Abstract
Trisomy 21 (Down syndrome, DS) is the main human genetic cause of intellectual disability (ID). Lejeune hypothesized that DS could be considered a metabolic disease, and we found that subjects with DS have a specific plasma and urinary metabolomic profile. In this work we confirmed the alteration of mitochondrial metabolism in DS and also investigated if metabolite levels are related to cognitive aspects of DS. We analyzed the metabolomic profiles of plasma samples from 129 subjects with DS and 46 healthy control (CTRL) subjects by 1H Nuclear Magnetic Resonance (NMR). Multivariate analysis of the NMR metabolomic profiles showed a clear discrimination (up to 94% accuracy) between the two groups. The univariate analysis revealed a significant alteration in 7 metabolites out of 28 assigned unambiguously. Correlations among the metabolite levels in DS and CTRL groups were separately investigated and statistically significant relationships appeared. On the contrary, statistically significant correlations among the NMR-detectable part of DS plasma metabolome and the different intelligence quotient ranges obtained by Griffiths-III or WPPSI-III tests were not found. Even if metabolic imbalance provides a clear discrimination between DS and CTRL groups, it appears that the investigated metabolomic profiles cannot be associated with the degree of ID.
Subject terms: Metabolomics, Genomics, Medical genetics, Neurodevelopmental disorders, Biochemistry, Chemistry
Introduction
Down syndrome (DS) is the most common chromosomal disorder described in humans1 and affects approximately 1 in every 1000–1100 newborns worldwide2. It is caused by the presence of human chromosome 21 in three copies in the cells of affected subjects. The main features of DS consist of intellectual disability (ID), cardiovascular defects and craniofacial dysmorphisms3.
After a systematic reanalysis of all described partial trisomy 21 cases, Pelleri et al.4 identified a highly restricted Down syndrome critical region (HR-DSCR) on human chromosome 21. This region is 34 kb long and is located on the distal 21q22.13. It is duplicated in all DS subjects while it is not duplicated in subjects without a diagnosis of DS5. To date there is no information about the role of this region (HR-DSCR), which does not include known genes but which is part of a very long intron of the isoform 2 of the KCNJ6 gene (https://www.ncbi.nlm.nih.gov/gene/3763)5.
Recently, Caracausi et al.6 conducted an analysis of the 1H Nuclear Magnetic Resonance (NMR)-detectable part of the metabolome in plasma and urine samples of 67 DS subjects and 29 healthy controls (CTRLs) for the first time. They found that DS samples have an altered metabolomic profile, in particular significantly modified levels of several metabolites were observed and attributed to mitochondrial dysmetabolism.
Lejeune had already hypothesized that DS can be considered a metabolic disease7, suggesting that a “blocked” mechanism might determine the level of ID severity, and that specific molecular protagonists of this complex mechanism might be identified.
Finding the possible relationship between metabolomic profile and ID in DS might be of immense value. Searching for the main altered gene products instead for the gene defects may change the point of view of the possible treatment of this condition.
ID is considered to be a significant limitation in both intellectual functioning and adaptive behavior. The mental development of children with DS shows a deceleration during the developmental period8 and their final intelligence quotient (IQ) ranges between 35 and 70, with a mean value of 509,10. However, since IQ is calculated with respect to the distribution of typically developing children of the same chronological age, IQ tends to decrease with age as an effect of the developmental deceleration characteristic of this syndrome. Moreover a high degree of interindividual variability has been demonstrated11.
Previous works indicated that DS is associated with a specific cognitive phenotype, characterized by impairments in speech and language12, with greater difficulties in expressive language than in receptive language, working memory13, as well as in executive functions13,14. Considering motor skills, subjects with DS have fine motor problems due to specific visuo-motor integration and eye-hand coordination problems, combined with slow movement15. On the other hand, non-verbal skills are less severely affected, though recent studies have shown a variable picture depending on which aspect of visuospatial cognition is considered16. Another area of relative strength is social functioning17.
In this work we performed the metabolomic analysis of plasma samples from a higher, almost doubled, number of subjects than in the previous work6. In addition, whenever possible, the cognitive data of DS subjects were collected, and statistical correlations between the metabolomic profiles and cognitive aspects were carried out. The aim of the present work was to confirm the previously observed changes in the metabolome, to identify if the metabolic imbalance makes a clear discrimination between DS and CTRL groups and to verify if specific metabolomic profiles can be associated with the degree of ID.
Results
Study design
Metabolomic data were obtained from a total of 129 subjects with DS (mean age ± standard deviation (SD) = 11.23 ± 6.64 years) and 46 CTRL subjects (mean age ± SD = 15.18 ± 7.99 years) chosen among DS siblings. Sex distribution was 51 female (F) and 78 male (M) among DS and 22 F and 24 M among CTRLs (Table 1). This sample size allowed us to obtain a statistical power of 0.92 (software G*Power, estimation for Wilcoxon-Mann-Whitney test, two tails, effect size d = 0.6, α = 0.05). The main features of the subjects enrolled in this study are described in Table 1. Data concerning age, IQ and medications at the time of the survey are reported in the Supplementary Dataset S1.
Table 1.
Number of Down syndrome (DS) and healthy control (CTRL) subjects.
| Patients | Total | Male | Female | Fasting | Non-Fasting |
|---|---|---|---|---|---|
| DS | 129 | 78 | 51 | 76 | 53 |
| CTRL | 46 | 24 | 22 | 35 | 11 |
As explained in our previous work6, because of the pediatric age of most of the subjects, it was not always possible to collect samples at a fasting state. Thus, to avoid that non-fasting conditions could alter the results, we performed multivariate and univariate analyses for two groups of subjects: the “all” group including fasting and non-fasting subjects, and the “fasting” group (76 DS and 35 CTRL).
With the aim of minimizing healthcare-induced stress, the metabolomic analyses were conducted on EDTA-plasma samples (as in the previous study6). This sample type is indeed suitable for multiple in vitro testing using different techniques, including the analysis of circulating microRNA18.
Plasma metabolome analysis
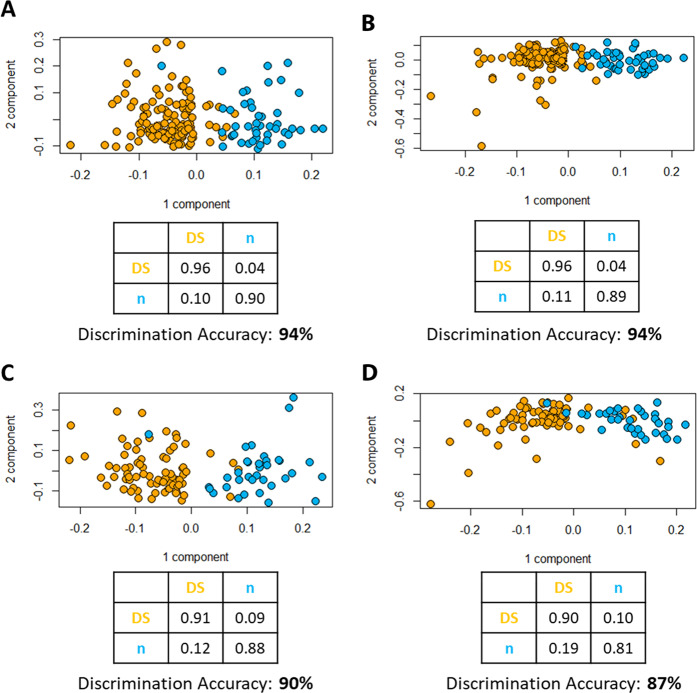
Partial Least Squares-Canonical analysis (PLS-CA) of all plasma samples discriminated DS and CTRL groups with an accuracy of 94% in both CPMG (Carr-Purcell-Meiboom-Gill) and NOESY (Nuclear Overhauser Effect Spectroscopy) spectra (Fig. 1A,B, respectively). Under fasting conditions, the discrimination accuracies were 90% in CPMG spectra (Fig. 1C) and 87% in NOESY spectra (Fig. 1D).
Figure 1.
PLS-CA analysis of all the plasma samples: (A) CPMG and (B) NOESY spectra. Score plot, each dot represents a different plasma sample. Orange dots: Down syndrome samples (DS, n = 129); blue dots: healthy controls (CTRL, n = 46). PLS-CA analysis of the fasting plasma samples: (C) CPMG and (D) NOESY spectra. Score plot, each dot represents a different plasma sample. Orange dots: Down syndrome samples (DS, n = 76); blue dots: healthy controls (CTRL, n = 35).
A random selection of 46 DS and 46 CTRL samples derived from all subjects, repeated 500 times, highlighted a discrimination between DS and CTRL groups with an average accuracy of 87% (p-value < 0.01) in CPMG spectra and 88% (p-value < 0.01) in NOESY spectra, Supplementary Figures S1A and S1B. Considering only fasting subjects, a casual selection of 35 DS and 35 CTRL samples, repeated 500 times, showed a discrimination between DS and CTRL groups with a mean accuracy of 87% in CPMG spectra and 86% in NOESY spectra (p-value < 0.01 in both analyses), Supplementary Figures S1C and S1D.
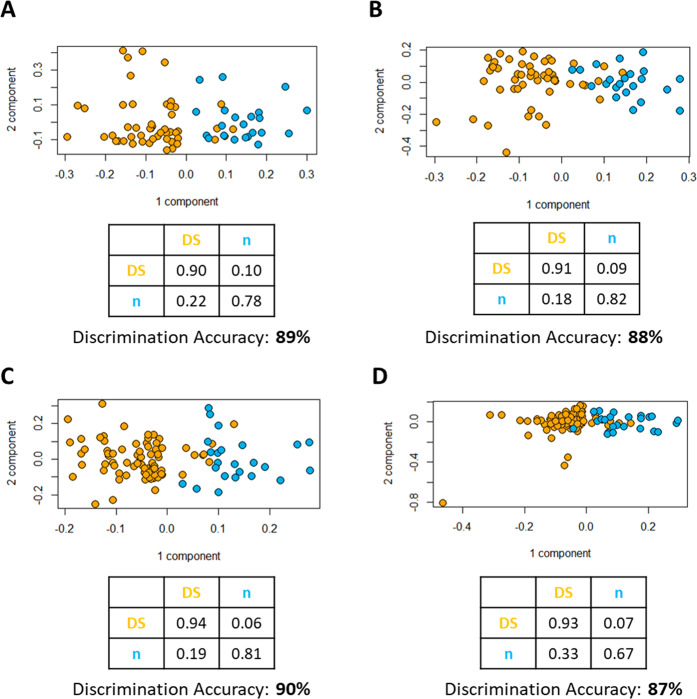
The same analyses were also performed with samples grouped by sex. In all-female samples, PLS-CA discriminated DS from CTRL groups with an accuracy of 89% (p-value < 0.01) in CPMG spectra (Fig. 2A), while in NOESY spectra (Fig. 2B) the discrimination accuracy was 88% (p-value < 0.01). Discrimination accuracy in all-male groups was 90% (p-value < 0.01) in CPMG spectra (Fig. 2C) and 87% (p-value < 0.01) in NOESY spectra (Fig. 2D). Considering only fasting female subjects, the discrimination accuracies were 84% and 85% (p-value < 0.01) in CPMG and NOESY spectra, respectively. Considering only fasting male subjects, the discrimination accuracy was 86% (p-value < 0.01) in both CPMG and NOESY spectra.
Figure 2.
PLS-CA analysis of all the plasma samples from female subjects: (A) CPMG and (B) NOESY spectra. Score plot, each dot represents a different plasma sample. Orange dots: Down syndrome samples (DS, n = 51); blue dots: healthy controls (CTRL, n = 22). PLS-CA analysis of all the plasma samples from male subjects: (C) CPMG and (D) NOESY spectra. Score plot, each dot represents a different plasma sample. Orange dots: Down syndrome samples (DS, n = 78); blue dots: healthy controls (CTRL, n = 24).
Among female samples, a random selection of 22 DS and 22 CTRLs, repeated 500 times, gave a discrimination between DS and CTRL groups with a mean accuracy of 82% in CPMG spectra (p-value < 0.01) and 85% in NOESY spectra (p-value < 0.01), Supplementary Figure S2A and S2B. Among male samples, a random selection of 24 DS and 24 CTRL, repeated 500 times, gave a discrimination between DS and CTRL groups with a mean accuracy of 80% in CPMG spectra (p-value < 0.01) and 76% in NOESY spectra (p-value < 0.01), Supplementary Figure S2C and S2D.
The signals of 28 metabolites were unambiguously assigned in all the NMR spectra (Supplementary Dataset S1). Their levels, together with the levels of 3 unknown signals (unk1, unk2 and unk3), were analyzed through univariate statistical analysis (Tables 2 and 3). Considering the samples from all subjects, independently from their fasting state, acetate, pyruvate, acetone, creatine, formate, acetoacetate, unk3 and succinate showed significantly increased levels in DS, with a DS/CTRL ratio > 1 whereas unk1, tyrosine, histidine and threonine showed significantly reduced levels in DS, with a DS/CTRL ratio < 1 (Table 2). Taking into account only fasting samples, the same metabolites were also found to be significantly increased or decreased in DS, with the exception of acetone, threonine and unk1, which resulted not significantly different in the two groups, and of methionine, which increases significantly in DS plasma (Table 3).
Table 2.
Univariate statistical analysis of all plasma samples (Down syndrome (DS), n = 129; healthy control (CTRL), n = 46).
| Metabolites | DS (median) | CTRL (median) | DS/CTRL median | p-value | p-value after FDR Correction |
| Leucine | 0.00344507 | 0.003570211 | 0.96 | 0.1372 | 0.2658 |
| Isoleucine | 0.00040417 | 0.000429518 | 0.94 | 0.0794 | 0.1797 |
| Valine | 0.00352026 | 0.003575671 | 0.98 | 0.2910 | 0.4748 |
| Alanine | 0.00475446 | 0.004815092 | 0.99 | 0.4072 | 0.5460 |
| Acetate | 0.00089010 | 0.000736686 | 1.21 | 0.0086 | 0.0443 |
| Pyruvate | 0.00222810 | 0.002017161 | 1.10 | 0.0110 | 0.0485 |
| Acetone | 0.00057313 | 0.000491021 | 1.17 | 0.0461 | 0.1190 |
| Glutamine | 0.00628499 | 0.006405399 | 0.98 | 0.8455 | 0.8455 |
| Citrate | 0.00041200 | 0.000393842 | 1.05 | 0.3132 | 0.4855 |
| Glycine | 0.00210159 | 0.002229647 | 0.94 | 0.4507 | 0.5589 |
| Creatine | 0.00115954 | 0.001007514 | 1.15 | 0.0002 | 0.0018 |
| Creatinine | 0.00041121 | 0.000458047 | 0.90 | 0.0812 | 0.1797 |
| Lactate | 0.00396610 | 0.003596582 | 1.10 | 0.1983 | 0.3616 |
| Glucose | 0.02785484 | 0.027973926 | 1.00 | 0.6730 | 0.7194 |
| Mannose | 7.778E-05 | 8.34062E-05 | 0.93 | 0.6267 | 0.7011 |
| Tyrosine | 0.00059000 | 0.000668668 | 0.88 | 0.0184 | 0.0651 |
| Histidine | 0.00031190 | 0.000349493 | 0.89 | 0.0189 | 0.0651 |
| Phenylalanine | 0.00035077 | 0.000352009 | 1.00 | 0.4818 | 0.5745 |
| Formate | 0.00020871 | 0.000183081 | 1.14 | 0.0085 | 0.0443 |
| Fumarate | 0.00001230 | 1.15222E-05 | 1.07 | 0.4227 | 0.5460 |
| Threonine | 0.00019038 | 0.000220489 | 0.86 | 0.0315 | 0.0880 |
| Lysine | 0.00047591 | 0.000466559 | 1.02 | 0.8111 | 0.8382 |
| Acetatoacetate | 0.00027010 | 0.000167440 | 1.61* | 0.0241 | 0.0746 |
| Methionine | 0.00079102 | 0.000732161 | 1.08 | 0.0924 | 0.1909 |
| unk1 | 0.00006890 | 8.40005E-05 | 0.82 | 0.0039 | 0.0305 |
| unk2 | 5.3121E-05 | 4.60515E-05 | 1.15 | 0.2154 | 0.3709 |
| unk3 | 0.00099389 | 0.000691254 | 1.44* | 0.0000001 | 0.0000009 |
| 3-hydroxybutyrate | 0.00041516 | 0.000380792 | 1.09 | 0.4091 | 0.5460 |
| Succinate | 0.00006360 | 4.63183E-05 | 1.37* | 0.000001 | 0.000009 |
| 2-hydroxybutyrate | 0.00010237 | 7.38223E-05 | 1.39* | 0.6333 | 0.7013 |
| Proline | 0.00025393 | 0.000246843 | 1.03 | 0.4188 | 0.5460 |
The table contains the list of metabolites analyzed with NMR. We reported both p-value of the univariate Wilcoxon test and p-value after FDR correction for each metabolite. Metabolites that show significant concentration differences in the two groups (p-value < 0.05) and/or show values in the interval next to 3:2 are reported in bold. *Values in the interval next to 3:2 (range 1.3–1.7).
Table 3.
Univariate statistical analysis of fasting samples (Down syndrome (DS), n = 76; healthy control (CTRL), n = 35).
| Metabolites | DS (median) | CTRL (median) | DS/CTRL median | p-value | p-value after FDR Correction |
| Leucine | 0.00361293 | 0.003694505 | 0.98 | 0.3591 | 0.4840 |
| Isoleucine | 0.00042202 | 0.000475091 | 0.89 | 0.1063 | 0.2059 |
| Valine | 0.00374662 | 0.003824309 | 0.98 | 0.5700 | 0.6796 |
| Alanine | 0.00483927 | 0.004903123 | 0.99 | 0.5277 | 0.6544 |
| Acetate | 0.00090934 | 0.000736206 | 1.24 | 0.0015 | 0.0137 |
| Pyruvate | 0.00226253 | 0.002109906 | 1.07 | 0.0085 | 0.0528 |
| Acetone | 0.00057523 | 0.000517008 | 1.11 | 0.1857 | 0.3199 |
| Glutamine | 0.00642759 | 0.006454857 | 1.00 | 0.8415 | 0.8696 |
| Citrate | 0.00042516 | 0.000385006 | 1.10 | 0.0996 | 0.2058 |
| Glycine | 0.00215948 | 0.002230915 | 0.97 | 0.6409 | 0.7358 |
| Creatine | 0.00114727 | 0.001000137 | 1.15 | 0.0018 | 0.0137 |
| Creatinine | 0.00045933 | 0.000489828 | 0.94 | 0.7342 | 0.7848 |
| Lactate | 0.00413212 | 0.003602912 | 1.15 | 0.0495 | 0.1395 |
| Glucose | 0.02830345 | 0.027789163 | 1.02 | 0.3054 | 0.4303 |
| Mannose | 0.00008250 | 8.55137E-05 | 0.96 | 0.6963 | 0.7709 |
| Tyrosine | 0.00060915 | 0.000681629 | 0.89 | 0.0166 | 0.0828 |
| Histidine | 0.00034121 | 0.000361783 | 0.94 | 0.0307 | 0.1030 |
| Phenylalanine | 0.00035893 | 0.000336898 | 1.07 | 0.4111 | 0.5311 |
| Formate | 0.00020937 | 0.00018 | 1.16 | 0.0187 | 0.0828 |
| Fumarate | 0.00001415 | 1.25068E-05 | 1.13 | 0.2377 | 0.3879 |
| Threonine | 0.00020752 | 0.000231226 | 0.90 | 0.0932 | 0.2058 |
| Lysine | 0.00049623 | 0.000488073 | 1.02 | 0.9873 | 0.9873 |
| Acetatoacetate | 0.00026706 | 0.000167242 | 1.60* | 0.0265 | 0.1029 |
| Methionine | 0.00082180 | 0.000730695 | 1.12 | 0.0332 | 0.1030 |
| unk1 | 0.00008045 | 8.6702E-05 | 0.93 | 0.0920 | 0.2058 |
| unk2 | 0.00005625 | 4.58131E-05 | 1.23 | 0.0957 | 0.2058 |
| unk3 | 0.00104660 | 0.000699813 | 1.50* | 0.0000016 | 0.000025 |
| 3-hydroxybutyrate | 0.00042926 | 0.000398342 | 1.08 | 0.2994 | 0.4303 |
| Succinate | 7.3119E-05 | 4.65665E-05 | 1.57* | 0.0000001 | 0.0000035 |
| 2-hydroxybutyrate | 0.00011189 | 7.75603E-05 | 1.44* | 0.1753 | 0.3196 |
| Proline | 0.00025412 | 0.000230208 | 1.10 | 0.2878 | 0.4303 |
The table contains the list of metabolites analyzed with NMR from samples taken from fasting patient. We reported both p-value of the univariate Wilcoxon test and p-value after FDR correction for each metabolite. Metabolites that show significant concentration differences in the two groups (p-value < 0.05) and/or show values in the interval next to 3:2 are reported in bold. *Values in the interval next to 3:2 (range 1.3–1.7).
Correlations between metabolites in DS and control groups
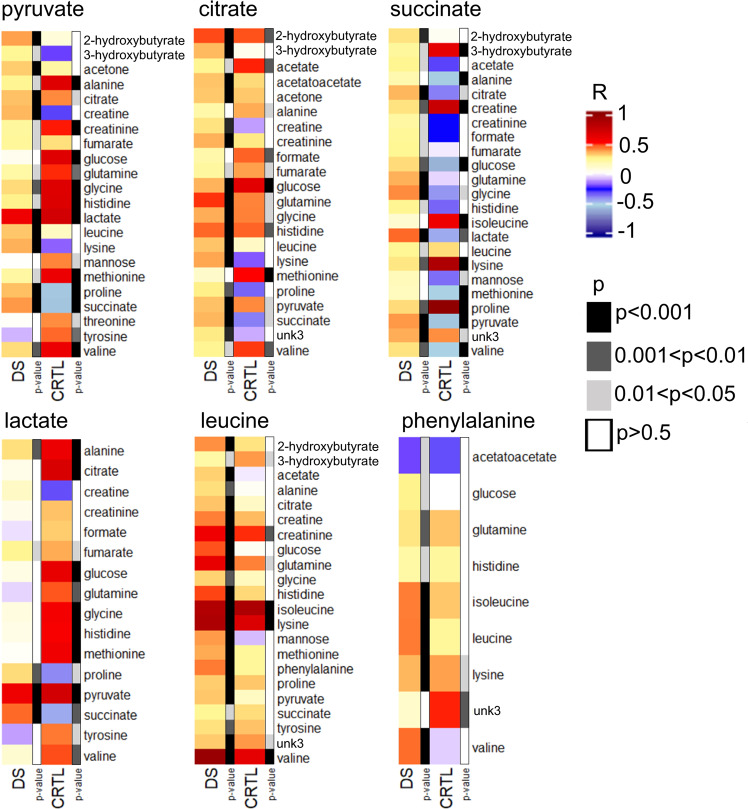
We compared the levels of each metabolite with those of all the others performing a series of correlation analyses in both DS and CTRL samples, separately. Partial correlations including “chronological age” as covariate were conducted (see “Statistical analysis” paragraph in the “Materials and methods” section). We considered only correlations with p-value after FDR (False Discovery Rate) < 0.05 and with a correlation coefficient r > 0.4 or < −0.4 to be statistically significant. We clustered the results into 7 main groups of metabolites whose levels have statistically significant correlations: the Krebs cycle metabolites (Supplementary Table 1); formate (Supplementary Table 2); ketone bodies (Supplementary Table 3); lactate, glucose and mannose (Supplementary Table 4); branched-chain amino acids (BCAA) (Supplementary Table 5); creatine and creatinine (Supplementary Table 6); amino acids (Supplementary Table 7). Interestingly, some correlations found in CTRL samples had no correspondence in DS samples and vice versa (Supplementary Tables 1, 2, 3, 4, 5, 6, 7; Fig. 3).
Figure 3.
Statistically significant correlations between the levels of some of the altered metabolites and all other detected metabolites. Heat maps of the correlations (r) of representative metabolites in both Down syndrome (DS) and healthy control (CTRL) samples, separately. The p-value (after FDR correction) of each correlation is also reported.
Correlations between metabolome and cognitive skills
We investigated if the NMR-detectable part of the plasma metabolome of subjects with DS contains signatures of the different IQ scores.
39 subjects with DS were evaluated through the WPPSI-III test (Supplementary Dataset S1). We created two main groups of DS patients on the basis of their IQ scores: one group included all DS subjects with an IQ > 40 (n = 13, mean age=125.69 months) and the other one included all DS subjects with an IQ ≤ 40 (n = 26, mean age=141.38 months). The difference between the two mean ages was not statistically significant by unpaired t-test (t = −1.694; p = 0.099). Using the PLS-CA model we obtained a discrimination accuracy of 74.5% between the plasma metabolomic profiles of the two groups, i.e. DS subjects with IQ > 40 vs. IQ ≤ 40 (Supplementary Figure S3). Additionally, we were not able to identify any metabolites with statistically significant concentration differences between the two groups.
22 subjects with DS were evaluated using the Griffiths-III test (Supplementary Dataset S1). Using the same statistical approach specified for the WPPSI-III test, we obtained a discrimination accuracy of 74.3% between the metabolomic profiles of DS subjects with IQ > 40 (n = 17, mean age=55.94 months) and those one with IQ ≤ 40 (n = 5, mean age=66.4 months). Again, the difference between the two mean ages was not statistically significant by unpaired t-test (t = −1.997; p = 0.06). Also in this case we were not able to identify any metabolites with statistically significant concentration differences (Supplementary Figure S4).
Furthermore, we performed a series of correlation analyses in order to investigate correlations between each metabolite level and the age equivalent (AE) scores collected from Griffiths-III and WPPSI-III tests from DS patients (see AE scores in the Supplementary Dataset S1 and Supplementary Tables 8 and 9). Partial correlations including “chronological age” as covariate were conducted (see “Statistical analysis” paragraph in the “Materials and methods” section). We were unable to obtain any statistically significant correlations considering p < 0.01 and r < −0.4 or r > 0.4 (Supplementary Tables 8 and 9).
Discussion
The aim of this study was to establish whether an alteration in metabolite levels may have an important role in DS. For this reason, we analyzed NMR spectra of plasma samples from patients with DS and their siblings, used as CTRL group, thus significantly increasing the number of subjects with respect to our previous work6 (from 67 to 129 DS subjects and from 29 to 46 CTRLs). First of all, we confirmed the presence of significantly different plasma metabolomic profiles of DS and CTRL groups and identified the metabolites with significantly different concentrations in the two groups. Then, we searched for correlations among metabolite levels in DS and CTRL samples separately and correlations between metabolite levels and cognitive data of DS subjects. Finally, we investigated whether the characteristic plasma metabolomic profile of DS subjects contains signatures of different grades of ID.
The discrimination accuracy between DS and CTRL groups is higher than in our previous analysis6, reaching 94% both with CPMG and NOESY spectra (Fig. 1A,B). If we only consider fasting subjects, the discrimination accuracy only slightly decreases to 90% with CPMG spectra and 87% with NOESY spectra (Fig. 1C,D). The decrease can be explained considering the reduction in the sample size using only fasting samples. Sex is not a confounding factor; in fact, our results show similar discrimination accuracy between DS and CTRL groups when considering females and males separately in both CPMG and NOESY spectra (again, a slight decrease in discrimination accuracy with respect to the complete group can be attributed to the reduction in the numbers of individuals). This is a further confirmation of our previous work6, where we demonstrated that both sex and age were not confounding factors for the discrimination between DS and CTRL metabolomic profiles.
The analysis of plasma samples provided the concentrations of 28 different metabolites involved in multiple metabolic pathways.
The univariate analysis confirmed the significant alterations of the levels of metabolites involved in processes related to mitochondrial metabolism in DS highlighted by Caracausi et al.6; the DS/CTRL ratio allowed us to understand when the metabolite level increases (DS/CTRL > 1) or decreases (DS/CTRL < 1) in DS vs CTRL samples. The analysis showed significantly increased concentrations (p-value after FDR correction < 0.05) of the following metabolites: acetate; pyruvate; succinate; formate and creatine. All of them are involved in energy production. Acetate is fundamental in supporting acetyl-coenzyme A metabolism and thus Krebs cycle progression and lipogenesis19. Pyruvate and succinate are produced before and during the Krebs cycle20. Formate is known to be necessary for the formylation of mitochondrial tRNAs21 and for the formylation of tetrahydrofolate during the folic acid cycle and the purine synthesis pathway. Finally creatine is used by muscles for the production of ATP22. Previous works demonstrated that measurable enzymes whose genes are located on Hsa21 adhere to the 3:2 overexpression model expected in trisomy 2123–27. Although the dependence of the levels of the metabolites on the enzyme concentration is complex and not easily interpretable, we point out that a ratio DS/CRTL near to 3:2 (1.5) or 2:3 (0.67) is observed for a certain set of molecules. In this framework, interesting data are the significant alterations of succinate in DS vs CTRL. Succinate levels increase in DS samples (p-value after FDR correction <0.0001 in both “all samples” and “fasting samples” groups) with a DS/CTRL ratio close to 3:2 (1.37 in “all samples” and 1.57 in “fasting samples” groups).
The alterations of succinate metabolism may play a role in the development of ID, but to date there is no strong evidence. However, it is already known that some enzymes which metabolize succinate are involved in developmental delay and brain injuries. In particular, the succinic semialdehyde dehydrogenase (SSADH) deficiency, caused by a mutation of the ALDH5A1 gene (6p22.3), determines an increase of γ-aminobutyric acid (GABA) and γ-hydroxybutyric acid levels. These alterations cause developmental delay, hypotonia, hyporeflexia, ataxia, neuropsychiatric problems, and epilepsy28.
Furthermore, alterations of activities for the Krebs cycle enzymes were also observed in some brain disorders such as Alzheimer’s disease29 and Huntington’s disease30. In both diseases, the increase of succinate dehydrogenase and malate dehydrogenase and the decrease of the pyruvate dehydrogenase complex, isocitrate dehydrogenase, and the alpha-ketoglutarate dehydrogenase complex were observed, suggesting that a mitochondrial alteration occurs. These results also suggest that measures to improve tricarboxylic acid cycle metabolism might lessen the effects of the diseases29,30.
In order to obtain more information about the alteration of the metabolic pathways in DS we compared the correlations (corrected by age) among the levels of metabolites in DS subjects and CTRL groups, separately.
Importantly, we observed that some correlations are strong/moderate (r > 0.4 or r < −0.4) and significant (p < 0.05) in CTRL samples but lose their significance or weaken their correlation in DS samples. These results are referred to metabolites involved in the Krebs cycle (pyruvate, citrate and succinate), formate and lactate and amino acids like alanine, threonine and tyrosine (Supplementary Table 1, 2, 4, 7; Fig. 3). On the contrary, some statistically significant correlations are found only in DS samples, like associations between phenylalanine and branched amino acids (Supplementary Table 7; Fig. 3). Moreover, the loss of correlations among strictly related metabolites (from a biochemical point of view) and the appearance of new correlation patterns in DS confirmed the dysregulation of some metabolic pathways. Accordingly, the inverse correlation between lactate and succinate characteristic of CTRL samples becomes direct in DS samples (Supplementary Table 1 and 4; Fig. 3).
These findings support the hypothesis of an altered metabolism in DS, strengthening the idea of the involvement of the Krebs cycle and of a few amino acids like leucine, phenylalanine, tyrosine and alanine. Leucine and phenylalanine are both involved in brain metabolism: the role of leucine as a nitrogen donor in the glutamine/glycine pathway may influence the function of some neurotransmitters31, while modifications in the phenylalanine pathway can alter dopamine levels32.
Even if metabolic imbalance clearly discriminates between DS and CTRL groups, it appears that specific metabolomic profiles cannot be associated with the degree of ID, as shown by both the low discrimination accuracies obtained when comparing the metabolomic profiles of DS patients with different IQ disability and the lack of correlation between the levels of the investigated metabolites and the scores obtained by cognitive tests (Supplementary Tables 8 and 9). It would be interesting to test if strengthening the statistical power of the models by increasing the number of patients in each range of ID could reveal such a correlation, as it would be of immense value in identifying critical points of metabolism as treatment targets. An alternative or complementary explanation for this result might be that, in accordance with the neuroconstructivist perspective on development, the effect of metabolome on patient’s skills might be mediated by environmental factors (such as family environment, school, early intervention, etc.), so further study is required here.
In addition, it is possible that metabolites critically altered in DS are not included among those we have investigated (i.e., fall below the NMR detection limit). This hypothesis should be further investigated by studying a larger number of metabolites using complementary analytical platforms with a better sensitivity with respect to NMR, like mass spectrometry33,34. By increasing the number of measured metabolites, extended metabolic network models could be developed and might provide deeper insight in the biochemical origin of cognitive impairment35,36.
Materials and methods
Ethics Statement
The study was approved by the independent Ethics Committee of the University Hospital St. Orsola-Malpighi Polyclinic, Bologna, Italy (study number: 39/2013/U/Tess). Informed written consent was obtained from all participants or their legal guardians. The patients, if over 18, or their legal guardians, signed an informed consent form for the collection of blood and clinical data before taking part in the study. All procedures were carried out in accordance with the Ethical Principles for Medical Research Involving Human Subjects of the Helsinki Declaration.
Case selection
All subjects were admitted to the Day Hospital of the Neonatology Unit, Sant’Orsola-Malpighi Polyclinic, Bologna, and this study was proposed in the context of the yearly routine follow-up provided for DS. Inclusion criteria for subjects with DS were diagnosis of DS with homogeneous or mosaic trisomy 21 and minimum age of 2 years.
A total of 175 subjects, between 3 and 37 years old, participated in the study including 129 patients with DS and 46 healthy CTRLs chosen among DS siblings and with no evidence of abnormal karyotype. 26 DS subjects had at least one sibling enrolled in the study. Blood samples, as well as clinical data, were collected during the follow-up visit.
For every collected sample, parents filled out a form with information about current fasting state, last meal, consumed medications.
Plasma sample preparation
Plasma samples were collected from all subjects enrolled in the study, according to standard procedures37–39.
Blood samples were collected in EDTA-coated blood collection tubes and stored at room temperature. They were treated within two hours of blood collection and every delayed treatment of the sample was recorded.
The samples were transferred to a new tube and centrifuged at 1,200 g for 10 min to separate corpuscular fraction from plasma. The plasma fraction was isolated and centrifuged for a second time at 800 g for 30 min. The supernatant was transferred to new tubes avoiding contact with pellets or with the bottom of the tube and then divided in aliquots of 400 μL. All plasma samples were rapidly stored in a − 80 °C freezer.
To avoid contamination all procedures were conducted carefully. The plasma samples were excluded from the analyses when the treatment of blood samples occurred more than two hours from their collection, or when evident contamination by residual erythrocytes at the end of the treatments occurred. Any anomalies like different plasma color or precipitates after centrifugation were noted and considered in further analysis.
NMR sample preparation, spectra processing and spectral analysis
NMR samples were prepared according to standard procedures6,40,41.
NMR spectra for all samples were acquired using a Bruker 600 MHz spectrometer (Bruker BioSpin) operating at 600.13 MHz proton Larmor frequency and equipped with a 5 mm PATXI 1H-13C-15N and 2H-decoupling probe including a z-axis gradient coil, an automatic tuning-matching (ATM) and an automatic and refrigerate sample changer (SampleJet, Bruker BioSpin). A BTO 2000 thermocouple served for temperature stabilization at the level of approximately 0.1 K of the sample. Before measurement, samples were kept inside the NMR probe head for 5 minutes for temperature equilibration at 310 K.
For each plasma sample, two monodimensional 1H-NMR spectra were acquired with water peak suppression and different pulse sequences that allowed the selective observation of different molecular components. The spectra were: 1) a standard NOESY42 using 32 scans, 98,304 data points, a spectral width of 18,028 Hz, an acquisition time of 2.7 s, a relaxation delay of 4 s and a mixing time of 0.1 s. This type of spectrum is made up of signals arising from low molecular weight molecules (metabolites) and signals arising from macromolecules such as lipoproteins and lipids; 2) a standard CPMG (Purcell) using 32 scans, 73,728 data points, a spectral width of 12,019 Hz and a relaxation delay of 4 s. This type of spectrum contains only the signals arising from low molecular weight molecules (metabolites).
Before applying Fourier transform, free induction decays were multiplied by an exponential function equivalent to a 0.3 Hz line-broadening factor. Transformed spectra were automatically corrected for phase and baseline distortions and calibrated to the glucose doubled at δ 5.24 ppm, using TopSpin 3.5 (Bruker BioSpin)6,40,41.
Cognitive data collection
Cognitive data were collected and processed for a total of 61 DS children/adolescents from 3 to 16 years old. Following an increasingly widespread procedure in the field of intellectual disability43, the cognitive level was assessed through tests for expected mental age rather than for chronological age. This approach provides a more sensitive measure that avoids floor effects.
Children from 3 to 6 years and 11 months were assessed using the Griffiths-III scale44, a play-oriented developmental test. Considering the DS cognitive profile, two scales were used for the purpose of this study: foundation of learning (scale A), which assesses different aspects of thinking; and language and communication (scale B), which measures overall language development, including expressive language, receptive language, and to a lesser extent, the use of language to communicate socially.
Children/adolescents from 7 to 16 years old were assessed using the WPPSI-III scale45 which consists of different subtests summarized in three principal indexes: Verbal, Non Verbal and Total.
For both tests, raw scores were registered and later converted into AE scores. However, since the AE scores do not take into account the subject’s age, every statistical analysis involving AE values has been corrected for chronological age.
Moreover, an IQ score was calculated as the ratio of the subject’s AE to his/her chronological age, multiplied by 100.
Statistical analysis
Multivariate analysis of the NMR data (aimed at analyzing the spectra as a whole) was performed on binned spectra. Each spectrum in the 10.00–0.2 ppm region was segmented into 0.02 ppm chemical shift bins and the corresponding spectral areas were integrated using the AMIX software (Bruker BioSpin). The presence of EDTA as anticoagulant gives rise to few NMR signals that are very intense and whose concentration levels can be slightly different among samples. Thus, together with the spectral region containing water signals (region:4.40–5.00 ppm), spectral regions including EDTA signals (regions: 2.53–2.60, 2.68–2.73, 3.07–3.24, 3.58–3.64 ppm) were excluded before integration to avoid the presence of potentially confounding factors in multivariate analyses41,46,47. The multivariate statistical analysis was performed using both CPMG and NOESY binned spectra.
Different kinds of multivariate statistical techniques were used on the obtained bins using R 3.0.2 in house scripts41.
Unsupervised Principal Component Analysis (PCA) was used to obtain a preliminary outlook of the data (visualization in reduced space, cluster detection, screening for outliers). Partial Least Squares (PLS) analysis was employed to perform supervised data reduction and classification between samples from healthy and diseased volunteers. Canonical analysis (CA) was used in combination with PLS to increase supervised data reduction and classification. The accuracy for classification was assessed by means of a Monte Carlo validation scheme: each dataset was randomly divided by 200 times into a training set (90% of the data) which was used to build the model and a test set (10% of the data) which was used to test the integrity of the model. The resulting confusion matrix was reported and its discrimination accuracy, specificity and sensitivity were estimated according to standard definitions. Each classification model was also validated using permutation test (n = 500) and the resulting p-value was reported.
Univariate analysis of the NMR data was performed on Fourier transformed and calibrated CPMG spectra. Metabolites, whose peaks in the spectra were well defined and resolved, were assigned and their levels analyzed. The assignment procedure was made up using an 1H-NMR spectra library of pure organic compounds, public databases, e.g. Human Metabolome Database48, storing reference 1H-NMR spectra of metabolites, spiking 1H-NMR experiments and using literature data49. The relative concentrations of the various metabolites were calculated by integrating the corresponding signals in the spectra50, using the AssureNMR Software (Bruker BioSpin) and a home-made tool for R Software.
The nonparametric Wilcoxon-Mann-Whitney test was used for the determination of the meaningful metabolites. Here, a p-value < 0.05 was considered statistically significant. Considering FDR, the p-value was corrected using the Benjamini-Hochberg formula and reported as pFDR51.
SPSS Statistics (IBM, Version 25 for Mac OS X) was used to perform partial correlation between the level of each metabolite and the levels of all the other metabolites checking for the effect of chronological age (at the moment of blood-collection). We considered r-value between 0.4 and 0.7 as moderate correlation and r > 0.7 as strong correlation52. Briefly, from the main Menu of the “SPSS Statistics” software, we selected “Analyze” and then “Correlate”; we chose “Partial...” and finally we inserted our data in the main box and inserted “Age at Blood Collection” in the “Controlling for” box. To obtain p-value after FDR correction, we created a file with all the p-values obtained from the previous analysis and, using JMP software (SAS Institute, Version 14), from the main menu we selected “Add-in”, then “False Discovery Rate P-value” command and finally inserted the p-value column in “PValue column”.
To analyze the correlations between metabolite levels and AE scores obtained from Griffiths-III and WPPSI-III tests, we performed a partial correlation checking for the effect of chronological age (at the moment of the cognitive test) using SPSS Statistics software (from the main software Menu we selected “Analyze” and then “Correlate”, then “Partial...” and finally we included our data in the main box and inserted “Age” in the “Controlling for” box).
To investigate the influence of the different IQ scores on the metabolomic profiles, we divided the metabolome results into two groups: those deriving from subjects with DS having taken the Griffiths-III test and those deriving from subjects with DS having taken the WPPSI-III test. We distinguished the subjects according to the IQ scores obtained from the two kinds of cognitive tests for both groups. It is known that the IQ scores have a mean of 100 and an SD of 15 and that a subject with an IQ < −2SD has an intellectual disability. All subjects with DS included in this study have an IQ < −2SD. To perform the multivariate statistical analysis between a significant number of metabolomic profiles for each kind of cognitive test, we decided to create two main groups of data: a group of metabolomic profiles from subjects with an IQ > 40 (between 2 and 4 SD below average) and a second group from subjects with an IQ ≤ 40 (more than 4 SD below average).
Supplementary information
Acknowledgements
The authors sincerely thank all the children and their families who participated in the study. We are very grateful to Donatella Pascai and Giuseppina Dibenedetto for their expert performance of blood draws. P.T., V.G. and C.Lu. acknowledge the support and the use of resources of Instruct-ERIC, a Landmark ESFRI project, and specifically the CERM/CIRMMP Italy Centre. As Bologna groups, we are profoundly grateful to the “Fondazione Umano Progresso”, Milano, for their continuous and fundamental support to our research on trisomy 21 and to this study. We are so truly grateful to Alberto Ospite and Diego Discepoli, as well as to Laura Buonfino, Stefano Marzeglia, and Marco Pasinetti, for the organization of various overwhelmingly successful fundraising events in London in support of our research as well as to all the donors contributing to these initiatives, as documented at the sites: https://www.justgiving.com/crowdfunding/genome21 or /genome21research. We wish to sincerely thank the following for their support to our research: Matteo and Elisa Mele; Giovanni Roncoroni; “Associazione Amicorum” and “Associazione + di 21 (piudi21) onlus”, Cassano Magnago (VA); “Comitato Arzdore di Dozza” (BO) and the Costa family; Enzo Iacchetti, “Nito Produzioni Srl”, Alessandro Pagnotti and Carmen Spagnolo; “Associazione Vola con Martin oltre il 21”, Mandello del Lario (LC); Rina Bini; Guido Marangoni; Stefano Martinz; Lauretta and Domenico Mele, with relatives and friends, in occasion of their 50th wedding anniversary. Very special thanks to “Progetto Pulcino Onlus” - Reggio Emilia, to its President Dr Cristiana Magnani and to Dr Massimiliano Iori for the valuable donation of the −80 °C freezer hosting our biobank. Our heartfelt thanks for their initiatives in support of our study, as well all the participants in these events, goes to: “Associazione Centro 21”, “Associazione Crescere Insieme”, “Coop. Sociale Cuore 21”, “Tenuta del Monsignore”, “La Prima Coccola”, “Luca Carni”, “Associazione Sergio Zavatta”, and friends of Giovanna Lelli (Rimini and province); “Parrocchie di Pieve Corleto e Basiago”, Faenza (RA); Luciano Perondi and Paolo Ciaroni for having created a series of car meetings called “Top Down”, devoted to spider cars; Facebook group “Sindrome di Down - up not down”, in particular to Jonatan Benvenuti and Antonella Loia, for promoting a fundraising campaign at https://it-it.facebook.com/donate/952905191722252/, as well as to Cristina Boldrini, Francesco Pezzi, and Illumia for supporting the related meeting in Bologna; fundraising initiative at “Fondazione Sant’Angelo”, Loverciano, Switzerland; Friends of Savigliano and of Flauto Magico onlus for the show of Guido Marangoni, 10-02-2019; “Compagnia dell’Idra”; Simone Bombardi and “Associazione Culturale Compagnia dell’Anello”, Forlì; “Parrocchia di Montespertoli” (Firenze). We are especially grateful to Medical Students Gerardo Lo Russo, Giovanni Lucertini, Andrea Pace, and Veronica Sandroni for their donations originated from the wonderful exhibition they designed about Dr Gian Carlo Rastelli (1933–1970), whose work led to cardiac surgery useful for children with trisomy 21. We are very grateful to all the other people that have very kindly contributed through individual donations to support this research, in particular: “Associazione Culturale SDR”, Castel Guelfo (BO); Martino Boni and Alma Shqalsi; Ernesto Bronzini; Giovanni Bubani; “Il Carro Soc. Coop. Soc.”, Paullo (MI); “Centro di Aiuto alla Vita Daniela Gulden”, Legnano (MI); Marcello Colombo and the group of whisky tasting: “Compagni di Classe 5a C di Anselmo”; Fam. Elena Gennari and Paolo D’Intino; Maddalena Gnappi; Carlo and Paola Gussoni; Vito La Monica; Matteo and Maddalena; parents of Luigi Fuoco; Maria Luisa Pensieri and Dante Costantini; Marina Peschiera; “Polisportiva Senigallia ASD”; Cesare Pozzoli and Paola Pasquin; Giuseppe and Nadia Rabaioli; “Scuola Primaria Bambini di Sarajevo”, Crespellano (BO); Guido Samarani and Carla Molinari Samarani; Antonella Zanotti and “Gruppo Arzdore”, Dozza (BO). V.G. thanks AIRC-FIRC (Fondazione Italiana per la Ricerca sul Cancro, Milan) for a 2-year fellowship for Italy (Project Code: 22576). The fellowships for A.P. and M.C. have been funded by donations from the Fondazione Umano Progresso and Matteo and Elisa Mele. The fellowships for F.A. and F.P., as well as the experimental activity of the Bologna group, have been funded by donations from several donors (acknowledged above).
Author contributions
F.A. performed the procedures necessary to treat and store the samples before the NMR analyses; V.G. acquired and assigned the NMR spectra and performed the statistical analyses; P.S., L.V., M.C.P. and P.T. conceived and designed the experiments; F.P., S.L., S.O. and R.V. administered the cognitive tests to the subjects with DS; C.Lo. and G.C. enrolled the subjects involved in the study; G.R., A.M. and A.F. collaborated for the enrollment of the subjects; E.C. and E.M. collaborated for the initial treatment of the samples; G.R., M.C., F.A., F.P., P.S. and A.P. contributed to data analysis; V.G., P.T. and C.Lu. drafted the NMR sections of the manuscript and contributed to the final discussion; M.C., G.R. and F.P. wrote the manuscript draft. M.C., A.P. and P.T. supervised the project. All authors critically reviewed and approved the final manuscript.
Data availability
The datasets generated and analyzed during the current study have been made available as Supplementary Dataset S1 (plasma metabolome and cognitive data).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Francesca Antonaros, Veronica Ghini and Francesca Pulina
Contributor Information
Paola Turano, Email: turano@cerm.unifi.it.
Allison Piovesan, Email: allison.piovesan2@unibo.it.
Supplementary information
is available for this paper at 10.1038/s41598-020-67195-z.
References
- 1.Lukowski AF, Milojevich HM, Eales L. Cognitive Functioning in Children with Down Syndrome: Current Knowledge and Future Directions. Adv. Child Dev. Behav. 2019;56:257–289. doi: 10.1016/bs.acdb.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 2.Lejeune J, Gauthier M, Turpin R. [Human chromosomes in tissue cultures] Comptes rendus hebdomadaires des seances de l’Academie des sciences. 1959;248:602–603. [PubMed] [Google Scholar]
- 3.Gardiner K, et al. Down syndrome: from understanding the neurobiology to therapy. J. Neurosci. 2010;30:14943–14945. doi: 10.1523/JNEUROSCI.3728-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pelleri MC, et al. Systematic reanalysis of partial trisomy 21 cases with or without Down syndrome suggests a small region on 21q22.13 as critical to the phenotype. Hum. Mol. Genet. 2016;25:2525–2538. doi: 10.1093/hmg/ddw116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pelleri MC, et al. Partial trisomy 21 map: Ten cases further supporting the highly restricted Down syndrome critical region (HR-DSCR) on human chromosome 21. Mol. Genet. Genomic Med. 2019;7:e797. doi: 10.1002/mgg3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caracausi M, et al. Plasma and urinary metabolomic profiles of Down syndrome correlate with alteration of mitochondrial metabolism. Sci. Rep. 2018;8:2977. doi: 10.1038/s41598-018-20834-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lejeune J. On the mechanism of mental deficiency in chromosomal diseases. Hereditas. 1977;86:9–14. doi: 10.1111/j.1601-5223.1977.tb01207.x. [DOI] [PubMed] [Google Scholar]
- 8.Volman MJ, Visser JJ, Lensvelt-Mulders GJ. Functional status in 5 to 7-year-old children with Down syndrome in relation to motor ability and performance mental ability. Disabil. Rehabil. 2007;29:25–31. doi: 10.1080/09638280600947617. [DOI] [PubMed] [Google Scholar]
- 9.Weijerman ME, de Winter JP. Clinical practice. The care of children with Down syndrome. Eur. J. Pediatr. 2010;169:1445–1452. doi: 10.1007/s00431-010-1253-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bull MJ. Health supervision for children with Down syndrome. Pediatrics. 2011;128:393–406. doi: 10.1542/peds.2011-1605. [DOI] [PubMed] [Google Scholar]
- 11.Karmiloff-Smith, A. et al. The importance of understanding individual differences in Down syndrome. F1000Research5 (2016). [DOI] [PMC free article] [PubMed]
- 12.Chapman RS, Hesketh LJ. Behavioral phenotype of individuals with Down syndrome. Ment Retard Dev Disabil Res Rev. 2000;6:84–95. doi: 10.1002/1098-2779(2000)6:2<84::AID-MRDD2>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 13.Lanfranchi S, Jerman O, Dal Pont E, Alberti A, Vianello R. Executive function in adolescents with Down Syndrome. J. Intellect. Disabil. Res. 2010;54:308–319. doi: 10.1111/j.1365-2788.2010.01262.x. [DOI] [PubMed] [Google Scholar]
- 14.Lee NR, et al. Caregiver report of executive functioning in a population-based sample of young children with Down syndrome. Am J Intellect Dev Disabil. 2011;116:290–304. doi: 10.1352/1944-7558-116.4.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marchal JP, et al. Growing up with Down syndrome: Development from 6 months to 10.7 years. Res. Dev. Disabil. 2016;59:437–450. doi: 10.1016/j.ridd.2016.09.019. [DOI] [PubMed] [Google Scholar]
- 16.Yang Y, Conners FA, Merrill EC. Visuo-spatial ability in individuals with Down syndrome: is it really a strength? Res. Dev. Disabil. 2014;35:1473–1500. doi: 10.1016/j.ridd.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fidler DJ. The Emerging Down Syndrome Behavioral Phenotype in Early Childhood - Implications for Practice. Infants & Young Children. 2005;18:86–103. doi: 10.1097/00001163-200507000-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salvi A, et al. Analysis of a nanoparticleenriched fraction of plasma reveals miRNA candidates for Down syndrome pathogenesis. Int. J. Mol. Med. 2019;43:2303–2318. doi: 10.3892/ijmm.2019.4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu X, et al. Acetate Production from Glucose and Coupling to Mitochondrial Metabolism in Mammals. Cell. 2018;175:502–513.e513. doi: 10.1016/j.cell.2018.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez-Reyes I, Chandel NS. Mitochondrial TCA cycle metabolites control physiology and disease. Nat Commun. 2020;11:102. doi: 10.1038/s41467-019-13668-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ducker GS, Rabinowitz JD. One-Carbon Metabolism in Health and Disease. Cell Metab. 2017;25:27–42. doi: 10.1016/j.cmet.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barclay CJ. Energy demand and supply in human skeletal muscle. J. Muscle Res. Cell Motil. 2017;38:143–155. doi: 10.1007/s10974-017-9467-7. [DOI] [PubMed] [Google Scholar]
- 23.Feaster WW, Kwok LW, Epstein CJ. Dosage effects for superoxide dismutase-1 in nucleated cells aneuploid for chromosome 21. Am. J. Hum. Genet. 1977;29:563–570. [PMC free article] [PubMed] [Google Scholar]
- 24.Chadefaux B, et al. Cystathionine beta synthase: gene dosage effect in trisomy 21. Biochem. Biophys. Res. Commun. 1985;128:40–44. doi: 10.1016/0006-291x(85)91641-9. [DOI] [PubMed] [Google Scholar]
- 25.Balcz, B., Kirchner, L., Cairns, N., Fountoulakis, M. & Lubec, G. Increased brain protein levels of carbonyl reductase and alcohol dehydrogenase in Down syndrome and Alzheimer’s disease. J. Neural Transm. Suppl., 193–201 (2001). [DOI] [PubMed]
- 26.Arai Y, Ijuin T, Takenawa T, Becker LE, Takashima S. Excessive expression of synaptojanin in brains with Down syndrome. Brain Dev. 2002;24:67–72. doi: 10.1016/s0387-7604(01)00405-3. [DOI] [PubMed] [Google Scholar]
- 27.Pelleri MC, et al. Integrated Quantitative Transcriptome Maps of Human Trisomy 21 Tissues and Cells. Front. Genet. 2018;9:125. doi: 10.3389/fgene.2018.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang P, et al. Clinical diagnosis and mutation analysis of four Chinese families with succinic semialdehyde dehydrogenase deficiency. BMC Med. Genet. 2019;20:88. doi: 10.1186/s12881-019-0821-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bubber P, Haroutunian V, Fisch G, Blass JP, Gibson GE. Mitochondrial abnormalities in Alzheimer brain: mechanistic implications. Ann. Neurol. 2005;57:695–703. doi: 10.1002/ana.20474. [DOI] [PubMed] [Google Scholar]
- 30.Naseri NN, et al. Novel Metabolic Abnormalities in the Tricarboxylic Acid Cycle in Peripheral Cells From Huntington’s Disease Patients. PLoS One. 2016;11:e0160384. doi: 10.1371/journal.pone.0160384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sperringer JE, Addington A, Hutson SM. Branched-Chain Amino Acids and Brain Metabolism. Neurochem. Res. 2017;42:1697–1709. doi: 10.1007/s11064-017-2261-5. [DOI] [PubMed] [Google Scholar]
- 32.Nelson, D. L., Cox, M. M. In Principles of Biochemistry (ed W. H. Freeman) Ch. 859–905, (macmillan education, 2017).
- 33.Gowda GA, Djukovic D. Overview of mass spectrometry-based metabolomics: opportunities and challenges. Methods Mol. Biol. 2014;1198:3–12. doi: 10.1007/978-1-4939-1258-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Powers RK, et al. Trisomy 21 activates the kynurenine pathway via increased dosage of interferon receptors. Nat Commun. 2019;10:4766. doi: 10.1038/s41467-019-12739-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schellenberger J, et al. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2.0. Nat. Protoc. 2011;6:1290–1307. doi: 10.1038/nprot.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ebrahim A, Lerman JA, Palsson BO, Hyduke DR. COBRApy: COnstraints-Based Reconstruction and Analysis for Python. BMC Syst. Biol. 2013;7:74. doi: 10.1186/1752-0509-7-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bernini P, et al. Standard operating procedures for pre-analytical handling of blood and urine for metabolomic studies and biobanks. J. Biomol. NMR. 2011;49:231–243. doi: 10.1007/s10858-011-9489-1. [DOI] [PubMed] [Google Scholar]
- 38.Ghini V, Quaglio D, Luchinat C, Turano P. NMR for sample quality assessment in metabolomics. N Biotechnol. 2019;52:25–34. doi: 10.1016/j.nbt.2019.04.004. [DOI] [PubMed] [Google Scholar]
- 39.CEN/TS. 16945 - Molecular in vitro diagnostic examinations — Specifications for pre-examination processes for metabolomics in urine, venous blood serum and plasma. (2016).
- 40.Takis, P. G., Ghini, V., Tenori, L., Turano, P. & Luchinat, C. Uniqueness of the NMR approach to metabolomics. TrAC Trends in Analytical Chemistry120 (2019).
- 41.Vignoli A, et al. High-Throughput Metabolomics by 1D NMR. Angew. Chem. Int. Ed. Engl. 2019;58:968–994. doi: 10.1002/anie.201804736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mckay RT. How the 1D-NOESY suppresses solvent signal in metabonomics NMR spectroscopy: an examination of the pulse sequence components and evolution. Concepts in Magnetic Resonance. 2011;38A:197–220. [Google Scholar]
- 43.Pulina, F., Vianello, R. & Lanfranchi, S. In State of the Art of Research on Down Syndrome Vol. 56 (ed S. Lanfranchi) 67–92 (Academic Press, 2019).
- 44.Green, E. et al. Griffiths scales of child development, third edition (Griffiths III) (2016).
- 45.Wechsler, D. W Preschool and Primary Scale of Intelligence-Third edition. WPPSI-III. (2002).
- 46.Beckonert O, et al. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat. Protoc. 2007;2:2692–2703. doi: 10.1038/nprot.2007.376. [DOI] [PubMed] [Google Scholar]
- 47.Emwas AH, et al. Recommended strategies for spectral processing and post-processing of 1D (1)H-NMR data of biofluids with a particular focus on urine. Metabolomics: Official journal of the Metabolomic Society. 2018;14:31. doi: 10.1007/s11306-018-1321-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wishart DS, et al. HMDB 3.0–The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41:D801–807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Psychogios N, et al. The human serum metabolome. PLoS One. 2011;6:e16957. doi: 10.1371/journal.pone.0016957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wishart DS. Quantitative metabolomics using NMR. TrAC Trends in Analytical Chemistry. 2008;27:228–237. [Google Scholar]
- 51.Benjamini Y, Hochberg Y. On the Adaptive Control of the False Discovery Rate in Multiple Testing With Independent Statistics. Journal of Educational and Behavioral Statistics. 2000;25:60–83. [Google Scholar]
- 52.Schober P, Boer C, Schwarte LA. Correlation Coefficients: Appropriate Use and Interpretation. Anesth. Analg. 2018;126:1763–1768. doi: 10.1213/ANE.0000000000002864. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed during the current study have been made available as Supplementary Dataset S1 (plasma metabolome and cognitive data).