Highlights
-
•
To summarize the blood CD4+ or CD8+ T cell count in COVID-19 patients.
-
•
The first meta-analysis on blood CD4+ or CD8+ T cell count between mild and severe COVID-19 patients.
-
•
The lower blood CD4+ or CD8+ T cell count could predict worse prognosis in COVID-19 patients.
Keywords: COVID-19, CD4+ T cells, CD8+ T cells
Abstract
This study mainly focused on the very serious COVID-19 epidemic situation at present and provided a new insight for the treatment and monitor of patients with COVID-19. Through this meta-analysis, we could draw a conclusion that less expression of blood CD4+ and CD8+ T cells count might reflect the severity of infection and often accompanied by a poor prognosis. Hence, we inferred blood CD4+ and CD8+ T cells count could be a promising biomarker for disease assessment and monitor of patients with COVID-19.
1. Introduction
The outbreak of Coronavirus disease (COVID-19) began in December 2019, which is a novel coronavirus (SARS-CoV-2) discovered by mankind. Up to 28th April 2020, more than 2.9 million cases were confirmed, died more than 200,000, manifested as global public health problem, and average mortality rate is about 6.9% [1]. There is growing evidence that the outcome of novel coronavirus's infection is closely related to the disorder of the immune system [2], [3]. Clinical experience in the treatment of patients with COVID-19 showed that the blood lymphocyte count decreased progressively in severe cases [4], [5], but still lacked sufficient evidence-based medical support until now. Hence, this study is aimed to investigate whether blood CD4+ or CD8+ T cells count is associated with severity of patients with COVID-19 using meta-analysis.
We conducted a literature search in PubMed,Scopus, Web of Science and CNKI database and retrieved the publications using the key words “(Coronavirus OR 2019 novel coronavirus pneumonia OR SARS-CoV-2 OR 2019-nCoV OR COVID-19 OR coronavirus 2019) AND (CD4 OR CD8)” without date(i.e. until April 28, 2020) or language restriction. The title, abstract and full text of all potentially studies were reviewed to identify the eligible studies, and disagreement was solved through discussion and consultation by authors. Finally, all the literatures reporting data on patients with severe (defined as needing to enter intensive care unit, using of mechanical ventilation or shock occurrence, etc.) or not-severe COVID-19 were included in our meta-analysis.
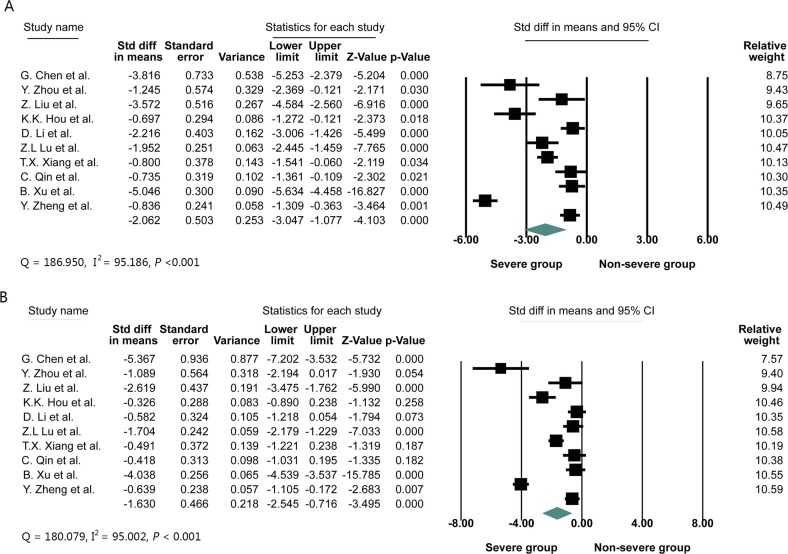
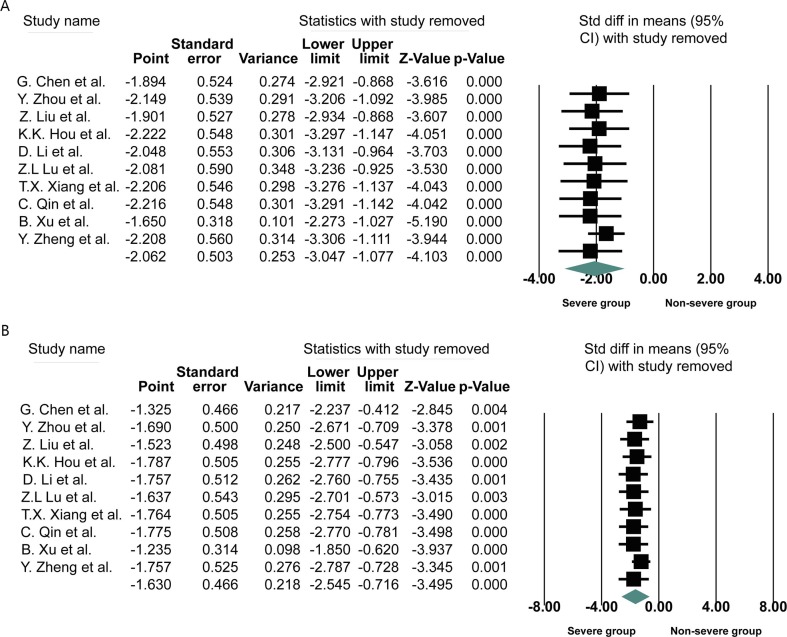
Ten studies were finally included in this meta-analysis, consisting of 643 COVID-19 cases, 277 of whom (43.08%) were severe cases. The primary characteristics of included COVID-19 patients was displayed in Table 1 . Due to the large differences in the mean values among the included studies and heterogeneity was observed, standardized mean difference (SMD) was adopted as the effect quantity in this meta-analysis. In this study, meta-analysis was performed using Comprehensive Meta Analysis Version 3.3 to calculate SMD and the 95% confidence interval (95% CI) of CD4+, CD8+ T cells count in COVID-19 patients with or without severe diseases. When the original study could not provide the mean and standard deviation, the mean and standard deviation of CD4+ and CD8+ T cells count were extrapolated according to the sample size, median and quartile range [6]. Eventually, The SMD values and other relevant indexes from ten studies were presented in Fig. 1 . The heterogeneity (I2 statistics) among the ten studies exceeded 50%, so random effect model was adopted. The pooled results revealed the blood CD4+ or CD8+ T cells count (CD4+: SMD = −2.062, 95%CI = −3.047 to −1.077; CD8+: SMD = −1.630, 95%CI = −2.545 to −0.716) was significantly lower in severe COVID-19 patients compared to non-severe group. Sensitivity analysis by removing one study in turn indicated the pooled result was robust and reliable (Fig. 2 ). The heterogeneity among included studies was high (CD4+: I2 = 95.186; CD8+: I2 = 95.002). To search for the source of heterogeneity, a subgroup analysis was conducted. Subgroup analysis pointed that severe COVID-19 patients with hypertension become the important source of heterogeneity. In addition, inherent variability in blood CD4+ and CD8+ T cells count among patients also possibly be a vital cause of heterogeneity. Furthermore, the Begg rank correction test and Egger linear regression did not support the existence of publication bias among the ten studies (Begg, P = 0.1074; Egger’s, P = 0.5268). It is well known that CD4+and CD8+T cells count are widely involved in the process of immune response, and virus-specific CD4+and CD8+ T cells often play a critical role in clearing virus by eliminating virus infected cells [7]. As in humans infected with coronavirus, CD4+ T cells can induce a protective immune response via IFN-γ expression and enhancement of CD8+ T cell and antibody responses [8]. SARS-CoV antigen is recognized and processed by antigen presenting cells to activate T cells. The activated virus specific effector T cells can elicit release of inflammatory cytokines (TNF-α, IL-6, IL-8, etc.) and chemokine (CXCL-1, CXCL-2, CCL-3, CCL-5, etc.), further inhibit the reproduction of the virus and play an antiviral effect [7]. Guo L et al. reported that the absolute count of CD3+T cells, CD3+CD4+T cells and CD3+CD8+T cells count in the dead pneumonic group are significantly lower than those in the survival group, suggesting that a large number of T cells are activated and depleted in the antiviral process, which is consistant to the result of our study [9].
Table 1.
The primary characteristics of 643 COVID-19 patients in the included studies.
| Study | Country | Males, n (%) |
Age, years |
No. Cases (severe cases) | Severe group CD4+ T cells count, ×106/L |
Non-severe group CD4+ T cells count,×106/L |
Severe CD8+ T cells count,×106/L |
Non-severe group CD8+ T cells count,×106/L |
||
|---|---|---|---|---|---|---|---|---|---|---|
| Severe group | Non-severe group | Severe group | Non-severe group | |||||||
| G. Chen et al. [10] | China | 10, (90.9%) | 7 (70.0%) | 61.0(56.5–66.0) | 52.0(42.8–56.0) | 21(11) | 177.5(104.0–249.8) | 381.5(255.0–451.0) | 89.0(61.5–130.3) | 254.0(183.3–312.8) |
| Y. Zhou et al. [11] | China | 0, (0.0%) | 6 (50.0%) | 42.2 ± 14.91 | 41.5 ± 14.31 | 17(5) | 377.2 ± 229.6 | 698.2 ± 267.4 | 147(116.0–446.0) | 364(111.0–799.0) |
| Z. Liu et al. [12] | China | – | – | – | – | 39(18) | 217.5(112.8–324.5) | 591(266.0–718.5) | 122.5(76.0–256.8) | 288(165.0–414.5) |
| K.K. Hou et al. [13] | China | – | – | – | – | 56(18) | 238.33 ± 142.18 | 453.39 ± 360.22 | 204.95 ± 154.72 | 263.50 ± 190.19 |
| D. Li et al. [14] | China | 12(54.5%) | 7(38.9) | 54.5(37–70) | 43(29–75) | 40(22) | 373(270–463) | 485(373–612) | 286(158–330) | 313(168–376) |
| Z.L Lu et al. [15] | China | 17(50.0%) | 17(25.4%) | 61.4 ± 17.24 | 41.0 ± 14.36 | 101(34) | 213.26 ± 118.89 | 663.57 ± 269.71 | 117.03 ± 73.61 | 417.27 ± 209.41 |
| T.X. Xiang et al. [16] | China | 8(88.9%) | 25(62.5) | 53.0 ± 14.0 | 40.6 ± 14.3 | 49(9) | 221.32 ± 181.65 | 352.75 ± 160.42 | 145.35 ± 89.24 | 201.00 ± 117.64 |
| C. Qin et al. [2] | China | – | – | – | – | 44(27) | 285.10 ± 168.0 | 420.5 ± 207.8 | 154.7 ± 116.5 | 201.9 ± 107.1 |
| B. Xu et al. [3] | China | 73(68.2%) | 30(37.5%) | – | 56.00 (44.00–67.00) | 187(107) | 223.38 ± 77.13 | 573.50 ± 57.375 | 132.66 ± 54.15 | 323.50 ± 36.00 |
| Y. Zheng et al. [17] | China | – | – | – | – | 89(26) | 273.92 ± 185.21 | 553.25 ± 377.81 | 202.31 ± 144.31 | 349.13 ± 256.50 |
Note: “–” represent Not available in the literature; Continuous variables were expressed using Median (range) or mean ± sd (Standard deviation) in this table.
Fig. 1.
Standardized mean difference (SMD) and 95% confidence interval (CI) of CD4+ (A) and CD8+ (B) T cells count in COVID-19 patients with or without severe disease.
Fig. 2.
Forest plot of sensitivity analysis of CD4+ (A) or CD8+ (B) T cells count in COVID-19 patients with or without severe disease. By removing each study in Turn.
To our knowledge, this is the first meta-analysis to comprehensively clarify the relationship between the levels of blood CD4+ and CD8+ T cells and the severity of COVID-19. This study suggests that the detection of blood CD4+ and CD8+ T cells count in patients with COVID-19 is helpful to evaluate the severity of COVID-19, and then treat severe patients as soon as possible. In the future, we probably use big data analysis to establish the reference interval of CD4+ and CD8+ T cell count in COVID-19 patients to distinguish between severe and non-severe patients and monitor the therapeutic effect of COVID-19.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cca.2020.06.040.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.W.H. Organization, Coronavirus disease (COVID-19) outbreak situation, 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019.
- 2.Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., Xie C., Ma K., Shang K., Wang W., Tian D.S. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu B., Fan C.Y., Wang A.L., Zou Y.L., Yu Y.H., He C., Xia W.G., Zhang J.X., Miao Q. Suppressed T cell-mediated immunity in patients with COVID- 19: A clinical retrospective study in Wuhan, China. J. Infect. 2020 doi: 10.1016/j.jinf.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. The Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hozo S.P., Djulbegovic B., Hozo I. Estimating the mean and variance from the median, range, and the size of a sample. BMC Med. Res. Methodol. 2005;5:13. doi: 10.1186/1471-2288-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Channappanavar R., Zhao J., Perlman S. T cell-mediated immune response to respiratory coronaviruses. Immunol. Res. 2014;59(1–3):118–128. doi: 10.1007/s12026-014-8534-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao J., Mangalam A.K., Channappanavar R., Fett C., Meyerholz D.K., Agnihothram S., Baric R.S., David C.S., Perlman S. Airway Memory CD4(+) T Cells Mediate Protective Immunity against Emerging Respiratory Coronaviruses. Immunity. 2016;44(6):1379–1391. doi: 10.1016/j.immuni.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo L., Wei D., Zhang X., Wu Y., Li Q., Zhou M., Qu J. Clinical Features Predicting Mortality Risk in Patients With Viral Pneumonia: The MuLBSTA Score. Front. Microbiol. 2019;10:2752. doi: 10.3389/fmicb.2019.02752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H., Zhang M., Wu S., Song J., Chen T., Han M., Li S., Luo X., Zhao J., Ning Q. Clinical and immunologic features in severe and moderate Coronavirus Disease 2019. J. Clin. Invest. 2020 doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Y., Zhang Z., Tian J., Xiong S. Risk factors associated with disease progression in a cohort of patients infected with the 2019 novel coronavirus. Ann. Palliat. Med. 2020;9(2):428–436. doi: 10.21037/apm.2020.03.26. [DOI] [PubMed] [Google Scholar]
- 12.Liu Z., Long W., Tu M., Chen S., Huang Y., Wang S., Zhou W., Chen D., Zhou L., Wang M., Wu M., Huang Q., Xu H., Zeng W., Guo L. Lymphocyte subset (CD4+, CD8+) counts reflect the severity of infection and predict the clinical outcomes in patients with COVID-19. J. Infect. 2020 doi: 10.1016/j.jinf.2020.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hou K., Zhang N., Li T., Zhou M., Shi X., Zhao G. CT features of corona virus disease 2019 (COVID-19) in different stages and its correlation neutrophil-lymphocyte ratio (NLR) and T lymphocyte subsets. Radiol. Pract. 2020;35(03):272–276. (In Chinese) [Google Scholar]
- 14.Li D., Wang M., He B., Xu Y., Zhou X., Li W., Lu W., Wan J. Laboratory test analysis of sixty-two COVID-19 patients. Med. J. Wuhan Univ. 2020:1–5. (In Chinese) [Google Scholar]
- 15.Lu Z., He R., Jiang W., Fan T., Geng Q. Clinical characteristics and immune function analysis of COVID-19. Med. J. Wuhan Univ. 2020;41(04):529–534. (In Chinese) [Google Scholar]
- 16.Xiang T., Liu J., Xu F., Chen N., Liu Y., Qian K., Zhang W. Analysis of clinical characteristics of 49 patients with coronavirus disease 2019 in Jiangxi. Chin. J. Respirat. Crit. Care Med. 2020;19(02):154–160. (In Chinese) [Google Scholar]
- 17.Zheng Y., Xu H., Yang M., Zeng Y., Chen H., Liu R., Li Q., Zhang N., Wang D. Epidemiological characteristics and clinical features of 32 critical and 67 noncritical cases of COVID-19 in Chengdu. J. Clin. Virol. 2020;127:104366. doi: 10.1016/j.jcv.2020.104366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.