Abstract
MicroRNAs are endogenous highly conserved short (~ 21 nucleotides) non-coding RNA molecules that play key roles in post-transcriptional gene regulation by translational inhibition or by target mRNA cleavage. In this report, using high stringent computational-based methods, a total of 101 putative miRNAs were identified from European sardine fish (Sardina pilchardus Walbaum, 1792). All the precursors of identified sardine miRNAs formed stable stem-loop structures and displayed high minimum free energy index (MFEI) values. For the experimental validation of the computationally predicted miRNAs, a tissue-specific quantitative study of eight randomly selected putative sardine miRNAs (spi-miR9, spi-miR26, spi-miR128, spi-miR129, spi-miR132, spi-miR212, spi-miR219, and spi-miR338) was performed in brain and liver and all the selected miRNAs were found to be overexpressed in brain tissue. Moreover, using RNAhybrid, a total of 83 potential target proteins of the characterized sardine miRNAs were identified those are involved in transcription, cellular development, defense mechanism, and various signaling pathways. To the best of our knowledge, this is the first report of sardine microRNAs and their targets.
Electronic supplementary material
The online version of this article (10.1007/s13205-020-02298-y) contains supplementary material, which is available to authorized users.
Keywords: Sardine (sardina pilchardus walbaum, 1792), microRNAs, Computational identification, qRT-PCR analysis, Minimum folding free energy index (MFEI)
Introduction
European sardine (Sardina pilchardus Walbaum, 1792), commonly known as European pilchard or simply sardine is one of the most abundant small pelagic fish in the world from the Clupeidae family that occurs mostly in the Atlantic Ocean and the Mediterranean Sea (Louro et al. 2019) and widely consumed by humans. Due to the richest and cheapest source of healthy omega-3 fatty acids, over the last fifty years, the global capture of the sardine has raised from 610,438 t in 1970 up to 1,281,391 t in 2016 representing a significant fish species for fisheries (https://www.fao.org/fishery/species/2910/en). Besides the commercial importance, sardine also plays a key role in the food chain of the marine ecosystem by connecting the first producers of energy to the top of the trophic chain reaching predators. Although European sardine is considered in a global scope assessment with a conservation status of least concern species in the International Union for Conservation of Nature and Natural Resources red list of threatened species (https://www.iucnredlist.org/species/198580/15542481), recently, marine biologists have warned that due to overfishing, pollution, habitat damage, climate change, and various diseases sardine population is declining rapidly worldwide facing the threat of extinction in near future (https://www.express.co.uk/news/nature/756417/Sardines-extinction-threat-overfishing-wiped-outAtlantic-fish-onservationists). Nevertheless, current advancement in molecular technologies greatly facilitates the use of genomics or transcriptomics knowledge to develop modern and rapid monitoring tools for endangered or threatened biodiversity.
MicroRNAs (miRNAs) are small endogenous ~ 21-nucleotide (nt) long non-coding regulatory RNA molecules that play a pivotal role in gene expression at the post-transcriptional level. It has been evidenced that miRNAs regulate a wide variety of biological processes such as cell cycle control, cell proliferation and differentiation, organ development, apoptosis, and stress response signaling in both animals and plants (Sun and Lai 2013; Paul et al. 2011, 2020). Moreover, highly tissue-specific expression patterns during embryogenesis suggest that microRNAs also play an important role in the differentiation and maintenance of tissue identity (Ribeiro et al. 2014). The evolutionarily conserved sequences of miRNAs across different species simplify the characterization process of new miRNA orthologues through computational based homology analysis (Sharma et al. 2019); however, in silico miRNA identification only based on sequence similarity generates false-positive results and hence other stringent parameters of the predicted miRNA precursors such as minimum folding free energy (MFE), sequence length, GC content, and the minimum folding free energy index (MFEI) are required to increase the prediction precision (Paul et al. 2018). Though, experimental validation of the predicted miRNAs is a crucial step to authenticate the prediction (Sharma et al. 2019).
Nonetheless, due to the rapid increment of the human population marine ecosystem are progressively exposing to numerous anthropogenic stressors resulting in a negative impact on biodiversity. This emphasizes the need to assess the effects of stressors on aquatic organisms so that the regulation could be tighter before irreversible damages occur to the marine communities. Thus, precise molecular approaches such as using microRNAs as a biomarker to study the effect of anthropogenic stressors to the aquatic communities could be an indicator for the policymaker to make their decision regarding the conservation of a species (Ikert and Craig 2020). Moreover, few miRNAs such as miR-9, miR-128, miR-129, miR132, and miR-219 are expressed in the brain providing a more effective knowledge of the environment, physiology, and for understanding the molecular mechanism involved in teleost fishes giving baseline information for commercial and conservation tasks (Subramanian et al. 2017; Xu et al. 2017; Bizuayehu and Babiak 2014). Nevertheless, since miRNAs play various regulatory roles and take part in a wide variety of biological processes it is important to exploit the recently published sardine genome information (GenBank accession UIGZ00000000; Louro et al. 2019) to gain a better understanding about the physiological role of miRNAs in sardine. Furthermore, recently, chemically modified antisense oligonucleotides (antimiRs), which sequester the mature miRNAs in competition with cellular target mRNAs leading to the functional inhibition of the miRNAs and de-repression of the direct targets, have been successfully employed in vivo, including in zebrafish (Stenvang et al. 2012). We believe that in near future current sardine miRNA information will help for the development of stress biomarkers as well as facilitate antimiR research to counteract biotic and abiotic stress-related disorders in sardine and other fishes. In summary, to increase knowledge about miRNAs and their functions in a commercially valuable popular fish sardine we aimed to characterize the unknown microRNAs and their targets in sardine and explore their tissue-specific expression pattern through a quantitative approach.
Material and methods
Computational prediction of sardine miRNA
For the in silico prediction of potential sardine miRNAs, two different reference sets of mature fish miRNA sequences were obtained from the miRbase miRNA database (https://www.mirbase.org/cgi-bin/browse.pl) and aligned with the whole genome sequence (WGS) of sardine. The reference set comprised a total of 889 mature miRNAs sequences including 373 mature sequences from vertebrate model fish Danio rerio (dre) or Zebrafish and 516 mature sequences from popular fish cod Gadus morhua (gmo). The alignment between the reference set of mature miRNAs and the WGS of sardine was done with the BLASTn tool and the sequences that showed the exact match were chosen manually. The potential precursor (pre-miRNA) sequences of nearly 400 nt (200 nt downstream and 200 nt upstream of the hit region from BLAST) were mined and sequences coding for proteins were eliminated. To check the reliability of the potential precursors, the secondary structures were predicted using the MFOLD web server (https://unafold.rna.albany.edu/?q=mfold). Since the stable secondary structure of the precursors is considered as one of the important factors to be a miRNA candidate some previously demonstrated strict filtering criteria were applied during secondary structure prediction such as: (1) the precursors must form a stem‐loop structure containing mature miRNA sequences within one arm (2) the potential miRNA sequences should not be positioned at the terminal loop of the hairpin structures, (3) mature miRNAs should have fewer than nine mismatches with the opposite miRNA* sequence, and (4) the predicted secondary structures must have low MFE and high MFEI values since it is required for distinguishing the miRNAs from other RNAs molecules (MFEIs of tRNAs, rRNAs or mRNAs candidates are 0.64, 0.59 and 0.62–0.66, respectively) (Zhang et al. 2006). The MFE or ΔG (-kcal/mol) values generated from the MFOLD web server of the stem-loop structures were used to calculate the MFEI values using the following formula:
Prediction of sardine miRNA targets and their functional annotation
The near precise complementarity between miRNAs and their target sequences enabled in silico prediction of potential target transcripts in sardine. In this report, the potential target transcripts of sardine miRNAs were initially predicted using the NCBI BLASTn program by subjecting the mature miRNA sequences as queries. The Reference RNA sequence database (rfseq_rna) of teleost fishes was chosen during the BLAST analysis. The mRNA sequences with ≥ 75% of query coverage as well as the percent identity were selected for further analysis by RNA-hybrid program (Krüger and Rehmsmeier 2006), and the parameters used are defined as follows: (1) no mismatches at 2–8 nt position (seed region) of mature miRNA with its complementary sequence, (2) only one G:U pairing in the seed region, and (3) no more than four gaps in miRNA from the 9 nt to 21 nt. To achieve a better comprehension functional annotation of the predicted targets was performed using the AmiGO2 platform (https://amigo.geneontology.org/amigo/dd_browse).
RNA extraction and tissue-specific miRNA expression analysis
Five frozen adult sardine fish samples size ranging from 19 to 23 cm were used for total RNA including small RNA extraction from liver and brain tissues using miRNeasy Mini Kit (Qiagen) and pooled separately for each tissue type. The quality and quantity of RNA samples were measured with Nanodrop One (Thermo Scientific, Wilmington, USA), and subsequently polyadenylated (using modified oligo dT primer) as well as reverse transcribed using mRQ Buffer (2X) and enzyme provided with Mir-X miRNA First-Stand Synthesis kit (Takara, Tokyo, Japan). In this study, 1 µg of total RNA (including small RNAs) was used for reverse transcription reaction. Randomly selected eight sardine microRNAs (spi-miR-9-3p, spi-miR-26a-5p, spi-miR-128-3p, spi-miR-129-1-3p, spi-miR132-3p, spi-miR-212, miR219-3p, spi-miR-338) were experimentally validated and their tissue-specific expression pattern in brain and liver was checked using Step One Real-Time PCR System (Applied Biosystems, Carlsbad, CA) and Mir-X miRNA TB Green qRT-PCR kit (Takara, Tokyo, Japan). The real-time qRT-PCR reaction was made in a volume of 12.5 µl containing 1X TB Green Advantage Premix, 1X ROX Dye, 0.2 µm each of forward and reverse primers, and 0.5 µl of cDNA. U6 was employed as an internal reference and each reaction was done in three technical replicates. The qRT-PCR program was as follows: initial denaturation for 10 s at 95 °C, then 45 cycles of denaturation for 5 s at 95 °C and annealing for 20 s at 60 °C. This cycle was followed by a melting curve analysis ranging from 55 to 95 °C, with temperature increasing steps of 0.5 °C every 10 s. Melting curves for each amplicon were observed carefully to confirm the specificity of the primers used. Finally, the relative fold change values were obtained using the comparative Ct method or Ct (2−ΔΔCT).
Results and discussion
Characterization of sardine miRNAs and their tissue-specific expression analysis
In this report using strict filtering criteria, a total of 101 potentially conserved sardine miRNAs were identified (Table 1). The majority of the identified sardine miRNAs were 22 nucleotides (nt) long while their precursors displayed great size variability ranging between 53 and 116 nt with an average of 62 nt (Table 1). Regarding the miRNA location, 64.4% of the putative sardine miRNAs were found located at the 3′ arm of the stem-loop precursors, while the remaining 35.6% were located at the 5′ arm. Moreover, 59% of the predicted sequences began with the uracil (U) nucleotide corroborating the study of Zhang et al. (2008) that miRNA mediated regulation is highly dependent on U existing at the initial position of the mature miRNA sequence. The content of guanine-cytosine (GC) of sardine miRNA precursors had an average of 44.90%. It is well known that low MFE values of the stem-loop precursors attain more stable miRNA predictions (Bonnet et al. 2004), in this study, quite low MFE values of the precursors varied from − 16.10 to − 46.80 with an average of − 25.50, was successfully achieved. Moreover, the MFEI scores ranged from 0.70 to 1.33 with a mean of 0.93 excluding the chance of being another small RNA. The predicted stem-loop secondary structures of sardine’s miRNA precursors pre-miRNA with higher MFEI values (top 20) were displayed in supplementary file 1.
Table 1.
Summary of the identified Sardina pilchardus (Walbaum, 1792) miRNAs
| Identified miRNAs | LM (nt) | Query miRNAs | miRNA sequences | Location | LP (nt) | GC % | MFEs | MFEI |
|---|---|---|---|---|---|---|---|---|
| spi-miR-1 | 22 | dre-miR-1 | UGGAAUGUAAAGAAGUAUGUAU | 3′ | 64 | 29.69 | − 24.00 | 1.26 |
| spi-let-7a | 22 | dre-let-7a | UGAGGUAGUAGGUUGUAUAGUU | 5′ | 70 | 37.14 | − 28.80 | 1.10 |
| spi-miR-9-3p | 21 | dre-miR-9-3p | UAAAGCUAGAUAACCGAAAGU | 3′ | 60 | 38.33 | − 26.70 | 1.16 |
| spi-miR-10a-5p | 22 | dre-miR-10a-5p | UACCCUGUAGAUCCGAAUUUGU | 5′ | 61 | 45.90 | − 21.60 | 0.77 |
| spi-miR-15a-3p | 22 | dre-miR-15a-3p | CAGGCCGUACUGUGCUGCGGCA | 3′ | 61 | 57.38 | − 29.60 | 0.84 |
| spi-miR-16a | 22 | dre-miR-16a | UAGCAGCACGUAAAUAUUGGUG | 5′ | 65 | 38.46 | − 22.70 | 0.90 |
| spi-miR-17a-2-3p | 23 | dre-miR-17a-2-3p | ACUGCAGUGGAGGCACUUCAAGC | 3′ | 63 | 47.62 | − 21.50 | 0.71 |
| spi-miR-19a-3p | 23 | dre-miR-19a-3p | UGUGCAAAUCUAUGCAAAACUGA | 3′ | 58 | 36.21 | − 23.60 | 1.12 |
| spi-miR-20a-3p | 22 | dre-miR-20a-3p | ACUGCAGUGUGAGCACUUGAAG | 3′ | 60 | 38.33 | − 24.70 | 1.07 |
| spi-miR-22a-3p | 22 | dre-miR-22a-3p | AAGCUGCCAGCUGAAGAACUGU | 3′ | 65 | 46.15 | − 32.50 | 1.08 |
| spi-miR-23a-3p | 22 | dre-miR-23a-3p | AUCACAUUGCCAGGGAUUUCCA | 3 | 60 | 45.26 | − 30.10 | 1.10 |
| spi-miR-24 | 22 | dre-miR-24 | UGGCUCAGUUCAGCAGGAACAG | 3′ | 58 | 56.9 | − 23.90 | 0.72 |
| spi-miR-25-3p | 22 | dre-miR-25-3p | CAUUGCACUUGUCUCGGUCUGA | 3′ | 62 | 54.84 | − 24.10 | 0.70 |
| spi-miR-26a-5p | 22 | dre-miR-26a-5p | UUCAAGUAAUCCAGGAUAGGCU | 5′ | 59 | 49.15 | − 27.00 | 0.93 |
| spi-miR-27a-3p | 22 | dre-miR-27a-3p | UUCACAGUGGCUAAGUUCCGCU | 3 | 67 | 48.19 | − 24.0 | 0.74 |
| spi-miR-29a | 22 | dre-miR-29a | UAGCACCAUUUGAAAUCGGUUA | 3′ | 62 | 45.73 | − 27.50 | 0.96 |
| spi-miR-30b | 22 | dre-miR-30b | UGUAAACAUCCUACACUCAGCU | 5′ | 57 | 49.12 | − 24.40 | 0.87 |
| spi-miR-31 | 22 | dre-miR-31 | UGGCAAGAUGUUGGCAUAGCUG | 5′ | 58 | 51.72 | − 27.00 | 0.90 |
| spi-miR-33b-5p | 21 | gmo-miR-33b-5p | GUGCAUUGUAGUUGCAUUGCA | 5′ | 57 | 43.86 | − 28.20 | 1.12 |
| spi-miR-34a | 22 | dre-miR-34a | UGGCAGUGUCUUAGCUGGUUGU | 5′ | 63 | 49.21 | − 28.30 | 0.91 |
| spi-miR-92a-3p | 22 | dre-miR-92a-3p | UAUUGCACUUGUCCCGGCCUGU | 3′ | 62 | 53.23 | − 35.50 | 1.07 |
| spi-miR93 | 22 | dre-miR93 | AAAAGUGCUGUUUGUGCAGGUA | 5′ | 59 | 47.46 | − 25.40 | 0.90 |
| spi-miR-96-3p | 22 | dre-miR-96-3p | CAAUUAUGUGUAGUGCCAAUAU | 3′ | 66 | 34.85 | − 25.70 | 1.11 |
| spi-miR-100–2-3p | 22 | dre-miR-100–2-3p | CAAGCUCGUGUCUAUAGGUAUG | 3′ | 59 | 49.29 | − 22.40 | 0.77 |
| spi-miR-101a | 22 | dre-miR-101a | UACAGUACUGUGAUAACUGAAG | 3′ | 61 | 40.98 | − 28.50 | 1.14 |
| spi-miR-103 | 23 | dre-miR-103 | AGCAGCAUUGUACAGGGCUAUGA | 3′ | 61 | 50.82 | − 25.30 | 0.81 |
| spi-miR-122 | 22 | dre-miR-122 | CAAACACCAUUGUCACACUCCA | 3′ | 58 | 41.67 | − 25.30 | 1.04 |
| spi-miR-124-3p | 22 | dre-miR-124-3p | UAAGGCACGCGGUGAAUGCCAA | 3′ | 61 | 45.90 | − 23.10 | 0.82 |
| spi-miR-125a | 22 | dre-miR-125a | UCCCUGAGACCCUUAACCUGUG | 5′ | 56 | 51.79 | − 25.80 | 0.88 |
| spi-miR-126a-3p | 21 | dre-miR-126a-3p | UCGUACCGUGAGUAAUAAUGC | 3′ | 59 | 42.37 | − 19.70 | 0.78 |
| spi-miR-128-3p | 22 | dre-miR-128-3p | UCACAGUGAACCGGUCUCUUUU | 3′ | 53 | 50.94 | − 19.50 | 0.72 |
| spi-miR-129–1-3p | 21 | dre-miR-129–1-3p | GAAGCCCUUACCCCAAAAAGU | 3′ | 65 | 51.54 | − 24.70 | 0.73 |
| spi-miR-130c-3p | 22 | dre-miR-130c-3p | CAGUGCAAUAUUAAAAGGGCAU | 3′ | 61 | 39.34 | − 25.60 | 1.06 |
| spi-miR132-3p | 22 | dre-miR132-3p | UAACAGUCUACAGCCAUGGUCG | 3′ | 63 | 52.38 | − 30.50 | 0.92 |
| spi-miR133a-2-5p | 21 | dre-miR133a-2-5p | AGCUGGUAAAAUGGAACCAAA | 5′ | 58 | 44.83 | − 22.00 | 0.84 |
| spi-miR-135a | 23 | dre-miR-135a | UAUGGCUUUUUAUUCCUAUGUGA | 5′ | 60 | 40.00 | − 24.10 | 1.00 |
| spi-miR-138–2-3p | 21 | dre-miR-138–2-3p | GCUUCUUCACAACACCAGGGU | 3′ | 62 | 56.45 | − 29.90 | 0.85 |
| spi-miR-140-3p | 23 | dre-miR-140-3p | UACCACAGGGUAGAACCACGGAC | 3′ | 66 | 53.03 | − 33.50 | 0.95 |
| spi-miR-142a-3p | 23 | dre-miR-142a-3p | UGUAGUGUUUCCUACUUUAUGG | 3′ | 60 | 41.67 | − 25.00 | 0.99 |
| spi-miR-143 | 21 | dre-miR-143 | UGAGAUGAAGCACUGUAGCUC | 3′ | 57 | 50.88 | − 26.60 | 0.91 |
| spi-miR-144-3p | 20 | dre-miR-144-3p | UACAGUAUAGAUGAUGUACU | 3′ | 59 | 30.51 | − 21.30 | 1.18 |
| spi-miR-145-3p | 22 | dre-miR-145-3p | GGAUUCCUGGAAAUACUGUUCU | 3′ | 63 | 47.62 | − 27.80 | 0.92 |
| spi-miR-146b | 22 | dre-miR-146b | UGAGAACUGAAUUCCAAGGGUG | 5′ | 58 | 48.28 | − 22.60 | 0.80 |
| spi-miR-148 | 22 | dre-miR-148 | UCAGUGCAUUACAGAACUUUGU | 3′ | 62 | 40.32 | − 24.20 | 0.96 |
| spi-miR-150 | 22 | dre-miR-150 | UCUCCCAAUCCUUGUACCAGUG | 5′ | 59 | 54.24 | − 31.80 | 0.99 |
| spi-miR-152-3p | 21 | gmo-miR-152-3p | CAAAGUUCUGUUAUGCACUGA | 5′ | 61 | 39.34 | − 21.90 | 0.91 |
| spi-miR-153a-3p | 22 | dre-miR-153a-3p | UUGCAUAGUCACAAAAGUGAUC | 3′ | 62 | 37.1 | − 24.40 | 1.06 |
| spi-miR-155 | 22 | dre-miR-155 | UUAAUGCUAAUCGUGAUAGGGG | 5′ | 59 | 35.59 | − 22.10 | 1.05 |
| spi-miR-183-5p | 23 | dre-miR-183-5p | UAUGGCACUGGUAGAAUUCACUG | 5′ | 60 | 40.00 | − 20.70 | 0.86 |
| spi-miR-184 | 22 | dre-miR-184 | UGGACGGAGAACUGAUAAGGGC | 3′ | 64 | 48.44 | − 22.60 | 0.72 |
| spi-miR-185a-5p | 23 | dre-miR-185a-5p | AACAUUCAACGCUGUCGGUGAGU | 5′ | 61 | 44.26 | − 20.40 | 0.75 |
| spi-miR-187 | 20 | dre-miR-187 | UCGUGUCUUGUGUUGCAGCC | 3′ | 60 | 63.33 | − 39.50 | 1.03 |
| spi-miR-190a | 22 | dre-miR-190a | ACCUAAUAUAUCAAACAUAUCA | 3′ | 57 | 26.32 | − 18.00 | 1.19 |
| spi-miR-192 | 21 | dre-miR-192 | AUGACCUAUGAAUUGACAGCC | 5′ | 61 | 47.54 | − 23.90 | 0.82 |
| spi-miR-193a-3p | 22 | dre-miR-193a-3p | AACUGGCCUACAAAGUCCCAGU | 3′ | 62 | 46.77 | − 23.40 | 0.80 |
| spi-miR-194a | 21 | dre-miR-194a | UGUAACAGCAACUCCAUGUGG | 5′ | 55 | 47.27 | − 27.90 | 1.07 |
| spi-miR-196a-5p | 22 | dre-miR-196a-5p | UAGGUAGUUUCAUGUUGUUGGG | 5′ | 60 | 36.67 | − 21.00 | 0.95 |
| spi-miR-199-3p | 22 | dre-miR-199-3p | UACAGUAGUCUGCACAUUGGUU | 3′ | 61 | 49.18 | − 23.90 | 0.79 |
| spi-miR-200a-3p | 22 | dre-miR-200a-3p | UAACACUGUCUGGUAACGAUGU | 3′ | 61 | 45.9 | − 23.90 | 0.85 |
| spi-miR-202-5p | 22 | dre-miR-202-5p | UUCCUAUGCAUAUACCUCUUUG | 5′ | 56 | 42.86 | − 22.60 | 0.94 |
| spi-miR-203a-3p | 22 | dre-miR-203a-3p | CAAGUGGUCCUAAACAUUUCAC | 5′ | 62 | 40.32 | − 18.20 | 0.72 |
| spi-miR-204-5p | 22 | dre-miR-204-5p | UUCCCUUUGUCAUCCUAUGCCU | 5′ | 56 | 53.57 | − 26.50 | 0.88 |
| spi-miR-205-5p | 22 | dre-miR-205-5p | UCCUUCAUUCCACCGGAGUCUG | 5′ | 57 | 45.61 | − 25.20 | 0.96 |
| spi-miR-206-3p | 22 | dre-miR-206-3p | UGGAAUGUAAGGAAGUGUGUGG | 3′ | 62 | 41.94 | − 30.20 | 1.16 |
| spi-miR-212 | 21 | dre-miR-212 | UAACAGUCUACAGUCAUGGCU | 3′ | 57 | 50.75 | − 24.50 | 0.84 |
| spi-miR-214 | 21 | dre-miR-214 | CUGCCUGUCUGUGCCUGCUGU | 5′ | 62 | 56.45 | − 36.80 | 1.05 |
| spi-miR-216a | 22 | dre-miR-216a | UAAUCUCAGCUGGCAACUGUGA | 5′ | 60 | 53.33 | − 26.00 | 0.81 |
| spi-miR-217 | 22 | dre-miR-217 | UACUGCAUCAGGAACUGAUUGG | 5′ | 59 | 42.37 | − 33.40 | 1.33 |
| spi-miR-218a | 22 | dre-miR-218a | UUGUGCUUGAUCUAACCAUGUG | 5′ | 63 | 55.56 | − 27.10 | 0.77 |
| spi-miR219-3p | 23 | dre-miR219-3p | GGAGUUGUGGAUGGACAUCACGC | 3′ | 68 | 47.06 | − 30.20 | 0.94 |
| spi-miR-221-3p | 23 | dre-miR-221-3p | AGCUACAUUGUCUGCUGGGUUUC | 3′ | 65 | 43.08 | − 29.60 | 1.05 |
| spi-miR-222a-3p | 24 | dre-miR-222a-3p | AGCUACAUCUGGCUACUGGGUCUC | 3′ | 64 | 50.00 | − 28.40 | 0.88 |
| spi-miR-223 | 21 | dre-miR-223 | UGUCAGUUUGUCAAAUACCCC | 3′ | 63 | 44.44 | − 24.60 | 0.87 |
| spi-miR-338 | 22 | dre-miR-338 | UCCAGCAUCAGUGAUUUUGUUG | 3′ | 61 | 44.26 | − 22.50 | 0.83 |
| spi-miR-363-3p | 22 | dre-miR-363-3p | AAUUGCACGGUAUCCAUCUGUA | 3′ | 76 | 34.21 | − 23.50 | 0.90 |
| spi-miR-365 | 22 | dre-miR-365 | UAAUGCCCCUAAAAAUCCUUAU | 3′ | 65 | 46.15 | − 28.40 | 0.94 |
| spi-miR-429a | 22 | dre-miR-429a | UAAUACUGUCUGGUAAUGCCGU | 3′ | 64 | 40.63 | − 26.90 | 1.03 |
| spi-miR-430a-3p | 22 | dre-miR-430a-3p | UAAGUGCUAUUUGUUGGGGUAG | 3′ | 61 | 44.26 | − 21.00 | 0.77 |
| spi-miR-454b | 22 | dre-miR-454b | UAGUGCAAUAUUGCUUAUAGGG | 3′ | 63 | 38.1 | − 23.10 | 0.96 |
| spi-miR-455-3p | 23 | dre-miR-455-3p | GUGUAUAUGCCCAUGGACUGCAU | 5′ | 62 | 53.23 | − 34.30 | 1.03 |
| spi-miR-456 | 22 | dre-miR-456 | CAGGCUGGUUAGAUGGUUGUCA | 3′ | 65 | 52.31 | − 28.80 | 0.84 |
| spi-miR-458-3p | 22 | dre-miR-458-3p | AUAGCUCUUUGAAUGGUACUGC | 3′ | 60 | 48.33 | − 31.60 | 1.08 |
| spi-miR-459-5p | 22 | dre-miR-459-5p | UCAGUAACAAGGAUUCAUCCUG | 5′ | 61 | 45.90 | − 25.50 | 0.91 |
| spi-miR-460-3p | 22 | dre-miR-460-3p | CACAGCGCAUACAAUGUGGAUG | 3′ | 61 | 44.26 | − 20.00 | 0.74 |
| spi-miR-462 | 22 | dre-miR-462 | UAACGGAACCCAUAAUGCAGCU | 5′ | 116 | 50.00 | − 46.80 | 0.80 |
| spi-miR-489 | 23 | dre-miR-489 | AGUGACAUCAUAUGUACGGCUGC | 3′ | 61 | 42.62 | − 27.20 | 1.04 |
| spi-miR-499-3p | 22 | dre-miR-499-3p | AACAUCACUUUAAGUCUGUGCU | 3′ | 62 | 38.71 | − 22.80 | 0.94 |
| spi-miR-722 | 24 | dre-miR-722 | UUUUUUGCAGAAACGUUUCAGAUU | 3′ | 67 | 31.34 | − 21.40 | 1.01 |
| spi-miR-724 | 22 | dre-miR-724 | AACAGUCGCAAAUUCCCUUUAA | 3′ | 58 | 37.93 | − 21.70 | 0.98 |
| spi-miR-726 | 21 | dre-miR-726 | UUCACUACUAGCAGAACUCGG | 3′ | 63 | 42.86 | − 19.20 | 0.71 |
| spi-miR-727-3p | 22 | dre-miR-727-3p | GUUGAGGCGAGUUGAAGACUUA | 3′ | 65 | 47.69 | − 22.50 | 0.72 |
| spi-miR-728 | 22 | dre-miR-728 | AUACUAAGUACACUACGUUUUC | 3′ | 66 | 33.33 | − 21.30 | 0.96 |
| spi-miR-729 | 24 | dre-miR-729 | CAUGGGUAUGAUACGACCUGGGUU | 3′ | 64 | 43.75 | − 29.20 | 1.04 |
| spi-miR-734-3p | 21 | gmo-miR-734-3p | UAAAUGCUGCAGAAUUGUGCU | 3′ | 60 | 38.33 | − 16.10 | 0.70 |
| spi-miR-737-3p | 21 | dre-miR-737-3p | AAUCAAAACCUAAAGAAAAUA | 3′ | 64 | 28.13 | − 20.70 | 1.14 |
| spi-miR-1788-3p | 21 | dre-miR-1788-3p | CAGGCAGCUAAAGCAAGUCUG | 3′ | 60 | 48.33 | − 29.20 | 1.00 |
| spi-miR-2187-3p | 22 | dre-miR-2187-3p | UUACAGGCUAUGCUAAUCUAUG | 3′ | 63 | 30.16 | − 23.80 | 1.25 |
| spi-miR-2188-5p | 21 | dre-miR-2188-5p | AAGGUCCAACCUCACAUGUCC | 5′ | 56 | 57.14 | − 26.30 | 0.82 |
| spi-miR-7552-5p | 22 | gmo-miR-7552-5p | UUACAAUUAAAGGAUAUUUCUU | 5′ | 60 | 30.00 | − 17.90 | 0.99 |
| spi-miR-8160b-5p | 21 | gmo-miR-8160b-5p | AGAAUAAUGCCAGCAGUCGGC | 5′ | 56 | 51.79 | − 22.50 | 0.77 |
| spi-miR-10545-5p | 22 | gmo-miR-10545-5p | UAAGUCUCACACCAGUGCAAAA | 5′ | 56 | 46.30 | − 19.60 | 0.75 |
LM length of miRNA, LP length of precursor, GC guanine-cytosine, MFE minimum folding free energy, MFEI minimum folding free energy index
Randomly selected eight sardine miRNAs (spi-miR9, spi-miR26, spi-miR128, spi-miR129, spi-miR132, spi-miR212, spi-miR219, and spi-miR338) were successfully validated in this study by qRT-PCR and their significant differential expression between brain and liver tissues was noticed. Interestingly, all the selected sardine miRNAs were overexpressed in the brain as compared to the liver. The expression of spi-miR338, spi-miR26, and spi-miR129 had the high fold changes of 109.13, 98.36, and 45.93, respectively, while spi-miR128 and spi-miR132 showed near similar fold changes of 23.50 and 20.35, respectively. However, spi-miR212, spi-miR129, and spi-miR9 exhibited the lowest fold changes with the values of 11.10, 7.00, and 6.23, respectively (Fig. 1). Since individual functions of those selected miRNAs are not well studied in teleost fish, we explored their function in other vertebrates, and it was revealed that all of them are brain-enriched miRNAs and participate in several neurological functions in other vertebrate species, including human. For example, miR9 was found to be one of the most highly expressed microRNAs in the developing and adult vertebrate brain and participate mainly in neural differentiation proliferation, differentiation, and cell migration (Coolen et al. 2013); while miR26 regulates neural stem cell development and targets brain-derived neurotrophic factor proteins involved in plasticity and synaptogenesis (Caputo et al. 2011). Similarly, miR129, miR212, and miR132 are also found to be involved in synaptic plasticity (Follert et al. 2014; Thangaleela et al. 2018); while miR219 was reported to endorse neural precursor cell differentiation (Murai et al. 2016). Likewise, brain enriched miR128 and miR338 are involved in neuronal cell migration and oligodendrocytes development, respectively (Evangelisti et al. 2009; Follert et al. 2014).
Fig. 1.
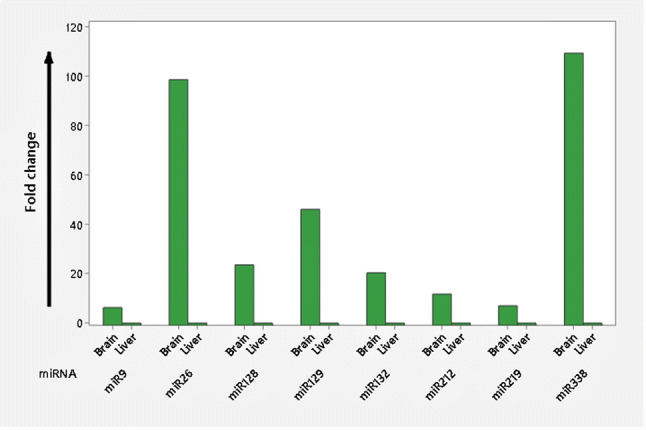
Graphical representation of the fold changes (differential expression pattern) of selected Sardina pilchardus (Walbaum, 1792) microRNAs (spi-miR9, spi-miR26, spi-miR128, spi-miR129, spi-miR132, spi-miR212, spi-miR219, and spi-miR338) between brain and liver tissue samples. U6 was chosen as an internal reference. All the miRNAs were found to be brain-specific and overexpressed in brain tissue. MicroRNA spi-miR338 was the highest expressed miRNAs among all followed by spi-miR26
Identification of potential target transcripts of putative sardine miRNAs
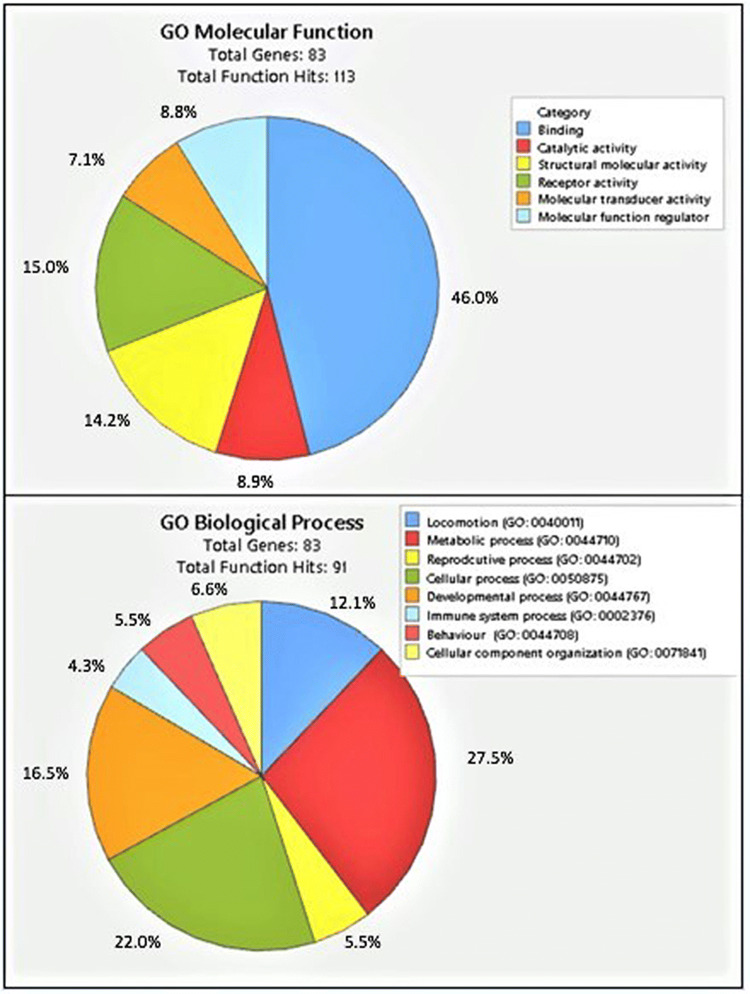
In this report, a total of 83 potential target transcripts of sardine miRNAs were identified, and among them, several microRNAs were found to target more than one transcript (Supplementary File 1). Most of the sardine miRNA targets identified in this study are involved in transcription, cellular development, defense mechanism, and signaling pathways (Supplementary file 1). Functional annotation by GO term enrichment analysis revealed that different sardine miRNA target proteins with molecular functions such as binding, transduction, and regulatory activity are involved in important biological processes such as cellular, developmental, metabolic, and reproductive processes (Fig. 2). Several experimental and computational studies have demonstrated that transcription factors are the major target molecules for various miRNAs (Barozai 2012) while for the proper functioning of the cells many miRNAs were reported to target signaling molecules (Hagen and Lai 2008). In this study important transcription factors targeted by sardine miRNAs include homeobox protein (spi-miR-10a-5p) that regulate gene expression and cell differentiation during early embryonic development and are involved in the regulation of patterns of anatomical development (morphogenesis) in both animals and plants (Bürglin and Affolter 2016); Wnt (spi-miR-22a-3p)-signaling cascade plays critical roles in embryonic patterning, cell fate determination, and tissue homeostasis (Van Noort and Clevers 2002); zinc finger proteins (spi-miR-30b, spi-miR-152-3p, and spi-miR-734-3p) are one of the most abundant groups of proteins and have a wide range of molecular functions including transcriptional regulation, ubiquitin-mediated protein degradation, actin targeting, DNA repair, cell migration, and numerous other processes (Cassandri et al. 2017); SRY-box 7 or SOX-7 (spi-miR-143) are transcription factors having critical roles in the regulation of diverse developmental processes in the animal kingdom and detected during embryonic development in many tissues, suggesting a role in differentiation and development (Takash 2001); and FEV (spi-miR-1788-3p), a member of one of the largest transcription factor family ETS. Among the important target signalling molecules Alpha kinase 2 (spi-miR-122), a member of Alpha kinase family are implicated in a large variety of cellular processes such as Mg2+ homeostasis, intracellular transport, cell migration, adhesion, and proliferation (Middelbeek et al. 2010); WD repeat-containing proteins (spi-miR-489) have critical roles in many biological functions such as signal transduction, transcription regulation and apoptosis (Li and Roberts. 2001); G protein-coupled receptor proteins (spi-miR-34a) are the largest family of membrane proteins and mediate most cellular responses to hormones and neurotransmitters, as well as being responsible for vision, olfaction and taste (Rosenbaum et al. 2009); Exportin 7 (spi-miR-146b) escorts multiple cytosolic proteins from the nucleus back into the cytoplasm, and thus may function to exclude numerous proteins that otherwise would interfere with gene expression if allowed to gather in the nucleus (Aksu et al. 2018); glutamate is the most abundant excitatory neurotransmitter in the vertebrate nervous system and one of the major functions of glutamate receptors (spi-miR-7552-5p) was found to be the modulation of synaptic plasticity, a property of the brain thought to be vital for memory and learning (Debanne et al. 2003); and Adenosine receptors (spi-let-7a and spi-miR-143) are reported to be involved in several key physiological processes, ranging from neuromodulation to immune regulation, and from vascular function to metabolic control (Chen et al. 2013) (Supplementary File 1).
Fig. 2.
Functional annotation of potential Sardina pilchardus (Walbaum, 1792) target transcripts. Most of the predicted targets were found to be involved in binding (46%) in the molecular function category, while in the biological process category most of the targets are involved in metabolic processes (27.5%)
Conclusion
It is well established that several conserved, as well as species-specific miRNAs, are very crucial for different biological and metabolic pathways in animals as well as they can be used as biomarkers to monitor the effect of different anthropogenic stressors to the aquatic communities, especially for the teleost fishes, and hence it is important to profile miRNAs in non-model commercially as well as ecologically important teleost fishes to get an indication whether their conservation is required or not. Moreover, a number of brain-specific miRNAs from different marine animals already provided baseline information for commercial and conservation tasks. In this study, for the first time, using homology-based computational analysis and strict filtering criteria, 101 conserved miRNAs and 83 corresponding targets were identified in sardine fish. Among the predicted miRNAs, eight randomly selected miRNAs (spi-miR9, spi-miR26, spi-miR128, spi-miR129, spi-miR132, spi-miR212, spi-miR219, and spi-miR338) were validated and their quantitative expression revealed that all of them are brain enriched miRNAs corroborating some previous reports. Among the predicted miRNA targets, numerous targets were found to be involved in transcription and signaling pathways. Nonetheless, identification of miRNAs and their targets is the crucial step to initiate a miRNA-related study in a non-model animal species. Additionally, in the near future, current miRNA documentation may help in the creation of direct antimiRs and de-repressing specific targets in vivo to neutralize abiotic and biotic stress disorders in sardines. Nevertheless, we believe that our current study will be useful for strengthening the research on miRNA-mediated metabolic control in sardine and other fishes.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Funding
None.
Data accessibility
The datasets during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Ashutosh Sharma, Email: asharma@tec.mx.
Sujay Paul, Email: spaul@tec.mx.
References
- Aksu M, Pleiner T, Karaca S, et al. Xpo7 is a broad-spectrum exportin and a nuclear import receptor. J Cell Biol. 2018;217:2329–2340. doi: 10.1083/jcb.201712013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barozai MYK. Identification and characterization of the microRNAs and their targets in Salmo salar. Gene. 2012;499:163–168. doi: 10.1016/j.gene.2012.03.006. [DOI] [PubMed] [Google Scholar]
- Bizuayehu TT, Babiak I. MircoRNA in teleost fishes. Genom Biol Evol. 2014;6(8):1911–1937. doi: 10.1093/gbe/evu151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet E, Wuyts J, Rouzé P, Van de Peer Y. Evidence that microRNA precursors, unlike other non-coding RNAs, have lower folding free energies than random sequences. Bioinformatics. 2004;20:2911–2917. doi: 10.1093/bioinformatics/bth374. [DOI] [PubMed] [Google Scholar]
- Bürglin TR, Affolter M. Homeodomain proteins: an update. Chromosoma. 2016;125:497–521. doi: 10.1007/s00412-015-0543-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputo V, Sinibaldi L, Fiorentino A, et al. Brain derived neurotrophic factor (BDNF) expression is regulated by microRNAs miR-26a and miR-26b allele-specific binding. PLoS ONE. 2011;6:12. doi: 10.1371/journal.pone.0028656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassandri M, Smirnov A, Novelli F, et al. Zinc-finger proteins in health and disease. Cell Death Discov. 2017;3:1–12. doi: 10.1038/cddiscovery.2017.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JF, Eltzschig HK, Fredholm BB. Adenosine receptors as drug targets-what are the challenges? Nat Rev Drug Discov. 2013;12:265–286. doi: 10.1038/nrd3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolen M, Katz S, Bally-Cuif L. miR-9: a versatile regulator of neurogenesis. Front Cell Neurosci. 2013;7:220. doi: 10.3389/fncel.2013.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debanne D, Daoudal G, Sourdet V, Russier M. Brain plasticity and ion channels. J Physiol Paris. 2003;97:403–414. doi: 10.1016/j.jphysparis.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Evangelisti C, Florian MC, Massimi I, et al. MiR-128 up-regulation inhibits Reelin and DCX expression and reduces neuroblastoma cell motility and invasiveness. FASEB J. 2009;23:4276–4287. doi: 10.1096/fj.09-134965. [DOI] [PubMed] [Google Scholar]
- Follert P, Cremer H, Béclin C. MicroRNAs in brain development and function: a matter of flexibility and stability. Front Mol Neurosci. 2014;7:5. doi: 10.3389/fnmol.2014.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen JW, Lai EC. MicroRNA control of cell-cell signaling during development and disease. Cell Cycle. 2008;7:2327–2332. doi: 10.4161/cc.6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homayooni B, Sahari MA, Barzegar M. Concentrations of omega-3 fatty acids from rainbow sardine fish oil by various methods. Int Food Res J. 2014;21:743. [Google Scholar]
- Ikert H, Craig PM. Chronic exposure to venlafaxine and increased water temperature reversibly alters microRNA in zebrafish gonads (Danio rerio) Comp Biochem Phys D. 2020;33:100634. doi: 10.1016/j.cbd.2019.100634. [DOI] [PubMed] [Google Scholar]
- Krüger J, Rehmsmeier M. RNAhybrid: MicroRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34:W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Roberts R. WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cell Mol Life Sci C. 2001;58:2085–2097. doi: 10.1007/PL00000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louro B, De Moro G, Garcia C, et al. A haplotype-resolved draft genome of the European sardine (Sardina pilchardus) Gigascience. 2019;8:giz059. doi: 10.1093/gigascience/giz059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middelbeek J, Clark K, Venselaar H, et al. The alpha-kinase family: an exceptional branch on the protein kinase tree. Cell Mol Life Sci. 2010;67:875–890. doi: 10.1007/s00018-009-0215-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai K, Sun G, Ye P, et al. The TLX-miR-219 cascade regulates neural stem cell proliferation in neurodevelopment and schizophrenia iPSC model. Nat Commun. 2016;7:1–15. doi: 10.1038/ncomms10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naya L, Paul S, Valdés-López O, et al. Regulation of copper homeostasis and biotic interactions by microRNA 398b in common bean. PLoS ONE. 2014;9:1. doi: 10.1371/journal.pone.0084416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Kundu A, Pal A. Identification and validation of conserved microRNAs along with their differential expression in roots of Vigna unguiculata grown under salt stress. Plant Cell Tissue Organ Cult. 2011;105:233–242. [Google Scholar]
- Paul SC, Sharma A, Mehta R, Paul S. Genome wide computational identification of Tuna (Thunnus orientalis) MicroRNAs and their targets. Ocean Sci J. 2018;53:727–734. [Google Scholar]
- Paul S, de la Fuente-Jiménez JL, Manriquez CG, Sharma A. Identification, characterization and expression analysis of passion fruit (Passiflora edulis) microRNAs. 3 Biotech. 2020;10:25. doi: 10.1007/s13205-019-2000-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro A, Schoof C, Izzotti A, et al. MicroRNAs: modulators of cell identity, and their applications in tissue engineering. MicroRNA. 2014;3:45–53. doi: 10.2174/2211536603666140522003539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum DM, Rasmussen SGF, Kobilka BK. The structure and function of G-protein-coupled receptors. Nature. 2009;459:356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Bejerano PIA, Maldonado IC, et al. Genome-wide computational prediction and experimental validation of quinoa (Chenopodium quinoa) micrornas. Can J Plant Sci. 2019;99:666–675. [Google Scholar]
- Stenvang J, Petri A, Lindow M, et al. Inhibition of microRNA function by antimiR oligonucleotides. Silence. 2012;3:1. doi: 10.1186/1758-907X-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian T, Shanmugapriya V, Mukilan M, Radhakrishnan K, Rajan KE. Alterations in mircroRNA-132/212 expression impairs fear memory in goldfish Cassarius auratus. Ann Neurosci. 2017;25:90–97. doi: 10.1159/000486842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K, Lai EC. Adult-specific functions of animal microRNAs. Nat Rev Genet. 2013;14:535–548. doi: 10.1038/nrg3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takash W. SOX7 transcription factor: sequence, chromosomal localisation, expression, transactivation and interference with Wnt signalling. Nucleic Acids Res. 2001;29:4274–4283. doi: 10.1093/nar/29.21.4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangaleela S, Shanmugapriya V, Mukilan M, et al. Alterations in MicroRNA-132/212 expression impairs fear memory in Goldfish Carassius auratus. Ann Neurosci. 2018;25:90–97. doi: 10.1159/000486842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Noort M, Clevers H. TCF transcription factors, mediators of Wnt-signaling in development and cancer. Dev Biol. 2002;244:1–8. doi: 10.1006/dbio.2001.0566. [DOI] [PubMed] [Google Scholar]
- Xu B, Zhang Y, Du XF, Zi HX, et al. Neurons secrete miR-132 containing exosomes to regulate brain vascular integrity. Cell Res. 2017;27:882–897. doi: 10.1038/cr.2017.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Pan X, Cobb GP, Anderson TA. Plant microRNA: a small regulatory molecule with big impact. Dev Biol. 2006;289:3–16. doi: 10.1016/j.ydbio.2005.10.036. [DOI] [PubMed] [Google Scholar]
- Zhang B, Pan X, Stellwag EJ. Identification of soybean microRNAs and their targets. Planta. 2008;229:161–182. doi: 10.1007/s00425-008-0818-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets during and/or analyzed during the current study are available from the corresponding author on reasonable request.