Abstract
Developmental dyslexia (DD) is a neurodevelopmental condition with complex genetic mechanisms. A number of candidate genes have been identified, some of which are linked to neuronal development and migration and to ciliary functions. However, expression and regulation of these genes in human brain development and neuronal differentiation remain uncharted. Here, we used human long-term self-renewing neuroepithelial stem (lt-NES, here termed NES) cells derived from human induced pluripotent stem cells to study neuronal differentiation in vitro. We characterized gene expression changes during differentiation by using RNA sequencing and validated dynamics for selected genes by qRT-PCR. Interestingly, we found that genes related to cilia were significantly enriched among upregulated genes during differentiation, including genes linked to ciliopathies with neurodevelopmental phenotypes. We confirmed the presence of primary cilia throughout neuronal differentiation. Focusing on dyslexia candidate genes, 33 out of 50 DD candidate genes were detected in NES cells by RNA sequencing, and seven candidate genes were upregulated during differentiation to neurons, including DYX1C1 (DNAAF4), a highly replicated DD candidate gene. Our results suggest a role of ciliary genes in differentiating neuronal cells and show that NES cells provide a relevant human neuronal model to study ciliary and DD candidate genes.
Electronic supplementary material
The online version of this article (10.1007/s12035-020-01905-6) contains supplementary material, which is available to authorized users.
Keywords: Cilia, Ciliopathies, Reading disorder, Human neuroepithelial stem cells, RNA sequencing
Introduction
Developmental dyslexia (DD) is a learning disorder with a neurodevelopmental origin. It is the most common learning disorder present in about 5–10% of the population. Early studies in postmortem human brains have suggested underlying neuronal migration anomalies [1]. DD is highly heritable and many candidate genes have been proposed [2] (Suppl. Table 1). Some of the most genetically replicated DD candidate genes (DCGs), namely DYX1C1, DCDC2, and KIAA0319, have been implied in neuronal development and migration in rodents, supporting the early studies in human brains [3–5]. Interestingly, the same genes have been shown independently to have a role in ciliary biology [6–11]. Cilia are present on mammalian neuronal cells at different stages of development—in progenitor cells and post-mitotic neurons [12]. At present, little is known about their function in neuronal development and homeostasis, and their roles in neuronal proliferation, migration, and maturation are just beginning to be elucidated [13]. Many ciliopathies show neurologic symptoms, and cilia have been linked to neuropsychiatric disorders [14–18].
In contrast to the vast literature on genetic studies of DCGs, few functional studies have been carried out. Some studies in animal model systems and cell lines have addressed the regulation and functions of DYX1C1, DCDC2, and KIAA0319 (reviewed in [19]). However, their function in human neuronal cells and cilia is still unsettled. While studies in model systems provide valuable insights, given the human-specific phenotype of DD, it is important to address DCG regulation in a human neuronal system. However, a systematic assessment of DCGs in human neuronal development is so far lacking. To study human-specific gene regulatory events and neurodevelopmental disorders, modeling human brain development in vitro derived from induced pluripotent stem cells (iPSCs)/embryonic stem cells (ESCs) combined with transcriptomic characterization has become a crucial tool [20]. Human long-term self-renewing neuroepithelial stem (lt-NES, here termed NES) cells derived from human iPSCs (hiPSCs) can mimic human neuronal development in vitro. They resemble neuroepithelial cells in vivo, self-renew in the presence of fibroblast growth factor (FGF) and epidermal growth factor (EGF), and can differentiate into neuronal and glial cells [21–23]. They have been used as a model for neurodevelopmental processes and disorders [24, 25].
Here, we sought to map gene expression changes during early human neuronal development in vitro with a focus on DCG regulation. We monitored gene expression throughout differentiation from NES cells to neuronal cells by RNA-sequencing (RNA-seq) on bulk RNA samples. In addition, we characterized specifically the dynamics of DCG expression.
Materials and Methods
Cell Culture
The ethical guidelines for derivation of cell line AF22 were described previously [21]. Reprogramming of human cells was permitted by Regional ethical committee Stockholm (Dnr 2012/208–31/3). The derivation and culturing of NES cells (line AF22, derived from a healthy female person) were described previously [21, 25]. Briefly, NES cells were cultured in DMEM/F12+Glutamax supplemented with penicillin (100 U/ml), streptomycin (100 μg/ml), N2 (1:100), B27 (1:1000), FGF (10 ng/ml) (all from Life Technologies, Thermo Fisher Scientific, Carlsbad, CA, USA), and EGF (10 ng/ml; Peprotech, Rocky Hill, NJ, USA) in a 5% CO2 incubator. Half of the medium was changed daily, and cells were passaged at a ratio of 1:3 upon confluency. Plates were pre-coated using poly-ornithine (0.1 mg/ml; Sigma-Aldrich, St. Louis, MO, USA) and laminin (2 μg/ml; Sigma-Aldrich, L2020). For differentiation, cells were plated in complete medium for 2 days, then medium was changed to medium without growth factors FGF and EGF. After 1 week, medium was changed to a 1:1 mixture of DMEM/F12+Glutamax and Neurobasal (Life Technologies) containing N2 (1:200) and B27 (1:100). During differentiation, half of the medium was changed every 2 to 3 days containing laminin (1:1000).
RNA Sequencing
Total RNA was extracted using NucleoSpin RNA kit or NucleoSpin Triprep kit (Macherey-Nagel, Düren, Germany) according to the supplier’s instructions. RNA concentration was measured using Nanodrop ND-1000 and Qubit (Thermo Fisher Scientific). RNA integrity was analyzed by Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Three biological replicates were collected for each time point in two independent experiments (except for day 14 in experiment 1). We applied the STRT (single-cell tagged reverse transcription) RNA-seq protocol [26, 27] on total bulk RNA samples with the following modifications: 10 ng of high-quality total RNA was converted into cDNA, amplified and converted to form an Illumina-compatible library. ERCC92 spike-in was used for quality control of sequenced samples and normalization of all the endogenous genes [28]. ERCC spike-in mixture was diluted 1000× with water, and 1 μl was added to reverse transcriptase cDNA master mix. In total, 25 PCR cycles were used: 15 for the first, full cDNA amplification and additional 10 to amplify and introduce sequencing-required motifs. Ready library was sequenced on three lanes of Illumina HiSeq 2000 instrument using 60 bp single reads.
RNA-seq data have been deposited in the ArrayExpress database at EMBL-EBI (https://www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-7128.
RNA-Seq Data Analysis
Data processing of the RNA-seq STRT library was performed using the STRTprep pipeline (https://github.com/shka/STRTprep/tree/v3dev [27]). Total raw reads were filtered and demultiplexed into individual samples based on the sample-specific barcodes, and redundant reads were filtered out based on UMI (unique molecular identifier) sequences. After barcodes were trimmed, processed reads were aligned to the hg19 human reference genome, synthetic ERCC92 spike-in sequences, and human ribosomal DNA unit (GenBank: U13369) using TopHat v2.0.12 [29]. Uniquely mapped reads within the 5′-UTR or 500 bp upstream of Refseq protein-coding genes were counted, and read counts were normalized by the sum of spike-in reads in each sample. Differential expression was analyzed using the R package SAMstrt [28] based on the combination of false discovery rate (FDR) < 0.01 and Benjamini-Hochberg-adjusted p value of the degree of variation < 0.05 [27]. The heatmap was generated using the Ward clustering method with the Spearman correlation distance matrix based on the log10-normalized expression levels of the 2516 differentially expressed genes between day 0 and day 35. The minimum value but non-zero (4.30e-05) was added to all the normalized expression levels to avoid the logarithm of zero. For the RNA-seq vs. qRT-PCR correlation plots, a constant of 0.00001 was added to all the mean values of DCX to avoid the logarithm of zero. Enrichment analysis of tissue-specific expression (UniProt UP_TISSUE) and gene ontology (GO) terms were performed using the DAVID web tool (http://david.abcc.ncifcrf.gov/) [30, 31]. GO graphs were displayed using GraphPad Prism 7 software. The gene set enrichment analysis (GSEA) was performed with GSEA v3.0 (http://software.broadinstitute.org/gsea/) using GSEAPreranked tool [32]. A total of 11,180 genes, which were expressed in at least three of the day 0 and day 35 samples, except spike-ins, were preranked according to the combination of FDR and fold change of their expression level. This gene list was then compared with the gene sets of ciliary genes and dyslexia-candidate genes. The gene set of ciliary genes was composed of 302 genes obtained from The SYSCILIA Gold Standard v1 [33]. The gene set of DCGs is shown in Suppl. Table 1. The gene set of DCGs was generated with a literature search in PubMed, including linkage and association studies and translocations and deletions co-segregating with DD, excluding CNVs (accession date: 29/5/2018). Since we chose an inclusive, exploratory approach, non-replicated genes were also included. GSEA plots were generated using ReplotGSEA.R (https://github.com/PeeperLab/Rtoolbox/blob/master/R/ReplotGSEA.R) with some modifications. For the time course analysis, 5976 genes with adjusted p value of the degree of variation < 0.05 were analyzed. Among them, a total of 2303 genes highly correlated between independent experiments (Pearson’s correlation coefficient > 0.7) were classified into 4 clusters, based on k-means clustering determined by X-Means algorithm using the R package Weka [34].
Publicly Available Transcriptome Data Analysis
Two publicly available cap analysis of gene expression (CAGE) datasets were downloaded from the FANTOM (Functional annotation of the mammalian genome) 5 database (http://fantom.gsc.riken.jp/5/data/; [35]), and an RNA-seq dataset was downloaded from Gene Expression Omnibus (GEO) database [36] under the accession number GSE99469 [37]. (1) Human iPSCs to neurons by CAGE: six replicates of day 0 and six replicates of day 18 samples from 2 iPS cell lines (one from newborn male fibroblasts and another from 12-week gestation female fibroblasts) were analyzed. Promoters with over 3 counts per million (cpm) in at least 6 samples were selected. (2) Human ESCs (hESCs) to cardiomyocytes by CAGE: three replicates of day 0 and three replicates of day 12 samples from hESCs were analyzed. Promoters with over 3 cpm in at least 3 samples were selected. (3) Human iPSCs to kidney organoids by RNA-seq: three replicates of day 0 and six replicates of day 18 samples from a healthy female human iPSC line were analyzed. Genes with over 3 cpm in at least 3 samples were selected.
The read counts were normalized using the RLE method, and significantly upregulated genes or promoters were identified using the R (version 3.6.0) package “edgeR” (version 3.26.5) based on Benjamini-Hochberg-adjusted p value < 0.01 along with the log2-fold change > 1 in the differentiated samples against the day 0 samples. Significantly upregulated genes or official gene symbols extracted from the significantly upregulated promoters were then annotated with DAVID web tool (http://david.abcc.ncifcrf.gov/) for the GO term enrichment analysis. GO graphs were displayed using GraphPad Prism 7 software.
Quantitative Real-Time PCR
cDNA was synthesized with Maxima First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) using 500 ng of RNA. cDNA was diluted 1:5 and 2 μl of diluted cDNA was used per reaction. The analysis was performed on a 7500 Fast Real-Time PCR system (Applied Biosystems, Thermo Fisher Scientific). The reagents used for RT-qPCR were TaqMan fast Universal PCR Master Mix (Thermo Fisher Scientific) and Taqman probes (BBS2: Hs00230400_m1; DCDC2: Hs00393203_m1; DYX1C1: Hs00370049_m1; GAPDH: Hs02758991_g1; IFT57 (ESRRBL1): Hs00215973_m1; KIAA0319: Hs00207788_m1; TCTN2: Hs00430714_m1; TMEM231: Hs00226008_m1; TUBA1A: Hs00362387_m1) or FastStart Universal SYBR Green Master (Roche Diagnostics, Mannheim, Germany) and SYBR green primers (DCX_F: TTG CTG GCT GAC CTG ACG CG; DCX_R: GCT GCT AGC CAA GGA CTG GGG; GAPDH_F: CCA CAT CGC TCA GAC ACC AT; GAPDH_R: GCG CCC AAT ACG ACC AAA T; MAP2_F: AGG CAG AGA CAC AGG TGC TT; MAP2_R: GGG TTT GCT CCT AGG GTT TC; TUBB3_F: CCT ACT GCA TCG ACA ACG AG; TUBB3_R: CGA TAC CAG GTG GTT GAG GT). GAPDH was used as a control to normalize expression levels [21, 24]. Expression was compared relative to day 0 using the ΔΔCt method and relative expression levels were displayed as 2^- ΔΔCt [38]. The data were analyzed and displayed using Microsoft Excel and GraphPad Prism 7 softwares.
Immunocytochemistry
Immunocytochemistry was performed as described before [9, 25]. Cells were grown on glass coverslips and differentiated during 0, 7, 14, 21, 28, or 35 days as described above. Cells were fixed either with ice-cold methanol after 45 min incubation on ice or with 4% formaldehyde (Sigma-Aldrich) at room temperature, permeabilized and blocked with 0.05% PBST with 5% horse serum. Samples were incubated with primary antibody (mouse anti-acetylated alpha-tubulin, 1:5000, T7451, Sigma-Aldrich, RRID:AB_609894; rabbit anti-PCNT, 1:1000, HPA019887, Atlas antibodies, RRID: AB_1855080) overnight at 4 °C and with secondary antibody (donkey anti-rabbit Alexa 488, 1:1000, Thermo Fisher Scientific, RRID:AB_2535792; donkey anti-mouse Alexa 568, 1:1000, Thermo Fisher Scientific, RRID:AB_2534013) for 1 h at room temperature, then counterstained with DRAQ5 (1:1000, Cell Signaling Technology, Cambridge, UK) for 10 min at room temperature or with DAPI (1:1000, Sigma-Aldrich) for 1 min at room temperature. Samples were embedded in Prolong Gold antifade reagent (Thermo Fisher Scientific). Images were acquired with a Nikon A1R Ti confocal (Nikon Instruments, Inc., Melville, NY, USA) with a Plan Apo λ 60 × NA 1.4 objective with z-stack imaging mode. Images were processed using Nikon NIS-elements version 4.51 (Laboratory Imaging/Nikon), DRAQ5 was pseudocolored in blue, LUT were applied, and z-stacks were collapsed to maximum intensity projections. Images were converted to 8-bit RGB and subsequently arranged using Adobe Illustrator CS6.
Results
NES Cell Differentiation, Quality Control, and Global Transcriptomics Analysis
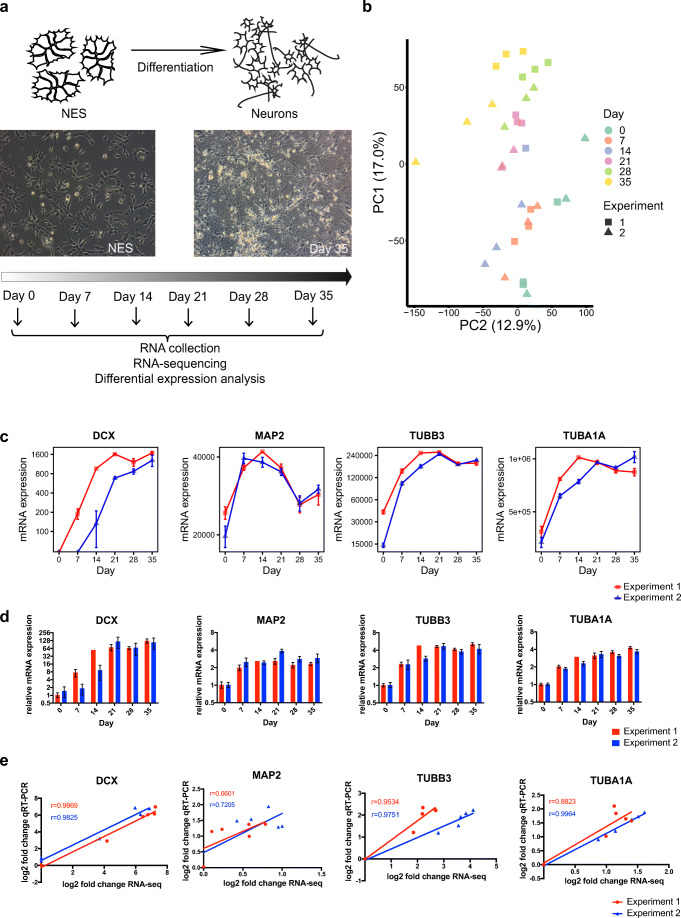
In order to model early human neuronal development, we used established protocols for NES cells (cell line AF22) [21]. We differentiated NES cells in two independent experiments towards neuronal cells during 35 days by undirected differentiation via removal of the growth factors EGF and FGF. To monitor gene expression at different stages of development, we sampled RNA at 0, 7, 14, 21, 28, and 35 days using three separately cultured replicates at each timepoint (Fig. 1a). We applied STRT RNA-seq on bulk RNA samples as a transcriptomics approach. On average, we obtained 3.7 million reads per sample, 90.6% of which were mapped to the human genome hg19 (Suppl. Table 2). First, we asked how similar the samples were according to their timepoints and experiment numbers to assess the robustness of our differentiation protocol. Sample classification by principal component analysis (PCA) of all detected genes showed that samples clustered together according to timepoints and across the two differentiation experiments (Fig. 1b).
Fig. 1.
Neuronal differentiation and quality control. a Two independent batches (Experiment 1 + Experiment 2) of NES cells were differentiated for 35 days, and RNA samples were collected at 0, 7, 14, 21, 28, and 35 days. b Principal component analysis of all the samples analyzed by RNA-seq. Dot colors represent differentiation days and dot shapes represent independent experiments. PC1: principal component 1, PC2: principal component 2. c Time course expression pattern of the neuronal markers DCX, MAP2, TUBB3, and TUBA1A measured by RNA-seq (red: Experiment1, blue: Experiment 2). Number of molecules calculated by multiplying the normalized read counts and spike-ins are shown. Mean ± SEM are displayed. d qRT-PCR showing upregulation of neuronal markers DCX, MAP2, TUBB3, and TUBA1A during differentiation. Mean ± SEM are displayed, red: Experiment 1, blue: Experiment 2. e Correlation plots of log2 fold change in RNA-seq vs. log2 fold change in qRT-PCR. r denotes Pearson correlation coefficient
To monitor neuronal differentiation, we tested the expression of neuronal markers. Indeed, we observed an upregulation of the established neuronal markers DCX, MAP2, TUBB3 [39], and TUBA1A (Fig. 1c). We confirmed the significant upregulation of these marker genes by using qRT-PCR (Fig. 1d, Suppl. Fig. 2). Comparison of the results revealed a high concordance between RNA-seq and qRT-PCR results (Fig. 1e). In summary, these results show that the experiment was reproducible and confirm the successful differentiation of NES cells to neuronal cells.
Characterization of Differentially Expressed Genes During Neuronal Differentiation
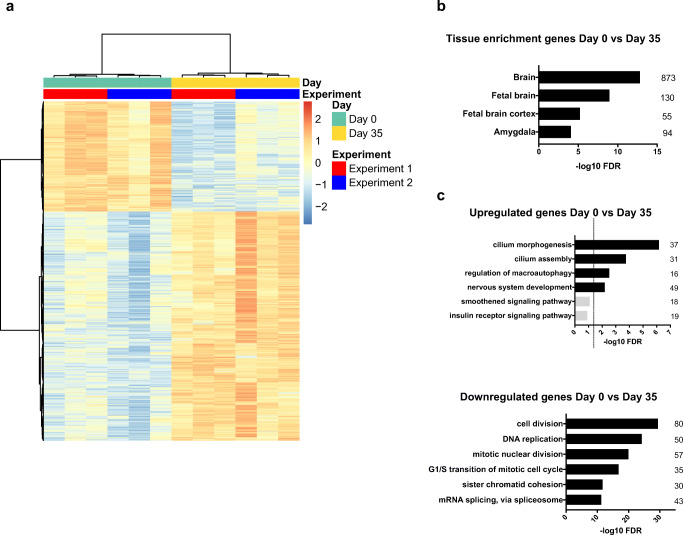
Next, we asked which genes were expressed in NES cells and which genes were differentially regulated by the maturation from NES cells to neurons. Based on the results of PCA analysis, we performed differential expression analysis between the extreme timepoints day 0 and day 35 to compare the most different expression profiles. We observed that 817 genes were significantly downregulated and 1699 genes were significantly upregulated between day 0 and day 35. Unsupervised hierarchical clustering revealed that day 0 samples clustered separately from day 35 samples and that samples clustered together depending on their respective experiment number (Fig. 2a). Enrichment analysis of the 1699 upregulated genes using UniProt tissue expression categories (UP_TISSUE) revealed that 873 genes overlapped with the brain category, 130 genes with the fetal brain category, 55 genes with the fetal brain cortex category, and 94 genes with the amygdala category, confirming the neuronal identity of the differentiated cell samples (Fig. 2b).
Fig. 2.
Differential gene expression analysis between day 0 and day 35. a Heatmap of differentially expressed genes between day 0 and day 35. All the genes at FDR < 0.01 are shown (817 downregulated, 1699 upregulated). b Enrichment analysis of the upregulated genes on UniProt tissue expression categories (UP_TISSUE). c Gene ontology term enrichment analysis of differentially expressed genes comparing day 0 and day 35. GO terms of six top ranked biological processes are shown. Terms with FDR < 0.05 are displayed in black and terms with FDR > 0.05 are displayed in gray. The dashed line indicates the limit of FDR = 0.05. The number of genes in each category is indicated on the right of the bars. FDR false discovery rate
To explore the function of the differentially regulated genes, we performed GO term enrichment analysis using the DAVID web tool [30, 31]. The six most significantly upregulated and downregulated GO terms are shown in Fig. 2c. As expected, we found neuronal development–related terms in the upregulated category and cell cycle–related terms in the downregulated category. Interestingly, we found cilia-related terms as the most significantly enriched upregulated terms (Fig. 2c). To make sure that the enrichment of ciliary genes is not a false positive signal due to a high number of tubulin genes upregulated in neurons, we examined the genes enriched in the cilium morphogenesis and cilium assembly categories. None of the genes in these categories were tubulin genes (Suppl. Table 3).
We then asked if the upregulation of cilia-related terms could also be observed in other neuronal cell lines. We performed GO analysis of the upregulated genes of iPSCs differentiated to neurons during 18 days using a publicly available cap analysis of gene expression (CAGE) dataset [35]. Indeed, we observed a strong enrichment of cilia-related GO terms, along with neuronal development-related GO terms (Suppl. Fig. 1a). Next, we asked if cilia gene upregulation can be observed as a general phenomenon in all cell types differentiating from PSCs and exiting the cell cycle. We therefore analyzed datasets from two other cell types differentiated from hESCs or hiPSCs during 12 and 18 days, respectively: cardiomyocytes [35] and kidney cells [37]. We observed some enrichment of cilia-related terms in kidney cells but not in cardiomyocytes (Suppl. Fig. 1b, c). In none of these tissues, the enrichment was as strong as for neurons. These results indicate that enrichment of cilia-related GO terms during differentiation could be specifically observed in neurons.
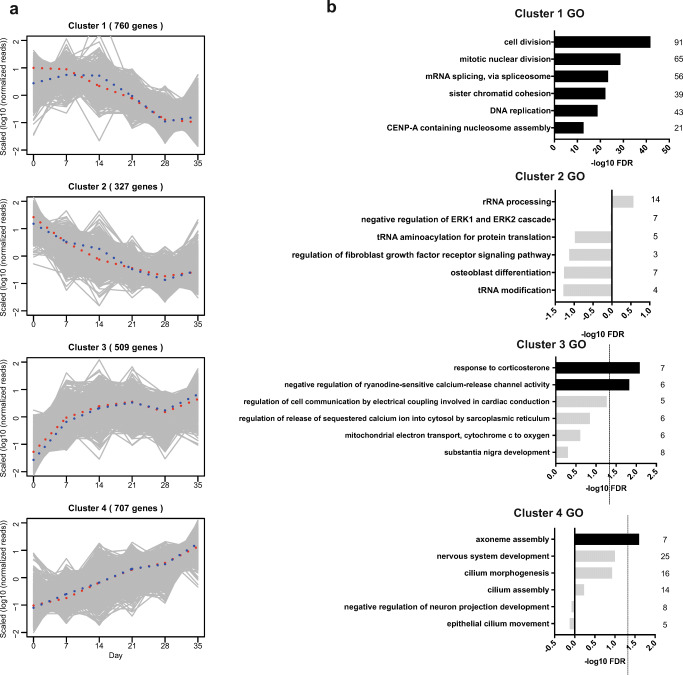
To further dissect the regulated gene classes, we performed clustering by time-course expression pattern analysis, revealing four different gene expression clusters (Fig. 3). Cluster 1 consisted of 760 genes, which are decreasing between day 7 and day 28 and cluster 2 comprised 327 monotonically decreasing genes. Cluster 3 contained 509 genes, which increase drastically in the beginning of differentiation, and cluster 4 consisted of 707 monotonically increasing genes (Fig. 3a). To determine the biological processes associated with the clusters, we performed GO term enrichment analysis. Cluster 1 was significantly enriched with cell cycle–related genes, further confirming the cell cycle exit of the cells after day 7 (Fig. 3b). Interestingly, cilia-related genes and nervous system development–related genes are in the same cluster (cluster 4), further indicating that they are correlated during neuronal differentiation (Fig. 3b). These results again suggest that ciliary genes might play a role during neuronal differentiation.
Fig. 3.
Clustering by time course gene expression analysis. a Clustering of all differentially expressed genes by time course expression pattern analysis resulting in clusters 1–4. b GO terms of biological processes associated to the clusters. GO terms of six top ranked biological processes are shown. Terms with FDR < 0.05 are displayed in black terms and terms with FDR > 0.05 are displayed in gray. The dashed line indicates the limit of FDR = 0.05. The number of genes in each category is indicated on the right of the bars. FDR false discovery rate
Genes Associated with Ciliopathies with Neurodevelopmental Phenotypes Are Upregulated During Neuronal Differentiation
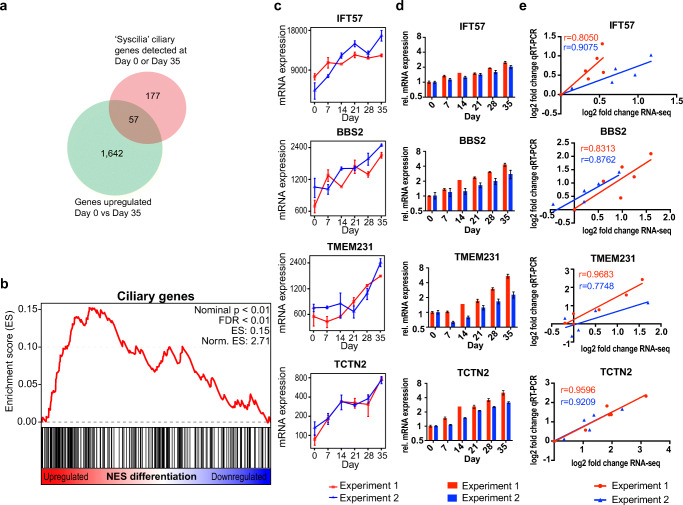
The occurrence of significant GO terms related to cilia incited us to investigate the ciliary genes more in detail. We used a conservative list of 302 bona fide ciliary genes (The SYSCILIA Gold Standard v1 [33]); the full list of ciliary genes is provided in Suppl. Table 4. We found that 234 of those ciliary genes were expressed either at day 0 or at day 35. By comparing day 0 to day 35, we observed that 57 ciliary genes out of 234 were among the 1699 short-listed upregulated genes at day 35 showing a significant enrichment of ciliary genes (p value = 0.0002; Fisher’s exact test) (Fig. 4a). Similarly, GSEA analysis ranking all 11,180 detected genes and comparing them to the 234 detected ciliary genes revealed a significant enrichment (FDR < 0.01) (Fig. 4b).
Fig. 4.
Time course expression pattern of ciliary genes. a Venn-diagram of all upregulated genes day 0 vs day 35 and all “Syscilia” ciliary genes detected at day 0 or 35. b Gene set enrichment analysis (GSEA) of “Syscilia” ciliary genes in all detected 11,180 genes. FDR false discovery rate, ES enrichment score. c RNA-seq time course progression plot of the ciliopathy-related genes IFT57, BBS2, TMEM231, TCTN2. d qRT-PCR showing upregulation of the dyslexia and ciliopathy-related genes IFT57, BBS2, TMEM231, TCTN2 during neuronal differentiation. e Correlation plots of log2 fold change in RNA-seq vs. log2 fold change in RT-qPCR. r denotes Pearson correlation coefficient. c, d Mean ± SEM are displayed. c, d, e red: Experiment 1, blue: Experiment 2. Norm. normalized, rel. relative
Many ciliopathies display neurodevelopmental phenotypes [40]. We therefore compared all the 57 upregulated ciliary genes to the known phenotypes in ciliopathies. Of the 57 upregulated ciliary genes between day 0 and day 35, 32 genes were associated with ciliopathies of which 15 display neurodevelopmental symptoms (Suppl. Table 5). Overall, these results point to a role of ciliary genes in neuronal differentiation.
The neurodevelopmental phenotypes present in many ciliopathies [14] and the enrichment in ciliary genes observed at day 35 prompted us to characterize the time course expression pattern of examples of upregulated ciliary genes that are mutated in ciliopathies with neurocognitive phenotypes. We examined the genes IFT57 (mutated in orofaciodigital syndrome (OFD, OMIM #311200)), BBS2 (Bardet-Biedl syndrome (BBS, OMIM #209900)), (TMEM231 (Meckel-Gruber syndrome (MKS, OMIM #249000)), TCTN2 (Joubert syndrome, (JBTS, OMIM #213300)) (Fig. 4c–e). Those genes were upregulated in the RNA-seq data (Fig. 4c) and qRT-PCR data (∆∆Ct day 0 vs day 35: p values IFT57, BBS2, TCTN2 < 0.0001; p value TMEM231 < 0.001 (Student’s t test) (Fig. 4d; Suppl. Fig. 2)). Correlation plots revealed a high concordance between RNA-seq and qRT-PCR results (Fig. 4e). These findings indicate that ciliopathy genes producing brain phenotypes when mutated are upregulated during human neuronal differentiation in vitro, suggesting human NES cells as a good model to study ciliopathies.
Individual Dyslexia Candidate Genes Are Upregulated During Neuronal Differentiation
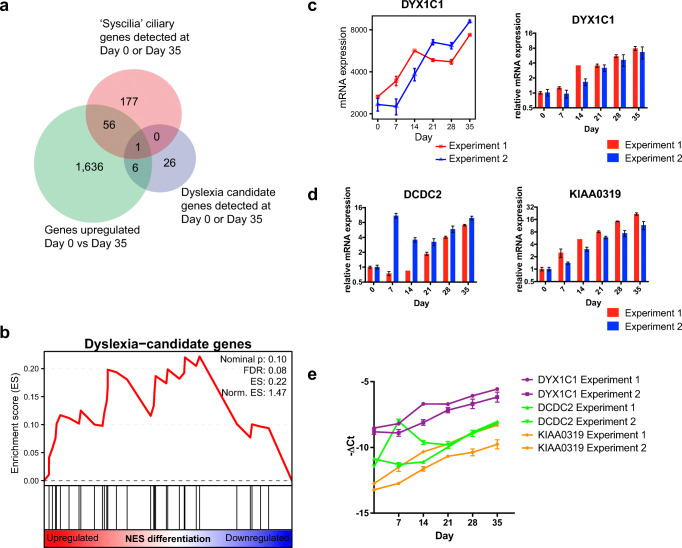
Next, we addressed the expression of DCGs. A number of DCGs have been shown to have a role in neuronal development and/or in cilia prompting us to ask whether they might have a role in NES cell differentiation [41, 42]. We therefore asked whether DCGs are expressed and differentially regulated during the differentiation from NES cells to neurons. While there is an abundant literature on genomic studies on DCGs, little is known on expression and function of these genes in human neuronal development. As our approach is exploratory, we chose to use an inclusive approach towards what constitutes a DCGs rather than focusing only on replicated genes that are few. We therefore used an inclusive list of 50 DCGs containing genes identified in families with dyslexia history as well as in specific population cohorts (Suppl. Table 1). Here, we found that thirty-three of these genes were expressed either at day 0 or at day 35 or both. Of those, seven DCGs were among the 1699 short-listed genes upregulated between day 0 and day 35 (CCDC136, COMT, DYX1C1, PRMT2, CCPG1, ZNF385D, GRIN2B) (Fig. 5a), indicating that there was no significant enrichment overall among the short-listed upregulated genes (p value = 0.33, Fisher’s exact test). Similarly, GSEA analysis ranking all 11,180 detected genes and comparing them to the 33 detected DCGs showed no significant enrichment (FDR = 0.08) (Fig. 5b). These findings lead to conclude that (1) many DCGs are expressed at some point during human neuronal differentiation; however, (2) there is no significant upregulation of DCGs as a group.
Fig. 5.
Time course expression pattern of dyslexia candidate genes. a Venn-diagram of all upregulated genes day 0 vs day 35, all DCGs detected at day 0 or 35 and all “Syscilia” ciliary genes detected at day 0 or 35. b Gene set enrichment analysis (GSEA) of DCGs in all detected 11,180 genes. FDR false discovery rate, ES enrichment score. cDYX1C1 expression measured by RNA-seq and qRT-PCR. d Relative expression of the DCGs DCDC2 and KIAA0319 assayed by qRT-PCR. e –ΔCt values by qRT-PCR of DYX1C1, DCDC2, and KIAA0319. c, d, e Mean ± SEM are displayed. c, d red: Experiment 1, blue: Experiment 2. Norm. normalized
Next, we examined the time course expression pattern of DCGs more in detail. For this purpose, we focused on a set of DCGs that has been highly replicated in genomic studies, namely DYX1C1, DCDC2, and KIAA0319 [43]. Interestingly, one of the most replicated DCGs—DYX1C1—was one of the most highly upregulated ciliary genes (Fig. 5c), which was confirmed by qRT-PCR (Fig. 5c; Suppl. Fig. 2) (p value ∆∆Ct day 0 vs day 35 < 0.0001; Student’s t test). Expression of the two other highly replicated DCGs, DCDC2 and KIAA0319, was not detected by RNA-seq, but their low-level expression was observed by qRT-PCR. Comparing their expression between day 0 and 35 by qRT-PCR revealed that they, too, were significantly upregulated during differentiation (p value ∆∆Ct day 0 vs day 35 < 0.0001; Student’s t test) (Fig. 5d; Suppl. Fig. 2). The Ct values for DCDC2 and KIAA0319 were much higher than the Ct values for DYX1C1 (Fig. 5e). These results underscore the importance of DYX1C1 in neuronal differentiation, whereas DCDC2 and KIAA0319 are expressed at a lower level or perhaps in specific cell types.
Primary Cilia Are Present Throughout Differentiation of NES Cells
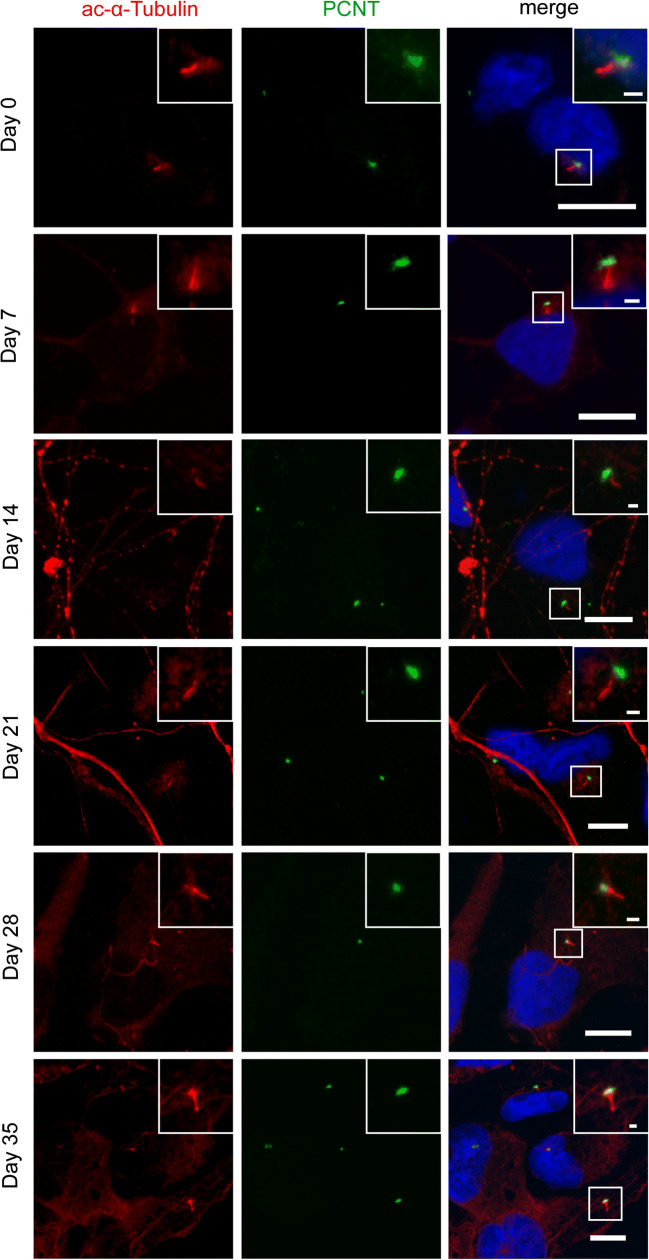
Based on our transcriptomics results and on earlier studies describing the presence of cilia on neural progenitor cells, we asked whether primary cilia are present on the cell surface during differentiation from NES cells to neuronal cells. We therefore stained NES cells and differentiating neurons for the ciliary marker acetylated-α-tubulin [44], in combination with the centrosomal marker pericentrin (PCNT) (Fig. 6). Interestingly, cilia were present on the cell surface during all examined differentiation stages. The cilia were located on the cell body. This finding confirms that NES cells display cilia throughout development.
Fig. 6.
Cilia are present throughout neuronal differentiation. NES cells were plated on glass coverslips and differentiated for the indicated number of days, then fixed and stained for the ciliary marker acetylated-α-Tubulin and the centrosomal marker pericentrin (PCNT). Scale bars = 10 μm, scale bars insets = 1 μm. Nuclei were counterstained with DAPI or DRAQ5
Discussion
Cilia are present on the surface of most mammalian cells and have important roles in processes such as cell to cell communication [45]. However, little is known about distribution and function of primary cilia in human neuronal cells and the mechanisms by which ciliary dysfunctions lead to neurodevelopmental defects are just beginning to emerge [46]. DD is a neuropsychiatric disorder frequently associated with ciliary genes [47].
Here, we studied gene expression changes in general and of DCGs in particular during human neuronal differentiation by combining long-term self-renewing NES cells with a transcriptomics approach. Surprisingly, we found that some of the most highly upregulated GO terms were related to cilia growth. The pronounced upregulation of ciliary genes observed may be explained by more cells exiting the cell cycle, entering G0, and undergoing ciliogenesis upon removal of growth factors. However, the specific enrichment of ciliary GO terms in neurons compared with other cell types suggests that ciliary genes have a more prominent role during neuronal differentiation in particular. The upregulation of ciliary genes during human neuronal differentiation in vitro has not been widely reported yet, but Van de Leemput et al. have observed a high enrichment of cilia-related GO terms in a human ESC neuronal differentiation model consistent with our results [48]. It is known that cilia play a role in neuronal development: In early brain development, the action of pathways such as SHH, WNT, PCP, and PDGF via the primary cilium are central to brain patterning and forebrain development [13]. Signaling via the cilium also plays a role in neuronal maturation [13]. Likely, cilia regulate the balance of neurogenesis via providing access to different signaling molecules in early and later stages of neuronal development [49]. In mouse ESCs, the ciliary genes Tmem67, Ahi1, and Ofd1 are crucial for neuronal differentiation [44, 50]. It is likely that cilia assume similar roles in the human brain.
We describe the presence of cilia on human NES and differentiating neuronal cells. This is in accordance with previous studies describing the occurrence of cilia in human neural progenitor cells (NPCs), differentiating cells in rosettes and organoids [51–53], hiPSC-derived neurons [54], and in the adult brain [55]. The hedgehog effector smoothened localizes to primary cilia in NPCs in maturing rosettes indicating that similar processes might be active in human neural cells as in rodents [51]. Cilia trigger differentiation of NPCs and might play a role in asymmetric cell division associated with neuronal differentiation [53]. The location of the cilium at the cell soma in our study is in accordance with previous observations, and it has been suggested that this localization optimizes the concentration of signaling molecules [56]. It remains to be determined whether ciliary length or number increases during differentiation.
For our study, we focused on the early neuronal development and therefore analyzed days 0 to 35. However, our data showed that the expression of most cilia genes did not yet plateau at 35 days. It would be highly interesting to analyze later timepoints, especially bearing in mind the late embryonic and postnatal growth and maturation of neuronal cilia described in rodents by Arellano and colleagues [57].
Many ciliopathies display neurodevelopmental symptoms such as mental retardation in JBTS, BBS, MKS, and OFD. More generally, 77 ciliary genes have been associated with neurodevelopmental or neurobehavioral defects [58]. In addition to their relevance in ciliopathies, ciliary genes have been associated with neuropsychiatric disorders [15–18]. One of the neurodevelopmental disorders frequently associated to ciliary genes is DD [42, 47]. It is remarkable that 33 genes out of an inclusive list of 50 DCGs were detected at day 0, day 35, or both, while seven genes were significantly upregulated during differentiation. The list of candidate genes was however not significantly enriched during neuronal differentiation using the stringent FDR cutoff of 0.05 in GSEA—yet, this does not exclude their possible roles as dyslexia candidates. As our approach was very inclusive, false-positive genes might dilute the result. It is also conceivable that these genes might function in different tissues—for example glial cells—or exert their impact on the brain in a paracrine mode. Indeed, we previously observed strong expression of DYX1C1 in glial cells, especially astrocytes, in the FANTOM5 expression database [35]. In addition, other lines of research have pointed into the direction that glial cells might play a role in DD. For example, certain polymorphisms in the dyslexia candidate genes DYX1C1, DCDC2, and KIAA0319 correlate with white matter density in the brain of children [59]. Recently, protocols to differentiate NES cells into the glial lineage have been developed, offering a way to address this question in the future [23, 60]. We further focused our analysis on a group of DCGs highly replicated in genetic studies and previously linked to neuronal development and cilia. Our results might facilitate to prioritize the candidates involved in neuronal differentiation. DYX1C1, the first identified DCG, has been replicated in many but not in all studies [61, 62]. It has previously been shown to act in neuronal migration [3, 63], learning, memory, and behavior [64] and later has been characterized as an axonemal dynein assembly factor [7]. Interestingly, the expression of DYX1C1 is highly upregulated during neuronal differentiation thereby differing strongly from the expression patterns of DNAAF1–3 that are absent or weakly expressed. These results suggest that DYX1C1 indeed has functions other than axonemal dynein assembly, consistent with previous reports on neuronal migration and development.
DCDC2 and KIAA0319—two DCGs replicated in many but not all studies—were previously associated with neuronal migration and cilia. Those genes were not detected in our RNA-seq approach, suggesting a low expression level. For DCDC2, this is consistent with human brain expression data from FANTOM5 and Allen Brain Atlas [35, 65]. Dcdc2 has been shown to have a role in neuronal migration and in behavior and learning [66] and localizes to rat hippocampal neuronal cilia [4, 8]. Certain variants in DCDC2 have been associated with gray and white matter changes [59, 67, 68]. Most likely, DCDC2 plays a role in neural development but may have a more restricted role than, e.g., DYX1C1, deserving further study.
Interestingly, while KIAA0319 is highly expressed in developing brain tissue [5, 65], it is lowly expressed in iPSC-derived neuronal cells in vitro [35], consistent with the observation in our in vitro model, implying that iPSC-derived neural cells might not be an ideal model to study KIAA0319. In vivo, KIAA0319 has been implicated in neuronal migration and axon regeneration [5, 69].Whether it is involved in cilia remains unclear—it has been detected as upregulated in ciliated tissues, and it is interesting to note that its protein possesses five PKD domains [11, 70, 71].
We here described the regulation of ciliary genes and investigated the expression of DCGs in a human neuronal in vitro model. The present findings should have implications for future work on ciliopathies and DD and prompt more studies on the neurodevelopmental roles of ciliary genes in human. Due to the immediate clinical relevance, a more thorough understanding of the functions of ciliary genes in human neurons is highly relevant. While our work provides a resource using a transcriptomics approach, it also sets the stage for future studies on patient-derived iPS/NES cells and modeling of neurodevelopmental disorders connected to cilia. While we do not provide evidence for a direct causal connection of ciliary genes and DCGs, we for the first time provide a systematic approach of their expression patterns in a human neuronal in vitro model and show that human NES cells provide a valid model to study ciliopathies or DCGs. Future functional studies should address the molecular mechanisms underlying the involvement of ciliary genes and DCGs in human brain development.
Electronic supplementary material
(DOC 697 kb)
Acknowledgments
We thank Ingegerd Fransson and Elisabet Einarsdottir for their help with sample handling and José Inzunza for providing cell laboratory facilities. We thank Kristiina Tammimies for critical reading of the manuscript.
Abbreviations
- BBS
Bardet-Biedl syndrome
- CAGE
Cap analysis of gene expression
- DAVID
Database for Annotation, Visualization and Integrated Discovery
- DCDC2
Doublecortin domain containing 2
- DCG
Dyslexia candidate gene
- DCX
Doublecortin
- DD
Developmental dyslexia
- DNAAF
Dynein axonemal assembly factor
- DYX1C1
Dyslexia susceptibility 1 candidate 1
- EGF
Epidermal growth factor
- FANTOM
Functional annotation of the mammalian genome
- FDR
False discovery rate
- FGF
Fibroblast growth factor
- GEO
Gene Expression Omnibus
- GO
Gene ontology
- GPCR
G protein coupled receptor
- GSEA
Gene set enrichment analysis
- hESC
Human embryonic stem cell
- hiPSC
Human induced pluripotent stem cell
- IFT57
Intraflagellar transport 57
- JBTS
Joubert syndrome
- MAP2
Microtubule-associated protein 2
- MKS
Meckel-Gruber syndrome
- NES cells
Long-term self-renewing neuroepithelial stem cells
- NPC
Neural progenitor cell
- OFD
Orofaciodigital syndrome
- PCA
Principal component analysis
- PCNT
Pericentrin
- STRT
Single cell tagged reverse transcription
- TCTN2
Tectonic family member 2
- TMEM231
Transmembrane protein 231
- TUBA1A
Tubulin α1α
- TUBB3
Tubulin β3
- UMI
Unique molecular identifier
Author Contributions
AB performed wet-lab experiments. AF provided NES cell line and expertise for NES cell culture. KK performed RNA sequencing methods. MY and SK, respectively, performed and supervised the bioinformatics analyses. AB and MY prepared figures and wrote the manuscript. AB, ITP, and JK conceived and ITP and JK supervised the project. All authors read and approved the final manuscript.
Funding Information
Open access funding provided by Karolinska Institutet. This study was supported by the Swedish Research Council (VR; JK), the Swedish Brain Foundation (Hjärnfonden; JK), and Sigrid Jusélius Foundation (JK). MY was supported by the Karolinska Institutet Research Foundation, the Scandinavia-Japan Sasakawa Foundation, the Japan Eye Bank Association, the Astellas Foundation for Research on Metabolic Disorders, and the Japan Society for the Promotion of Science Overseas Research Fellowships. This study was performed, in part, at the Live Cell Imaging Unit/Nikon Center of Excellence at the KI Department of Biosciences and Nutrition, which is supported by grants from the Knut and Alice Wallenberg Foundation, the Swedish Research Council, the KI Center for Innovative Medicine, and the Jonasson donation to the School of Technology and Health, Royal Institute of Technology. The computations were performed on resources provided by the Swedish National Infrastructure for Computing (SNIC) through the Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) under Project b2014069.
Compliance with Ethical Standards
Ethics Statement
The ethical guidelines for derivation of cell line AF22 were described previously [21]. Reprogramming of human cells was permitted by Regional ethical committee Stockholm (Dnr 2012/208–31/3).
Conflict of Interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Andrea Bieder and Masahito Yoshihara contributed equally to this work.
Contributor Information
Andrea Bieder, Email: andrea.bieder@ki.se.
Juha Kere, Email: juha.kere@ki.se.
References
- 1.Galaburda AM, Sherman GF, Rosen GD, Aboitiz F, Geschwind N. Developmental dyslexia: four consecutive patients with cortical anomalies. Ann Neurol. 1985;18(2):222–233. doi: 10.1002/ana.410180210. [DOI] [PubMed] [Google Scholar]
- 2.Kere J. The molecular genetics and neurobiology of developmental dyslexia as model of a complex phenotype. Biochem Biophys Res Commun. 2014;452(2):236–243. doi: 10.1016/j.bbrc.2014.07.102. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y, Paramasivam M, Thomas A, Bai J, Kaminen-Ahola N, Kere J, Voskuil J, Rosen GD, Galaburda AM, Loturco JJ. DYX1C1 functions in neuronal migration in developing neocortex. Neuroscience. 2006;143(2):515–522. doi: 10.1016/j.neuroscience.2006.08.022. [DOI] [PubMed] [Google Scholar]
- 4.Meng H, Smith SD, Hager K, Held M, Liu J, Olson RK, Pennington BF, DeFries JC, Gelernter J, O'Reilly-Pol T, Somlo S, Skudlarski P, Shaywitz SE, Shaywitz BA, Marchione K, Wang Y, Paramasivam M, LoTurco JJ, Page GP, Gruen JR. DCDC2 is associated with reading disability and modulates neuronal development in the brain. Proc Natl Acad Sci U S A. 2005;102(47):17053–17058. doi: 10.1073/pnas.0508591102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paracchini S, Thomas A, Castro S, Lai C, Paramasivam M, Wang Y, Keating BJ, Taylor JM, Hacking DF, Scerri T, Francks C, Richardson AJ, Wade-Martins R, Stein JF, Knight JC, Copp AJ, Loturco J, Monaco AP. The chromosome 6p22 haplotype associated with dyslexia reduces the expression of KIAA0319, a novel gene involved in neuronal migration. Hum Mol Genet. 2006;15(10):1659–1666. doi: 10.1093/hmg/ddl089. [DOI] [PubMed] [Google Scholar]
- 6.Chandrasekar G, Vesterlund L, Hultenby K, Tapia-Paez I, Kere J. The zebrafish orthologue of the dyslexia candidate gene DYX1C1 is essential for cilia growth and function. PLoS One. 2013;8(5):e63123. doi: 10.1371/journal.pone.0063123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tarkar A, Loges NT, Slagle CE, Francis R, Dougherty GW, Tamayo JV, Shook B, Cantino M et al (2013) DYX1C1 is required for axonemal dynein assembly and ciliary motility. 45(9):995–1003. 10.1038/ng.2707 [DOI] [PMC free article] [PubMed]
- 8.Massinen S, Hokkanen ME, Matsson H, Tammimies K, Tapia-Paez I, Dahlstrom-Heuser V, Kuja-Panula J, Burghoorn J, Jeppsson KE, Swoboda P, Peyrard-Janvid M, Toftgard R, Castren E, Kere J. Increased expression of the dyslexia candidate gene DCDC2 affects length and signaling of primary cilia in neurons. PLoS One. 2011;6(6):e20580. doi: 10.1371/journal.pone.0020580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tammimies K, Bieder A, Lauter G, Sugiaman-Trapman D, Torchet R, Hokkanen ME, Burghoorn J, Castren E, Kere J, Tapia-Paez I, Swoboda P. Ciliary dyslexia candidate genes DYX1C1 and DCDC2 are regulated by Regulatory Factor X (RFX) transcription factors through X-box promoter motifs. FASEB J. 2016;30(10):3578–3587. doi: 10.1096/fj.201500124RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schueler M, Braun DA, Chandrasekar G, Gee HY, Klasson TD, Halbritter J, Bieder A, Porath JD, Airik R, Zhou W, LoTurco JJ, Che A, Otto EA, Bockenhauer D, Sebire NJ, Honzik T, Harris PC, Koon SJ, Gunay-Aygun M, Saunier S, Zerres K, Bruechle NO, Drenth JP, Pelletier L, Tapia-Paez I, Lifton RP, Giles RH, Kere J, Hildebrandt F. DCDC2 mutations cause a renal-hepatic ciliopathy by disrupting Wnt signaling. Am J Hum Genet. 2015;96(1):81–92. doi: 10.1016/j.ajhg.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoh RA, Stowe TR, Turk E, Stearns T. Transcriptional program of ciliated epithelial cells reveals new cilium and centrosome components and links to human disease. PLoS One. 2012;7(12):e52166. doi: 10.1371/journal.pone.0052166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuchs JL, Schwark HD. Neuronal primary cilia: A review. Cell Biol Int. 2004;28(2):111–118. doi: 10.1016/j.cellbi.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 13.Sarkisian MR, Guadiana SM. Influences of primary cilia on cortical morphogenesis and neuronal subtype maturation. Neuroscientist. 2015;21(2):136–151. doi: 10.1177/1073858414531074. [DOI] [PubMed] [Google Scholar]
- 14.Valente EM, Rosti RO, Gibbs E, Gleeson JG. Primary cilia in neurodevelopmental disorders. Nat Rev Neurol. 2014;10(1):27–36. doi: 10.1038/nrneurol.2013.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trulioff A, Ermakov A, Malashichev Y (2017) Primary cilia as a possible link between left-right asymmetry and neurodevelopmental diseases. Genes 8(2). 10.3390/genes8020048 [DOI] [PMC free article] [PubMed]
- 16.Marley A, von Zastrow M. A simple cell-based assay reveals that diverse neuropsychiatric risk genes converge on primary cilia. PLoS One. 2012;7(10):e46647. doi: 10.1371/journal.pone.0046647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Munoz-Estrada J, Lora-Castellanos A, Meza I, Alarcon Elizalde S, Benitez-King G. Primary cilia formation is diminished in schizophrenia and bipolar disorder: a possible marker for these psychiatric diseases. Schizophr Res. 2018;195:412–420. doi: 10.1016/j.schres.2017.08.055. [DOI] [PubMed] [Google Scholar]
- 18.Alvarez Retuerto AI, Cantor RM, Gleeson JG, Ustaszewska A, Schackwitz WS, Pennacchio LA, Geschwind DH. Association of common variants in the Joubert syndrome gene (AHI1) with autism. Hum Mol Genet. 2008;17(24):3887–3896. doi: 10.1093/hmg/ddn291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guidi LG, Velayos-Baeza A, Martinez-Garay I, Monaco AP, Paracchini S, Bishop DVM, Molnar Z. The neuronal migration hypothesis of dyslexia: a critical evaluation 30 years on. Eur J Neurosci. 2018;48(10):3212–3233. doi: 10.1111/ejn.14149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin M, Lachman HM, Zheng D. Transcriptomics analysis of iPSC-derived neurons and modeling of neuropsychiatric disorders. Mol Cell Neurosci. 2016;73:32–42. doi: 10.1016/j.mcn.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falk A, Koch P, Kesavan J, Takashima Y, Ladewig J, Alexander M, Wiskow O, Tailor J, Trotter M, Pollard S, Smith A, Brustle O. Capture of neuroepithelial-like stem cells from pluripotent stem cells provides a versatile system for in vitro production of human neurons. PLoS One. 2012;7(1):e29597. doi: 10.1371/journal.pone.0029597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tailor J, Kittappa R, Leto K, Gates M, Borel M, Paulsen O, Spitzer S, Karadottir RT, Rossi F, Falk A, Smith A. Stem cells expanded from the human embryonic hindbrain stably retain regional specification and high neurogenic potency. J Neurosci. 2013;33(30):12407–12422. doi: 10.1523/jneurosci.0130-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lundin A, Delsing L, Clausen M, Ricchiuto P, Sanchez J, Sabirsh A, Ding M, Synnergren J, Zetterberg H, Brolen G, Hicks R, Herland A, Falk A. Human iPS-derived astroglia from a stable neural precursor state show improved functionality compared with conventional astrocytic models. Stem cell reports. 2018;10(3):1030–1045. doi: 10.1016/j.stemcr.2018.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shahsavani M, Pronk RJ, Falk R, Lam M, Moslem M, Linker SB, Salma J, Day K, Schuster J, Anderlid BM, Dahl N, Gage FH, Falk A. An in vitro model of lissencephaly: expanding the role of DCX during neurogenesis. Mol Psychiatry. 2017;23:1674–1684. doi: 10.1038/mp.2017.175. [DOI] [PubMed] [Google Scholar]
- 25.Yu NY, Bieder A, Raman A, Mileti E, Katayama S, Einarsdottir E, Fredholm BB, Falk A, Tapia-Paez I, Daub CO, Kere J. Acute doses of caffeine shift nervous system cell expression profiles toward promotion of neuronal projection growth. Sci Rep. 2017;7(1):11458. doi: 10.1038/s41598-017-11574-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Islam S, Kjallquist U, Moliner A, Zajac P, Fan JB, Lonnerberg P, Linnarsson S. Highly multiplexed and strand-specific single-cell RNA 5′ end sequencing. Nat Protoc. 2012;7(5):813–828. doi: 10.1038/nprot.2012.022. [DOI] [PubMed] [Google Scholar]
- 27.Krjutskov K, Katayama S, Saare M, Vera-Rodriguez M, Lubenets D, Samuel K, Laisk-Podar T, Teder H, Einarsdottir E, Salumets A, Kere J. Single-cell transcriptome analysis of endometrial tissue. Hum Reprod. 2016;31(4):844–853. doi: 10.1093/humrep/dew008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katayama S, Tohonen V, Linnarsson S, Kere J. SAMstrt: statistical test for differential expression in single-cell transcriptome with spike-in normalization. Bioinformatics. 2013;29(22):2943–2945. doi: 10.1093/bioinformatics/btt511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang d W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang d W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 32.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Dam TJ, Wheway G, Slaats GG, Huynen MA, Giles RH. The SYSCILIA gold standard (SCGSv1) of known ciliary components and its applications within a systems biology consortium. Cilia. 2013;2(1):7. doi: 10.1186/2046-2530-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hornik K, Buchta C, Zeileis A. Open-source machine learning: R meets Weka. Comput Stat. 2009;24(2):225–232. doi: 10.1007/s00180-008-0119-7. [DOI] [Google Scholar]
- 35.Forrest AR, Kawaji H, Rehli M, Baillie JK, de Hoon MJ, Haberle V, Lassmann T, Kulakovskiy IV, Lizio M, Itoh M, Andersson R, Mungall CJ, Meehan TF, Schmeier S, Bertin N, Jorgensen M, Dimont E, Arner E, Schmidl C, Schaefer U, Medvedeva YA, Plessy C, Vitezic M, Severin J, Semple C, Ishizu Y, Young RS, Francescatto M, Alam I, Albanese D, Altschuler GM, Arakawa T, Archer JA, Arner P, Babina M, Rennie S, Balwierz PJ, Beckhouse AG, Pradhan-Bhatt S, Blake JA, Blumenthal A, Bodega B, Bonetti A, Briggs J, Brombacher F, Burroughs AM, Califano A, Cannistraci CV, Carbajo D, Chen Y, Chierici M, Ciani Y, Clevers HC, Dalla E, Davis CA, Detmar M, Diehl AD, Dohi T, Drablos F, Edge AS, Edinger M, Ekwall K, Endoh M, Enomoto H, Fagiolini M, Fairbairn L, Fang H, Farach-Carson MC, Faulkner GJ, Favorov AV, Fisher ME, Frith MC, Fujita R, Fukuda S, Furlanello C, Furino M, Furusawa J, Geijtenbeek TB, Gibson AP, Gingeras T, Goldowitz D, Gough J, Guhl S, Guler R, Gustincich S, Ha TJ, Hamaguchi M, Hara M, Harbers M, Harshbarger J, Hasegawa A, Hasegawa Y, Hashimoto T, Herlyn M, Hitchens KJ, Ho Sui SJ, Hofmann OM, Hoof I, Hori F, Huminiecki L, Iida K, Ikawa T, Jankovic BR, Jia H, Joshi A, Jurman G, Kaczkowski B, Kai C, Kaida K, Kaiho A, Kajiyama K, Kanamori-Katayama M, Kasianov AS, Kasukawa T, Katayama S, Kato S, Kawaguchi S, Kawamoto H, Kawamura YI, Kawashima T, Kempfle JS, Kenna TJ, Kere J, Khachigian LM, Kitamura T, Klinken SP, Knox AJ, Kojima M, Kojima S, Kondo N, Koseki H, Koyasu S, Krampitz S, Kubosaki A, Kwon AT, Laros JF, Lee W, Lennartsson A, Li K, Lilje B, Lipovich L, Mackay-Sim A, Manabe R, Mar JC, Marchand B, Mathelier A, Mejhert N, Meynert A, Mizuno Y, de Lima Morais DA, Morikawa H, Morimoto M, Moro K, Motakis E, Motohashi H, Mummery CL, Murata M, Nagao-Sato S, Nakachi Y, Nakahara F, Nakamura T, Nakamura Y, Nakazato K, van Nimwegen E, Ninomiya N, Nishiyori H, Noma S, Noma S, Noazaki T, Ogishima S, Ohkura N, Ohimiya H, Ohno H, Ohshima M, Okada-Hatakeyama M, Okazaki Y, Orlando V, Ovchinnikov DA, Pain A, Passier R, Patrikakis M, Persson H, Piazza S, Prendergast JG, Rackham OJ, Ramilowski JA, Rashid M, Ravasi T, Rizzu P, Roncador M, Roy S, Rye MB, Saijyo E, Sajantila A, Saka A, Sakaguchi S, Sakai M, Sato H, Savvi S, Saxena A, Schneider C, Schultes EA, Schulze-Tanzil GG, Schwegmann A, Sengstag T, Sheng G, Shimoji H, Shimoni Y, Shin JW, Simon C, Sugiyama D, Sugiyama T, Suzuki M, Suzuki N, Swoboda RK, t Hoen PA, Tagami M, Takahashi N, Takai J, Tanaka H, Tatsukawa H, Tatum Z, Thompson M, Toyodo H, Toyoda T, Valen E, van de Wetering M, van den Berg LM, Verado R, Vijayan D, Vorontsov IE, Wasserman WW, Watanabe S, Wells CA, Winteringham LN, Wolvetang E, Wood EJ, Yamaguchi Y, Yamamoto M, Yoneda M, Yonekura Y, Yoshida S, Zabierowski SE, Zhang PG, Zhao X, Zucchelli S, Summers KM, Suzuki H, Daub CO, Kawai J, Heutink P, Hide W, Freeman TC, Lenhard B, Bajic VB, Taylor MS, Makeev VJ, Sandelin A, Hume DA, Carninci P, Hayashizaki Y. A promoter-level mammalian expression atlas. Nature. 2014;507(7493):462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phipson B, Er PX, Combes AN, Forbes TA, Howden SE, Zappia L, Yen HJ, Lawlor KT, Hale LJ, Sun J, Wolvetang E, Takasato M, Oshlack A, Little MH. Evaluation of variability in human kidney organoids. Nat Methods. 2019;16(1):79–87. doi: 10.1038/s41592-018-0253-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 39.Telias M, Ben-Yosef D. Modeling neurodevelopmental disorders using human pluripotent stem cells. Stem Cell Rev Rep. 2014;10(4):494–511. doi: 10.1007/s12015-014-9507-2. [DOI] [PubMed] [Google Scholar]
- 40.Braun DA, Hildebrandt F (2017) Ciliopathies. Cold Spring Harb Perspect Biol 9(3). 10.1101/cshperspect.a028191 [DOI] [PMC free article] [PubMed]
- 41.Gabel LA, Gibson CJ, Gruen JR, LoTurco JJ. Progress towards a cellular neurobiology of reading disability. Neurobiol Dis. 2010;38(2):173–180. doi: 10.1016/j.nbd.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lauter G, Swoboda P, Tapia-Páez I. Cilia in brain development and disease. In: Goggolidou P, editor. Cilia: development and disease. Boca Raton: CRC Press; 2018. pp. 1–35. [Google Scholar]
- 43.Carrion-Castillo A, Franke B, Fisher SE. Molecular genetics of dyslexia: An overview. Dyslexia. 2013;19(4):214–240. doi: 10.1002/dys.1464. [DOI] [PubMed] [Google Scholar]
- 44.Romani S, Illi B, De Mori R, Savino M, Gleeson JG, Valente EM. The ciliary proteins Meckelin and Jouberin are required for retinoic acid-dependent neural differentiation of mouse embryonic stem cells. Differentiation. 2014;87(3–4):134–146. doi: 10.1016/j.diff.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 2010;11(5):331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee JH, Gleeson JG. The role of primary cilia in neuronal function. Neurobiol Dis. 2010;38(2):167–172. doi: 10.1016/j.nbd.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paracchini S, Diaz R, Stein J. Advances in dyslexia genetics-new insights into the role of brain asymmetries. Adv Genet. 2016;96:53–97. doi: 10.1016/bs.adgen.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 48.van de Leemput J, Boles NC, Kiehl TR, Corneo B, Lederman P, Menon V, Lee C, Martinez RA, Levi BP, Thompson CL, Yao S, Kaykas A, Temple S, Fasano CA. CORTECON: a temporal transcriptome analysis of in vitro human cerebral cortex development from human embryonic stem cells. Neuron. 2014;83(1):51–68. doi: 10.1016/j.neuron.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 49.Lepanto P, Badano JL, Zolessi FR. Neuron's little helper: the role of primary cilia in neurogenesis. Neurogenesis. 2016;3(1):e1253363. doi: 10.1080/23262133.2016.1253363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hunkapiller J, Singla V, Seol A, Reiter JF. The ciliogenic protein oral-facial-digital 1 regulates the neuronal differentiation of embryonic stem cells. Stem Cells Dev. 2011;20(5):831–841. doi: 10.1089/scd.2010.0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Banda E, McKinsey A, Germain N, Carter J, Anderson NC, Grabel L. Cell polarity and neurogenesis in embryonic stem cell-derived neural rosettes. Stem Cells Dev. 2015;24(8):1022–1033. doi: 10.1089/scd.2014.0415. [DOI] [PubMed] [Google Scholar]
- 52.He Q, Wang G, Wakade S, Dasgupta S, Dinkins M, Kong JN, Spassieva SD, Bieberich E. Primary cilia in stem cells and neural progenitors are regulated by neutral sphingomyelinase 2 and ceramide. Mol Biol Cell. 2014;25(11):1715–1729. doi: 10.1091/mbc.E13-12-0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gabriel E, Wason A, Ramani A, Gooi LM, Keller P, Pozniakovsky A, Poser I, Noack F, Telugu NS, Calegari F, Saric T, Hescheler J, Hyman AA, Gottardo M, Callaini G, Alkuraya FS, Gopalakrishnan J. CPAP promotes timely cilium disassembly to maintain neural progenitor pool. EMBO J. 2016;35(8):803–819. doi: 10.15252/embj.201593679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miki D, Kobayashi Y, Okada T, Miyamoto T, Takei N, Sekino Y, Koganezawa N, Shirao T, Saito Y. Characterization of functional primary cilia in human induced pluripotent stem cell-derived neurons. Neurochem Res. 2019;44(7):1736–1744. doi: 10.1007/s11064-019-02806-4. [DOI] [PubMed] [Google Scholar]
- 55.Mandl L, Megele R. Primary cilia in normal human neocortical neurons. Zeitschrift fur mikroskopisch-anatomische Forschung. 1989;103(3):425–430. [PubMed] [Google Scholar]
- 56.Lancaster MA, Gleeson JG. The primary cilium as a cellular signaling center: lessons from disease. Curr Opin Genet Dev. 2009;19(3):220–229. doi: 10.1016/j.gde.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arellano JI, Guadiana SM, Breunig JJ, Rakic P, Sarkisian MR. Development and distribution of neuronal cilia in mouse neocortex. J Comp Neurol. 2012;520(4):848–873. doi: 10.1002/cne.22793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo J, Higginbotham H, Li J, Nichols J, Hirt J, Ghukasyan V, Anton ES. Developmental disruptions underlying brain abnormalities in ciliopathies. Nat Commun. 2015;6:7857. doi: 10.1038/ncomms8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Darki F, Peyrard-Janvid M, Matsson H, Kere J, Klingberg T. Three dyslexia susceptibility genes, DYX1C1, DCDC2, and KIAA0319, affect temporo-parietal white matter structure. Biol Psychiatry. 2012;72(8):671–676. doi: 10.1016/j.biopsych.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 60.Lam M, Sanosaka T, Lundin A, Imaizumi K, Etal D, Karlsson FH, Clausen M, Cairns J, Hicks R, Kohyama J, Kele M, Okano H, Falk A. Single-cell study of neural stem cells derived from human iPSCs reveals distinct progenitor populations with neurogenic and gliogenic potential. Genes Cells. 2019;24(12):836–847. doi: 10.1111/gtc.12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dahdouh F, Anthoni H, Tapia-Paez I, Peyrard-Janvid M, Schulte-Korne G, Warnke A, Remschmidt H, Ziegler A, Kere J, Muller-Myhsok B, Nothen MM, Schumacher J, Zucchelli M. Further evidence for DYX1C1 as a susceptibility factor for dyslexia. Psychiatr Genet. 2009;19(2):59–63. doi: 10.1097/YPG.0b013e32832080e1. [DOI] [PubMed] [Google Scholar]
- 62.Tran C, Gagnon F, Wigg KG, Feng Y, Gomez L, Cate-Carter TD, Kerr EN, Field LL, Kaplan BJ, Lovett MW, Barr CL. A family-based association analysis and meta-analysis of the reading disabilities candidate gene DYX1C1. Am J Med Genet B Neuropsychiatr Genet. 2013;162b(2):146–156. doi: 10.1002/ajmg.b.32123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tammimies K, Vitezic M, Matsson H, Le Guyader S, Burglin TR, Ohman T, Stromblad S, Daub CO, Nyman TA, Kere J, Tapia-Paez I. Molecular networks of DYX1C1 gene show connection to neuronal migration genes and cytoskeletal proteins. Biol Psychiatry. 2013;73(6):583–590. doi: 10.1016/j.biopsych.2012.08.012. [DOI] [PubMed] [Google Scholar]
- 64.Threlkeld SW, McClure MM, Bai J, Wang Y, LoTurco JJ, Rosen GD, Fitch RH. Developmental disruptions and behavioral impairments in rats following in utero RNAi of Dyx1c1. Brain Res Bull. 2007;71(5):508–514. doi: 10.1016/j.brainresbull.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, Szafer A, Ebbert A, Riley ZL, Royall JJ, Aiona K, Arnold JM, Bennet C, Bertagnolli D, Brouner K, Butler S, Caldejon S, Carey A, Cuhaciyan C, Dalley RA, Dee N, Dolbeare TA, Facer BA, Feng D, Fliss TP, Gee G, Goldy J, Gourley L, Gregor BW, Gu G, Howard RE, Jochim JM, Kuan CL, Lau C, Lee CK, Lee F, Lemon TA, Lesnar P, McMurray B, Mastan N, Mosqueda N, Naluai-Cecchini T, Ngo NK, Nyhus J, Oldre A, Olson E, Parente J, Parker PD, Parry SE, Stevens A, Pletikos M, Reding M, Roll K, Sandman D, Sarreal M, Shapouri S, Shapovalova NV, Shen EH, Sjoquist N, Slaughterbeck CR, Smith M, Sodt AJ, Williams D, Zollei L, Fischl B, Gerstein MB, Geschwind DH, Glass IA, Hawrylycz MJ, Hevner RF, Huang H, Jones AR, Knowles JA, Levitt P, Phillips JW, Sestan N, Wohnoutka P, Dang C, Bernard A, Hohmann JG, Lein ES. Transcriptional landscape of the prenatal human brain. Nature. 2014;508(7495):199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gabel LA, Marin I, LoTurco JJ, Che A, Murphy C, Manglani M, Kass S. Mutation of the dyslexia-associated gene Dcdc2 impairs LTM and visuo-spatial performance in mice. Genes Brain Behav. 2011;10(8):868–875. doi: 10.1111/j.1601-183X.2011.00727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Darki F, Peyrard-Janvid M, Matsson H, Kere J, Klingberg T. DCDC2 polymorphism is associated with left temporoparietal gray and white matter structures during development. J Neurosci. 2014;34(43):14455–14462. doi: 10.1523/jneurosci.1216-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marino C, Scifo P, Della Rosa PA, Mascheretti S, Facoetti A, Lorusso ML, Giorda R, Consonni M, Falini A, Molteni M, Gruen JR, Perani D. The DCDC2/intron 2 deletion and white matter disorganization: focus on developmental dyslexia. Cortex. 2014;57:227–243. doi: 10.1016/j.cortex.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Franquinho F, Nogueira-Rodrigues J, Duarte JM, Esteves SS, Carter-Su C, Monaco AP, Molnar Z, Velayos-Baeza A, Brites P, Sousa MM. The dyslexia-susceptibility protein KIAA0319 inhibits axon growth through Smad2 signaling. Cereb Cortex. 2017;27(3):1732–1747. doi: 10.1093/cercor/bhx023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ivliev AE, t Hoen PA, van Roon-Mom WM, Peters DJ, Sergeeva MG. Exploring the transcriptome of ciliated cells using in silico dissection of human tissues. PLoS One. 2012;7(4):e35618. doi: 10.1371/journal.pone.0035618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Velayos-Baeza A, Toma C, Paracchini S, Monaco AP. The dyslexia-associated gene KIAA0319 encodes highly N- and O-glycosylated plasma membrane and secreted isoforms. Hum Mol Genet. 2008;17(6):859–871. doi: 10.1093/hmg/ddm358. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 697 kb)