.36 | 0.29 | −0.93 | 0.22 | 1.53 | 0.22 |
| Days on antibiotics | 0.30 | 0.63 | −0.96 | 1.55 | 0.22 | 0.64 |
| Feeding type | 4.01 | 2.36 | −0.68 | 8.70 | 2.88 | 0.09 |
| Sepsis | −8.80 | 9.63 | −27.95 | 10.34 | 0.84 | 0.36 |
| Respiratory distress syndrome | 4.01 | 5.09 | −6.11 | 14.13 | 0.62 | 0.43 |
| Chorioamnionitis | 1.90 | 0.76 | 0.38 | 3.41 | 6.19 | 0.02 |
| Vaginal delivery | −3.82 | 5.09 | −13.94 | 6.30 | 0.56 | 0.46 |
| Multiple birtd | 3.84 | 5.25 | −6.59 | 14.27 | 0.54 | 0.47 |
| Blood transfusion | 0.46 | 4.52 | −8.52 | 9.45 | 0.01 | 0.92 |
| Hct (%) | 1.39 | 0.46 | 0.48 | 2.31 | 9.10 | <0.01 |
All variables are set as fixed effects
Stool microbiome and anemia:
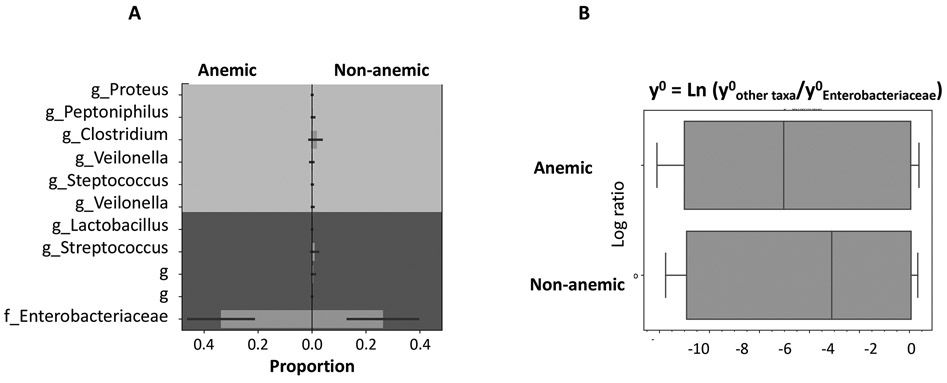
The anemic group had lower median percentages of Firmicutes (p < 0.001) and Clostridium (p = 0.002) and higher median percentages of Proteobacteria (p = 0.001) and Escherichia (p < 0.001) (Mann Whitney U test). The balance tree analysis showed higher proportion of Enterobacteriaceae, the main family in Proteobacteria phylum, and lower proportion of Clostridium in anemic infants (Figure 2A). This increase in Enterobacteriaceae proportion was independent of the shifting of other taxa because the Enterobacteriaceae counts were increased in relative to other bacterial counts in the anemic group (Figure 2B).
Figure 2: Bacterial balance of Enterobacteriaceae in relative to important genera between anemic and non-anemic groups.
A) Proportion diagram from the balance tree analysis shows higher proportion of Enterobacteriaceae (the major family in Proteobacteria phylum) and lower proportion of Clostridium in anemic group. B) Boxplots represent the accumulative natural log transformation of the ratios of major bacterial genera and Enterobacteriaceae; the smaller log values in anemic group represent higher Enterobacteriaceae counts in relative to the counts of other taxa.
In comparing the microbiome characteristics over postnatal age intervals, the microbiomes were similar between the two groups during the first 2 weeks. At 2–4 weeks, the median percentages of Bacteroidetes (p = 0.018) and Proteobacteria (p =0.036) were higher while those of Firmicutes (p = 0.012) and Clostridium (p = 0.002) were lower in the anemic group (Mann Whitney U test). At 4–8 weeks, the means of ESV and Shannon index (which represents bacterial richness and evenness) (p<0.05) (student t-test), the median percentages of Firmicutes (p = 0.001), Staphylococcus (p = 0.001), Clostridium (p = 0.035) and Veillonella (p = 0.006) were significantly lower while those of Proteobacteria (p = 0.025) and Escherichia (p = 0.001) were higher in the anemic group (Mann Whitney U test).
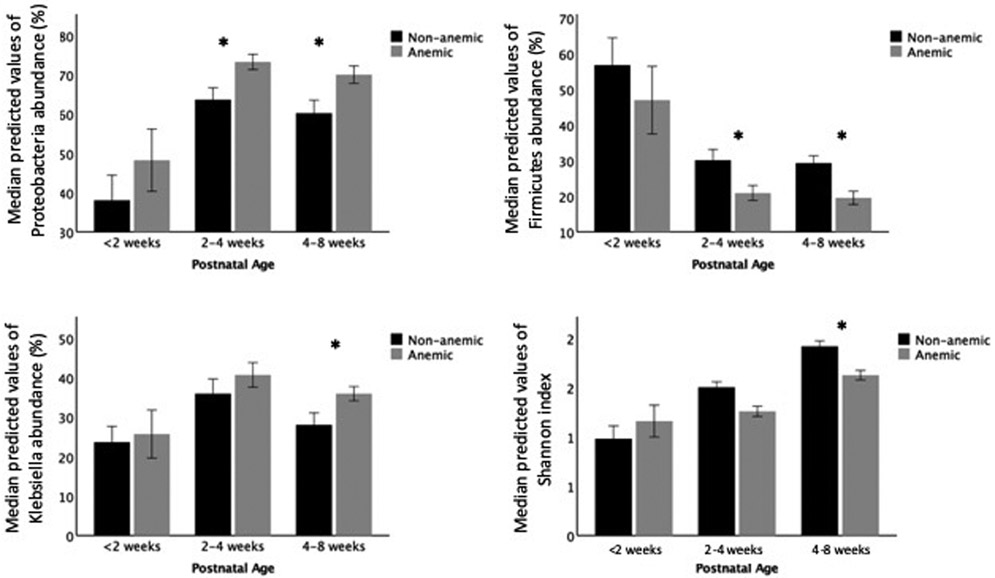
The linear mixed-effects model showed significant differences in bacterial compositions between the two groups. Over the first 8 weeks postnatal, the anemic group had higher percentages of Proteobacteria (B=14.10, F =6.77, p = 0.010) and Klebsiella (B=13.55, F=5.19, p=0.023) and lower in Enterococcus (B=−4.94, F=4.58, p=0.033). There were no significant differences in the microbiome composition before 2 weeks between the anemic and non-anemic groups in this cohort. After 2 weeks, the anemic group had higher percentage of Proteobacteria (2–4 weeks: B= 17.38, F = 5.09, p = 0.026; 4–8 weeks: B= 21.61, F = 10.80, p = 0.001) and Klebsiella (4–8 weeks: B= 24.51, F =8.33, p = 0.005), and lower Firmicutes percentage (2–4 weeks: B= −14.73, F =4.07, p = 0.046; 4–8 weeks: B= −10.29, F =4.16, p = 0.043) and lower Shannon index (4–8 weeks: B=−0.37, F=4.29, p=0.040) (Figure 3).
Figure 3: Anemic VLBW infants develop different stool microbiome over time.
A linear mixed-effects model shows anemic VLBW infants (Hct <30%) are colonized with higher percentages of Proteobacteria and Klebsiella, lower in Firmicutes, and lower Shannon index over time. The bar graphs show the median and 95th CI of the predicted values of bacterial percentages by groups at three postnatal age intervals. The model controls for birth gestational age, postnatal age, days on antibiotics, history of sepsis, feeding types, respiratory distress syndrome, chorioamnionitis, vaginal birth, multiple birth, and blood transfusion history (*p<0.05).
We further explored the differences in microbiomes based on the anemia severity. The severely anemic group had lower alpha diversity, both numbers of ESVs (B=−2.92, F=6.23, p=0.013) and Shannon index (B=−0.43, F=6.80, p=0.010), and higher percentage of Proteobacteria (B= 16.92, F=5.24, p=0.023) than the non-anemic group in the linear mixed model. The mildly anemic group was only different from the non-anemic group in higher abundance of Proteobacteria (B=12.92, F=4.22, p=0.041).
Discussion
Our study is the first to investigate the temporal changes in the gut bacterial colonization of VLBW infants in relation to the development of anemia. Hct level was associated positively with Firmicutes abundance and negatively with Proteobacteria after controlling for clinical factors such as birth gestational age, postnatal age, antibiotic days, and history of blood transfusion. VLBW infants who developed anemia, with a recorded Hct<30%, exhibited a diverging colonization pattern toward proteobacteria dominant intestinal dysbiosis compared to those who did not develop anemia. The anemic infants had higher proportions of Proteobacteria and Klebsiella, and lower proportion of Firmicutes and lower bacterial species diversity after the first postnatal month. These characteristics are similar to those of the intestinal microbiome that is observed prior to NEC development (8, 11). The changes in the gut microbiome was associated with the severity of anemia. These findings suggested that intestinal microbiome is an important characteristic of anemia and may provide an explanation for the observed relationship between severe anemia and the risk of NEC (24).
Our study had limitations secondary to using 16S rRNA next generation sequencing and the multiple confounding factors from studying preterm infants. Each variable region of 16S gene favors identification of certain bacteria genera over others and the selection of variable region can affect the bacterial percentages and alpha diversity (25). Bacterial relative abundances do not translate into absolute bacterial mass (26). Using relative abundances to compare the changes in bacterial presence favors the higher abundances and is less accurate in the lower abundances. Therefore, our findings should be compared to studies using similar 16S variable regions and reporting the changes in overall bacterial compositions. A multilevel mixed model, which does not require the assumptions of homogeneity of regression slopes and independence of errors and tolerates missing data, was used to minimize known and unknown confounders in our clinical data set.
Anemia is more common in infants born with lower gestational ages or those with more complicated NICU courses and we considered whether those factors may have driven the microbiome findings. Our study reduced the effects of clinical and infant characteristics on the developments of anemia and dysbiosis by mixed model and controlling for multiple confounding factors. More premature and sicker infants are often exposed to invasive procedures and instrumentations, prolonged and frequent antibiotics, delayed feeding or more days without enteral feeds. These factors can affect the overall bacterial diversity and composition and the colonization succession (22). However, the extent to which these factors individually and together influence the microbiome development is not well understood. In our cohort, the similarity in intestinal microbiome between the anemia and non-anemia groups during the first postnatal month suggested that the differences in perinatal factors and in the early antibiotic exposure were less likely to influence the subsequent differences in their microbiomes. The changes toward a dysbiotic microbiome together with a decline in Hct levels in anemic infants, warrant further investigations, especially in light of the findings in preclinical models (5, 6).
Although the study could not be powered to assess NEC as an outcome variable, the Proteobacteria-dominant intestinal dysbiosis observed in our preterm infants who developed anemia has a biologically plausible link to clinical NEC. Proteobacteria dominance has been reported to promote inflammation and weaken the epithelial barriers (27, 28). Gram-negative pathogens from the Proteobacteria phylum produce pro-inflammatory lipopolysaccharides (27, 29, 30). Inflammation further favors the growth of aerobic and facultative aerobic bacteria with reduced cellular capacity for beta oxidation, a necessary process in anaerobic conditions (31). The overgrowth of Proteobacteria can suppress the proper growth of commensal obligate anaerobes. Lower colonization of commensal bacteria reduces the production of short chain fatty acids such as butyrate (32). Butyrate promotes maturation and proliferation of colonocytes and it is also a critical mediator of colonic inflammatory responses (33) (34). The interplays between dysbiosis and inflammation along with the weakened gut barrier in the presence of pathogen overgrowth are important in the development of anemia-associated diseases of premature infants and therefore, they should be a research priority.
It has become more evident that anemia is a potential inflammatory mediator in NEC development. The hallmarks of NEC are intestinal inflammation and leakage, and abnormal gut bacterial composition. A previous study describes an inverse relationship between mucosal inflammation and Hct at the time of transfusion (3). Hyung et al. report that intestinal mucosal injury, measured by urine fatty acid binding protein levels, occurs prior to packed red blood cell transfusions in anemic preterm infants (4). A recent animal model demonstrates the anemic intestines are infiltrated with macrophages and the severity of transfusion-associated NEC increases with the severity and duration of anemia (6). Our anemic infants had the changes in gut microbial community that are similar to the previously reported signature microbiome changes that precede NEC onset (8, 35). Clinical studies establish that pathogenic gram-negative bacteria like Klebsiella and Escherichia are pro-inflammatory to the intestine (36, 37). Collectively, these findings indicate a potential microbiological process that supports previously published findings that anemia is related to risk of NEC (24).
This study supported an association between anemia and intestinal dysbiosis in preterm infants and raised the question of the order of their occurrences. A recent publication shows that the severity of anemia in VLBW infants correlates with inflammatory cytokine, IFN-gamma, level in serum (5). On the other hand, a pre-clinical murine model shows that anemia induces significant gut mucosal hypoxia (5). The anemia induced inflammation and hypoxia can play opposite roles in the growth of facultative anaerobic Proteobacteria (31). A randomized study is not ethical in human subjects, but a case control prospective study can provide valuable information on the directionality of anemia and dysbiosis relationship.
In conclusion, the Hct level and anemia were associated with the intestinal microbiome in VLBW infants. Anemic infants developed significantly greater abundance of Proteobacteria and less bacterial diversity, characteristics of dysbiotic microbiome, compared to non-anemic infants, after a month old. The timing of this divergence coincides with the usual onset of anemia associated NEC in VLBW infants. The development of Proteobacteria dominant intestinal dysbiosis in the presence of anemia can lead to plausible explanations for how inflammation and NEC develop in anemic VLBW infants. This study can provide supporting evidence for future investigations on anemia associated diseases, such as NEC, and the threshold of transfusion for anemia.
Acknowledgement
We thank the research nurses, Judy Zaritt and Marcia Kneusel, for their valuable contributions with recruitment, data and stool collection; Tampa General Hospital NICU nurses for stool collection; laboratory technician, Bradley Kane, for his assistance with sample processing; Dr. Michael Georgieff for scientific inputs and editing; and Dr. Jane Carver for editing the manuscript.
Funding Sources: R01 NR015446 (PI Groer), T32GM007281 (ALY)
Footnotes
Disclosure Statement: The authors have no conflicts of interest to declare.
Data sharing: Our 16S sequence data are uploaded and available to public at this website: https://www.ncbi.nlm.nih.gov/sra/?term=SRP136661
References
- 1.Paul DA, Mackley A, Novitsky A, Zhao Y, Brooks A, Locke RG. Increased odds of necrotizing enterocolitis after transfusion of red blood cells in premature infants. Pediatrics. 2011;127(4):635–41. [DOI] [PubMed] [Google Scholar]
- 2.Singh R, Visintainer PF, Frantz ID 3rd, Shah BL, Meyer KM, Favila SA, et al. Association of necrotizing enterocolitis with anemia and packed red blood cell transfusions in preterm infants. J Perinatol. 2011;31(3):176–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ho TT, Groer MW, Luciano AA, Schwartz A, Ji M, Miladinovic BS, et al. Red blood cell transfusions increase fecal calprotectin levels in premature infants. J Perinatol. 2015;35(10):837–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hyung N, Campwala I, Boskovic DS, Slater L, Asmerom Y, Holden MS, et al. The relationship of red blood cell transfusion to intestinal mucosal injury in premature infants. J Pediatr Surg. 2016. [DOI] [PubMed] [Google Scholar]
- 5.Arthur CM, Nalbant D, Feldman HA, Saeedi BJ, Matthews J, Robinson BS, et al. Anemia induces gut inflammation and injury in an animal model of preterm infants. Transfusion. 2019;59(4):1233–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MohanKumar K, Namachivayam K, Song T, Jake Cha B, Slate A, Hendrickson JE, et al. A murine neonatal model of necrotizing enterocolitis caused by anemia and red blood cell transfusions. Nat Commun. 2019;10(1):3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marin T, Patel RM, Roback JD, Stowell SR, Guo Y, Easley K, et al. Does red blood cell irradiation and/or anemia trigger intestinal injury in premature infants with birth weight </= 1250 g? An observational birth cohort study. BMC Pediatr. 2018;18(1):270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pammi M, Cope J, Tarr PI, Warner BB, Morrow AL, Mai V, et al. Intestinal dysbiosis in preterm infants preceding necrotizing enterocolitis: a systematic review and meta-analysis. Microbiome. 2017;5(1):31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stecher B The Roles of Inflammation, Nutrient Availability and the Commensal Microbiota in Enteric Pathogen Infection. Microbiol Spectr. 2015;3(3). [DOI] [PubMed] [Google Scholar]
- 10.Groer MW, Gregory KE, Louis-Jacques A, Thibeau S, Walker WA. The very low birth weight infant microbiome and childhood health. Birth Defects Res C Embryo Today. 2015;105(4):252–64. [DOI] [PubMed] [Google Scholar]
- 11.Mai V, Young CM, Ukhanova M, Wang X, Sun Y, Casella G, et al. Fecal microbiota in premature infants prior to necrotizing enterocolitis. PLoS One. 2011;6(6):e20647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richardson DK, Corcoran JD, Escobar GJ, Lee SK. SNAP-II and SNAPPE-II: Simplified newborn illness severity and mortality risk scores. J Pediatr. 2001;138(1):92–100. [DOI] [PubMed] [Google Scholar]
- 13.Walters W, Hyde ER, Berg-Lyons D, Ackermann G, Humphrey G, Parada A, et al. Improved Bacterial 16S rRNA Gene (V4 and V4–5) and Fungal Internal Transcribed Spacer Marker Gene Primers for Microbial Community Surveys. mSystems. 2016;1(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6(8):1621–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nature methods. 2016;13(7):581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morton JT, Sanders J, Quinn RA, McDonald D, Gonzalez A, Vazquez-Baeza Y, et al. Balance Trees Reveal Microbial Niche Differentiation. mSystems. 2017;2(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yi Q, Panzarella T. Estimating sample size for tests on trends across repeated measurements with missing data based on the interaction term in a mixed model. Control Clin Trials. 2002;23(5):481–96. [DOI] [PubMed] [Google Scholar]
- 18.Gatsonis C, Sampson AR. Multiple correlation: exact power and sample size calculations. Psychol Bull. 1989;106(3):516–24. [DOI] [PubMed] [Google Scholar]
- 19.Jopling J, Henry E, Wiedmeier SE, Christensen RD. Reference ranges for hematocrit and blood hemoglobin concentration during the neonatal period: data from a multihospital health care system. Pediatrics. 2009;123(2):e333–7. [DOI] [PubMed] [Google Scholar]
- 20.Howarth C, Banerjee J, Aladangady N. Red Blood Cell Transfusion in Preterm Infants: Current Evidence and Controversies. Neonatology. 2018;114(1):7–16. [DOI] [PubMed] [Google Scholar]
- 21.Omar RZ, Wright EM, Turner RM, Thompson SG. Analysing repeated measurements data: a practical comparison of methods. Stat Med. 1999;18(13):1587–603. [DOI] [PubMed] [Google Scholar]
- 22.Korpela K, Blakstad EW, Moltu SJ, Strommen K, Nakstad B, Ronnestad AE, et al. Intestinal microbiota development and gestational age in preterm neonates. Sci Rep. 2018;8(1):2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim HY. Statistical notes for clinical researchers: assessing normal distribution (2) using skewness and kurtosis. Restor Dent Endod. 2013;38(1):52–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patel RM, Knezevic A, Shenvi N, Hinkes M, Keene S, Roback JD, et al. Association of Red Blood Cell Transfusion, Anemia, and Necrotizing Enterocolitis in Very Low-Birth-Weight Infants. JAMA. 2016;315(9):889–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen J, Ryu E, Hathcock M, Ballman K, Chia N, Olson JE, et al. Impact of demographics on human gut microbial diversity in a US Midwest population. PeerJ. 2016;4:e1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allaband C, McDonald D, Vazquez-Baeza Y, Minich JJ, Tripathi A, Brenner DA, et al. Microbiome 101: Studying, Analyzing, and Interpreting Gut Microbiome Data for Clinicians. Clin Gastroenterol Hepatol. 2019;17(2):218–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zimmermann MB, Chassard C, Rohner F, N’Goran E K, Nindjin C, Dostal A, et al. The effects of iron fortification on the gut microbiota in African children: a randomized controlled trial in Cote d’Ivoire. Am J Clin Nutr. 2010;92(6):1406–15. [DOI] [PubMed] [Google Scholar]
- 28.Roxas JL, Koutsouris A, Bellmeyer A, Tesfay S, Royan S, Falzari K, et al. Enterohemorrhagic E. coli alters murine intestinal epithelial tight junction protein expression and barrier function in a Shiga toxin independent manner. Lab Invest. 2010;90(8):1152–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shreiner AB, Kao JY, Young VB. The gut microbiome in health and in disease. Curr Opin Gastroenterol. 2015;31(1):69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo S, Guo Y, Ergun A, Lu L, Walker WA, Ganguli K. Secreted Metabolites of Bifidobacterium infantis and Lactobacillus acidophilus Protect Immature Human Enterocytes from IL-1beta-Induced Inflammation: A Transcription Profiling Analysis. PLoS One. 2015;10(4):e0124549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hughes ER, Winter MG, Duerkop BA, Spiga L, Furtado de Carvalho T, Zhu W, et al. Microbial Respiration and Formate Oxidation as Metabolic Signatures of Inflammation-Associated Dysbiosis. Cell Host Microbe. 2017;21(2):208–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gibson MK, Wang B, Ahmadi S, Burnham CA, Tarr PI, Warner BB, et al. Developmental dynamics of the preterm infant gut microbiota and antibiotic resistome. Nat Microbiol. 2016;1:16024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rivera-Chavez F, Lopez CA, Baumler AJ. Oxygen as a driver of gut dysbiosis. Free Radic Biol Med. 2017;105:93–101. [DOI] [PubMed] [Google Scholar]
- 34.Rizzatti G, Lopetuso LR, Gibiino G, Binda C, Gasbarrini A. Proteobacteria: A Common Factor in Human Diseases. Biomed Res Int. 2017;2017:9351507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McMurtry VE, Gupta RW, Tran L, Blanchard EEt, Penn D, Taylor CM, et al. Bacterial diversity and Clostridia abundance decrease with increasing severity of necrotizing enterocolitis. Microbiome. 2015;3:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vigsnaes LK, Brynskov J, Steenholdt C, Wilcks A, Licht TR. Gram-negative bacteria account for main differences between faecal microbiota from patients with ulcerative colitis and healthy controls. Benef Microbes. 2012;3(4):287–97. [DOI] [PubMed] [Google Scholar]
- 37.Ho TTB, Groer MW, Kane B, Yee AL, Torres BA, Gilbert JA, et al. Enteric dysbiosis and fecal calprotectin expression in premature infants. Pediatr Res. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]